Abstract
Alcoholic liver disease (ALD) is a prominent cause of morbidity and mortality in the United States. Alterations in protein folding occur in numerous disease states, including ALD. The endoplasmic reticulum (ER) is the primary site of post-translational modifications (PTM) within the cell. Glycosylation, the most abundant PTM, affects protein stability, structure, localization and activity. Decreases in hepatic glycosylation machinery have been observed in rodent models of ALD, but specific protein targets have not been identified. Utilizing two-dimensional gel electrophoresis and liquid chromatography tandem mass spectrometry, glycoproteins were identified in hepatic microsomal fractions from control and ethanol-fed mice. This study reports for the first time a global decrease in ER glycosylation. Additionally, the identification of 30 glycoproteins within this fraction elucidates pathway-specific alterations in ALD impaired glycosylation. Among the identified proteins, triacylglycerol hydrolase (TGH) is positively affected by glycosylation, showing increased activity following the addition of sugar moieties. Impaired TGH activity is associated with increased cellular storage of lipids and provides a potential mechanism for the observed pathologies associated with ALD.
Keywords: Alcoholic liver disease, glycoproteomics, UPR, ER stress, 4-HNE, DAVID
Introduction
The endoplasmic reticulum (ER) is the primary site for the synthesis, folding and modification of proteins within the cell.1 Due to a large number of co- and post-translational modifications (PTMs), it is estimated that nearly 30% of newly translated proteins fail to reach their properly folded state.2 By far the most abundant PTM is the attachment of carbohydrate groups to Asn, Ser and Thr residues.3 These glycan modifications affect protein folding, structure, localization, half-life and enzymatic activity.3 Altered protein glycosylation has been implicated in various disease states, including diabetes, leukocyte adhesion deficiency, hepatitis B virus infection, fatty liver and alcoholic liver disease (ALD).4, 5
As the major cause of liver injury, ALD remains a prominent cause of both morbidity and mortality in the Western world, affecting nearly 2 million people annually in the United States alone.6 The damaging effects of ethanol are, in part, due to its oxidative metabolism, where the generation of reactive oxygen species (ROS) remains unbalanced with decreased antioxidant defenses. Under situations of chronic ethanol consumption, rate-limiting ethanol metabolizing enzymes become saturated, leading to the generation of toxic concentrations of acetaldehyde. As a cellular defense, induction of cytochrome P450 2E1 (Cyp2E1) becomes necessary to oxidize high concentrations of ethanol.7-9 This induction, however, remains a double-edged sword, where increased Cyp2E1 results in significant exacerbation of ROS production, leading to substantial oxidative stress. This results in enhanced lipid peroxidation and generation of pathologic concentrations of the lipid peroxidation products 4-hydroxynonenal (4-HNE) and 4-oxononenal (4-ONE).10 These reactive aldehydes covalently adduct Cys > His >Lys residues, potentially altering protein structure, localization and activity.11 Additionally, protein adduction by reactive aldehydes is proposed to play a key role in the progression of diseases associated with sustained oxidative stress, most notably, ALD.12-14
Among the most prevalent pathologic responses to chronic ethanol consumption is alcoholic fatty liver, or steatosis.7, 15, 16 Steatosis is marked by dysregulation of lipid metabolism, resulting in increased synthesis of fatty acids, increased levels of lipogenic enzymes and decreased fatty acid oxidation.16 Although steatosis remains the most predictable outcome, the precise mechanisms behind the onset and progression of ALD are largely unknown. Compounding the complications of this disease, numerous cellular derangements have been implicated in ALD, including, the induction of Cyp2E1, depletion of cellular antioxidants, mitochondrial dysregulation, increased lipid peroxidation and induction of the ER stress response, and subsequent unfolded protein response (UPR) signaling.2, 7
Glycosylation plays a key role in the folding of nascent proteins, where inhibition of this PTM may lead to enhanced UPR induction.17 Numerous reports have implicated the UPR following ethanol-mediated liver damage, however, the precise mechanisms and proteins involved have not been characterized.1, 2, 18-20 Interestingly, changes in protein glycosylation have been observed on numerous occasions following chronic ethanol consumption.21, 22 Clinically, glycosylation of serum proteins is used as a biomarker to detect liver damage following ethanol ingestion. Specifically, carbohydrate-deficient transferrin (CDT) and haptoglobin are two of the most recognized clinical markers for detecting chronic ethanol-mediated liver damage.4 The purpose of the present study was to investigate protein glycosylation in a rodent model of ALD, with the goal of identifying protein targets of glycosylation within the ER, potentially providing a mechanism for the observed UPR signaling following chronic ethanol consumption.
In this study, we demonstrate an apparent global decrease in the ER liver glycoproteome with the progression of chronic ethanol ingestion. Changes in glycosylation were observed using two-dimensional gel electrophoresis and altered proteins were then identified using mass spectrometry. Bioinformatic analyses were conducted and multiple ER-dependent processes were implicated following changes in protein glycosylation. Our data suggest an important role for ER glycosylation in the pathogenesis of ALD.
Experimental Procedures
Animal Model
All procedures involving animals were approved by the Institutional Animal Care and Use Committee of the University of Colorado and were performed in accordance with published National Institutes of Health guidelines. Male C57/BL6J mice (12 per group) were utilized for the analysis and characterization of ethanol-mediated liver damage. Briefly, mice were fed a modified Lieber-DeCarli liquid based-diet (Bio-Serv, Frenchtown, NJ) for 6 weeks. The diet consisted of 44% fat-derived calories, 16% protein-derived calories and the remaining balance being comprised of either carbohydrate or ethanol-derived calories. Ethanol-fed mice began the study on a diet consisting of 2% (v/v) ethanol, with the ethanol-derived calories increasing by 1% on a weekly basis until sacrifice; week 6 consisted of 6% ethanol (v/v) or 31.8% ethanol-derived calories. Pair-fed animals were each matched to an ethanol-fed mouse where ethanol-derived calories were replaced with calories from a carbohydrate source (maltose-dextrin). Food consumption was monitored daily and body weights were measured weekly. Upon completion of the study, animals were anesthetized via intraperitoneal injection of sodium pentobarbital and euthanized via exsanguination. Blood was collected from the inferior vena cava and plasma was separated through centrifugation. Livers were excised, weighed, and frozen for biochemical characterization, fixed in formalin for immunohistochemical analysis, or subjected to differential centrifugation using a sucrose gradient for subcellular fractionation as previously described.23
Biochemical Analysis
Plasma collected from the inferior vena cava was assayed for alanine aminotransferase (ALT) activity (Diagnostic Chemicals Limited, Oxford, CT). Liver triglycerides were measured using a 2:1 chloroform:methanol extract of liver homogenate using a kit from Diagnostic Chemicals Limited as previously reported.13 Protein concentrations were determined using a BCA Protein Assay from Pierce (Rockford, IL) or a modified Lowry Protein Assay from Bio-Rad (Hercules, CA).
