Abstract
Biochemical regulatory networks governing diverse cellular processes such as stress-response, differentiation and cell cycle often contain coupled feedback loops. We aim to understand how features of feedback architecture, such as the number of loops, the sign of the loops and the type of their coupling, affect network dynamical performance. Specifically, we investigate how bistability range, maximum open-loop gain and switching times of a network with transcriptional positive feedback are affected by additive or multiplicative coupling with another positive or negative feedback loop. We show that a network’s bistability range is positively correlated with its maximum open-loop gain and that both the quantities depend on the sign of the feedback loops and the type of feedback coupling. Moreover, we find that the addition of positive feedback could decrease the bistability range if we control the basal level in the signal-response curves of the two systems. Furthermore, the addition of negative feedback has the capacity to increase the bistability range if its dissociation constant is much lower than that of the positive feedback. We also find that addition of a positive feedback to a bistable network increases robustness of its bistability range, whereas addition of a negative feedback decreases it. Finally, we show that the switching time for a transition from a high to a low steady state increases with the effective fold change in gene regulation. In summary, we show that the effect of coupled feedback loops on the bistability range and switching times depends on the underlying mechanistic details.
Keywords: feedback, coupling, bistability, cooperativity, open-loop gain, switching time, robustness
1. Introduction
Coupled positive and negative feedback loops are common features of biochemical networks [1–3]. Such loops have been identified in regulatory pathways that govern diverse cellular processes, such as stress-response [4], galactose signaling [5] and the cell cycle [6]. The dynamic consequences of feedback in simple genetic circuits were theoretically analyzed by Savageau [7–9]. Since then, these predictions have been extended and verified experimentally. It was demonstrated that negative feedback is likely to result in faster transient responses [10], robustness against the fluctuation of parameters [11] and dynamical stability [12]. On the other hand, positive feedback is likely to result in slower response times [13], increased noise [14] and possibly bistability [15].
Network bistability is defined as the ability of a system to exhibit two different stable steady states for the same biochemical conditions. For a system to be bistable, the underlying circuit must possess positive feedback along with some type of non-linear behavior [16]. Previously, it was shown that the stability properties of a positive-feedback system can be deduced by analyzing the behavior of the open-loop system for which the feedback is blocked but the system is subject to tunable input instead. [17]. As long as the open-loop system is monotone (lacks negative feedback loops; excluding self-feedback) and displays sigmoidal input-output curve, the closed system is guaranteed to be bistable for some range of feedback strengths [17]. Furthermore, information about closed system’s stability is determined by analyzing the slope of the steady state input-output curve of the open-loop system at fixed points. For such analysis, it is often useful to analyze the slope of this curve in log-log coordinates. This is a local quantity defined for a given input signal and is referred to as open-loop gain (LG). Previously, we have shown that the necessary condition for bistability in a network depends on the maximum value of LG (mLG) – a global quantity computed over the range of signals [18].
The simplest transcriptional regulatory network that can exhibit bistability is an autoregulatory circuit, in which a transcription factor (TF) positively regulates the production of its own mRNA. Such single feedback autoregulatory circuits play a crucial role in several cellular processes, such as stress-response [19], differentiation [20] and cell cycle [21]. These observations raise the following question - if a circuit with a single positive feedback can mediate diverse cellular responses, then why do some biological systems have more than one feedback? Several answers have been suggested by past studies, some of which we briefly discuss here. First, duplicate feedback loops provide robustness against perturbations simply via redundancy of their effect [22]. For example, a genetic mutation or fluctuation disabling one of the feedback loops may not drastically affect the network performance if there is another loop. Second, the presence of fast and slow positive feedback loops can result in a “dual time” monostable or bistable switch that is both rapidly inducible and resistant to noise during signaling [23, 24]. Third, positive feedback loops can interact to increase effective cooperativity thereby increasing the range of conditions at which bistability is observed [25]. Fourth, coupled positive feedback loops can give rise to multistable decision switches that can robustly discriminate differences in strength, duration and timing of signal [26]. On the other hand, interlinked positive and negative feedback loops offer flexibility in the network’s dynamical performance as modulating their relative strengths enables transitions between bistable and oscillatory regime [27, 28]. The presence of a positive feedback in an oscillatory network (designed primarily using negative feedback loops) increases the robustness of the oscillations and allows for greater tunability [29]. The presence of a negative feedback in a bistable network produces faster reversible transitions and transient switching responses [30].
Here, we focus on how a single transcriptional positive feedback interacts with another positive or negative feedback loop to affect dynamical properties of a bistable network. Hereafter, the interaction between two feedback loops is referred to as ‘coupling’ and depending on the number of promoters in the genetic architecture it can be classified into additive (two promoters with one operator site each) or multiplicative (one promoter with two operator sites). Previously, it was shown that additive coupling between positive feedback loops always increases the bistability range and switching times [31]. In contrast, additive coupling between a positive and a negative feedback decreases the bistability range [28]. However, these results are based on a limited choice of parameter values and did not explicitly consider the possibility of multiplicative coupling. In this work, we investigate how the bistability range and the switching time of a network with an autoregulatory positive feedback are affected by both additive and multiplicative coupling with another positive or negative feedback loop. In general, we aim to understand how the features of feedback architecture, such as the number of loops, the sign of the loops and the type of their coupling affect a network’s dynamical performance.
2. Mathematical model for networks with coupled feedback loops
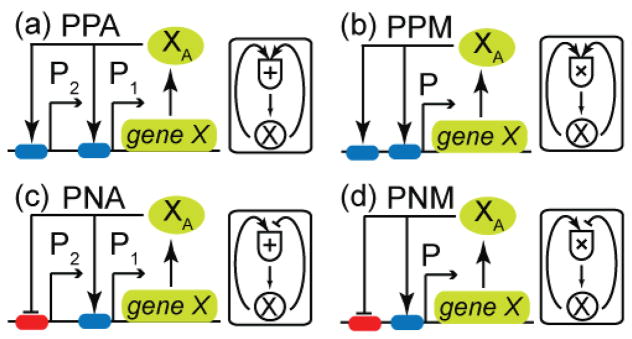
The type of coupling between feedback loops is a consequence of the underlying genetic architecture and as discussed below depends on the number of promoters. Here we consider four different genetic architectures which result in additively or multiplicatively coupled positive/positive and positive/negative feedback loops (figure 1). We built ordinary differential equation based models for these networks assuming mRNA concentrations are at quasi-steady state to understand the effects of the number of loops, the sign of the loops and the type of their coupling. The resulting equations are given below along with some key attributes, while detailed derivations are presented in the Supplementary Information.
Figure 1.
Genetic architectures and corresponding transcriptional logic depicting the coupling of feedback loops. (a) PPA: a gene network depicting the additive coupling of two positive feedback loops. The transcription of gene X is controlled from two independent promoters, P1 and P2, both of which possess upstream operator sites for the TF XA. Bound TF upregulates transcription from the two promoters, thereby giving rise to two positive feedback loops. (b) PPM: a gene network depicting multiplicative coupling of two positive feedback loops. The transcription of gene X is controlled from one promoter P. TF XA regulates transcription by binding to the operator sites upstream of the promoter, thereby resulting in two positive feedback loops such as in the case of additive coupling. (c) PNA: a gene network depicting additive coupling of a positive and a negative feedback loop. The transcription of gene X is controlled from two independent promoters P1 and P2, both of which possess upstream operator sites for the TF XA. Bound TF upregulates and downregulates the transcription from the promoters P1 and P2, respectively, thereby giving rise to positive and negative feedback loops. (d) PNM: a gene network depicting multiplicative coupling of a positive and a negative feedback loop. The transcription of gene X is controlled by one promoter P. TF XA regulates transcription by binding to the operator sites upstream of the promoter, thereby resulting in positive and negative feedback loops similar to the case of additive coupling.
