Abstract
STUDY QUESTION
Is there a contribution of the minor allele at the KRAS single nucleotide polymorphism (SNP) rs61764370 in the let-7 microRNA-binding site to endometriosis risk?
SUMMARY ANSWER
We found no evidence for association between endometriosis risk and rs61764370 or any other SNPs in KRAS.
WHAT IS KNOWN ALREADY
The rs61764370 SNP in the 3′ untranslated region of the KRAS gene is predicted to disrupt a complementary binding site (LCS6) for the let-7 microRNA, and was recently reported to be at a high frequency (31%) in 132 women of varying ancestry with endometriosis compared with frequencies in a database of population controls (up to 7.6% depending on ancestry), suggesting a strong effect of this KRAS SNP in the aetiology of endometriosis.
STUDY DESIGN, SIZE AND DURATION
This was a case–control study with a total of 11 206 subjects. The study was performed between February 2012 and July 2012.
PARTICIPANTS/MATERIALS, SETTING AND METHODS
We first investigated a possible association between common markers in KRAS and endometriosis risk from our genome-wide association (GWA) data in 3194 surgically confirmed endometriosis cases and 7060 controls of European ancestry. Although rs61764370 was not genotyped on the GWA arrays, five SNPs typed in the study were highly correlated with this variant. The rs61764370 and two SNPs highly correlated with rs61764370 were then genotyped in 933 endometriosis cases and 952 controls using the Sequenom MassARRAY platform.
MAIN RESULTS AND THE ROLE OF CHANCE
There was no evidence for an association between rs61764370 and endometriosis risk P = 0.411 and odds ratio = 1.10 (95% confidence intervals: 0.88–1.36). We also found no evidence for an association between the highly correlated SNP rs17387019 and endometriosis. Their minor allele frequencies in cases and controls were of 0.087–0.091 similar to the population frequency reported previously for this variant in controls. Analyses of endometriosis cases with revised American Fertility Society stage III/IV disease also showed no evidence for an association between these SNPs and endometriosis risk.
LIMITATIONS AND REASONS FOR CAUTION
The GWA and genotyped data sets were not independent since individuals and cases from some families overlap. Controls in our GWA study were not screened for endometriosis.
WIDER IMPLICATIONS OF THE FINDINGS
The key SNP, rs61764370, was genotyped in a subset of samples. Our results do not support the suggestion that carrying the minor allele at rs61764370 contributes to a significant number of endometriosis cases and rs61764370 is, therefore, unlikely to be a useful marker of endometriosis risk.
STUDY FUNDING/COMPETING INTEREST(S)
The research was funded by grants from the Australian National Health and Medical Research Council and Wellcome Trust. None of the authors has competing interests for the study.
Keywords: endometriosis, KRAS, microRNA, genetic association
Introduction
Endometriosis is a common, chronic gynaecological disease characterized by the development of functional endometrial tissue outside the uterus. The disease is an important contributor to pelvic pain and subfertility. It affects 7–10% of women of reproductive age and up to 50% of women with infertility (Treloar et al., 1999; Giudice, 2010). Medical treatment options are limited and surgical intervention may not prevent recurrence (Rogers et al., 2009; Giudice, 2010).
Genetic factors contribute to endometriosis and it is inherited as a complex genetic trait (Kennedy, 1999; Treloar et al., 1999; Stefanson et al., 2002; Simpson and Bischoff, 2003). Numerous candidate gene studies have reported association for markers in candidate genes and endometriosis susceptibility, but results have generally not been replicated in subsequent studies (Simpson and Bischoff, 2003; Bischoff and Simpson, 2004; Guo, 2005; Montgomery et al., 2008). This lack of success is likely due to a number of reasons, including differences in disease definitions, case/control population sampling issues and the lack of power in small-scale studies, which detect only variants with large effects and are prone to detection of false-positive results (Zondervan et al., 2002).
Two genome-wide association (GWA) studies have reported significant associations with endometriosis risk in three genomic regions on chromosomes 9p21, 7p15.2 and 1p36 (Uno et al., 2010; Painter et al., 2011). The regions showed strong evidence for association with disease risk and were replicated in independent data sets, demonstrating that the studies had the power to detect common variants contributing to disease risk. However, effect size estimates were small [odds ratios (ORs) ≤1.22] and the proportion of variance explained by the key single nucleotide polymorphism (SNP) on chromosome 7p15.2 (rs12700667) was 0.36, or 0.69% of the estimated 51% heritability of endometriosis (Painter et al., 2011).
