Abstract
Cell fate decisions during haematopoiesis are governed by lineage-specific transcription factors, such as RUNX1, SCL/TAL1, FLI1 and C/EBP family members. To gain insight into how these transcription factors regulate the activation of haematopoietic genes during embryonic development, we measured the genome-wide dynamics of transcription factor assembly on their target genes during the RUNX1-dependent transition from haemogenic endothelium (HE) to haematopoietic progenitors. Using a Runx1−/− embryonic stem cell differentiation model expressing an inducible Runx1 gene, we show that in the absence of RUNX1, haematopoietic genes bind SCL/TAL1, FLI1 and C/EBPβ and that this early priming is required for correct temporal expression of the myeloid master regulator PU.1 and its downstream targets. After induction, RUNX1 binds to numerous de novo sites, initiating a local increase in histone acetylation and rapid global alterations in the binding patterns of SCL/TAL1 and FLI1. The acquisition of haematopoietic fate controlled by Runx1 therefore does not represent the establishment of a new regulatory layer on top of a pre-existing HE program but instead entails global reorganization of lineage-specific transcription factor assemblies.
Keywords: cell fate decisions, endothelial–haematopoietic transition, haematopoiesis, RUNX1, transcriptional programming of chromatin
Introduction
During embryogenesis blood cells arise from the mesoderm via haemangioblast cells, a cell type capable of generating both endothelial and haematopoietic cells (Kennedy et al, 1997; Choi et al, 1998; Huber et al, 2004). These cells differentiate into the haemogenic endothelium (HE) which then undergoes a transition from an adherent, epithelial-like cell into a non-adherent blood cell precursor, a process called the endothelial–haematopoietic transition (Chen et al, 2009; Eilken et al, 2009; Lancrin et al, 2009; Kissa and Herbomel, 2010). Each of these differentiation steps is controlled by sequence-specific DNA-binding proteins. Mice lacking the ETS-factor family member FLI1 die from defects in blood vessel formation and show multiple haematopoietic defects (Spyropoulos et al, 2000), indicating that this factor is involved in the regulation of endothelial and haematopoietic gene expression programs. Mice with a deletion of the Tal1 gene that encodes the transcription factor SCL/TAL1 fail to generate any haematopoietic precursors and develop vascular defects (Robb et al, 1995; Porcher et al, 1996; Visvader et al, 1998). Using mouse embryonic stem (ES) cell differentiation as a model, it was later shown that SCL/TAL1 is required to form the HE (D'Souza et al, 2005; Lancrin et al, 2009). The same study together with experiments using conditional knockout mice demonstrated that the actual endothelial–haematopoietic transition is dependent on the presence of the transcription factor RUNX1 (Chen et al, 2009, 2011; Eilken et al, 2009; Lancrin et al, 2009). The development of mature blood cells from haematopoietic precursors requires additional transcription factors and includes the ETS-family member PU.1 whose balanced expression is required for correct myeloid and lymphoid development (McKercher et al, 1996; Scott et al, 1997; Rosenbauer et al, 2006; Leddin et al, 2011). Last, but not least, C/EBP family members are critical for myelopoiesis. C/EBPα regulates the formation of granulocyte-macrophage precursor cells and fulfills important roles in haematopoietic stem cell (HSC) maintenance (Zhang et al, 2004; Bereshchenko et al, 2009) and C/EBPβ is involved in the control of macrophage function (Tanaka et al, 1995). Mice lacking both factors die early in embryogenesis (Begay et al, 2004).
Transcription factors determine cell fate decisions by controlling the correct temporal sequence of lineage-specific gene activation or repression. This notion was emphasized by experiments in Drosophila that combined the determination of global binding patterns of transcription factors throughout a developmental pathway with mathematical modelling. This demonstrated that such knowledge is sufficient to predict the developmental activity of cis-regulatory elements (Zinzen et al, 2009; Junion et al, 2012). While the phenotypic and developmental consequences of the removal of each of the described transcription factors are well described, little is known of what drives their dynamic and specific assembly on their target genes during the process of haematopoietic specification. Recent genome-wide analyses of transcription factor binding have given insights into the complexities of combinations of transcription factors targeting lineage-specific genes (Wilson et al, 2010). However, other studies revealed that the same transcription factors are able to interact with different partners in different cell types and display different binding patterns (Pencovich et al, 2010; Hannah et al, 2011; Trompouki et al, 2011). Little is known about whether similar transitions govern the onset of haematopoiesis and how differential transcription factor assembly relates to cell fate decisions.
Here, we addressed these questions by employing a well-established in vitro model of haematopoiesis, the differentiation of ES cells into haematopoietic precursor cells (Choi et al, 1998; Sroczynska et al, 2009b). This system provides a traceable model to gain mechanistic insights into mechanistic the regulation of early events in embryonic haematopoiesis that are difficult to study in the mouse. Using an ES cell line that expresses an inducible form of RUNX1 in a Runx1−/− genetic background (Lancrin et al, 2009) we measured the dynamic assembly of specific transcription factor complexes containing C/EBPβ, SCL/TAL1 and FLI1 during the RUNX1-dependent endothelial-to-haematopoietic transition, both at the genome-wide and at the gene-specific level. We find that in the absence of RUNX1 in the HE C/EBPβ, SCL/TAL1 and FLI1 bind to specific cis-regulatory elements of haematopoiesis-specific genes and genes involved in changing cell shape. RUNX1 binds to the same genes and its induction leads to a rapid shift in the binding of SCL/TAL1 and FLI1, which is complete in multipotent precursor cells. Our data are consistent with a model by which the expression of RUNX1 in the HE orchestrates the assembly of a haematopoiesis-specific global pattern of transcription factor binding.
Results
Coordinated and hierarchical expression of haematopoietic regulator genes during ES cell differentiation
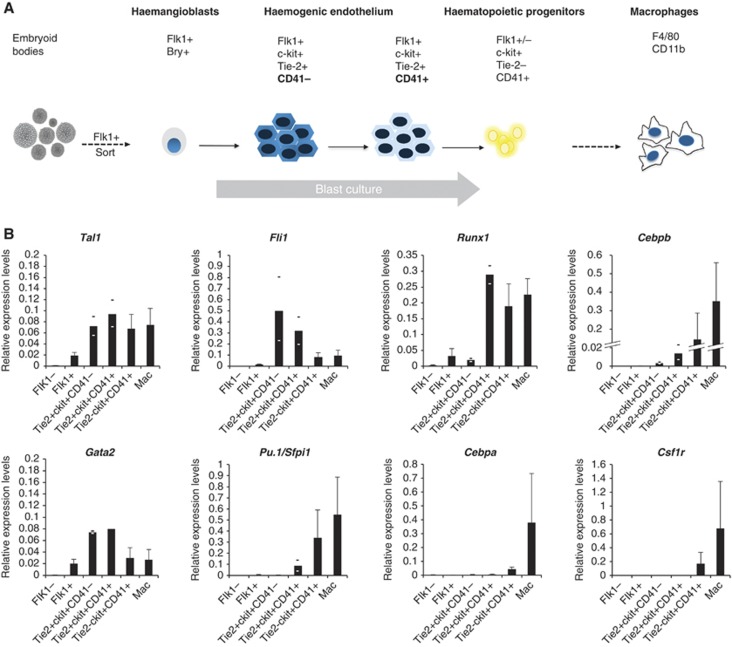
We first measured the accurate levels and the kinetics of expression of genes encoding transcription factors important for early haematopoiesis in purified cells. We differentiated ES cells and sampled developing cells at different time points by purifying them according to their surface marker profile using fluorescence activated cell sorting (FACS) as outlined in Figure 1A and Supplementary Figure 1A. We used an ES cell line carrying a GFP gene in the mesoderm-specific Brachyury locus which allowed for the purification of haemangioblasts by sorting cells for GFP expression and the expression of Flk-1, a marker for endothelial cells (Fehling et al, 2003). The next step in differentiation is the first stage of HE development which is defined by the expression of Tie-2 and c-kit, and a low level of Runx1 mRNA. In the second stage of HE development, Runx1 is upregulated and CD41 appears on the surface (Lancrin et al, 2009; Sroczynska et al, 2009a). Fully committed haematopoietic precursor cells express high levels of c-kit and CD41. All analysed genes showed a highly dynamic expression pattern (Figure 1B; Supplementary Figure 1B) whereby Cebpb was already expressed very early from the HE stage onwards but the gene encoding the myeloid master regulator PU.1 (Pu.1/Sfpi1) was only expressed after the onset of Runx1 expression (Hoogenkamp et al, 2009). The expression of Cebpa followed that of Pu.1. Late myeloid genes such as the PU.1 target gene Csf1r were only detected after PU.1 was expressed at high levels confirming previous cell line studies (Krysinska et al, 2007).
Figure 1.
Time course of expression of haematopoietic regulator genes during ES cell differentiation. (A) Schematic outline of the haematopoietic differentiation model. Haemangioblast (Flk1+, Bry+) cells are sorted from embryoid bodies on day 0 and during a 4-day blast culture through the stage of the haemogenic endothelium give rise to haematopoietic progenitors, which can be differentiated into macrophages (CD11b+, F4/80+). Relevant surface markers of sorted cell populations are indicated. (B) Relative expression analysis of haematopoietic regulator genes in purified cells from all stages of differentiation. Flk1− cells served as control and experiments were carried out at least in triplicates where STDEV was applied, otherwise the average of two biological duplicates and the respective values are shown.
Taken together, these data show a highly dynamic expression pattern of haematopoietic regulator genes whereby different upstream regulators independently and hierarchically feed into the expression of their downstream targets.
