Abstract
Premature senescence, a key strategy used to suppress carcinogenesis, can be driven by p53/p21 proteins in response to various stresses. Here, we demonstrate that Wig1 plays a critical role in this process through regulation of p21 mRNA stability. Wig1 controls the association of Argonaute2 (Ago2), a central component of the RNA-induced silencing complex (RISC), with target p21 mRNA via binding of the stem-loop structure near the microRNA (miRNA) target site. Depletion of Wig1 prohibited miRNA-mediated p21 mRNA decay and resulted in premature senescence. Wig1 plays an essential role in cell proliferation, as demonstrated in tumour xenografts in mice, and Wig1 and p21 mRNA levels are inversely correlated in human normal and cancer tissues. Together, our data indicate a novel role of Wig1 in RISC target accessibility, which is a key step in RNA-mediated gene silencing. In addition, these findings indicate that fine-tuning of p21 levels by Wig1 is essential for the prevention of cellular senescence.
Keywords: cellular senescence, p21 mRNA stability, RISC recruitment, RNA-binding protein, Wig1
Introduction
Cellular senescence was originally defined by the phenotype of human fibroblasts undergoing replicative exhaustion in vitro (Hayflick and Moorhead, 1961). Although this phenotype represents a stable state of cell-cycle arrest, cellular senescence is prematurely triggered in response to diverse forms of cellular damage or stress. Because cellular senescence limits the proliferative potential of premalignant cells, this process is thought to be a key strategy for the suppression of carcinogenesis (Schmitt et al, 2002; Chen et al, 2005; Lee et al, 2010). Premature senescence can be triggered through two complementary pathways, which involve the p53/p21 and p16/retinoblastoma (pRb) tumour suppressor proteins. First, p21 (also known as Cip1/Waf1/CDKN1A), which is a direct inhibitor of cyclin/cyclin-dependent kinase (CDK) complexes, is an important player that induces cellular senescence in response to DNA damage or oncogene imbalance (Brugarolas et al, 1995; Deng et al, 1995; Wang et al, 1999). The expression of p21 is strictly regulated at the transcriptional level through p53-dependent and/or -independent mechanisms and also at the post-transcriptional and post-translational levels through mechanisms involving mRNA stability, subcellular localization, and/or protein stability (Sheikh et al, 1994; Gartel and Tyner, 1999; Abbas and Dutta, 2009). Recently, post-transcriptional control through a microRNA (miRNA)-mediated mRNA silencing mechanism has been implicated as an important mechanism of p21 regulation (Wu et al, 2010).
The miRNAs comprise a group of short (typically ∼22 nucleotides), non-coding RNAs that suppress the expression of protein-coding genes by mRNA decay and/or suppress translation through guiding the ribonucleoprotein RNA-induced silencing complex (RISC), which contains the Argonaute (Ago) proteins (Bartel, 2009). RISC-loaded miRNAs recognize target sites in the 3′-untranslated regions (UTRs) of their target mRNAs (Kawamata and Tomari, 2010). The most critical requirement for target recognition is complementary base pairing between the target site and the 5′ region of the miRNA, the so-called canonical ‘seed’ region (Grimson et al, 2007; Bartel, 2009). For post-transcriptional control, modulation of miRNA function through miRNA biogenesis, localization, and degradation is a key process. In addition, miRNA activity is enhanced or hindered by RNA-binding proteins (RBPs; van Kouwenhove et al, 2011). For example, HuR, an Elav-like protein, binds to the 3′UTR of cationic amino-acid transporter 1 (CAT1) mRNA and relieves miR-122-mediated repression (Bhattacharyya et al, 2006). On the other hand, HuR facilitates c-Myc repression by recruiting let-7-loaded RICS via an association with the c-Myc 3′UTR that neighbours a let-7-binding site (Kim et al, 2009a). Thus, RBPs play fundamental roles in post-transcriptional control, which is regulated by different processes of mRNA metabolism and translation (Kim et al, 2009b). Since miRNA target recognition is a key process for RISC functions, the RBPs governing RISC-loaded miRNA recruitment to its target are waiting to be uncovered (Wiemer, 2007; Kawamata and Tomari, 2010; van Kouwenhove et al, 2011).
Wig1 (wild-type p53-induced gene 1; official gene symbol is ZMAT3, zinc finger, matrin-type 3) encodes a Cys2His2-type zinc finger protein that contains three zinc fingers (ZFs) and a nuclear localization signal (NLS) (Israeli et al, 1997; Varmeh-Ziaie et al, 1997). Wig1 was identified as a p53-responsive gene and is highly conserved from fish to humans, especially within the ZFs, which are almost completely conserved (Wilhelm et al, 2002; Hellborg and Wiman, 2004). Wig1 interacts with heterogeneous nuclear ribonucleoprotein (hnRNP) A2/B1 and RNA helicase A (RHA) via dsRNA binding and regulates p53 mRNA stability through the protection of deadenylation (Méndez Vidal et al, 2006; Prahl et al, 2008; Vilborg et al, 2009). Despite several previous studies, the function and biological significance of Wig1 remain nebulous.
In this study, we show that Wig1 depletion induces premature senescence due to p21 mRNA stabilization. Wig1 preferentially interacts with the stem-loop structure of the 3′UTR that neighbours miRNA target sites to determine the target accessibility of RISC. Our data describe a novel role for Wig1 in modulating miRNA-mediated mRNA decay.
Results
Wig1 depletion induces premature senescence
Wig1 has been identified as a p53-responsive gene and has been suggested to play a role in the p53-dependent stress response (Israeli et al, 1997; Hellborg et al, 2001); however, its biological significance has yet to be elucidated. To investigate the biological function of Wig1, we first examined the influence of Wig1 depletion on cell growth, cellular morphology, and apoptosis. MCF7 breast cancer cells, which were treated with an siRNA against Wig1 (Wig Si), exhibited decreased cell proliferation and clonogenic ability (Figure 1A–C). Interestingly, Wig1 depletion induced typical senescence-associated phenotypic changes: large and flattened morphology, positive senescence-associated β-galactosidase (SA-β-Gal) staining, and G1 cell-cycle arrest (Figure 1D–F). No significant increase in the subG1 phase was observed (Figure 1F). Wig1 depletion contributed to the gradual decrease in phospho-pRb and the increase in p21, and such changes were sustained regardless of p53 activation (Figure 1G). The premature senescent phenotypes were observed up to 10 days after the depletion of Wig1 (Supplementary Figure S1). We confirmed a lack of off-target effects of Wig1 Si in p21 accumulation and induction of premature senescence (Supplementary Figure S2). Wig1 depletion also resulted in premature senescence phenotypes including p21 induction in other cell types, such as H460 lung carcinoma cells, U2OS osteosarcoma cells, MCF10A normal mammary epithelial cells, and primary human diploid fibroblasts (Supplementary Figures S3 and S4). Overexpression of siRNA-resistant Wig1 resulted in rescue of the abovementioned premature senescent phenotypes observed following the depletion of Wig 1 (Supplementary Figure S5). These data demonstrate that Wig1 depletion induces p21 accumulation and hypophosphorylation of pRb and ultimately leads to premature senescence in both normal and cancer cells.
Figure 1.
Depletion of Wig1 in MCF7 cancer cells induces premature senescence via G1 arrest. (A) Growth retardation of either control siRNA (Con Si)- or Wig1 siRNA (Wig1 Si)-transfected MCF7 cells. Viable cell numbers were determined using a trypan blue exclusion assay at the indicated times after transfection with 100 nM Wig1 Si (in box). (B) A BrdU incorporation assay was conducted in Wig1 Si-transfected MCF7 cells at the indicated times. Values are expressed as the relative BrdU incorporation in Wig Si-transfected cells compared to Con Si-transfected cells. (C) Colony-forming ability of Wig1 Si-transfected cells. Cells were fixed and stained 7 days after siRNA transfection. (D) Morphologic changes of Wig1 Si-transfected MCF7 cells. (E) SA-β-Gal positivity was evaluated after siRNA transfection (left panel). Percentages of SA-β-Gal-positive cells are plotted (right panel). (F) FACS analysis of cell-cycle distribution in Wig1-depleted cells. Cells were harvested at the indicated times after Wig1 Si transfection. (G) Molecular status of phospho-pRB, p53, and p21 in Wig1-depleted cells. The cells were harvested at the indicated times after siRNA transfection and subjected to immunoblotting. Actin serves as a loading control. Each bar represents mean±standard deviation (s.d.) from three independent experiments. An asterisk indicates a significant difference, P<0.05.
