Abstract
Autophagy is a conserved process for the bulk degradation of cytoplasmic material. Triggering of autophagy results in the formation of double membrane-bound vesicles termed autophagosomes. The conserved Atg5–Atg12/Atg16 complex is essential for autophagosome formation. Here, we show that the yeast Atg5–Atg12/Atg16 complex directly binds membranes. Membrane binding is mediated by Atg5, inhibited by Atg12 and activated by Atg16. In a fully reconstituted system using giant unilamellar vesicles and recombinant proteins, we reveal that all components of the complex are required for efficient promotion of Atg8 conjugation to phosphatidylethanolamine and are able to assign precise functions to all of its components during this process. In addition, we report that in vitro the Atg5–Atg12/Atg16 complex is able to tether membranes independently of Atg8. Furthermore, we show that membrane binding by Atg5 is downstream of its recruitment to the pre-autophagosomal structure but is essential for autophagy and cytoplasm-to-vacuole transport at a stage preceding Atg8 conjugation and vesicle closure. Our findings provide important insights into the mechanism of action of the Atg5–Atg12/Atg16 complex during autophagosome formation.
Keywords: autophagy, autophagosome, Atg5, Atg8, Atg16
Introduction
Macroautophagy (hereafter autophagy) is a process mediating the bulk degradation of intracellular material. Autophagy is conserved in all eukaryotes and has been shown to promote the survival of cells and thus the organism when exposed to various stresses. During starvation portions of the cell’s cytoplasm are degraded in order to recycle essential building blocks such as amino acids. Autophagy additionally mediates the degradation of protein aggregates, intracellular pathogens and damaged or surplus organelles such as mitochondria and peroxisomes (Xie and Klionsky, 2007; Mizushima et al, 2008; Nakatogawa et al, 2009). Autophagy is thus a major contributor to cellular homeostasis.
Triggering of autophagy results in the de novo formation of double membrane-bound organelles called autophagosomes (Kraft and Martens, 2012). Small membrane structures named isolation membranes or phagophores are early precursors to autophagosomes. Isolation membranes are cup-shaped double membrane-bound structures that expand and gradually enclose cytoplasmic cargo. The isolation membranes then close giving rise to completed autophagosomes. Autophagosomes subsequently fuse with either the endo-lysosomal compartment (in higher eukaryotes) or the vacuole (in yeast) within which the inner autophagosomal membrane and the captured cytoplasmic cargo are degraded (Orsi et al, 2010).
In complex eukaryotes, most autophagosomes appear to be generated from, or are very close to, the endoplasmic reticulum (ER; Axe et al, 2008; Hayashi-Nishino et al, 2009; Ylä-Anttila et al, 2009). However, mitochondria (Hailey et al, 2010), the plasma membrane (Ravikumar et al, 2010; Moreau et al, 2011) and the Golgi (Young et al, 2006; Geng et al, 2010; Ohashi and Munro, 2010; Tooze and Yoshimori, 2010; van der Vaart et al, 2010) have also been reported to contribute membranes for the generation of autophagosomes. In yeast, autophagosomes appear to be generated at the pre-autophagosomal structure (PAS) localized close to the vacuole (Suzuki et al, 2001; Suzuki and Ohsumi, 2010). It has recently been shown that Golgi-derived, Atg9-positive membrane structures relocate to a site close to the vacuole in response to autophagy induction (Mari et al, 2010; Yamamoto et al, 2012). Collectively, these structures likely contribute to the PAS and must somehow undergo fusion and reorganization in order to generate the isolation membrane and eventually the autophagosome.
At the PAS, several proteins required for autophagosome formation localize in a hierarchical manner (Suzuki et al, 2001; Suzuki and Ohsumi, 2010). Among these are components of two ubiquitin-like conjugation systems (Mizushima et al, 1998; Ichimura et al, 2000; Suzuki et al, 2001). The first of these systems entails the conjugation of the ubiquitin-like protein Atg8 to the headgroup of the membrane lipid phosphatidylethanolamine (PE) via a C-terminal glycine (G116). This modification renders the otherwise soluble Atg8 membrane bound. Atg8 conjugation to PE requires the activity of several enzymes. Atg8 is initially synthesized with a C-terminal arginine (R117) that masks the penultimate G116. R117 is removed by the protease Atg4 allowing Atg8 to be transferred to the E1-like enzyme Atg7. Atg7 transfers Atg8 to the E2-like enzyme Atg3 that subsequently transfers Atg8 to the membrane lipid PE. Atg8–PE has been shown to recruit cytoplasmic cargo to the isolation membrane and so ensures its incorporation into autophagosomes. In addition, Atg8–PE has been proposed to mediate membrane tethering and fusion and thus to assist isolation membrane expansion by the promotion of vesicular carrier fusion (Nakatogawa et al, 2007). Membrane fusion and tethering activity have also been demonstrated for the mammalian Atg8-like proteins LC3 and GATE-16 (Weidberg et al, 2011). However, while for yeast Atg8 the tethering activity has been confirmed the fusion activity has recently been questioned (Nair et al, 2011). In mammalian cells, functional inhibition of the Atg8 conjugation system results in isolation membranes that apparently fail to close (Fujita et al, 2008a). This suggests that Atg8–PE is essential for a rather late step of autophagosome formation.
The second ubiquitin-like conjugation system functioning during autophagosome formation is the Atg12 system (Mizushima et al, 1998). Here, Atg7 activates the ubiquitin-like protein Atg12 and transfers it to Atg10. Finally, Atg10 covalently links the C-terminal glycine residue of Atg12 to a lysine residue of Atg5 (K149 of S. cerevisiae Atg5). This reaction appears to be irreversible and a major fraction of both Atg12 and Atg5 exists in the conjugated form (Mizushima et al, 1998; Kuma et al, 2002). The Atg5–Atg12 conjugate associates non-covalently with Atg16 (Mizushima et al, 1999; Kuma et al, 2002). Atg16 is required for the localization of the Atg5–Atg12 conjugate to the PAS in yeast and isolation membranes in higher eukaryotes (Suzuki et al, 2001; Fujita et al, 2008b). The Atg5–Atg12 conjugate has been shown to promote Atg8–PE formation in a manner analogous to the function of E3 enzymes during classical ubiquitin conjugation reactions (Hanada et al, 2007). Atg12 has been reported to bind to Atg3 (Tanida et al, 2002; Fujita et al, 2008b). Since the Atg5–Atg12 conjugate has also been shown to weakly associate with membranes by crosslinking experiments it has been proposed that Atg5–Atg12 recruits Atg3 (conjugated to Atg8) to the membrane thereby facilitating the transfer of Atg8 from Atg3 to membrane-localized PE (Hanada et al, 2007). Atg16 was apparently not required for this E3-like activity (Hanada et al, 2007) although Atg16 is required for the localization of the Atg5–Atg12 conjugate to isolation membranes and the PAS in vivo (Suzuki et al, 2001; Fujita et al, 2008b). Whether the Atg12 conjugation system has any functions in addition to the promotion of Atg8–PE conjugation is unclear. However, in yeast (Abeliovich et al, 1999, 2000; Suzuki et al, 2001) as well as in mammalian cells (Mizushima et al, 2001; Fujita et al, 2008a) autophagosome formation is more heavily impaired by inactivation of the Atg12 conjugation system than the Atg8 conjugation system suggesting additional roles for this conjugation system.
The molecular mechanisms underlying the generation of isolation membranes and their subsequent expansion and closure into autophagosomes are poorly understood. Furthermore, the mechanism of action of the Atg5–Atg12/Atg16 complex remains enigmatic. In order to gain further insights into the biochemical activities of the Atg5–Atg12/Atg16 complex, we developed a reconstituted system using recombinant proteins and giant unilamellar vesicles (GUVs). Here, we describe the regulation and lipid requirements of membrane binding by the Atg5–Atg12/Atg16 complex. We also show that efficient promotion of Atg8–PE attachment requires the association of Atg5–Atg12 with Atg16. Furthermore, we report that in vitro the Atg5–Atg12/Atg16 complex efficiently tethers membranes in an Atg16-dependent manner. Thus, the Atg5–Atg12/Atg16 complex may contribute to the initial tethering of vesicular precursors during autophagosome formation in addition to its Atg8–PE promoting activity. We further identify the membrane binding site in Atg5 and show that membrane binding by Atg5 is essential for both, autophagy and the cytoplasm-to-vacuole-transport (Cvt) pathway at a step preceding Atg8–PE conjugation and vesicle closure but not for the initial recruitment of the Atg5–Atg12/Atg16 complex to the PAS.
Results
Membrane binding by the Atg5–Atg12/Atg16 complex
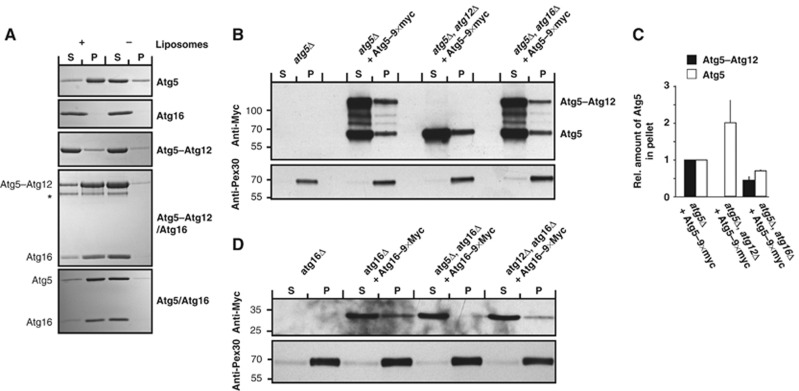
The Atg5–Atg12 conjugate is localized to the PAS in an Atg16-dependent manner (Suzuki et al, 2001). In order to gain insights into the regulation of the recruitment of the Atg5–Atg12 conjugate by Atg16, we (co-)expressed and purified Atg5, Atg16, the Atg5–Atg12 conjugate and the Atg5/Atg16 and Atg5–Atg12/Atg16 complexes from E. coli. We first analysed membrane binding by these proteins in a liposome co-sedimentation assay using highly charged Folch liposomes (Figure 1A). We found that Atg5 alone bound efficiently to these liposomes whereas Atg16 did not show any detectable binding (Figure 1A), consistent with previous cell fractionation experiments (Mizushima et al, 1999; George et al, 2000). Interestingly, compared to Atg5 alone the Atg5–Atg12 conjugate showed severely reduced membrane binding suggesting that Atg12 inhibits membrane binding by Atg5. In contrast, association of Atg5–Atg12 with Atg16 restored membrane binding. These findings suggest that complex formation of Atg5–Atg12 with Atg16 unmasks a membrane-binding site in Atg5. Consistent with this, a complex consisting of Atg5 and Atg16 showed membrane binding to a similar degree as the whole Atg5–Atg12/Atg16 complex. We were unable to test membrane binding by Atg12 in isolation, as we did not succeed in purifying soluble Atg12 alone.
