Abstract
N-methyl-D-aspartate receptors (NMDARs) are critical to synaptogenesis, neural circuitry and higher cognitive functions such as learning and memory. A hallmark feature of NMDARs is an early postnatal developmental switch from primarily GluN2B- to GluN2A-containing. Although the switch in phenotype has been an area of intense interest for two decades, the mechanisms that trigger it, and the link between experience and the switch are unclear. Here we show a novel role for the transcriptional repressor REST in the developmental switch of synaptic NMDARs. REST is activated at a critical window of time and acts via epigenetic remodeling to repress grin2b expression and properties at rat hippocampal synapses. Knockdown of REST in vivo prevented the decline in GluN2B and developmental switch in NMDARs. Notably, maternal deprivation impaired REST activation and acquisition of the mature NMDAR phenotype. Thus, REST is essential for experience-dependent fine-tuning of genes involved in synaptic plasticity.
INTRODUCTION
NMDARs are hetero-tetramers typically composed of GluN1 and GluN2 subunits, and the precise subunit composition determines NMDAR functional properties1, 2. A developmental switch in subunit composition from primarily GluN2B- to GluN2A-containing occurs during postnatal development3–7. This developmental switch is significant in that GluN2B expression can restrict synaptic incorporation of AMPARs8, 9, reduce the threshold for and enhance the magnitude of long-term potentiation (LTP)6, 10, 11, and promote hippocampal-dependent learning12, plasticity-induced spine growth10, and dendritic patterning critical to information processing13. Moreover, GluN2B-containing NMDARs exhibit slower decay times1, carry more Ca2+ current per unit charge7 and preferentially tether to the plasticity protein CaMKII14. These findings highlight the importance of maintaining correct GluN2A/GluN2B levels in adults. However, the molecular mechanisms controlling the long-term switch in NMDAR subunit composition during brain development remain unclear.
Epigenetic modification of chromatin is a key regulator of gene expression in virtually all tissues including the brain, and provides a mechanism through which neuronal activity and early experience in life can modify brain development15, 16. An attractive scenario is that during postnatal development, experience triggers chromatin remodeling and transcriptional repression of grin2b, the gene encoding the NMDAR subunit GluN2B. The Repressor element 1 silencing transcription factor (REST); also known as Neuron-restrictive silencer factor (NRSF) is a gene-silencing factor that is widely expressed during embryogenesis in pluripotent stem cells and neural progenitors, where it acts via epigenetic remodeling to silence a large array of coding and noncoding neuronal genes important to synaptic function including grin2b17–21. REST binds RE1 (also known as NRSE), a 23 base pair motif within the promoter of target genes, and acts as a platform that recruits co-repressors, which promote chromatin remodeling via modifications of core histone proteins and DNA22–24. Initially thought to function as a master negative regulator of genes involved in neurogenesis and neuronal differentiation, loss of REST is critical to acquisition of the neuronal phenotype18, 19. However, a role for REST in synaptic function in mature neurons under physiological conditions is, as yet, unknown.
The subunit composition of synaptic NMDARs is regulated acutely, on the order of minutes or hours, in response to neuronal activity10, 25–28 and in a long-lasting manner by early postnatal experience3, 29–33. The present study was undertaken to investigate molecular mechanisms underlying the long-term switch in GluN2 subunits at hippocampal synapses during normal postnatal development and in response to early-life experience. We show that REST is activated and recruited to the GluN2B promoter in differentiated neurons and is critical to the enduring physiological switch in synaptic NMDARs phenotype at dentate gyrus granule cell synapses observed in vivo during normal postnatal development. We further show that adverse experience in early life in the form of maternal deprivation disrupts activation of REST and the switch in synaptic NMDARs. Thus, REST is essential for experience-dependent fine-tuning of genes involved in synaptic activity and plasticity via epigenetic mechanisms.
RESULTS
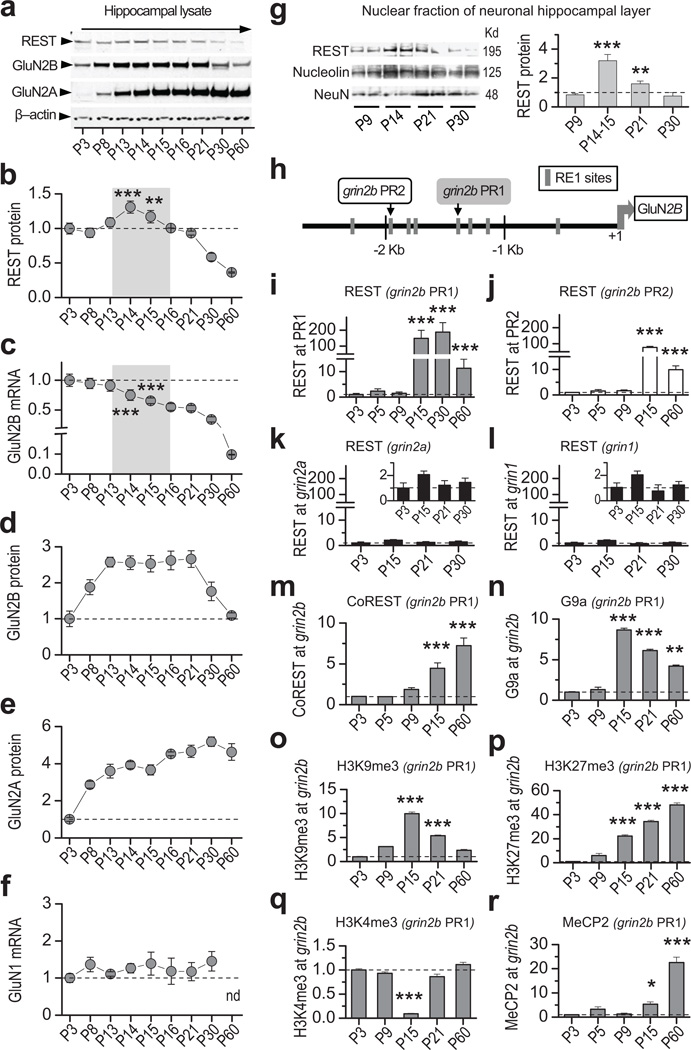
To examine a possible role for REST in the developmental switch of NMDAR subunit expression, we examined postnatal expression of REST, GluN2A, and GluN2B in the hippocampus of neonatal rats. A transient, but marked increase in REST abundance occurred at postnatal day 14–15 (P14–15; Fig 1a, b; see scanned membrane in Supplementary Fig. 1a) coinciding with a long-term decline in GluN2B mRNA expression (Fig. 1c). GluN2B protein abundance remained essentially constant from P8 until P21, after which it declined by ~2.5-fold to a level that persisted until adulthood (Fig. 1a, d). GluN2A protein was barely detectable at P3, and progressively increased by nearly 5-fold by P30, a level that persisted until P60 (Fig. 1a, e; Supplementary Table 1). Whereas GluN2B and GluN2A are developmentally regulated, GluN1 mRNA expression is unaltered postnatally (Fig. 1f). To determine whether the observed increase in REST at P14–15 occurs primarily in the nuclear fraction of neurons, we microdissected the cell body layer of the hippocampus at indicated ages and extracted the nuclear fraction. REST abundance in nuclei increased by more than 3-fold (vs. 0.3-fold in whole hippocampal lysates) at P14–15 relative to P9 (Fig. 1g; Supplementary Fig. 1b; Supplementary Table 2). These findings document a marked increase in the transcriptional repressor REST in the nuclei of hippocampal neurons.
Figure 1. REST increases transiently, is recruited to and coincides with epigenetic marks of repression at the grin2b promoter during rat hippocampal postnatal development.
a, Representative Western blots of whole hippocampal lysates showing that REST increases, GluN2B declines, and GluN2A increases during postnatal development (see full-length blot in Supplementary Fig. 1a). b, Time course showing that REST protein increases transiently at P14–15 (n = 5). c, GluN2B mRNA exhibits a long-term decline during postnatal development, assessed by RT-qPCR. The decline was highly significant from P15 through P60 (vs. P3; n = 5). d,e, Time course showing that whereas GluN2B protein declines after P21 (n = 6), GluN2A protein increases markedly from P8 to P16 and remains high up to P60 (n = 3). Data were normalized to corresponding values at P3. f, GluN1 mRNA is not altered during postnatal development (n = 6). g, Representative Western blot (left panel) and summary data (right panel) show REST protein expression in the nuclear fraction of the hippocampal cell body layer, which is enriched for neurons. Note that REST abundance in the neuronal nuclear fraction increases strikingly by P14–15 (n = 4). Data were normalized to corresponding data at P9. See full-length blot in Supplementary Fig. 12). h, Map of the rat grin2b gene indicating location of RE1 motifs contained within the proximal (PR1; gray box) and distal (PR2; white box) regions of the grin2b promoter probed by chromatin immunoprecipitation (ChIP). i,j, REST occupancy is markedly enriched at the grin2b proximal (PR1, grey bars) (n = 6) and distal PR2 (white bars) (n = 3) promoters at P15 but declines by P60. k,l, REST is not enriched at RE1 sites within the grin2a (n = 9), nor grin1 (n = 6) promoters. Inset, same data depicted with expanded y-axis. m,n, CoREST (n = 3) and G9a (n = 3) are enriched at PR1 by P15. o,p, Increase in H3K9me3 (n = 3) and H3K27me3 (n = 6) (marks of repression) at P15. q, Decrease in trimethylation of H3K4 (n = 3) (mark of active transcription), at PR1. r, MeCP2 occupancy is enriched at grin2b PR1 by P15 with a sharp increase by P60 (n = 3). All samples were normalized to input and to corresponding values at P3. Summary data represent the mean ± s.e.m. *p < 0.05; **p < 0.01; ***p < 0.001.