SCL/TAL1, FLI1 and C/EBPβ associate with a large number of genes in the HE in the absence of RUNX1
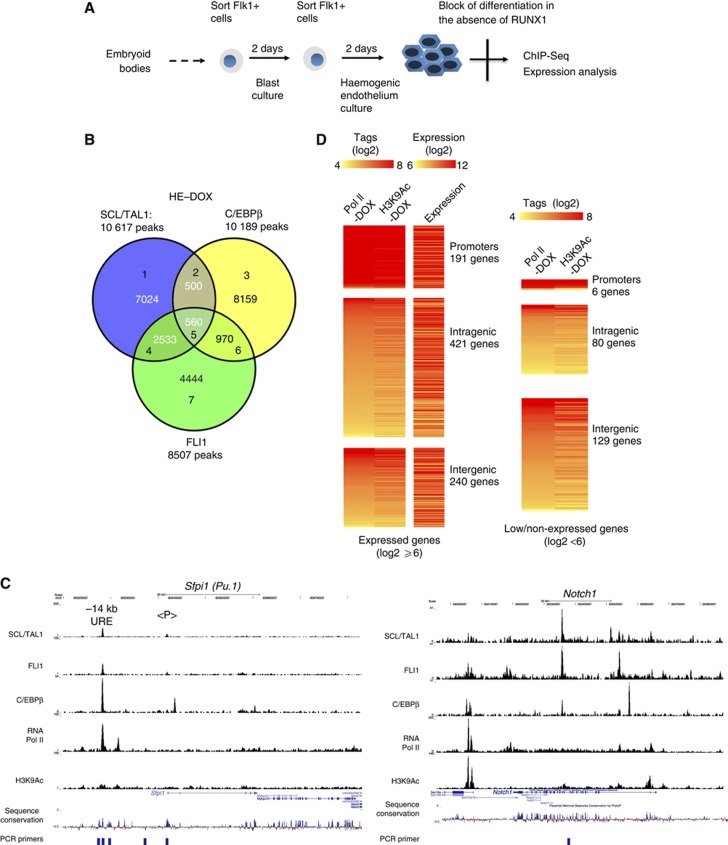
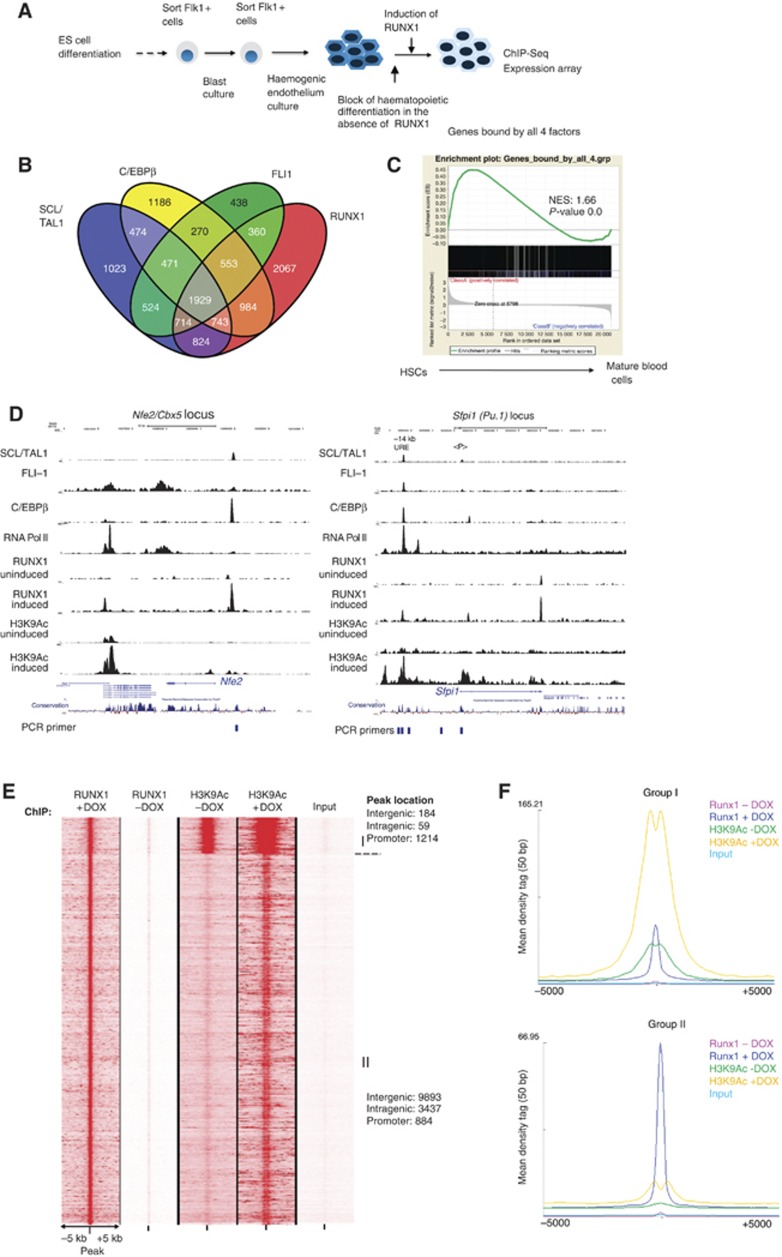
The onset of Tal1 and Fli1 expression does not depend on RUNX1 but Runx1 is a target of FLI1 (Nottingham et al, 2007) and its expression follows FLI1 upregulation (Figure 1B). The first stage of the HE is therefore a transient cell population and it is difficult to obtain enough cells for global analyses aimed at distinguishing the SCL/TAL1 and FLI1 regulatory circuits from those governed by RUNX1. To gain detailed insights into the molecular defects of haematopoiesis in the absence of RUNX1, we used an ES cell line that expressed an inducible form of RUNX1 in a Runx1−/− genetic background (iRUNX1) (Hoogenkamp et al, 2009; Lancrin et al, 2009). We differentiated these cells using a step-wise culture system that allows detailed studies of the endothelial–haematopoietic transition (outlined in Figure 2A and Supplementary Figure 2A) and where the order of expression of haematopoietic genes is faithfully preserved (Lancrin et al, 2009, see also Supplementary Figure 6I). Without induction, these cells are blocked in their differentiation and accumulate at the first HE stage. We therefore determined the genome-wide binding sites of SCL/TAL1 and FLI1 in the iRUNX1 cell line in the absence of induction. We also followed up on our finding of the very early C/EBPβ expression prior to the appearance of C/EBPα expression and determined whether this factor was bound to its target genes at this stage. To link transcription factor binding data with active chromatin formation, we determined the binding of RNA-Polymerase II (RNA Pol II) and measured histone H3 lysine 9 acetylation by chromatin immunoprecipitation (ChIP) sequencing. We found 10 617 binding sites for SCL/TAL1, 8507 binding sites for FLI1 and 10 189 peaks for C/EBPβ (Figure 2B; Supplementary Table 1). Examples of binding patterns at specific genes are shown in Figure 2C and Supplementary Figure 2B. ChIP-sequencing data were validated by manual analysis of different genes (Supplementary Figure 2E). We also performed ChIP assays in cells derived from Runx1−/− ES cells to demonstrate that factor binding was not an artifact of leakiness of the inducible system. Also here all three factors as well as RNA-Pol II interacted with an essential enhancer element (3′ upstream regulatory element (URE)) of the Pu.1 (Sfpi1) locus that binds the three factors as well as RUNX1 (Supplementary Figure 2D).
Figure 2.
Genome-wide SCL/TAL1, FLI1 and C/EBPβ binding in the haemogenic endothelium. (A) Schematic overview of iRUNX1 differentiation. In RUNX1-deficient iRUNX1 cells a two-step protocol was used, including a second Flk1 sort during blast culture to isolate haemogenic endothelium cells, which are arrested at that stage. (B) Venn diagram demonstrating the intersection of SCL/TAL1, C/EBPβ and FLI1 peaks. (C) UCSC genome browser screenshots showing the binding pattern of transcription factors, RNA-Pol II and H3K9Ac at Sfpi1 (Pu.1) and Notch1 and depicting unmanipulated aligned reads at each chromosomal location. (D) Heat maps showing the distribution of RNA-Pol II and H3K9 acetylation around SCL/TAL1 (left), FLI1 (middle) and C/EBPβ (right) binding sites within promoter/intragenic and intergenic regions. RNA Pol II and H3K9Ac levels for each promoter/intragenic and intergenic division were counted using a 1-kb window flanking the peak summits.
In the uninduced HE, SCL/TAL1 and FLI1 peaks showed some overlap (Figure 2B) but very few peaks were bound by all three factors (Figure 2B; Supplementary Figure 2C). We subdivided these peaks according to their genomic location (promoters, intergenic peaks and intragenic peaks) and ranked them according to the RNA-Pol II tag count around the respective factor binding sites. We also plotted the H3K9Ac levels at the corresponding positions (Figure 2D with manual validations shown in Supplementary Figure 2E). This analysis showed that binding of all factors was correlated with high levels of histone acetylation. Only a minority of peaks for each factor resided in promoters, while most binding sites were in intergenic or intragenic regions. A large number of these sites were co-occupied with RNA-Pol II and we observed a direct correlation between RNA Pol II and H3K9Ac levels at these peaks.