p21 plays an essential role in Wig1 depletion-mediated premature senescence
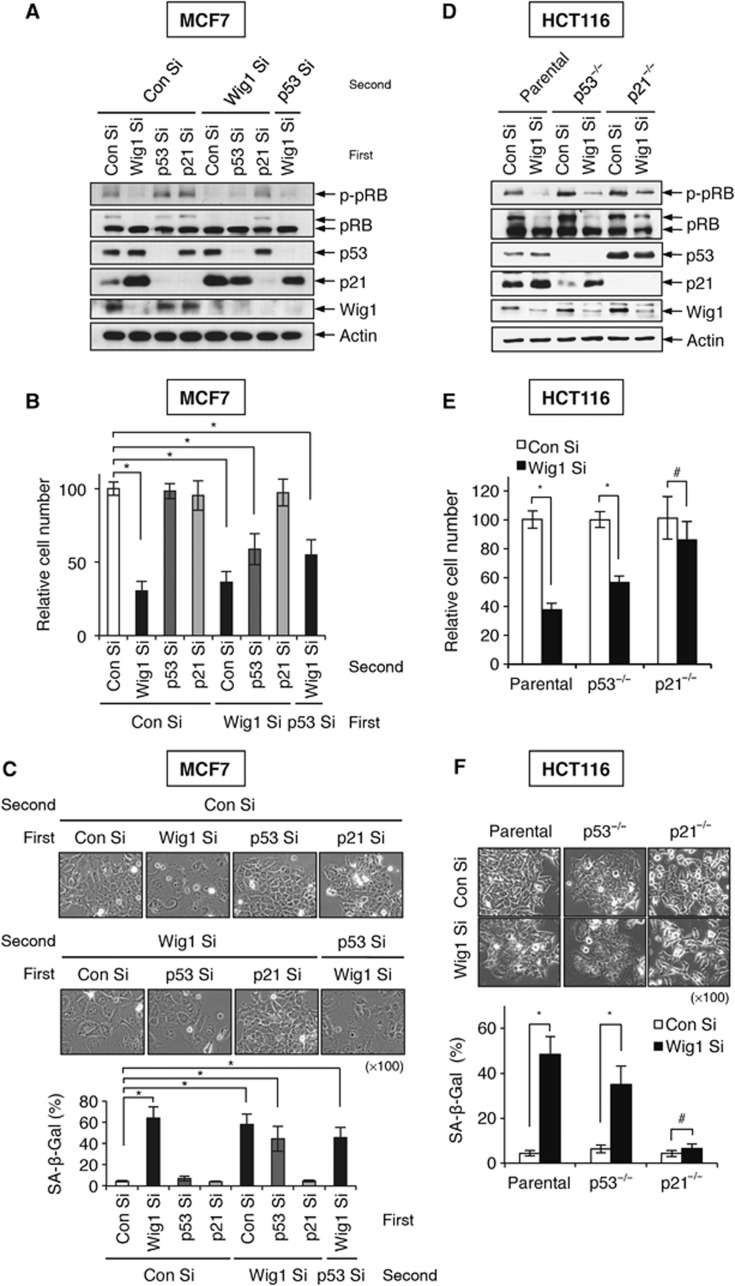
Next, we investigated the involvement of p53 and p21 in premature senescence induced by Wig1 depletion. Wig1 depletion resulted in p21 accumulation and decreased cell proliferation in p53-depleted MCF7 cells (Figure 2A and B). Analysis of SA-β-Gal staining demonstrated that Wig1 depletion induced premature senescence even in the absence of p53 (Figure 2C). In contrast, p21 depletion abrogated Wig1 depletion-mediated premature senescence (Figure 2A–C). To obtain additional evidence, we examined Wig1 depletion-mediated premature senescence in HCT116parental, HCT116p53−/−, and HCT116p21−/− isogenic cell lines. In accordance with HCT116parental cells, HCT116p53−/− cells exhibited increased p21 levels, limited cell proliferation, and positivity of SA-β-Gal staining in the absence of Wig1 (Figure 2D–F), whereas Wig1 depletion did not induce premature senescence in HCT116p21−/− cells, which exhibited no significant alterations in cell growth rate and SA-β-Gal positivity (Figure 2D–F). In the absence of p53, however, Wig1 depletion-mediated premature senescence was lessened due to a decreased level of p21 as compared to cells with intact p53 (Figure 2). These results indicate that p21 accumulation is indispensible for the Wig1 depletion-mediated premature senescence.
Figure 2.
p21 is essential for the induction of Wig1 depletion-mediated premature senescence that occurs in a p53-independent manner. (A) Western blot analysis in siRNA-transfected MCF7 cells. Cells were transfected with Con Si, Wig1 Si, p53 siRNA (p53 Si), or p21 siRNA (p21 Si) 24 h prior to transfection with Con Si, Wig1 Si, or p53 Si as indicated. After a 2-day incubation, cells were subjected to immunoblot analysis. Actin serves as a loading control. (B) Viable cells were counted using a trypan blue exclusion assay 4 days after double transfections as in (A). Cell numbers in Wig1 Si-, p53 Si-, and p21 Si-transfected cells are shown as a relative value compared to Con Si-transfected cells. (C) Cellular morphology and SA-β-Gal positivity analyses were performed 4 days after double transfections as in (A). (D) Western blot analysis of Wig1 Si-transfected HCT116 parental, p53−/−, and p21−/− isogenic cell lines. The cell lines were incubated for 2 days after transfection with either Con Si or Wig1 Si and then subjected to immunoblotting. (E) Viable cells were counted using a trypan blue exclusion assay 4 days after Wig1 Si transfection of HCT116 isogenic cell lines, and cell counts in Wig Si-transfected cells are presented as a relative value compared to each Con Si-transfected isogenic cell line. (F) Cellular morphology and SA-β-Gal positivity were determined 4 days after Wig1 Si transfection. Each bar represents mean±s.d. from three independent experiments. An * or # indicates either P<0.05 or P>0.05, respectively.
Wig1 modulates p21 levels via the regulation of p21 mRNA stability
To define the regulatory mechanism of p21 accumulation in Wig1-depleted cells, we examined the transcriptional activity, mRNA stability, and protein stability of p21. No significant difference was found in p21 promoter activity, which was measured using a luciferase reporter containing the p21 promoter (WWP-Luc), in Wig1 Si- and control siRNA (Con Si)- transfected MCF7 cells (Figure 3A). To determine whether Wig1 regulates p21 expression at the post-translational level, we treated cells with cycloheximide (CHX), an inhibitor of protein biosynthesis. Western blot analysis revealed that the p21 protein degradation rate was not affected by Wig1 depletion (Figure 3B). Next, to examine the change in p21 mRNA levels following Wig1 depletion, we performed quantitative real-time reverse transcription-PCR (qRT–PCR). The p21 mRNA levels were inversely correlated with Wig1 mRNA levels (Figure 3C). To explore the decay rate of the p21 transcript, actinomycin D (ActD) chase and qRT–PCR assays were performed. As shown in Figure 3D, the half-life of the p21 transcript was significantly increased due to Wig1 depletion (3.29 versus 5.34 h). To obtain additional evidence of p21 regulation by Wig1, the levels of p21 mRNA and protein were measured in Flag-tagged Wig1 (Flag–Wig1)-overexpressed and in Wig1-depleted cells. Overexpression and depletion of Wig 1 downregulated and upregulated p21, respectively, at both the mRNA and protein levels (Figure 3E). These results demonstrate that Wig1 regulates the p21 protein level via regulation of p21 mRNA decay.
Figure 3.
Wig1 plays a role in the regulation of p21 mRNA stability. (A) p21 promoter activity. Firefly luciferase activities were measured in MCF7 cells 2 days after co-transfection with a luciferase vector containing the p21 promoter and Renilla luciferase pRL-CMV (RL) reference plasmid prior to Wig1 Si transfection (upper). Cells exposed to 6 Gy of ionizing radiation (IR) were used as a positive control (PC). Purified proteins were subjected to immunoblotting (lower). (B) Wig1 depletion-mediated p21 protein decay in the presence of cycloheximide (CHX). MCF7 cells were treated with 40 μg/ml CHX following Wig1 Si transfection and then harvested at the indicated times for immunoblot analysis (upper). The normalized band intensities were evaluated with ImageJ software (lower). (C) Relative p21 mRNA levels in Wig1-depleted cells. Purified RNA harvested at the indicated times after Wig1 Si transfection was subjected to qRT–PCR using p21- or Wig1-specific primers. The data were normalized to actin mRNA levels. (D) Actinomycin D (ActD) chase experiment. Twenty-four hours after transfection with Wig1 Si (in box), 5 μg/ml ActD was added to cells for the indicated times. Purified RNA was subjected to qRT–PCR with p21-specific primers. (E) Effects of Wig1 overexpression on p21 mRNA and protein levels. RNA was isolated from cells transfected with either p3xFlag-empty vector (p3xFlag–EV) or p3xFlag–Wig1 in combination with either Con Si or Wig1 Si as indicated. Then, qRT–PCR was performed using p21-specific primers. Relative mRNA levels were normalized to actin mRNA (upper). Proteins were subjected to immunoblot analysis (lower). Each bar represents mean±s.d. from three independent experiments. t1/2 indicates the estimated half-lives of p21 protein or mRNA after Wig1 Si transfection. An * or # indicates either P<0.05 or P>0.05, respectively.