Immunohistochemistry
Following excision, livers were sectioned and placed in 10% neutral buffered formalin for 8 hours. Samples were then processed, imbedded in paraffin and mounted to slides by Colorado HistoPrep (Fort Collins, CO). One pair of slides was then stained with hematoxylin and eosin (H&E) for histological characterization while the remaining slides were subject to de-paraffinization and rehydration for immunohistochemical characterization. 4-HNE modified proteins were stained using a custom antibody generated in rabbit directed against 4-HNE-modified keyhole limpet hemocyanin. Sections were developed using a VECTASTAIN ABC Elite kit (Vector Laboratories, Burlingame, CA) and counterstained using hematoxylin. Quantification of 4-HNE staining was achieved using 3 independent 400x fields from 6 control and ethanol-fed mice. Protocols were adapted from Sampey et al. (2007).12
In Gel Staining of Glycosylated Proteins
Microsomal fractions isolated from the livers of both control and ethanol-fed mice were subjected to two-dimensional gel electrophoresis on IPG strips (pH 3-11) and separated on either 11 or 17cm gels (Bio-Rad, Hercules, CA) in duplicate. Proteins were then fixed overnight in the gels with a solution containing 50% methanol and 5% acetic acid in ddH2O. One gel was then stained for glycoproteins using the Pro-Q Emerald 300 Glycoprotein Gel Stain Kit following manufacturer’s instructions (Molecular Probes, Eugene, OR) and visualized using a GE Healthcare Ettan DIGE Imager (excitation wavelength, 390nm; emission wavelength, 595nm). The second gel was stained for total protein using a SilverQuest Silver Stain kit (Invitrogen, Carlsbad, CA). Gels were then overlaid and glycosylated proteins were picked from the Silver stained gel for identification. To ensure that the observed alterations in glycosylation were not due to decreased protein expression, additional gels were run and stained with coomassie-blue for total protein expression.
LCMS/MS Identification of Altered Protein Glycosylation
Differences in protein glycosylation were visualized using the Pro-Q Emerald 300 Glycoprotein Gel Stain Kit applied to the 17cm 2D-SDS-PAGE gels described above and were aligned to the corresponding spots on Silver stained companion gels. Glycosylated proteins were excised from the gels, digested with sequencing-grade trypsin (Promega, Madison, WI) in 50 mM ammonium bicarbonate overnight at 37°C and the resulting peptides prepared for analysis by LC-MS/MS as previously described.24 Peptide samples were introduced using an electrospray (ESI) source into an Agilent SL ion trap mass spectrometer (Palo Alto, CA) with an Agilent 1100 capillary HPLC system and a 1.0 × 150 mm Jupiter Proteo 90 Å column (Phenomenex, Torrance, CA). The flow rate was 5 L/min of 0.1% aqueous formic acid (solvent A) and 0.1% formic acid in acetonitrile (solvent B). Mobile-phase composition was held at 2% solvent B for 5 min and then increased to 40% using a linear gradient over 40 min. The instrument was operated under tandem MS (MS/MS) conditions with helium as the collision gas. Compound lists of peptide fragment ions were generated using Chemfinder (Agilent) and exported as Mascot Generic Format files and searched against the comprehensive Mass Spectrometry Database (MSDB) obtained from ftp.ncbi.nih.gov/repository/MSDB via the Mascot search engine (v 2.1.04, www.matrixscience.com). Searches resulted in the assignment of a UniProt (Universal Protein Resource http://www.uniprot.org/) accession number which was used for subsequent analysis. Of the 38 spots identified, four were not associated with a UniProt accession number (spots 28, 29, 34, 35), while in four instances, two spots were associated with the same accession number (O35490 (spots 31, 33), P16460 (spots 30, 34), Q8VCT4 (spots 13, 22), Q8VCU1 (spots 12, 16)). Once these features had been accounted for 30 unique UniProt accession numbers remained and formed the basis of further analysis.
Western Blotting
Microsomal protein (5.0μg) was subjected to standard SDS-PAGE and transferred to a Hybond-P membrane (GE Healthcare, Buckinghamshire, UK). Membranes were then blocked for 30 minutes with a tris-buffered saline solution containing 1% Tween-20 (TBST) and 5% non-fat dry milk (NFDM). Membranes were then probed with primary antibodies directed against Grp78/Grp94 (Stressgen, Ann Arbor, MI), PDI (Sigma-Aldrich, St. Louis, MO), Calreticulin (Abcam, Cambridge, MA) and Carboxylesterase 3 (ProteinTech Group, Chicago, IL). Following 3 washes with TBST, a horseradish peroxidase conjugated secondary (Jackson Labs, Bar Harbor, ME) was applied and membranes were developed using ECL-Plus Reagent from GE Healthcare. Chemiluminescence was then visualized using a Storm 860 scanner from Molecular Dynamics (Sunnyvale, CA). As a reliable loading control does not exist for microsomal proteins, a BCA assay was conducted prior to all western blotting to ensure equal protein load.
Prediction of N- and O-Linked Glycosylation using NetNGlyc 1.0 and NetOGlyc 3.1
Due to the immense diversity in possible glyco-modifications, detection of in vivo glycosylation is extremely difficult and unlikely; because of this challenge, multiple prediction servers have been established as tools to predict glycosylation sites on proteins based on their amino acid sequence, predicted secondary structures and surface accessibility of residues. Amino acid sequences from each accession number were entered in to NetNGlyc 1.0 and NetOGlyc 3.1 for the prediction of both N- and O-linked glycosylation sites. These servers employ a neural network analysis for the prediction of potentially glycosylated sites for a given protein. For N-linked glycosylation predictions, all Asn-Xaa-Ser/Thr motifs (where Xaa is not Pro) were considered, and are reported first. Positive sites were identified using default settings (glycosylation potential > 0.5), and are reported in parenthesis (for more information on prediction software, see http://www.cbs.dtu.dk/services/NetNGlyc/abstract.php).
Due to the magnitude of possible sites for O-linked glycosylation, the potential for false positive scoring does increase, however, NetOGly 3.1 has been shown to correctly predict 76% of positive sites, and 93% of negative sites for completely new proteins. This neural network utilizes 2 separate scores; the G-score is the score from the best general predictor, whereas the I-score is the score from the best isolated site predictor. For the analyses reported here, all predicted Ser and Thr residues were reported based on default settings (G-score > 0.5 or I-score > 0.5). For information on the prediction software used, and a more detailed description of the scoring systems utilized, see Julenius et al. 2005 25.
Identification and Enrichment of Functionally Important Terms
The Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7 [http://david.abcc.ncifcrf.gov]26 was used to identify biologically enriched themes and terms within the protein list of interest. The set of 30 unique UniProt accession numbers were submitted to DAVID for analysis; background and species identified as Mus musculus. DAVID correctly matched all 30 UniProt accessions. O35490 was identified as having two corresponding DAVID records (ENSMUSG00000074768 and ENSMUSG00000069324/ ENSMUSG00000082293) both of which were included for completeness.
Gene Functional classification, Functional Annotation Enrichment and Functional Annotation clustering were all undertaken using DAVIDs default parameters and stringency measures. Functional annotation enrichment was also undertaken utilizing default settings to identify annotation sources. DAVID utilizes functional annotations from a variety of sources, including Gene Ontology (GO) (biological processes, molecular function, cellular component), disease databases (OMIM), protein domain categories (InterPro, SMART), pathway databases (KEGG, BioCarta), and functional categories (Up Seq Feature, Sp Pir Keywords). Significance was determined with slight modifications as recommended by the DAVID authors as follows: enrichment scores ≥ 2.0, fold enrichment > 2.0, Benajmini corrected p < 0.05.26
Statistical Analysis
Statistical analysis and generation of graphs was performed using GraphPad Prism 4.02 (GraphPad Software, San Diego, CA). Differences between control and ethanol-fed mice were assessed using a paired Student’s t test. Differences were considered significant if p < 0.05.