REST binds RE1/NRSE sites within promoters of target genes23, including grin2b20, 21. To evaluate whether the increase in REST abundance at P14–15 coincides with REST enrichment at grin2b, we examined REST occupancy at two RE1 sites contained within the proximal (PR1) and distal (PR2) regions of the grin2b promoter (Fig. 1h; Supplementary Fig. 2a, b) by means of chromatin immunoprecipitation (ChIP). Of note, the mouse grin2b promoter has multiple RE1 motifs which act synergistically to enhance the binding affinity for REST20. REST was highly enriched at PR1 (Fig. 1i), and PR2 (Fig. 1j; Supplementary Fig. 3c, d and Supplementary Table 3) at P15, directly preceding the decline in GluN2B mRNA expression. Because REST occupancy was greater at PR1 than at PR2 at all ages examined, we focused on PR1 in subsequent experiments. REST occupancy at the grin2b promoter persisted as late as P60, consistent with a role for REST in long-term transcriptional repression of GluN2B. In contrast, REST was not enriched at exon 4 of the grin2b gene (Supplementary Fig. 4a), or at the promoter of the actin gene, both of which lack RE1 sites (Supplementary Fig. 4b). Moreover, REST was not enriched at RE1 sites within the grin2a (GluN2A; Fig. 1k), grin1 (GluN1; Fig. 1l), or gria2 (GluA2) gene promoters (Supplementary Fig. 4c). Collectively, these findings indicate target specificity of REST in mature, differentiated neurons.
Co-assembly of REST with the co-factor CoREST, enhances the ability of REST to repress genes34 and can effect long-term gene silencing22. Recruitment of CoREST to the grin2b promoter was first evident at P15, and was greatest at P60, the latest age examined (Fig. 1m). The REST corepressor complex promotes post-translational modifications of core histone proteins and methylation of cytosines within DNA18. G9a, a site-specific histone methyltransferase which confers methyl moieties to H3K9, is associated with REST repression35, and was highly enriched at the grin2b promoter (~9-fold) at P15 (Fig. 1n), after which it gradually decreased to ~6-fold at P21 and to ~4-fold at P60, the latest time point examined. As is the case of other transcription factors, the binding affinity of REST to RE1, and its ability to repress target genes is enhanced by co-existence of chromatin repressive marks36. Trimethylation of core histone 3 at lysine 9 (H3K9me3), a functional readout of G9a and a mark of transcriptional repression, transiently increased ~10-fold at the grin2b promoter by P15, and 5-fold by P21, after which it declined to levels near to those observed at P3 (Fig. 1o). In contrast, trimethylation of H3 at lysine 27 (H3K27me3), also a mark of gene silencing, increased dramatically (~20-fold) at the grin2b promoter at P15, but continued to rise in occupancy (~50-fold) as late as P60 (Fig. 1p; see Supplementary Fig 3f and Supplementary Table 4). Thus, although initially both repressive marks are likely acting in concert, the long-term decline in GluN2B expression is associated with the long-term repressive mark H3K27me3. The striking increase in repressive marks was accompanied by a marked and transient reduction in trimethylation of H3 at lysine 4 (H3K4me3), a mark of active gene transcription, to almost undetectable levels at P15, followed by a significant rise at P21 to the higher levels observed at earlier ages (Fig. 1q; Supplementary Fig 3e). At P21 and as late as P60, the latest time point examined, the grin2b promoter exhibited a bivalent pattern of epigenetic marks in which marks of repression (H3K27me3, H3K9me3) colocalize with marks of active gene transcription (H3K4me3); under these conditions, genes will be repressed yet at the same time poised for activation37. Methyl CpG binding protein 2 (MeCP2), a protein that reads epigenetic marks and functions as a transcriptional repressor15, was enriched at the grin2b promoter by ~5 fold at P15, and by ~20-fold at P60 (Fig. 1r; Supplementary Table 3). Thus, recruitment of REST to the grin2b promoter and epigenetic marks of repression directly precede the long term decline in GluN2B subunit.
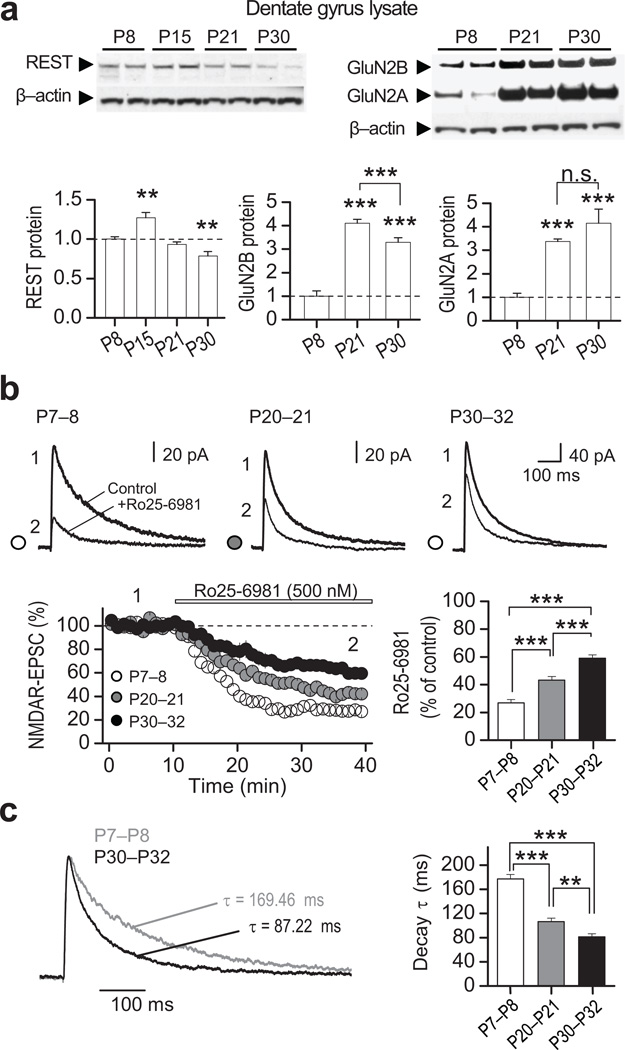
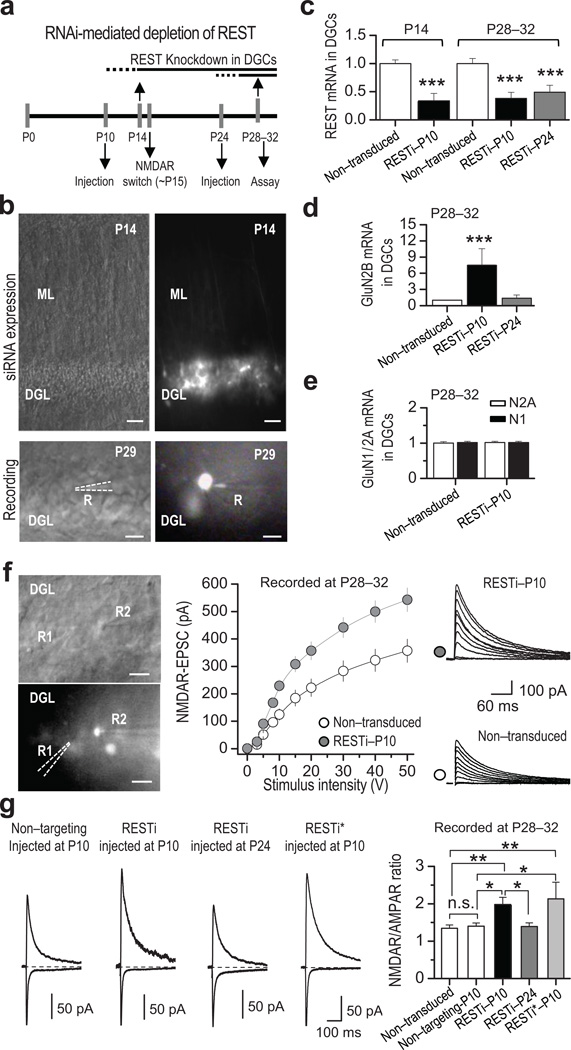
To examine a possible causal role for REST in the long-term decline in GluN2B expression, we focused on the dentate gyrus (DG), a key relay station in the hippocampal formation where a transient increase in REST precedes the developmental switch in NMDAR subunit composition (Fig. 2a and Supplementary Table 5). Pharmacologically-isolated NMDAR-mediated EPSCs (see Methods) exhibited both a progressive decrease in sensitivity to the GluN2B-selective antagonist Ro25–6981 (0.5 µM) (Fig. 2b), and faster EPSC decay kinetics with age (Fig. 2c). These findings suggest that in DG, as in other brain regions1, 4, 30, there is a developmental switch in synaptic NMDAR phenotype from primarily GluN2B- to primarily GluN2A-containing. We injected synthetic RNAi targeted to REST (RESTi)38 or nontargeting RNAi (does not target any known vertebrate gene; Supplementary Fig. 5) directly into the DG of rat pups by means of the lentivirus expression system at either P10, an age prior to the initial decline in GluN2B, or at P24 (Fig. 3a), after the initial decline in GluN2B (see Fig. 1a,d) and switch in NMDAR phenotype (Fig. 2b,c) have started. RESTi delivered at P10 was abundantly expressed in dentate granule cells (DGCs) by P14, as assessed by the intensity of green fluorescence (Fig. 3b, top panels). Injection of RESTi at P10 decreased the levels of REST mRNA assessed at P14 and P28–32 (Fig. 3c). While RESTi injected at P10 elicited increased the levels of GluN2B mRNA by more than 7-fold (Fig. 3d), it had little or no effect on GluN2A mRNA, nor GluN1 mRNA (Fig. 3e), assessed by qPCR of clusters of GFP-positive DGCs expressing RESTi at P28–32 (GFP positive neurons), relative to non-transduced control cells. This observation is consistent with the concept that REST regulates a subset of "transcriptionally responsive" genes in postnatal neurons, as recently reported for the chloride transporter KCC2 in cortical neurons in vitro39. Whereas RESTi delivered at P24 decreased REST mRNA expression (Fig. 3c), it did not significantly alter GluN2B mRNA, assessed at P28–31 (Fig. 3d; Supplementary Table 6), indicating that reducing REST after the switch has been triggered does not reverse the decline in grin2b transcription.
