Abstract
The centriole is a conserved microtubule-based organelle essential for both centrosome formation and cilium biogenesis. Five conserved proteins for centriole duplication have been identified. Two of them, SAS-5 and SAS-6, physically interact with each other and are codependent for their targeting to procentrioles. However, it remains unclear how these two proteins interact at the molecular level. Here, we demonstrate that the short SAS-5 C-terminal domain (residues 390–404) specifically binds to a narrow central region (residues 275–288) of the SAS-6 coiled coil. This was supported by the crystal structure of the SAS-6 coiled-coil domain (CCD), which, together with mutagenesis studies, indicated that the association is mediated by synergistic hydrophobic and electrostatic interactions. The crystal structure also shows a periodic charge pattern along the SAS-6 CCD, which gives rise to an anti-parallel tetramer. Overall, our findings establish the molecular basis of the specific interaction between SAS-5 and SAS-6, and suggest that both proteins individually adopt an oligomeric conformation that is disrupted upon the formation of the hetero-complex to facilitate the correct assembly of the nine-fold symmetric centriole.
Keywords: centriole, crystallography, SAS-5, SAS-6, structure
Introduction
Centrioles are self-replicating cylindrical organelles with a unique nine-fold symmetry. They are 0.1–0.5 μm long, 0.1–0.2 μm in diameter, and are essential for assembling centrosomes and cilia in eukaryotic cells (Azimzadeh and Marshall, 2010). Defects in centrosome and cilium assembly or function have been linked to a broad spectrum of human diseases (Nigg and Raff, 2009; Bettencourt-Dias et al, 2011). Centrioles are usually composed of a cartwheel structure and nine triplets of microtubules (MTs) (Preble et al, 2000; Marshall, 2001). Exceptions are singlets in centrioles of Caenorhabditis elegans sperm cells and early embryos (Delattre and Gonczy, 2004). Centrioles in C. elegans show a double-layered central tube, which is likely related to the cartwheel structure seen in other species (Pelletier et al, 2006). Studies in a number of research groups have uncovered five C. elegans centriolar proteins, namely the polo-like kinase ZYG-1 (O’Connell et al, 2001), and the four coiled-coil-containing proteins, SPD-2 (Kemp et al, 2004; Pelletier et al, 2004), SAS-4 (Kirkham et al, 2003; Leidel and Gonczy, 2003), SAS-5 (Dammermann et al, 2004; Delattre et al, 2004), and SAS-6 (Dammermann et al, 2004; Leidel and Gonczy, 2005). Homologues of these proteins have been identified in flies and humans (Hung et al, 2000; Andersen et al, 2003; Bettencourt-Dias et al, 2005; Habedanck et al, 2005; Leidel et al, 2005; Basto et al, 2006; Stevens et al, 2010a; Tang et al, 2011). Recently, it was reported that the protein phosphatase PP2A also plays an important role in centriole assembly by regulating SAS-5 recruitment and/or maintaining the structural stability of SAS-5 and ZYG-1 (Song et al, 2011; Kitagawa et al, 2011a). In green algae and humans, Cep135/Bld10 also participates in the formation of the cartwheel structure during centriole assembly (Hiraki et al, 2007; Mottier-Pavie and Megraw, 2009).
RNAi and mating-based assays in C. elegans have shown that centriole duplication is a multistep process, with the five centriolar proteins being recruited in a sequential manner (Delattre et al, 2006; Pelletier et al, 2006; Dammermann et al, 2008). First, SPD-2 is brought close to the mother centriole. The kinase ZYG-1, which is required for the subsequent recruitment of the SAS-5/SAS-6 complex, is then incorporated into the nascent daughter centriole. SAS-5 and SAS-6 together form the initial central tube. Subsequently, SAS-4 is recruited to build an outer layer of the central tube. Finally, nine singlet MTs are assembled around the central tube to generate a daughter centriole that is identical to the mother.
The nine-spoked cartwheel is the first assembled structure during centriole duplication in many organisms, and SAS-6 has been previously shown to ensure the nine-fold symmetry of centrioles in green alga (Nakazawa et al, 2007). Recently, crystal structures of the N-terminal head group of SAS-6 from several organisms have been determined, which suggested that SAS-6 could self-associate in vitro into assemblies akin to the central hub of the cartwheel (van Breugel et al, 2011; Kitagawa et al, 2011b). However, whether SAS-6 alone could faithfully drive the formation of the strict nine-fold symmetry of centrioles is still a matter of some debate (Cottee et al, 2011). Indeed, modelling the oligomeric assembly of SAS-6 using the crystal structures of the Chlamydomonas reinhardtii and Danio rerio SAS-6 head groups results in spirals of different orientations or a flat ring that fits into an eight-fold symmetry (Cottee et al, 2011). Furthermore, the oligomeric structure of recombinant Drosophila SAS-6 (DSAS-6) is different from the in vivo structure of centrioles (Gopalakrishnan et al, 2010). Similarly, overexpression of DSAS-6 in Drosophila embryos resulted in de novo formation of irregular tube-like structures that can be much larger than the centriole; interestingly, overexpression of other centriolar proteins such as SAS-4 and the polo-like kinase SAK also generates such tube-like structures (Peel et al, 2007). Intriguingly, although the binding affinity between the head groups of C. elegans SAS-6 is comparable to that of C. reinhardtii, H. sapiens, and D. rerio SAS-6, recombinant C. elegans SAS-6 alone does not form a cartwheel-like structure similar to that of non-nematode SAS-6 proteins (Pelletier et al, 2006; van Breugel et al, 2011; Kitagawa et al, 2011b). Notably, the head group interaction of different SAS-6 proteins (dissociation constant (Kd)∼60–110 μM) is relatively weak and has been thought unlikely to be the driving force for forming the nine-fold symmetry (Cottee et al, 2011). Taken together, these data suggest that faithful duplication of the strict nine-fold symmetric centrioles likely requires other symmetry-ensuring factors.
It was shown previously that, although overexpression of DSAS-6 alone resulted in an irregular tube-like structure, co-overexpression of DSAS-6 with Ana2, the putative Drosophila orthologue of SAS-5, generated a highly ordered tubular structure, the SAS tubule. This structure looked the same as the in vivo cartwheel structure, suggesting that Ana2 assists SAS-6 in Drosophila centriole assembly (Stevens et al, 2010b). Similarly, STIL, the putative vertebrate orthologue of SAS-5, regulates centrosome integrity (Castiel et al, 2011), and depletion of either SAS-6 or STIL made the other protein fail to target to the procentriole, indicating that SAS-6 and STIL in vertebrates are mutually dependent for centriolar localization (Tang et al, 2011; Arquint et al, 2012; Vulprecht et al, 2012). Earlier experiments carried out in worms revealed that SAS-5 and SAS-6 physically interact with each other for their codependent centriolar localization and that centriole duplication failed in embryos with a sas-5-mutant that fails to interact with SAS-6, indicating that SAS-5 works synergistically with SAS-6 in C. elegans centriole assembly (Leidel et al, 2005). Altogether, these findings suggest that the SAS-5/Ana2/STIL family of proteins is likely the extra factor needed for SAS-6 to generate the nine-fold symmetry of centrioles.
It was previously reported that SAS-5 binds to the SAS-6 coiled coil and that SAS-6 fails to interact with the sas-5(t2079) mutant, which corresponds to a C-terminal single-residue mutation of SAS-5, R397C (Leidel et al, 2005). Studies of centriole duplication in C. elegans indicate that the SAS-5/SAS-6-based central tube is the first observable structure in procentrioles, which grows wider upon the recruitment of SAS-4 (Pelletier et al, 2006). However, it remains elusive what the molecular mechanism of SAS-5 and SAS-6 interaction is and how the central tube of C. elegans centrioles is formed.
In this study, we determine the crystal structure of the C. elegans SAS-6 coiled coil and establish the molecular basis for SAS-5/SAS-6 interaction using structure-based mutagenesis studies. We further show that purified SAS-5/SAS-6 complex assembles into semicircular or arc-like structures with a diameter similar to that of the central tube of C. elegans centrioles and that their interaction disrupts the autoinhibitory oligomerization of both SAS-5 and SAS-6. Based on our findings and previous reports, we propose a molecular mechanism for centriole duplication in C. elegans.
Results
The C-terminal domain of SAS-5 is both necessary and sufficient for interaction with SAS-6
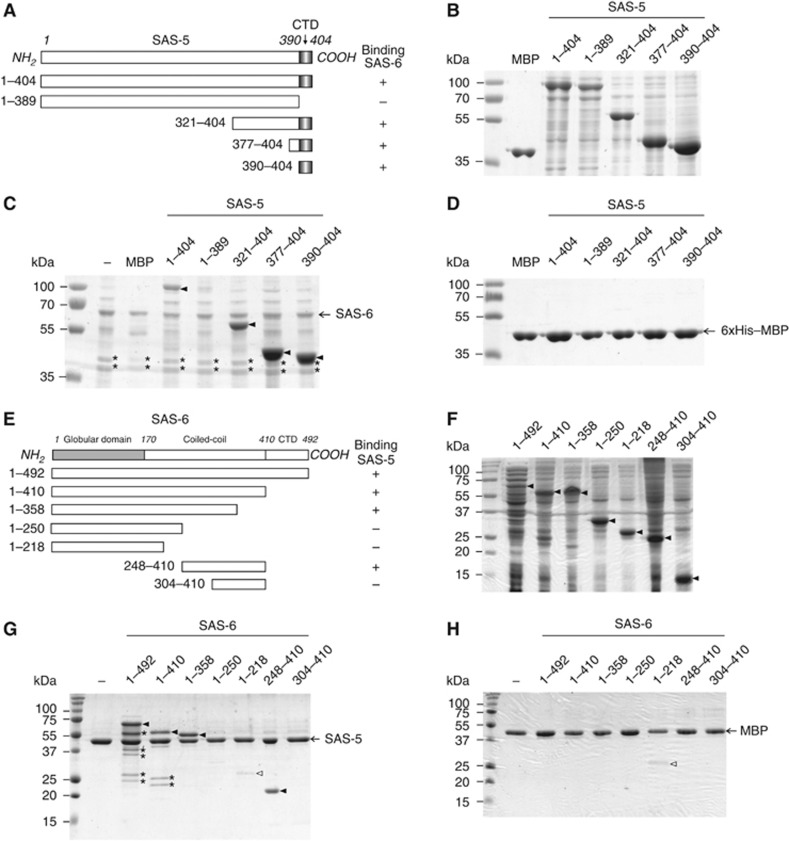
Previous studies using yeast two-hybrid assays showed that the interaction between SAS-6 and SAS-5 was undetectable when using SAS-5 corresponding to the sas-5(t2079) mutant allele (Leidel et al, 2005). Since this mutation (R397→C) is located close to the C-terminus of SAS-5 and as the last 15 residues of SAS-5 are predicted to form an α helix, we wondered whether this C-terminal helix alone is sufficient for binding SAS-6. To test this, we carried out in vitro pull-down experiments using five truncations of SAS-5 in addition to the full-length protein (Figure 1A). In order to increase yield and to better visualize the smaller fragments, we added maltose-binding protein (MBP, Mw∼42 kDa) as a fusion tag to the N-termini of all six constructs (Figure 1B). Our in vitro pull-down results showed that the SAS-5 CTD (residues 390–404) is both necessary and sufficient to bind SAS-6 (Figure 1C). In a control experiment, we used MBP-loaded Ni-NTA beads to pull down SAS-5, which shows no bound SAS-5 (Figure 1D), indicating that the determined interaction between SAS-5 and SAS-6 is specific. Notably, during purification of SAS-6 we consistently observed two degraded fragments on SDS–PAGE gels (Figure 1C, asterisks). Using N-terminal amino-acid analysis, we found that the two fragments correspond to sequences starting at residues 225 and 239, respectively. This indicates that this neck region of SAS-6 (in reference to the head group and the coiled-coil tail), approximately spanning residues 220–240, is flexible and prone to proteolysis.