Wig1 binds to the 3′UTR of p21 mRNA through ZF domains 1 and 2
Because Wig1 is a ZF protein that contains an unusual dsRNA-binding domain (Méndez Vidal et al, 2006), we investigated whether Wig1 directly binds to p21 mRNA using ribonucleoprotein immunoprecipitation (RNP-IP) and a semiquantitative (sq) RT–PCR assay. As shown in Figure 4A, Wig1 associated with the p21 mRNA and reduced its level in Wig1-overexpressing MCF7 cells. In general, cis-acting determinants located in the 3′UTR regulate the stability of mRNA by association with specific proteins (Guhaniyogi and Brewer, 2001). Therefore, we hypothesized that Wig1 binds to the 3′UTR of p21 mRNA to regulate its stability. To test this hypothesis, we used a reporter system containing pFL-p21-3′UTRs that express the N-terminal ORF of firefly luciferase (FL) mRNA and contain the full-length 3′UTR (nt 1–1531) derived from p21 mRNA (Figure 4B, upper panel). The cells were simultaneously transfected with three plasmids: (1) p3xFlag–empty vector (EV) or p3xFlag–Wig1, (2) pFL-EV or pFL-p21-3′UTR as an mRNA reporter plasmid, and (3) Renilla luciferase-expressing vector pRL-CMV (pRL) as a reference plasmid. Whole cell lysates were subjected to RNP-IP with anti-Flag M2 affinity gel, and RNA was isolated from immunoprecipitates. The target FL and reference RL mRNA was amplified using [32P]-α-dNTP and quantified using a radioisotope-imaging system (Figure 4B, lower panel). Indeed, Wig1 overexpression led to a decrease in reporter mRNA levels, and Wig1 directly bound to the p21 3′UTR.
Figure 4.
Wig1 binds to the 3′UTR of the p21 mRNA through zinc finger domains 1 and 2. (A) Ribonucleoprotein immunoprecipitation (RNP-IP) assay. MCF7 cells were harvested 2 days after transfection with either p3xFlag–EV or p3xFlag–Wig1 (Flag–Wig1), and then an RNP-IP assay using anti-Flag M2 affinity gel was performed. Immunoprecipitated proteins were then subjected to immunoblotting using anti-Flag antibody (upper). Isolated total RNA was radiolabelled with RT–PCR using p21-specific primers (lower), and band intensities were quantified using imaging software. The four leftmost lanes represent two-fold serial dilutions of RNA and demonstrate that the RT–PCR is semiquantitative. (B) Interaction of Wig1 and p21 3′UTR reporter. Schematic representations of pFL-EV and pFL-p21-3′UTR, which encodes the N-terminal portion of firefly luciferase with the full-length 3′UTR of p21 (upper). HEK 293T cells were transfected with either pFL-EV or pFL-p21-3′UTR in combination with p3xFlag–EV or p3xFlag–Wig1. Cells were also co-transfected with pRL-CMV (pRL) as a reference plasmid. Cells were harvested 2 days after transfection and subjected to RNP-IP using anti-Flag M2 affinity gel. Purified RNA from IP was radiolabelled using RT–PCR using RL- and FL-specific primers, and products were subjected to PAGE. The p21 3′UTR reporter mRNA levels were quantified using imaging software, and data are presented as mean±s.d. from three independent experiments (lower). (C) Schematic representations of the p3xFlag–Wig1-wild-type (wt), p3xFlag–Wig1-mutant 1 (mut1), and p3xFlag–Wig1-mutant 2 (mut2) overexpression plasmids. ZF, zinc finger; NLS, nuclear localization signal. (D) Immunoblotting and qRT–PCR. MCF7 cells were transfected with Wig1 Si prior to transfection with pFL-p21-3′UTR in combination with p3xFlag–EV, p3xFlag–Wig1-wt, p3xFlag–Wig1-mut1, or p3xFlag–Wig1-mut2. The cells were also co-transfected with pRL-CMV as a reference plasmid. Whole cell lysates and RNA were isolated 2 days after transfection. Endogenous Wig1 depletion and recombinant Wig1 overexpression were assessed using immunoblot analysis (upper). The qRT–PCR analysis shows the relative levels of FL-p21-3′UTR reporter mRNA (lower). Each bar represents mean±s.d. from three independent experiments. An * or # indicates either P<0.05 or P>0.05, respectively. (E) Endogenous p21 protein levels in cells overexpressing recombinant Wig1. MCF7 cells were harvested 2 days after transfection with p3xFlag–EV, p3xFlag–Wig1-wt, p3xFlag–Wig1-mut1, or p3xFlag–Wig1-mut2 and subjected to immunoblotting.
To identify the regions of Wig1 involved in binding to the p21 3′UTR, we constructed deletion mutants of Wig1 that lack either ZF3 (mut1) or ZF1 and ZF2 (mut2; Figure 4C). The effects of Wig1-mut1 and Wig1-mut2 on reporter mRNA levels were investigated following Wig1 depletion in MCF7 cells that expressed the p21 3′UTR reporter (Figure 4D). Because Wig1 Si targeted the 3′UTR of its own mRNA, Wig1 overexpression vectors that lack a 3′UTR sequence were resistant to Wig1 Si treatment. The qRT–PCR assay revealed that Wig1-wt and Wig1-mut1 reverted the p21 3′UTR reporter mRNA levels that were increased due to Wig1 depletion to levels similar to those of Con Si-transfected cells (bars 4 and 5 versus bar 1, Figure 4D, lower panel), whereas Flag–Wig1-mut2 failed to rescue these reporter mRNA levels (bar 6 versus bar 1, Figure 4D, lower panel). The importance of the Wig1 ZF1 and ZF2 region in the regulation of p21 was confirmed by western blot analysis (Figure 4E). Wig1-wt and Wig1-mut1 decreased endogenous p21 protein levels, whereas Wig1-mut2 did not. These results indicate that Wig1 regulates p21 mRNA levels and p21 protein levels via its ZF1 and ZF2 domains.
Wig1 is critical for the recruitment of Ago2 to p21 mRNA 3′UTR
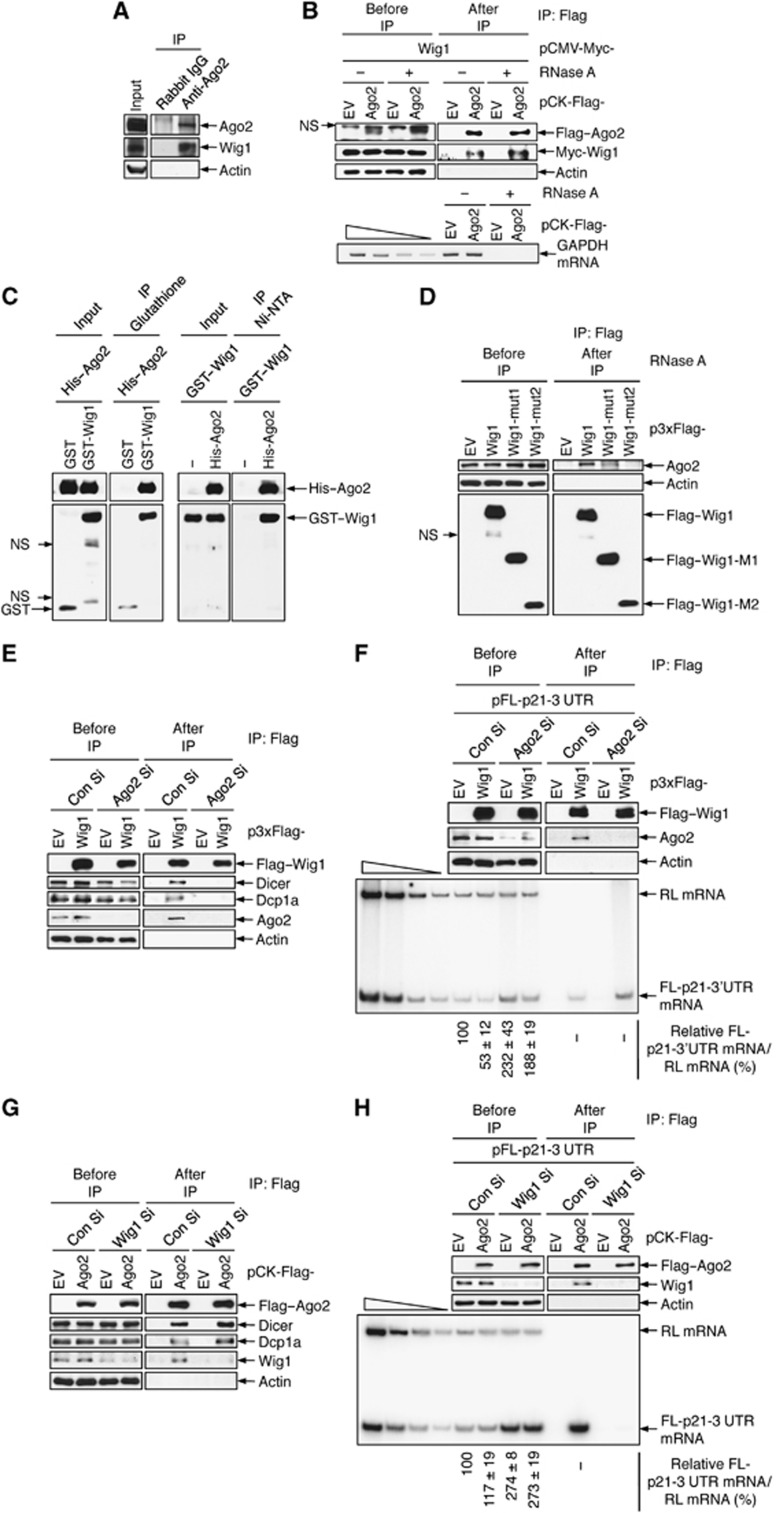
RHA, which was recently reported to interact with Wig1 via dsRNA, directly binds to RISC components, including Ago2 (Robb and Rana, 2007; Prahl et al, 2008). Thus, we hypothesized that Wig1 may participate in miRNA-mediated mRNA silencing. To test this hypothesis, we performed IP-immunoblot assays with anti-Ago2 antibody and found that endogenous Wig1 protein was physically associated with Ago2 protein (Figure 5A). Moreover, Ago2 interacted with Wig1 in an RNase A-insensitive manner in HEK 293T cells that overexpressed Wig1 and Ago2 (Figure 5B). The direct binding of Wig1 to Ago2 was confirmed by an in vitro binding assay with polyhistidine-tagged Ago2 (His–Ago2) and glutathione S-transferase-tagged Wig1 (GST–Wig1) proteins (Figure 5C). We then tested the binding capacity of Wig1 mutants to Ago2 protein and found that Ago2 was associated with Wig1-wt and Wig1-mut1 but not with Wig1-mut2 (Figure 5D). This finding is consistent with our data showing that Wig1 regulates p21 mRNA levels via its ZF1 and ZF2 domains (Figure 4D).