Results
Biochemical Characterization of Animal Feeding
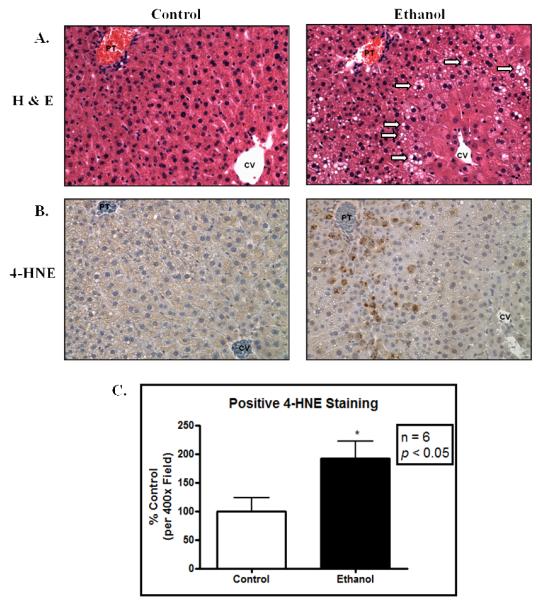
Following 6 weeks of chronic ethanol consumption, mice were sacrificed and subjected to various biochemical and histological analyses to assess the extent of liver damage. Data presented in Table 1 show that ethanol consuming mice displayed a significant decrease in body weight gain and significant increases in liver/body weight ratio, reflecting a state of hepatomegaly as typically observed following chronic ethanol consumption. To assess the extent of liver damage, plasma ALT activities were determined; ethanol-fed mice displayed nearly a 3-fold increase in serum ALT activity as compared to pair-fed, isocaloric control mice. Further characterization revealed nearly a 2-fold increase in liver triglycerides, indicative of ethanol-mediated steatosis (Table 1). As shown in Figure 1, H&E staining revealed significant lipid accumulation in the ethanol-fed mice. Using antibodies directed against 4-HNE modified proteins, immunohistochemical analysis reveals approximately a 2-fold increase in staining in the ethanol-fed livers, indicating a state of sustained oxidative stress. Taken together, these data demonstrate significant liver damage, consistent with early-stage ALD. This allowed for further investigation using this model into the analysis of glycosylation and glycoprotein status in the ethanol-fed mouse liver.
Table 1.
Biochemical characterization of ALD. Chronic ethanol consumption leads to a decrease in body weight gain as well as a significant increase in liver/body weight. Liver damage is also indicated by a significant increase in ALT activity in serum. Steatosis was achieved as indicated by a marked increase in liver triglycerides. Statistical significance was determined between the control and ethanol-fed mice as follows: n = 12;
| Parameter | Control | Ethanol |
|---|---|---|
| Body Weight (g) | 31.017 ± 0.752 | 25.783 ± 0.647† |
| Liver/Body Weight | 0.038 ± 0.001 | 0.044 ± 0.001† |
| ALT (U/L) | 18.411 ± 4.612 | 42.985 ± 4.523# |
| Triglycerides (mmol/L/mg tissue) | 0.146 ± 0.021 | 0.246 ± 0.037* |
p < 0.05;
p < 0.01;
p < 0.001.
Figure 1.
Chronic ethanol ingestion results in hepatic lipid accumulation and marked oxidative stress. (A.) H&E stained liver (400x) displays significant lipid accumulation following ethanol ingestion. Areas of significant lipid accumulation are designated by arrows; (B.) Immunohistochemical staining for 4-HNE shows increased staining in the ethanol-fed mice. (C.) Quantification of 4-HNE immunopositive staining (n = 6 pairs; p < 0.05). PT, portal triad; CV, central vein.
Apparent Reduction in Glycosylation of Microsomal Proteins Following Chronic Ethanol Consumption
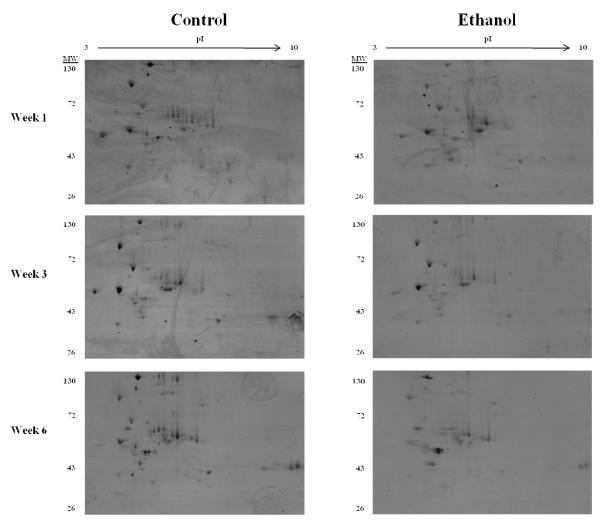
Microsomal fractions isolated from the livers of control and mice ingesting ethanol for 1, 3 and 6 weeks were analyzed for glycoprotein status using 2-dimensional gel electrophoresis. First dimension isoelectric focusing (IEF) was performed with microsomal proteins (250μg) using 11cm, pH 3-10 IPG strips. IPG strips were then resolved in the second dimension using 10% acrylamide gels and stained for glycosylation as mentioned. As shown in Figure 2, a gradual reduction in glycoprotein staining was observed as ethanol consumption progressed, with the greatest decrease occurring between the week 6 control- and ethanol-fed mice. These results were independent of overall protein expression, which showed no significant alterations as determined by coomassie-blue staining (data not shown). Reports have indicated a concern regarding the composition of the Lieber-DeCarli diet, and the observed alterations on glycosylation machinery potentially being a result of a decrease in carbohydrate-derived calories. Using a “carbohydrate compensated chronic-ethanol” group, this concern was addressed by Ghosh et al. in 1997, who showed no significant differences in glycosylation machinery regardless of the caloric source.21 In an effort to identify novel mechanisms behind the progression of ALD, a glycoproteomic approach was undertaken and remained the focus of this study.
Figure 2.
Chronic ethanol ingestion results in an apparent progressive decrease in glycosylation of microsomal proteins. 2-dimensional gel electrophoresis using a pI 3-10 non-linear IPG strip and 10% acrylamide gel was performed using microsomal fractions isolated from livers of both control and ethanol-fed mice. Gels were then stained with the Pro-Q Emerald 300 Glycoprotein Gel Stain Kit and visualized on an Ettan DIGE Imager.
Identification of Altered Microsomal Proteins Using LC-MS/MS
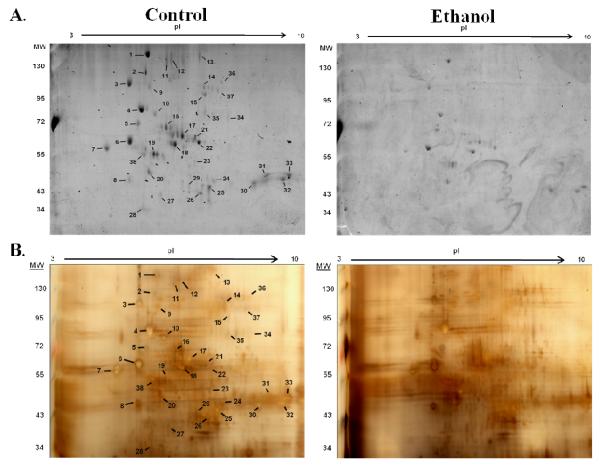
Microsomal samples obtained from the week 6 time point, were run in duplicate on 17cm gels to further increase resolution in the 50kD range; one gel was stained with the aforementioned glycoprotein staining, and one gel was Silver stained. Following excision of glycosylated proteins, gel spots were destained and digested with trypsin. Digested peptides were then analyzed using LC-MS/MS for protein identification. As shown in Figure 3, 38 spots were picked for identification; it was observed that numerous proteins resolved over a range of pI’s, resulting in redundant identifications. This was observed with carboxylesterase 3 (spot 22), liver carboxylesterase 31 (spot 17) and alpha-1-antitrypsin 1-1 (spot 19). Of the 38 spots chosen, 4 did not include a significant MOWSE score (spots 28, 29, 34, 35) and 4 were recognized as the same protein as a previous identification (spots 16, 20, 32 and 33). This yielded 30 unique proteins which were independently identified as glycosylated using our techniques. A summary of proteins identified, along with MS/MS characterization is presented in Table 2.
Figure 3.