Figure 1.
The C-terminal domain of SAS-5 interacts with the central part of the SAS-6 coiled coil. (A) Deletion constructs of SAS-5 used for in vitro binding assays with SAS-6. CTD, carboxy-terminal domain. Numbers indicate amino-acid positions or ranges. The right column shows a summary of the binding results in (C). (B) Purified MBP and soluble fractions of MBP-tagged SAS-5 proteins for in vitro pull-down assays. (C) In vitro pull-down results of SAS-5 proteins using Ni-NTA bound full-length SAS-6 as the bait. MBP is used as a negative control for detecting tag-dependent binding. SAS-5 proteins specifically pulled down by SAS-6 are indicated with arrowheads. Marked by asterisks are the two degradation products of SAS-6. (D) No non-specific interaction to the resin or the MBP tag was detected. (E) Truncation constructs of SAS-6 used for in vitro binding assays with the SAS-5 CTD. The right column shows the summary of the binding results in (G). (F) Soluble fractions of SAS-6 proteins used in the in vitro pull-down assays. Arrowheads indicate the target proteins. (G) In vitro pull-down results of SAS-6 proteins using amylose beads preloaded with the MBP-tagged SAS-5 CTD as the bait. Filled arrowheads indicate SAS-6 proteins pulled down by SAS-5. An empty arrowhead indicates the MBP-dependent non-specific binding of the construct containing residues 1–218 of SAS-6, which is comparable to what is seen in the control experiment in (H). Asterisks indicate the degradation products of SAS-6. (H) Control experiments to show no non-specific binding of SAS-6 proteins to the MBP tag.
The C-terminal domain of SAS-5 binds specifically to the central part of the SAS-6 coiled coil
It was previously reported that SAS-5 binds specifically to the coiled-coil domain (CCD) of SAS-6 (residues 180–415) (Leidel et al, 2005; Boxem et al, 2008). Our results in Figure 1C show that only the last 15 residues of the SAS-5 CTD are required for SAS-6 interaction. As this CTD is much smaller in size compared with the SAS-6 coiled coil, we anticipated that only a small segment of the SAS-6 coiled coil would be involved in the interaction. To locate the SAS-5-binding site, we generated six truncations of SAS-6 (Figure 1E), which were all expressed and soluble in solution (Figure 1F). We then carried out in vitro binding assays using amylose beads preloaded with the MBP-tagged SAS-5 CTD to pull down SAS-6 protein. The SAS-5 CTD specifically bound to the central region of the SAS-6 coiled coil, spanning residues 248–303 (Figure 1G). In a control assay we used MBP alone as the bait to pull down SAS-6 and no significant binding was detected (Figure 1H), showing that the interaction between the SAS-5 CTD and the SAS-6 coiled coil is specific. In a reciprocal binding experiment, we used Ni-NTA bound SAS-6 constructs to pull down the MBP-tagged SAS-5 CTD, which further confirms that SAS-5 binds to the same region of the SAS-6 coiled coil (Supplementary Figure S1).
Crystal structure of the SAS-6 CCD reveals an electrostatic periodicity along the coiled coil
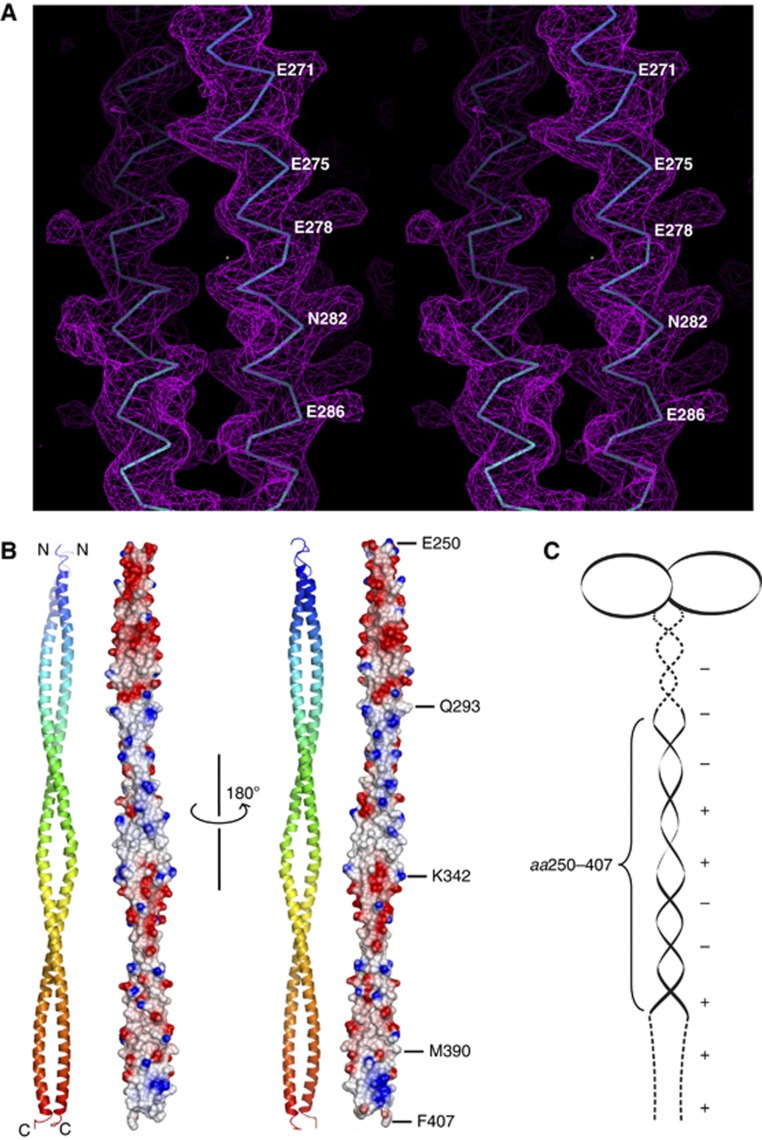
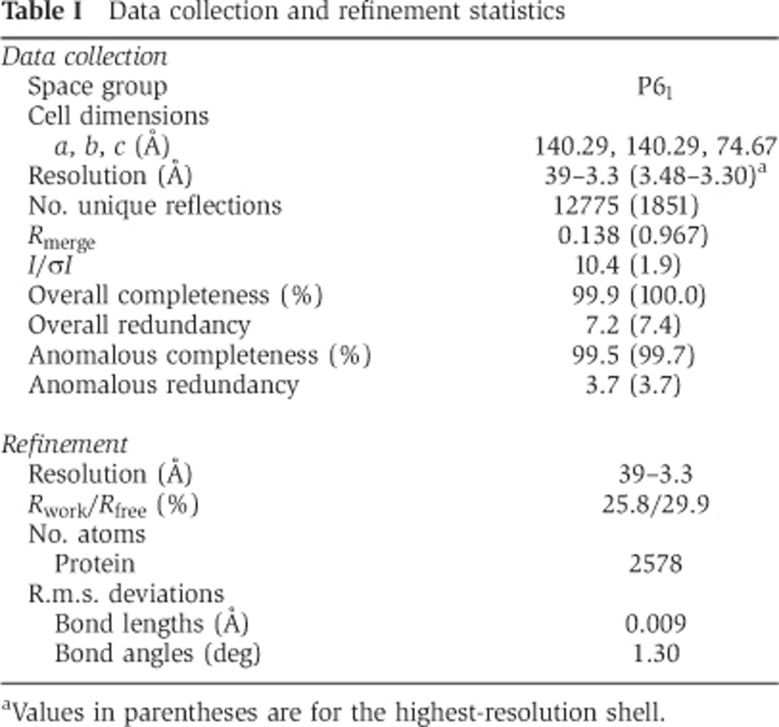
To investigate the interaction between SAS-5 and SAS-6 at the molecular level, we determined the crystal structure of the SAS-6 CCD (residues 248–410). This segment contains the SAS-5-binding site mapped above. The structure was determined to 3.3-Å resolution (Table I; Figure 2A). In the structure, each of the two chains is folded into a continuous α helix spanning residues 250–407. The two helices form a parallel coiled coil extending to 230 Å in length. Interestingly, an electrostatic surface plot indicated that the SAS-6 CCD exhibits a periodically charged pattern along the coiled coil—the segments spanning residues 250–293 and 343–390 are predominantly negatively charged, and residues 294–342 and 391–407 are mainly positively charged (Figure 2B). Sequence alignment of SAS-6 proteins from three different Caenorhabditis species indicated that the surface of the coiled-coil region preceding the CCD, spanning residues 220–250, is also negatively charged (Supplementary Figure S2), whereas the C-terminal part of SAS-6 (residues 410–492) contains five conserved lysine/arginine residues and is positively charged (Supplementary Figure S3). Previously published crystal structures of SAS-6 proteins have shown that the head group together with the N-terminal part of the CCD of SAS-6 forms a dimer (van Breugel et al, 2011; Kitagawa et al, 2011b). Taken together, we conclude that C. elegans SAS-6 folds into a tadpole-like structure with an alternating charge distribution along its coiled-coil tail (Figure 2C).