Figure 2. Transient increase in REST precedes the switch in NMDAR phenotype in rat dentate gyrus.
a, Representative immunoblots (top) from microdissected dentate gyrus tissue at indicated postnatal (P) ages, showing the developmental expression for REST, GluN2A, GluN2B and β-actin (see full-length blots in Supplementary Fig. 12). All values were normalized relative to P8. Quantification (bottom) showing that, as in the hippocampal formation (see Fig. 1), REST expression in dentate gyrus is transiently increased at P15 (bottom left panel), the NMDAR subunits GluN2B declines (bottom middle panel) and GluN2A increases (bottom right panel) (n = 4). b, Averaged sample traces of NMDAR-EPSCs (top) before and after bath application of the selective GluN2B antagonist Ro25–6981 (500 nM) at indicated postnatal ages (P7–8, P20–21, P30–32). Time course and summary data (bottom) showing that sensitivity to Ro25–6981 decreases progressively with age (P7–8, 2 animals/7 cells; P20–21, 2 animals/8 cells; and P30–32, 2 animals/6 cells). Averaged sample traces were taken at times indicated by numbers on the summary plot. c, Normalized NMDAR-EPSCs traces at different postnatal ages and summary data showing that the decay kinetics of NMDARs increases progressively with age. Summary data represent the mean ± s.e.m. **p<0.01; ***p<0.001; n.s., not significant.
Figure 3. RNAi-mediated depletion of REST increases GluN2B mRNA and alters NMDAR properties.
a, Diagram illustrating RNAi injection and time course of assays (qRT-PCR or patch recording). Lentiviral mediated RNAi directed against REST (RESTi) and nontargeting RNAi were delivered by means of sterotaxic injection into the hippocampus of rats at P10 or P24, and evaluated at indicated time points. b, Differential interference contrast (DIC) and GFP fluorescence images showing viral-mediated transduction of DGCs at P14 (top), and a patched DGC at P29 (bottom). Scale bars = 50 µm (top panel) and 20 µm (bottom panel). DGL, dentate granule cell layer; ML, molecular layer; R, recording pipette. c, RESTi delivered at P10 (RESTi-P10) effectively knocks down REST by P14 (n = 3), as assessed by qPCR. Delivery of RESTi at P10 and P24 (RESTi-P24) knocked-down REST measured at P28–32 (n = 3) in dentate granule cells (DGCs). d, RESTi injected at P10, but not P24, significantly increased GluN2B mRNA(n = 3) measured at P28–32. e, RESTi delivered at P10 does not exert any effect on GluN2A (n = 3) or GluN1 (n = 3) mRNA expression measured at P28–32. f, Representative DIC (top left panel) and fluorescence (bottom left panel) images showing simultaneous whole-cell recordings from RESTi-P10 expressing (GFP+) and non-transduced control (GFP−) DGCs. R1 and R2 indicate the recording pipettes from GFP− and GFP+ neurons, respectively. Scale bars = 20 µm. Summary data (middle panel) and representative traces (right panels), showing that delivery of RESTi at P10 (gray circles; 6 animals/13 cells) increases NMDAR-mediated transmission relative to that of neighboring non-transduced (control) neurons (white circles; 6 animals/13 cells). g, Representative traces (left) and summary data (right) showing that the NMDAR/AMPAR ratio is higher in DGCs expressing REST RNAi delivered at P10 (6 animals/23 cells) but not at P24 (6 animals/18 cells) relative to non-transduced controls (6 animals/29 cells) or controls expressing nontargeting RNAi neurons (6 animals/18 cells). A second lentiviral mediated RNAi targeted to a different sequence in REST (RESTi*) also increased the NMDAR/AMPAR ratio (2 animals/7 cells). Summary data represent the mean ± s.e.m. *p < 0.05; **p < 0.01; ***p < 0.001; n.s., not significant.
To determine whether the increase in GluN2B mRNA expression induced by RESTi is functionally significant, we monitored pharmacologically-isolated, NMDAR-EPSCs onto DGC synapses (see Methods). We first analyzed the input/output function of NMDAR-mediated transmission in RESTi expressing neurons relative to neighboring, non-transduced control cells in the same hippocampal slice. RESTi injected at P10 significantly increased NMDAR transmission (Fig. 3f; Supplementary Table 7). Consistent with this observation, in animals injected at P10, but not at P24, neurons expressing RESTi exhibited a higher NMDAR/AMPAR ratio than did neurons expressing nontargeting RNAi (Fig. 3b, bottom panels; Fig. 3g). Moreover, another synthetic RNAi that targets a different sequence within the REST gene (called RESTi*, see Methods) also increased the NMDAR/AMPAR ratio (Fig. 3g). In contrast, the frequency and amplitude of AMPAR-mediated miniature EPSCs, the paired-pulse ratio of evoked EPSCs, and I/V curves for AMPAR-EPSCs and NMDAR-EPSCs were unchanged in RESTi expressing neurons (Supplementary Figs. 6, 7), strongly suggesting that RESTi does not detectably alter glutamate release probability, quantal size of AMPAR-mediated synaptic transmission or rectification properties of synaptic AMPARs or NMDARs at DGC synapses. Together, these results suggest that knocking down REST prevents the normal developmental decline in GluN2B mRNA and presumably protein, thereby selectively increasing NMDAR- (but not AMPAR-) mediated transmission.
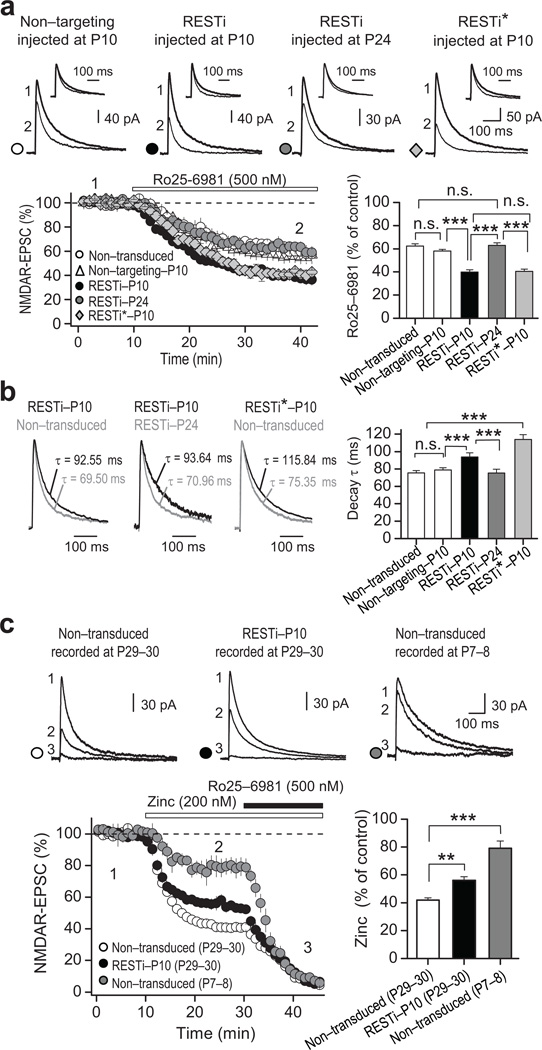
We next examined the impact of RESTi on the properties of NMDAR-mediated transmission. In animals injected with either RESTi or RESTi* at P10, NMDAR-EPSCs recorded at P28–32 exhibited a greater sensitivity to the GluN2B-selective antagonist Ro25–6981 (0.5 µM; Fig. 4a) and slower decay kinetics, relative to NMDAR-EPSCs recorded from non-transduced DGCs or DGCs expressing nontargeting RNAi (Fig. 4b; Supplementary Table 8). These observations are consistent with an increase in synaptic GluN2B-containing receptors5, 40. In contrast, neurons injected with RESTi at P24 showed no detectable change in sensitivity of NMDAR-EPSCs to Ro25–6981 (Fig. 4a) or decay kinetics (Fig. 4b) relative to non-transduced DGCs, consistent with the notion that REST acts within a critical time window during development to promote epigenetic marks of repression that contribute to the switch in synaptic NMDAR properties. To examine the impact of RESTi injected at P10 on GluN2A-containing NMDARs we assessed the sensitivity of NMDAR-EPSCs to Zn2+, which in the nanomolar range, preferentially inhibits GluN2A- vs. GluN2B-containing NMDARs (see Methods). To validate the selectivity of Zn2+ for GluN2A-containing NMDARs, we recorded NMDAR-EPSCs at an older age (P29–30), when synaptic NMDARs are primarily GluN2A-containing, and at a younger age (P7–8), when the contribution of GluN2A-containing receptors is minimal. The reduction in NMDAR-EPSC amplitude produced by Zn2+ was marked at P29–30 but greatly diminished at P7–8 (Fig. 4c; Supplementary Table 8). NMDAR-EPSCs recorded from RESTi-P10 neurons at P29–30 exhibited significantly less sensitivity to 200 nM Zn2+ (see Methods and Supplementary Fig. 8) than NMDAR-EPSCs recorded from non-transduced DGCs (Fig. 4c; p<0.001). These results demonstrate that REST regulates the GluN2B/GluN2A expression ratio and significantly contributes to the switch in synaptic NMDAR phenotype from primarily GluN2A- to GluN2B-containing during postnatal development.