Figure 1.
Membrane binding by Atg5–Atg12/Atg16 complex and its components. (A) Coomassie-stained gels showing the results of liposome co-sedimentation assays using Folch liposomes and the indicated proteins. Liposome binding allows the protein to be pelleted. The star indicates a degradation product of the Atg5–Atg12 conjugate. (B, D) Yeast cells of the indicated genotype were transformed with plasmids coding for Atg5–9 × Myc and Atg16–9 × Myc, respectively, and subjected to fractionation experiments. The proteins were detected by anti-Myc western blotting. The presence of the protein in the pellet fraction indicated membrane binding. (C) Quantification showing the relative amounts of Atg5–9 × Myc and Atg5–9 × Myc–Atg12 in the pellet fractions relative to Pex30. The amount of Atg5–9 × Myc and Atg5–9 × Myc–Atg12 in atg5Δ cells was set to 1. See Supplementary Figure 1 for graph based on non-normed data. The quantification is based on three independent experiments and the averages and the standard deviations are shown. The numbers next to the blots indicate the molecular weight in kDa. P, pellet; S, supernatant. Figure source data can be found with the Supplementary data.
To test whether our in vitro results reflect the situation in vivo, we determined the presence of Atg5 and Atg16 in the membrane fractions of different deletion strains (Figure 1B–D; Supplementary Figure 1). Consistent with our in vitro results we found that the presence of Atg5–Atg12 in the membrane fraction was reduced in yeast cells lacking Atg16 (Figure 1B and C). Atg12 was not required for the presence of Atg5 in the membrane fraction and additionally, in the absence of Atg12 the amount of Atg5 in the pellet fraction was increased (Figure 1B and C). Furthermore, Atg16 was present in the membrane fraction in cells lacking Atg12 but not in cells lacking Atg5 (Figure 1D).
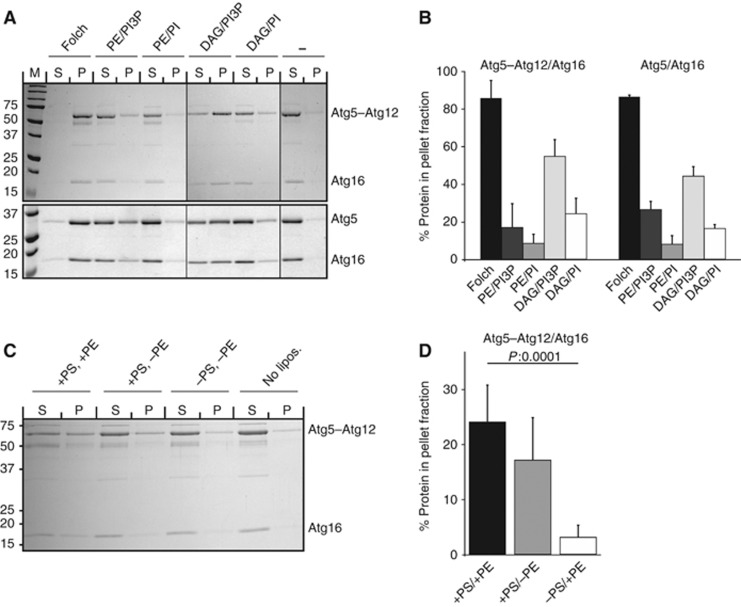
In order to further characterize membrane binding by the Atg5–Atg12/Atg16 complex, we used liposomes with varying lipid compositions (Figure 2). Atg5–Atg12/Atg16 and Atg5/Atg16 showed strong binding to Folch liposomes (Figures 1A, 2A and B). Binding to liposomes with a more physiological lipid composition (PE/PI) was considerably weaker compared to Folch liposomes but interestingly the presence of PI3P slightly enhanced binding (compare PE/PI and PE/PI3P in Figure 2A and B). The fact that Atg5–Atg12/Atg16 showed much stronger binding to the highly charged Folch liposomes compared to liposomes of more physiological composition suggested that binding is charge dependent. Consistently, binding of Atg5–Atg12/Atg16 to liposomes containing no PS was very weak (Figure 2C and D). The positive effect of PI3P on membrane binding by Atg5–Atg12/Atg16 and Atg5/Atg16 is likely due to rather unspecific charge effects since binding to PI3P-containing liposomes was rather weak and not strictly dependent on PI3P. Interestingly, we also found that the presence of PE had a positive effect on membrane binding (Figure 2C and D). We reasoned that Atg5–Atg12/Atg16 may insert some part of itself into the membrane upon binding and that the small headgroup of PE may aid this insertion. In order to test this hypothesis, we substituted PE with diacylglycerol (DAG), which is devoid of any headgroup and should aid membrane insertion even more than PE. Indeed, binding of the Atg5–Atg12/Atg16 complex to DAG-containing liposomes was considerably stronger compared to PE-containing liposomes (Figure 2A and B). Consistent with the results obtained for the PE-containing liposomes, we detected stronger binding upon addition of PI3P. In summary, we conclude that binding of Atg5–Atg12/Atg16 to membranes requires both charge and hydrophobic insertion.
Figure 2.
The lipid requirements for membrane binding by the Atg5–Atg12/Atg16 complex. (A) Coomassie-stained gels of liposome co-sedimentation assays using either Atg5–Atg12/Atg16 (upper panel) or Atg5/Atg16 (lower panel) and liposomes with the indicated lipid composition (Folch: Folch lipids, PE/PI3P: 40% POPC, 35% POPS, 20% POPE, 5% PI3P, PE/PI: 40% POPC, 35% POPS, 20% POPE, 5% PI, DAG/PI3P: 40% POPC, 35% POPS, 20% DAG, 5% PI3P, DAG/PI: 40% POPC, 35% POPS, 20% DAG, 5% PI). Note that the Atg5–Atg12/Atg16 and the Atg5/Atg16 complexes show almost identical binding behaviours. (B) Quantification showing the amounts of Atg5–Atg12/Atg16 and Atg5/Atg16 in the pellet fractions. The amount of protein in the pellet fraction in the absence of liposomes (background) was subtracted from the amount of protein in the pellet fractions in the presence of liposomes. Data were obtained from three independent experiments, one of which is shown in (A). The Atg5–Atg12 and Atg5 bands were used for quantification. (C) Coomassie-stained gel of a liposome co-sedimentation assay showing the lipid requirements of the Atg5–Atg12/Atg16 complex. Liposomes were composed of 40% POPC, 35% POPS, 20% POPE and 5% PI3P. In the liposomes containing no POPE or POPS, these lipids were replaced with POPC. (D) Quantification of Atg5–Atg12/Atg16 in the pellet fraction in liposome co-sedimentation, one of which is shown in (C). Data were obtained from six independent experiments. The Atg5–Atg12 was used for quantification. The average value is shown and the error bars represent the standard deviation. P-values were calculated using Student’s t-test. The numbers next to the gels indicate the molecular weight in kDa. DAG, diacylglycerol; P, pellet; PI, phosphatidylinositol; PI3P, phosphatidylinositol-3-phosphate; (PO)PC, (1-palmitoyl-2-oleoyl-) phosphatidylcholine; (PO)PE, (1-palmitoyl-2-oleoyl-) phosphatidylethanolamine; (PO)PS, (1-palmitoyl-2-oleoyl-) phosphatidylserine; S, supernatant. Figure source data can be found with the Supplementary data.
Membrane recruitment of Atg3 by the Atg5–Atg12/Atg16 complex
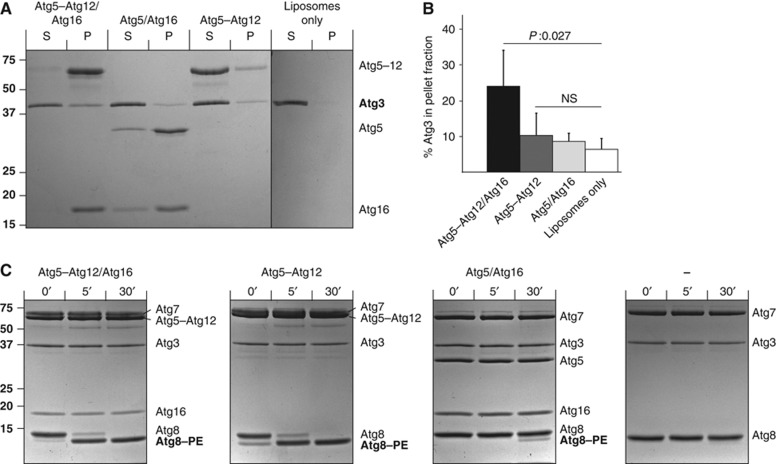
Atg5–Atg12 has been shown to possess an E3 ubiquitin ligase-like activity, promoting the transfer of Atg8 from Atg3 to membrane localized PE (Hanada et al, 2007). Since we found that the Atg5–Atg12/Atg16 complex binds efficiently to liposomes while the Atg5–Atg12 conjugate shows rather poor binding (Figure 1A) we investigated whether the Atg5–Atg12/Atg16 complex recruits Atg3 more efficiently to membranes than the Atg5–Atg12 conjugate. To this end, we incubated Folch liposomes with Atg3 in the presence of the Atg5–Atg12/Atg16 complex, the Atg5/Atg16 complex or the Atg5–Atg12 conjugate. Indeed, we found that in the presence of Atg5–Atg12/Atg16 the amount of Atg3 in the pellet fraction was about three-fold higher compared to liposomes only (Figure 3A and B). In agreement with very weak membrane binding by the Atg5–Atg12 conjugate, the amount of Atg3 in the pellet fraction in its presence was only slightly above background. Consistent with the finding that Atg3 interacts with Atg12 (Tanida et al, 2002), membrane recruitment of Atg3 by the Atg5–Atg12/Atg16 complex was Atg12 dependent as the Atg5/Atg16 complex was very inefficient in recruiting Atg3 to the membrane despite the fact that it efficiently bound to liposomes (Figure 3A and B).