Figure 5.
Wig1 is essential for recruitment of RNA-inducing silencing complex (RISC) to the p21 mRNA 3′UTR. (A) Protein–protein interactions between Wig1 and Ago2. Endogenous Ago2 protein was immunoprecipitated from MCF7 cell lysates with polyclonal anti-Ago2 antibody and then subjected to immunoblotting using anti-Ago2 and anti-Wig1 antibodies as indicated. (B) Physical interaction between Wig1 and Ago2. HEK 293T cell lysates were isolated after co-transfection with pCK-Flag–Ago2 and pCMV-Myc–Wig1 and immunoprecipitated using anti-Flag M2 affinity gel. RNase A was added to half of each sample prior to immunoprecipitation. Immunoblotting was performed to detect the protein interaction (upper). GAPDH mRNA was analysed using RT–PCR to demonstrate that RNase A digestion was complete (lower). (C) In vitro binding assay of polyhistidine-tagged Ago2 (His–Ago2) and glutathione S transferase-tagged Wig1 (GST–Wig1) proteins. Equal amounts of recombinant His–Ago2 proteins were precipitated with glutathione beads via bound GST or GST–Wig1 proteins. Precipitates were then subjected to immunoblotting with anti-His and anti-GST antibodies as indicated (left). Conversely, the GST–Wig1 proteins were precipitated with Ni-NTA agarose beads via bound His–Ago2 proteins and then subjected to immunoblot analysis (right). (D) Interaction of Ago2 with Wig1 mutant proteins. Lysates from Flag–Wig1-wt-, Flag–Wig1-mut1-, or Flag–Wig1-mut2-expressing HEK 293T cells were subjected to immunoprecipitation using anti-Flag M2 affinity gel followed by immunoblotting with anti-Ago2 antibody as indicated. (E, G) Association of either Wig1 or Ago2 with RISC. Ago2- or Wig1-depleted HEK 293T cells were transfected with either p3xFlag–Wig1 or pCK-Flag–Ago2, respectively. Proteins were immunoprecipitated using anti-Flag M2 affinity gel. Western blot analysis was used to detect the association of the specified proteins. (F) Interaction of Wig1 with the p21 3′UTR in the absence of Ago2. Ago2-depleted HEK 293T cells were harvested 2 days after transfection with pFL-p21-3′UTR in combination with either p3xFlag–EV or p3xFlag–Wig1 and pRL as a reference plasmid, and then an RNP-IP assay was performed with anti-Flag M2 affinity gel. Proteins were subjected to immunoblotting using anti-Flag and anti-Ago2 antibodies as indicated (upper). Total RNA isolated from IP was radiolabelled using RT–PCR with RL- and FL-specific primers and subjected to PAGE (lower). (H) No interaction of Ago2 with the p21 3′UTR was observed in the absence of Wig1. Wig1-depleted HEK 293T cells were harvested 2 days after transfection of pFL-p21-3′UTR in combination with either pCK-Flag–EV or pCK-Flag–Ago2 and pRL as a reference plasmid and then subjected to RNP-IP as described in (F). The four leftmost lanes represent two-fold serial dilutions of RNA and demonstrate that the RT–PCR is semiquantitative. The p21 3′UTR reporter mRNA levels were quantified by imaging software, and data are presented as mean±s.d. from three independent experiments (bottom in F and H). NS indicates non-specific bands.
RISC assembles with multiple silencing factors, including small RNA duplexes and Dicer as well as Ago proteins, and requires a decapping complex, which includes Dcp1 and Dcp2, to silence mRNA (Lee et al, 2006; Eulalio et al, 2008). We determined whether Wig1 associates with other players of the RISC-mediated silencing machinery. Indeed, Wig1 associated with Ago2, Dicer, and Dcp1a in Con Si-transfected HEK 293T cells (Figure 5E and G). In Ago2-depleted cells, Wig1 did not associate with Dicer or Dcp1a (Figure 5E). In contrast, Ago2 interacted with Dicer and Dcp1a in the Wig1-depleted cells (Figure 5G). Next, we performed RNP-IP and sqRT–PCR analysis of the p21 3′UTR reporter in order to determine whether binding of Ago2 to the 3′UTR of p21 mRNA is dependent on Wig1 and vice versa. Depletion of Ago2 had no effect on binding of Wig1 to the p21 3′UTR (Figure 5F); however, Ago2 failed to associate with the p21 3′UTR in Wig1-depleted cells (Figure 5H). Altogether, these results demonstrate that Wig1 binding to p21 mRNA is essential for the recruitment of RISC, while Wig1 is not directly involved in RISC assembly per se.
Wig1 governs RISC accessibility through binding of RNA secondary structure that neighbours the miRNA target site
Because we found that Wig1 participates in the decay of p21 mRNA by guiding RISC recruitment to its 3′UTR, we next sought to identify miRNAs that are involved in p21 mRNA process. A search of the mRNA database at TargetScan (http://www.targetscan.org; version 5.2) retrieved hsa-miR-106b and hsa-let-7a as candidates involved in the targeting of RISC to the p21 3′UTR sequence, as the seed regions of these miRNAs are perfectly complementary to this 3′UTR sequence (Figure 6A). Wig1 depletion did not significantly affect mature miR-106 and let-7a miRNA levels (Supplementary Figure S6). Thus, we studied the potential roles of miR-106b and let-7a in Wig1-dependent p21 mRNA destabilization.
Figure 6.