2.1 Additively coupled positive/positive feedback loops (PPA-network)
Figure 1(a) depicts a scenario in which the transcription of gene X is controlled from two independent promoters, P1 and P2. Post-translationally, an activating signal converts the protein X into an active form XA. This form functions as a TF and positively regulates transcription of its own gene by binding to the operator sites upstream of the two promoters, thereby resulting in two positive feedback loops. An example of such a regulatory architecture is the phoPR operon in Bacillus subtilis whose transcription is controlled by two σA-dependent promoters both regulated by TF PhoP~P [32]. We built a detailed biochemical model for this network and found that the coupling between the feedback loops results in an additive relationship between them. The following equations between the normalized concentrations of the active transcription factor (xA) and the total protein (xT) display the additive effect:
| (1) |
| (2) |
These equations are in dimensionless form -- concentrations are normalized with the dissociation constant of TF binding to the first operator site, whereas dimensionless time τ is obtained by normalizing time with the protein degradation rate. All parameters in the above equations are positive and have a clear meaning: b is the combined basal protein synthesis rate from two promoters, f1 and f2 are the fold changes in protein synthesis resulting from each operator site, n1 and n2 are the respective Hill coefficients for the two positive feedback loops (number of molecules of XA required to activate each operator site), and l is the ratio of the TF dissociation constants of the second site to that of the first site. For simplicity, the activation of TF is modeled with a linear equation (2), which expresses the concentration of active TF as a fraction of the concentration of total protein. This linear relation corresponds to first order activation and deactivation reactions (see Supplementary Information). Biologically, this can occur when activation or deactivation is achieved via binding to a small molecule present in excess or via unsaturated phosphorylation-dephosphorylation reactions. The parameter γ represents the fraction of the total TF that is active and thus has a value between 0 and 1. Henceforth, it will be considered as the activation signal for the network.
2.2 Multiplicatively coupled positive/positive feedback loops (PPM-network)
Figure 1(b) depicts an alternate scenario in which the transcription of gene X is controlled by one promoter P with two operator sites that bind an active form XA and positively regulate transcription rate. Consequently, there are two positive feedback loops, similar to section 2.1 but with different coupling. In B. subtilis TF DegU~P controls its own transcription in a similar fashion by to two operator sites upstream of its promoter [33]. We built a detailed biochemical model for this network with one promoter. Under the two assumptions stated below we found that the coupling between the feedback loops results in a multiplicative relationship between them. First, we assume TF molecules binding to different operator sites do not interact, i.e. bind non-cooperatively. This assumption leads to an additive free energy of binding and as a result the dissociation constant of the TF molecules from the two operator sites is the product of the dissociation constants from each individual operator site. Second assumption is that TF molecules bound to different operator sites interact with RNA polymerase independently and, as a result, the fold change in the rate of protein synthesis from the two operator sites is the product of the fold changes from the individual operator sites. Under these assumptions, equation (1) is replaced with the following equation, which displays the multiplicative effect:
| (3) |
We assume the same mechanism for linear activation as in equation (2). All parameters have the same meanings as in section 2.1, except b, which is now the basal protein synthesis rate from single promoter.
Note that equation (3) is derived assuming non-cooperative binding of TFs. However, cooperative binding of TFs at different operator sites is possible in the case of coupled positive/positive feedback loops. For example, in bacteriophage λ the CI repressor positively regulates its own transcription from promoter PRM by cooperatively binding to the operator sites OR1 and OR2 [34]. Therefore, we also developed an alternate framework which takes cooperativity into account; however, our main conclusions do not depend on it (see Supplementary Information).
2.3 Additively coupled positive/negative feedback loops (PNA-network)
Figure 1(c) depicts a scenario in which the transcription of gene X is controlled from two independent promoters, P1 and P2. The two-promoter situation is similar to figure 1(a); however, in this case, the active form XA regulates the transcription both positively and negatively from the promoters P1 and P2, respectively, thereby resulting in positive and negative feedback loops. For example, in B. subtilis TF Spo0A~P positively and negatively regulates its own transcription through σH and σA-dependent promoters by binding to different operator sites [35]. As in section 2.1, we built a detailed biochemical model for this network and found that the coupling between the two feedback loops results in an additive relationship between them, as depicted in the following equation:
| (4) |
We assume the same mechanism for linear activation as in equation (2). Compared to section 2.1, some of the parameters now have slightly different meanings, as noted. f2 is now the fold change reduction in protein synthesis, n2 is the Hill coefficient for the negative feedback. All parameters are positive. Additionally, to ensure that the rate of change of protein (dxT/dτ) in the above equation remains positive in the limit of non-saturated positive feedback and saturated negative feedback (l ≪ xA ≪ 1), we constrain f2 to be less than one.
2.4 Multiplicatively coupled positive/negative feedback loops (PNM-network)
Figure 1(d) depicts a scenario in which the transcription of gene X is controlled from one promoter P. The one promoter situation is similar to figure 1(b); however, in this case, the active form XA both positively and negatively regulates its own transcription by binding to operator sites that are upstream of the promoter, thereby resulting in positive and negative feedback loops. In E. coli PapB autoregulates itself in a similar manner – it activates transcription by binding to an upstream operator site, while it represses transcription by binding to another which includes the position of transcription initiation by RNA polymerase [36]. As in section 2.2, we built a detailed biochemical model for this network, to find that the coupling between the two feedback loops results in a multiplicative relationship between them, as depicted in the following equation:
| (5) |
We assume the same mechanism for linear activation as in equation (2), and all of the parameters have the same meaning as in section 2.3.
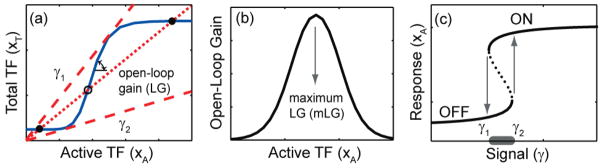
Note that substituting f2 = 0 in equation (1) (or in equations (3)–(5)) removes the second positive or negative feedback, whereby the system reduces to a single positive feedback network (P-network). The bistability properties of this network are depicted in figure 2. Steady states of the network are determined by the intersections between the system’s nullclines: open-loop transcriptional transfer function (production term in equation (1) after substituting f2 = 0, solid curve) and the dotted line (equation (2)), depicting the input signal (figure 2(a)). The two dashed lines with slopes γ1−1 and γ2−1 represent the boundaries of the bistability range. Any signal inside this range (dotted line in figure 2(a)) will result in three intersections with the transcriptional transfer function, indicating the existence of two stable steady states and an unstable steady state. LG in figure 2(a) refers to the slope of the transcriptional transfer function in the log-log coordinates. For a sigmoidal transcriptional transfer function (as in figure 2(a)), LG depends on the given input signal through the concentration of TF XA and attains its maximum value in the interval γ1 <γ < γ2 (figure 2(b)). Figure 2(c) shows the steady-state signal-response curve (one-parameter bifurcation diagram) containing a range of signals for which two different steady states are possible. This curve consists of three branches: two of the branches represent the stable steady states (solid lines), and the intermediate dotted branch represents the unstable steady state (dotted line). At the boundaries of the bistability range, the steady-state response of the system discontinuously jumps from one state to the other (see arrows in figure 2(c)).