Table 1. Data collection and refinement statistics.
aValues in parentheses are for the highest-resolution shell.
Figure 2.
Crystal structure of the SAS-6 CCD. (A) Stereo view of a representative portion of the 2Fo–Fc experimental electron density map covering residues 268–290 (contoured at 2.0 σ). For clarity, only the main chains of the final model are shown. (B) Ribbon diagram and electrostatic surface plot of the SAS-6 CCD structure. Residues at the boundaries of differently charged segments are indicated. (C) Schematic representation of the SAS-6 dimer. Dashed lines indicate the regions lacking a known structure. Positive and negative charges along the coiled coil and in the C-terminal domain are depicted as ‘+’ and ‘−’, respectively.
Association of SAS-5 and SAS-6 is based on synergistic hydrophobic and electrostatic interactions
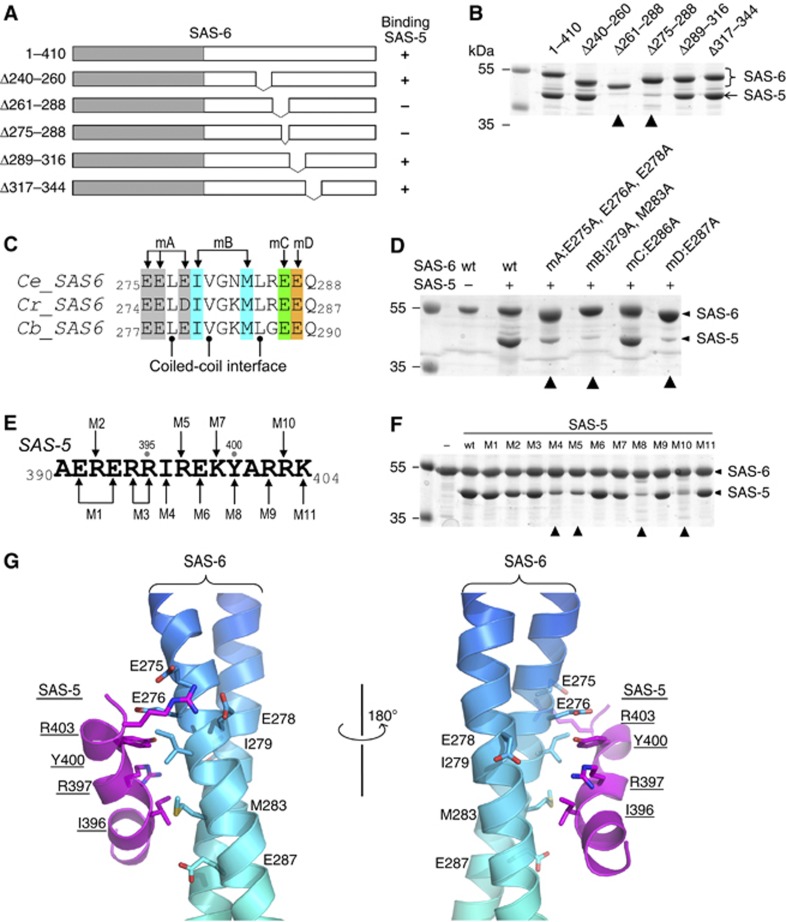
To further narrow down the specific binding site of SAS-5 on SAS-6, we generated multiple structure-based deletion constructs of SAS-6 in the coiled-coil region covering residues 248–303 that is essential for their interaction (Figure 3A). To avoid disrupting or distorting the coiled-coil structure, each deletion removed n × 7 residues (n=2, 3, or 4) to maintain the register of the heptad repeats of the coiled coil. We also generated a deletion outside of this region, spanning residues 317–344, as a control to show that a partial deletion of the SAS-6 coiled coil did not affect its folding or the binding ability of the neighbouring region to SAS-5. Using in vitro pull-down assays, we determined that the region containing residues 275–288 of SAS-6 is essential for SAS-5 binding (Figure 3B).
Figure 3.
Association of the SAS-5 CTD and the SAS-6 CCD is mediated by synergistic hydrophobic and electrostatic interactions. (A) Schematic of SAS-6 deletion constructs. The right column summarizes the interaction results in (B). (B) In vitro pull-down results of MBP-tagged SAS-5 CTD using Ni-NTA bound SAS-6 as the bait. The two deletions of SAS-6 that failed to pull down SAS-5 are indicated with arrowheads. (C) Sequence alignment of the SAS-5-binding site from three Caenorhabditis species. Ce, Caenorhabditis elegans; Cr, C. remanei; Cb, C. briggsae. Mutations of the four groups of conserved, solvent-exposed residues (to alanines) are highlighted in different colours. (D) Coomassie stained SDS–PAGE gel showing the result of in vitro pull-down of SAS-5 by wild-type (wt) and the four mutations of SAS-6. All mutations except for mC failed to interact with SAS-5. (E) Sequence of the SAS-5 CTD. Eleven mutations are indicated as M1–M11. (F) Coomassie stained SDS–PAGE gel showing the results of in vitro pull-down of wild-type or mutants of the SAS-5 CTD by SAS-6. The four mutations that show a drastic decrease of binding to SAS-5 are indicated with arrowheads. (G) Docking the SAS-5 CTD to its binding site on the SAS-6 CCD by ClusPro 2.0 (Kozakov et al, 2010). Side chains of the residues that participate in the interaction are shown.
To determine which individual residues are directly involved in the interaction, we generated four structure-based mutations of all residues in this 14-residue segment that are conserved among Caenorhabditis species but are not parts of the coiled-coil interface (Figure 3C, mA, mB, mC, and mD). Results of the in vitro pull-down experiments indicated that mutations of either the central hydrophobic residues (mB: I279A+M283A) or the flanking negatively charged residues (mA: E275A+E276A+E278A, mD: E287A) nearly completely abolished the SAS-5/SAS-6 interaction. In contrast, mutation of another charged residue in the same region (mC: E286A) seemed not to affect the interaction (Figure 3D). The influence of the mutations on the SAS-5/SAS-6 interaction was further measured by isothermal titration calorimetry (ITC) assays. These indicated that each of the three mutations (mA, mB, and mD) completely abolished the interaction between SAS-5 and SAS-6, whereas the mutation mC only slightly reduced the binding affinity (Supplementary Figure S4A). Given that all these mutated residues are solvent exposed, as seen in the crystal structure, this suggests that the interaction between SAS-5 and SAS-6 is structure-based rather than a non-specific electrostatic interaction.
We showed above that the SAS-5 CTD (residues 390–404) is responsible for interacting with SAS-6 (Figure 1C). To further identify which residues in this region are directly involved in the interaction with SAS-6, we generated 11 mutations in the SAS-5 CTD that substituted each non-alanine residue with an alanine, except for the residue R397, which was replaced by a cysteine as in the previously reported sas-5(t2079) mutant (Leidel et al, 2005) (Figure 3E). All SAS-5 CTD mutations were fused to the C-terminus of MBP to facilitate the visualization of the proteins on SDS–PAGE gels. We then carried out in vitro binding assays using Ni-NTA bound 6 × His–SAS-6 (residues 1–410) to pull down SAS-5. While most of the mutations did not affect the amount of SAS-5 pulled down, four of them (M4/I396A, M5/R397C, M8/Y400A, and M10/R403A) drastically reduced the interaction between SAS-6 and SAS-5 (Figure 3F). To quantitate the influence of the mutations on the interaction, we again carried out ITC experiments using individually purified proteins. None of the four mutations that could not be pulled down by SAS-6 showed a measurable Kd (Supplementary Figure S4B).
In summary, we have identified the residues on both SAS-5 and SAS-6 that are directly involved in the interaction of these two centriolar proteins. These data, together with the crystal structure of the SAS-6 CCD and the predicted helical structure of the SAS-5 CTD, allowed us to generate a docking model for the interaction between the two proteins (Figure 3G). The docking was carried out by ClusPro 2.0 (Kozakov et al, 2010) using the crystal structure of the SAS-6 CCD as the receptor and a theoretical helical model of the SAS-5 CTD as the ligand. Multiple predicted interaction models were generated (Supplementary Figure S5). In a representative docked model, the helix of the SAS-5 CTD was placed nearly perpendicular onto the SAS-6 coiled coil. This arrangement allowed both the hydrophobic interactions between the central two pairs of non-polar residues (SAS-6: I279/M283 versus SAS-5: I396/Y400) and the electrostatic interactions between the flanking oppositely charged residues (SAS-6: E275/E276/E278 versus SAS-5: R403). However, the distance between E287 of SAS-6 and R397 of SAS-5 seems too far for establishing an electrostatic interaction. One possibility is that the side chains of these two charged residues form salt bridges with the backbone of the opposite molecule. Alternatively, the SAS-6 coiled coil might bend at the interaction site to maximize the intermolecular contacts, which has been seen in other interactions between a helix and a coiled coil (Sibanda et al, 2001). Given the nearly symmetric arrangement of the SAS-6 coiled coil and the stoichiometry of the complex (SAS-6 dimer: SAS-5=1:1. See the ITC results in Supplementary Figure S4), it suggests that SAS-5 only binds to one of the two interacting sites on the SAS-6 coiled coil, which implies that binding of SAS-5 to one site either occludes the other site from binding SAS-5 or disrupts the structural symmetry by inducing local conformational changes of the SAS-6 coiled coil.
Mutations in the SAS-6 CCD disrupt centriolar recruitment and function in centriole assembly in C. elegans embryos
Previous work has shown that SAS-5 does not localize and centriole assembly fails in C. elegans embryos carrying a mutation in the C-terminal domain of SAS-5 disrupting its interaction with SAS-6 (Delattre et al, 2004). Similar results would be expected with SAS-6 if we disrupt its interaction with SAS-5, given the co-dependence of SAS-5 and SAS-6 for each other’s recruitment (Leidel et al, 2005; Dammermann et al, 2008). To determine whether this prediction holds true and to confirm our in vitro data, we set out to examine this question in C. elegans.