Recently, the minor allele at a SNP (rs61764370) within the 3′ untranslated region (UTR) of the Kirsten rat sarcoma viral oncogene homolog (KRAS) gene was reported to be at a high frequency (31%) in women with endometriosis, much higher than frequencies reported in unselected population samples (Grechukhina et al., 2012). KRAS is a strong candidate gene for endometriosis. The activation of Kras induced peritoneal endometriosis in 47% of mice, and 100% of mice developed benign endometriosis-like lesions within the ovarian surface epithelium (Dinulescu et al., 2005). Endometriosis-like lesions also developed and survived over a prolonged period in wild-type mice after transplanting endometrial tissue from mice with the activated Kras mutation (Cheng et al., 2011). The rs61764370 SNP is predicted to disrupt a complementary binding site (LCS6) for the let-7 microRNA (miRNA) (Johnson et al., 2005; Esquela-Kerscher and Slack, 2006). The minor allele at rs61764370 was shown to alter mRNA and protein expression levels in cultured endometrial stromal cells (Grechukhina et al., 2012), and stromal cells from women carrying the variant allele also showed increased proliferation and invasion. The authors concluded that carrying the minor allele at the rs61764370 variant might account for up to one-third of endometriosis cases (Grechukhina et al., 2012).
Previous studies have examined an association between variants in KRAS and endometriosis risk. Mutational screening of KRAS identified KRAS mutations in endometrioid carcinoma, but not in endometriosis lesions (Amemiya et al., 2004). Genotyping 32 tagging SNPs across KRAS in 958 endometriosis cases and 959 controls showed no evidence for common variants contributing to disease risk in a study that had 80% power to detect a risk variant with a minor allele frequency (MAF) of 5% at a genotype relative risk of 1.7 (Zhao et al., 2006). We would have expected the association signal for rs61764370 to have been detected by correlation with tagging SNPs (Zhao et al., 2006) or in the GWA studies (Uno et al., 2010; Painter et al., 2011).
We therefore re-examined evidence for an association between common markers in KRAS and endometriosis risk from our GWA data (Painter et al., 2011) and genotyped rs61764370 in endometriosis cases and controls.
Materials and Methods
Study samples
We examined data from our previous GWA study including 3194 endometriosis cases and 7060 controls, with 2270 endometriosis cases from Australia (QIMR, Australian data set) and 924 cases from the UK (Oxford, UK data set) (Painter et al., 2011). The QIMR samples were unrelated cases (one individual per family) drawn from 3908 affected women from multiple case families and single cases previously recruited into our endometriosis study. Women included in the study completed questionnaires, provided a blood sample and gave access to their medical records, allowing retrospective confirmation of diagnosis for affected individuals (Treloar et al., 2005; Painter et al., 2011). UK cases included unrelated cases drawn from 245 families with ≥2 affected sisters with surgically confirmed endometriosis (Treloar et al., 2002), and 785 singleton cases. We obtained medical records for all the cases and diagnosis of endometriosis was confirmed from surgical records in 100% of cases in Australia and 97% of cases in the UK (Painter et al., 2011). Disease severity was retrospectively assessed using the revised American Fertility Society (rAFS) classification system (American Fertility Society, 1985); A total of n = 1686 (52.7%) were diagnosed with stage A disease (rAFS I/II or some ovarian disease with few adhesions), n = 1364 (42.7%) had stage B (stage III/IV disease) and n = 144 (4.6%) had an unknown stage. In both the Australian and UK data sets, affected women completed a self-report questionnaire including questions about fertility history and pain symptoms; however, as questions were phrased differently, sub-phenotype analysis was limited to data from the largest case data set (QIMR, 2078 cases).
The controls consisted of 1870 individuals recruited by QIMR and a further 5194 individuals provided by the Wellcome Trust Case Control Consortium 2 (WTCCC2) (Painter et al., 2011). Ethics approval for the studies was obtained from the QIMR Human Ethics Research Committee, the Australian Twin Registry and the Oxford regional multi-centre and local research ethics committees. All the participants gave informed consent prior to testing.