Figure 4. REST knockdown prevents acquisition of the mature NMDAR phenotype.
a, Representative NMDAR-EPSCs traces (top panel) and summary data (bottom panel) showing an increase in Ro25–6981-mediated inhibition of NMDAR-EPSCs in DGCs from animals injected at P10 with RESTi (RESTi-P10; 4 animals/12 cells) or RESTi* (RESTi*; 2 animals/6 cells), but not at P24 (RESTi-P24; 4 animals/12 cells), relative to non-transduced controls (3 animals/7 cells) or cells expressing nontargeting RNAi neurons (4 animals/11 cells). Inserts in top panel show normalized current before and after Ro25–6981-mediated inhibition. b, Normalized current (left panels) and summary data (right panel) showing an increase in decay kinetics of NMDAR-EPSCs from animals injected at P10 with RESTi or RESTi*, but not P24, compared to non-transduced control DGCs. c, Sample traces (top panel), time course (bottom left) and summary data (bottom right) showing Zn2+ (200 nM)-dependent inhibition of NMDAR-EPSCs. Note that Zn2+ inhibition is high in DGCs from non-transduced older (P29–30) animals (left), which express primarily GluN2A-containing NMDARs (8 animals/14 cells), and is diminished in older animals injected with RESTi at P10 (center, 8 animals/16 cells). Zn2+ inhibition is diminished in DGCs from non-transduced younger animals (P7–8) animals (right), which express primarily GluN2B-containing NMDARs (2 animals/7 cells). In all panels, averaged sample traces were taken at times indicated in the summary data. Summary data show the mean ± s.e.m. ** p<0.01; *** p<0.001; n.s., not significant.
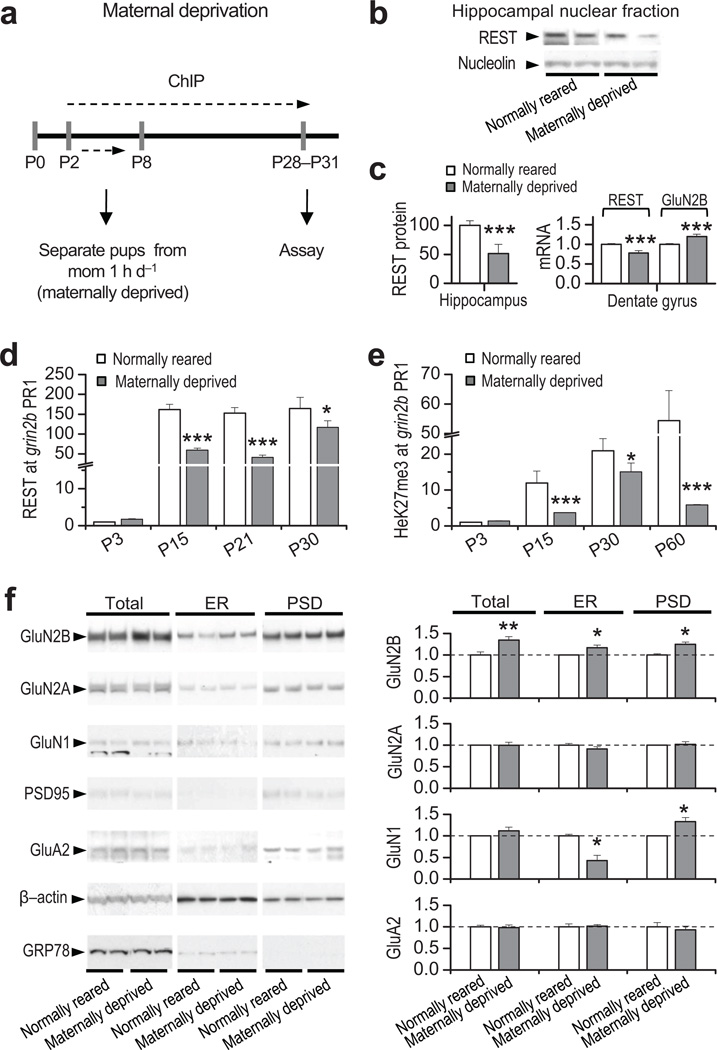
Epigenetic modifications of chromatin reflect environmental influences that are not “hard-wired” into the DNA sequence15, 16. Given that REST acts via epigenetic remodeling to promote the developmental switch in NMDAR phenotype, we reasoned that experience might influence activation of REST and the long-term decline in GluN2B expression. Maternal deprivation during the first week of postnatal life has a profound and enduring impact on hippocampal development41 and NMDAR subunit composition in the hippocampus31. We first assessed whether adverse experience early in life in the form of maternal deprivation could influence REST and REST-dependent silencing of grin2b. Animals subjected to maternal deprivation (Fig. 5a, see Methods) showed a marked reduction in REST protein in the nuclear fraction of the hippocampus assessed by Western analysis (Fig. 5b, 5c left panel), a decrease in REST mRNA (Fig. 5c, right panel), and increase in GluN2B mRNA expression in DG, assessed by qRT-PCR at P30 (Fig. 5c, right panel). In contrast, GluN2A, GluN1, and GluA2 mRNA expression were unaltered in DG at P30, indicating target specificity of REST (Supplementary Fig. 9; Supplementary Table 9). Moreover, maternally-deprived rats exhibited a marked reduction in REST (Fig. 5d), and H3K27me3 (Fig. 5e) occupancy at the grin2b promoter in DG, relative to age-matched, normally-reared rats (Supplementary Table 10). To examine the impact of maternal deprivation on NMDAR subunit composition at synapses, we assessed levels of receptor subunits in endoplasmic reticulum (ER) and postsynaptic density (PSD) from normally-reared and maternally-deprived pups at P30 (see Methods). GluN2B was markedly increased in both ER and PSD fractions of maternally-deprived vs. normally-reared animals (Fig. 5f). Whereas total cellular abundance of GluN1 protein was unchanged, GluN1 protein was decreased in the ER fraction and increased in the PSD fraction (Fig. 5d), consistent with forward trafficking of GluN1 from the ER to the PSD in the hippocampus of maternally-deprived rats. The increase in synaptic GluN1 likely reflects assembly and synaptic incorporation of new GluN1/GluN2B receptors. In contrast, GluN2A was unchanged in total cell lysates, ER and PSD fractions from maternally-deprived vs. normal-reared rats (Fig. 5d; Supplementary Table 9), consistent with no change in occupancy of REST at grin2a in maternally-deprived animals (Supplementary 11; Table 10). These findings show that maternal deprivation during the first week of life interferes with activation of REST and occupancy of epigenetic marks of repression, and prevents the decline in synaptic GluN2B (but not the rise in GluN2A) during postnatal development.
Figure 5. Maternal deprivation disrupts the increase in REST, epigenetic remodeling and decrease in GluN2B during postnatal development.
a, Diagram illustrating the maternal deprivation paradigm. b, Representative Western blot of hippocampal nuclear fraction. c, Summary plot (left panel) showing a robust decrease in REST protein expression in the hippocampus of maternally-deprived vs. normally-reared pups (n = 3). In the dentate gyrus, whereas REST mRNA decreases (n = 3), GluN2B mRNA increases (right panel; n = 3). d, Abundance of REST and e, H3K27me3 at the grin2b promoter are decreased relative to age-matched, normally-reared rats (n = 3). f, Representative Western blots (left panel) and summary data (right panel), showing that whereas maternal deprivation increases GluN2B protein in total lysate, endoplasmic reticulum (ER) and postsynaptic density (PSD) fractions, GluN2A and GluA2 are unchanged in all fractions. GluN1 was reduced in the ER fraction and increased in the PSD in the hippocampus from maternally-deprived pups vs. normally-reared pups. All samples were assessed at P28–31 (n = 3). Total lysate samples were normalized to β-actin, ER samples were normalized to GRP78, an ER marker, and PSD samples were normalized to PSD-95, which is not altered after maternally-deprived (see Supplementary Table 9 and full length blots in Supplementary Fig. 12).