Wig1 binds to the miR-106b recognition site and enhances the accessibility of RISC. (A) Schematic representation of the candidate binding sites of miRNAs in the 3′UTR via the TargetScan database. (B) Effect of miRNAs on the p21 3′UTR reporter in Wig1-depleted MCF7 cells. Cells were harvested 2 days after transfection with either pFL-EV or pFL-p21-3′UTR, 100 nM of each miRNA candidate, and pRL-CMV as a reference plasmid following Wig1 Si transfection. Total RNA was subjected to qRT–PCR to quantitate FL-3′UTR mRNA abundance (upper). Whole cell lysates were subjected to immunoblot analysis. The p21 protein levels were determined by quantification of band intensities (lower). (C) Effects of miRNA on the p21 3′UTR reporter in the presence of Wig1 overexpression. Cells were harvested 2 days after transfection with the p21 3′UTR reporter and miRNA mimics as in (B) following transfection with either p3xFlag–EV or p3xFlag–Wig1. Total RNA was subjected to qRT–PCR to quantitate FL-3′UTR mRNA (upper). Whole cell lysates were subjected to immunoblotting, and p21 protein levels were quantitated as described in (B) (lower). (D) Interaction of miR-106b with p21 3′UTR reporter mRNA in the absence of Wig1. Cells expressing the p21 3′UTR reporter were transfected with biotinylated miR-106b in combination with Wig1 Si, and then lysates were subjected to precipitation using streptavidin-conjugated beads. RNA was purified from precipitates, and qRT–PCR was performed. (E) Cloning strategy of the miR-106b target region in the p21 3′UTR. Three stem-loop structures in the p21 3′UTR (spanning nt 366–488) were placed in the pFL-p21-3′UTR. pFL-p21-3′UTR-F1 contains both nt 366–384 and nt 464–488 including the miR-106 target site. The pFL-p21-3′UTR-F2 and pFL-p21-3′UTR-F3 contain nt 387–426 and nt 427–454, respectively. The pFL-p21-3′UTR (366–488)-ΔmiR is a mutant in which the miR-106 target site in p21 3′UTR (nt 366–488) was deleted. The base-pair probability is indicated in the colour key. (F) The Wig1-interacting region in p21 3′UTR spanning nt 366–488. HEK 293T cells that overexpressed Flag-tagged Wig1 were harvested 2 days after transfection with pFL-EV, pFL-p21-3′UTR-F1, pFL-p21-3′UTR-F2, pFL-p21-3′UTR-F3, or pFL-p21-3′UTR (366–488) in combination with the pRL-CMV reference plasmid. An RNP-IP assay using anti-Flag M2 affinity gel was performed. Immunoblot analysis using anti-Flag antibody (upper). RNA isolated from immunoprecipitates was radiolabelled using RT–PCR with RL- and FL-specific primers, and the products were subjected to PAGE. The four leftmost lanes represent two-fold serial dilutions of RNA and demonstrate that the RT–PCR is semiquantitative (lower). (G) Levels of p21 3′UTR reporters that contain stem-loop structures in the p21 3′UTR nt 366–488 in Wig1-depleted MCF7 cells. Following Wig1 Si transfection, the cells were transfected with the p21 3′UTR reporter vector as in (F), and cells were then harvested after 2 days. Isolated RNA was subjected to qRT–PCR using RL- and FL-specific primers. (H) Effects of deletion of the miR-106b target site in the p21 3′UTR (nt 366–488) in Wig1-depleted MCF7 cells. Cells were transfected with Wig1 Si and then pFL-p21-3′UTR (366–488) or pFL-p21-3′UTR (366–488)-ΔmiR in combination with the pRL-CMV reference plasmid. Cells were harvested 2 days after transfection, and then isolated RNA was subjected to qRT–PCR using RL- and FL-specific primers. Knockdown effects of Wig1 are shown in boxes for (G) and (H). The intensity values of qRT–PCR represent mean±s.d. from three independent experiments. Amplified RNA intensities before IP were quantified, and data are presented as mean±s.d. from three independent experiments (bottom in D and F). An * or # indicates either P<0.05 or P>0.05, respectively.
First, we performed qRT–PCR of the p21 reporter mRNA after transfection with either an miR-106b or let-7a mimic in Wig1-depleted MCF7 cells (Figure 6B, upper panel). Transfection of miRNA mimics decreased p21 3′UTR reporter mRNA in Con Si-transfected cells; however, this decrease was not observed in Wig1-depleted cells. On the contrary, Wig1 overexpression augmented the suppression of the p21 3′UTR reporter in response to transfection of either of the miRNA mimics (Figure 6C, upper panel). Overexpression of anti-miR for either miR-106b or let-7a together with Wig1 had no effect on p21 3′UTR reporter mRNA levels (Supplementary Figure S7, upper panel). Consistent with the results of the p21 3′UTR reporter system, both Wig1 depletion and overexpression contributed to the regulation of p21 protein levels in concert with miR-106b or let-7a; however, these effects were not observed in the presence of anti-miR (Figure 6B and C; Supplementary Figure S7, lower panels). To test whether Wig1 affects the interaction between miR-106b and the p21 mRNA 3′UTR, we analysed p21 3′UTR reporter after precipitation of biotinylated miR-106b with streptavidin-conjugated beads. We found that miR-106b did not bind to p21 mRNA 3′UTR in the absence of Wig1 (Figure 6D); however, Ago2-bound miR-106b levels were not affected by the absence of Wig1 (Supplementary Figure S8).
Using the RNA secondary structure prediction software (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi; Vienna RNA package version 2.0.0), we selected p21 3′UTR stem-loop structures that were predicted to be recognized by miR-106b or let-7a (Supplementary Figure S9A). To examine the possibility that Wig1 associates with secondary structures in the vicinity of the miRNA recognition sites, we cloned the p21 3′UTR regions that are recognized by miR-106b (nt 366–488 and nt 1035–1191) or let-7a (nt 840–993) into the pFL plasmid (Supplementary Figure S9B). The full-length p21 3′UTR (nt 1–1531) served as a positive control. The p21 3′UTR region that spans nt 601–683, which is predicted to be recognized by miRNAs with low conservation, was used as a negative control. The qRT–PCR analysis revealed that the levels of the reporter mRNA including the miR-106b target site were upregulated by Wig1 depletion and were downregulated by Wig1 overexpression (Supplementary Figure S9C and D). Next, we dissected the stem-loop structures of the 3′UTR nt 366–488 into fragments: fragment 1 (F1), which contained the miR-106b target site, F2, and F3 (Figure 6E). RNP-IP and radiolabelled sqRT–PCR analysis revealed that FL-p21-3′UTR-F1 mRNA was present in association with Flag–Wig1 and was stabilized by Wig1 depletion, whereas neither p21-3′UTR-F2 mRNA nor p21-3′UTR-F3 mRNA was detected (Figure 6F and G). When the miR-106b target site was deleted from the stem-loop structures containing nt 366–488 (Figure 6E), the mRNA levels of FL-p21-3′UTR (366–488)-ΔmiR were not regulated in the Wig1-depleted cells (Figure 6H). Furthermore, the levels of p21-3′UTR-F1 mRNA were decreased by overexpression of either Wig1-wt or Wig1-mut1 due to associations between p21-3′UTR-F1 mRNA and Wig1-wt or Wig1-mut1; however, neither regulation nor association of p21-3′UTR-F1 mRNA with Wig1-mut2 was detected (Supplementary Figure S10). Taken together, these results indicate that Wig1 binds to the p21 3′UTR stem-loop structure neighbouring the miR-106b recognition site and enhances the accessibility of miRNA-loaded Ago2 to the target site, ultimately facilitating p21 mRNA silencing.
Wig1 depletion retards tumour growth in a murine xenograft model, and Wig1 and p21 mRNA levels are inversely correlated in human cancer tissues
To explore the biological significance of our in vitro observations, we examined the role of Wig1 in a murine xenograft model. In experiments using the in vivo Atelocollagen (KOKEN Co. Ltd., Tokyo, Japan) delivery system for siRNA, the growth rate of xenografted tumours was significantly retarded in Wig1 Si-administered mice during the observation period (Figure 7A, left panel). Western blot analysis of tumour tissues confirmed that Wig1 depletion induced p21 accumulation in a p53-independent manner (Figure 7A, right panel). Furthermore, we assessed the mRNA levels of Wig1 and p21 in lung cancer patient tissues. We collected cancerous and corresponding normal tissue samples from 33 lung cancer patients who were diagnosed with adenocarcinoma, bronchioalveolar carcinoma, or squamous cell carcinoma. The relative mRNA expression levels were analysed using qRT–PCR (Figure 7B). Cancer tissues exhibited higher Wig1 mRNA levels than the normal tissues (1.00±0.13 versus 3.14±0.54, P=0.0004; data are presented as mean±s.e.m.), whereas p21 mRNA levels were lower in cancer tissues than in normal tissues (1.15±0.42 versus 0.05±0.01, P=0.0004). The correlation between Wig1 and p21 mRNA in tumours and normal tissues as a normalized expression ratio was assessed using the Pearson correlation coefficient, and the R-value indicated a negative correlation between Wig1 and p21 mRNA expression (R=−0.2798). Plotting of the Wig1 and p21 mRNA levels from each individual patient showed distinct clustering between normal and cancer tissues (Figure 7C). Collectively, these data indicate that Wig1 depletion suppresses the growth of xenografted tumours in vivo as well, and upregulation of Wig1 may be, at least in part, involved in human lung cancer development via suppression of p21.
Figure 7.
Wig1 depletion in a murine xenograft model and Wig1 and p21 mRNA levels in human cancer tissues. (A) H460 cells (1 × 106) were injected subcutaneously into the femurs of nude mice. When tumours reached 50 mm3, 200 μl AteloGene gel containing 20 μM of Wig1 Si was injected to encompass the whole tumour mass. Tumour size was then measured at the indicated times. Error bars represent s.d. (n≥3) (left). Xenograft tissue lysates at 5 days after siRNA injection were subjected to immunoblot analysis (right). An asterisk indicates a significant difference, P<0.05. (B) Quantitative RT–PCR analysis of Wig1 and p21 mRNA in patient tissues. RNA was purified from lung cancer tissues and corresponding normal counterparts from 33 patients and subjected to qRT–PCR using Wig1- and p21-specific primers. Expression levels were normalized to actin mRNA. The correlation between the relative levels of Wig1 and p21 mRNA was analysed by tumour versus normal counterpart expression ratio of either Wig1 or p21 mRNA using the Pearson correlation coefficient. Error bars represent standard error of the mean (s.e.m.). (C) In vivo correlation between Wig1 and p21 mRNA expression in each patient tissue. Expression levels in either normal or tumour tissues were clustered as either solid or dotted lines, respectively. Normalized Wig1 and p21 mRNA levels from each individual sample were plotted on logarithmic scales. (D) Proposed model for Wig1-dependent target recognition of RISC. Wig1 is a critical determinant for the decision between proliferation and cellular senescence via fine-tuning of p21 mRNA levels through facilitation of the accessibility of RISC to its target. In the presence of Wig1, RISC is effectively recruited to the target p21 mRNA, modulates its decay, and finally results in cell-cycle progression. In the case of Wig1 depletion, p21 mRNA is stabilized, leading to the onset of cellular senescence.