Figure 3.
Atg3 recruitment to membranes is dependent on Atg16 and Atg12. (A) Coomassie-stained gels showing co-sedimentation assays using Folch liposomes and the indicated proteins. Atg5–Atg12/Atg16 was most efficient in recruiting Atg3 to the pellet fraction. (B) Quantification of Atg3 in the pellet fraction in the presence of the indicated proteins. The quantification is based on three independent experiments. (C) Coomassie-stained gels showing the results of Atg8–PE conjugation assays using the indicated proteins and E. coli lipids derived liposomes. Atg8–PE conjugation was detected as characteristic band shift as the conjugated form runs faster on urea-SDS gels. The numbers above the gels indicate the time in minutes. Note that at least under these conditions almost no Atg8–PE conjugation was detected in the absence of the Atg5–Atg12 conjugate or the Atg5–Atg12/Atg16 complex. The numbers next to the gels indicate the molecular weight in kDa. The averages and the error bars representing the standard deviations are shown. P-values were calculated using Student’s t-test. NS, non-significant; P, pellet; S, supernatant. Figure source data can be found with the Supplementary data.
Promotion of Atg8–PE conjugation in the small unilamellar vesicle assay requires Atg12 but not Atg16
The fact that the Atg5–Atg12/Atg16 complex more efficiently recruited Atg3 to the membrane compared to the Atg5–Atg12 conjugate is consistent with the requirement for Atg16 for efficient Atg8–PE generation in vivo (Suzuki et al, 2001; Matsushita et al, 2007). We therefore tested promotion of Atg8–PE conjugation in an in vitro assay in the presence of Atg5–Atg12/Atg16, Atg5–Atg12 and Atg5/Atg16 using small unilamellar vesicles (SUVs; Hanada et al, 2007). We could reproduce substantial stimulation of Atg8–PE conjugation by both the Atg5–Atg12/Atg16 complex and the Atg5–Atg12 conjugate (Figure 3C) confirming previous results (Hanada et al, 2007). Conjugation in the absence of the Atg5–Atg12/Atg16 or the Atg5–Atg12 conjugate was so inefficient that it was undetectable under these conditions (Figure 3C). In stark contrast, Atg8–PE conjugation was almost complete after 5 min in the presence of Atg5–Atg12/Atg16 or Atg5–Atg12 and even occurred during initial mixing of the components, hence the weak band at the 0-min time point. Promotion of Atg8–PE conjugation was largely Atg12 dependent as the Atg5/Atg16 complex almost entirely lacked its promoting activity. Interestingly, promotion of Atg8–PE conjugation was not completely abolished by the absence of Atg12 as a weak Atg8–PE band was detectable after 30 min of incubation with Atg5/Atg16 (Figure 3C). The molecular basis for this is currently unknown. In conclusion, promotion of Atg8–PE conjugation is largely Atg16 dependent in vivo (Suzuki et al, 2001; Matsushita et al, 2007) but not in the in vitro assay employing SUVs (Figure 3; Hanada et al, 2007).
Promotion of Atg8–PE conjugation is Atg16 dependent in a GUV assay
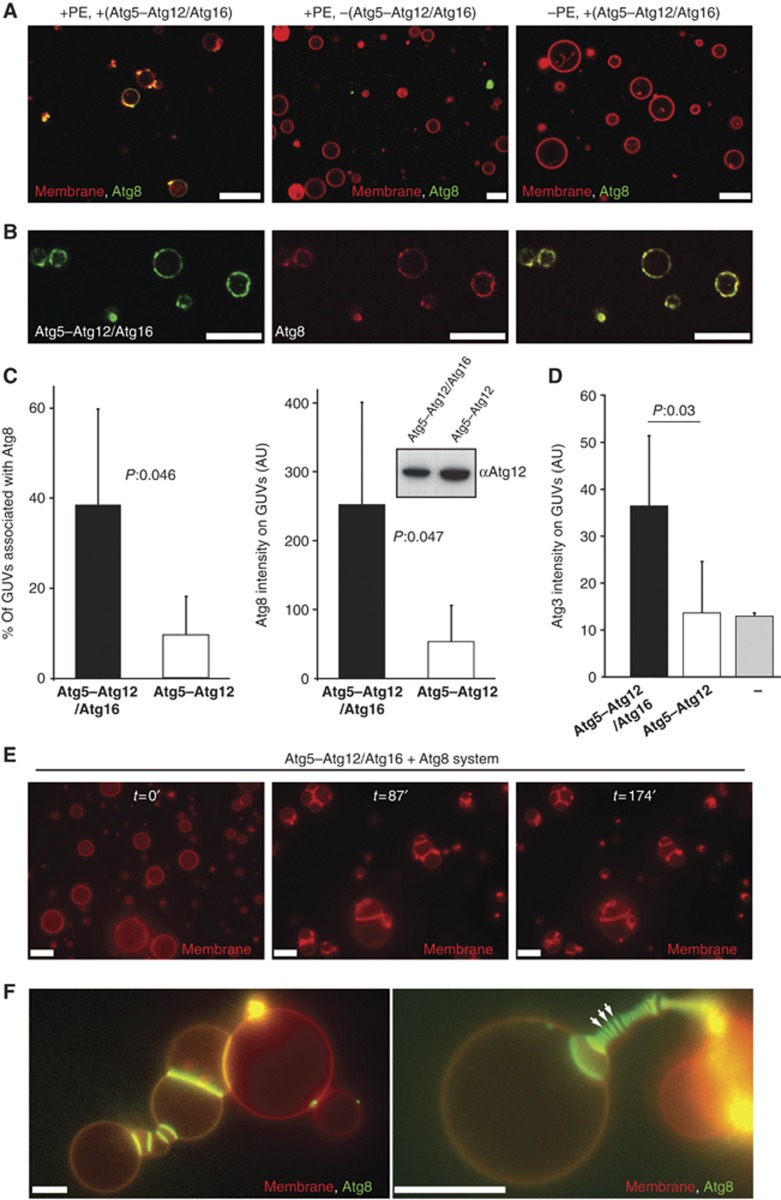
The in vitro assay using SUVs does thus not recapitulate the requirement of Atg16 for efficient Atg8–PE conjugation in vivo (Suzuki et al, 2001; Matsushita et al, 2007). The reason for this may be the high concentration of SUVs in the assay and thus a high probability of collision between the vesicle and Atg3–Atg8 and/or the high curvature of the SUVs. We therefore developed an assay employing GUVs, which are visible by light microscopy (Figure 4). In this assay, membranes are far less abundant and less curved. In order to detect Atg8 in the GUV assay, we purified eGFP–Atg8 and mCherry–Atg8 fusion proteins from E. coli. Both eGFP–Atg8 and mCherry–Atg8 were efficiently conjugated to PE in the in vitro assay using SUVs (Supplementary Figure 1). In the presence of Atg3, Atg7 and the Atg5–Atg12/Atg16 complex, eGFP–Atg8 became localized to the GUVs (Figure 4A). We interpret this localization to GUVs as conjugation to PE. Atg8–PE conjugation was strictly dependent on the presence of the Atg5–Atg12/Atg16 complex and PE in the GUV membrane (Figure 4A). In order to detect the Atg5–Atg12/Atg16 complex on the GUV membrane, we expressed and purified Atg16 C-terminally fused to eGFP and used this fusion protein for complex formation with the Atg5–Atg12 conjugate. The Atg5–Atg12/Atg16–eGFP complex bound efficiently to membranes (Supplementary Figure 1). Moreover, the Atg5–Atg12/Atg16–eGFP complex colocalized with mCherry–Atg8 on the same GUVs (Figure 4B). Having established that the GUV assay reproduces these crucial in vivo properties of the system we asked if complex formation of the Atg5–Atg12 conjugate with Atg16 was required for Atg8–PE conjugation. Interestingly, in the GUV assay Atg5–Atg12/Atg16 was much more efficient in promoting Atg8–PE conjugation than the Atg5–Atg12 conjugate (Figure 4C). Both the number of Atg8-associated GUVs and the intensity of eGFP–Atg8 fluorescence on the GUVs were considerably higher in the presence of the Atg5–Atg12/Atg16 complex. This correlated with the ability of the Atg5–Atg12/Atg16 complex to recruit Atg3 to the surface of GUVs (Figure 4D). In contrast, Atg5–Atg12 did not promote Atg3 recruitment to the GUV membrane (Figure 4D). Thus, membrane localization of the Atg5–Atg12 conjugate by complex formation with Atg16 becomes crucial for membrane recruitment of Atg8 if the membranes are limiting and/or less highly curved, offering an explanation for the strict dependence of Atg8–PE conjugation on Atg16 in vivo (Suzuki et al, 2001; Fujita et al, 2008b). We did not observe any membrane recruitment of Atg8 in the presence of Atg5 and the Atg5/Atg16 complex.
Figure 4.