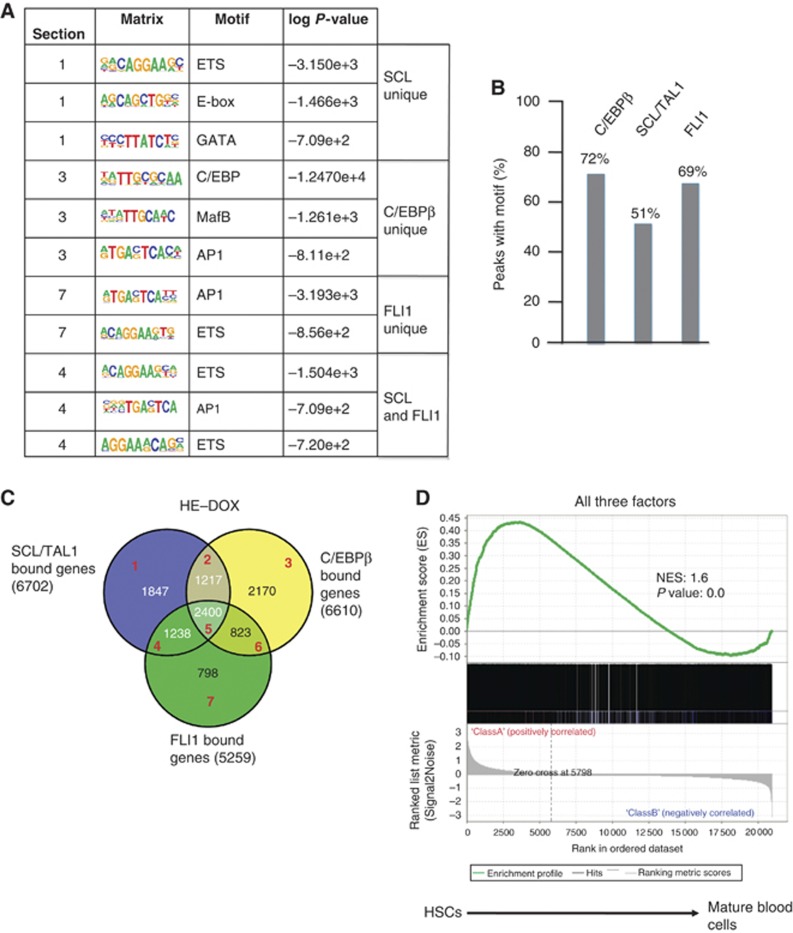
Unbiased de novo motif discovery with peak regions for the three factors (Figure 3A) showed that SCL/TAL1 unique peaks were mostly associated with E-box motifs, as expected, but also with motifs for ETS- and GATA family members, a combination that has been associated with cis-regulatory elements active in HSCs (Pimanda et al, 2007). C/EBPβ bound peaks associated with binding sites for inducible factors such as AP1. This colocalization with AP1 consensus sequences was also seen with FLI1 peaks and peaks overlapping SCL/TAL1 and FLI1. In all, 69% of all FLI1 bound peaks also contained an ETS consensus sequence and the C/EBP consensus sequence was found in 72% of all C/EBPβ bound peaks, but only about half of the SCL/TAL1 peaks showed an E-box motif (Figure 3B).
Figure 3.
Sequence motifs and genes associated with SCL/TAL1, C/EBPβ and FLI1 binding. (A) Unbiased binding motif search underlying unique SCL/TAL1, C/EBPβ or FLI1 peaks and those overlapping between SCL/TAL1 and FLI1. (B) Frequency of the corresponding binding site motifs within ChIP-sequencing peaks. (C) Venn diagram showing the overlap of genes bound by the three factors (2400) and the sections analysed by GSEA (red numbers, Supplementary Figure 3A). (D) Gene set enrichment analysis (GSEA) of genes bound by all three factors, SCL/TAL1, FLI1 and C/EBPβ comparing these genes with genes expressed in haematopoietic stem cells (HSCs) and differentiated mature blood cells.
In summary, we show (i) that SCL/TAL1, FLI1 and also C/EBPβ bind to a large number of cis-regulatory elements at the first stage of the HE and (ii) that many distal elements binding these factors are also bound by RNA Polymerase II and marked by H3K9 acetylation.
In the HE SCL/TAL1, FLI1 and C/EBPβ associate with genes regulating haematopoiesis and cell shape
We next annotated peaks with nearby genes (Figure 3C) and identified 2400 genes that were bound by all three factors (Supplementary Table 3). We first performed a gene set enrichment analysis (GSEA) comparing the genes bound by different combinations of the three factors with microarray expression data obtained from differentiating murine haematopoietic cells, including HSCs (Chambers et al, 2007; Figure 3D; Supplementary Figure 3A). Although many haematopoietic genes were contained in the overlap set between FLI1 and SCL/TAL1 bound genes, the gene set that was bound by all three factors showed the highest enrichment score for HSC genes (Figure 3D). Genes bound by the three factors included haematopoietic transcriptional regulator genes such as Notch1, Pu.1, Nfe2 and Erg as well as cytokine and cytokine receptor genes (Supplementary Table 3). In the analysis of the physiological and system developmental function of this gene set, haematopoiesis featured prominently (Supplementary Figure 3B), together with a high representation of genes involved in the regulation of tissue morphology. The latter was also confirmed by a KEGG pathway analysis, which showed that genes bound by the three factors were highly enriched in the focal adhesion pathway (Supplementary Figure 3C, marked by red asterisks). This included genes such as Flnb (Filamin B), Fak (focal adhesion kinase), Rock2 and Bcar1, all of which were differentially regulated after the expression of RUNX1 (Supplementary Figure 3D). This pathway is important for the endothelial–haematopoietic transition (Yue et al, 2012), demonstrating that besides binding to genes important for haematopoiesis, SCL/TAL1, FLI1 and C/EBPβ bind to genes involved in regulating the morphological alterations occurring during this process.
To examine how factor binding, RNA-Pol II occupancy, histone acetylation and genomic location correlated with gene activity, we also measured the expression of genes in the uninduced HE using expression microarrays (Supplementary Figure 3E) and plotted expression levels alongside RNA-Pol II occupancy and H3K9Ac. This showed that the majority of the genes were expressed irrespective of the genomic location of the factor binding sites. We then subdivided this gene set into a high expression set (13 876 genes) and a set with low or absent expression (8167 genes), according to absolute expression levels using a threshold value of (log2 ≤6). KEGG pathway analysis of the high expression set binding all three factors (1908 genes) demonstrated that also in this data set the pathways involved in regulating the actin cytoskeleton (P=5.15 × 10−8) and focal adhesion (P=1.17 × 10−9) were highly enriched. To correlate factor binding with expression levels, we examined the enrichment of high expression genes in sets of genes binding each combination of one, two or all three transcription factors. All these lists show significant enrichment (Supplementary Figure 3F). The set of genes binding all three factors showed the most significant enrichment of high expression genes (1908 out of 2300, P=5.86 × 10−77). However, within this gene set, there was little correlation between the number of binding sites for any of the three factors and actual gene expression levels (Pearson coefficient 0.1).
FLI1 binding at the HE stage is required for the correct timing of Pu.1 gene expression
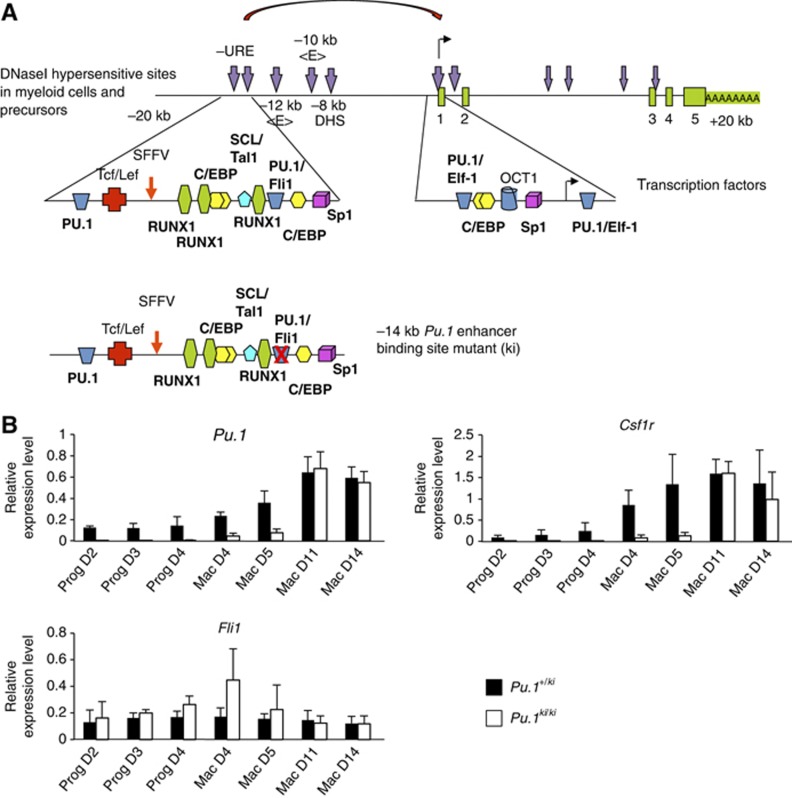
Our experiments demonstrate that the formation of the first stage of the HE is associated with a high level of SCL/TAL1 and FLI1 expression as well as low levels of RUNX1 and C/EBPβ. This is followed by the further upregulation of RUNX1 at the second stage of the HE. The Pu.1 locus was bound by SCL/TAL1, FLI1 and C/EBPβ at the first stage of the HE. We showed previously that a low level of RUNX1 is required to prime Pu.1 chromatin at the haemangioblast stage when FLI1 expression is low/absent (Hoogenkamp et al, 2009). The elimination of the RUNX1 binding site in the Pu.1 URE leads to an inactivation of the enhancer and a dramatic reduction in Pu.1 mRNA levels (Huang et al, 2008). To test whether not only RUNX1 binding, but also early binding by FLI1 is required for the correct timing of Pu.1 expression, we generated an ES cell line where the FLI1 binding site in the URE was mutated in both alleles (Pu.1ki/ki, Figure 4A), but was otherwise normal. Functional effects of this mutation on the development of myeloid cells in mice will be described elsewhere (Staber, P, and Tenen DRG, submitted). ES cells heterozygous for the mutated allele (Pu.1+/ki) served as a control. Both cell lines were differentiated and purified Flk1+ cells were subjected first to blast culture and precursors were isolated which were subjected to macrophage differentiation culture over a period of 14 days. We measured Pu.1 expression at different time points. In mature macrophages, Pu.1 expression levels were identical between control and Pu.1ki/ki cells, and precursor cell formation was unaffected (Supplementary Figure 4). However, in Pu.1ki/ki cells the onset of expression of Pu.1 was delayed (Figure 4B), while Fli1 expression remained the same. Expression of the PU.1 target gene Csf1r was delayed even further, indicating that the delay of expression of PU.1 had a knock-on effect on the expression of downstream genes. Pu.1 contains other enhancer elements at −12 and −10 kb, which depend on PU.1 expression and are activated in macrophages (Leddin et al, 2011), and which are likely to compensate for the defect in the URE. This result shows that the FLI1 binding site is required for the correct developmental timing of expression of at least two genes essential for myelopoiesis and macrophage differentiation.