Discussion
In the present study, we elucidated the molecular mechanism involved in Wig1 modulation of cell fate between proliferation and senescence. Wig1 binds to stem-loop structures neighbouring the miRNA target site in the 3′UTR of p21 mRNA and facilitates RISC recruitment to suppress p21 expression. Thus, Wig1 plays an essential role in tuning p21 expression levels and ultimately in preventing the onset of cellular senescence.
Cellular senescence was initially described as finite proliferative capacity that results from telomere shortening in normal human cells (Hayflick and Moorhead, 1961). In contrast, cancer cells were believed to have lost the ability for senescence due to their limitless replicative potential. Cellular senescence, however, has recently begun to be described as a tumour suppression mechanism as potent as apoptosis (Ventura et al, 2007; Wu et al, 2007; Byun et al, 2009; Lee et al, 2011). One of the distinct molecular features of senescent cells is elevated expression of cell-cycle inhibitors. The CDK inhibitor p21 plays a crucial role in stress-induced cellular senescence via transcriptional upregulation (Kramer et al, 2001; Zhang et al, 2005); however, accumulating evidence suggests that post-transcriptional and post-translational regulation is also important for p21 expression and activity (Jung et al, 2010). Indeed, dysregulation of p21 accelerates tumour formation, suggesting that p21 expression and activity must be tightly regulated by multiple mechanisms (Brugarolas et al, 1995; Deng et al, 1995). In addition, induction of p21 expression or p21 stabilization promotes cellular senescence (Jascur et al, 2005; Vigneron et al, 2005). Recently, regulation of p21 mRNA by various miRNAs and RBPs has begun to be explored. Our results provide strong evidence to support a molecular mechanism that involves tight regulation of p21 by the RBP Wig1 and miRNAs at the post-transcriptional level to allow cell proliferation. With respect to a clinical application for these findings, we demonstrated that regulation of Wig1 levels is important for determining final cell fate, either premature senescence or cell proliferation, in various cancer cell lines and in cancer patient tissues. Vilborg et al (2009) reported that Wig1 depletion resulted in a decrease in p53 protein levels through the regulation of p53 mRNA stability; however, we observed no correlation between Wig1 and p53 in various cancer cells, including U2OS cells, and in normal cells (Supplementary Figure S11; also in Figure 1 and Supplementary Figures S3 and S5). Our findings were consistent with the recent report by Sedaghat et al (2012).
The miRNA-mediated mRNA decay plays a central role in the regulation of gene expression in eukaryotic cells (Hutvágner et al, 2001; Lee et al, 2003; Bartel, 2009). Here, we showed that Wig1 overexpression facilitated miR-106b- and let-7a-mediated degradation of p21 reporter mRNA following transfection of miRNA mimics (Figure 6C). In addition, Wig1 depletion suppressed reporter mRNA decay in miRNA mimic-transfected cells (Figure 6B). In the absence of let-7a mimic transfection, however, the effect of Wig1 on FL-p21-3′UTR (840–993) reporter mRNA levels was barely detectable (Supplementary Figure S9C and D), perhaps due to the low endogenous level of let-7a compared to that of miR106b (Supplementary Figure S12). Since miR-106b and let-7a target sites likely form a stem-loop structure due to considerable base-pairing probabilities, we further defined the exact role of this stem-loop structure of the p21 3′UTR in mRNA decay by Wig1. We found that Wig1 preferentially associated with stem-loop fragments containing the miR-106b target site in order to regulate the reporter mRNA levels (Figure 6F and G).
Although several proteins modulate the efficacy of miRNA biogenesis, target recognition of miRNAs is based on base-pairing interactions via thermodynamic asymmetry and the 5′-nucleotide identity between the miRNA ‘seed’ sequence and the complementary nucleotides in the 3′UTR of target mRNAs (van Kouwenhove et al, 2011). Recently, several reports have suggested that an alternative regulatory mechanism is involved in influencing miRNA activity, since most miRNAs pair imperfectly with their cognate mRNAs (Jing et al, 2005; Kedde et al, 2007; Kedde et al, 2010; van Kouwenhove et al, 2011). RBPs have been proposed to be key components in the determination of miRNA function. Indeed, alterations of RBP expression and activity could affect many physiological and developmental processes (van Kouwenhove et al, 2011). Although >500 human RBPs have been identified, the mechanisms of action of only a few have been elucidated. Therefore, as RBPs play an essential role in RNA metabolism, functional studies of various classes of RBPs should be the focus of more attention (Höck et al, 2007; Landthaler et al, 2008).
If an miRNA target site is located in a stem-loop structure, then the site may be concealed by the secondary structure to prevent pairing with the miRNA. Thus, RBPs may affect target site accessibility by facilitating an open structure via association with the secondary structure of the mRNA and may, thereby, modulate gene expression regulation by miRNAs (Brodersen and Voinnet, 2009). Pumilio-1 (PUM1) binds to the 3′UTR of p27 and induces a local change in mRNA structure to favour association with miR-221 and miR-222 (Kedde et al, 2010). The AU-rich element (ARE) binding protein, tristetraprolin (TTP), is also required in miR16-mediated ARE-RNA decay (Jing et al, 2005). Furthermore, competitive binding of RBP and miRNA to the target sites may lead to modulation of miRNA-mediated mRNA decay. Studies have demonstrated that hnRNP L, Dnd1, and RBM38 relieve miRNA-mediated mRNA repression because the target site overlaps with a binding site for an RBP (Kedde et al, 2007; Jafarifar et al, 2011; Léveillé et al, 2011). In this study, we proposed possible mechanisms for the regulation of p21 expression at the post-transcriptional level by Wig1 (Figure 7D). Wig1 may dictate miRNA-mediated mRNA decay through the regulation of RISC accessibility to its target site via relaxation of the mRNA secondary structure and/or recruitment of miRNA-loaded RISC by direct association with the Ago2 protein. Our results not only provide evidence for a novel contribution to the prevention of premature senescence onset but also suggest the molecular architecture to shed light on the target recognition mechanism of miRNA-containing RISCs. Furthermore, we demonstrated that Wig1 depletion inhibited tumour progression in a murine xenograft model and provided evidence of an inverse correlation between Wig1 and p21 mRNAs in human normal and cancer tissues, indicating that Wig1 may play a critical role in tumour progression via its regulation of p21 (Figure 7A–C). Since dysregulation of a cell-cycle regulatory protein may contribute to malignant transformation, we suggest that fine-tuning of cell-cycle regulators via the coordination between miRNAs and the RBP Wig1 may offer a therapeutic and prognostic strategy for cancer treatment. We observed that levels of Wig1 and p21 were not significantly altered during the replicative senescence of human diploid fibroblasts (Supplementary Figure S13), suggesting that these molecules may not be critical executioners in the process of replicative senescence.
Recently, co-expression of miRNAs and their target genes was documented, suggesting that these miRNAs may, indeed, be involved in negative transcriptional co-regulation circuits, which are termed as miRNA-mediated feed-forward loops (Tsang et al, 2007). This circuit may have the potential to provide fine-tuning and maintenance of protein steady state, although the functional significance of the circuit has not been fully characterized. If a temporal gap in the activation of the target gene and the miRNA exists, then the circuit may be utilized for either the delayed shutdown of the regulated genes or buffering for noisy fluctuations (Shalgi et al, 2007). Although both miR-17/92 and E2F are transcriptionally activated by c-Myc, a tight regulatory connection is involved as miR-17/92 represses E2F (O'Donnell et al, 2005). Interestingly, we found that Wig1 induced by p53 negatively regulated another p53-responsive gene, p21. Under normal conditions, the basal levels of Wig1 negatively regulate p21 to prevent the onset of cellular senescence (Figure 1; Supplementary Figure S3). Under stressed conditions, activated p53 induces Wig1 expression as well as p21 expression, and this induced Wig1 functions as a guardian to regulate the induced p21 level (Supplementary Figure S14A and B). Summarily, our findings provide evidence that a TF controls the transcription of both a target mRNA and the RBP involved in regulation of the decay of the same target mRNA, providing more precise regulation via a combined transcriptional/post-transcriptional regulatory network (Supplementary Figure S14C).