Figure 2.
Dynamical characteristics of a bistable P-network. (a) Steady states of the network are determined by the intersections between the transcriptional transfer function (solid curve) and the dotted line depicting the input signal. Dashed lines parameterized by signals γ1 and γ2 represent the boundaries of the bistability range. The boundaries of the bistability range are inverse of the slopes of the dashed lines. The dotted line represents a signal inside the bistability range and results in three intersections with the transcriptional transfer function, corresponding to two stable steady states (filled circles) and an unstable steady state (empty circle). (b) The slope of the transcriptional transfer function represents the system’s open-loop gain which depends on the input signal through the concentration of TF XA and attains its maximum value in the interval γ1 < γ < γ2. (c) The steady-state signal-response curve shows a range of signals (horizontal bar) for which two different steady-state responses are possible. At the boundaries of the bistability range, the steady-state response of the system discontinuously jumps from one state to the other (arrows). The two solid curves represent the stable steady states (OFF and ON), which are separated by the unstable steady state (dotted curve).
Although the above models in sections 2.1–2.4 are developed for different variants of an autoregulatory network with a single TF, they can be extended to include multiple TFs. For example, let us assume that the first positive feedback in the scenarios discussed above is mediated through another gene Y, such that genes X and Y mutually activate each other’s transcription. In this case, gene Y serves as an intermediate node, which increases the dimensionality of the equations at hand. Nevertheless, this scenario can be reduced to the form of equations discussed above by assuming a quasi-steady state approximation for . This assumption will allow us to substitute TF YA as a function of TF XA in the differential equation governing xT, thereby reducing the system to one dimension. The quasi-steady state approximation discussed above may not always be applicable for additional TFs, but is sometimes useful for example when the second TF has a faster degradation rate. Moreover, this assumption does not affect the condition for bistability (LG > 1) which has been derived for systems with indirect feedback loops and is hence applicable for networks with multiple TFs [17].
Methods
We sampled 10,000 parameter sets (See Supplementary Information for details) for which the following networks were simultaneously bistable: i) P, PPA and PPM, and ii) P, PNA and PNM. For each parameter set, we used the command-line bifurcation package MATCONT [37] to obtain the bifurcation diagram. Subsequently, MATLAB was used to analyze the sampling data and to numerically compute bistability range, mLG, effective fold change and switching times. Bistability range is defined as the ratio between the higher (γ2) and lower (γ1) thresholds of the signal at which the system, respectively, switches from an OFF state to an ON state, and vice versa. LG is defined as the slope of transcriptional transfer function at steady state (xT = U(xA)) in log-log coordinates ( ), while mLG is the maximum value of this slope. Effective fold change is defined as the ratio of the maximum and basal total protein levels. The effective fold change for P-network is equal to the fold change in protein synthesis due to the feedback, i.e. f1, but in the case of remaining four networks it will be a function of the two fold changes f1 and f2. A network’s switching time from an OFF to an ON state is defined as the time that is taken to make the transition between the states when there is a step increase in the activating signal (γ) from 0 to 1. Similarly, the switching time from an ON to an OFF state is defined for the reverse transition.
We adopt the methodology of [38] to compute a network’s robustness score which represents the fraction of sampled parameter sets that result in bistability range greater than 5. The mean and standard deviation for each network’s robustness score was obtained using bootstrapping techniques as follows. For each network, the distribution of bistability ranges (original dataset obtained after parameter sampling) was used to produce 1000 resamples of the original dataset using bootstrp function in MATLAB. Each resample is created by random sampling with replacement from the original dataset, and its size is equal to the size of the original dataset (10,000 in this case). Next, for each resample the robustness score was computed as described above. Finally, the robustness scores from 1000 resamples were used to calculate the mean and standard deviation of a network’s robustness score. As an alternate robustness measure, we also compute the sensitivity of bistability range to fold change f1, the common parameter across all networks, . Sensitivity for P- network was computed analytically using its bistability range expression derived in Supplementary Information. For other networks MATCONT was used to obtain bistability ranges when f1 was varied by ±10% and ± 20% while other parameters are kept constant. These values were used to calculate the first derivative in the sensitivity expression using the five-point center difference formula and thereby compute the sensitivity.
3. Results
3.1 Bistability range positively correlates with maximum open-loop gain
The deciding role played by mLG in determining the necessary condition for bistability [18] led us to analyze whether it also controls the bistability range – the range of signal levels over which a system displays two stable states (see figure 2). First, we examine P-network (case f2 = 0 discussed in section 2.1) and analytically compute the expressions for mLG and bistability range (see Supplementary Information). We find that both of these quantities increase with the Hill coefficient (n1) and the fold change in protein synthesis (f1) (figures S1(a) and (b)). This confirms our initial supposition that bistability range is controlled by mLG.
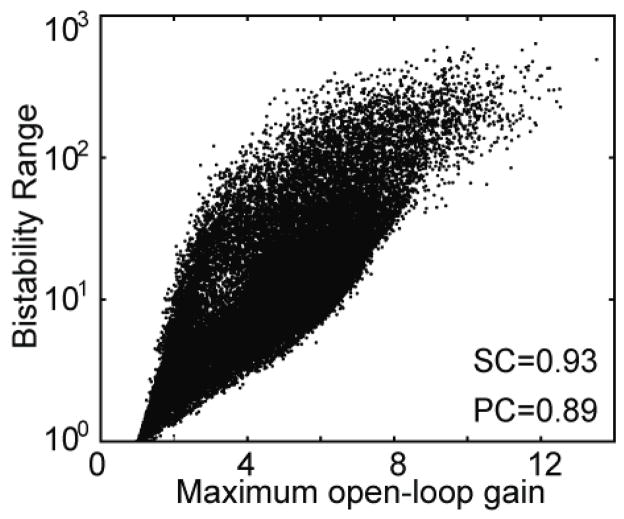
Subsequently, we extend our analysis to include PPA, PPM, PNA and PNM networks, which are discussed in sections 2.1–2.4. We numerically compute bistability ranges and mLGs for these networks because their model equations are analytically intractable. We sample parameter sets for all five networks (see Methods for details), and for each parameter set, we numerically compute the two aforementioned quantities. Figure 3 shows the result of this analysis, which leads us to conclude that mLG and bistability range of a network are highly correlated (Spearman correlation coefficient=0.93, Pearson correlation coefficient=0.89). The reason for this high correlation becomes clear by analyzing a typical transcriptional transfer function, as shown in figure 2(a). The steepest slope of this transfer function in log-log coordinates represents the mLG, which governs the separation between the two tangents (dashed lines) that determine a network’s bistability range. Therefore, an increase in mLG increases the separation between the two tangents, eventually resulting in a larger bistability range.
Figure 3.
Bistability range is positively correlated with maximum open-loop gain. Parameter sets were sampled for P, PPA, PPM, PNA and PNM bistable networks, and the corresponding bistability ranges and maximum open-loop gains were computed (see Methods for details). High Spearman and Pearson correlation coefficients demonstrate a strong correlation between a network’s bistability range and its maximum open-loop gain.