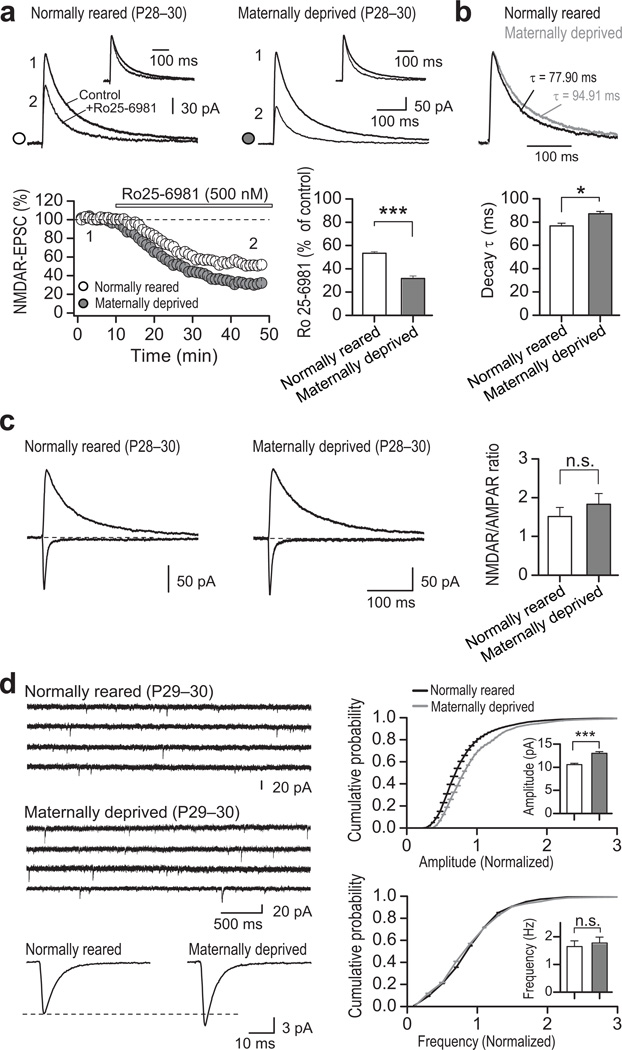
To evaluate the functional consequences of maternal deprivation on NMDAR-mediated synaptic transmission, we recorded NMDAR-EPSCs at synapses onto DGCs in slices from maternally deprived pups and normally-reared littermates at P28–31. Consistent with an increase in synaptic GluN2B, with little or no change in GluN2A (Fig. 5f, Supplementary Fig. 9), NMDAR-EPSCs of maternally-deprived rats exhibited enhanced sensitivity to the GluN2B-selective antagonist Ro25–6981 (0.5 µM; Fig. 6a) and slower EPSC decay kinetics (Fig. 6b, Supplementary Table 11). In contrast, the paired-pulse ratio of evoked EPSCs (Supplementary Fig. 10) and NMDAR/AMPAR ratios (Fig. 6c) were unchanged. Intriguingly, maternal deprivation induced a modest increase in the quantal amplitude of AMPAR-mediated transmission –as indicated by changes in miniature EPSC (mEPSC) amplitude but not frequency– (Fig. 6d, Supplementary Table 11), which could account for the lack of change in NMDAR/AMPAR ratio. While the increase in mEPSC amplitude might be due to compensatory changes in AMPAR number or function, we did not detect a change in GluA2 protein level (Fig. 5d; Supplementary Table 9). Neither REST abundance at gria2 (Supplementary Fig. 11; Supplementary Table 10), nor GluA2 mRNA (Supplementary Fig. 9; Supplementary Table 10) were changed at P30. Thus, while the mechanism underlying the increase in AMPAR-mediated transmission in maternally-deprived animals remains unclear, it is unlikely due to REST-dependent transcriptional regulation of GluA2. Together, these findings strongly suggest that maternal deprivation, known to be associated with aberrant behavior42, impairs activation of REST and the switch in NMDAR subunit composition during development, likely due to the inability of low levels of REST to repress transcription of grin2b.
Figure 6. Maternal deprivation impairs acquisition of the mature NMDAR phenotype at synapses.
a, Representative traces (top panel) and summary data (bottom panel) showing that inhibition of NMDAR-EPSCs by Ro25–6981 was greater in neurons from maternally-deprived (4 animals/13 cells) vs. normally-reared pups (4 animals/14 cells) assessed at P28–30. Averaged sample traces were taken at times indicated by numbers on the summary plot. b, Normalized NMDAR-EPSCs (top panel) and summary data (bottom panel) showing that DGCs from maternally-deprived pups display a small, albeit significant decrease in NMDAR-EPSC decay kinetics (4 animals/18 cells) vs. control pups (4 animals/16 cells). c, Representative traces (left and center panels) and summary data (right panel) showing that the NMDAR/AMPAR ratio was unchanged in DGCs from maternally-deprived (4 animals/16 cells) vs. control pups (4 animals/18 cells). d, Representative traces showing AMPAR-mEPSC activity in control (top left) and DGCs from maternally-deprived animals (middle left). Average AMPAR-mEPSC traces (bottom left), and cumulative probability plots of amplitude (top right) and frequency (bottom right) showing a significant increase in the amplitude (p<0.001), but not frequency (p=0.098) of AMPAR-mEPSCs in DGCs from maternally-deprived (3 animals/15 cells) vs. control pups (3 animals/14 cells).
DISCUSSION
The present study demonstrates a critical and previously unrecognized role for experience-dependent, epigenetic remodeling of synaptic NMDARs during hippocampal development. Our results indicate that REST is essential to the developmental switch in GluN2A/GluN2B ratio at hippocampal synapses in vivo. REST orchestrates epigenetic remodeling of the grin2b promoter and long-lasting transcriptional repression of GluN2B early in postnatal development. While previous studies have shown that acute neuronal activity can regulate NMDAR subunit composition on the order of minutes or hours10, 25–28, these studies have not addressed the enduring physiological switch in synaptic NMDARs observed in vivo during postnatal development. Whereas studies of the switch in NMDARs during postnatal development have focused on the increase in GluN2A at synapses of visual cortex3, 33, we demonstrate an essential role for the decline in GluN2B expression at hippocampal synapses. A prevailing view has been that REST serves as a master regulator of neuronal gene expression in pluripotent stem cells and neural progenitors. To our knowledge, our study is the first demonstration that REST-dependent epigenetic modifications alter properties of a synaptic protein in differentiated neurons and that REST can regulate synaptic function. We further show that adverse early life experience in the form of maternal deprivation disrupts activation of REST, alters the epigenetic landscape at the grin2b promoter, and prevents acquisition of the mature NMDAR phenotype at hippocampal synapses. Our finding that the switch is mediated by experience-dependent mechanisms in vivo supports a novel and previously unappreciated role for REST as a link between experience and synaptic function.
REST exhibits both target and temporal specificity
While genome wide and cell line studies reveal that only ~13% of predicted RE1 sites actually bind REST, there is little information on mechanisms underlying target specificity and affinity in neurons in vivo. Our studies show that REST exhibits target specificity in differentiated neurons during postnatal development. Whereas REST is enriched at the grin2b promoter, it is not enriched at grin1, grin2a or gria2 promoter, which contain known RE1 sites. Some of these findings are not entirely surprising, as GluN1 mRNA expression in the hippocampus is not developmentally regulated (see Fig. 1d), and GluN2A mRNA expression is highly increased during development40. Also, our finding that REST is not enriched at grin2a in differentiated neurons is consistent with findings of others that grin2a is not a functional target of REST in mammalian cells23, 43. The observation that REST is not recruited to the gria2 promoter during early postnatal development is in contrast to findings that gria2 is a known functional target of REST in mammalian cells44 and in CA1 neurons in response to ischemic insults in vivo38. Thus, target specificity of REST may vary with cell type and context.
Although not addressed in the present study, the long-term increase in GluN2A expression during postnatal development is likely regulated at least in part by REST-independent transcriptional activation, as for example CREB43, and/or post-transcriptional mechanisms such as translation and receptor trafficking. It is interesting that REST knockdown enhances NMDAR-mediated transmission (Fig. 3f,g). Such an increase is presumably due to the selective REST-dependent increase in GluN2B expression at the synapse (Figs. 3d, 4a,b), consistent with the notion that availability of GluN2 subunits determines the total number of functional NMDARs45. Our results reinforce the notion that although REST can repress thousands of genes, other supporting mechanisms may be necessary to enable REST-dependent repression, as for example changes in the epigenetic landscape at the gene promoter. An open question is what mechanism turns on REST expression in differentiated neurons at P15–17. Recent studies indicate that a fundamental mechanism by which REST is regulated is at the level of protein stability and degradation. REST protein stability is bidirectionally regulated by β-TrCP-dependent, ubiquitin-based proteasomal degradation46, 47 and HAUSP-dependent deubiquitination, leading to enhanced protein stability48. One possibility is that early life experience regulates REST expression through these degradative mechanisms.
We also report that the temporal context in which REST modulates grin2b is restricted to a critical window of time during postnatal development associated with heightened plasticity. Whereas in vivo knockdown of REST at P10, an age prior to the decline in GluN2B, derepresses GluN2B and disrupts the change in synaptic NMDAR properties at hippocampal synapses, knockdown of REST at P24, an age after the decline in GluN2B has begun, is ineffective. Moreover, once REST has triggered grin2b repression at P14–15, corepressors such as CoREST and MeCP2, and/or chromatin repressive marks persist at the grin2b promoter at high levels at older ages, thereby maintaining GluN2B at low levels. This would, in turn, make acute REST knockdown ineffective at older ages (e.g. >P24). Alternatively, once the switch in NMDAR phenotype has occurred, it is difficult to reverse. Recruitment of REST to the grin2b promoter is coincident with a striking alteration in the chromatin landscape. At P15 the grin2b promoter undergoes a dramatic change in which an increase in marks of stable gene repression (H3K9me3 and H3K27me3)49 is coincident with a decline in the activation mark H3K4me3. Notably, the decrease in H3K4me3 is transient, in that by P21 (and at P60), it is increased and coincides with a persistent enrichment in H3K27me3. These findings document a bivalent pattern of epigenetic marks involving both “repressive” and “activating” chromatin modifications at the grin2b promoter in early postnatal life, possibly maintaining GluN2B in a repressed state, yet poised for activation37. To our knowledge, this is the first study showing a bivalent pattern of epigenetic marks at the gene promoter of a synaptic protein in fully-differentiated neurons.
Maternal deprivation disrupts epigenetic remodeling and the switch in NMDAR phenotype
Epigenetic modifications are associated with experience-driven chromatin modifications that lead to long-lasting changes in gene transcription16, 49. While it is well-established that experience can regulate the GluN2A/GluN2B ratio at central synapses3, 29–33, the link between experience and synaptic function remains unclear. Our results clearly demonstrate a role for experience-dependent epigenetic remodeling, leading to a synaptic modification in vivo. We show that early in life the NMDAR subunit GluN2B is modulated by REST-dependent epigenetic processes involving histone methylation. Although not addressed by the present study, our results do not preclude the possibility that DNA methylation is also altered. Experience in the form of maternal deprivation (a paradigm known to disrupt the developmental switch in NMDAR phenotype31), prevents activation of REST and epigenetic remodeling of the grin2b promoter, silencing of GluN2B and the switch in synaptic NMDAR phenotype. Consistent with these findings, it was recently shown that maternal deprivation impairs REST function, resulting in enhanced vulnerability to stress in rats42. Maternal deprivation is also associated with an increase in anxiety and alcohol preference, impaired maternal care, and diminished spatial navigation learning50, but the mechanisms underlying these abnormal behaviors remain poorly understood. While it is clear that altered GluN2B expression highly impacts synaptic function and plasticity8–10, 12, 13, further studies are necessary to understand how disrupting repression of GluN2B during early postnatal development can impact behavioral deficits and pathological disorders associated with dysregulation of GluN2B-containing receptors.