2-dimensional gel electrophoresis was performed using a pI 3-10 non-linear IPG strip and a 17cm, 8% polyacrylamide gel. (A.) Microsomal fractions isolated from livers of control and ethanol-fed mice were stained for glycosylation. (B.) Paired gels were Silver Stained and spots were picked from the control gel, as staining was more intense. Gel spots (indicated) were then subjected to tryptic digest and analyzed using LC-MS/MS for protein identification.
Table 2.
Proteins identified as glycosylated using glycoproteomics in pair-fed control mice.
| Spot | Protein | Swiss-Prot | Calc MW | Calc PI | Peptides Matched |
Sequence Coverage (%) |
MOWSE Score |
|---|---|---|---|---|---|---|---|
| 1 | Hypoxia Upregulated Protein 1 (Grp170) | Q9JKR6 | 111181 | 5.12 | 35 | 27 | 224 |
| 2 | Serine Protease Inhibitor | P07759 | 46850 | 5.05 | 5 | 17 | 106 |
| 3 | Endoplasmin (Grp94) | P08113 | 92418 | 4.74 | 39 | 28 | 291 |
| 4 | 78 kDa glucose-regulated protein (Grp78) | P20029 | 72377 | 5.07 | 64 | 46 | 542 |
| 5 | Liver Carboxylesterase N | P23953 | 61133 | 5.1 | 6 | 11 | 45 |
| 6 | Protein disulfide-isomerase | P09103 | 57108 | 4.79 | 56 | 62 | 335 |
| 7 | Calreticulin | P14211 | 47965 | 4.33 | 14 | 18 | 99 |
| 8 | 40S ribosomal protein SA | P14206 | 32817 | 4.8 | 11 | 25 | 253 |
| 9 | Transitional endoplasmic reticulum ATPase | Q01853 | 89266 | 5.14 | 33 | 37 | 286 |
| 10 | Heat shock cognate 71 kDa protein | P63017 | 70827 | 5.37 | 41 | 43 | 405 |
| 11 | Glutamate [NMDA] receptor subunit zeta-1 | P35438 | 105414 | 8.99 | 1 | 0 | 40 |
| 12 | Liver carboxylesterase 31-like | Q8VCU1 | 62966 | 5.65 | 5 | 8 | 70 |
| 13 | Carboxylesterase 3 | Q8VCT4 | 61749 | 6.17 | 9 | 14 | 72 |
| 14 | Glycogen phosphorylase, liver | Q9ET01 | 97369 | 6.63 | 21 | 20 | 87 |
| 15 | GDH/6PGL endoplasmic bifunctional protein | Q8CFX1 | 88855 | 6.4 | 16 | 17 | 156 |
| 16 | Liver carboxylesterase 31-like | Q8VCU1 | 62966 | 5.65 | 8 | 11 | 102 |
| 17 | Liver carboxylesterase 31 | Q63880 | 63277 | 5.78 | 18 | 21 | 131 |
| 18 | Protein disulfide-isomerase A3 | P27773 | 56643 | 5.88 | 40 | 46 | 316 |
| 19 | Alpha-1-antitrypsin 1-1 | P07758 | 45974 | 5.44 | 7 | 13 | 82 |
| 20 | Actin, cytoplasmic 1 | P60710 | 41710 | 5.29 | 10 | 24 | 156 |
| 21 | Probable carboxypeptidase PM20D1 | Q8C165 | 55628 | 5.99 | 10 | 23 | 94 |
| 22 | Carboxylesterase 3 | Q8VCT4 | 61749 | 6.17 | 24 | 34 | 223 |
| 23 | Alpha-enolase | P17182 | 47111 | 6.37 | 15 | 33 | 82 |
| 24 | 4-hydroxyphenylpyruvate dioxygenase | P49429 | 45026 | 6.58 | 18 | 44 | 232 |
| 25 | Fumarylacetoacetase | P35505 | 46074 | 6.92 | 8 | 19 | 130 |
| 26 | Arginase-1 | Q61176 | 34786 | 6.51 | 21 | 43 | 282 |
| 27 | Peroxiredoxin-2 | Q61171 | 21765 | 5.2 | 2 | 9 | 38 |
| 28 | No Score | - | - | - | - | - | - |
| 29 | No Score | - | - | - | - | - | - |
| 30 | Argininosuccinate synthase | P16460 | 46555 | 8.36 | 28 | 27 | 183 |
| 31 | Betaine-homocysteine S-methyltransferase 1 | O35490 | 44992 | 8.01 | 23 | 44 | 199 |
| 32 | Argininosuccinate synthase | P16460 | 46555 | 8.36 | 28 | 29 | 192 |
| 33 | Betaine-homocysteine S-methyltransferase 1 | O35490 | 44992 | 8.01 | 30 | 45 | 326 |
| 34 | No Score | - | - | - | - | - | - |
| 35 | No Score | - | - | - | - | - | - |
| 36 | C-1-tetrahydrofolate synthase, cytoplasmic | Q922D8 | 101192 | 6.68 | 29 | 30 | 174 |
| 37 | Elongation factor 2 | P58252 | 95253 | 6.41 | 21 | 25 | 199 |
| 38 | ATP synthase subunit beta, mitochondrial | P56480 | 56265 | 5.19 | 22 | 37 | 372 |
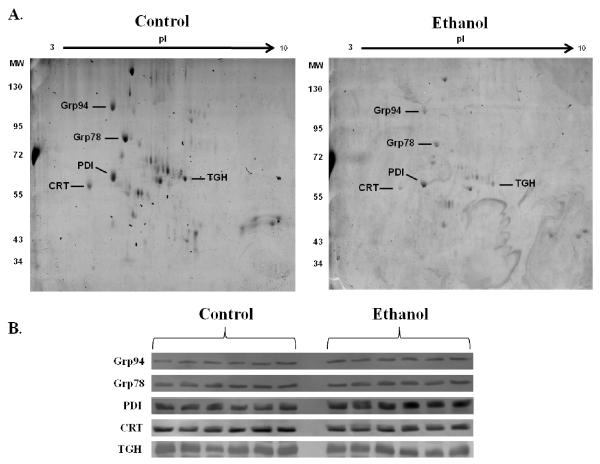
To confirm that the observed fluctuations in microsomal glycosylation were not due to altered protein expression, western blotting was conducted. As shown in Figure 4, no difference in expression of the prototypic ER proteins Grp94 (spot 3), Grp78 (spot 4), PDI (spot 6), Calreticulin (spot 7) and Carboxylesterase 3 (spot 13) was observed. Although additional targets were identified, these 5 proteins displayed a large change in overall glycosylation staining from week 3 to week 6 and were thus chosen for analysis of protein expression.
Figure 4.
Immunoblotting reveals no change in protein expression of identified targets of glycosylation. (A.) Glycoproteins analyzed via western blotting in (B.) are indicated for reference on 2-dimensional gels. (B.) Western blotting was performed using microsomal fractions from control and ethanol-fed mice for protein expression of Glucose Regulated Protein 94 (Grp94), Grp78, Protein Disulfide Isomerase (PDI), Calreticulin (CRT) and Carboxylesterase 3 (TGH). No change in expression was observed in these proteins.