Materials and methods
Cell culture
MCF7, HEK 293T, U2OS, and HDF cells were cultured in DMEM (PAA Laboratories GmbH). H460 cells were cultured in RPMI 1640 (WelGENE). HCT116 parental, p53−/−, and p21−/− isogenic cell lines were cultured in McCoy’s 5A medium (WelGENE). Cells were supplemented with 10% FBS (Lonza Group Ltd.) and 1% penicillin and streptomycin solution (WelGENE) at 37°C in a 5% CO2 incubator.
RNA interference and plasmid transfection
Cells were transfected with siRNA duplexes or miRNA mimics using Lipofectamine RNAiMAX (Invitrogen Corp.), and transfection of plasmids was carried out using Lipofectamine 2000 reagent (Invitrogen Corp.) according to manufacturer’s instructions. The siRNAs used in this study are listed in Supplementary Table I.
Immunoblot analysis
Immunoblotting was performed as described (Lee et al, 2011), and the antibodies used in this study are listed in Supplementary Table II.
Analysis of p21 mRNA stability and protein half-life
Wig1 Si-transfected cells were treated with 5 μg/ml actinomycin D (Sigma-Aldrich) for various periods of time (0–4 h). Total RNA from each time point was purified from the cells, and quantitative real-time PCR was performed. To analyse protein half-life, Wig1 Si-transfected cells were treated with 40 μg/ml CHX (Sigma-Aldrich Co. LLC.) for the indicated periods of time (0–6 h). Cellular extracts obtained at each time point were subjected to immunoblotting using the appropriate antibodies. The intensities of the bands normalized with actin were determined using the ImageJ software (http://rsb.info.nih.gov/ij/) from the National Institutes of Health (USA).
sqRT–PCR and quantitative real-time PCR
sqRT–PCR was performed by radioisotope-labelling PCR and polyacrylamide gel electrophoresis (PAGE). qRT–PCR was performed in a DNAEngine Thermal Cycler with a Chromo4 Real-Time PCR Detector (Bio-Rad Laboratories). Primers used in this study are given in Supplementary Table III.
Immunoprecipitation
Lysates from cells that overexpress Flag-tagged Wig1 were immunoprecipitated with anti-Flag® M2 affinity gel (Sigma-Aldrich Co. LLC.). Total RNA isolated from these immunoprecipitates was subjected to sqRT–PCR.
In vitro binding assay
In vitro binding assays were performed using recombinant Flag-tagged Wig1 (Novus Biologicals, Inc.) and polyhistidine-tagged Ago2 (Sino Biological Inc.). After incubation of either GST or GST–Wig1 in combination with His–Ago2 in binding buffer (10 mM Tris–HCl (pH 8.0), 150 mM NaCl, 10% glycerol, 0.1% BSA, 0.1% Triton X-100) at 4°C for 2 h, samples were incubated with either glutathione Sepharose 4B (GE Healthcare Bio-Sciences AB) or Ni-NTA agarose resin (Qiagen GmbH) for 1.5 h. After adsorption, the resin was washed five times with binding buffer, and then bound proteins were resolved by 10% SDS–PAGE and subjected to immunoblot using either His probe (Santa Cruz Biotechnology) or HRP-conjugated anti-GST antibody (GE Healthcare Bio-Sciences AB).
Biotin precipitation
Wig1 Si-transfected HEK 293T cells were transfected with the pFL-p21-3′UTR reporter plasmid and biotinyl-miR-106b (Bioneer Inc.). In addition, pRL-CMV was used as a reference plasmid. After 2 days of incubation, the cells were rinsed and sonicated in NET-2 buffer as described above for IP. Yeast tRNA-coated streptavidin beads (Sigma-Aldrich Co. LLC.) were added to cell lysates and incubated for 3 h at 4°C. The beads were then washed 5–7 times with NET-2 buffer. RNA was isolated from the beads, and the mRNA associated with the biotinylated miR-106b was amplified using RT–PCR with FL- and RL-specific primers as described above for sqRT–PCR.
Establishment of xenograft tumours and siRNA transfection using AteloGene® gel
A single-cell suspension of H460 cells (1 × 106) was injected subcutaneously into lateral hind legs of 6-week-old BALB/c nude mice (n=6). Average tumour length was determined as (L × W2)/2 with measurements of tumour length (L) and width (W) taken with a caliper. When the tumour reached an average volume of ∼50 mm3 according to this formula, a Wig1 Si mixture with AteloGene® Local Use (KOKEN Co. Ltd.) was injected so as to wrap up the whole tumour mass (Takei et al, 2004). Tumour volumes were determined daily after the siRNA gel injection. The mice were sacrificed on days 5 and 7 after xenograft injection, and tumours were removed for further analysis. All animal protocols and studies were conducted in accordance with the guidelines of the Institutional Animal Care and Use Committee of the Korea Institute Radiological and Medical Sciences (IRB Approval No. KIRAMS 2012-2).
Human tissue collection and qRT–PCR analysis
This study involving human tissues was approved by the Institutional Review Board of Korea Institute of Radiological and Medical Sciences (IRB Approval No. KIRAMS 2010-0526) and the Asan Medical Center Institutional Review Board (IRB Approval No. K-1007-002-067). Snap-frozen tissues from lung tissues were obtained from patients with lung cancer that had been diagnosed as lung adenocarcinoma, squamous cell carcinoma, or bronchioloalveolar carcinoma. Samples were stored in liquid nitrogen. To measure Wig1 and p21 mRNA expression in human specimens, total RNA was isolated and subjected to reverse transcription using SuperScript III Reverse Transcriptase (Invitrogen Corp.) according to manufacturer’s recommendations. Then, qRT–PCR was performed using the Chromo4 Real-Time PCR Detection System (Bio-Rad Laboratories). The relative fold-change in RNA expression was calculated using the 2−ΔΔCt method, where the average of ΔCt values for the amplicon of interest was normalized to that of an endogenous control gene (β-actin), and compared with normal counterpart specimens.
Statistical analysis
Differences between the various experimental groups were calculated using Student’s two-tailed t-test. P-values of <0.05 were considered as significant. The correlation between Wig1 and p21 mRNA levels in tumours versus normal tissues was assessed based on the tumour/normal expression ratio of relative mRNA levels of each target using Pearson’s correlation coefficient analysis.
Supplementary Material
Acknowledgments
Primary human diploid fibroblasts and Ago2 overexpressing vector were kindly provided by Professor Kyung A Cho at Chonnam National University and Professor V Narry Kim at Seoul National University, respectively. We thank Professor Yoon Ki Kim and Professor Young Sik Lee at Korea University, Dr Kyung S Lee at National Cancer Institute, NIH, and Professor Edward KL Chan at University of Florida for critical review of the manuscript. This work was supported by the Nuclear Research and Development Program of the National Research Foundation funded by the Korean Government (MEST).