Figure 4.
The FLI1 binding site in the URE is required for the correct timing of Pu.1 upregulation. (A) Upper panel: Map of regulatory elements of the Pu.1 gene. Transcription factor binding sites of the −14 kb upstream regulatory element (URE) and promoter are highlighted in the enlarged detailed map. DNase I hypersensitive sites are marked by vertical arrows and the transcription start site by a horizontal arrow. Lower panel: Section of the −14 kb URE containing a mutation in the FLI1 binding site. (B) Relative expression analysis during the ES cell to macrophage differentiation in heterozygous Pu.1+/ki cells (control: black bars) and the homozygous Pu.1ki/ki cells (white bars). Experiments represent the mean values of three biological replicates.
SCL/TAL1 and FLI1 binding profiles change during development
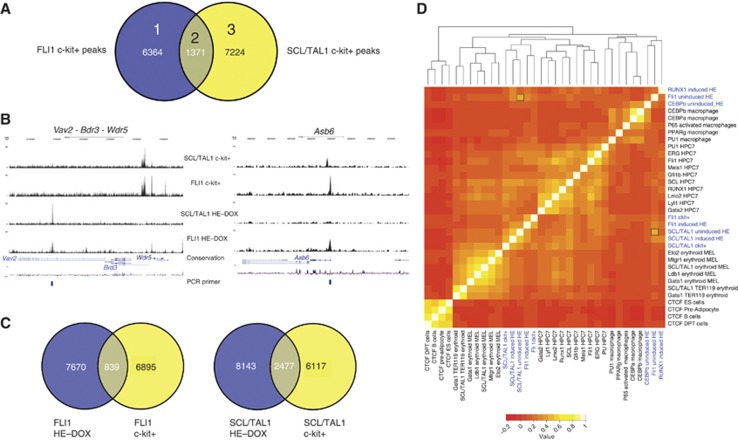
The combinatorial binding of haematopoietic regulators such as RUNX1, SCL/TAL1 and FLI1 and others specifies cis-regulatory elements active in haematopoietic cells (Wilson et al, 2010). However, our analysis indicates that while haematopoietic genes were bound by SCL/TAL1, FLI1 and C/EBPβ in the HE, these commonly did not bind together within a 400-bp window. We therefore considered the possibility that they changed position during the specification of haematopoietic cells. To this end we determined the binding sites of FLI1 and SCL/TAL1 in wild-type c-kit+/CD41+/Tie2− progenitor cells (c-kit+ cells) and obtained 7735 and 8205 peaks, respectively (Figure 5A) and 1371 joint peaks. The comparison of the overlap of peak regions from the uninduced HE and c-kit+ cells for the two factors showed that only a proportion of these regions were shared (Figure 5C) and the pattern for many individual genes was different (examples are shown in Figure 5B and Supplementary Figure 5A, with manual validations depicted in Supplementary Figure 5B). This was most pronounced for FLI1 where only a few binding sites were shared between the two cell types. In addition, the sequence motifs associated with FLI1 and SCL/TAL1 in c-kit+ cells differed from those in the HE (Supplementary Figure 5C). Most notable was a lack of enrichment of AP1-associated consensus sequences (compare with Figure 3A). Instead, the pattern resembled that of cis elements active in haematopoietic cells (Wilson et al, 2010), with GATA and RUNX1 binding sites featuring prominently. Again, most FLI1 peaks contained an ETS consensus motif (93%) and more than half of the SCL peaks featured E-box motifs (58%) (Supplementary Figure 5D).
Figure 5.
FLI1 and SCL/TAL1 binding profiles in haematopoietic c-kit+ precursor cells. (A) Venn diagram showing the intersection of ChIP-sequencing data of FLI1 and SCL/TAL1 in c-kit+/CD41+/Tie2− (c-kit+) wild-type progenitor cells prepared on day 4 of blast differentiation. (B) UCSC genome browser screenshots providing examples of genes where the binding pattern of FLI1 and SCL/TAL1 changes during differentiation from the uninduced haemogenic endothelium (HE) to c-kit+ progenitors. (C) Venn diagrams showing the overlap of peaks of SCL/TAL1 (left) and FLI1 (right) between the uninduced HE and wild-type c-kit+ progenitors. (D) Hierarchical clustering of transcription factor binding sequences in different cell types (data sets from this study are highlighted in blue). Based on similar binding patterns of the different ChIP-seq data, a correlation matrix was generated and sequence similarities are displayed after hierarchical clustering and using a colour code.
The redistribution of FLI1 and SCL/TAL1 towards a haematopoietic binding pattern was also indicated when examining genes bound by FLI1, SCL/TAL1 and both factors. However, while the peak overlap was relatively low (Figure 5A), around 50% of genes were bound by both factors (Supplementary Figure 5E, top panel). This was also observed when comparing genes bound by FLI1 and SCL/TAL1, respectively, in the HE and c-kit+ cells (Supplementary Figure 5E, middle and bottom panels). GSEA of genes bound by FLI1, SCL/TAL1 and both factors showed that the genes bound by both factors had the highest enrichment score for HSC-specific genes (Supplementary Figure 5F; Supplementary Table 3). This suggests that much of the redistribution of factors during differentiation occurs within one gene locus or in its vicinity.
To substantiate this finding, we clustered binding sites according to their sequence similarity and compared them to a compendium of other, previously published ChIP-sequencing data sets (Figure 5D). This type of analysis has demonstrated that binding sites for tissue-specific transcription factors often cluster according to the cell types in which the factor was analysed, in contrast to CTCF whose binding sites in different cell types are invariant (Hannah et al, 2011). Our analysis showed that DNA sequences within peaks bound by SCL/TAL1 in the HE and in c-kit+ cells relate to those obtained from ChIP data from the haematopoietic precursor cell line HPC7 (Wilson et al, 2010), although there were still sequence similarities with FLI1 bound peaks in the uninduced HE, as indicated by a signal in the heat map (see marked grid locations). In contrast, FLI1 bound sequences in the uninduced HE clustered with those bound by C/EBPβ whereas in the c-kit+ cells they clustered with SCL/TAL1 (Figure 5D). C/EBPβ sites in the uninduced HE clustered with C/EBPβ and C/EBPα sites in macrophages, suggesting that the macrophage-type C/EBPβ occupancy pattern is established early in development.
Taken together, our results demonstrate that the differentiation from HE into haematopoietic precursor cells is associated with a redistribution of SCL/TAL1 and FLI1 to new binding sites.
RUNX1 binds to genes that are previously bound by SCL/TAL1, FLI1 and C/EBPβ
We next addressed the question of what caused this shift in binding site patterns. RUNX1 is absolutely required to drive the differentiation of the HE into haematopoietic cells and for the activation of important haematopoietic regulator genes, such as Pu.1 (Sfpi1) (Chen et al, 2009; Hoogenkamp et al, 2009; Lancrin et al, 2009). We therefore investigated the role of RUNX1 in this shift using the iRUNX1 system. To this end, we induced RUNX1 expression in the HE by overnight treatment with low levels of doxycycline as described (Hoogenkamp et al, 2009) and measured global RUNX1 binding sites (Figure 6A; Supplementary Figure 6A). To test the effect of RUNX1 induction on active chromatin formation, we also measured H3K9Ac. Uninduced cells served as control. We obtained 15 669 RUNX1 peaks (Supplementary Table 1, examples are shown in Figure 6D). In the heat map, RUNX1 binding sites clustered with those of FLI1 and C/EBPβ sites in the uninduced HE (Figure 5D).
Figure 6.