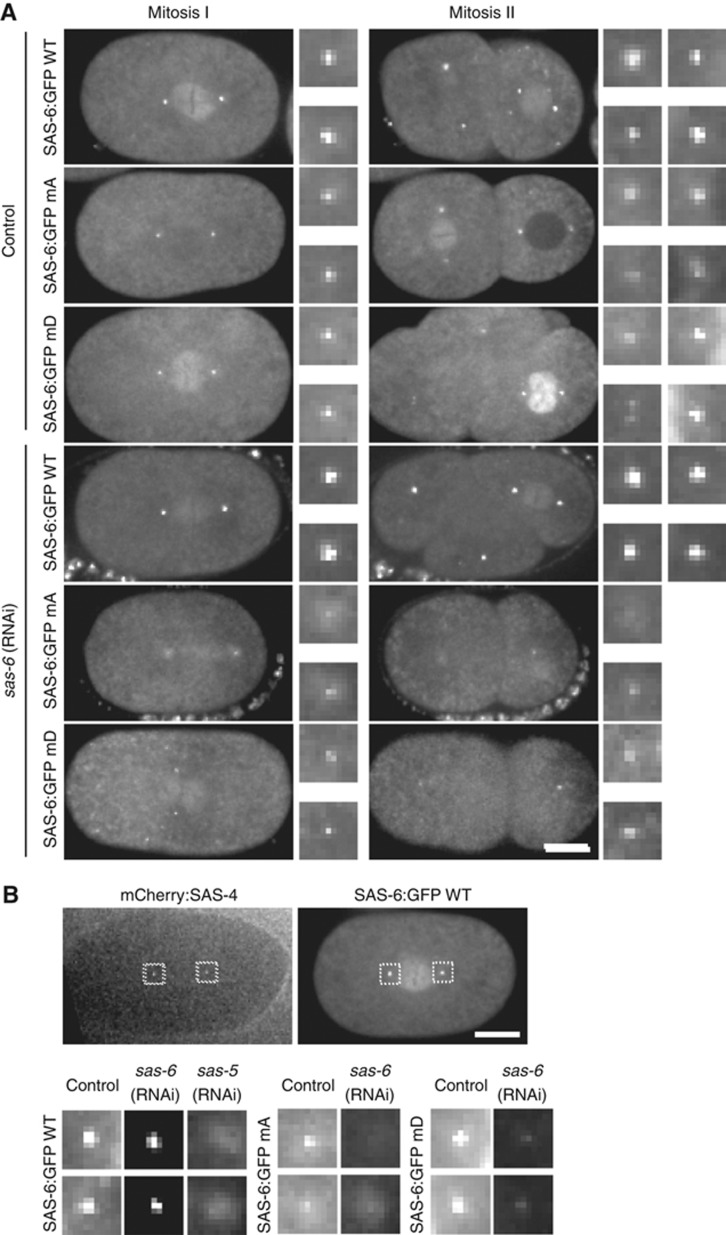
Re-encoded GFP transgenes have been used successfully in C. elegans to confirm functionality following depletion of the endogenous protein by RNAi (Dammermann et al, 2008). However, the intrinsic variabilty in germline expression associated with traditional methods of transformation, notably ballistic bombardment, made comparisons of different mutant isoforms unreliable. The recently established method of Mos1 transposon-mediated insertion at a defined integration site eliminates that variability by providing a fixed chromosomal context for transgene expression (Frokjaer-Jensen et al, 2008). Any differences in phenotype can therefore be confidently assigned to the mutations introduced prior to transformation. We first generated a wild-type SAS-6:GFP fusion under the control of its own promoter and regulatory elements. This fusion was found to be fully functional in restoring centriole assembly and rescuing embryo viability following RNAi-mediated depletion of endogenous SAS-6 (Figure 4A), as well as in the context of a putative null mutant deleting most of the SAS-6 open reading frame (sas-6(ok2554); data not shown).
Figure 4.
In vivo analysis of SAS-6 mutants mA and mD in C. elegans embryos. (A) GFP fusions to wild-type SAS-6 and mutants mA and mD localize to centrioles. However, only wild-type SAS-6:GFP can sustain centriole assembly and consequently spindle bipolarity following depletion of endogenous SAS-6 by isoform-specific RNAi. All wild-type SAS-6:GFP embryos displayed bipolar second divisions following depletion of endogenous SAS-6, all mutant embryos monopolar second divisions. N=10/9 embryos wild-type, 8/22 embryos mA, and 11/17 embryos mD (control/RNAi). Note that GFP signal in RNAi-depleted mutant embryos reflects sperm centriole-associated SAS-6:GFP unaffected by depletion. (B) Centriolar recruitment of SAS-6 requires interaction with SAS-5. Mating was used to introduce mCherry:SAS-4-labelled centrioles into each SAS-6:GFP strain, thereby marking the site of centriole assembly. All embryos are in late prophase-metaphase of the first embryonic division. SAS-6 was detected in wild-type SAS-6:GFP strain following depletion of endogenous SAS-6, but severely diminished in SAS-6 mutants as well as following depletion of SAS-5 in wild-type worms. Scale bar is 10 μm. Insets are magnified × 3. Images in (B) are scaled identically across all strains and conditions to allow cross-comparison.
We next generated constructs carrying each of the three sets of mutations that disrupted interaction with SAS-5 in vitro (mA, mB, and mD) and used these for C. elegans transformation. For mA and mD we obtained several independent strains with identical behaviour. We were unable to generate a strain carrying an integration of mB. This may be due to technical limitations or reflect differences in the ability of C. elegans to tolerate the two classes of mutations (mA, mD charged versus mB hydrophobic). Interestingly, SAS-6 mutants mA and mD both localized to centrioles, at levels not appreciably different from wild-type SAS-6, in control embryos (Figure 4A and B). However, neither mutant could sustain centriole assembly following depletion of the endogenous protein by RNAi, with the characteristic monopolar second division phenotype observed in all embryos (Figure 4A). Note that under the standard RNAi regimen, sperm centrioles are unaffected by RNAi-mediated depletion, and these do remain GFP:SAS-6 positive, even for the two SAS-6 mutants.
To specifically assess recruitment to the site of new centriole assembly, mating can be used to introduce unlabelled sperm centrioles (Kirkham et al, 2003; Dammermann et al, 2004) or centrioles marked with mCherry:SAS-4 (Dammermann et al, 2008), the latter being particularly useful for live assays. Since SAS-6 stably incorporates into centrioles (Dammermann et al, 2004; Leidel et al, 2005), any GFP signal seen under mating conditions reflects new recruitment, rather than exchange of sperm centriole-associated SAS-6 with the cytoplasmic pool. Under these conditions, SAS-6 recruitment was nearly completely abolished for both mutants when endogenous SAS-6 was depleted, similar to what was seen with wild-type SAS-6 following depletion of SAS-5 (Figure 4B). Thus, the residues mutated in mA and mD are indeed critical for SAS-6 recruitment and function in centriole assembly, and the severity of the phenotype mirrors that of depleting SAS-5 itself.
SAS-6 molecules form an anti-parallel tetramer through the electrostatic interactions of their CCDs
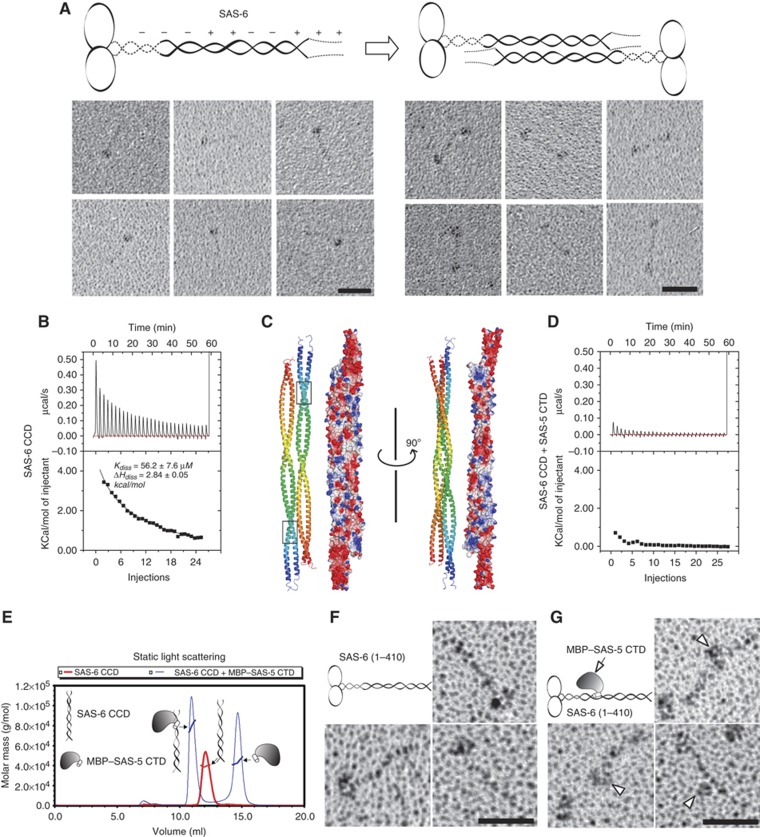
Our rotary metal shadowing electron microscopy studies of purified recombinant SAS-6 protein showed that although many of the observed particles are tadpole-like structures as reported previously (Kitagawa et al, 2011b), a significant fraction of them (∼20%) are dumbbell-like structures with a central rod measuring 35–45 nm in length (Figure 5A). As shown in Figure 2B, there is an alternating charge distribution along the SAS-6 CCD, which we suspected may dictate further self-association of SAS-6 dimers along their coiled coils. To test this hypothesis, we carried out a dilution ITC experiment that has been used successfully for analysing the dissociation equilibrium of other proteins (Lovatt et al, 1996). In this assay, a series of small aliquots of concentrated SAS-6 CCD were injected into a large volume of buffer, which generated a sequence of endothermic heat pulses, characteristic of molecular dissociation (Figure 5B, upper panel). It has been reported that the Kd of the SAS-6 coiled-coil dimer is ∼0.9 μM (Kitagawa et al, 2011b). The fit dissociation curve had a Kdiss of 56.2±7.6 μM and ΔHdiss of 2.84±0.05 kcal/mol, which suggests a dimer-tetramer equilibrium (Figure 5B, lower panel). The tetrameric association of the SAS-6 CCD is apparently much weaker than the coiled-coil dimer and thus may not always survive the grid preparation for rotary metal shadowing as the coiled-coil dimer does. This explains why the dumbbell-like tetramer structure was not as frequently observed as the tadpole-like structure of the SAS-6 dimer.