GWA association data
Samples from the GWA study were genotyped on Illumina genotyping platforms. We reviewed a total of 97 common SNPs (MAF range from 2.4 to 47.7%) located between 150 kb upstream and 150 kb downstream of KRAS in all the endometriosis cases and in stage III/IV cases compared with the controls. The KRAS variant rs61764370 was not present on the chips, so we searched for correlated SNPs using the SNP Annotation and Proxy Search (SNAP) software available through the Broad Institute (http://www.broadinstitute.org/mpg/snap/ldsearch.php). Linkage disequilibrium (LD) is the non-random association of alleles at different loci and indicates to what extent the SNPs are correlated in a particular ethnic population. Using the 1000 Genome pilot 1 data generated on 179 individuals including 60 of European origin (‘CEU’), the SNAP program compared the genotypes of SNPs within this region and identified those present on Illumina Human 610 and 1 M chips highly correlated (in strong LD) with rs61764370. We identified five SNPs typed that were strongly correlated with rs61764370 (r2 > 0.5) (Table I) and reviewed the association results previously calculated in our GWA study (Painter et al., 2011). The effect of SNP genotype on the distributions of women with stage B disease or who answered ‘Yes’ to questions on subfertility and pain were tested by Pearson’s chi-squared test.
Table I.
Results of the tests of association for SNPs correlated with rs61764370 in all endometriosis and stage B disease.
| SNPa | Position | r2 with rs61764370* | A1 | A2 | Allele frequency in cases | Allele frequency in controls | Chi-square | P-value | OR (95% CIs) |
|---|---|---|---|---|---|---|---|---|---|
| All Cases | |||||||||
| rs859141 | 25113291 | 0.789 | G | A | 0.090 | 0.098 | 3.346 | 0.067 | 0.90 (0.82–1.02) |
| rs7303889 | 25146242 | 0.571 | C | A | 0.173 | 0.179 | 0.877 | 0.350 | 0.96 (0.89–1.04) |
| rs17387019 | 25155526 | 1.000 | G | A | 0.087 | 0.091 | 1.053 | 0.305 | 0.95 (0.85–1.05) |
| rs17388893 | 25285132 | 0.568 | A | C | 0.060 | 0.064 | 0.790 | 0.375 | 0.95 (0.84–1.07) |
| rs17329975 | 25290239 | 0.568 | C | T | 0.060 | 0.064 | 1.145 | 0.285 | 0.93 (0.83–1.06) |
| Stage B Cases | |||||||||
| rs859141 | 25113291 | 0.789 | G | A | 0.094 | 0.098 | 0.441 | 0.506 | 0.95 (0.82–1.10) |
| rs7303889 | 25146242 | 0.571 | C | A | 0.179 | 0.179 | 0.000 | 0.984 | 0.99 (0.90–1.11) |
| rs17387019 | 25155526 | 1.000 | G | A | 0.089 | 0.091 | 0.080 | 0.777 | 0.98 (0.85–1.13) |
| rs17388893 | 25285132 | 0.568 | A | C | 0.062 | 0.064 | 0.067 | 0.795 | 0.98 (0.83–1.16) |
| rs17329975 | 25290239 | 0.568 | C | T | 0.061 | 0.064 | 0.302 | 0.583 | 0.95 (0.80–1.13) |
r2: A measure of LD or non-random association for the observed frequencies of alleles at two markers measured as the square of the correlation coefficient.
LD was assessed using the SNAP proxy search program (http://www.broadinstitute.org/mpg/snap/ldsearch.php).
A1, reference allele; A2, alternative allele.
aSNPs present on Illumina HumanHap610-Quad BeadChip and Illumina Human1M-DuoChip that are highly correlated with SNP rs61764370 in 1000 Genomes Pilot 1 data of 60 Utah residents with European ancestry (CEU) individuals.
Genotyping
To confirm results from the GWA data, we genotyped rs61764370 and two correlated SNPs in the same sample of 958 endometriosis cases and 959 unrelated controls examined in our previous KRAS study (Zhao et al., 2006) using the Sequenom MassARRAY Genomics Platform. SNPs were genotyped as part of multiplex assays designed using the Sequenom MassARRAY Assay Design software (version 3.0: Sequenom Inc.) and samples were genotyped using standard methods (Zhao et al., 2007, 2008). Departures from Hardy-Weinberg equilibrium and statistical analysis for association between SNPs and endometriosis risk were tested using the PLINK (http://pngu.mgh.harvard.edu/~purcell/plink/). Quality control of case samples from the GWA data analyses had identified case families with varying degrees of non-Caucasian ancestry. These were removed from the analyses of this data set and genotype data from 933 cases and 952 controls were included in the association analyses.