Predicted Glycosylation Sites on Identified Proteins
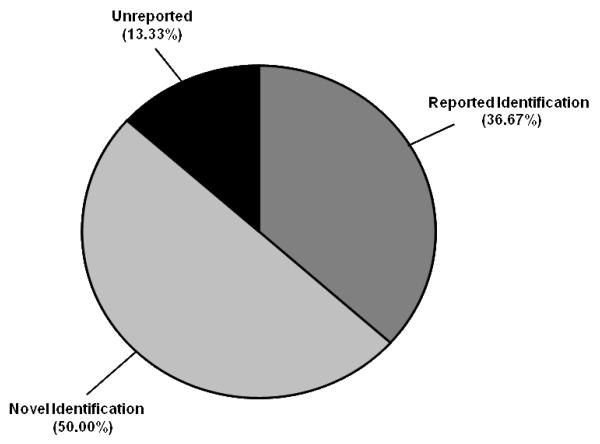
Protein glycosylation is widely variable, differing in the type (e.g. N-linked or O-linked), composition (e.g. N-acetylglucosamines, mannose, galactose, fucose) and chain length.4, 5 O-linked glycosylation predominately occurs on both Ser and Thr residues, however, no consensus sequence is reported. Although a consensus sequence for N-linked glycosylation (Asn-Xaa-Ser/Thr, where Xaa is not Pro) does exist, this does not guarantee that a protein will undergo this type of modification. To further investigate our findings, we utilized both N- and O-linked web-based glycosylation servers. This freeware examines protein sequences and predicts potentially glycosylated residues based on their surface accessibility, secondary structure and distance constraints.25, 27 The proteins identified by LC-MS/MS were submitted and the results are presented in Table 3. Of the 30 unique proteins identified, 22 contained the signature Asn-Xaa-Ser/Thr motif, with 21 of these proteins identified as positive for N-linked glycosylation sites; 19 proteins contained positive scores for O-linked glycosylation. As shown in Figure 5, only 11 of the proteins identified have been previously reported to be glycosylated (36.67%); our studies resulted in 15 unique and novel glycosylated proteins (50.0%); 4 of these proteins did not fall under either category and are designated “Unreported” (13.33%).
Table 3.
Identified glycoproteins and their predicted glycosylation sites using NetNGlyc and NetOGlyc servers. “Reported Glycosylation” indicates proteins known to be glycosylated in the UniProt database, reference numbers are in brackets. “Predicted N-Glycosylation Sites” indicates the number of Asn-Xaa-Ser/Thr motifs present with positive scores (glycosylation potential > 0.5) reported from NetNGlyc in parenthesis. “Predicted O-Glycosylation Sites” indicates all positive scores (G- or I-score > 0.5) associated with either Ser or Thr residues as reported by NetOGlyc. Redundant proteins were removed.
| Spot | Protein | Reported Glycosylation [Ref.] |
Predicted N-Glycosylation Sites (positive scores) |
Predicted O- Glycosylation Sites |
|---|---|---|---|---|
| 1 | Hypoxia Upregulated Protein 1 (Grp170) | X [45] | 9 (5) | 2 |
| 2 | Serine Protease Inhibitor | X [46] | 4 (2) | 0 |
| 3 | Endoplasmin (Grp94) | X [45] | 6 (5) | 2 |
| 4 | Glucose Regulated Protein 78 (Grp78) | 0 (0) | 4 | |
| 5 | Liver Carboxylesterase N | X [47] | 5 (5) | 3 |
| 6 | Protein disulfide-isomerase A1 | 0 | 0 | |
| 7 | Calreticulin | 1 (0) | 0 | |
| 8 | 40S ribosomal protein SA | 0 | 10 | |
| 9 | Transitional endoplasmic reticulum ATPase | 4 (1) | 1 | |
| 10 | Heat shock cognate 71 kDa protein | 7(5) | 3 | |
| 11 | Glutamate [NMDA] receptor subunit zeta-1 | X [48] | 11 (9) | 0 |
| 12 | Liver carboxylesterase 31-like | X [49] | 4 (3) | 2 |
| 13 | Carboxylesterase 3 | X [49] | 2 (2) | 2 |
| 14 | Glycogen phosphorylase, liver | 6 (5) | 0 | |
| 15 | GDH/6PGL endoplasmic bifunctional protein | X [50]* | 3 (2) | 0 |
| 17 | Liver carboxylesterase 31 | X [49] | 3 (2) | 2 |
| 18 | Protein disulfide-isomerase A3 | 0 | 0 | |
| 19 | Alpha-1-antitrypsin 1-1 | X [47] | 3 (3) | 1 |
| 20 | Actin, cytoplasmic 1 | 1 (1) | 1 | |
| 21 | Probable carboxypeptidase PM20D1 | X [50]** | 4 (2) | 1 |
| 23 | Alpha-enolase | 2 (2) | 0 | |
| 24 | 4-hydroxyphenylpyruvate dioxygenase | 2 (2) | 1 | |
| 25 | Fumarylacetoacetase | 1 (1) | 1 | |
| 26 | Arginase-1 | 1 (1) | 1 | |
| 27 | Peroxiredoxin-2 | 0 (0) | 2 | |
| 30 | Argininosuccinate synthase | 0 (0) | 3 | |
| 31 | Betaine-homocysteine S-methyltransferase 1 | 0 (0) | 0 | |
| 36 | C-1-tetrahydrofolate synthase, cytoplasmic | 2 (1) | 3 | |
| 37 | Elongation factor 2 | 5 (2) | 0 | |
| 38 | ATP synthase subunit beta, mitochondrial | 0 (0) | 0 |
Identified in human samples;
Human isoforms identified.
Figure 5.
Depiction of identified glycoproteins in murine microsomal fractions from liver extracts. “Reported Identification” designates all proteins identified that have been reported to be glycosylated in the UniProt database. “Novel Identification” designates all proteins identified that are predicted to be glycosylated that have not been previously reported in the literature. “Unreported” designates all proteins that are unreported and not predicted to be glycosylated.
Categorization of Glycosylated Microsomal Proteins Using DAVID
To further investigate the observed alterations in glycosylation, a bioinformatics approach was taken to elucidate potential ER-dependent mechanisms of ethanol-induced liver injury. The 30 proteins identified were subjected to DAVID analysis and categorized based on functional annotation enrichment. Given that all of the proteins analyzed were isolated from the microsomal fractions of murine liver, localization and tissue specificity terms were excluded from analysis. Table 4 provides a summary of these results based on their associated molecular processes, presenting both fold enrichment (values > 2) and a Benjamini corrected p-value (p < 0.05). A total of 27 different annotation terms associated with the 30 identified proteins of interest achieved both statistical and enrichment significance. Relevant to our model, processes involving protein folding, esterase and stress response pathways were highly enriched.
Table 4.
Functional enrichment analysis using DAVID. Only processes with an enrichment factor (fold > 2)a and Benjamini corrected p-value (p < 0.05)a were reported.