The large variability in figure 3 is a consequence of different effective fold changes (ratio of maximal and basal total protein levels) for parameter sets that result in the same mLG. Since the effective fold change controls the height of the transcriptional transfer function, it also plays a role in determining the points of tangency that describe the bistability range. Thus, between two parameter sets with same mLG, the set with a larger effective fold change will lead to a higher bistability range as it will further separate the two tangents. Consequently, the variability in the sampling results can be explained by the fact that parameter sets with same mLG could result in different bistability ranges for different fold-change values. Therefore, we decided to check whether the bistability range also depends on the effective fold change. We use the data obtained from parameter sampling to compute the effective fold change for each of the five networks. Surprisingly, we find that the bistability range is somewhat weakly correlated with the effective fold change, as shown by the low Spearman and Pearson correlation coefficients (figure S2).
3.2 Network’s open-loop gain is determined by the sign of feedback loops and the type of coupling
The above analysis suggests that mLG is an important property of a bistable network as it not only determines the possibility of bistability but also determines the bistability range. LG can sometimes be computed independently of the choice of parameters and transcriptional transfer functions. For example, let the following equation describe the rate of change of the total protein in the case of coupled feedback loops:
| (6) |
For additive coupling
| (7) |
whereas for multiplicative coupling
| (8) |
here F(xA) and G(xA) represent general transcriptional transfer functions for the two feedback loops. Using the formula for LG defined in Methods we obtain the following expressions for LGs for the two types of coupling:
| (9) |
| (10) |
where and are the LGs from the first and second feedback, respectively, and represents the ratio between the amount of total protein produced from the first feedback loop and the coupled feedback loops.
Because the expressions in equations (9) and (10) are general, they can be converted to the LGs for the five networks discussed in section 2, after substituting corresponding F(xA) and G(xA) from equations (1), (3)–(5). Through this analysis, we expect to unravel the effects of the sign of feedback loops and the type of coupling on a network’s LG.
1. Open-loop gain for PPA-network
The LG for PPA-network (LGPPA) is given by equation (9), where and are, respectively, the transcriptional transfer functions (defined in equation (1)) for the first and second positive feedback loops and ω<1. As LGPPA is a weighted average of two LGs, it lies between those two quantities. If LGF > LGG, then equation (9) leads us to conclude that additive coupling with a second feedback that has a smaller LG (LGG) decreases the LG of P-network (LGP) which corresponds to the limiting case G = 0 and ω = 1 in equation (9) :
| (11) |
The opposite is true in the case LGF < LGG. Thus, if second feedback’s LG is smaller than that of the first one, then additive coupling between the two feedback loops always result in a smaller LG compared to a single positive feedback network.
2. Open-loop gain for PPM-network
The LG for PPM-network (LGPPM) is given by equation (10), in which F (defined in section 3.2.1) and are, respectively, the transcriptional transfer functions (defined in equation (3)) for the first and second positive feedback loops. Comparing equations (10) and (11) shows that LGPPM > LGF. Hence, we conclude that multiplicative coupling of positive feedback loops always results in a larger LG compared to a single positive feedback network.
3. Open-loop gain for PNA-network
The LG for PNA-network (LGPNA) is given by equation (9), in which F (defined in section 3.2.1) and are, respectively, the transcriptional transfer functions (defined in equation (4)) for the positive and negative feedback loops and ω > 1 (since G < 0). Subtracting equation (11) from equation (9), we obtain LGPNA − LGP = (ω−1)(LGF − LGG). Now, if LGF > LGG, then we conclude that additive coupling with a negative feedback that has a smaller LG (LGG) increases the LG of P-network (LGP). The opposite is true in the case LGF < LGG. Thus, if the negative feedback’s LG is smaller than that of the positive feedback, then additive coupling between the two feedback loops always results in a larger LG compared to a single positive feedback network.
4. Open-loop gain for PNM-network
The LG for PNM-network (LGPNM) is given by equation (10), in which F (defined in section 3.2.1) and are, respectively, the transcriptional transfer functions (defined in equation (5)) for the positive and negative feedback loops. However, LGG < 0 because . Hence, equation (10) results in LGPNM < LGF, which allows us to conclude that multiplicative coupling of positive/negative feedback loops always results in a smaller LG compared to a single positive feedback network.
To summarize, we combine the conclusions from sections 3.2.1–3.2.4 in the following inequality, which depicts the relationship between the LGs for the five networks:
| (12) |
In order to increase the LG of P-network, we can add a i) positive feedback that is multiplicatively coupled to the first one, or ii) positive feedback that is additively coupled and has an LG larger than the first one, or iii) negative feedback that is additively coupled and has an LG smaller than the first one. On the other hand, decrease in the LG could be achieved by replacing positive feedback by a negative feedback (in i and ii) and vice versa (in iii). We note that the theoretical results obtained in this section are independent of parameter values and actual functional forms of F and G, and are therefore quite general in nature. Additionally, we numerically compute mLG for each parameter set during the parameter sampling discussed above. The sampling results (figure S3) are in accord with the theoretical results summarized in the above inequality.
3.3 Network’s bistability range is determined by the sign of feedback loops and the type of coupling
The preceding sections revealed that a network’s bistability range is highly correlated with mLG, which itself depends on the sign of feedback loops and the type of coupling. Hence, we expect that the bistability range also depends on these two network features. We therefore expand on the previous studies [28, 31] to investigate the relationship between the bistability range and the features of the feedback architecture over a wide range of parameter values. To this end, we sample parameter sets for all five networks (see Methods for details). In contrast with previous studies [28, 31], we include a non-zero basal protein production rate, explore the effects of multiplicative coupling and also consider distinct dissociation constants of TF binding for the different operator sites.
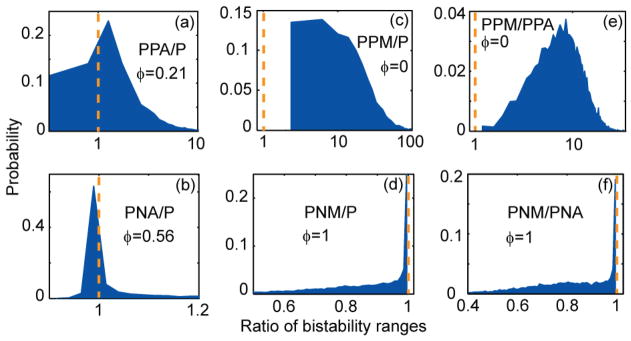
To understand how coupled feedback loops control the bistability range, we compare bistability ranges in networks with i) one feedback versus two feedback loops, ii) additive versus multiplicative coupling, and iii) positive versus negative feedback loops. Sampling results for these comparisons are presented in figure 4 and are discussed below.
Figure 4.
Comparison of bistability ranges for networks with different feedback architecture. Panels represent histograms of the ratio of the bistability ranges for networks (a) PPA and P, (b) PNA and P, (c) PPM and P, (d) PNM and P, (e) PPM and PPA, (f) PNM and PNA. φ represents the fraction of sampled parameter sets for which the ratio is less than one. (c)–(f) Histograms lying entirely on the right or left of the dashed line (for which ratio=1)indicate that one of the networks has a larger bistability range.(a)–(b) Depending on parameters, either of the networks can achieve a larger bistability range.