In summary, our data provide evidence for a unique mechanism underlying the switch in synaptic NMDAR phenotype in vivo. These findings add a new dimension to our understanding of how synaptic proteins and function are influenced by experience and epigenetic mechanisms early in life, which can have long-term consequences. Given that the developmental switch in synaptic NMDAR phenotype occurs widely throughout the forebrain and has important physiological consequences, REST-dependent epigenetic remodeling of synaptic NMDARs during brain development may be a widespread and enduring mechanism to adjust synaptic function. NMDARs are critical to synaptogenesis, formation of neural circuitry and to higher cognitive functions such as learning and memory, and their dysregulation is implicated in neuropsychiatric disorders such as schizophrenia, stroke, Parkinson's disease and Huntington's disease. Thus, our findings have important ramifications for NMDARs not only during normal brain development but possibly in a large number of brain diseases and disorders.
METHODS
Animals
All animals were treated in accordance with the principles and procedures of the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals. Protocols were approved by the Institutional Animal Care and Use Committee of the Albert Einstein College of Medicine. Sprague Dawley rats (Charles River Laboratories, Inc.) were maintained in a temperature- and light-controlled environment with a 12:12 h light/dark cycle. For maternal deprivation, rat pups were isolated from the dam, nest, and siblings for a period of 1 h once per day over P2–8 as described previously31. Animals of both isolated and non-isolated groups received equal amounts of handling. Pups in the isolation treatment group were placed in individual plastic cups (9 cm diameter) and maintained at warm temperatures using a heat lamp. At the end of the isolation period, pups were returned to the nest with the dam.
Western blots
Western blot analysis was performed as described38. In brief, hippocampi were rapidly dissected snap frozen in liquid nitrogen. Proteins were extracted in lysis buffer (Tris-HCl (pH 7.40, 25 mM), NaCl (150 mM), EDTA (pH 8.0, 1 mM), SDS (0.1%), Na deoxycholate (0.5%), and a 1% cocktail of protease inhibitors (Sigma)). To enrich for the nuclear fractions, samples were gently homogenized in homogenization buffer [HEPES (5 mM), MgCl2 (1 mM), EGTA (100 mM), sucrose (0.32 M), 1% cocktail of protease inhibitors (Sigma)] and centrifuged at 3200 rpm × 10 min (4°C). Nuclear pellet was isolated, lysed and sonicated. Equal aliquots of protein (40–80 µg) were subjected to SDS-PAGE, transferred to a membrane and processed for incubation with antibody. Band densities were normalized to β-actin. Mean band densities for samples from experimental animals were normalized to the corresponding samples from control animals as indicated. For REST expression, a band just below 195 Kd was detected and verified to be REST by its absence in hippocampal neuron cultures expressing RESTi by means of lentiviral transduction (Supplementary Fig. 1a), and in hippocampal extracts loaded alongside Hela nuclear extract (Supplementary Fig. 1b), known to express REST at high levels.
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were performed as previously described38. Transverse slices of dorsal hippocampus (1 mm) were immersed in 1% formaldehyde (12 min, room temperature) to cross-link histone proteins to DNA. The cross-linking reaction was stopped by addition of glycine (final concentration, 0.125 M). Samples were lysed and sonicated to afford chromosomal DNA in the range of 0.6–1 kb. Pre-immunoprecipitated chromatin (“input”, 100 µl) was saved. Samples were precleared and immunoprecipitated with antibody (15 µg). Immunocomplexes were collected on protein A agarose beads or Dynabeads and eluted. Samples of DNA were treated with proteinase K and purified by means of the PCR purification kit (Qiagen). Abundance of a protein at RE1 sites within gene promoter regions was quantified by real time PCR using a TaqMan probe directed to grin2b (Supplementary Figure 3), gria2 (forward: TGTGTGTATGTGTGTGTATGTGTGT; reverse: CCCTCTCTCGAGCTCTCTCTCT), grin2a (forward: TCCGGAGTGGAACAGAAAGC; reverse: CTCATCCAGCCCCATGCT) and grin1 (forward: TCCCTGCTTCCTCTCTTGGA; reverse: AATGACTGCTGGGAGCAAGAC). The real time PCR ChIP data was analyzed using the software tool REST (http://www.REST.de.com), and normalized to “input” samples.
PSD and ER homogenate preparation
PSD fractions were prepared from rat brains. Hippocampi were dissected from control and maternally-deprived animals, and immediately snap frozen in liquid nitrogen. Hippocampi were homogenized in HEPES buffered sucrose (HBS) and centrifuged at 800 × g for 10 min. The pellet containing the nuclear fraction (P1) was assessed for REST expression. The supernatant was centrifuged at 12,000 × g for 15 min. The resulting supernatant (S2) was spun to obtain a pellet containing enriched ER fraction (140,000 × g for 120 min). The pellet (P2) was washed in HBS and centrifuged at 12,000 × g for 15 min, lysed via hypo-osmotic shock in ice cold water, and adjusted to 4 mM HEPES. Sample was spun at 25,000 × g for 20 min and the resulting pellet (P3) was resuspended in HBS and layered in a sucrose gradient (1.2 M sucrose:1.0 M sucrose: 0.8 M sucrose). Samples were centrifuged at 150,000 × g for 120 min, and the synaptic plasma membrane isolated. Pellet was resuspended in 50 mM HEPES plus 0.5% Triton X–100, kept at 4°C for 15 min. Samples were centrifuged at 32,000 × g for 20 min, and the resulting pellet (P4), which contained the PSD fraction was assessed using western blotting.
Antibodies used
Mouse anti-GluN2B (05–920), rabbit anti-GluN2B (06–600), rabbit anti-GluN2A (07–632), anti-GluN1 (05–432), anti-REST (07–579), anti-CoREST (07–455), anti-G9a (07–551), anti-H3K9me3 (07–442), anti-H3K27me3 (07–449), anti-H3K4me3 (07–473), anti-MeCP2 (07–013), and anti-PSD95 (AB9708) were obtained from Upstate Biotechnology/Millipore. Rabbit anti-GluR2 (556341) was purchased from Pharmigen. Rabbit anti-GRP78 (RB10478) was obtained from Neomarkers/Labvision Corp. Mouse anti-β-actin (A5316) was purchased from Sigma.
RT–qPCR
Hippocampi were dissected, and snap frozen in liquid nitrogen. RNA was extracted by means of the PureLink micro-to-midi total RNA purification system (Invitrogen) and treated with DNase I (Invitrogen). Aliquots of RNA (1 µg) were reverse-transcribed to cDNA with Superscript III Reverse Transcriptase (Invitrogen). For RT-qPCR, GFP-positive DGCs were suctioned with a patch pipette from acute slices maintained in artificial cerebral-spinal fluid (ACSF). RNA was extracted by means of the Dynabeads mRNA direct micro kit (Invitrogen). The entire reaction was reverse-transcribed to cDNA with Superscript III Reverse Transcriptase. Quantitative real-time PCR (qPCR) was performed with TaqMan probes (Applied Biosystems) for REST (Rn01413149_s1), GluN2A (Rn00561341_m1), GluN2B (Rn00561352_m1), GluN1 (Rn00433800_m1), and GluA2 (Rn00568514_m1); HPRT (hypoxanthine phosphoribosyl transferase) (Ref: Rn01527838_g1) and Gapdh (Rn99999916_s1) served as an endogenous reference. Reactions were run in triplicate in a StepOnePlus real-time PCR system (Applied Biosystems). mRNA abundance was calculated by means of the comparative Ct method at a threshold of 0.02. Data was analyzed using the software tool REST (http://www.REST.de.com), on the basis of the group mean for the target transcript, versus reference HPRT transcript or GAPDH transcript.
Generation and titer of lentiviral vectors
Lentiviral vectors encoding REST RNAi were generated and evaluated for specificity and efficacy of REST knockdown as described38. In this study we used two RNAi antisense sequences targeted to REST: RESTi (AACAAACTCTCTCAGTTAC), and RESTi* (AATGGAAGCACTGCTGTTT). A non-transducing generic control which has the same number of bases as RESTi, but does not target any known vertebrate gene was purchases from Applied Biosystems. For lentivirus-mediated silencing of REST in DGCs of intact animals, high-titer vesicular stomatitis virus (VSV)-pseudotyped lentiviral stocks were produced in HEK-293T cells. In brief, HEK-293T cells were transfected with lentiviral transfer construct pRRL.PPT.hCMV.eGFP.REST-miR.Wpre, or pRRL.PPT.hCMV.eGFP.control-miR.Wpre, and packaging constructs pMDLg/pRRE and pRSV-REV and pMD2.G envelope protein construct by means of calcium phosphate. Titers were determined by infecting HEK-293T cells with serial dilutions of concentrated lentivirus. eGFP fluorescence was evaluated by flow cytometry (FACSCalibur, Becton Dickinson Immunocytometry Systems) at 48 h after transduction; titers were 1x109 transducing units (TU) per ml. The ability of REST-RNAi to knockdown REST, was evaluated by Western blot analysis 14 d after transduction of hippocampal neurons at DIV21. REST miR3 was effective in knockdown of REST (Supplementary Fig. 5). Nontargeting control-miR had no effect on expression of REST.