| Term | Fold Enrichment | p-Value | Annotation Source |
|---|---|---|---|
| Arginine metabolic process | 167.75 | 2.71E-02 | GO Biological Process, GO:0006525 |
| Heat shock protein | 70 161.48 | 5.67E-03 | InterPro, IPR013126 |
| Methionine biosynthetic process | 150.98 | 2.26E-02 | GO Biological Process, GO:0009086 |
| Methionine metabolic process | 137.25 | 2.07E-02 | GO Biological Process, GO:0006555 |
| Heat shock protein 70, conserved site | 136.64 | 5.35E-03 | InterPro, IPR018181 |
| Active site:Acyl-ester intermediate | 114.44 | 1.56E-02 | Up Seq Feature |
| Chaperone HSP70 | 108.96 | 8.18E-03 | PIR Superfamily, PIRSF002581 |
| Sulfur amino acid biosynthetic process | 107.84 | 2.28E-02 | GO Biological Process, GO:0000097 |
| Carboxylesterase type B, active site | 98.68 | 7.82E-03 | InterPro, IPR019826 |
| Aspartate family amino acid biosynthetic process | 94.36 | 2.25E-02 | GO Biological Process, GO:0009067 |
| Short sequence motif:Prevents secretion from ER | 81.46 | 9.27E-12 | Up Seq Feature |
| Carboxylesterase type B, conserved site | 74.01 | 1.12E-02 | InterPro, IPR019819 |
| Aspartate family amino acid metabolic process | 68.63 | 3.79E-02 | GO Biological Process, GO:0009066 |
| Sulfur amino acid metabolic process | 65.64 | 3.73E-02 | GO Biological Process, GO:0000096 |
| Serine esterase | 57.59 | 1.28E-02 | Sp Pir Keyword |
| Endoplasmic reticulum, targeting sequence | 55.08 | 3.74E-03 | InterPro, IPR000886 |
| Stress response | 52.90 | 1.76E-03 | Sp Pir Keyword |
| Thioredoxin-like | 49.34 | 2.09E-02 | InterPro, IPR017936 |
| Redox-active center | 45.78 | 1.82E-02 | Sp Pir Keyword |
| Cellular amino acid biosynthetic process | 45.75 | 3.36E-02 | GO Biological Process, GO:0008652 |
| Amine biosynthetic process | 27.58 | 2.19E-02 | GO Biological Process, GO:0009309 |
| Chaperone | 19.84 | 2.44E-03 | Sp Pir Keyword |
| Nitrogen compound biosynthetic process | 10.00 | 2.03E-02 | Go Biological Process, GO:0044271 |
| Atp-binding | 4.16 | 9.79E-03 | Sp Pir Keyword |
| Nucleotide-binding | 4.01 | 3.14E-03 | Sp Pir Keyword |
| Hydrolase | 3.97 | 6.24E-03 | Sp Pir Keyword |
| Acetylation | 3.84 | 2.23E-04 | Sp Pir Keyword |
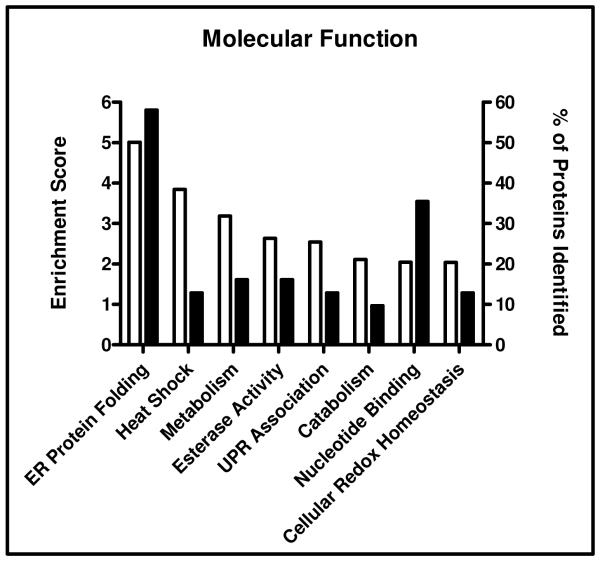
Utilizing functional annotation clustering analysis, proteins were categorized based on molecular function (Figure 6). As predicted, terms associated with ER protein folding (enrichment factor 5.01), heat shock response (3.84), UPR (2.54) and cellular redox homeostasis (2.04) all had significant enrichment factors, including up to 58% of the total proteins identified (terms associated with each cluster, as well as UniProt accession assignments, may be found in Supplemental Table 1). Interestingly, 16% of the proteins identified were annotated with terms associated with esterase activity (enrichment factor 2.63). This suggests a potential novel mechanism for the lipid accumulation typically observed in ALD. Taken together, these analyses provide crucial information regarding glycosylation and its role in the pathogenesis of ALD.
Figure 6.
Functional annotation clustering analysis using DAVID. The identified proteins were clustered based on molecular functions. Enrichment scores ≥ 2.0 were reported and are depicted by the open bars on the left y-axis. Total percentage of proteins identified in each category is represented by the closed bars in the right y-axis.
Discussion
Protein glycosylation is the most abundant and structurally diverse PTM occurring in eukaroytes.3, 25 Due to the enormous diversity in both chain length and sugar composition for an individual protein, detection of in vivo glyco-modifications by mass spectrometry is extremely difficult and heavily reliant on costly instrumentation with high mass accuracy.28-31 As research into PTMs has gained considerable attention in recent decades, methodology and tools for the analysis of glycoproteins has made research in this area more accessible. Although more specific techniques for glycosylation have evolved (i.e. lectin affinity), hydrazide chemistry remains the primary approach for the analysis of global glycosylation in complex samples.28, 32 In the past decade, commercial assays have become readily available, which couple hydrazide chemistry with highly sensitive fluorescent detection.33 Using these techniques, in conjunction with two-dimensional gel electrophoresis, our results suggest a marked diminution in protein glycosylation within the ER following chronic ethanol consumption. These fluctuations are likely to contribute significantly to the pathologies associated with ALD.
Preliminary studies by Marinari et al. provided a potential link between glycosylation and diseases of oxidative stress.34 These studies investigated the glycosylation enzymes galactosyltransferase and sialyltransferase and their susceptibility to aldehyde modification. Following incubation with physiologically relevant concentrations of 4-HNE (100μM), a 41 and 59% decrease in activity of galactosyltransferase and sialyltransferase activities was observed, respectively. Sustained ingestion of ethanol is known to result in pathologic concentrations of 4-HNE, altering both protein activity and structure.12-14 The rodent model for ALD used in this study resulted in a 2-fold increase in 4-HNE immunopositive proteins, further suggesting a role for this aldehyde in the course of disease progression. Although alterations in glycosylation machinery occur in ALD, the mechanisms behind these fluctuations are unknown.21, 22, 35 These data provide a potential mechanism by which sustained ethanol consumption, and its oxidative metabolism, may lead to the observed decrease in ER glycosylation.
Oxidative stress is a known consequence of chronic ethanol ingestion.8-10 The cause-effect behind this phenomenon, however, remains a topic of debate. In the past decade, UPR signaling has been implicated in various disease states, including ALD, however, the factors behind its initiation are unknown.1, 2, 36 A link between protein folding and situations of sustained oxidative stress has been intricately outlined in numerous reports.37-40 It has been documented that chronic oxidative stress may lead to UPR activation, while induction of UPR-associated proteins leads to enhanced ROS production .38 This cycling effect between oxidative stress and UPR induction outlines a mechanism that may be described as a “double-edge sword”. In regard to ALD progression, a direct link between UPR activation and oxidative stress has not been established. Following 6 weeks of ethanol consumption, significant immunohistochemical staining of 4-HNE modified proteins is present, indicating sustained oxidative stress. These mice also display a decrease in ER-resident protein glycosylation, suggesting a possible initiating factor for UPR activation.
Steatosis is a pathologic response to chronic ethanol consumption and currently, mechanisms behind this dysregulation in lipid homeostasis are not clear. The UPR has been implicated in the course of ALD progression, showing activation of multiple proteins, including sterol-response element binding proteins (SREBP).18, 36 Activation of SREBPs has been shown to lead to significant accumulation of triglycerides, resulting in steatosis. Tunicamycin, a pharmacological inhibitor of N-linked glycosylation, induces a potent and rapid ER stress response, resulting in numerous UPR-dependent pathways, including SREBP activation.41 Using our 6 week rodent model of early-stage ALD, a decrease in overall glycosylation were observed in liver microsomal fractions. These findings demonstrate for the first time, a progressive decrease in glycosylation, which coincides with histological analysis confirming the initiating stages of liver damage associated with ALD. Although other processes are likely implicated, alterations in glycosylation may provide a mechanism by which the UPR is induced and provide a novel area for therapeutic intervention.