Additive coupling with another positive or negative feedback can either increase or decrease P- network’s bistability range. Figures 4(a) and 4(b) present comparisons between the bistability ranges of a single positive feedback network and additively coupled positive/positive or positive/negative feedback loops. Histograms show that, irrespective of the sign of second feedback, additive coupling has the capacity to both increase and decrease the bistability range of P- network. This is in contrast with previous research which showed that the second positive or negative feedback respectively increases or decreases the bistability range. We discuss counterexamples to this original claim in the next section.
Multiplicative coupling with another positive or negative feedback always increases or decreases P-network’s bistability range. Figures 4(c) and 4(d) present comparisons between the bistability ranges of a single positive feedback network and multiplicatively coupled positive/positive or positive/negative feedback loops.. Histograms show that multiplicative coupling always increases the bistability range of P-network if the second feedback is positive, and decreases the bistability range if the second feedback is negative.
PPM-network has larger bistability range, whereas PNM-network has smaller bistability range, compared to analogous additively coupled system. Figures 4(e) and 4(f) present comparisons between the bistability ranges of additively and multiplicatively coupled positive/positive or positive/negative feedback loops. Histograms show that, when the coupling between two positive feedback loops is changed from additive to multiplicative, the bistability range increases. However, the same modification in the case of coupled positive/negative feedback loops decreases the bistability range.
We summarize the conclusions in the following inequality, which governs the relationship between the bistability ranges for the five networks:
| (13) |
Comparing equations (13) and (12) further confirms that bistability range and mLG are correlated. Thus, if we aim to increase or decrease P-network’s bistability range, we can add a multiplicatively coupled positive or negative feedback, respectively. Additive coupling can also increase or decrease the bistability range by employing the mechanisms discussed below.
3.4 Positive feedback can decrease, while negative feedback can increase bistability range
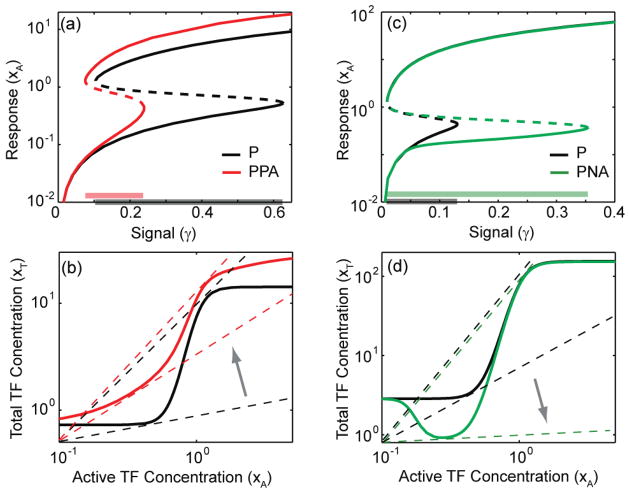
Parameter sampling shows that in contrast to previous findings [28, 31] adding a second positive feedback to P-network can decrease the bistability range, whereas adding a negative feedback can increase the bistability range (figure 5). In figure 5(a), the steady-state signal-response curves for P and PPA networks reveal that the latter has a smaller bistability range (compare corresponding horizontal bars on the x axis). This result can be explained by inspecting the transcriptional transfer functions in figure 5(c), which show that mLG for PPA-network is smaller than that of P-network (compare the maximum slopes of the two curves). The reduction in mLG causes the higher signal boundary of the bistability range to move significantly (see arrow), thereby resulting in a smaller bistability range.
Figure 5.
Positive feedback can decrease, while negative feedback can increase the bistability range. Steady-state signal-response curves for networks (a) P and PPA, and (c) P and PNA show that the addition of a positive and negative feedback has the capacity to, respectively, decrease and increase the bistability range (compare corresponding horizontal bars on the x-axis). (b) Steady-state curves (similar to figure 2(a)) for panel (a) show that maximum open-loop gain for PPA-network is smaller than that of P-network. This causes the higher threshold (γ2 in figure 2(a)) to decrease significantly (see arrow; the slope of the dashed line is the inverse of the threshold), resulting in a smaller bistability range. (d) Steady-state curves for panel (c) show that PNA-network has a non-monotonic response. This causes the higher to increase significantly (see arrow), resulting in a larger bistability range. Parameters (panels (a) and (b)): b = 0.73, f1 = 18.52, f2 = 20.48, n1 = 7.42, n2 = 1.65, l = 2.06. Parameters (panels (c) and (d)): b = 2.86, f1 = 53.2, f2 = 0.69, n1 = 6.89, n2 = 9.68, l = 0.17.
In figure 5(b), the steady-state signal-response curves for P and PNA networks reveal that the latter has a larger bistability range (compare corresponding horizontal bars on the x axis). This result is also explained by inspecting the corresponding transcriptional transfer functions in figure 5(d). The presence of negative feedback in the PNA-network results in a non-monotonic response in the transcriptional transfer function. This response is produced because the dissociation constant of TF binding for the negative feedback operator site is much lower than that the positive feedback operator site. Correspondingly, parameter l, the ratio between the two dissociation constants, is much smaller than 1 (l = 0.17). The non-monotonic response causes the higher signal level boundary of the bistability range to move significantly (see arrow), thereby resulting in a larger bistability range.
3.5 Coupling of feedback loops affects robustness of network’s bistability range
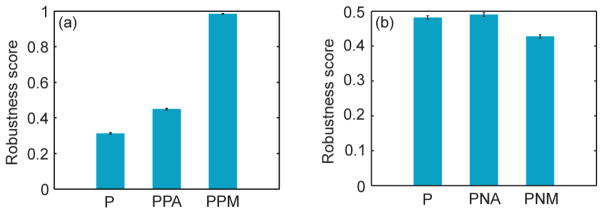
Although the results summarized in equations (12) and (13) describe important relationships between the bistability ranges and LGs for the five networks, these equations do not reveal if the network with coupled feedback loops has any performance advantage over a single positive feedback network. For example, single positive feedback can achieve very large bistability range and mLG if it has sufficiently high fold change and hill coefficient (figures S1(a) and (b)), and does not necessarily need a multiplicatively coupled second positive feedback. To understand possible performance advantages of multiple feedback loops, we adopt the methodology of [38] to quantify how strongly bistable a network is by computing robustness scores. These scores represent the fraction of sampled parameter sets that result in robust network bistability (bistability range greater than 5). We find that an additively coupled second positive feedback as in the PPA-network increases the robustness score as compared to P-network(figure 6(a)). Moreover, changing the type of coupling to multiplicative (PPM-network) further increases the robustness score (figure 6(a)). On the other hand, a multiplicatively coupled negative feedback as in the PNM-network decreases the robustness score as compared to P-network. (figure 6(b)). However, an additively coupled negative feedback (PNM-network) does not affect its robustness score (figure 6(b)).
Figure 6.
Positive feedback increases while negative feedback decreases robustness of network’s bistability range. Robustness scores represent the fraction of sampled parameter sets that result in robust network bistability (bistability range > 5, see Methods for details). (a) An additively coupled second positive feedback increases the robustness of P-network. Changing the type of coupling to multiplicative further increases the robustness. (b) An additively coupled negative feedback does not affect the robustness of P-network, whereas a multiplicatively coupled negative feedback decreases it.