Figure 5.
C. elegans SAS-6 alone forms an anti-parallel tetramer, whereas binding of SAS-5 disrupts the tetrameric association of SAS-6. (A) Schematic model and the rotary metal shadowing electron micrographs of recombinant SAS-6. Scale bars: 30 nm. (B) Experimental and integrated dilution ITC curves for the SAS-6 CCD (residues 248–410). (C) Docking of the SAS-6 CCD self-association by the automated protein docking program ClusPro 2.0 (Kozakov et al, 2010). Both ribbon diagrams and electrostatic surface plots are shown. Boxed are the SAS-5-binding sites on the SAS-6 CCD. (D) Experimental and integrated dilution ITC curves for SAS-6 CCD+SAS-5 CTD. (E) SLS analysis of the complex of the SAS-6 CCD and the MBP–SAS-5 CTD. The SAS-6 CCD by itself forms a dimer (Mw ∼38 kDa), whereas mixing it with the MBP–SAS-5 CTD (molar ratio=1:1.5) gave rise to a hetero-trimer (Mw ∼80 kDa). Mw of the MBP–SAS-5 CTD is 44 kDa. (F) Rotary metal shadowing electron micrographs of SAS-6 (residues 1–410). Scale bar: 30 nm. (G) Rotary shadowing electron micrographs of SAS-6 in complex with MBP-tagged SAS-5 CTD. Arrowheads indicate the bound SAS-5 on the SAS-6 coiled coil. Scale bar: 30 nm.
To better understand the self-association of the SAS-6 CCD, we subjected our solved crystal structure of the SAS-6 CCD to the ClusPro 2.0 protein–protein docking server (Kozakov et al, 2010). Docking results suggested an anti-parallel interaction of the coiled coils, with the opposite charges complementing each other in each segment (Figure 5C). The fully extended central rod of the anti-parallel SAS-6 tetramer based on this docking model is calculated to be ∼45-nm long, which is in agreement with the length of the rod in the dumbbell-like structure seen in the electron micrographs (Figure 5A). The length variation of the central rods in the dumbbell-like structures is likely due to the flexible region at the N-terminal part of the coiled coil as shown above (Figure 1C, asterisks). These data altogether suggest that C. elegans SAS-6 forms a dumbbell-like tetramer through the anti-parallel association of the coiled coils. Notably, the SAS-5-binding sites on the SAS-6 CCD are obscured in the anti-parallel SAS-6 tetramer (Figure 5C, boxes).
Binding of SAS-5 both disrupts the tetrameric association of the SAS-6 CCD and promotes the formation of an ordered structure reminiscent of the central tube of C. elegans centrioles
We found that SAS-5 binds specifically to a segment of the SAS-6 CCD that is part of the periodic charge region being obscured in the anti-parallel tetramer (Figure 5C, boxes). Moreover, the interaction between SAS-5 and SAS-6 (Kd∼2 μM) is over one order of magnitude stronger than the self-association of the SAS-6 CCD tetramer (Kd∼56 μM). Therefore, binding of SAS-5 should potentially disrupt SAS-6 self association. To test this, we carried out a dilution ITC assay similar to that for the SAS-6 CCD alone but with 1.5 × fold (molar ratio) of the SAS-5 CTD supplemented to both the injections and the buffer. The presence of the SAS-5 CTD in the buffer prevents the dissociation of the SAS-5/SAS-6 complex. However, in case that the anti-parallel self-association of SAS-6 is not affected by SAS-5 binding, we would still observe the dissociation of the tetramer of the SAS-6 CCD to dimers. As shown in Figure 5D, no endothermic heat pulses were observed, which was in contrast to the strong dissociation signal for the SAS-6 CCD (Figure 5B). This suggests that binding of SAS-5 either stabilizes the SAS-6 CCD tetramer or disrupts the tetrametric association of the SAS-6 CCD. As shown in Figure 5E, analysis of the complex by static light scattering (SLS) indicated that the complex is a hetero-trimer (Mw ∼80 kDa; Mws of the SAS-6 CCD monomer and the MBP–SAS-5 CTD are 19 and 44 kDa, respectively). Furthermore, examination by rotary metal shadowing electron microscopy indicated that while SAS-6 (residues 1–410) alone forms a tadpole-like structure (Figure 5F), the complex shows a bound MBP-tagged SAS-5 CTD in the middle of the SAS-6 tail (Figure 5G). Therefore, it is the latter case that accounts for the loss of endothermic heat pulses in the ITC. Moreover, examination using dynamic light scattering also indicates that the SAS-5 CTD shifts the equilibrium between two species of the SAS-6 CCD into one species when mixing the two proteins in a stoichiometric 1:1 ratio (Supplementary Figure S6). Therefore, binding of SAS-5 disrupts the tetrameric association of the SAS-6 CCD.
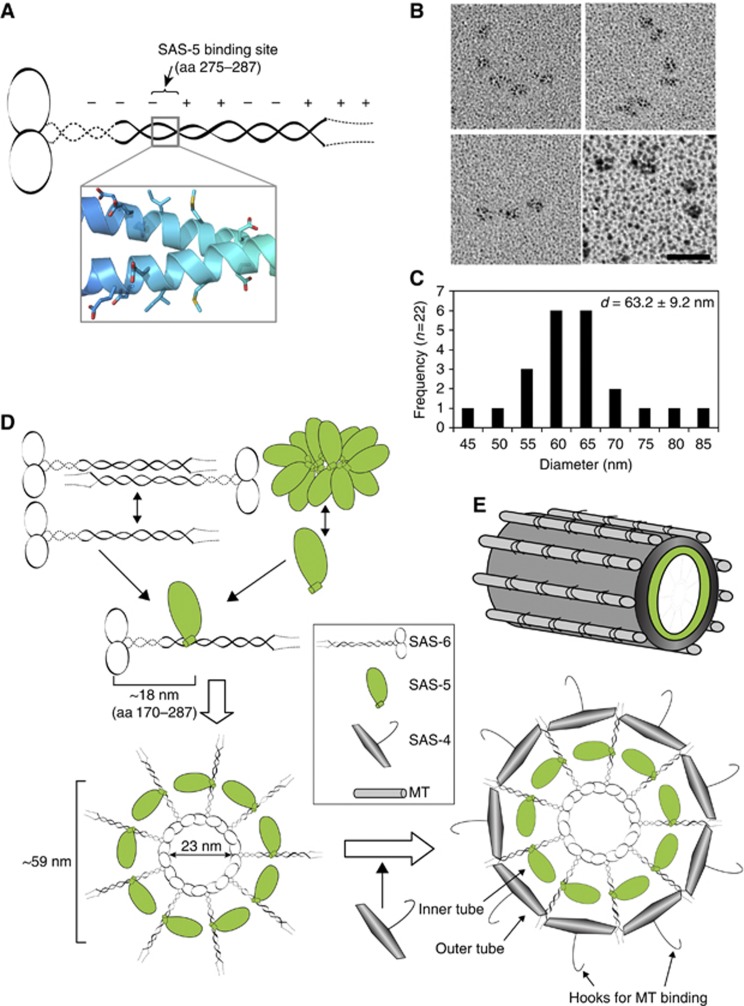
As seen in Figure 4, mutations of SAS-6 residues involved in SAS-5 interaction disrupt SAS-6 centriolar recruitment comparably to what occurs in SAS-5 depletion, which is consistent with previous report that SAS-5 and SAS-6 are mutually dependent for their centrosome localization (Pelletier et al, 2006). Observation of the tadpole- and dumbbell-like structures (Figure 5A) rather than the cartwheel structure of recombinant C. elegans SAS-6 suggests that SAS-5 may play a critical role for establishing the nine-fold symmetry of C. elegans centroles. To find this out, we used rotary metal shadowing electron microscopy to examine the purified complex of recombinant SAS-6 and MBP-tagged SAS-5. Interestingly, we repeatedly observed curved structures (n=22) with particles of the same size arranged with a similar distance in between (Figure 6B). These particles are 2–3 times larger than the SAS-6 head group shown in Figure 5A and are likely SAS-5 molecules. The mean diameter of the fit rings of these structures is 63.2±9.2 nm (Figure 6C), which is in good agreement with the 60-nm diameter of the inner layer of the central tube of C. elegans centrioles (Pelletier et al, 2006).
Figure 6.
The SAS-5/SAS-6 complex forms curved structures similar to the central tube of C. elegans centrioles. (A) Schematic of the SAS-6 dimer and the nearly symmetric arrangement of the residues involved in SAS-5 binding. (B) Rotary metal shadowing electron micrographs of the full-length SAS-5-/SAS-6 complex. Scale bar: 30 nm. (C) Histogram representation of mean diameters of the rings measured from circle structures. The majority of the circle structures have a diameter of 60–65 nm, which is in good agreement with that of the central tube of C. elegans centrioles. (D) Hypothetical mechanism of recruitment of SAS-5, SAS-6, and SAS-4. (E) Schematic illustration of the C. elegans centriole. The inner layer of the central tube is formed by SAS-5, whereas the outer layer is built by SAS-4, which has a hook-like structure that serves to recruit the singlet MTs.
To find out whether SAS-5 alone can form such structures, we tried to purify the MBP–SAS-5 with a C-terminal 6 × His-tag. However, we could not purify the protein using Ni-NTA resin (data not shown), suggesting that the His-tag is inaccessible either due to being shielded by a neighbouring structure or being buried in aggregates. Using size exclusion chromatography and negative staining electron microscopy, we found that SAS-5 forms large aggregates (Supplementary Figure S7). Interestingly, the C-terminus of SAS-5 became accessible when mixing with SAS-6 as demonstrated by the success in the pull-down of SAS-5 by Ni-NTA bound SAS-6 (Figure 1C), and the complex shows semicircle and arc-like structures (Figure 6B). This indicates that binding to SAS-6 indeed releases SAS-5 from its aggregates. Overall, our data suggest that while SAS-6 and SAS-5 individually form an oligomeric conformation, together they can assemble into a highly ordered structure resembling the central tube of the C. elegans centrioles.