Identification of genome-wide RUNX1 binding sites in the HE. (A) Experimental strategy for iRUNX ES cell differentiation. (B) Four-way Venn diagram of genes bound by RUNX1, SCL/TAL1, FLI1 and C/EBPβ. (C) GSEA of this group of genes. (D) Screenshots from the UCSC genome browser showing genes binding SCL/TAL1, FLI1 C/EBPβ and RNA Pol II in the uninduced HE. RUNX1 binding and H3K9Ac are shown before and after induction of RUNX1. (E) Heat map showing two groups of RUNX1 bound sequences and corresponding H3K9Ac levels before and after RUNX1 induction. ChIP enrichment is shown 5 kb upstream and downstream of the peak centre. (F) Integration of the sequence enrichment of each group into a density plot clearly demonstrating RUNX1 binding to pre-existing sites of histone acetylation and de novo sites.
Again, de novo RUNX1 binding sites in the HE did not always overlap with the binding sites for SCL/TAL1, FLI1 and C/EBPβ (Supplementary Figure 6B, with manual validations for different genes shown in Supplementary Figure 6C). Pu.1 (Sfpi1) is one of only a few genes where all factors bind together to a single cis-regulatory element, the 3′ URE (Figure 6D; Supplementary Figure 6D). However, the gene set bound by RUNX1 was a subset of that bound by SCL/TAL1, FLI1 and C/EBPβ (Figure 6B) and also showed the highest enrichment factor for HSC-specific genes (Figure 6C). In all, 61% of all RUNX1 peaks contained a RUNX1 consensus sequence. The main motifs associated with RUNX1 sites were ETS binding sites and AP1 consensus sequences (Supplementary Figure 6E).
We next examined the pattern of RUNX1 induced genes using expression microarrays (Supplementary Figure 6F and G). In all, 1179 genes responded to overnight RUNX1 induction and 64.2% (431/671) of the upregulated and 46.2% (229/508) of the downregulated genes were direct RUNX1 targets. In all, 16% (188) of these genes were bound by all four factors (Supplementary Figure 6H). To examine how important haematopoietic regulator genes respond to RUNX1 induction, we measured the expression of a number of these genes in Flk-1+ cells, and in the HE in the presence and absence of RUNX1 (Supplementary Figure 6I). Despite containing RUNX1 binding sites (genes marked in black), genes known to be involved in the regulation of haematopoiesis such as Fli1, Tal1, Elf1 and Etv6 did not respond to RUNX1 induction. The known RUNX1 target genes Pu.1 (Sfpi1) and the regulator of erythropoiesis Nfe2 (Huang et al, 2008; Wang et al, 2010) were upregulated by RUNX1, while others such as Erg and Notch1 were downregulated. The latter result uncovered an interesting potential feedback loop, as it has been proposed that NOTCH1 is involved in upregulating Runx1 expression (Nakagawa et al, 2006; Wang et al, 2010). Cebpa did not respond to RUNX1 induction, and neither did the known RUNX1 target Csf1r. As shown previously (Hoogenkamp et al, 2009), Csf1r cannot be bound by RUNX1 at this developmental stage (Supplementary Figure 6D).
In summary, our data demonstrate that RUNX1 binds to haematopoietic genes that have previously bound SCL/TAL1, FLI1 and C/EBPβ, but they also show that these factors do not direct RUNX1 to specific sequences.
RUNX1 induction activates additional distal cis-regulatory elements
To test whether RUNX1 preferentially associated with pre-existing sites of active chromatin or nucleated transcription factor assembly at new sites, we integrated RUNX1 binding data with H3K9Ac data. The heat map in Figure 6E shows that RUNX1 binding sites cluster into two groups. Group I (Figure 6F, upper panel) consisted mostly of promoter sequences and was marked by pre-existing H3K9Ac surrounding the peak, indicating that RUNX1 bound to pre-existing transcription factor complexes that were flanked by acetylated histones. The binding of RUNX1 strongly increased H3K9Ac at these sites and beyond. Group II (Figure 6F, bottom panel) represents mostly promoter distal sites of de novo binding that have very low or absent H3K9Ac, indicating that RUNX1 binding does not commonly require high levels of active chromatin marks. After RUNX1 binding, H3K9Ac was strongly increased at these sites and the pattern indicated the formation of a new factor complex that was flanked by acetylated histones.
Taken together, our data show that (i) the binding of RUNX1 does not require pre-existing histone H3K9 acetylation, but binding strongly increases local H3K9Ac levels and (ii) the vast majority of de novo RUNX1 binding sites consist of intergenic and intragenic sites that are likely to be enhancers.
RUNX1 association leads to a rapid redistribution of SCL/TAL1 and FLI1 binding
After establishing the RUNX1 binding pattern in the HE, we tested the hypothesis that the induction of RUNX1 initiated the shift in the binding pattern of SCL/TAL1 and FLI1 during the development of haematopoietic precursor cells. We therefore measured the genome-wide association of these two factors in HE cells after overnight induction of RUNX1 and compared them to the pattern obtained with c-kit+ cells. We obtained 10 395 SCL/TAL1 peaks and 5577 FLI1 peaks in HE cells after RUNX1 induction (Figure 7A; Supplementary Table 1). Examples for specific genes are depicted in Figure 7B and Supplementary Figure 7A and demonstrate a striking complexity of alterations of binding patterns mediated by RUNX1 induction whereby the actual formation of non-adherent precursor cells does not yet take place. Note also that a subset of peaks overlap with each other in multiple individual experiments thus emphasizing the specificity of our data. For example, the Sox17 locus that encodes a transcription factor whose expression distinguishes fetal from adult HSCs (Kim et al, 2007) was strongly downregulated after RUNX1 binding (Figure 7B; Supplementary Figure 6I). While in uninduced and induced cells SCL/TAL1 binding at the promoter was maintained, it increased FLI1 binding after induction and all peaks were finally lost in c-kit+ cells. At another downregulated gene (Erg) encoding a transcription factor important for haematopoiesis (Supplementary Figure 6I), RUNX1 binding at the promoter caused a reduction in H3K9Ac, SCL/TAL1 binding was shifted and was lost in c-kit+ cells (Supplementary Figure 7A and B). The comparison of the overlap between SCL/TAL1 and FLI1 binding sites between non-induced and induced HE cells as well as c-kit+ cells demonstrated a steady loss of peak colocalization with differentiation (HE−DOX→HE+DOX→c-kit+ cells) that was most pronounced with FLI1 (Figure 7A). The latter was also confirmed by clustering analysis (Figure 5D). While the induction of RUNX1 did not cause a major change in the position of SCL/TAL1 binding sites in the heat map, it caused FLI1 binding sequences to now cluster with those of c-kit+ cells. Again, the overlap between associated genes was higher than that of peaks (Supplementary Figure 7C). To further confirm the association with SCL/TAL1 and FLI1 with different sequences, we performed an unbiased motif analysis of binding sites for each factor unique for HE−DOX and HE+DOX. These analyses demonstrate that after RUNX1 induction both factors now associate with sequences that were highly enriched for juxtaposed RUNX1 binding sites, but not before (Figure 7C).
Figure 7.
Redistribution of SCL/TAL1 and FLI1 binding after RUNX1 induction. (A) Venn diagrams intersecting SCL/TAL1 peaks and FLI1 peaks in the uninduced HE, the induced HE and in c-kit+ cells. (B) UCSC genome browser screenshot showing the Sox17 locus exemplifying the movement of different transcription factors after RUNX1 induction in the different cell types. (C) Induction of RUNX1 leads to a shift in the binding of SCL/TAL1 and FLI1 into genomic regions containing RUNX1 motifs. Unbiased motif analysis of genomic regions uniquely bound by the two transcription factors in uninduced or induced iRUNX1 cells.
Two potential scenarios could explain the redistribution of SCL/TAL1 and FLI1. One possibility would be that RUNX1 binding could either displace SCL/TL1 and FLI1 from their binding sites, alternatively it could attract them to new locations. To distinguish between these alternatives, we examined the distances between SCL/TAL1 and FLI1 peaks and the next RUNX1 peak within a 400-bp window at the genomic coordinates. SCL/TAL1 and FLI1 peaks were subdivided into peaks that were gained or lost within 400 bp of a RUNX1 peak on RUNX1 induction (Figure 8A, Supplementary Figure 8A). In both cases, the number of gained peaks within 400 bp of a new RUNX1 peak was significantly greater than expected by chance and many peaks were in close proximity (Figure 8B), providing evidence for RUNX1 attracting SCL/TAL1 and FLI1 to nearby sites. The number of peaks lost after RUNX1 induction was smaller than that of gained peaks. In the case of SCL/TAL1, the occurrence of such peaks was no more than would be expected by chance. However, in the case of FLI1, there was statistical evidence that RUNX1 may displace it from its binding sites (Figure 8B), suggesting that in different contexts RUNX1 binding may either displace FLI1 or attract it. Unbiased motif analysis of these sequences showed the presence of RUNX1 motifs in the majority of each peak population (up to 75%), demonstrating the specificity of our analysis (Figure 8C). Moreover, 72% of new FLI1 sites also contained an ETS motif whereas only 37% of new SCL/TAL1 sites contained an E-Box motif, similar to the pattern described in a haematopoietic precursor cell line (Wilson et al, 2010).
Figure 8.