Results
We identified five SNPs present on Illumina HumanHap610-Quad and 1M-Duo Chips that were highly correlated with the SNP rs61764370 genotyped in the Grechukhina et al. (2012) study. These SNPs were in strong LD with rs61764370 (r2≥ 0.568, Table I) and were all located within 140 kb of the 3′UTR for KRAS. One SNP rs17387019 was reported to be in perfect (r2 = 1) LD with rs61764370. There was no evidence for an association between this SNP and endometriosis risk (Table I) with P = 0.305 [OR = 0.95; 95% confidence intervals (CIs): 0.85–1.05]. The MAF for this SNP in our GWA data was 0.087 in the cases, consistent with published estimates in other Caucasian populations (Hapmap-CEU and EUR—interim phase 1–1000 Genomes) (Chin et al., 2008).
There was no evidence for any association between endometriosis risk and other correlated SNPs, including rs859141, which was in high LD with rs61764370 (r2 = 0.789) (Table I) with P = 0.067 and OR = 0.90 (95% CIs: 0.82–1.02). The latter SNP also has an MAF of 0.090. In addition, we checked all other common SNPs genotyped across the region, from 150 kb upstream to 150 kb downstream of KRAS in our GWAS data. There was no evidence for an association between any of the genotyped SNPs in the KRAS region and endometriosis risk (Fig. 1).
Figure 1.
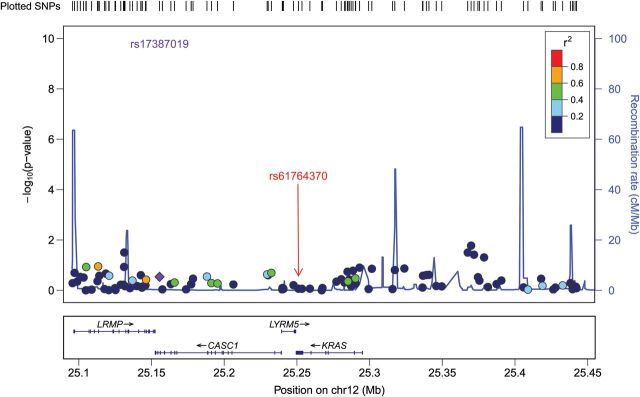
Evidence for association (−log10 P-values, Y-axis) between genotyped SNPs and endometriosis across the chromosome 12 region ∼150 kb from the 3′UTR KRAS variant (rs61764370). In this Locus Zoom association plot SNP rs17387019 is represented by a purple diamond (http://csg.sph.umich.edu/locuszoom/) and is perfectly correlated with the 3′UTR KRAS variant rs61764370. The red arrow shows the position of the 3′UTR KRAS variant rs61764370. Other SNPs are colour coded according to the strength of their correlation (LD) with rs17387019 (measured by r2).
As cases in Grechukhina et al.'s analysis included 89% with a rAFS stage III/IV diagnosis, we restricted our analysis to cases diagnosed with rAFS stage III/IV (stage B) disease, and the results showed that the minor allele at each correlated SNP was slightly lower in the cases compared with the controls (Table I). We also found no evidence for an association between endometriosis risk and any correlated SNPs (Table I). We further analysed the genotypic association between SNP rs17387019 and sub-phenotypes of endometriosis collected in the Australian data set (see methods). There was no evidence that genotypes at rs17387019 influenced the proportions of women with stage B disease, infertility or pain (Table II).
Table II.