Discussion
Accumulating data indicate that SAS-5 and its putative orthologues, Ana2 in flies and STIL in vertebrates, work cooperatively with SAS-6 in centriole formation (Stevens et al, 2010a, 2010b; Tang et al, 2011; Arquint et al, 2012; Vulprecht et al, 2012). To determine how SAS-5 assists SAS-6 in centriole assembly, we first need to know how the two proteins interact. Here, we demonstrate that the short SAS-5 CTD specifically interacts with a very narrow segment of the SAS-6 coiled coil. We have also solved the crystal structure of the SAS-6 CCD that contains the binding site of SAS-5 and further used structure-based mutagenesis studies to identify the residues on both proteins that are directly involved in their interaction. Interestingly, we found that the interaction is mediated by synergistic hydrophobic and electrostatic interactions of multiple residues on either protein. Single-residue mutation analyses showed that mutating any one of these residues completely abolished the interaction. We further showed that the recombinant SAS-5/SAS-6 complex could potentially form semicircular or arc-like structures (Figure 6B). How can one put this into the context of centriole duplication?
Unlike the clearly visible cartwheel structure in non-nematode centrioles, centriole duplication in C. elegans begins with a 60-nm central tube dependent on SAS-5/SAS-6. This central tube grows wider at the pronuclear migration stage when SAS-4 is recruited, which coincides with the emergence of SAS-4 derived hook-like structures around the outer wall at positions where MT assembly occurs (Pelletier et al, 2006). Consistently, SAS-4 homologues in flies and humans also localize to the outer wall of centrioles and are essential for recruiting MTs and pericentriolar materials (Kohlmaier et al, 2009; Tang et al, 2009; Gopalakrishnan et al, 2011). In this study, we show that SAS-6 forms an anti-parallel tetramer whereas SAS-5 aggregates. Crystal structure of the SAS-6 CCD reveals a periodic charge distribution with the SAS-5-binding site in the centre of the coiled coil (Figure 6A). We also discovered that binding of the SAS-5 CTD to the SAS-6 coiled coil both releases SAS-5 from its aggregates and prevents the tetrameric association of SAS-6. This would allow the efficient interaction between the SAS-6 head groups. Our electron microscopy studies confirmed that SAS-5 binds to the central region of the SAS-6 coiled coil (Figure 5G) and demonstrated that the recombinant SAS-5/SAS-6 complex assembles into arc-like structures with an average diameter of 63.2±9.2 nm for the corresponding rings (Figure 6B and C; Supplementary Figure S8). We therefore believe that the emerging 60-nm wide central tube in the procentriole is formed by circularly arranged SAS-5 molecules bound onto the coiled coils of SAS-6 (Figure 6D). SAS-5 and SAS-6 together assemble into an unstable tubular structure, whereas loading of SAS-4 stabilizes this tube (now becomes the inner layer) by generating an outer wall with protruding hook-like appendages that serve to recruit the nine singlet MTs (Figure 6E).
Apart from disrupting the tetrameric association of SAS-6, SAS-5 may also play a more active role in centriole assembly. It was recently revealed that the Trichonympha basal body has a cartwheel structure with spokes from two neighbouring layers merging into a single bundle at the position ∼20 nm away from the central hub (Guichard et al, 2012). However, it remains mysterious how this merge occurs. Given that the merging points around the central hub would encircle a 62-nm ring (d=20 nm × 2+22 nm), we speculate that SAS-5 and its functional orthologues might mediate this merge. Notably, SAS-5 was shown to self-associate in a reported domain-based interactome network in C. elegans (Boxem et al, 2008). The self-association of SAS-5 could bridge spokes of neighbouring layers so that the nine-fold symmetric rings are stacked onto one another to form the cylindrical structure. The other possibility for SAS-5 to play an active role is that binding of SAS-5 may induce conformational changes in the SAS-6 coiled coil that subsequently facilitate centriole assembly. The crystal structure of the SAS-6 CCD indicates that each SAS-6 coiled-coil contains two almost symmetrically arranged SAS-5-binding sites (Figure 6A). However, results of both ITC and SLS experiments indicate that only one SAS-5 molecule could bind to the SAS-6 dimer (Figure 5E; Supplementary Figure S4). One reason for this might be that binding of SAS-5 to one site sterically blocks the other site from recruiting another SAS-5. Alternatively, SAS-5 binding may disrupt the structural symmetry of SAS-6 CCD by inducing a kink of the SAS-6 coiled coil as was seen in the DNA ligase IV-induced bending of the Xrcc4 coiled coil (Sibanda et al, 2001). Such a local conformational change may facilitate SAS-6 directed centriole assembly.
Why were only arc-like structures but not full circles seen in the EM images? Since ZYG-1 is required for the recruitment of SAS-5/SAS-6, it is likely that ZYG-1 plays a structural role in the assembly of the central tube. It was reported previously that ZYG-1 phosphorylates SAS-6 and this phosphorylation is crucial for centriole duplication in vivo (Kitagawa et al, 2009). The phosphorylation site, serine 123, is located in a long flexible loop (disordered in the crystal structure) next to the dimerization interface of the SAS-6 head group (Kitagawa et al, 2011b). It is conceivable that phosphorylation by ZYG-1 might strengthen the head group interaction of SAS-6 by providing an electrostatic interaction between the phospho-group and a positively charged surface patch on the opposite molecule (Supplementary Figure S9). This hypothesis is not only consistent with the observation that ZYG-1-dependent phosphorylation of SAS-6 is needed for both central tube formation and maintenance of SAS-6 at the central tube (Kitagawa et al, 2009), but would also explain why we only observed arc-like structures of the SAS-5/SAS-6 complex in vitro but not closed rings. In the future, it will be interesting to examine whether adding ZYG-1 to the recombinant SAS-5/SAS-6 complex or co-expressing the three proteins could stimulate the formation of a ring-like structure.
It was shown previously that SAS-5 failed to localize to centrioles in a mutant sas-5 (t2033) corresponding to a single amino-acid substitution (R397C) in SAS-5 (Delattre et al, 2004). This substitution disrupts SAS-5 and SAS-6 interaction as demonstrated in a yeast two-hybrid assay (Leidel et al, 2005). Using in vitro pull-down and ITC assays, here we showed that the R397C substitution in SAS-5 completely abolishes its interaction with SAS-6 (Figure 3F). Thus, SAS-5 seems to shuttle to procentrioles through the specific interaction of its CTD with SAS-6. Given that ZYG-1-dependent recruitment of SAS-6 failed when SAS-5 was depleted (Figure 4B), SAS-5 could help SAS-6 target to the procentriole by forming with SAS-6 a specific recognition site for ZYG-1. Indeed, the combination of conserved charges on the SAS-5 CTD and its binding site on the SAS-6 CCD may provide a unique recognition site for ZYG-1 binding (Figures 3G and 6A). Notably, although the SAS-5 CTD is sufficient to bind SAS-6, the N-terminal part of SAS-5 seems to ensure the fidelity of the interaction. As shown in Figure 1C, while full-length SAS-5 was pulled down stoichiometrically relative to SAS-6, considerably more protein was pulled down for all the three N-terminal truncations of SAS-5. This disproportionate interaction occurs only when using full-length SAS-6 but not for SAS-6 lacking the C-terminal disordered tail (residues 411–492) (Figure 3D and F). Therefore, we conclude that the N-terminal domain of SAS-5 (residues 1–389) prevents the non-specific interaction between the SAS-5 CTD and the unstructured SAS-6 C-terminal tail. Consistently, most of the N-terminal part of SAS-5 is conserved in the three Caenorhabditis species. These conserved residues may confer on SAS-5 the ability to regulate its interaction with SAS-6.
What does our finding of SAS-5/SAS-6 interaction imply for the mechanisms of centriole formation in other organisms? Homologues of SAS-6 and putative orthologues of SAS-5 have been identified in flies and vertebrates, which are DSAS-6/hSAS-6 and Ana2/STIL, respectively. Crystal structures of several SAS-6 proteins show that the head group of SAS-6 has a conserved fold that mediates the intermolecular interaction in SAS-6 oligomeric assembly, implying that the mechanism of centriole biogenesis may be conserved through evolution. While structural segmentation of SAS-6 family of proteins is easy to define, domain arrangements of SAS-5/Ana2/STIL are very vague because of the lack of distinct motif structures. An ∼90-residue region toward the C-terminus of the SAS-5 family of proteins, the STAN motif, was suggested to be important for their function (Stevens et al, 2010a). However, while the STAN motif is modestly conserved between Ana2 and STIL (31% sequence identity), it is very divergent in SAS-5. Recently, a second conserved motif called TIM was identified, which is located at the extreme C-terminus of STIL, Ana2, and SAS-5 (Arquint et al, 2012) and includes the SAS-5 CTD. Interestingly, the corresponding sequences in STIL and Ana2 are also predicted to form a helix and two of the four key residues that we found to be essential for SAS-5 interaction with SAS-6 are also conserved in Ana2 and STIL (Supplementary Figure S10A and B). Ana2 has previously been reported to also interact with SAS-6 in Drosophila (Stevens et al, 2010a). While the binding sites have not been precisely defined, as with C. elegans SAS-5 and SAS-6, the interaction appears to involve the C-terminus of Ana2 and part of the coiled coil of DSas-6. Given the poor sequence homology among SAS-5, Ana2, and STIL, confirming and defining the interaction between Ana2/STIL and SAS-6 would be a powerful argument for an orthology relationship between these three proteins.