SCL/TAL1 and FLI1 move towards RUNX1 peaks after induction. (A) Total number and the percentage of peaks that disappear or emerge upon RUNX1 induction. (B) Frequency of distances between SCL/TAL1 or FLI1 peaks and the corresponding nearest RUNX1 binding sites for peaks that were lost (left panel) and peaks that were gained (right panel) after DOX induction. Both SCL/TAL1 and FLI1 gained peak overlaps with RUNX1 peaks were found to be significant (indicated by two asterisks), with Z scores of 107.6 and 34.9, respectively. We found no evidence that RUNX displaced SCL/TAL (Z=0.54), however, it had a weak propensity to replace FLI-1 near its binding sites (Z=24.7, one asterisk). (C) Unbiased motif analysis of SCL/TAL1 and FLI1 bound regions near RUNX1 sites that were either gained or lost after RUNX1 binding, demonstrating the presence of RUNX1 motifs in each analysed peak population. The analysed peak populations are depicted in Supplementary Figure 8. (D) Experimental strategy for withdrawal experiments. (E) Expression (upper panels) and factor binding (lower panels) before and after RUNX1 induction and withdrawal. Amplicons containing a RUNX1 binding site are marked by an asterisk. Error bars indicate the standard deviation between three independent differentiation and ChIP experiments.
To further substantiate the finding that RUNX1 was directly responsible for the shift in transcription factor binding, we took advantage of the full temporal control afforded by the inducible system, and examined whether withdrawal of RUNX1 following an overnight pulse of induction influenced the binding of SCL/TAL1 and FLI1 as well. We therefore measured binding of RUNX1, FLI-1 and SCL/TAL1 before and after RUNX1 withdrawal and at binding sites known to change binding patterns dependent on RUNX1 by manual ChIP to be able to perform accurate quantification (Figure 8E; Supplementary Figure 8B). We also measured expression and compared upregulated and downregulated genes by real-time PCR. At most downregulated genes (Erg, Vav2, Evi1, Notch1, Ets1), the withdrawal of RUNX1 also altered the binding of the other factors and expression went back up. This was independent of the presence of a nearby RUNX1 binding site (genes with a RUNX1 binding site at the relevant amplicons are indicated by a red asterisk). At another downregulated gene, Sox17, the pattern was different, as this gene contained a stable RUNX1 complex that did not disappear within 24 h of withdrawal. Upregulated RUNX1 target genes showed a similar complex picture. While at Nfe2, Itga2b (CD41), AI467606 and Gfi1 the binding of SCL/TAL1 and/or FLI1 diminished following RUNX1 withdrawal, this was not true for Pu.1. These experiments therefore show a direct requirement for the presence of RUNX1 for the observed reorganization of SCL/TAL1 and FLI1 binding patterns, and also indicates preferential retention of RUNX1 protein at a subset of target regions following the withdrawal of doxycycline induction, reminiscent of recently described CTCF binding dynamics following shRNA mediated knockdown (Schmidt et al, 2012).
RUNX1 is required for the interaction between transcription factor complexes at specific genes (Collins et al, 2011; Levantini et al, 2011). Our data suggest that RUNX1 may directly interact with SCL/TAL1 and FLI1 to redirect their genomic location. To show that RUNX1 forms complexes with these factors in the HE, we performed co-immunoprecipitation experiments using nuclear extracts from induced HE cells (Supplementary Figure 8C) and showed that this was indeed the case.
In summary, the data presented here demonstrate (i) that haematopoietic genes are primed by the binding of SCL/TAL1, FLI1 and C/EBPβ in a pattern that is specific for the HE, (ii) that priming is required for the correct temporal regulation of specific genes involved in the regulation of myeloid differentiation and (iii) that RUNX1 redirects the binding pattern of SCL/TAL1 and FLI1 towards that of haematopoietic cells.
Discussion
Haematopoietic genes are primed in the HE
In the absence of RUNX1 many regulator genes such as Tal1, Fli1, Lmo2 or Cebpb are already expressed and many haematopoietic genes are bound by C/EBPβ, SCL/TAL1 and FLI1 at this stage. We also show that at the first stage of the HE SCL/TAL1, FLI1 and C/EBPβ largely bind to different sites, but are already associated with the same genomic neighbourhood. It has been suggested that coordinately regulated genes occupy similar areas within the nucleus and that this colocalization is mediated by specific transcription factors (Schoenfelder et al, 2010). This would explain the presence of RNA-Pol II at distal and intergenic elements and suggests that such a binding pattern may be part of the priming mechanism for haematopoietic genes in the HE. Such genes would already reside in the appropriate transcription factory facilitating low-level expression and would be poised for rapid activation in haematopoietic cells. This is also consistent with our gene expression data. It will be very interesting to examine whether such elements also give rise to transcripts with specific functions in development as suggested in Kowalczyk et al (2012). As exemplified by Sox17, the same mechanism may also allow the rapid inactivation of genes not expressed in haematopoietic cells by incoming repressors.
We suggest that the coordinated activation/upregulation of developmentally regulated genes depends on such priming events. While it is difficult to show this in a genome-wide fashion, in the case of Pu.1 we can unambiguously demonstrate that priming prior to gene activation is essential for the correct temporal regulation of expression. Here, the inactivation of an early cis-regulatory element by a binding site mutation does not abolish expression, but only delays it until late cis elements are activated and compensate for the reduction of activity of the URE. This activation involves an auto-regulatory loop depending on high levels of PU.1 expression (Pimanda et al, 2007; Leddin et al, 2011), which provides a mechanistic explanation for the delay. Such compensatory action may ensure that important genes are always expressed and differentiation is not completely blocked. Studies with mice in which the RUNX1 binding site at the Pu.1 3′URE was mutated or where the URE was deleted demonstrated that progenitor cells expressing low levels of PU.1 reside longer in the self-renewal stage until PU.1 levels rise high enough to drive differentiation. Such mice are prone to develop leukaemia (Rosenbauer et al, 2004; Huang et al, 2008).
SCL/TAL1 and FLI1 binding patterns shift in response to RUNX1 expression
In early myeloid precursor cells, haematopoietic genes are bound by transcription factors known to be important for the development of blood cells, including SCL/TAL1, RUNX1, FLI1 and GATA2 which colocalize on the same cis-regulatory elements. The motif composition of these elements reflects this notion (Wilson et al, 2010). However, the factor binding pattern in the HE prior to the expression of RUNX1 is different. SCL/TAL1 and FLI1 binding sites mostly do not overlap and a predominant motif occurring at their binding sites is an AP1 consensus site, while GATA and RUNX motifs are not enriched. Endothelial cells contain large numbers of functional binding sites for AP1 (Linnemann et al, 2011), suggesting that this factor is part of the binding signature specific for endothelial cells and our experiments suggest that this is also true for the HE. In c-kit+ precursor cells, SCL/TAL1 and FLI1 associate with motifs for haematopoietic transcription factors such as GATA, RUNX and ETS family members and in the clustering analysis now cluster together with sequences binding these factors in haematopoietic cells (Wilson et al, 2010; Hannah et al, 2011). RUNX1 induction causes a rapid shift of the transcription factor binding pattern towards that observed in c-kit+ precursor cells in the absence of overt precursor formation. Moreover, RUNX1 initiates changes in the epigenome, including the formation of new transcription factor complexes with a concomitant increase in histone acetylation at a large number of newly formed cis elements. A significant fraction of these elements now bind SCL/TAL1 and FLI1 in close proximity to RUNX1. RUNX1 is required to mediate long-distance interactions between distant cis-regulatory elements (Collins et al, 2011; Levantini et al, 2011) and from our analysis it is likely that this is the main mechanism by which RUNX1 attracts SCL/TAL1 and FLI1. The fact that we find a physical interaction of RUNX1 with each of these factors in the induced HE supports this idea. Another mechanism operating later in development may be the RUNX1-dependent induction of other transcription factors, notably PU.1. Many ETS factors such as PU.1 and FLI1 share binding sites, as exemplified by the Pu.1 5′ URE (Hoogenkamp et al, 2007), and it is possible that PU.1 displaces FLI1 at some of them once it is expressed at high enough levels and/or provides new interaction platforms for existing factors.
RUNX1 has previously shown to be expendable in haematopoietic precursor cells once they have formed from the HE (Chen et al, 2009). However, our precisely timed withdrawal studies indicate that immediately after RUNX1 induction transcription factor complex assembly at many genes is still flexible, but not at all of them. Our experiments therefore suggest a dynamic interplay between RUNX1 and other transcription factors that dictates the half-life of transcription factor complexes and their dependency on RUNX1. How the different complexes are stabilized, how this translates into cell differentiation and how RUNX1 levels come into play will be a fascinating topic for further investigations.
In summary, the experiments described here highlight a role of RUNX1 in orchestrating the formation of a haematopoiesis-specific transcription factor binding pattern at the onset of haematopoietic development. Our study provides a wealth of data for more in depth mechanistic studies on transcriptional programming by RUNX1 and its downstream effector genes.
Materials and methods
ES cell culture
ES cells were maintained on primary embryonic fibroblasts in DMEM (high glucose) (Sigma D5648), 15% FCS, 1 mM Sodium pyruvate, 50 units/ml Penicillin and 50 μg/ml Streptomycin (Pen/Strep), 1 mM glutamine, 0.15 mM MTG, 25 mM Hepes buffer, 103 U/ml ESGRO® (Millipore), 1 × non-essential amino acids (Sigma). Prior to differentiation, the ES cells were grown without fibroblast feeder cells for two passages on gelatinized tissue culture-treated plates.
ES cell differentiation
A single cell suspension of ES cells was transferred into IVD media (IMDM with 15% FCS, Pen/Strep, 1 mM glutamine, 0.15 mM MTG, 0.18 mg/ml human transferrin (Roche 652202) and 50 μg/ml ascorbic acid) on 15 cm low adherence bacteriological plates (Sterilin) at a concentration of 2.5 × 104/ml. After 3.25 days, embryoid bodies were collected and dissociated by digestion with 1 × trypsin/EDTA. The cell suspension was resuspended in IMDM+20% FCS. Flk-1+ (CD309) cells were isolated using a biotinylated Flk-1 antibody (eBioscience 13-5821) used at 5 μl per 107 cells for 10 min on ice, followed by 2 × wash with MACS buffer (PBS+5% BSA and 0.5 mM EDTA). Cells bound by the antibody were then isolated using antibiotin beads and MACS LS columns (Miltenyi Biotec) according to manufacturer’s instructions.