Reconstitution of Atg5–Atg12/Atg16 and Atg8 on GUVs. (A) Confocal images showing eGFP–Atg8 recruitment to GUVs is dependent on the Atg5–Atg12/Atg16 complex and PE. The membrane was labelled by incorporation of rhodamine-PE. (B) Single confocal section showing colocalization of Atg5–Atg12/Atg16–eGFP and mCherry–Atg8 on GUVs. (C) Quantification of GUV-associated eGFP–Atg8 in the presence of the Atg5–Atg12/Atg16 complex and the Atg5–Atg12 conjugate. The first graph shows the percentage of GUVs associated with eGFP–Atg8 fluorescence. The second graph shows the intensity of eGFP–Atg8 on the GUVs. The quantification is based on four independent experiments. Inset: anti-Atg12 western blot using the rabbit anti-Atg12 antiserum showing the amounts of Atg5–Atg12/Atg16 complex and Atg5–Atg12 conjugate in one of the assays used for the quantification shown in (C). (D) Quantification of membrane recruitment of Atg3–mCherry by the Atg5–Atg12/Atg16 complex, the Atg5–Atg12 conjugate and a buffer control. The graph is based on two independent experiments. (E) Widefield images showing individual frames of a time series of GUVs incubated with Atg3, Atg7, Atg8 and the Atg5–Atg12/Atg16 complex (see Materials and methods for details). The membrane was labelled by incorporation of rhodamine-PE. The time is indicated in minutes. (F) Widefield images of GUVs incubated with Atg3, Atg7, eGFP–Atg8 and the Atg5–Atg12/Atg16 complex for 35 min (left) or 70 min (right). The membrane was labelled by incorporation of rhodamine-PE. Note the massive accumulation of eGFP–Atg8 at the interface between individual GUVs. The arrows point to some of these interfaces. For the quantifications, the averages and standard deviations are shown. P-values were calculated using Student’s t-test. Scale bars: 10 μm. Figure source data can be found with the Supplementary data.
PE-conjugated yeast Atg8 has been reported to mediate membrane tethering and hemifusion of vesicles containing high amounts of PE (Nakatogawa et al, 2007). Similarly, membrane tethering and fusion activity have also been demonstrated for the mammalian Atg8-like proteins LC3 and GATE-16 (Weidberg et al, 2011). However, the hemifusion activity of Atg8 at physiological PE concentrations has recently been questioned by another study (Nair et al, 2011). The mammalian Atg8 homologues LC3 and GATE-16 have been shown to mediate membrane fusion when anchored to the membrane by lipidation (Weidberg et al, 2011). In our GUV assay, we found that in the presence of Atg5–Atg12/Atg16 and the Atg8 conjugation system composed of Atg3, Atg7 and Atg8, GUVs underwent extensive clustering (Figure 4E). Atg8 massively accumulated at sites of GUV–GUV contact (Figure 4F). This suggests that the contact sites are not hemifusion diaphragms, since steric constraints would not allow Atg8 to accumulate there (Chernomordik and Kozlov, 2005). We did not find any evidence for massive fusion of GUVs although we cannot exclude that some fusion occurred in our system.
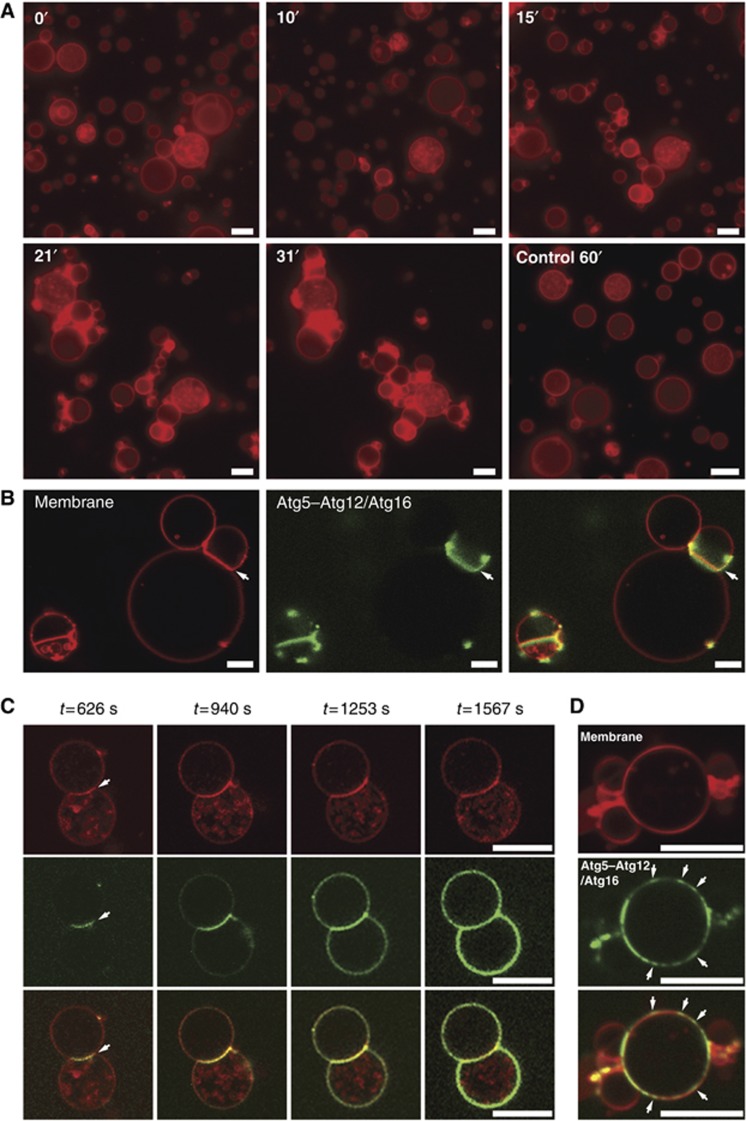
The Atg5–Atg12/Atg16 complex is sufficient for GUV tethering
In the course of these experiments, we also studied the effect of Atg5–Atg12/Atg16 on the GUVs. Strikingly, we noticed that the addition of Atg5–Atg12/Atg16 resulted in massive clustering of GUVs even in the absence of the Atg8 conjugation system (Figure 5A). When we imaged the Atg5–Atg12/Atg16–eGFP complex on the GUV membranes we noticed that it initially bound at the interface between clustered GUVs (Figure 5B). From the GUV–GUV interfaces the Atg5–Atg12/Atg16–eGFP signal grew eventually covering the complete GUV (Figure 5C). We found the same behaviour when we imaged mCherry and monomeric eGFP-tagged versions of the Atg5–Atg12/Atg16 complex. In addition, we noticed apparent cooperativity with respect to GUV binding by the complex since the Atg5–Atg12/Atg16 complex bound some GUVs a few minutes after addition of the protein. The complex quickly covered these GUVs whereas others were bound at a much later time point. In addition we frequently observed that the Atg5–Atg12/Atg16 complex did not homogenously cover the GUV membrane but was concentrated in small mobile assemblies (Figure 5D; Supplementary Movie 1). We also noted that the Atg5–Atg12/Atg16 complex bound much faster to small vesicles that were smaller than the resolution limit than to the large GUVs.
Figure 5.
Membrane tethering by the Atg5–Atg12/Atg16 complex. (A) Frames of widefield images taken from a time series of rhodamine-PE labelled GUVs in the presence of the Atg5–Atg12/Atg16 complex. For the control condition buffer but no protein was added. Note the increase in clustered GUVs over time. The numbers indicate the time in minutes. (B) Confocal images of rhodamine-PE labelled GUVs incubated with Atg5–Atg12/Atg16–eGFP complex. Note the accumulation of the complex at the interface between GUVs (arrows). (C) Individual confocal frames of a time series of rhodamine-PE labelled GUVs incubated with Atg5–Atg12/Atg16–eGFP. The arrows point to the GUV–GUV interface where the protein initially accumulated. (D) Confocal images of rhodamine-PE labelled GUVs incubated with Atg5–Atg12/Atg16–eGFP. The arrows point to the concentrations of Atg5–Atg12/Atg16 on the GUV membrane. Scale bars: 10 μm (A–C) and 3 μm (D).
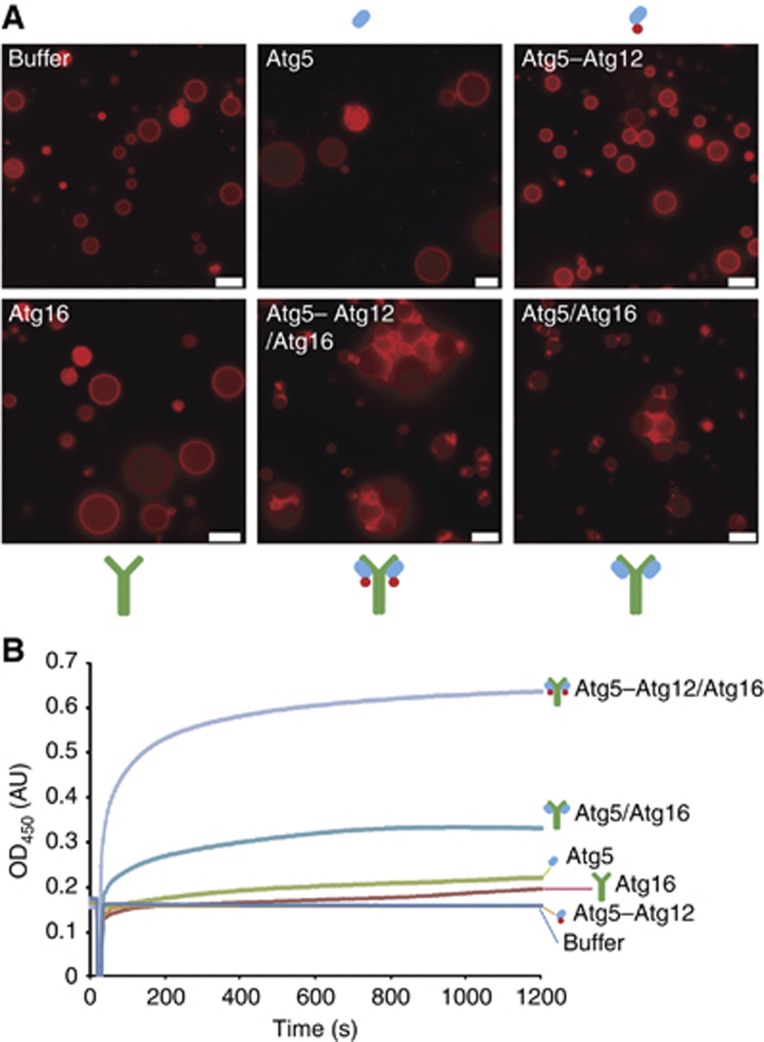
Subunit requirement for GUV tethering by the Atg5–Atg12/Atg16 complex
Next, we asked what the molecular basis for membrane clustering by the Atg5–Atg12/Atg16 could be. We therefore imaged GUVs in the presence of Atg5, Atg16, the Atg5–Atg12 conjugate, the Atg5–Atg12/Atg16 complex or the Atg5/Atg16 complex (Figure 6A). While the Atg5–Atg12/Atg16 and Atg5/Atg16 complexes efficiently clustered GUVs Atg5, Atg5–Atg12 and Atg16 did not induce any detectable clustering. To corroborate these findings, we determined the ability of the above proteins and complexes to cluster SUVs using a turbidity assay (Figure 6B). The results we obtained with the turbidity assay correlated well with our results we obtained using the GUV system. Thus while Atg5, Atg16 and Atg5–Atg12 failed to cluster SUVs the Atg5–Atg12/Atg16 complex efficiently clustered SUVs. Furthermore, also the Atg5/Atg16 complex clustered SUVs. Interestingly, its clustering activity was markedly reduced compared to the Atg5–Atg12/Atg16 complex, suggesting that Atg12 enhances the clustering activity of the complex. Atg16 has been reported to form multimeric complexes with Atg5–Atg12 (Kuma et al, 2002) and thus Atg16-dependent dimerization or multimerization of the membrane binding site provided by Atg5 may be required for membrane clustering.