Using full-length SAS-5 and SAS-6 proteins, we observed semicircle and arc-like structures with an average extrapolated diameter of ∼63 nm (Figure 6B), which is close to the dimensions of the inner part of the central tube seen in electron micrographs of C. elegans embryos (Pelletier et al, 2006). Furthermore, such circles could accommodate ∼8–10 globular structures, consistent with the characteristic nine-fold symmetry of centrioles. Intriguingly, in the rotary metal shadowing micrographs of the full-length SAS-5/SAS-6 complex, we did not observe the 23-nm central hub or the coiled-coil spokes as that formed by C. reinhardtii and D. rerio SAS-6 (van Breugel et al, 2011; Kitagawa et al, 2011b), which is notably consistent with the missing cartwheel structure in C. elegans centrioles in vivo (Pelletier et al, 2006). However, using the same experimental setup, we could observe the coiled-coil tail of SAS-6 when using the residues 1–410 of SAS-6 alone (Figure 5F) or in complex with the MBP-tagged SAS-5 CTD (Figure 5G). It is worth to note though that, similarly to what was observed in the electron micrographs of the full-length SAS-6/SAS-5 complex, no rod-like structures in the complex of SAS-6(aa1–410)/MBP–SAS-5-CTD could be observed upon cross-linking by glutaradehyde (0.05% (v/v), incubated with 0.1 mg/ml protein for 5min at 22°C). While the invisible coiled-coil structure in the truncated complex is likely an artefact generated by cross-linking, we speculate that the lack of the cartwheel structure for the full-length SAS-5/SAS-6 complex might arise from some structural modulations of SAS-6 upon SAS-5 binding. Interestingly, we found that although the majority of the C. elegans SAS-6 coiled coil is well folded as seen in the crystal structure, the N-terminal region (approximately residues 220–240) of the SAS-6 coiled coil is sensitive to proteolysis and seems flexible (Figure 1C, asterisks). It needs to be investigated whether the flexibility of this region accounts for the invisible hub structure in C. elegans centrioles.
In summary, our findings uncover the specific interaction between SAS-5 and SAS-6 and provide a possible explanation for the double-layered central tube structure in C. elegans centrioles. The data further confirm a role for SAS-5 in assisting SAS-6 to determine the nine-fold symmetry of centrioles and suggest a possible mechanism of the regulation in C. elegans centriole assembly. Our results also provide hints for SAS-6 and Ana2/STIL interaction in other organisms and may have general relevance for future studies.
Materials and methods
Cloning, protein expression, and purification
Sequences encoding full-length C. elegans SAS-6 (residues 1–492) and SAS-5 (residues 1–404) were amplified by PCR from cDNA and cloned, respectively, into pET-29a (Novagen) and a custom vector KiM5α that adds an N-terminal MBP tag to the target protein. Truncations of SAS-6 were cloned into pET-15b (Novagen), which provides an N-terminal 6 × His tag cleavable by thrombin. Truncations of SAS-5 were cloned in a similar manner to full-length SAS-5. Deletions and point mutations were generated by the QuickChange Kit (Stratagene) and confirmed by DNA sequencing.
All recombinant proteins were expressed in Escherichia coli BL21 (DE3) cells. The cells were grown at 37°C. At an OD600 of 0.6–0.8, the cells were cold shocked on ice for 10 min and then shifted to 18°C. Protein induction was done overnight with 0.5 mM of isopropyl-β-d-thiogalactopyranoside (IPTG). The cells were harvested and resuspended in cold lysis buffer (20 mM Tris–HCl (pH 8), 300 mM NaCl, 20 mM imidazole, and 5% glycerol). The cells were broken by the EmulsiFlex-C3 homogenizer (Avestin) and the lysate was cleared by centrifugation at 30 000 g for 30 min. The supernatant was filtered through a 0.4-μm filter and loaded onto a Ni-HiTrap column (GE Healthcare) pre-equilibrated in the same lysis buffer. The column was washed with 5 × column volume (cv) of lysis buffer, and bound protein was eluted by a liner gradient concentration of imidazole (20–500 mM, 10 × cv) in the lysis buffer. The N-terminal 6 × His tag was removed by incubation with 2% (w/w) of thrombin overnight at 4°C. The protein was concentrated and further purified with a Superdex-200 16/60 column (GE Healthcare) pre-equilibrated with 20 mM Tris–HCl (pH 8), 50 mM NaCl and 5% glycerol. The protein was concentrated to 10 mg/ml, divided into aliquots and stored at −80°C.
Selenomethionine(SeMet)-substituted SAS-6 CCD (residues 248–410) for crystallization was expressed using M9 minimal medium supplemented with all amino acids (2 mg/ml) except for methionine. Prior to induction, L-SeMet was added to 80 mg/l, and additional threonine, lysine, phenylalanine, leucine, isoleucine, and valine were added to inhibit the methionine biosynthetic pathway (Doublie, 1997). The SeMet-protein was purified as described above, except for the addition of 15 mM β-mercaptoethanol (b-ME) for Ni-HiTrap purification and 10 mM dithiothreitol (DTT) for gel filtration.
Crystallization and data collection
The SAS-6 CCD (residues 248–410) was crystallized at 4°C by the hanging drop method against a reservoir solution containing 0.1 M tri-sodium citrate (pH 5.6), 10% (w/v) PEG 4000 and 10% (v/v) isopropanol. Rod-shaped crystals appeared in 2 days and reached the maximal size of ∼0.03 × 0.03 × 0.5 mm after 1 week. The crystals belong to space group P61 (a=b=140.29 Å, c=74.67 Å). For harvesting, crystals were soaked in the same reservoir solution augmented with increasing concentrations of glycerol (final concentration 20% [v/v]), loop mounted, and flash frozen in liquid nitrogen. Diffraction data to 3.3-Å resolution was collected at the beamline ID23-1 at the European Synchrotron Radiation Facility (ESRF). A complete and highly redundant data set at the anomalous peak of Se (λ=0.9792 Å) was collected.
Structure determination and model docking
Data were integrated using iMosflm (Battye et al, 2011) and scaled using the program SCALA (Evans, 2006). Selenium sites were located and experimental maps were calculated using AutoSol in the software suite Phenix (Terwilliger et al, 2009). Models were built using the program COOT (Emsley and Cowtan, 2004), and refinement carried out using CNS (Brunger et al, 1998) to final Rwork of 0.258 and Rfree of 0.299.
For modelling of the SAS-6 coiled-coil tetramer and the SAS-6/SAS-5 complex, we submitted our solved crystal structure of the SAS-6 CCD and a theoretical helical model the SAS-5 CTD to the web-based ClusPro 2.0 docking server (http://cluspro.bu.edu/), which filters docked conformations with good surface and charge complementarity and ranks them based on their clustering properties. The docking was carried out with default parameters.
Pull-down assays
Small aliquots (50 μl of beads) of 6 × His-tagged full-length or truncated SAS-6 proteins bound to Ni-NTA beads (QIAGEN) were used to pull down MBP-tagged SAS-5 protein from crude cell lysate. Afterwards, the beads were washed using 5 × cv of lysis buffer supplemented with 0.1% Triton X-100 to remove contaminants. After boiling for 2 min in 1 × SDS loading buffer, the proteins were separated on an SDS–PAGE gel and stained with Coomassie Brilliant Blue G250 (Sigma-Aldrich). In a reciprocal binding experiment, we loaded MBP-tagged SAS-5 CTD onto amylose beads and then used these beads to pull down SAS-6 proteins. Subsequent wash and examination were carried out in the same way as the Ni-NTA pull-down. As a negative control to show that SAS-5 proteins did not non-specifically bind to Ni-NTA beads and SAS-6 did not bind to MBP and/or the amylose beads, mock experiments were carried out, in which we used Ni-NTA bound 6 × His-tagged MBP to pull down SAS-5 or MBP alone on amylose beads to pull down SAS-6.
Electron microscopy
Purified full-length SAS-6 and SAS-6 (residues 1–410), either alone or in complex with MBP-tagged SAS-5 CTD, were prepared at 0.05–0.1 mg/ml in 100 mM ammonium bicarbonate (pH 7.5), 30% (v/v) glycerol. The samples were sprayed onto freshly cleaved mica chips. After drying in a Bal-Tec MED020 high vacuum coater (Leica Microsystems) for at least 6 h, the chips were rotary shadowed with 0.7 nm platinum/carbon at an elevation angle of 4 degree for SAS-6 and 7 degree for SAS-5/SAS-6 complex and with carbon at a tilted angle of 45 degree. Electron micrographs were taken on an FEI Morgagni 268D transmission electron microscope operated at 80 kV equipped with a 11-megapixel CCD camera. Images were examined and analysed using ImageJ (http://imagej.nih.gov/ij/).
Accession code
Coordinates and structure factors have been deposited in the Protein Data Bank (PDB) under accession code 4GKW.
Supplementary Material
Acknowledgments
We are grateful to J Lesigang for technical help and V Feng for assisting the characterization of SAS-5/SAS-6 interaction during her visit in the summer of 2010. We thank the staff at ID23-1 of the ESRF for their help with data collection. We also thank G Resch and M Brandstetter for their help in preparing rotary metal shadowing grids and E Shimanovskaya for recording electron micrographs. We greatly appreciate B Morriswood and G Warren for thoroughly reading the manuscript. This work was supported by funding from the MFPL to GD and AD and Grants P23440-B20 and P24296-B20 from the Austrian Science Fund (FWF) to GD and AD, respectively. GC is a DOC-fFORTE fellow of the Austrian Academy of Sciences (ÖAW).