Author contributions: BCK, HCL, JJL, CMC, DKK, JCL, YGK, and JSL designed the experiments and analysed the data. BCK and HCL performed the experiments. CMC and DKK collected the patient tissue samples. BCK and JSL wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abbas T, Dutta A (2009) p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer 9: 400–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W (2006) Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 125: 1111–1124 [DOI] [PubMed] [Google Scholar]
- Brodersen P, Voinnet O (2009) Revisiting the principles of microRNA target recognition and mode of action. Nat Rev Mol Cell Biol 10: 141–148 [DOI] [PubMed] [Google Scholar]
- Brugarolas J, Chandrasekaran C, Gordon JI, Beach D, Jacks T, Hannon GJ (1995) Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature 377: 552–557 [DOI] [PubMed] [Google Scholar]
- Byun HO, Han NK, Lee HJ, Kim KB, Ko YG, Yoon G, Lee YS, Hong SI, Lee JS (2009) Cathepsin D and eukaryotic translation elongation factor 1 as promising markers of cellular senescence. Cancer Res 69: 4638–4647 [DOI] [PubMed] [Google Scholar]
- Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, Koutcher JA, Scher HI, Ludwig T, Gerald W, Cordon-Cardo C, Pandolfi PP (2005) Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature 436: 725–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C, Zhang P, Harper JW, Elledge SJ, Leder P (1995) Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell 82: 675–684 [DOI] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Izaurralde E (2008) GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat Struct Mol Biol 15: 346–353 [DOI] [PubMed] [Google Scholar]
- Gartel AL, Tyner AL (1999) Transcriptional regulation of the p21WAF1/CIP1 gene. Exp Cell Res 246: 280–289 [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP (2007) MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 27: 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guhaniyogi J, Brewer G (2001) Regulation of mRNA stability in mammalian cells. Gene 265: 11–23 [DOI] [PubMed] [Google Scholar]
- Hayflick L, Moorhead PS (1961) The serial cultivation of human diploid cell strains. Exp Cell Res 25: 585–621 [DOI] [PubMed] [Google Scholar]
- Hellborg F, Qian W, Méndez-Vidal C, Asker C, Kost-Alimova M, Wilhelm M, Imreh S, Wiman KG (2001) Human wig-1, a p53 target gene that encodes a growth inhibitory zinc finger protein. Oncogene 20: 5466–5474 [DOI] [PubMed] [Google Scholar]
- Hellborg F, Wiman KG (2004) The p53-induced Wig-1 zinc finger protein is highly conserved from fish to man. Int J Oncol 24: 1559–1564 [PubMed] [Google Scholar]
- Höck J, Weinmann L, Ender C, Rüdel S, Kremmer E, Raabe M, Urlaub H, Meister G (2007) Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep 8: 1052–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, Zamore PD (2001) A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293: 834–838 [DOI] [PubMed] [Google Scholar]
- Israeli D, Tessler E, Haupt Y, Elkeles A, Wilder S, Amson R, Telerman A, Oren M (1997) A novel p53-inducible gene, PAG608, encodes a nuclear zinc finger protein whose overexpression promotes apoptosis. EMBO J 16: 4384–4392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafarifar F, Yao P, Eswarappa SM, Fox PL (2011) Repression of VEGFA by CA-rich element-binding microRNAs is modulated by hnRNP L. EMBO J 30: 1324–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jascur T, Brickner H, Salles-Passador I, Barbier V, El Khissiin A, Smith B, Fotedar R, Fotedar A (2005) Regulation of p21WAF1/CIP1 stability by WISp39, a Hsp90 binding TPR protein. Mol Cell 17: 237–249 [DOI] [PubMed] [Google Scholar]
- Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, Di Padova F, Lin SC, Gram H, Han J (2005) Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell 120: 623–634 [DOI] [PubMed] [Google Scholar]
- Jung YS, Qian Y, Chen X (2010) Examination of the expanding pathways for the regulation of p21 expression and activity. Cell Signal 22: 1003–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata T, Tomari Y (2010) Making RISC. Trends Biochem Sci 35: 368–376 [DOI] [PubMed] [Google Scholar]
- Kedde M, Strasser MJ, Boldajipour B, Oude Vrielink JA, Slanchev K, le Sage C, Nagel R, Voorhoeve PM, van Duijse J, Ørom UA, Lund AH, Perrakis A, Raz E, Agami R (2007) RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell 131: 1273–1286 [DOI] [PubMed] [Google Scholar]
- Kedde M, van Kouwenhove M, Zwart W, Oude Vrielink JA, Elkon R, Agami R (2010) A Pumilio-induced RNA structure switch in p27-3′ UTR controls miR-221 and miR-222 accessibility. Nat Cell Biol 12: 1014–1020 [DOI] [PubMed] [Google Scholar]
- Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M (2009a) HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev 23: 1743–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MY, Hur J, Jeong S (2009b) Emerging roles of RNA and RNA-binding protein network in cancer cells. BMB Rep 42: 125–130 [DOI] [PubMed] [Google Scholar]
- Kramer DL, Chang BD, Chen Y, Diegelman P, Alm K, Black AR, Roninson IB, Porter CW (2001) Polyamine depletion in human melanoma cells leads to G1 arrest associated with induction of p21WAF1/CIP1/SDI1, changes in the expression of p21-regulated genes, and a senescence-like phenotype. Cancer Res 61: 7754–7762 [PubMed] [Google Scholar]
- Landthaler M, Gaidatzis D, Rothballer A, Chen PY, Soll SJ, Dinic L, Ojo T, Hafner M, Zavolan M, Tuschl T (2008) Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA 14: 2580–2596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JJ, Kim BC, Park MJ, Lee YS, Kim YN, Lee BL, Lee JS (2011) PTEN status switches cell fate between premature senescence and apoptosis in glioma exposed to ionizing radiation. Cell Death Differ 18: 666–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JJ, Lee JH, Ko YG, Hong SI, Lee JS (2010) Prevention of premature senescence requires JNK regulation of Bcl-2 and reactive oxygen species. Oncogene 29: 561–575 [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, Kim VN (2003) The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415–419 [DOI] [PubMed] [Google Scholar]
- Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN (2006) The role of PACT in the RNA silencing pathway. EMBO J 25: 522–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léveillé N, Elkon R, Davalos V, Manoharan V, Hollingworth D, Oude Vrielink J, le Sage C, Melo CA, Horlings HM, Wesseling J, Ule J, Esteller M, Ramos A, Agami R (2011) Selective inhibition of microRNA accessibility by RBM38 is required for p53 activity. Nat Commun 2: 513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez Vidal C, Prahl M, Wiman KG (2006) The p53-induced Wig-1 protein binds double-stranded RNAs with structural characteristics of siRNAs and miRNAs. FEBS Lett 580: 4401–4408 [DOI] [PubMed] [Google Scholar]
- O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT (2005) c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435: 839–843 [DOI] [PubMed] [Google Scholar]
- Prahl M, Vilborg A, Palmberg C, Jörnvall H, Asker C, Wiman KG (2008) The p53 target protein Wig-1 binds hnRNP A2/B1 and RNA Helicase A via RNA. FEBS Lett 582: 2173–2177 [DOI] [PubMed] [Google Scholar]
- Robb GB, Rana TM (2007) RNA helicase A interacts with RISC in human cells and functions in RISC loading. Mol Cell 26: 523–537 [DOI] [PubMed] [Google Scholar]
- Sedaghat Y, Mazur C, Sabripour M, Hung G, Monica BP (2012) Genomic analysis of wig-1 pathways. PLoS ONE 7: e29429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt CA, Fridman JS, Yang M, Lee S, Baranov E, Hoffman RM, Lowe SW (2002) A senescence program controlled by p53 and p16INK4a contributes to the outcome of cancer therapy. Cell 109: 335–346 [DOI] [PubMed] [Google Scholar]
- Shalgi R, Lieber D, Oren M, Pilpel Y (2007) Global and local architecture of the mammalian microRNA-transcription factor regulatory network. PLoS Comput Biol 3: e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh MS, Li XS, Chen JC, Shao ZM, Ordonez JV, Fontana JA (1994) Mechanisms of regulation of WAF1/Cip1 gene expression in human breast carcinoma: role of p53-dependent and independent signal transduction pathways. Oncogene 9: 3407–3415 [PubMed] [Google Scholar]
- Takei Y, Kadomatsu K, Yuzawa Y, Matsuo S, Muramatsu T (2004) A small interfering RNA targeting vascular endothelial growth factor as cancer therapeutics. Cancer Res 64: 3365–3370 [DOI] [PubMed] [Google Scholar]
- Tsang J, Zhu J, van Oudenaarden A (2007) MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell 26: 753–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kouwenhove M, Kedde M, Agami R (2011) MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat Rev Cancer 11: 644–656 [DOI] [PubMed] [Google Scholar]
- Varmeh-Ziaie S, Okan I, Wang Y, Magnusson KP, Warthoe P, Strauss M, Wiman KG (1997) Wig-1, a new p53-induced gene encoding a zinc finger protein. Oncogene 15: 2699–2704 [DOI] [PubMed] [Google Scholar]
- Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T (2007) Restoration of p53 function leads to tumour regression in vivo. Nature 445: 661–665 [DOI] [PubMed] [Google Scholar]
- Vigneron A, Roninson IB, Gamelin E, Coqueret O (2005) Src inhibits adriamycin-induced senescence and G2 checkpoint arrest by blocking the induction of p21waf1. Cancer Res 65: 8927–8935 [DOI] [PubMed] [Google Scholar]
- Vilborg A, Glahder JA, Wilhelm MT, Bersani C, Corcoran M, Mahmoudi S, Rosenstierne M, Grandér D, Farnebo M, Norrild B, Wiman KG (2009) The p53 target Wig-1 regulates p53 mRNA stability through an AU-rich element. Proc Natl Acad Sci USA 106: 15756–15761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Blandino G, Givol D (1999) Induced p21waf expression in H1299 cell line promotes cell senescence and protects against cytotoxic effect of radiation and doxorubicin. Oncogene 18: 2643–2649 [DOI] [PubMed] [Google Scholar]
- Wiemer EA (2007) The role of microRNAs in cancer: no small matter. Eur J Cancer 43: 1529–1544 [DOI] [PubMed] [Google Scholar]
- Wilhelm MT, Méndez-Vidal C, Wiman KG (2002) Identification of functional p53-binding motifs in the mouse wig-1 promoter. FEBS Lett 524: 69–72 [DOI] [PubMed] [Google Scholar]
- Wu CH, van Riggelen J, Yetil A, Fan AC, Bachireddy P, Felsher DW (2007) Cellular senescence is an important mechanism of tumor regression upon c-Myc inactivation. Proc Natl Acad Sci USA 104: 13028–13033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Huang S, Ding J, Zhao Y, Liang L, Liu T, Zhan R, He X (2010) Multiple microRNAs modulate p21Cip1/Waf1 expression by directly targeting its 3′ untranslated region. Oncogene 29: 2302–2308 [DOI] [PubMed] [Google Scholar]
- Zhang X, Li J, Sejas DP, Pang Q (2005) The ATM/p53/p21 pathway influences cell fate decision between apoptosis and senescence in reoxygenated hematopoietic progenitor cells. J Biol Chem 280: 19635–19640 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.