Genotype at rs17387019 in the Australian cases and the number (and proportions) with moderate/severe (stage B) disease or who answered ‘Yes’ to questions on subfertility or pain.
| Genotype at rs17387019 |
P-value | |||
|---|---|---|---|---|
| AA | AG | GG | ||
| Stage B | 7 (0.39) | 149 (0.42) | 754 (0.39) | 0.74 |
| Subfertilitya | 5 (0.29) | 129 (0.37) | 717 (0.38) | 0.64 |
| Menstrual painb | 17 (1.00) | 332 (0.94) | 1729 (0.91) | 0.13 |
| Pelvic painc | 16 (0.94) | 294 (0.83) | 1540 (0.82) | 0.40 |
| Emergency treatmentd | 10 (0.58) | 156 (0.45) | 792 (0.42) | 0.30 |
| Dysperuniae | 11 (0.69) | 276 (0.79) | 1433 (0.77) | 0.48 |
The number of women who answered 'Yes' to the following questions:
aHave you tried for 12 months or more on any occasion to conceive without success?
bHave you EVER experienced severe menstrual pain?
cHave you EVER experienced severe pelvic pain?
dHave you ever had to seek emergency treatment because of pain?
eHave you ever experienced pain during sexual intercourse?
To directly examine the association between rs61764370 and endometriosis risk, we genotyped this SNP, together with rs17387019 and rs859141 in our subset of endometriosis cases with a family history of disease and controls used in previous studies. There was no evidence for association between rs61764370 and endometriosis risk (Table III) with P = 0.411 and OR = 1.10 (95% CIs: 0.88–1.36). The MAF for this SNP was 0.099 in endometriosis cases. In agreement with the GWA data, we did not find any evidence for association between endometriosis risk and rs17387019 and rs859141 (Table III). These two SNPs were also in high LD with the KRAS SNP rs61764370 in our data set (r2 = 0.858 and 0.537, respectively), although the estimates were slightly lower than in the 1000 Genome pilot 1 data in our larger sample.
Table III.
Results of the tests of association for rs61764370 and SNPs correlated with rs61764370 in multiplex genotyping of endometriosis cases and controls.
| SNP | Position | r2 with rs61764370 | A1 | A2 | Allele frequency in cases (n = 933) | Allele frequency in controls (n = 952) | Chi-square | P-value | OR (95% CIs) |
|---|---|---|---|---|---|---|---|---|---|
| rs61764370 | 25360224 | G | T | 0.099 | 0.091 | 0.677 | 0.411 | 1.10 (0.88–1.36) | |
| rs17387019 | 25155526 | 0.858 | A | G | 0.089 | 0.082 | 0.509 | 0.476 | 1.09 (0.86–1.37) |
| rs859141 | 2511329 | 0.537 | T | C | 0.097 | 0.090 | 0.493 | 0.483 | 1.08 (0.87–1.35) |
LD was assessed using the PLINK program which was run on our multiplex genotyping data set.
Discussion
Ras proteins have an important role as binary molecular switches in controlling signal transduction pathways. In mouse models, the activation of Kras triggers the development of tissue resembling endometriosis and endometrioid ovarian cancer (Dinulescu et al., 2005; Cheng et al., 2011). In women with endometriosis, the frequency of the minor allele at rs61764370 in the 3′ UTR of KRAS gene was reported to be 31% (41/132 cases), a much higher frequency than expected in the general population (Grechukhina et al., 2012). The authors concluded that rs61764370 is a marker of endometriosis risk, potentially contributing to nearly one-third of all endometriosis cases and that it provides a novel method for diagnosis (Grechukhina et al., 2012). However, the results from our analyses in two large data sets provide no support for an association between rs61764370 and endometriosis risk.
The KRAS polymorphism rs61764370 was not present on the Illumina 610 chip used for our GWAS. We therefore examined LD between rs61764370 and the genotyped SNPs. SNP rs17387019 which was typed on the chips is in strong LD with rs61764370 and genotyping of this SNP provides almost complete information on the other. We also identified four other SNPs typed on the Illumina 610 chip and in moderate to high LD (r2 > 0.57) with the KRAS 3′UTR SNP. There was no evidence for an association with endometriosis for any of these correlated SNPs; allele frequencies in the cases and the controls were similar in our GWA sample including 3194 endometriosis cases.
Since rs61764370 was not on the Illumina chips, we genotyped this SNP in a sample of endometriosis cases with a family history of disease. The studies were not independent since individual cases and some families overlap between the two sample sets. There was no evidence for an association as the allele frequency for rs61764370 in our genotyped sample was 0.099 for the cases and 0.091 for the controls, similar to the population frequency reported previously for this variant (Grechukhina et al., 2012). The results also agree with those of our previous study, where we found no association between common variants in KRAS and endometriosis risk from genotyping 32 tagging SNPs across the KRAS gene (Zhao et al., 2006). Analysis in our GWA data for 97 SNPs located within the region from 150 kb upstream and downstream of KRAS gene showed no evidence for different allele frequencies between the cases and the controls and no association signal between any SNP in KRAS and endometriosis risk.