Figure 6.
Membrane tethering is Atg16 dependent. (A) Widefield images of rhodamine-PE labelled GUVs 30 min after incubation with the indicated proteins. Note that GUVs cluster only in the presence of the Atg5–Atg12/Atg16 and Atg5/Atg16 complexes. (B) SUV aggregation assay using Folch liposomes and the indicated proteins. The proteins were added after a short delay. The symbols above, below the images or next to the curves represent Atg5 (blue oval), Atg12 (red circle) and Atg16 (green Y). The overall shape of the Atg5–Atg12/Atg16 complex is according to the structural model of the complex as proposed in Fujioka et al (2010). Scale bars: 10 μm.
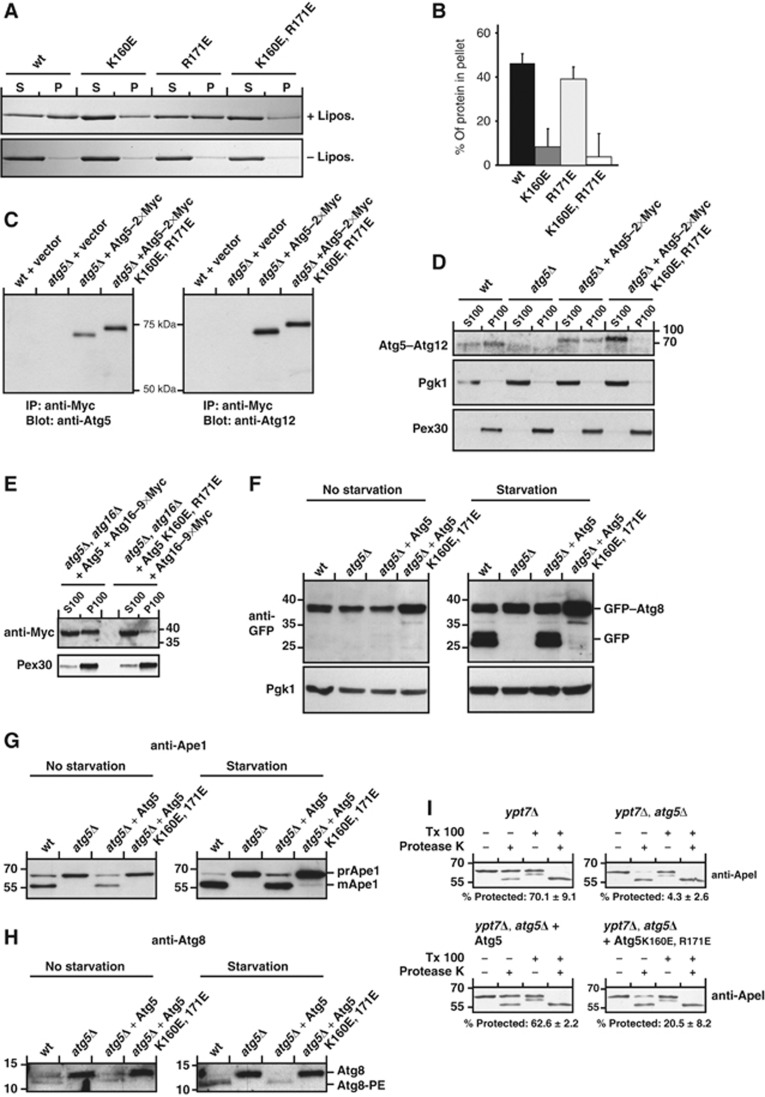
Identification of the membrane-binding site in Atg5
In order to be able to test if Atg5-mediated membrane binding by the Atg5–Atg12/Atg16 complex is required for autophagosome formation in vivo, we set out to identify the membrane-binding site in Atg5. Since Atg12 inhibits membrane binding by Atg5 (Figure 1) we hypothesized that the membrane-binding site is located close to K149, which is conjugated to Atg12. In addition, membrane binding by Atg5 is dependent on negatively charged lipids (Figure 2). We therefore mutated positively charged residues in vicinity of K149 to glutamate (E) (Supplementary Figure 2). Atg5 alone showed considerable background pelleting in the absence of liposomes in our co-sedimentation assays (Figure 1). In order to accurately test individual mutants for loss of membrane binding, we therefore tested the Atg5 mutants in complex with an N-terminal region of Atg16 (amino acids 1–46) (Figure 7A and B), which has a stabilizing effect (Matsushita et al, 2007). We also tested the Atg5 mutants in complex with full-length Atg16 (Supplementary Figure 2). Two mutations, K160E and R171E, reduced membrane binding (Figure 7A and B; Supplementary Figure 2). Mutation of K160 to E had a larger effect than the R171E mutation. When the two mutations were co-introduced into Atg5 membrane binding was severely reduced (Figure 7A and B; Supplementary Figure 2). The Atg5 K160E, R171E double mutant was still efficiently conjugated to Atg12 in vitro (Supplementary Figure 3) and bound to Atg16. Also in vivo the Atg5 K160E, R171E double mutant was expressed at levels comparable to the wild-type protein and was efficiently conjugated to Atg12 (Figure 7C; Supplementary Figure 3). The addition of Myc tags to the C-terminus of Atg5 did not interfere with its ability to restore Ape1 processing in Atg5-deficient yeast cells (Supplementary Figure 3).
Figure 7.
Membrane binding by Atg5 is essential for autophagy and the Cvt pathway in vivo. (A) Liposome co-sedimentation assay using Folch liposomes and the indicated Atg5 proteins stabilized with an N-terminal fragment of Atg16 (amino acids 1–46). (B) Quantification based on three co-sedimentation assays as shown in (A). The amount of protein pelleted in the absence of liposomes was subtracted. The averages and the standard deviations are shown. (C) Yeast cells were transformed with the indicated expression constructs and Atg5–2 × Myc was immunoprecipitated with an anti-Myc antibody. The precipitated protein was subjected to anti-Atg5 and anti-Atg12 western blotting. The substitution of R171 and K160 to glutamate caused the mutant protein to run higher. The same phenomenon was observed for the recombinant protein (Supplementary Figure 3). (D) Anti-Atg12 western blot of cell fractions prepared from rapamycin-treated yeast cells. The anti-Pgk1 and anti-Pex30 served as control for the cytosolic and membrane fractions, respectively. (E) Anti-Myc and anti-Pex30 western blots of yeast cell fractions from cells of the indicated genotype and expressing Atg16–9 × Myc and Atg5 or Atg5 K160E, R171E. (F) Starvation assay with the indicated yeast strains and the indicated rescue constructs. GFP was detected by western blotting. Pgk1 served as loading control. (G) Same assay as shown in (F) but using an anti-Ape1 antiserum for western blotting. (H) Western blots of yeast cell lysates using an anti-Atg8 antiserum. (I) Ape1 protease protection assay with the indicated yeast strains. The percentages below the blots indicate the percent of protected ApeI in the +protease K, −Triton X-100 lanes and are the average of three independent experiments±the standard deviation. The numbers next to the blots indicate the molecular weight in kDa. Figure source data can be found with the Supplementary data.
Membrane binding by Atg5 is required for autophagy and the Cvt pathway
In order to test if the Atg5 K160E, R171E mutant also showed reduced membrane binding in vivo we fractionated membranes from rapamycin-treated yeast cells by separating the cytosol (S100) from the membrane fraction (P100) by ultracentrifugation. While the endogenous and the introduced wild-type Atg5–Atg12 conjugates were present in the membrane fractions the Atg5 K160E, R171E mutant showed a severely reduced presence in the pellet fraction (Figure 7D; Supplementary Figure 3). In addition, also when we expressed 9 × Myc-tagged versions of wild type and K160E, R171E Atg5 we found reduced membrane binding by the mutant conjugate (Supplementary Figure 3). Consistent with our results that Atg5 is required for the recruitment of Atg16 to the membrane (Figure 1), the presence of Atg16 in the membrane fraction was severely reduced in cells expressing the Atg5 K160E, R171E mutant compared to the wild-type Atg5 protein (Figure 7E). To analyse if membrane binding by Atg5 is required for autophagy and the Cvt pathway in vivo, we expressed Atg5 K160E, R171E in Atg5-deficient S. cerevisiae and monitored autophagy by the GFP–Atg8 degradation assay (Figure 7F; Klionsky et al, 2007). While a plasmid encoding wild-type Atg5 efficiently restored the lack of GFP–Atg8 degradation in Atg5-deficient cells the Atg5 K160E, R171E mutant completely failed to restore GFP–Atg8 degradation (Figure 7F). Furthermore, the GFP signal accumulated in the vacuole in yeast cells expressing wild-type Atg5 while it stayed in the cytoplasm in cells expressing Atg5 K160E, R171E (Supplementary Figure 4), suggesting that membrane binding by Atg5 is essential for autophagy.