Stereotaxic injection of LV-REST-miR constructs
For in vivo experiments LV-REST-miRNA was delivered by stereotaxic injection into the hippocampus of live rats at postnatal day 10 (P10), or 24 (P24) (Fig. 3a). Animals were placed in a stereotaxic frame, and anesthetized with isoflurane. Concentrated viral solution (1.5 µl) was injected into the right hippocampus at a flow rate of 0.2 µl/min. The injection site was defined by the following coordinates: at P10, 3.4 mm posterior to bregma, 3.2 mm lateral to bregma, 3.1 mm ventral from dura; at P24, 3.9 mm posterior to bregma, 3.4 mm lateral to bregma, 3.5 mm ventral from dura. The needle was left in place for an additional 3 min and gently withdrawn. To verify region-specific delivery of the virus, we assessed eGFP fluorescence in brain sections 4 d after injection. eGFP-positive neurons were prominent in dentate granule cell layer (Fig. 3b, top panels)
Hippocampal slice preparation
Acute transverse hippocampal slices (290–300 µm thick) were prepared from Sprague Dawley rats at different postnatal days (from P7 to P31). Briefly, the hippocampi were isolated and cut in an extracellular solution containing (in mM): 215 sucrose, 2.5 KCl, 20 glucose, 26 NaHCO3, 1.6 NaH2PO4, 1 CaCl2, 4 MgCl2 and 4 MgSO4. Thirty minutes post sectioning, the cutting medium was gradually switched to the extracellular artificial cerebrospinal (ACSF) recording solution containing: 124 NaCl, 2.5 KCl, 26 NaHCO3, 1 NaH2PO4, 2.5 CaCl2, 1.3 MgSO4 and 10 glucose. All solutions were equilibrated with 95% O2 and 5% CO2 (pH 7.4). Slices were incubated for at least 30 min in the ASCF solution prior to recordings.
Electrophysiology
All hippocampal slices were visualized using infrared differential interference contrast (DIC) and nontargeting GFP-control and RESTi dentate granule cells (DGCs) were visualized using fluorescence videomicroscopy on a Nikon eclipse E600FN microscope. All experiments, except where indicated, were performed at 28 ± 1 °C in a submersion-type recording chamber perfused at ~1–2 ml/min with ASCF supplemented with the GABAA receptor antagonist picrotoxin (PTX; 100 µM) and the AMPA/Kainate receptor antagonist NBQX (10 µM). Whole-cell patch-clamp recordings using a Multiclamp 700A amplifier (Molecular Devices, Sunnyvale, CA) were made from DGCs voltage clamped at −60 and +40 mV using patch-type pipette electrodes (~3–4 MΩ) containing (in mM): 131 Cs-Gluconate, 8 NaCl, 1 CaCl2, 10 EGTA, 10 glucose, 10 HEPES; pH 7.2, 292 mmol/kg.
Series resistance (~12–25 MΩ) was monitored throughout the experiment with a −5 mV, 80 ms voltage step, and cells that exhibited significant change in series resistance (>20%) were excluded from analysis. To stimulate excitatory synaptic inputs, a monopolar stimulating patch-type pipette was filled with ACSF and placed in the middle third of the molecular layer to stimulate medial perforant path (MPP) inputs. Excitatory postsynaptic currents (EPSCs) were elicited at 20 s intervals, filtered at 2.2 kHz, and acquired at 5 kHz using a custom-made software written in Igor Pro 4.09A (Wavemetrics, Inc., Lake Oswego, OR, USA). Miniature excitatory currents (mEPSCs) were recorded during 10 minutes at 32 °C from DGCs voltage clamped at –60 mV in ASCF containing 100 µM PTX and 1 µM tetrodotoxin (TTX). mEPSCs were identified using a minimal threshold amplitude (≥ 5 pA) and analyzed using the mini-analysis software Synaptosoft (Synaptosoft, Inc., Fort Lee, NJ). For Zn2+ experiments, 10 mM Tricine (N-[tris(hydroxymethyl)methyl]glycine) was added to the ACSF to buffer Zn2+ as described previously51. In the Tricine-ACSF solution the NaHCO3 was increased up to 27 mM and the pH was adjusted with NaOH. Importantly, Tricine-ACSF does not affect NMDAR-mediated basal synaptic transmission (Supplementary Figure 8). A total extracellular-free Zn2+ concentration of 200 nM was estimated by adding 40 µM ZnCl2 to the Tricine-ACSF. NMDAR-EPSC decay kinetic was fit with a single exponential function using OriginPro 7.0 software (OriginLab Corporation, Northampton, MA). Both RESTi and maternal deprivation experiments and analyses were done in a blind fashion.
Pharmacological agents (e.g. Ro 25–6981) were bath applied after establishment of a stable baseline (~10 min), and their effects were measured after responses reached a new steady state (typically >15 min). Drugs were obtained from Sigma-Aldrich (Zn2+, PTX), Tocris Bioscience (Ro 25–6981) and ASCENT (NBQX). Stock solutions were prepared in water and DMSO (PTX, NBQX) and added to the ACSF as needed. Total DMSO in the ACSF was maintained at <0.1%.
Statistical analysis
For RT-qPCR, qPCR and ChIP experiments, statistical analysis, was done using the “relative gene expression approach: REST” program (http://www.REST.de.com), which uses “Pfaffl analysis method” in the evaluation of relative real-time RT-PCR52. For Western band densities, One-way ANOVA, followed by Dunnett’s test was performed for developmental blots normalized to postnatal 3 (P3) and for cellular fractionation experiments. For REST expression in neuron-enriched nuclear fractions, One-way ANOVA followed by Bonferroni test, was performed. For electrophysiological results, statistical analysis was performed using One-way ANOVA in OriginPro 7.0 and 8.6 software (OriginLab Corporation, Northampton, MA). All One-way ANOVA were performed at the p<0.05 significance level. Cumulative probability plots of mEPSC were compared using the Kolmogorov-Smirnov test. All illustrated traces are averages of 20–31 responses. Unless otherwise stated, in the figures, *p < 0.05, **p < 0.01, and ***p < 0.001. All data are presented as mean ± s.e.m.
Supplementary Material
Acknowledgments
This work was supported by NIH grants to RSZ (R01 NS46742 and NS20752) and PEC (R01 MH081935) and generous support from the F.M. Kirby Foundation. A.E.C. was supported by a Ruth L Kirschstein Award from the National Institute of Neurological Disorders and Stroke (F32NS071821). RSZ is the F.M. Kirby Professor of Neural Repair and Protection. We thank Dr. Kyung-Min Noh for providing the REST RNAi lentiviral construct, and Dr. Anne Etgen and all the members of the Zukin and Castillo Labs for their constructive comments on the data and the manuscript.
Footnotes
Author contributions: A.R.R designed and performed all the biochemical experiments and analyzed the results. M.J.C performed biochemical experiments. A.E.C designed and performed all the electrophysiological experiments and analyzed the results. A.R.R, A.E.C., P.E.C. and R.S.Z interpreted all the results and wrote the paper.