Blast culture and the differentiation of HE and haematopoietic precursor cells have been described in Lancrin et al (2009) and Sroczynska et al (2009b). A detailed description of these methods can be found in Supplementary Materials and Methods.
Chromatin immunoprecipitation
Differentiated ES cells were crosslinked for 10 min at room temperature with 1% formaldehyde (Pierce, Rockford, IL, USA) and quenched with 1/10 volume 2 M glycine. Nuclei were essentially prepared as described previously (Lefevre et al, 2003). For a more detailed description of the ChIP, see Supplementary Materials and Methods. Antibodies used for ChIP were C/EBPβ Santa Cruz SC-150, FLI1 Santa Cruz SC-356, PU.1 Santa Cruz SC-352, RUNX1 Abcam ab23980, SCL/TAL1 Santa Cruz SC-12984, RNA-Pol II ab817, H3K9Ac ab4441, H3 ab1791 and HA Sigma H6908. Precipitated material was subjected to library preparation and run on an Illumina genome analyzer. A detailed protocol for manual library preparation can be found in Supplementary Methods.
mRNA expression analysis
RNA was extracted from cells using TRIzol® (Invitrogen) according to manufacturer’s instruction. First-strand cDNA synthesis was carried out using M-MLV reverse transcriptase (Invitrogen) according to manufacturer’s instructions using 1–2 μg of RNA. Real-time PCR was carried out using ABI SYBR® green master mix with 2 μl of diluted cDNA and 0.25 μM primer per 10 μl reaction on an ABI 7500 machine. The analysis was carried out at least on biological triplicates measured in duplicate. See Supplementary Table 2 for primer sequences. Microarray expression experiments were performed on whole mouse gene expression microarrays (60 K, Agilent) using three independent biological replicates showing Pearson correlation coefficients between 0.977 and 1.00.
Co-immunoprecipitation assay
Inducible RUNX1 (iRUNX1) ES cells were differentiated into HE and induced overnight with 0.3 μg/ml doxycyclin. Approximately 1.2 × 107 cells were lysed with buffer P (300 mM NaCl, 20 mM HEPES pH 7.5, 0.5 mM EDTA, 0.1% Triton X-100, 0.5 mM sodium vanadate, 2 mM NaF, 2 mM DTT, 0.1 mM PMSF, proteinase inhibitor mix). The extract was diluted 1:1 with buffer P without 300 mM sodium chloride and then incubated with Protein A/G dynabeads previously coupled with 2 μg of the corresponding antibodies or a non-specific control antibody overnight. After washing with buffer P, the immunoprecipitated proteins were eluted by incubation with non-reducing SDS sample buffer. Inputs and precipitated samples were analysed by western blot using an HA-specific antibody. The antibodies used for immunoprecipitation and detection were the same as those used for ChIP. Control antibodies were normal rabbit IgG sc-2027 and normal goat IgG sc-2028.
Generation of homozygous Pu.1 knock-in mutant ES cells
Pu.1 site in Pu.1 URE mutant knock-in (KI) heterozygous ES cells containing one copy of neo (clone C4853 kindly provided by Dan Tenen) were plated at 5 × 105 cells per Ø 10 cm plate of mitomycin-C-treated STO feeder cells. Cells were incubated overnight before changing the ES culture media/LIF to media plus 2.0 mg/ml G418. Surviving colonies were picked after 2 weeks, expanded on new STO feeder cells in the presence of 2 mg/ml G418 and frozen stocks made. An aliquot of each clone was grown on 0.1% gelatin-coated tissue culture flasks for DNA extraction in order to perform PCR genotyping. The primers used for the PU.1 site mut-KI were mA/mP/KI/sense: TGTTTCGAGAACCGAAGGGAATGAC and mA/mP/KI/antisense: AGCTGGACCGGAGGATCTGCGGAAC; PCR products (WT 400 and KI 500 bp) were separated on a 1% agarose gel.
Data analysis
A detailed description of data analyses can be found in Supplementary Materials.
Data accession numbers
All high-throughput data have been deposited at GEO (accession number GSE40235).
Supplementary Material
Acknowledgments
This work was funded by a BBSRC strategic LoLa grant to CB, BG, VK, GL and DW, the MRC (CB and DW), a grant from the European Community (FP6 Integrated Project EuTRACC, grant no. LSHG-CT-2007-037445; CB), the NIH (NIH R01 CA118316, R01 HL112719; DGT), the NIHR Cambridge Biomedical Research Centre grant (RH), Leukaemia & Lymphoma Research (BG) and Cancer Research UK (GL, VK). We thank A Joshi, E Diamanti and L Wernisch for advice on generating heat map displays and Gang Huang and Pu Zhang (Harvard Stem Cell Institute) for Pu.1ki/+ ES cells.
Author contributions: ML, RI, DC, DM, LN and MW performed experiments, MLAL, SAA, VMS and DWR analysed data, DGT, VK and GL provided essential materials, helped writing the paper and designed experiments. CB and BG designed the study and CB wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Begay V, Smink J, Leutz A (2004) Essential requirement of CCAAT/enhancer binding proteins in embryogenesis. Mol Cell Biol 24: 9744–9751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bereshchenko O, Mancini E, Moore S, Bilbao D, Mansson R, Luc S, Grover A, Jacobsen SE, Bryder D, Nerlov C (2009) Hematopoietic stem cell expansion precedes the generation of committed myeloid leukemia-initiating cells in C/EBPalpha mutant AML. Cancer Cell 16: 390–400 [DOI] [PubMed] [Google Scholar]
- Chambers SM, Boles NC, Lin KY, Tierney MP, Bowman TV, Bradfute SB, Chen AJ, Merchant AA, Sirin O, Weksberg DC, Merchant MG, Fisk CJ, Shaw CA, Goodell MA (2007) Hematopoietic fingerprints: an expression database of stem cells and their progeny. Cell Stem Cell 1: 578–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MJ, Li Y, De Obaldia ME, Yang Q, Yzaguirre AD, Yamada-Inagawa T, Vink CS, Bhandoola A, Dzierzak E, Speck NA (2011) Erythroid/Myeloid progenitors and hematopoietic stem cells originate from distinct populations of endothelial cells. Cell Stem Cell 9: 541–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MJ, Yokomizo T, Zeigler BM, Dzierzak E, Speck NA (2009) Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature 457: 887–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Kennedy M, Kazarov A, Papadimitriou JC, Keller G (1998) A common precursor for hematopoietic and endothelial cells. Development 125: 725–732 [DOI] [PubMed] [Google Scholar]
- Collins A, Hewitt SL, Chaumeil J, Sellars M, Micsinai M, Alline J, Parisi F, Nora EP, Bolland DJ, Corcoran AE, Kluger Y, Bosselut R, Ellmeier W, Chong MM, Littman DR, Skok JA (2011) RUNX transcription factor-mediated association of Cd4 and Cd8 enables coordinate gene regulation. Immunity 34: 303–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza SL, Elefanty AG, Keller G (2005) SCL/Tal-1 is essential for hematopoietic commitment of the hemangioblast but not for its development. Blood 105: 3862–3870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilken HM, Nishikawa S, Schroeder T (2009) Continuous single-cell imaging of blood generation from haemogenic endothelium. Nature 457: 896–900 [DOI] [PubMed] [Google Scholar]
- Fehling HJ, Lacaud G, Kubo A, Kennedy M, Robertson S, Keller G, Kouskoff V (2003) Tracking mesoderm induction and its specification to the hemangioblast during embryonic stem cell differentiation. Development 130: 4217–4227 [DOI] [PubMed] [Google Scholar]
- Hannah R, Joshi A, Wilson NK, Kinston S, Gottgens B (2011) A compendium of genome-wide hematopoietic transcription factor maps supports the identification of gene regulatory control mechanisms. Exp Hematol 39: 531–541 [DOI] [PubMed] [Google Scholar]
- Hoogenkamp M, Krysinska H, Ingram R, Huang G, Barlow R, Clarke D, Ebralidze A, Zhang P, Tagoh H, Cockerill PN, Tenen DG, Bonifer C (2007) The Pu.1 locus is differentially regulated at the level of chromatin structure and noncoding transcription by alternate mechanisms at distinct developmental stages of hematopoiesis. Mol Cell Biol 27: 7425–7438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogenkamp M, Lichtinger M, Krysinska H, Lancrin C, Clarke D, Williamson A, Mazzarella L, Ingram R, Jorgensen H, Fisher A, Tenen DG, Kouskoff V, Lacaud G, Bonifer C (2009) Early chromatin unfolding by RUNX1: a molecular explanation for differential requirements during specification versus maintenance of the hematopoietic gene expression program. Blood 114: 299–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Zhang P, Hirai H, Elf S, Yan X, Chen Z, Koschmieder S, Okuno Y, Dayaram T, Growney JD, Shivdasani RA, Gilliland DG, Speck NA, Nimer SD, Tenen DG (2008) PU.1 is a major downstream target of AML1 (RUNX1) in adult mouse hematopoiesis. Nat Genet 40: 51–60 [DOI] [PubMed] [Google Scholar]
- Huber TL, Kouskoff V, Fehling HJ, Palis J, Keller G (2004) Haemangioblast commitment is initiated in the primitive streak of the mouse embryo. Nature 432: 625–630 [DOI] [PubMed] [Google Scholar]