Of 132 women with endometriosis typed for the KRAS SNP rs61764370 by Grechukhina et al. (2012), 89% (n = 117) of the cases were assigned stage III/IV disease. When we limited our analysis to cases with stage B (III/IV) disease, our data set showed no evidence of any association between endometriosis risk and SNPs highly correlated with rs61764370 [proxy SNP rs17387019: OR = 0.98 (95% CIs: 0.85–1.13)]. MAFs in stage B cases were slightly lower in the cases compared with the controls, very similar to those in all endometriosis cases and our Australian and UK control population samples. Taking these results together, our data provide no support for the minor allele of rs61764370 conferring risk of endometriosis.
Our analysis of KRAS variants is based on a sample of 2270 endometriosis cases recruited in Australia and 924 cases recruited in the UK (Painter et al., 2011). The diagnosis of endometriosis was confirmed from surgical records in 100% of cases in Australia and 97% of cases in the UK (Painter et al., 2011). The controls in our GWAS were not screened for endometriosis. However, Grechukhina et al. used data from public databases for control allele frequency information and it is also unlikely that these individuals were screened for disease.
In summary, we find no evidence for an association between risk of endometriosis and variants in KRAS including the rs61764370 SNP in the let-7 miRNA-binding site in the 3′UTR of KRAS. Our results do not address the physiological consequences of carrying the variant allele in the let-7 miRNA-binding site. However, our study provides no support for suggestions that carrying the minor allele of rs61764370 contributes to a significant number of endometriosis cases and rs61764370 is unlikely to be a useful marker of endometriosis risk.
Authors’ roles
H.T.T.L, G.W.M., and D.R.N.: Study design and manuscript preparation; S.K., G.W.M., D.R.N., J.N.P., S.A.T., and K.T.Z.: GWAS data and clinical phenotyping; B.C.: Replication genotyping; H.T.T.L., G.W.M. D.R.N., J.N.P.: Data analysis; B.C., H.T.T.L, S.K., G.W.M., D.R.N., J.N.P., S.A.T., and K.T.Z.: Revision for critical content and approval of the final version of the manuscript.
Funding
The QIMR Study was supported by grants from the National Health and Medical Research Council (NHMRC) of Australia (241944, 339462, 389927, 389875, 389891, 389892, 389938, 443036, 442915, 442981, 496610, 496739 552485 and 552498), the Cooperative Research Centre for Discovery of Genes for Common Human Diseases (CRC), Cerylid Biosciences (Melbourne) and donations from N. Hawkins and S. Hawkins. D.R.N. was supported by the NHMRC Fellowship (339462 and 613674) and the ARC Future Fellowship (FT0991022) schemes and G.W.M. (339446, 619667) was supported by the NHMRC Fellowships Scheme. The work was supported by a grant from the Wellcome Trust (WT084766/Z/08/Z) and makes use of WTCCC2 control data generated by the Wellcome Trust Case-Control Consortium. A full list of the investigators who contributed to the generation of these data is available at http://www.wtccc.org.uk. Funding for the WTCCC project was provided by the Wellcome Trust under awards 076113 and 085475. S.K. is supported by the Oxford Partnership Comprehensive Biomedical Research Centre with funding from the Department of Health NIHR Biomedical Research Centres funding scheme. K.T.Z. is supported by a Wellcome Trust Research Career Development Fellowship (WT085235/Z/08/Z). Funding to pay the Open Access publication charges for this article was provided by the Wellcome Trust.
Conflict of interest
None declared.