REFERENCES
- 1.Cull-Candy SG, Leszkiewicz DN. Role of distinct NMDA receptor subtypes at central synapses. Sci STKE. 2004;2004:re16. doi: 10.1126/stke.2552004re16. [DOI] [PubMed] [Google Scholar]
- 2.Paoletti P, Neyton J. NMDA receptor subunits: function and pharmacology. Curr Opin Pharmacol. 2007;7:39–47. doi: 10.1016/j.coph.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 3.Quinlan EM, Olstein DH, Bear MF. Bidirectional, experience-dependent regulation of N-methyl-D-aspartate receptor subunit composition in the rat visual cortex during postnatal development. Proc Natl Acad Sci U S A. 1999;96:12876–12880. doi: 10.1073/pnas.96.22.12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sheng M, Cummings J, Roldan LA, Jan YN, Jan LY. Changing subunit composition of heteromeric NMDA receptors during development of rat cortex. Nature. 1994;368:144–147. doi: 10.1038/368144a0. [DOI] [PubMed] [Google Scholar]
- 5.Williams K, Russell SL, Shen YM, Molinoff PB. Developmental switch in the expression of NMDA receptors occurs in vivo and in vitro. Neuron. 1993;10:267–278. doi: 10.1016/0896-6273(93)90317-k. [DOI] [PubMed] [Google Scholar]
- 6.Yashiro K, Philpot BD. Regulation of NMDA receptor subunit expression and its implications for LTD, LTP, and metaplasticity. Neuropharmacology. 2008;55:1081–1094. doi: 10.1016/j.neuropharm.2008.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sobczyk A, Scheuss V, Svoboda K. NMDA receptor subunit-dependent [Ca2+] signaling in individual hippocampal dendritic spines. J Neurosci. 2005;25:6037–6046. doi: 10.1523/JNEUROSCI.1221-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hall BJ, Ripley B, Ghosh A. NR2B signaling regulates the development of synaptic AMPA receptor current. J Neurosci. 2007;27:13446–13456. doi: 10.1523/JNEUROSCI.3793-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gray JA, et al. Distinct modes of AMPA receptor suppression at developing synapses by GluN2A and GluN2B: single-cell NMDA receptor subunit deletion in vivo. Neuron. 2011;71:1085–1101. doi: 10.1016/j.neuron.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee MC, Yasuda R, Ehlers MD. Metaplasticity at single glutamatergic synapses. Neuron. 2010;66:859–870. doi: 10.1016/j.neuron.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu Z, et al. Metaplastic regulation of long-term potentiation/long-term depression threshold by activity-dependent changes of NR2A/NR2B ratio. J Neurosci. 2009;29:8764–8773. doi: 10.1523/JNEUROSCI.1014-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.von Engelhardt J, et al. Contribution of hippocampal and extra-hippocampal NR2B-containing NMDA receptors to performance on spatial learning tasks. Neuron. 2008;60:846–860. doi: 10.1016/j.neuron.2008.09.039. [DOI] [PubMed] [Google Scholar]
- 13.Espinosa JS, Wheeler DG, Tsien RW, Luo L. Uncoupling dendrite growth and patterning: single-cell knockout analysis of NMDA receptor 2B. Neuron. 2009;62:205–217. doi: 10.1016/j.neuron.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barria A, Malinow R. NMDA receptor subunit composition controls synaptic plasticity by regulating binding to CaMKII. Neuron. 2005;48:289–301. doi: 10.1016/j.neuron.2005.08.034. [DOI] [PubMed] [Google Scholar]
- 15.Borrelli E, Nestler EJ, Allis CD, Sassone-Corsi P. Decoding the epigenetic language of neuronal plasticity. Neuron. 2008;60:961–974. doi: 10.1016/j.neuron.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levenson JM, Sweatt JD. Epigenetic mechanisms in memory formation. Nat Rev Neurosci. 2005;6:108–118. doi: 10.1038/nrn1604. [DOI] [PubMed] [Google Scholar]
- 17.Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 18.Ballas N, Mandel G. The many faces of REST oversee epigenetic programming of neuronal genes. Curr Opin Neurobiol. 2005;15:500–506. doi: 10.1016/j.conb.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 19.Schoenherr CJ, Anderson DJ. The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science. 1995;267:1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- 20.Qiang M, Rani CS, Ticku MK. Neuron-restrictive silencer factor regulates the N-methyl-D-aspartate receptor 2B subunit gene in basal and ethanol-induced gene expression in fetal cortical neurons. Mol Pharmacol. 2005;67:2115–2125. doi: 10.1124/mol.104.010751. [DOI] [PubMed] [Google Scholar]
- 21.Sasner M, Buonanno A. Distinct N-methyl-D-aspartate receptor 2B subunit gene sequences confer neural and developmental specific expression. J Biol Chem. 1996;271:21316–21322. doi: 10.1074/jbc.271.35.21316. [DOI] [PubMed] [Google Scholar]
- 22.Andres ME, et al. CoREST: a functional corepressor required for regulation of neural-specific gene expression. Proc Natl Acad Sci U S A. 1999;96:9873–9878. doi: 10.1073/pnas.96.17.9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bruce AW, et al. Genome-wide analysis of repressor element 1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) target genes. Proc Natl Acad Sci U S A. 2004;101:10458–10463. doi: 10.1073/pnas.0401827101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ooi L, Wood IC. Chromatin crosstalk in development and disease: lessons from REST. Nat Rev Genet. 2007;8:544–554. doi: 10.1038/nrg2100. [DOI] [PubMed] [Google Scholar]
- 25.Barria A, Malinow R. Subunit-specific NMDA receptor trafficking to synapses. Neuron. 2002;35:345–353. doi: 10.1016/s0896-6273(02)00776-6. [DOI] [PubMed] [Google Scholar]
- 26.Bellone C, Nicoll RA. Rapid bidirectional switching of synaptic NMDA receptors. Neuron. 2007;55:779–785. doi: 10.1016/j.neuron.2007.07.035. [DOI] [PubMed] [Google Scholar]
- 27.Matta JA, Ashby MC, Sanz-Clemente A, Roche KW, Isaac JT. mGluR5 and NMDA receptors drive the experience- and activity-dependent NMDA receptor NR2B to NR2A subunit switch. Neuron. 2011;70:339–351. doi: 10.1016/j.neuron.2011.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanz-Clemente A, Matta JA, Isaac JT, Roche KW. Casein kinase 2 regulates the NR2 subunit composition of synaptic NMDA receptors. Neuron. 2010;67:984–996. doi: 10.1016/j.neuron.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mierau SB, Meredith RM, Upton AL, Paulsen O. Dissociation of experience-dependent and -independent changes in excitatory synaptic transmission during development of barrel cortex. Proc Natl Acad Sci U S A. 2004;101:15518–15523. doi: 10.1073/pnas.0402916101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carmignoto G, Vicini S. Activity-dependent decrease in NMDA receptor responses during development of the visual cortex. Science. 1992;258:1007–1011. doi: 10.1126/science.1279803. [DOI] [PubMed] [Google Scholar]
- 31.Ku HY, Huang YF, Chao PH, Huang CC, Hsu KS. Neonatal isolation delays the developmental decline of long-term depression in the CA1 region of rat hippocampus. Neuropsychopharmacology. 2008;33:2847–2859. doi: 10.1038/npp.2008.36. [DOI] [PubMed] [Google Scholar]
- 32.Nase G, Weishaupt J, Stern P, Singer W, Monyer H. Genetic and epigenetic regulation of NMDA receptor expression in the rat visual cortex. Eur J Neurosci. 1999;11:4320–4326. doi: 10.1046/j.1460-9568.1999.00859.x. [DOI] [PubMed] [Google Scholar]
- 33.Philpot BD, Sekhar AK, Shouval HZ, Bear MF. Visual experience and deprivation bidirectionally modify the composition and function of NMDA receptors in visual cortex. Neuron. 2001;29:157–169. doi: 10.1016/s0896-6273(01)00187-8. [DOI] [PubMed] [Google Scholar]
- 34.Yu HB, Johnson R, Kunarso G, Stanton LW. Coassembly of REST and its cofactors at sites of gene repression in embryonic stem cells. Genome Res. 2011;21:1284–1293. doi: 10.1101/gr.114488.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roopra A, Qazi R, Schoenike B, Daley TJ, Morrison JF. Localized domains of G9a-mediated histone methylation are required for silencing of neuronal genes. Mol Cell. 2004;14:727–738. doi: 10.1016/j.molcel.2004.05.026. [DOI] [PubMed] [Google Scholar]
- 36.Bruce AW, et al. Functional diversity for REST (NRSF) is defined by in vivo binding affinity hierarchies at the DNA sequence level. Genome Res. 2009;19:994–1005. doi: 10.1101/gr.089086.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 38.Noh KM, et al. Repressor element-1 silencing transcription factor (REST)-dependent epigenetic remodeling is critical to ischemia-induced neuronal death. Proc Natl Acad Sci U S A. 2012;109:E962–E971. doi: 10.1073/pnas.1121568109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yeo M, Berglund K, Augustine G, Liedtke W. Novel repression of Kcc2 transcription by REST-RE-1 controls developmental switch in neuronal chloride. J Neurosci. 2009;29:14652–14662. doi: 10.1523/JNEUROSCI.2934-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Monyer H, Burnashev N, Laurie DJ, Sakmann B, Seeburg PH. Developmental and regional expression in the rat brain and functional properties of four NMDA receptors. Neuron. 1994;12:529–540. doi: 10.1016/0896-6273(94)90210-0. [DOI] [PubMed] [Google Scholar]
- 41.Liu D, Diorio J, Day JC, Francis DD, Meaney MJ. Maternal care, hippocampal synaptogenesis and cognitive development in rats. Nat Neurosci. 2000;3:799–806. doi: 10.1038/77702. [DOI] [PubMed] [Google Scholar]
- 42.Uchida S, et al. Early life stress enhances behavioral vulnerability to stress through the activation of REST4-mediated gene transcription in the medial prefrontal cortex of rodents. J Neurosci. 2010;30:15007–15018. doi: 10.1523/JNEUROSCI.1436-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Desai A, Turetsky D, Vasudevan K, Buonanno A. Analysis of transcriptional regulatory sequences of the N-methyl-D-aspartate receptor 2A subunit gene in cultured cortical neurons and transgenic mice. J Biol Chem. 2002;277:46374–46384. doi: 10.1074/jbc.M203032200. [DOI] [PubMed] [Google Scholar]
- 44.Huang Y, Myers SJ, Dingledine R. Transcriptional repression by REST: recruitment of Sin3A and histone deacetylase to neuronal genes. Nat Neurosci. 1999;2:867–872. doi: 10.1038/13165. [DOI] [PubMed] [Google Scholar]
- 45.Prybylowski K, et al. Relationship between availability of NMDA receptor subunits and their expression at the synapse. J Neurosci. 2002;22:8902–8910. doi: 10.1523/JNEUROSCI.22-20-08902.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guardavaccaro D, et al. Control of chromosome stability by the beta-TrCP-REST-Mad2 axis. Nature. 2008;452:365–369. doi: 10.1038/nature06641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Westbrook TF, et al. SCFbeta-TRCP controls oncogenic transformation and neural differentiation through REST degradation. Nature. 2008;452:370–374. doi: 10.1038/nature06780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang Z, et al. Deubiquitylase HAUSP stabilizes REST and promotes maintenance of neural progenitor cells. Nat Cell Biol. 2011;13:142–152. doi: 10.1038/ncb2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meaney MJ, Ferguson-Smith AC. Epigenetic regulation of the neural transcriptome: the meaning of the marks. Nat Neurosci. 2010;13:1313–1318. doi: 10.1038/nn1110-1313. [DOI] [PubMed] [Google Scholar]
- 50.Huot RL, Thrivikraman KV, Meaney MJ, Plotsky PM. Development of adult ethanol preference and anxiety as a consequence of neonatal maternal separation in Long Evans rats and reversal with antidepressant treatment. Psychopharmacology (Berl) 2001;158:366–373. doi: 10.1007/s002130100701. [DOI] [PubMed] [Google Scholar]
- 51.Paoletti P, Ascher P, Neyton J. High-affinity zinc inhibition of NMDA NR1-NR2A receptors. J Neurosci. 1997;17:5711–5725. doi: 10.1523/JNEUROSCI.17-15-05711.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.