- Junion G, Spivakov M, Girardot C, Braun M, Gustafson EH, Birney E, Furlong EE (2012) A transcription factor collective defines cardiac cell fate and reflects lineage history. Cell 148: 473–486 [DOI] [PubMed] [Google Scholar]
- Kennedy M, Firpo M, Choi K, Wall C, Robertson S, Kabrun N, Keller G (1997) A common precursor for primitive erythropoiesis and definitive haematopoiesis. Nature 386: 488–493 [DOI] [PubMed] [Google Scholar]
- Kim I, Saunders TL, Morrison SJ (2007) Sox17 dependence distinguishes the transcriptional regulation of fetal from adult hematopoietic stem cells. Cell 130: 470–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissa K, Herbomel P (2010) Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature 464: 112–115 [DOI] [PubMed] [Google Scholar]
- Kowalczyk MS, Higgs DR, Gingeras TR (2012) Molecular biology: RNA discrimination. Nature 482: 310–311 [DOI] [PubMed] [Google Scholar]
- Krysinska H, Hoogenkamp M, Ingram R, Wilson N, Tagoh H, Laslo P, Singh H, Bonifer C (2007) A two-step, PU.1-dependent mechanism for developmentally regulated chromatin remodeling and transcription of the c-fms gene. Mol Cell Biol 27: 878–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancrin C, Sroczynska P, Stephenson C, Allen T, Kouskoff V, Lacaud G (2009) The haemangioblast generates haematopoietic cells through a haemogenic endothelium stage. Nature 457: 892–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leddin M, Perrod C, Hoogenkamp M, Ghani S, Assi S, Heinz S, Wilson NK, Follows G, Schonheit J, Vockentanz L, Mosammam AM, Chen W, Tenen DG, Westhead DR, Gottgens B, Bonifer C, Rosenbauer F (2011) Two distinct auto-regulatory loops operate at the PU.1 locus in B cells and myeloid cells. Blood 117: 2827–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefevre P, Melnik S, Wilson N, Riggs AD, Bonifer C (2003) Developmentally regulated recruitment of transcription factors and chromatin modification activities to chicken lysozyme cis-regulatory elements in vivo. Mol Cell Biol 23: 4386–4400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levantini E, Lee S, Radomska HS, Hetherington C, Alberich-Jorda M, Amabile G, Zhang P, Huang G, Zhang DE, Ebralidze AK, Bonifer C, Okuno Y, Gottgens B, Tenen DG (2011) RUNX1 regulates the CD34 gene in haematopoietic stem cells by mediating interactions with a distal regulatory element. EMBO J 30: 4059–4070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnemann AK, O’Geen H, Keles S, Farnham PJ, Bresnick EH (2011) Genetic framework for GATA factor function in vascular biology. Proc Natl Acad Sci USA 108: 13641–13646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKercher SR, Torbett BE, Anderson KL, Henkel GW, Vestal DJ, Baribault H, Klemsz M, Feeney AJ, Wu GE, Paige CJ, Maki RA (1996) Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J 15: 5647–5658 [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M, Ichikawa M, Kumano K, Goyama S, Kawazu M, Asai T, Ogawa S, Kurokawa M, Chiba S (2006) AML1/Runx1 rescues Notch1-null mutation-induced deficiency of para-aortic splanchnopleural hematopoiesis. Blood 108: 3329–3334 [DOI] [PubMed] [Google Scholar]
- Nottingham WT, Jarratt A, Burgess M, Speck CL, Cheng JF, Prabhakar S, Rubin EM, Li PS, Sloane-Stanley J, Kong ASJ, de Bruijn MF (2007) Runx1-mediated hematopoietic stem-cell emergence is controlled by a Gata/Ets/SCL-regulated enhancer. Blood 110: 4188–4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pencovich N, Jaschek R, Tanay A, Groner Y (2010) Dynamic combinatorial interactions of RUNX1 and cooperating partners regulates megakaryocytic differentiation in cell line models. Blood 117: e1–14 [DOI] [PubMed] [Google Scholar]
- Pimanda JE, Ottersbach K, Knezevic K, Kinston S, Chan WY, Wilson NK, Landry JR, Wood AD, Kolb-Kokocinski A, Green AR, Tannahill D, Lacaud G, Kouskoff V, Gottgens B (2007) Gata2, Fli1, and Scl form a recursively wired gene-regulatory circuit during early hematopoietic development. Proc Natl Acad Sci USA 104: 17692–17697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcher C, Swat W, Rockwell K, Fujiwara Y, Alt FW, Orkin SH (1996) The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell 86: 47–57 [DOI] [PubMed] [Google Scholar]
- Robb L, Lyons I, Li R, Hartley L, Kontgen F, Harvey RP, Metcalf D, Begley CG (1995) Absence of yolk sac hematopoiesis from mice with a targeted disruption of the scl gene. Proc Natl Acad Sci USA 92: 7075–7079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbauer F, Owens BM, Yu L, Tumang JR, Steidl U, Kutok JL, Clayton LK, Wagner K, Scheller M, Iwasaki H, Liu C, Hackanson B, Akashi K, Leutz A, Rothstein TL, Plass C, Tenen DG (2006) Lymphoid cell growth and transformation are suppressed by a key regulatory element of the gene encoding PU.1. Nat Genet 38: 27–37 [DOI] [PubMed] [Google Scholar]
- Rosenbauer F, Wagner K, Kutok JL, Iwasaki H, Le Beau MM, Okuno Y, Akashi K, Fiering S, Tenen DG (2004) Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU.1. Nat Genet 36: 624–630 [DOI] [PubMed] [Google Scholar]
- Schmidt D, Schwalie PC, Wilson MD, Ballester B, Gonçalves A, Kutter C, Brown GD, Marshall A, Flicek P, Odom DT (2012) Waves of retrotransposon expansion remodel genome organization and CTCF binding in multiple mammalian lineages. Cell 148: 335–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder S, Sexton T, Chakalove L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, Eskiw CH, Luo Y, Wei CL, Ruan Y, Bieker JJ, Fraser P (2010) Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet 42: 53–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott EW, Fisher RC, Olson MC, Kehrli EW, Simon MC, Singh H (1997) PU.1 functions in a cell-autonomous manner to control the differentiation of multipotential lymphoid-myeloid progenitors. Immunity 6: 437–447 [DOI] [PubMed] [Google Scholar]
- Spyropoulos DD, Pharr PN, Lavenburg KR, Jackers P, Papas TS, Ogawa M, Watson DK (2000) Hemorrhage, impaired hematopoiesis, and lethality in mouse embryos carrying a targeted disruption of the Fli1 transcription factor. Mol Cell Biol 20: 5643–5652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sroczynska P, Lancrin C, Kouskoff V, Lacaud G (2009a) The differential activities of Runx1 promoters define milestones during embryonic hematopoiesis. Blood 114: 5279–5289 [DOI] [PubMed] [Google Scholar]
- Sroczynska P, Lancrin C, Pearson S, Kouskoff V, Lacaud G (2009b) In vitro differentiation of mouse embryonic stem cells as a model of early hematopoietic development. Methods Mol Biol 538: 1–18 [DOI] [PubMed] [Google Scholar]
- Tanaka T, Akira S, Yoshida K, Umemoto M, Yoneda Y, Shirafuji N, Fujiwara H, Suematsu S, Yoshida N, Kishimoto T (1995) Targeted disruption of the NF-IL6 gene discloses its essential role in bacteria killing and tumor cytotoxicity by macrophages. Cell 80: 353–361 [DOI] [PubMed] [Google Scholar]
- Trompouki E, Bowman TV, Lawton LN, Fan ZP, Wu DC, DiBiase A, Martin CS, Cech JN, Sessa AK, Leblanc JL, Li P, Durand EM, Mosimann C, Heffner GC, Daley GQ, Paulson RF, Young RA, Zon LI (2011) Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell 147: 577–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader JE, Fujiwara Y, Orkin SH (1998) Unsuspected role for the T-cell leukemia protein SCL/Tal-1 in vascular development. Genes Dev 12: 473–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Schwemmers S, Hexner EO, Pahl HL (2010) AML1 is overexpressed in patients with myeloproliferative neoplasms and mediates JAK2V617F-independent overexpression of NF-E2. Blood 116: 254–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson NK, Foster SD, Wang X, Knezevic K, Schutte J, Kaimakis P, Chilarska PM, Kinston S, Ouwehand WH, Dzierzak E, Pimanda JE, de Bruijn MF, Gottgens B (2010) Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell 7: 532–544 [DOI] [PubMed] [Google Scholar]
- Yue R, Li H, Liu H, Li Y, Wei B, Gao G, Jin Y, Liu T, Wei L, Du J, Pei G (2012) Thrombin receptor regulates hematopoiesis and endothelial-to-hematopoietic transition. Dev Cell 22: 1092–1100 [DOI] [PubMed] [Google Scholar]
- Zhang P, Iwasaki-Arai J, Iwasaki H, Fenyus ML, Dayaram T, Owens BM, Shigematsu H, Levantini E, Huettner CS, Lekstrom-Himes JA, Akashi K, Tenen DG (2004) Enhancement of hematopoietic stem cell repopulating capacity and self-renewal in the absence of the transcription factor C/EBP alpha. Immunity 21: 853–863 [DOI] [PubMed] [Google Scholar]
- Zinzen RP, Girardot C, Gagneur J, Braun M, Furlong EE (2009) Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature 462: 65–70 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.