The measure of robustness described above was based on the number of sampled parameter sets, some with very different bistability ranges. In addition, we were interested in knowing the differences between networks with single and coupled feedback loops for a particular parameter set that resulted in the same bistability range. Specifically we asked the question – what is the advantage of a network with coupled feedback loops if we can achieve the same bistability properties with a single positive feedback network? To answer this we first selected all the parameter sets for which the bistability range and mLG of P-network are equal to that of the network with coupled feedback loops. We did not include PPM-network in this analysis as we could not find any parameter set in our sampling results which matched the above criteria. For the PPA, PNA and PNM network we calculated the sensitivity of bistability range to fold change f1 (see Methods for details). We find that irrespective of the sign of feedback adding a second loop to P-network can reduce this sensitivity (figure S4). Thus, in networks with coupled feedback loops bistability range is more robust to parameter variation.
3.6 Examining the dependence of switching times on number, sign and coupling of feedback loops
In a monostable system, positive feedback results in slower response times, whereas negative feedback leads to faster response times [10, 39] (response time is defined as the time it takes to reach half-way between the initial and final protein levels). A more interesting dynamical property to study in the case of a bistable system is the time that it takes for the system to switch between the two steady states when exposed to a step increase or decrease in the signal. In light of the dependence of response times for monostable systems on the sign of feedback loops [10, 39], we expect that the addition of another positive or negative feedback to P-network may cause the network’s switching times to increase or decrease, respectively.
OFF to ON switching time displays no clear dependence on the sign of feedback loops and the type of coupling
Parameter sampling results show that any of the four networks with coupled feedback loops can have faster or slower switching time compared to a single positive feedback network (figure S5). Thus, OFF to ON switching time displays no clear dependence on the sign of feedback loops and the type of coupling. The variability in switching times for any of the five networks is a consequence of different OFF to ON switching thresholds for different parameter sets. Notably, for a bistable system, the switching time depends on the signal strength which in turn determines the distance between the input signal and the switching threshold [40]. For example, if a bistable system is in the OFF state and we provide two step-increases in the signal γL and γH > γL such that both cross the switching threshold (γ2, see figure 2(c)), then the signal closer to the threshold (γL in this case) will result in slower dynamics.
ON to OFF switching time is determined by the network’s effective fold change
A step decrease in the signal from 1 to 0 instantly makes the active TF concentration zero (see equation (2)). This reduces the production term in the differential equations (1), (3)–(5) to basal synthesis level, which is same for all five networks. As a result, during the transition, the protein production term remains constant and does not depend on the active TF concentration. Moreover, since the protein degradation rate is identical in all five networks, the switching time depends only on the pre-transition (ON state) total protein levels. Thus, larger the effective fold change for a network, greater is the separation between the ON and OFF states and, hence, slower are the switching times. Effective fold changes are in turn affected by the sign of feedback loops and the type of coupling. Consequently, ON to OFF switching time depends on network’s feedback structure, but through a combined parameter --effective fold change.
Not surprisingly, parameter sampling results show that ON to OFF switching time depends on the sign of feedback loops and the type of coupling (figure S6). As compared to P-network, PPA and PPM networks always increases the switching time, whereas PNA and PNM networks always decreases the switching time. As expected ON to OFF switching time is highly correlated with the effective fold change (figure S7). Subsequently, we compute the analytical expression for ON to OFF switching time and show that it is determined only by the effective fold change (See Supplementary Information). The effective fold changes for the five networks are related as follows:
| (14) |
Thus, based on analytical expression and sampling results ON to OFF switching times for the five networks are related as follows:
| (15) |
4. Discussion
Complex cellular behavior is a result of interactions between the biochemical molecules that mediate responses to external stimuli. Therefore, to understand cellular behavior, one must examine the network of interactions and their dynamical properties. This investigation is further simplified by focusing on network motifs, i.e., a set of recurring regulatory interactions [41]. The most commonly studied network motif is a feedback loop for which dynamical properties in isolation are well known. Feedback loops are often coupled together suggesting that these networks may possess a performance advantage over networks with single feedback loops. In this work, we analyze how bistability range and switching times of a network with a transcriptional positive feedback are affected by additive or multiplicative coupling with another positive or negative feedback loop.
We show that mLG is highly correlated with the bistability range. Hence, experimental perturbations that increase mLG of a bistable network will, in general, lead to an increase in the bistability range. Moreover, a system’s stability properties could be inferred by analyzing LGs [17]. For example, in figure 2(a), the stability of intersection points of transcriptional transfer function (solid curve) and dotted line depicting input signal can be determined by comparing the LGs of the two curves at those points. If the product of the LGs of the two curves is less than one, then the intersection point is stable (filled circles); otherwise, it is unstable (empty circle). A possible experimental manipulation facilitating LG measurement for the transcriptional transfer function involves replacing the native copy of a gene under positive feedback by a transcriptional-fusion reporter, and integrating it somewhere else on the chromosome under the control of an inducible promoter. In such a synthetic construct, we can measure the promoter activity as fluorescence readout at different inducer (signal) levels. Finally, the slope of the log-log plot between the fluorescence readout and signal will give LGs. Thus, without a detailed understanding of the biochemical mechanism that produces the input-output curve, we have some information about the system’s stability and its bistability range. This type of analysis could prove to be useful, because very rarely detailed mechanisms associated with a biochemical network are available. Additionally, we analytically show that the LG and the bistability range for increase with increases in the fold change and with increases in the Hill coefficient. Our results have general implications in synthetic biology, as a useful guide on which parameters can be tuned experimentally for the optimization of a bistable switch.
Earlier models of coupled positive feedback loops have focused only on additive coupling [31]. In the case of additive coupling the network’s LG is a weighted average of the individual LGs of the two loops. Thus, adding a second feedback whose LG is smaller than that of the first feedback will reduce the network’s LG, which in turn reduces the bistability range (figure 5(a)). We discovered this special scenario, which was not observed in previous studies, because we considered a non-zero basal protein production rate. Setting the basal production rate to zero constrains the system’s lower steady state to be zero; as a result, the higher threshold (γ2 in figure 2(c)) is at infinity. Because of this approximation, only the lower threshold (γ1 in figure 2(c)) was used to compare the bistability ranges in the previous analysis. Moreover, additive coupling with a positive feedback always increases the total protein levels (see equation (1)), and consequently its transcriptional transfer function always lies above that of a single positive feedback network (see figure 5(b)). As a result, the slope of the left tangent to this transfer function increases. Since the lower threshold is the inverse of the slope, it decreases in the case of (see figure 5(b)). Not surprisingly, a zero basal production rate in the previous analysis resulted in a larger bistability range for PPA-network, as compared to P-network. In the case of multiplicative coupling (which has not been considered earlier), the individual LGs resulting from the two loops add, as a result of which the network’s overall LG always increases due to the addition of a second feedback. This increase results in a larger mLG which causes the lower threshold to decrease. Hence, a multiplicatively coupled second positive feedback always increases the bistability range. We also examined the effects of incorporating positive or negative cooperativity in the PPM-network and found that cooperativity does not change our main conclusion – PPM-network has a larger LG and bistability range compared to P and PPA networks (figures S8(a–d) and S9(a–d)). In conclusion the type of feedback coupling plays a crucial role in determining bistability range.