Author contributions: GD and RQ designed the experiments and performed data analyses. MML contributed the original wild-type SAS-6:GFP strain. GC and AD conducted in vivo analysis presented in Figure 4 as well as their discussion within the text. RQ conducted all other experiments. GD supervised the project and wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M (2003) Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426: 570–574 [DOI] [PubMed] [Google Scholar]
- Arquint C, Sonnen KF, Stierhof YD, Nigg EA (2012) Cell-cycle-regulated expression of STIL controls centriole number in human cells. J Cell Sci 125: 1342–1352 [DOI] [PubMed] [Google Scholar]
- Azimzadeh J, Marshall WF (2010) Building the centriole. Curr Biol 20: R816–R825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto R, Lau J, Vinogradova T, Gardiol A, Woods CG, Khodjakov A, Raff JW (2006) Flies without centrioles. Cell 125: 1375–1386 [DOI] [PubMed] [Google Scholar]
- Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG (2011) iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr 67: 271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Hildebrandt F, Pellman D, Woods G, Godinho SA (2011) Centrosomes and cilia in human disease. Trends Genet 27: 307–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Rodrigues-Martins A, Carpenter L, Riparbelli M, Lehmann L, Gatt MK, Carmo N, Balloux F, Callaini G, Glover DM (2005) SAK/PLK4 is required for centriole duplication and flagella development. Curr Biol 15: 2199–2207 [DOI] [PubMed] [Google Scholar]
- Boxem M, Maliga Z, Klitgord N, Li N, Lemmens I, Mana M, de Lichtervelde L, Mul JD, van de Peut D, Devos M, Simonis N, Yildirim MA, Cokol M, Kao HL, de Smet AS, Wang H, Schlaitz AL, Hao T, Milstein S, Fan C et al. (2008) A protein domain-based interactome network for C. elegans early embryogenesis. Cell 134: 534–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Castiel A, Danieli MM, David A, Moshkovitz S, Aplan PD, Kirsch IR, Brandeis M, Kramer A, Izraeli S (2011) The Stil protein regulates centrosome integrity and mitosis through suppression of Chfr. J Cell Sci 124: 532–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottee MA, Raff JW, Lea SM, Roque H (2011) SAS-6 oligomerization: the key to the centriole? Nat Chem Biol 7: 650–653 [DOI] [PubMed] [Google Scholar]
- Dammermann A, Maddox PS, Desai A, Oegema K (2008) SAS-4 is recruited to a dynamic structure in newly forming centrioles that is stabilized by the gamma-tubulin-mediated addition of centriolar microtubules. J Cell Biol 180: 771–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammermann A, Muller-Reichert T, Pelletier L, Habermann B, Desai A, Oegema K (2004) Centriole assembly requires both centriolar and pericentriolar material proteins. Dev Cell 7: 815–829 [DOI] [PubMed] [Google Scholar]
- Delattre M, Canard C, Gonczy P (2006) Sequential protein recruitment in C. elegans centriole formation. Curr Biol 16: 1844–1849 [DOI] [PubMed] [Google Scholar]
- Delattre M, Gonczy P (2004) The arithmetic of centrosome biogenesis. J Cell Sci 117: 1619–1630 [DOI] [PubMed] [Google Scholar]
- Delattre M, Leidel S, Wani K, Baumer K, Bamat J, Schnabel H, Feichtinger R, Schnabel R, Gonczy P (2004) Centriolar SAS-5 is required for centrosome duplication in C. elegans. Nat Cell Biol 6: 656–664 [DOI] [PubMed] [Google Scholar]
- Doublie S (1997) Preparation of selenomethionyl proteins for phase determination. Methods Enzymol 276: 523–530 [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Evans P (2006) Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr 62: 72–82 [DOI] [PubMed] [Google Scholar]
- Frokjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, Grunnet M, Jorgensen EM (2008) Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet 40: 1375–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan J, Guichard P, Smith AH, Schwarz H, Agard DA, Marco S, Avidor-Reiss T (2010) Self-assembling SAS-6 multimer is a core centriole building block. J Biol Chem 285: 8759–8770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan J, Mennella V, Blachon S, Zhai B, Smith AH, Megraw TL, Nicastro D, Gygi SP, Agard DA, Avidor-Reiss T (2011) Sas-4 provides a scaffold for cytoplasmic complexes and tethers them in a centrosome. Nat Commun 2: 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichard P, Desfosses A, Maheshwari A, Hachet V, Dietrich C, Brune A, Ishikawa T, Sachse C, Gonczy P (2012) Cartwheel architecture of Trichonympha basal body. Science 337: 553. [DOI] [PubMed] [Google Scholar]
- Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA (2005) The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol 7: 1140–1146 [DOI] [PubMed] [Google Scholar]
- Hiraki M, Nakazawa Y, Kamiya R, Hirono M (2007) Bld10p constitutes the cartwheel-spoke tip and stabilizes the 9-fold symmetry of the centriole. Curr Biol 17: 1778–1783 [DOI] [PubMed] [Google Scholar]
- Hung LY, Tang CJ, Tang TK (2000) Protein 4.1 R-135 interacts with a novel centrosomal protein (CPAP) which is associated with the gamma-tubulin complex. Mol Cell Biol 20: 7813–7825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp CA, Kopish KR, Zipperlen P, Ahringer J, O'Connell KF (2004) Centrosome maturation and duplication in C. elegans require the coiled-coil protein SPD-2. Dev Cell 6: 511–523 [DOI] [PubMed] [Google Scholar]
- Kirkham M, Muller-Reichert T, Oegema K, Grill S, Hyman AA (2003) SAS-4 is a C. elegans centriolar protein that controls centrosome size. Cell 112: 575–587 [DOI] [PubMed] [Google Scholar]
- Kitagawa D, Busso C, Fluckiger I, Gonczy P (2009) Phosphorylation of SAS-6 by ZYG-1 is critical for centriole formation in C. elegans embryos. Dev Cell 17: 900–907 [DOI] [PubMed] [Google Scholar]
- Kitagawa D, Fluckiger I, Polanowska J, Keller D, Reboul J, Gonczy P (2011a) PP2A phosphatase acts upon SAS-5 to ensure centriole formation in C. elegans embryos. Dev Cell 20: 550–562 [DOI] [PubMed] [Google Scholar]
- Kitagawa D, Vakonakis I, Olieric N, Hilbert M, Keller D, Olieric V, Bortfeld M, Erat MC, Fluckiger I, Gonczy P, Steinmetz MO (2011b) Structural basis of the 9-fold symmetry of centrioles. Cell 144: 364–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlmaier G, Loncarek J, Meng X, McEwen BF, Mogensen MM, Spektor A, Dynlacht BD, Khodjakov A, Gonczy P (2009) Overly long centrioles and defective cell division upon excess of the SAS-4-related protein CPAP. Curr Biol 19: 1012–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozakov D, Hall DR, Beglov D, Brenke R, Comeau SR, Shen Y, Li K, Zheng J, Vakili P, Paschalidis ICh., Vajda S (2010) Achieving reliability and high accuracy in automated protein docking: ClusPro, PIPER, SDU, and stability analysis in CAPRI rounds 13-19. Proteins 78: 3124–3130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidel S, Delattre M, Cerutti L, Baumer K, Gonczy P (2005) SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat Cell Biol 7: 115–125 [DOI] [PubMed] [Google Scholar]
- Leidel S, Gonczy P (2003) SAS-4 is essential for centrosome duplication in C elegans and is recruited to daughter centrioles once per cell cycle. Dev Cell 4: 431–439 [DOI] [PubMed] [Google Scholar]
- Leidel S, Gonczy P (2005) Centrosome duplication and nematodes: recent insights from an old relationship. Dev Cell 9: 317–325 [DOI] [PubMed] [Google Scholar]
- Lovatt M, Cooper A, Camilleri P (1996) Energetics of cyclodextrin-induced dissociation of insulin. Eur Biophys J 24: 354–357 [DOI] [PubMed] [Google Scholar]
- Marshall WF (2001) Centrioles take center stage. Curr Biol 11: R487–R496 [DOI] [PubMed] [Google Scholar]
- Mottier-Pavie V, Megraw TL (2009) Drosophila bld10 is a centriolar protein that regulates centriole, basal body, and motile cilium assembly. Mol Biol Cell 20: 2605–2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa Y, Hiraki M, Kamiya R, Hirono M (2007) SAS-6 is a cartwheel protein that establishes the 9-fold symmetry of the centriole. Curr Biol 17: 2169–2174 [DOI] [PubMed] [Google Scholar]
- Nigg EA, Raff JW (2009) Centrioles, centrosomes, and cilia in health and disease. Cell 139: 663–678 [DOI] [PubMed] [Google Scholar]
- O'Connell KF, Caron C, Kopish KR, Hurd DD, Kemphues KJ, Li Y, White JG (2001) The C. elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell 105: 547–558 [DOI] [PubMed] [Google Scholar]
- Peel N, Stevens NR, Basto R, Raff JW (2007) Overexpressing centriole-replication proteins in vivo induces centriole overduplication and de novo formation. Curr Biol 17: 834–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier L, O’Toole E, Schwager A, Hyman AA, Muller-Reichert T (2006) Centriole assembly in Caenorhabditis elegans. Nature 444: 619–623 [DOI] [PubMed] [Google Scholar]
- Pelletier L, Ozlu N, Hannak E, Cowan C, Habermann B, Ruer M, Muller-Reichert T, Hyman AA (2004) The Caenorhabditis elegans centrosomal protein SPD-2 is required for both pericentriolar material recruitment and centriole duplication. Curr Biol 14: 863–873 [DOI] [PubMed] [Google Scholar]
- Preble AM, Giddings TM Jr, Dutcher SK (2000) Basal bodies and centrioles: their function and structure. Curr Top Dev Biol 49: 207–233 [DOI] [PubMed] [Google Scholar]
- Sibanda BL, Critchlow SE, Begun J, Pei XY, Jackson SP, Blundell TL, Pellegrini L (2001) Crystal structure of an Xrcc4-DNA ligase IV complex. Nat Struct Biol 8: 1015–1019 [DOI] [PubMed] [Google Scholar]
- Song MH, Liu Y, Anderson DE, Jahng WJ, O’Connell KF (2011) Protein phosphatase 2A-SUR-6/B55 regulates centriole duplication in C. elegans by controlling the levels of centriole assembly factors. Dev Cell 20: 563–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens NR, Dobbelaere J, Brunk K, Franz A, Raff JW (2010a) Drosophila Ana2 is a conserved centriole duplication factor. J Cell Biol 188: 313–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens NR, Roque H, Raff JW (2010b) DSas-6 and Ana2 coassemble into tubules to promote centriole duplication and engagement. Dev Cell 19: 913–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang CJ, Fu RH, Wu KS, Hsu WB, Tang TK (2009) CPAP is a cell-cycle regulated protein that controls centriole length. Nat Cell Biol 11: 825–831 [DOI] [PubMed] [Google Scholar]
- Tang CJ, Lin SY, Hsu WB, Lin YN, Wu CT, Lin YC, Chang CW, Wu KS, Tang TK (2011) The human microcephaly protein STIL interacts with CPAP and is required for procentriole formation. EMBO J 30: 4790–4804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Adams PD, Read RJ, McCoy AJ, Moriarty NW, Grosse-Kunstleve RW, Afonine PV, Zwart PH, Hung LW (2009) Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr D Biol Crystallogr 65: 582–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Breugel M, Hirono M, Andreeva A, Yanagisawa HA, Yamaguchi S, Nakazawa Y, Morgner N, Petrovich M, Ebong IO, Robinson CV, Johnson CM, Veprintsev D, Zuber B (2011) Structures of SAS-6 suggest its organization in centrioles. Science 331: 1196–1199 [DOI] [PubMed] [Google Scholar]
- Vulprecht J, David A, Tibelius A, Castiel A, Konotop G, Liu F, Bestvater F, Raab MS, Zentgraf H, Izraeli S, Krämer A (2012) STIL is required for centriole duplication in human cells. J Cell Sci 125: Part 5 1353–1362 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.