The present glycoproteomic analysis has lead to the identification of 30 glycosylated proteins in an animal model of ALD. A progressive decrease in glycosylation was also observed with these proteins and may play a role in the observed pathologies of steatotic ALD. Among proteins identified, liver carboxylesterase 3 (also known as triacylglycerol hydrolase, TGH) has been implicated in intracellular lipid metabolism, through its function in the hydrolysis of triacylglycerols (TG).42 Inhibition of this enzyme results in decreased very low density lipoprotein (VLDL) secretion, leading to increased intracellular accumulation.43 Kroetz et al. demonstrated altered enzymatic activity of liver carboxylesterases based on their N-glycosylation status. The esterase activities were determined to be positively regulated by overall glycosylation status, leading to nearly a 75% reduction in activity when glycosylation was removed.44
Our rodent model for ALD demonstrates an apparent global decrease in overall ER glycosylation. Four unique liver carboxylesterases, including TGH, were determined to be altered in our disease model, indicating a potential mechanism for the observed steatosis following chronic ethanol consumption. Although the enzymatic processes behind altered glycosylation in ALD have been noted, this phenomenon has yet to be evaluated in the context of disease progression.21, 22 The identification of 30 unique protein-targets within the ER provides new avenues for research in the field of ALD; however, further investigation into these proteins is necessary to fully elucidate their role in altered lipid metabolism and the pathogenesis of ALD.
Supplementary Material
Acknowledgement
Studies were supported by the National Institutes of Health/National Institutes of Alcoholism and Alcohol Abuse under grant numbers R37AA09300 (DRP), R01DK074487-01 (DRP) and F31AA018606-01 (JJG). HT is supported by NIH R01 grants LM009254-04 (LEH) and LM008111-05 (LEH).
Footnotes
Supporting Information Available: Supporting table includes annotations associated with each functional gene cluster, enrichment scores, assigned UniProt accession numbers and Benjamini corrected p-values. This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Kaplowitz N, Than TA, Shinohara M, Ji C. Endoplasmic reticulum stress and liver injury. Semin Liver Dis. 2007;27(4):367–77. doi: 10.1055/s-2007-991513. [DOI] [PubMed] [Google Scholar]
- 2.Ji C. Dissection of endoplasmic reticulum stress signaling in alcoholic and non-alcoholic liver injury. J Gastroenterol Hepatol. 2008;23(Suppl 1):S16–24. doi: 10.1111/j.1440-1746.2007.05276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seitz O. Glycopeptide synthesis and the effects of glycosylation on protein structure and activity. Chembiochem. 2000;1(4):214–46. doi: 10.1002/1439-7633(20001117)1:4<214::AID-CBIC214>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 4.Blomme B, Van Steenkiste C, Callewaert N, Van Vlierberghe H. Alteration of protein glycosylation in liver diseases. J Hepatol. 2009;50(3):592–603. doi: 10.1016/j.jhep.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 5.Spiro RG. Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology. 2002;12(4):43R–56R. doi: 10.1093/glycob/12.4.43r. [DOI] [PubMed] [Google Scholar]
- 6.Seitz HK, Lieber CS, Stickel F, Salaspuro M, Schlemmer HP, Horie Y. Alcoholic liver disease: from pathophysiology to therapy. Alcohol Clin Exp Res. 2005;29(7):1276–81. doi: 10.1097/01.alc.0000171896.37022.f7. [DOI] [PubMed] [Google Scholar]
- 7.Arteel G, Marsano L, Mendez C, Bentley F, McClain CJ. Advances in alcoholic liver disease. Best Pract Res Clin Gastroenterol. 2003;17(4):625–47. doi: 10.1016/s1521-6918(03)00053-2. [DOI] [PubMed] [Google Scholar]
- 8.Cederbaum AI, Lu Y, Wu D. Role of oxidative stress in alcohol-induced liver injury. Arch Toxicol. 2009;83(6):519–48. doi: 10.1007/s00204-009-0432-0. [DOI] [PubMed] [Google Scholar]
- 9.Koch OR, Pani G, Borrello S, Colavitti R, Cravero A, Farre S, Galeotti T. Oxidative stress and antioxidant defenses in ethanol-induced cell injury. Mol Aspects Med. 2004;25(1-2):191–8. doi: 10.1016/j.mam.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 10.Dey A, Cederbaum AI. Alcohol and oxidative liver injury. Hepatology. 2006;43(2 Suppl 1):S63–74. doi: 10.1002/hep.20957. [DOI] [PubMed] [Google Scholar]
- 11.Sayre LM, Lin D, Yuan Q, Zhu X, Tang X. Protein adducts generated from products of lipid oxidation: focus on HNE and one. Drug Metab Rev. 2006;38(4):651–75. doi: 10.1080/03602530600959508. [DOI] [PubMed] [Google Scholar]
- 12.Sampey BP, Stewart BJ, Petersen DR. Ethanol-induced modulation of hepatocellular extracellular signal-regulated kinase-1/2 activity via 4-hydroxynonenal. J Biol Chem. 2007;282(3):1925–37. doi: 10.1074/jbc.M610602200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roede JR, Orlicky DJ, Fisher AB, Petersen DR. Overexpression of peroxiredoxin 6 does not prevent ethanol-mediated oxidative stress and may play a role in hepatic lipid accumulation. J Pharmacol Exp Ther. 2009;330(1):79–88. doi: 10.1124/jpet.109.152983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carbone DL, Doorn JA, Kiebler Z, Petersen DR. Cysteine modification by lipid peroxidation products inhibits protein disulfide isomerase. Chem Res Toxicol. 2005;18(8):1324–31. doi: 10.1021/tx050078z. [DOI] [PubMed] [Google Scholar]
- 15.Lucey MR. Management of alcoholic liver disease. Clin Liver Dis. 2009;13(2):267–75. doi: 10.1016/j.cld.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 16.You M, Crabb DW. Recent advances in alcoholic liver disease II. Minireview: molecular mechanisms of alcoholic fatty liver. Am J Physiol Gastrointest Liver Physiol. 2004;287(1):G1–6. doi: 10.1152/ajpgi.00056.2004. [DOI] [PubMed] [Google Scholar]
- 17.Malhotra JD, Kaufman RJ. The endoplasmic reticulum and the unfolded protein response. Semin Cell Dev Biol. 2007;18(6):716–31. doi: 10.1016/j.semcdb.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ji C, Kaplowitz N. Betaine decreases hyperhomocysteinemia, endoplasmic reticulum stress, and liver injury in alcohol-fed mice. Gastroenterology. 2003;124(5):1488–99. doi: 10.1016/s0016-5085(03)00276-2. [DOI] [PubMed] [Google Scholar]
- 19.Ji C, Kaplowitz N. Hyperhomocysteinemia, endoplasmic reticulum stress, and alcoholic liver injury. World J Gastroenterol. 2004;10(12):1699–708. doi: 10.3748/wjg.v10.i12.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshida H. ER stress and diseases. FEBS J. 2007;274(3):630–58. doi: 10.1111/j.1742-4658.2007.05639.x. [DOI] [PubMed] [Google Scholar]
- 21.Ghosh P, Lakshman MR. Chronic ethanol induced impairment of hepatic glycosylation machinery in rat is independent of dietary carbohydrate. Alcohol Clin Exp Res. 1997;21(1):76–81. [PubMed] [Google Scholar]
- 22.Ghosh P, Liu QH, Lakshman MR. Long-term ethanol exposure impairs glycosylation of both N- and O-glycosylated proteins in rat liver. Metabolism. 1995;44(7):890–8. doi: 10.1016/0026-0495(95)90242-2. [DOI] [PubMed] [Google Scholar]
- 23.Roede JR, Stewart BJ, Petersen DR. Decreased expression of peroxiredoxin 6 in a mouse model of ethanol consumption. Free Radic Biol Med. 2008;45(11):1551–8. doi: 10.1016/j.freeradbiomed.2008.08.032. [DOI] [PubMed] [Google Scholar]