To test if the Cvt pathway was still active in cells expressing the Atg5 K160E, R171E mutant, we monitored Ape1 processing in these cells (Figure 7G). Ape1 is synthesized in the cytosol with an N-terminal propeptide (prApe1) and is transported into the vacuole by its inclusion into double membrane-bound vesicles called Cvt vesicles. Within the vacuole the propeptide of Ape1 is removed. The formation of Cvt vesicles is dependent on the autophagic core machinery including Atg5 (Harding et al, 1995; Mizushima et al, 1998). In the presence of nutrients, about 50–75% of the Ape1 protein is localized in the vacuole and thus lacks the N-terminal propeptide. During starvation, Ape1 is delivered to the vacuole by autophagy and is almost exclusively localized within the vacuole. The Atg5 K160E, R171E mutant almost completely failed to restore the Ape1 processing defect in Atg5-deficient yeast both in the presence of nutrients and during starvation (Figure 7G). Autophagy and the Cvt pathway appear to be blocked at a relatively early step since Atg8 conjugation to PE was severely reduced in the Atg5 K160E, R171E expressing cells (Figure 7H). To corroborate this result, we conducted Ape1 protection assays. Consistent with a requirement for membrane binding by Atg5 before autophagosome completion, Ape1 was substantially less protected from protease K digestion in cells expressing the Atg5 K160E, R171E mutant compared to wild-type Atg5 (Figure 7I).
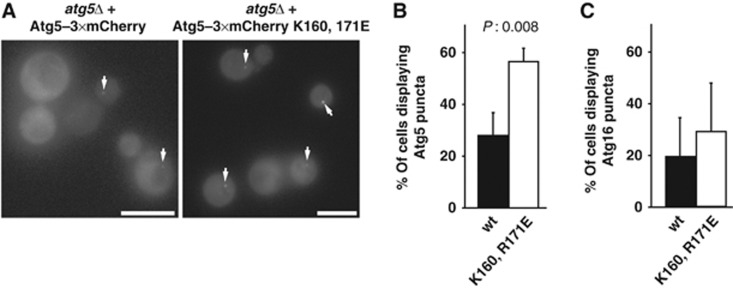
Interestingly, despite the loss of the Atg5 K160E, R171E mutant from the membrane fraction (Figure 7D; Supplementary Figure 3) the mutant protein was still localized to the PAS (Figure 8A and B; Supplementary Figure 5). Compared to the wild-type Atg5–3 × mCherry protein almost twice as many cells displayed a detectable dot-like localization in cells treated with rapamycin (Figure 8B). A similar though statistically not significant tendency was also observed for Atg16–3 × mCherry in cells expressing wild-type Atg5 and the Atg5 K160E, R171E mutant, respectively (Figure 8C; Supplementary Figure 5). We also noticed that the signal at the PAS tended to be more intense for the Atg5 K160E, R171E mutant. The Atg5 K160E, R171E-positive dots colocalized with ApeI-GFP, suggesting that the Atg5 K160E, R171E-positive dots represent the PAS (Supplementary Figure 5). These results suggest that the initial recruitment of the Atg5–Atg12/Atg16 complex occurs independent of Atg5 membrane binding, most probably via another protein.
Figure 8.
Localization of wild-type Atg5 and Atg5 K160E, R171E to the PAS. (A) Yeast cells of the indicated genotype expressing Atg5–3 × mCherry or Atg5 K160E, R171E–3 × mCherry were treated with rapamycin and imaged using a Deltavision microscope. Maximum projections of z-stacks are shown. (B) Graph showing the percentage of mCherry-positive cells that display Atg5–3 × mCherry dots. In total, 3 independent experiments were conducted and >400 cells were counted. (C) Graph showing the percentage of cells that display Atg16–3 × mCherry dots. In total, 3 independent experiments were conducted and >750 cells were counted. The graphs show the averages and the error bars representing the standard deviations. P-values were calculated using Student’s t-test. Scale bars: 5 μm.
Discussion
The mechanisms underlying autophagosome formation are only poorly understood even though >30 Atg genes have been reported to play key roles at various stages during autophagy (Nakatogawa et al, 2009). In particular, two ubiquitin-like conjugation systems, based around the ubiquitin-like proteins Atg8 and Atg12, have central roles during the formation of isolation membranes and autophagosomes (Mizushima et al, 1998; Ichimura et al, 2000). To date, eight Atg proteins have been reported to belong to these two ubiquitin-like conjugation systems (Geng and Klionsky, 2008). Despite their apparent importance during autophagosome formation, their mechanism of action is only incompletely understood. This is mainly because little is known about their biochemical activities. This is particularly true for the Atg12 conjugation system. The Atg5–Atg12/Atg16 complex has been observed to localize to isolation membranes (Suzuki and Ohsumi, 2010) and when individual components of this complex are deleted autophagosomes fail to form (Mizushima et al, 1998, 1999, 2001; Hanada et al, 2007; Suzuki and Ohsumi, 2010). The Atg5–Atg12 conjugate and the Atg5–Atg12/Atg16 complex have been reported to facilitate the conjugation of Atg8 to PE (Hanada et al, 2007). This, however, may not be the only activity the Atg5–Atg12/Atg16 complex possesses, since autophagosome formation is more severely impaired when the Atg5–Atg12/Atg16 complex is removed compared to inactivation of the Atg8 conjugation system (Abeliovich et al, 1999, 2000; Mizushima et al, 2001; Fujita et al, 2008a).
Direct membrane binding by Atg5
To unravel the biochemical properties of the Atg5–Atg12/Atg16 complex, we studied the complex in reconstituted systems using recombinant proteins. We found that Atg5 alone is capable of binding to membranes. Atg12 conjugation largely inhibits membrane binding by Atg5. Since the Atg5–Atg12 conjugation is covalent and constitutive (Mizushima et al, 1998; Kuma et al, 2002), this inhibition could contribute to the regulation of membrane binding by Atg5 in vivo. Upon binding of the Atg5–Atg12 conjugate to Atg16, Atg5 regains its ability to associate with membranes. We can currently not rule out that Atg12 also contributes to membrane binding within the Atg5–Atg12/Atg16 complex since we were not successful in purifying soluble Atg12 for membrane binding analyses. However, the fact that the Atg5 K160E, R171E mutant showed severely reduced membrane binding in our fractionation experiment suggests that the contribution of Atg12 to membrane binding in vivo is minor. Association with Atg16 may trigger membrane binding by the Atg5–Atg12 conjugate in vivo and thus allows it to promote the subsequent stages of autophagosome formation. The fact that the Atg5–Atg12 conjugate and Atg16 were found in a complex even in the absence of any pro-autophagic stimulus suggests that an additional level of regulation may be involved (Kuma et al, 2002) possibly mediated by another PAS localized factor (Figure 8). Interestingly, we found some preference of the Atg5–Atg12/Atg16 complex for PI3P-containing membranes. PI3-Kinase activity is essential for autophagosome formation and thus local production of PI3P on autophagosome precursors may contribute to membrane recruitment of the Atg5–Atg12/Atg16 complex (Kihara et al, 2001). Membrane binding by Atg5 is essential for its function in vivo because the Atg5 K160E, R171E mutant that showed severely reduced membrane binding in vitro was unable to support autophagy and the Cvt pathway.
Interestingly, we found that loss of membrane binding by Atg5 did not interfere with its localization to the PAS (Figures 7 and 8). In fact, the Atg5 K160E, R171E mutant displayed an even enhanced PAS localization. Thus, an upstream factor that still remains to be identified may be required for the initial recruitment of the Atg5–Atg12/Atg16 complex to the PAS while membrane binding by Atg5 is a downstream requirement for the promotion of the subsequent steps of autophagosome formation. The enhanced localization of the Atg5 K160E, R171E mutant to the PAS may be explained by the arrest of autophagosome formation downstream of its recruitment, whereas the wild-type protein may show fluctuating fluorescence at the PAS reflecting different stages of autophagosome biogenesis. Such fluctuations have been seen for Atg8 (Geng et al, 2008). The factor that recruits the Atg5–Atg12/Atg16 complex to the PAS remains to be identified but the recruitment of components of the Atg5–Atg12/Atg16 complex by another protein has an interesting precedence in mammalian cells where TECPR1 has been demonstrated to recruit the Atg5–Atg12 conjugate to autolysosomes (Chen et al, 2012).
Atg16 is required for efficient promotion of Atg8–PE conjugation under physiological conditions
The Atg5–Atg12 conjugate has been reported to dramatically facilitate Atg8 conjugation to PE in an in vitro assay using SUVs (Hanada et al, 2007). Complex formation of the Atg5–Atg12 conjugate with Atg16 did not further enhance promotion of Atg8–PE generation in this assay (Figure 3; Hanada et al, 2007) despite the fact that Atg16 is essential for Atg8–PE conjugation in vivo (Suzuki et al, 2001). Since the Atg5–Atg12/Atg16 complex showed much stronger membrane binding than the Atg5–Atg12 conjugate we analysed membrane recruitment of Atg3 by the two proteins. Indeed, the Atg5–Atg12/Atg16 complex was more efficient in recruiting Atg3 to membranes than the Atg5–Atg12 conjugate. Atg3 recruitment to membranes was dependent on Atg12. It was therefore initially surprising to find that the Atg5–Atg12 conjugate was equally efficient in promoting Atg8–PE conjugation. However, the in vitro assay employing SUVs contains a high concentration of highly mobile vesicles facilitating the collision of Atg3–Atg8 with membranes. Thus, membrane recruitment of Atg8-loaded Atg3 might not be rate limiting in this system. In addition, SUVs are highly curved which might additionally facilitate the conjugation reaction. The fact that the conjugate still promoted Atg8–PE conjugation suggests either that the weak membrane binding activity of the Atg5–Atg12 conjugate is sufficient in this system or that Atg5–Atg12 binding to Atg3 has a stimulatory effect independent of membrane recruitment.
We therefore tested membrane recruitment of Atg8 and by implication promotion of Atg8–PE conjugation in a reconstituted assay employing GUVs. In this system, the membranes are far less abundant, less mobile and have a much lower curvature. In the GUV system, the Atg5–Atg12/Atg16 complex was much more efficient in promoting Atg8 recruitment to the membrane than the Atg5–Atg12 conjugate offering an explanation for why Atg16 is essential for Atg8–PE generation in vivo (Suzuki et al, 2001). Thus, we are able to assign functions to all components of the Atg5–Atg12/Atg16 complex during Atg8–PE conjugation: Atg5 binds to membranes, Atg16 triggers membrane binding by Atg5 due to the release of its inhibition of membrane binding by Atg12 and Atg12 recruits Atg3 to the membrane once localized there by Atg5 within the Atg5–Atg12/Atg16 complex.