We also studied the effect of adding a negative feedback on the bistability range of a single positive feedback network. Past findings show that an additively coupled negative feedback always decreases the bistability range [28]. This decrease was attributed to the increase in the lower threshold, while the higher threshold was not affected significantly. The increase in the lower threshold is explained by inspecting the transcriptional transfer function for PNA-network, which is always below than that of P-network (see equation (4) and figure 5(d)). This downward shift in the transfer function causes the lower threshold to increase. Hence, if the higher threshold is not affected significantly, then an additively coupled negative feedback always decreases the bistability range because of an increase in the lower threshold. However, parameter sampling reveals that an additively coupled negative feedback can increase the bistability range (figure 5(c)). This happens when the TF dissociation constant for the negative operator site is much lower than that of the positive feedback site, as a result of which the transcriptional transfer function becomes non-monotonic. The non-monotonic response decreases the slope of the right tangent (corresponding to the higher threshold, figure 5(d)), which in turn increases the higher threshold (figure 5(c)). If the increase in the higher threshold is greater than the increase in the lower threshold, the net effect is an increase in the bistability range. Thus, in contrast with past findings, negative feedback has the capacity to increase the bistability range when it is additively coupled to a positive feedback. On the other hand, negative feedback always decreases the bistability range when it is multiplicatively coupled to a positive feedback as it always reduces the mLG which in turn increases the lower threshold. Multiplicative coupling can also produce a non-monotonic response; however, it scales both the thresholds by the same factor. For this reason, bistability range, defined as the ratio of the thresholds, is not affected as the common factor gets cancelled out during division.
Our analysis shows that, irrespective of the sign of the second feedback (first feedback being positive), additive coupling has the capacity to both increase and decrease a network’s LG and bistability range. In contrast, the effects produced by multiplicative coupling on the two quantities depend on the sign of the second feedback. In this sense, multiplicative coupling is more restrictive because the second positive feedback will only increase the LG and bistability range, while adding a negative feedback instead will decrease them. We speculate that, owing to its flexibility, additive coupling could be selected for during evolution. In fact, an earlier study notes that OR regulation logic is more common than AND logic in a coupled feedback system [31], where OR and AND logic closely resemble additive and multiplicative coupling. On the other hand, we found that multiplicatively coupling of positive feedback loops increases the robustness of network’s bistability range as compared to additive coupling (figure 6(a)). In contrast, in the case of coupled positive/negative feedback loops multiplicative coupling decreases the robustness compared to additive coupling (figure 6(b)).
The differential equation-based modeling used in our analysis represents a deterministic approximation that is only capable of describing systemic quantities averaged over a large population of cells. However, many important dynamic responses are observed at the single-cell level, where fluctuations in the number of molecules present at low concentrations as well as the random nature of binding kinetics at the promoter produce stochastic effects [42]. These effects result in large variability in the switching times of bistable networks. A recent study showed that, at large effective fold changes, the switching times for an autoregulatory network computed using a stochastic model deviates strongly from the deterministic network, and a significant slowdown is predicted [43]. Thus, it would be interesting to explore the effects of the features of feedback architecture studied here in a stochastic framework especially with regards to switching times.
The bistability generating mechanism that we have analyzed in this work is based on purely transcriptional interactions, such as the presence of positive feedback loops and cooperativity resulting from multimerization of TFs [44]. There also exist post-translational mechanisms that can generate bistability, such as multi-site activation of a substrate, cascading of phosphorylation steps and sequestration of a protein by its specific antagonist [44]. In the presence of more complicated post-translational modifications, the assumption of a linear activation (equation (2)) may not hold and will require a more rigorous analysis of the corresponding interactions. This scenario could simplify the analysis of transcriptional interactions as high cooperativity in TF binding may no longer be essential in generating bistability. The crucial role played by post-translational interactions has been further emphasized by a recent study that showed that a purely transcriptional network is the most fragile in generating ultrasensitive and bistable responses [38]. It was concluded that hybrid networks that consist of both transcriptional and enzymatic reactions were the most robust.
In conclusion, we show that the specific effect of coupled feedback loops on open-loop gain, bistability range and switching times depends on the underlying mechanistic details. Our results serve as a useful guide and provide information about various parameters, such as the number of feedback loops, the sign of the loops and the type of coupling, which could be tuned experimentally to construct a bistable switch.
Supplementary Material
Acknowledgments
This work was partially supported by 2009 John S Dunn Collaborative research award and by the grants MCB-0920463 from the National Science Foundation and R01-GM096189-01 from the National Institutes of Health.
References
- 1.Kim D, Kwon YK, Cho KH. Coupled positive and negative feedback circuits form an essential building block of cellular signaling pathways. Bioessays. 2007;29:85–90. doi: 10.1002/bies.20511. [DOI] [PubMed] [Google Scholar]
- 2.Brandman O, Meyer T. Feedback Loops Shape Cellular Signals in Space and Time. Science. 2008;322:390–5. doi: 10.1126/science.1160617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Freeman M. Feedback control of intercellular signalling in development. Nature. 2000;408:313–9. doi: 10.1038/35042500. [DOI] [PubMed] [Google Scholar]
- 4.Manganelli R, Provvedi R. An integrated regulatory network including two positive feedback loops to modulate the activity of sigma(E) in mycobacteria. Mol Microbiol. 2010;75:538–42. doi: 10.1111/j.1365-2958.2009.07009.x. [DOI] [PubMed] [Google Scholar]
- 5.Acar M, Becskei A, van Oudenaarden A. Enhancement of cellular memory by reducing stochastic transitions. Nature. 2005;435:228–32. doi: 10.1038/nature03524. [DOI] [PubMed] [Google Scholar]
- 6.Pomerening JR, Kim SY, Ferrell JE., Jr Systems-level dissection of the cell-cycle oscillator: bypassing positive feedback produces damped oscillations. Cell. 2005;122:565–78. doi: 10.1016/j.cell.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 7.Savageau MA. Comparison of classical and autogenous systems of regulation in inducible operons. Nature. 1974;252:546–9. doi: 10.1038/252546a0. [DOI] [PubMed] [Google Scholar]
- 8.Savageau MA. Biochemical systems analysis : a study of function and design in molecular biology. Reading, Mass: Addison-Wesley; 1976. [Google Scholar]
- 9.Wall ME, Hlavacek WS, Savageau MA. Design of gene circuits: lessons from bacteria. Nat Rev Genet. 2004;5:34–42. doi: 10.1038/nrg1244. [DOI] [PubMed] [Google Scholar]