- 24.Roede JR, Carbone DL, Doorn JA, Kirichenko OV, Reigan P, Petersen DR. In vitro and in silico characterization of peroxiredoxin 6 modified by 4-hydroxynonenal and 4-oxononenal. Chem Res Toxicol. 2008;21(12):2289–99. doi: 10.1021/tx800244u. [DOI] [PubMed] [Google Scholar]
- 25.Julenius K, Molgaard A, Gupta R, Brunak S. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology. 2005;15(2):153–64. doi: 10.1093/glycob/cwh151. [DOI] [PubMed] [Google Scholar]
- 26.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 27.Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4(6):1633–49. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- 28.Cao J, Shen C, Wang H, Shen H, Chen Y, Nie A, Yan G, Lu H, Liu Y, Yang P. Identification of N-glycosylation sites on secreted proteins of human hepatocellular carcinoma cells with a complementary proteomics approach. J Proteome Res. 2009;8(2):662–72. doi: 10.1021/pr800826u. [DOI] [PubMed] [Google Scholar]
- 29.Liu T, Qian WJ, Gritsenko MA, Camp DG, 2nd, Monroe ME, Moore RJ, Smith RD. Human plasma N-glycoproteome analysis by immunoaffinity subtraction, hydrazide chemistry, and mass spectrometry. J Proteome Res. 2005;4(6):2070–80. doi: 10.1021/pr0502065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lattova E, Tomanek B, Bartusik D, Perreault H. N-glycomic changes in human breast carcinoma MCF-7 and T-lymphoblastoid cells after treatment with herceptin and herceptin/Lipoplex. J Proteome Res. 9(3):1533–40. doi: 10.1021/pr9010266. [DOI] [PubMed] [Google Scholar]
- 31.Dell A, Morris HR. Glycoprotein structure determination by mass spectrometry. Science. 2001;291(5512):2351–6. doi: 10.1126/science.1058890. [DOI] [PubMed] [Google Scholar]
- 32.Klement E, Lipinszki Z, Kupihar Z, Udvardy A, Medzihradszky KF. Enrichment of O-GlcNAc modified proteins by the periodate oxidation-hydrazide resin capture approach. J Proteome Res. 9(5):2200–6. doi: 10.1021/pr900984h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hart C, Schulenberg B, Steinberg TH, Leung WY, Patton WF. Detection of glycoproteins in polyacrylamide gels and on electroblots using Pro-Q Emerald 488 dye, a fluorescent periodate Schiff-base stain. Electrophoresis. 2003;24(4):588–98. doi: 10.1002/elps.200390069. [DOI] [PubMed] [Google Scholar]
- 34.Marinari UM, Pronzato MA, Cottalasso D, Rolla C, Biasi F, Poli G, Nanni G, Dianzani MU. Inhibition of liver Golgi glycosylation activities by carbonyl products of lipid peroxidation. Free Radic Res Commun. 1987;3(1-5):319–24. doi: 10.3109/10715768709069799. [DOI] [PubMed] [Google Scholar]
- 35.Gong M, Castillo L, Redman RS, Garige M, Hirsch K, Azuine M, Amdur RL, Seth D, Haber PS, Lakshman MR. Down-regulation of liver Galbeta1, 4GlcNAc alpha2, 6-sialyltransferase gene by ethanol significantly correlates with alcoholic steatosis in humans. Metabolism. 2008;57(12):1663–8. doi: 10.1016/j.metabol.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 36.Ji C, Mehrian-Shai R, Chan C, Hsu YH, Kaplowitz N. Role of CHOP in hepatic apoptosis in the murine model of intragastric ethanol feeding. Alcohol Clin Exp Res. 2005;29(8):1496–503. doi: 10.1097/01.alc.0000174691.03751.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Csala M, Margittai E, Banhegyi G. Redox control of endoplasmic reticulum function. Antioxid Redox Signal. 13(1):77–108. doi: 10.1089/ars.2009.2529. [DOI] [PubMed] [Google Scholar]
- 38.Malhotra JD, Kaufman RJ. Endoplasmic reticulum stress and oxidative stress: a vicious cycle or a double-edged sword? Antioxid Redox Signal. 2007;9(12):2277–93. doi: 10.1089/ars.2007.1782. [DOI] [PubMed] [Google Scholar]
- 39.Malhotra JD, Miao H, Zhang K, Wolfson A, Pennathur S, Pipe SW, Kaufman RJ. Antioxidants reduce endoplasmic reticulum stress and improve protein secretion. Proc Natl Acad Sci U S A. 2008;105(47):18525–30. doi: 10.1073/pnas.0809677105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santos CX, Tanaka LY, Wosniak J, Laurindo FR. Mechanisms and implications of reactive oxygen species generation during the unfolded protein response: roles of endoplasmic reticulum oxidoreductases, mitochondrial electron transport, and NADPH oxidase. Antioxid Redox Signal. 2009;11(10):2409–27. doi: 10.1089/ars.2009.2625. [DOI] [PubMed] [Google Scholar]
- 41.Yamamoto K, Takahara K, Oyadomari S, Okada T, Sato T, Harada A, Mori K. Induction of liver steatosis and lipid droplet formation in ATF6alpha-knockout mice burdened with pharmacological endoplasmic reticulum stress. Mol Biol Cell. 21(17):2975–86. doi: 10.1091/mbc.E09-02-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dolinsky VW, Gilham D, Alam M, Vance DE, Lehner R. Triacylglycerol hydrolase: role in intracellular lipid metabolism. Cell Mol Life Sci. 2004;61(13):1633–51. doi: 10.1007/s00018-004-3426-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gilham D, Ho S, Rasouli M, Martres P, Vance DE, Lehner R. Inhibitors of hepatic microsomal triacylglycerol hydrolase decrease very low density lipoprotein secretion. FASEB J. 2003;17(12):1685–7. doi: 10.1096/fj.02-0728fje. [DOI] [PubMed] [Google Scholar]
- 44.Kroetz DL, McBride OW, Gonzalez FJ. Glycosylation-dependent activity of baculovirus-expressed human liver carboxylesterases: cDNA cloning and characterization of two highly similar enzyme forms. Biochemistry. 1993;32(43):11606–17. doi: 10.1021/bi00094a018. [DOI] [PubMed] [Google Scholar]
- 45.Wollscheid B, Bausch-Fluck D, Henderson C, O’Brien R, Bibel M, Schiess R, Aebersold R, Watts JD. Mass-spectrometric identification and relative quantification of N-linked cell surface glycoproteins. Nat Biotechnol. 2009;27(4):378–86. doi: 10.1038/nbt.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ghesquiere B, Van Damme J, Martens L, Vandekerckhove J, Gevaert K. Proteome-wide characterization of N-glycosylation events by diagonal chromatography. J Proteome Res. 2006;5(9):2438–47. doi: 10.1021/pr060186m. [DOI] [PubMed] [Google Scholar]
- 47.Bernhard OK, Kapp EA, Simpson RJ. Enhanced analysis of the mouse plasma proteome using cysteine-containing tryptic glycopeptides. J Proteome Res. 2007;6(3):987–95. doi: 10.1021/pr0604559. [DOI] [PubMed] [Google Scholar]
- 48.Kawamoto S, Uchino S, Hattori S, Hamajima K, Mishina M, Nakajima-Iijima S, Okuda K. Expression and characterization of the zeta 1 subunit of the N-methyl-D-aspartate (NMDA) receptor channel in a baculovirus system. Brain Res Mol Brain Res. 1995;30(1):137–48. doi: 10.1016/0169-328x(95)00005-d. [DOI] [PubMed] [Google Scholar]
- 49.Ostasiewicz P, Zielinska DF, Mann M, Wisniewski JR. Proteome, phosphoproteome, and N-glycoproteome are quantitatively preserved in formalin-fixed paraffin-embedded tissue and analyzable by high-resolution mass spectrometry. J Proteome Res. 9(7):3688–700. doi: 10.1021/pr100234w. [DOI] [PubMed] [Google Scholar]
- 50.Chen R, Jiang X, Sun D, Han G, Wang F, Ye M, Wang L, Zou H. Glycoproteomics analysis of human liver tissue by combination of multiple enzyme digestion and hydrazide chemistry. J Proteome Res. 2009;8(2):651–61. doi: 10.1021/pr8008012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.