When incubated with the Atg5–Atg12/Atg16 complex and the Atg8 conjugation system (Atg3, Atg7, Atg8, MgCl2 and ATP) we observed massive clustering of GUVs. The interfaces between the GUVs sometimes extended so extensively that the individual GUVs became massively deformed and flattened. Since we have not directly measured fusion in our system we cannot rule out that fusion occurred. However, consistent with a recent report employing SUVs in a bulk assay (Nair et al, 2011) we did not detect any evidence for massive fusion in our GUV system.
Homotypic tethering of membranes depends on Atg16
During the course of our experiments designed to analyse Atg8 recruitment to GUVs we observed that the Atg5–Atg12/Atg16 complex itself was sufficient to induce clustering of GUVs. Clustering of GUVs was not absolutely dependent on the presence of Atg12 as the Atg5/Atg16 complex was sufficient to induce GUV aggregation, however, to a lesser extent. In contrast, Atg5 alone did not induce clustering. We obtained very similar results with an aggregation assay employing SUVs. In this assay, it was apparent that the absence of Atg12 reduced the clustering activity of the complex. This suggests that Atg12 might weakly but directly contribute to membrane binding, activate membrane binding by the Atg5–Atg12/Atg16 complex in an indirect manner or promote some higher order assemblies of the complex that in turn may be more efficient in clustering vesicles.
The C-terminal domain of Atg16 consists of a coiled-coil region and crystallized as a homodimer (Fujioka et al, 2010). Since Atg5 directly binds to the N-terminus of Atg16 (Matsushita et al, 2007) it is therefore likely that at least two copies of Atg5 are present in the Atg5–Atg12/Atg16 complex (Mizushima et al, 1999; Kuma et al, 2002). The most likely explanation for the clustering of GUVs in our system is thus that one Atg5 molecule binds to one membrane and the other Atg5 molecule to the other. Interestingly, the coiled-coil domain of Atg16 is essential for autophagosome formation (Fujioka et al, 2010) and one of its functions may be the dimerization of the Atg5-localized membrane binding site. Consistently, dimerization or oligomerization of Atg16 is required for autophagy in vivo (Kuma et al, 2002). The interface between the two membranes appears to be a preferred localization of the Atg5–Atg12/Atg16 complex as we often observed massive accumulation of the Atg5–Atg12/Atg16 complex in the interface between GUVs before the rest of the GUV was covered. We frequently noticed that the Atg5–Atg12/Atg16 signal grew from the interface between two GUVs around the rest of the GUV. In addition, the Atg5–Atg12/Atg16 complex did often form highly mobile foci. We also noticed that the Atg5–Atg12/Atg16 complex bound strongly to individual GUVs rather than exhibiting a uniform level of binding across the whole GUV population. This observation suggests some sort of cooperativity with respect to membrane binding and would be consistent with oligomerization of the Atg5–Atg12/Atg16 complex on the membrane, at least in vitro.
The fact that membrane tethering of the Atg5–Atg12/Atg16 complex is independent of the Atg8 conjugation system might explain why inactivation of the Atg5–Atg12/Atg16 complex affects autophagosome formation more severely than inactivation of the Atg8 conjugation system (Abeliovich et al, 1999, 2000; Mizushima et al, 2001; Fujita et al, 2008a) although we have so far no definitive evidence for this clustering activity in vivo. It has recently been shown that an Atg9-containing tubulovesicular compartment relocalizes to a site close to the vacuole and likely represents the PAS (Mari et al, 2010). Also, individual Atg9-positive vesicles have been seen to coalesce at the PAS (Yamamoto et al, 2012). Furthermore, SNAREs are required for the formation of Atg9-positive tubulovesicular isolation membrane precursors and thus isolation membranes in yeast (Nair et al, 2011). In mammalian cells, SNARE-dependent fusion of Atg16-positive vesicular precursors has been reported (Moreau et al, 2011). The Atg5–Atg12/Atg16 complex may thus contribute to the homotypic tethering of these precursors in order to allow SNARE-depended fusion to occur.
Materials and methods
Antibodies
The rabbit anti-Atg12 antiserum (NB600-603) was from Novus Biologicals, Cambridge, UK. Monoclonal anti-Atg5 (8B12-3B11) and anti-Atg12 (7B11-1F9) antibodies were raised in mouse against the recombinantly expressed and purified S. cerevisiae Atg5–Atg12 conjugate and were selected for specific recognition of the conjugate as well as the respective individual proteins. The rabbit polyclonal anti-Ape1 and the rabbit polyclonal anti-Atg8 antibodies were a kind gift from Claudine Kraft. Anti-Pex30 was a kind gift of Cecile Brocard. Anti-GFP antibodies were purchased from Roche. Anti-Pgk1 was purchased from Invitrogen and anti Penta-His from Qiagen. The anti-Myc tag antibody (clone 4A6) is available from Millipore.
Protein expression and purification
All proteins were expressed in E. coli Rosetta pLySS cells. Atg3 was expressed as N-terminal GST fusion protein from pGEX-4T1. Atg7 was expressed with an N-terminal His tag from pOPTHrsTEV. Atg8 lacking the C-terminal arginine was expressed from pOPC. GST-tagged Atg16 was expressed from pOPTG. Atg5 was expressed with an N-terminal His tag from pOPTH. eGFP–Atg8, monomeric-eGFP–Atg8 and mCherry–Atg8 (all lacking the C-terminal arginine) and monomeric-eGFP–Atg16 were expressed from pETDuet-1. The monomeric-eGFP versions were produced by inserting a A206K mutation into the open reading frame of eGFP derived from pEGFP-N1. The Atg5–Atg12 conjugate was purified by co-expression of His-tagged Atg5 and Atg12 from pETDuet-1 with Atg10 and Atg7 from pCOLADuet-1. The Atg5–Atg12/Atg16 complex was purified by co-expression of His-tagged Atg5 and Atg12 from pETDuet-1 with Atg10 and Atg7 from pCOLADuet-1 and Atg16 from pCDFDuet-1.
See Supplementary data for detailed protocols.
Liposome co-sedimentation assays
Lipids were mixed, dried under argon stream and desiccated. Buffer (150 mM NaCl, 50 mM HEPES pH 7.5, 1 mM DTT) was added to give a final concentration of 1 mg/ml lipids and sonicated. 5 μg protein was incubated with 0.5 mg/ml liposomes for 30 min at room temperature. The reactions were centrifuged at 180 000 g at 22°C, supernatants and pellets were separated and equal amounts were run on 12% SDS/polyacrylamide gels. For details, see Supplementary data.
In vitro Atg8–PE conjugation reactions using SUVs
E. coli lipids (total extracts, Avanti Inc. #100500) were used to prepare liposomes at a final concentration of 1 mg/ml in 50 mM HEPES pH 7.5, 150 mM NaCl. Liposomes were prepared as described for the liposome co-sedimentation assays. The final concentration of lipids in the assays was 0.8 mg/ml. Atg3, Atg7, Atg5–Atg12, Atg5/Atg16 and Atg5–Atg12/Atg16 were used at a final concentration of 1 μM whereas Atg8 was used at a final concentration of 5 μM. ATP was used at a final concentration of 2 mM and MgCl2 at a final concentration of 1 mM. The reactions were done at 30°C and stopped by the addition of SDS loading buffer. The reactions were run on 16% gels containing 6 M urea.
Preparation of GUVs
GUVs were prepared by electroformation. The electroformation was conducted either at 24°C or at 60°C dependent on the phase transition temperature of the lipids. The GUVs were diluted in 15 mM HEPES pH 7.5, 135 mM NaCl.
Proteins were used at the following final concentrations: Atg3 and Atg7: 80 nM, Atg8, eGFP–Atg8, monomeric-eGFP–Atg8, mCherry–Atg8, Atg5, Atg5–Atg12, Atg5/Atg16, Atg5/Atg16–monomeric-eGFP, Atg5–Atg12/Atg16-monomeric–eGFP, Atg5–Atg12/Atg16–eGFP: 400 nM. MgCl2 was added to a final concentration of 0.5 mM and ATP was added to a final concentration of 1 mM.
See Supplementary data for details.
Microscopy
Confocal images were acquired using a Zeiss LSM 510 or Visitron spinning disc microscope with × 63 (NA 1.4) or × 100 (NA 1.45) oil objectives. Widefield images were obtained on an Olympus CellˆR imaging station with × 40 or × 60 oil objectives (NA 1.4) or a Zeiss Observer Z1 inverted fluorescent microscope with a × 63 (NA 1.4) oil objective or a Deltavision using a × 100 (NA 1.4) oil objective. Images were processed and analysed using the ImageJ software.
Yeast strains and constructs
For yeast strains used in this study, please see Supplementary Table 1. Plasmids for yeast expression were produced by amplification of Atg5 with its endogenous promoter from genomic DNA and cloning to pRS316. Mutations were introduced by QuikChange mutagenesis (Stratagene). Atg5–Myc contained Myc tags fused to its C-terminus and was expressed from its endogenous promoter from pRS416. GFP–Atg8 was expressed from pRS315-GFP–Atg8 described in Kraft et al (2008). Atg5–3 × mCherry and Atg5 K160E, R171E–3 × mCherry were expressed from pRS316, under the control of the endogenous promoter and cyc1 terminator. Atg16–3 × mCherry was expressed from pRS315. Yeast strains were derivatives of the BY4741 background, kindly provided by Claudine Kraft.
Yeast manipulations
Standard methods were used for cell growth and manipulation. Yeast were grown in selective medium to log phase and subjected to nitrogen starvation for 5 h. Whole cell lysates were prepared by a trichloroacetic acid extraction. Proteins were separated by SDS/PAGE and subjected to western blotting. Alternatively, for the detection of Atg5 and Atg12, native cell lysates were prepared by glass bead disruption in lysis buffer supplemented with protease inhibitors. Atg5–2 × Myc was immunoprecipitated with an anti-Myc tag antibody (clone 4A6) covalently crosslinked to protein A sepharose beads (GE Healthcare). Immunoprecipitated proteins were analysed by 11% SDS–PAGE/western blot probed with the monoclonal Atg5 or Atg12 antibody. For detailed information of used strains and protocols, see Supplementary data.