- 10.Rosenfeld N, Elowitz MB, Alon U. Negative autoregulation speeds the response times of transcription networks. J Mol Biol. 2002;323:785–93. doi: 10.1016/s0022-2836(02)00994-4. [DOI] [PubMed] [Google Scholar]
- 11.Nevozhay D, Adams RM, Murphy KF, Josic K, Balazsi G. Negative autoregulation linearizes the dose-response and suppresses the heterogeneity of gene expression. Proc Natl Acad Sci U S A. 2009;106:5123–8. doi: 10.1073/pnas.0809901106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Becskei A, Serrano L. Engineering stability in gene networks by autoregulation. Nature. 2000;405:590–3. doi: 10.1038/35014651. [DOI] [PubMed] [Google Scholar]
- 13.Igoshin OA, Alves R, Savageau MA. Hysteretic and graded responses in bacterial two-component signal transduction. Mol Microbiol. 2008;68:1196–215. doi: 10.1111/j.1365-2958.2008.06221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Austin DW, Allen MS, McCollum JM, Dar RD, Wilgus JR, Sayler GS, Samatova NF, Cox CD, Simpson ML. Gene network shaping of inherent noise spectra. Nature. 2006;439:608–11. doi: 10.1038/nature04194. [DOI] [PubMed] [Google Scholar]
- 15.Xiong W, Ferrell JE., Jr A positive-feedback-based bistable ‘memory module’ that governs a cell fate decision. Nature. 2003;426:460–5. doi: 10.1038/nature02089. [DOI] [PubMed] [Google Scholar]
- 16.Ferrell JE., Jr Self-perpetuating states in signal transduction: positive feedback, double-negative feedback and bistability. Curr Opin Cell Biol. 2002;14:140–8. doi: 10.1016/s0955-0674(02)00314-9. [DOI] [PubMed] [Google Scholar]
- 17.Angeli D, Ferrell JE, Jr, Sontag ED. Detection of multistability, bifurcations, and hysteresis in a large class of biological positive-feedback systems. Proc Natl Acad Sci U S A. 2004;101:1822–7. doi: 10.1073/pnas.0308265100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tiwari A, Balazsi G, Gennaro ML, Igoshin OA. The interplay of multiple feedback loops with post-translational kinetics results in bistability of mycobacterial stress response. Phys Biol. 2010;7:036005. doi: 10.1088/1478-3975/7/3/036005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goulian M. Two-component signaling circuit structure and properties. Curr Opin Microbiol. 2010;13:184–9. doi: 10.1016/j.mib.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Narula J, Smith AM, Gottgens B, Igoshin OA. Modeling reveals bistability and low-pass filtering in the network module determining blood stem cell fate. PLoS Comput Biol. 2010;6:e1000771. doi: 10.1371/journal.pcbi.1000771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm-2:autoregulatory feedback loop. Genes Dev. 1993;7:1126–32. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 22.Kwon YK, Cho KH. Quantitative analysis of robustness and fragility in biological networks based on feedback dynamics. Bioinformatics. 2008;24:987–94. doi: 10.1093/bioinformatics/btn060. [DOI] [PubMed] [Google Scholar]
- 23.Brandman O, Ferrell JE, Jr, Li R, Meyer T. Interlinked fast and slow positive feedback loops drive reliable cell decisions. Science. 2005;310:496–8. doi: 10.1126/science.1113834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang XP, Cheng Z, Liu F, Wang W. Linking fast and slow positive feedback loops creates an optimal bistable switch in cell signaling. Phys Rev E Stat Nonlin Soft Matter Phys. 2007;76:031924. doi: 10.1103/PhysRevE.76.031924. [DOI] [PubMed] [Google Scholar]
- 25.Chang DE, Leung S, Atkinson MR, Reifler A, Forger D, Ninfa AJ. Building biological memory by linking positive feedback loops. Proc Natl Acad Sci U S A. 2009;107:175–80. doi: 10.1073/pnas.0908314107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guantes R, Poyatos JF. Multistable decision switches for flexible control of epigenetic differentiation. PLoS Comput Biol. 2008;4:e1000235. doi: 10.1371/journal.pcbi.1000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song H, Smolen P, Av-Ron E, Baxter DA, Byrne JH. Dynamics of a minimal model of interlocked positive and negative feedback loops of transcriptional regulation by cAMP-response element binding proteins. Biophys J. 2007;92:3407–24. doi: 10.1529/biophysj.106.096891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tian XJ, Zhang XP, Liu F, Wang W. Interlinking positive and negative feedback loops creates a tunable motif in gene regulatory networks. Phys Rev E Stat Nonlin Soft Matter Phys. 2009;80:011926. doi: 10.1103/PhysRevE.80.011926. [DOI] [PubMed] [Google Scholar]
- 29.Stricker J, Cookson S, Bennett MR, Mather WH, Tsimring LS, Hasty J. A fast, robust and tunable synthetic gene oscillator. Nature. 2008;456:516–9. doi: 10.1038/nature07389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pfeuty B, Kaneko K. The combination of positive and negative feedback loops confers exquisite flexibility to biochemical switches. Phys Biol. 2009;6:046013. doi: 10.1088/1478-3975/6/4/046013. [DOI] [PubMed] [Google Scholar]
- 31.Kim JR, Yoon Y, Cho KH. Coupled feedback loops form dynamic motifs of cellular networks. Biophys J. 2008;94:359–65. doi: 10.1529/biophysj.107.105106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paul S, Birkey S, Liu W, Hulett FM. Autoinduction of Bacillus subtilis phoPR operon transcription results from enhanced transcription from EsigmaA- and EsigmaE-responsive promoters by phosphorylated PhoP. J Bacteriol. 2004;186:4262–75. doi: 10.1128/JB.186.13.4262-4275.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogura M, Tsukahara K. Autoregulation of the Bacillus subtilis response regulator gene degU is coupled with the proteolysis of DegU-P by ClpCP. Mol Microbiol. 2010;75:1244–59. doi: 10.1111/j.1365-2958.2010.07047.x. [DOI] [PubMed] [Google Scholar]
- 34.Meyer BJ, Maurer R, Ptashne M. Gene-Regulation at the Right Operator (or) of Bacteriophage-Lambda. 2. Or1, Or2, and Or3 - Their Roles in Mediating the Effects of Repressor and Cro. J Mol Biol. 1980;139:163–94. doi: 10.1016/0022-2836(80)90303-4. [DOI] [PubMed] [Google Scholar]
- 35.Chastanet A, Losick R. Just-in-time control of Spo0A synthesis in Bacillus subtilis by multiple regulatory mechanisms. J Bacteriol. 2011;193:6366–74. doi: 10.1128/JB.06057-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Forsman K, Goransson M, Uhlin BE. Autoregulation and multiple DNA interactions by a transcriptional regulatory protein in E. coli pili biogenesis. EMBO J. 1989;8:1271–7. doi: 10.1002/j.1460-2075.1989.tb03501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dhooge A, Govaerts W, Kuznetsov, Yu A. MATCONT: A MATLAB package for numerical bifurcation analysis of ODEs. ACM Transactions on Mathematical Software. 2003;29:141–64. [Google Scholar]
- 38.Shah NA, Sarkar CA. Robust network topologies for generating switch-like cellular responses. PLoS Comput Biol. 2011;7:e1002085. doi: 10.1371/journal.pcbi.1002085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maeda YT, Sano M. Regulatory dynamics of synthetic gene networks with positive feedback. J Mol Biol. 2006;359:1107–24. doi: 10.1016/j.jmb.2006.03.064. [DOI] [PubMed] [Google Scholar]
- 40.Fontich E, Sardanyes J. General scaling law in the saddle-node bifurcation: a complex phase space study. J Phys A: Math Theor. 2008;41:015102. [Google Scholar]
- 41.Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8:450–61. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- 42.Kaern M, Elston TC, Blake WJ, Collins JJ. Stochasticity in gene expression: from theories to phenotypes. Nat Rev Genet. 2005;6:451–64. doi: 10.1038/nrg1615. [DOI] [PubMed] [Google Scholar]
- 43.Hermsen R, Erickson DW, Hwa T. Speed, Sensitivity, and Bistability in Auto-activating Signaling Circuits. PLoS Comput Biol. 2011;7:e1002265. doi: 10.1371/journal.pcbi.1002265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tiwari A, Ray JC, Narula J, Igoshin OA. Bistable responses in bacterial genetic networks: designs and dynamical consequences. Math Biosci. 2011;231:76–89. doi: 10.1016/j.mbs.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.