Cell fractionation
Yeast cells were grown overnight in selection medium, resuspended in YPD, grown to log phase and treated with 220 nM rapamycin for 2 h. Cells were harvested and spheroplasts were prepared using zymolyase. The spheroplasts were resuspended in a hypotonic buffer. After non-lysed cells were removed by low speed centrifugation, the lysate was spun at a 100 000 g. Proteins in the supernatant were precipitated with TCA and dissolved in urea loading buffer (ULB). Equal amounts of the supernatants and pellets were run on 4–20% gradient or 12% SDS gels and subjected to western blotting using the anti-Atg12 (7B11-1F9), anti-Myc (4A6) antibody, anti-Pex30 or anti-Pgk1 antibody.
Proteinase protection assay.
Yeast cells starved for 4 h in SD-N were washed and incubated with 2-OD/ml DTT buffer (10 mM DTT, 10 mM Tris pH 9.4; 15 min, 30°C), resuspended in 10-OD/ml SP buffer (1 M sorbitol, 20 mM PIPES pH 6.8) and spheroplasted by Zymolyase-20T (MP Biomedicals) treatment. Spheroplasts were collected by centrifugation (2000 g for 5 min) and gently lysed with osmotic lysis buffer (200 mM sorbitol, 20 mM PIPES pH 6.8, 5 mM MgCl2) to preserve organelle-enclosed particles. After two 500 g centrifugation steps to remove unbroken cells, the resulting total lysate was further separated into a 5000 g supernatant and a pellet fraction. The 5000 g pellet was resuspended in osmotic lysis buffer with or without proteinase K (50 μg/ml, AppliChem) and 0.2% Triton X-100. After 30 min incubation on ice, samples were precipitated with TCA and analysed by western blotting with an anti-Ape1antiserum.
Statistical methods
The P-values were calculated using an unpaired two-tailed Student’s t-test. A P-value of <0.05 was considered as significant.
Accession numbers
Accession numbers are Atg3: NP_014404; Atg5: NP_015176.1; Atg7: NP_012041.1; Atg8: NP_009475.1; Atg10: NP_013058.1; Atg12: NP_009776.1; Atg16: NP_013882.1.
Supplementary Material
Acknowledgments
We are grateful to Cecile Brocard for providing the anti-Pex30 antiserum and Lars Demmel for technical help. We also thank Graham Warren, Justyna Sawa-Makarska and Brooke Morriswood for critically reading the manuscript. Funding by the University of Vienna is gratefully acknowledged. The research leading to these results has received funding from the European Research Council under the European Community’s Seventh Framework Programme (FP7/2007-2013)/ ERC grant agreement No. 260304. This work was also supported by a grant from the Austrian Science Foundation to E.O. (FWF_L_697-B20).
Author contributions: JR and MW contributed equally. SM conceived and supervised the study and wrote the manuscript. JR, MW, II, SS and SM conducted experiments. All authors contributed to data analysis, interpretation and commented on the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abeliovich H, Darsow T, Emr SD (1999) Cytoplasm to vacuole trafficking of aminopeptidase I requires a t-SNARE-Sec1p complex composed of Tlg2p and Vps45p. EMBO J 18: 6005–6016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeliovich H, Dunn WA Jr, Kim J, Klionsky DJ (2000) Dissection of autophagosome biogenesis into distinct nucleation and expansion steps. J Cell Biol 151: 1025–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT (2008) Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 182: 685–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Fan W, Lu Y, Ding X, Chen S, Zhong Q (2012) A mammalian autophagosome maturation mechanism mediated by TECPR1 and the Atg12-Atg5 conjugate. Mol Cell 45: 629–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernomordik LV, Kozlov MM (2005) Membrane hemifusion: crossing a chasm in two leaps. Cell 123: 375–382 [DOI] [PubMed] [Google Scholar]
- Fujioka Y, Noda NN, Nakatogawa H, Ohsumi Y, Inagaki F (2010) Dimeric coiled-coil structure of Saccharomyces cerevisiae Atg16 and its functional significance in autophagy. J Biol Chem 285: 1508–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Hayashi-Nishino M, Fukumoto H, Omori H, Yamamoto A, Noda T, Yoshimori T (2008a) An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol Biol Cell 19: 4651–4659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T (2008b) The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell 19: 2092–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng J, Baba M, Nair U, Klionsky DJ (2008) Quantitative analysis of autophagy-related protein stoichiometry by fluorescence microscopy. J Cell Biol 182: 129–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng J, Klionsky DJ (2008) The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. EMBO Rep 9: 859–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng J, Nair U, Yasumura-Yorimitsu K, Klionsky DJ (2010) Post-Golgi sec proteins are required for autophagy in Saccharomyces cerevisiae. Mol Biol Cell 21: 2257–2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George MD, Baba M, Scott SV, Mizushima N, Garrison BS, Ohsumi Y, Klionsky DJ (2000) Apg5p functions in the sequestration step in the cytoplasm-to-vacuole targeting and macroautophagy pathways. Mol Biol Cell 11: 969–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailey D, Rambold A, Satpute-Krishnan P, Mitra K, Sougrat R, Kim P, Lippincott-Schwartz J (2010) Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell 141: 656–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, Ohsumi Y (2007) The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem 282: 37298–37302 [DOI] [PubMed] [Google Scholar]
- Harding TM, Morano KA, Scott SV, Klionsky DJ (1995) Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J Cell Biol 131: 591–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A (2009) A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol 11: 1433–1437 [DOI] [PubMed] [Google Scholar]
- Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, Noda T, Ohsumi Y (2000) A ubiquitin-like system mediates protein lipidation. Nature 408: 488–492 [DOI] [PubMed] [Google Scholar]
- Kihara A, Noda T, Ishihara N, Ohsumi Y (2001) Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol 152: 519–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Cuervo AM, Seglen PO (2007) Methods for monitoring autophagy from yeast to human. Autophagy 3: 181–206 [DOI] [PubMed] [Google Scholar]
- Kraft C, Deplazes A, Sohrmann M, Peter M (2008) Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol 10: 602–610 [DOI] [PubMed] [Google Scholar]
- Kraft C, Martens S (2012) Mechanisms and regulation of autophagosome formation. Curr Opin Cell Biol 24: 496–501 [DOI] [PubMed] [Google Scholar]
- Kuma A, Mizushima N, Ishihara N, Ohsumi Y (2002) Formation of the ∼350-kDa Apg12-Apg5·Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. J Biol Chem 277: 18619–18625 [DOI] [PubMed] [Google Scholar]
- Mari M, Griffith J, Rieter E, Krishnappa L, Klionsky DJ, Reggiori F (2010) An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J Cell Biol 190: 1005–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita M, Suzuki NN, Obara K, Fujioka Y, Ohsumi Y, Inagaki F (2007) Structure of Atg5·Atg16, a complex essential for autophagy. J Biol Chem 282: 6763–6772 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ (2008) Autophagy fights disease through cellular self-digestion. Nature 451: 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Noda T, Ohsumi Y (1999) Apg16p is required for the function of the Apg12p-Apg5p conjugate in the yeast autophagy pathway. EMBO J 18: 3888–3896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y (1998) A protein conjugation system essential for autophagy. Nature 395: 395–398 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T (2001) Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol 152: 657–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau K, Ravikumar B, Renna M, Puri C, Rubinsztein David C (2011) Autophagosome precursor maturation requires homotypic fusion. Cell 146: 303–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair U, Jotwani A, Geng J, Gammoh N, Richerson D, Yen W-L, Griffith J, Nag S, Wang K, Moss T, Baba M, McNew James A, Jiang X, Reggiori F, Melia Thomas J, Klionsky Daniel J (2011) SNARE proteins are required for macroautophagy. Cell 146: 290–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H, Ichimura Y, Ohsumi Y (2007) Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130: 165–178 [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y (2009) Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol 10: 458–467 [DOI] [PubMed] [Google Scholar]
- Ohashi Y, Munro S (2010) Membrane delivery to the yeast autophagosome from the golgi-endosomal system. Mol Biol Cell 21: 3998–4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsi A, Polson HEJ, Tooze SA (2010) Membrane trafficking events that partake in autophagy. Curr Opin Cell Biol 22: 150–156 [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC (2010) Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol 12: 747–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y (2001) The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J 20: 5971–5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Ohsumi Y (2010) Current knowledge of the pre-autophagosomal structure (PAS). FEBS Lett 584: 1280–1286 [DOI] [PubMed] [Google Scholar]
- Tanida I, Tanida-Miyake E, Komatsu M, Ueno T, Kominami E (2002) Human Apg3p/Aut1p homologue is an authentic E2 enzyme for multiple substrates, GATE-16, GABARAP, and MAP-LC3, and facilitates the conjugation of hApg12p to hApg5p. J Biol Chem 277: 13739–13744 [DOI] [PubMed] [Google Scholar]
- Tooze SA, Yoshimori T (2010) The origin of the autophagosomal membrane. Nat Cell Biol 12: 831–835 [DOI] [PubMed] [Google Scholar]
- van der Vaart A, Griffith J, Reggiori F (2010) Exit from the Golgi is required for the expansion of the autophagosomal phagophore in yeast Saccharomyces cerevisiae. Mol Biol Cell 21: 2270–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg H, Shpilka T, Shvets E, Abada A, Shimron F, Elazar Z (2011) LC3 and GATE-16 N termini mediate membrane fusion processes required for autophagosome biogenesis. Dev Cell 20: 444–454 [DOI] [PubMed] [Google Scholar]
- Xie Z, Klionsky DJ (2007) Autophagosome formation: core machinery and adaptations. Nat Cell Biol 9: 1102–1109 [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Kakuta S, Watanabe TM, Kitamura A, Sekito T, Kondo-Kakuta C, Ichikawa R, Kinjo M, Ohsumi Y (2012) Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol 198: 219–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylä-Anttila P, Vihinen H, Jokitalo E, Eskelinen E-L (2009) 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy 5: 1180–1185 [DOI] [PubMed] [Google Scholar]
- Young AR, Chan EY, Hu XW, Kochl R, Crawshaw SG, High S, Hailey DW, Lippincott-Schwartz J, Tooze SA (2006) Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci 119: 3888–3900 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.