Acknowledgements
We acknowledge with appreciation all the women who participated in the QIMR and OXEGENE studies. We thank Endometriosis Associations for supporting the study recruitment and the many hospital directors and staff, gynaecologists, general practitioners and pathology services in Australia and the UK who provided assistance with confirmation of diagnoses. We thank S. Nicolaides and the Queensland Medical Laboratory for pro bono collection and delivery of blood samples and other pathology services for assistance with blood collection. We thank A.K. Henders, L. Wallace, B. Haddon, D. Smyth, H. Beeby, and O. Zheng for project and database management, sample processing and genotyping. We thank Brisbane gynaecologist D.T. O'Connor for confirmation of diagnosis and staging of disease from clinical records of many cases, including 251 in these analyses. We are grateful to the many research assistants and interviewers for assistance with the studies contributing to the QIMR collection. We thank L. Cotton, L. Pope, G. Chalk and G. Farmer (University of Oxford). We also thank P. Koninckx (Leuven, Belgium), M. Sillem (Heidelberg, Germany), C.O'Herlihy and M. Wingfield (Dublin, Ireland), M. Moen (Trondheim, Norway), L. Adamyan (Moscow, Russia), E. McVeigh (Oxford, UK), C. Sutton (Guildford, UK), D. Adamson (Palo Alto, California, USA) and R. Batt (Buffalo, New York, USA) for providing diagnostic confirmation.
References
- Amemiya S, Sekizawa A, Otsuka J, Tachikawa T, Saito H, Okai T. Malignant transformation of endometriosis and genetic alterations of K-ras and microsatellite instability. Int J Gynecol Obstet. 2004;86:371–376. doi: 10.1016/j.ijgo.2004.04.036. [DOI] [PubMed] [Google Scholar]
- Bischoff F, Simpson JL. Genetics of endometriosis: heritability and candidate genes. Best Prac Res Clin Obstet Gynaecol. 2004;18:219–232. doi: 10.1016/j.bpobgyn.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Cheng CW, Licence D, Cook E, Luo FJ, Arends MJ, Smith SK, Print CG, Charnock-Jones DS. Activation of mutated K-ras in donor endometrial epithelium and stroma promotes lesion growth in an intact immunocompetent murine model of endometriosis. J Pathol. 2011;224:261–269. doi: 10.1002/path.2852. [DOI] [PubMed] [Google Scholar]
- Chin LJ, Ratner E, Leng SG, Zhai RH, Nallur S, Babar I, Muller RU, Straka E, Su L, Burki EA, et al. A SNP in a let-7 microRNA complementary site in the KRAS 3 untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68:8535–8540. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinulescu DM, Ince TA, Quade BJ, Shafer SA, Crowley D, Jacks T. Role of K-ras and Pten in the development of mouse models of endometriosis and endometrioid ovarian cancer. Nat Med. 2005;11:63–70. doi: 10.1038/nm1173. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Giudice LC. Endometriosis. N Engl J Med. 2010;362:2389–2398. doi: 10.1056/NEJMcp1000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grechukhina O, Petracco R, Popkhadze S, Massasa E, Paranjape T, Chan E, Flores I, Weidhaas JB, Taylor HS. A polymorphism in a let-7 microRNA binding site of KRAS in women with endometriosis. EMBO Mol Med. 2012;4:206–217. doi: 10.1002/emmm.201100200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo SW. Glutathione S-transferases M1/T1 gene polymorphisms and endometriosis: a meta-analysis of genetic association studies. Mol Hum Reprod. 2005;11:729–743. doi: 10.1093/molehr/gah206. [DOI] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 MicroRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Kennedy S. The genetics of endometriosis. Eur J Obstet and Gynecol Reprod Biol. 1999;82:129–133. doi: 10.1016/s0301-2115(98)00213-9. [DOI] [PubMed] [Google Scholar]
- Montgomery GW, Nyholt DR, Zhao ZZ, Treloar SA, Painter JN, Missmer SA, Kennedy SH, Zondervan KT. The search for genes contributing to endometriosis risk. Hum Reprod Update. 2008;14:447–457. doi: 10.1093/humupd/dmn016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter JN, Anderson CA, Nyholt DR, Macgregor S, Lin JH, Lee SH, Lambert A, Zhao ZZ, Roseman F, Guo Q, et al. Genome-wide association study identifies a locus at 7p15.2 associated with endometriosis. Nat Genet. 2011;43:51–54. doi: 10.1038/ng.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers PAW, D'Hooghe TM, Fazleabas A, Gargett CE, Giudice LC, Montgomery GW, Rombauts L, Salamonsen LA, Zondervan KT. Priorities for endometriosis research: recommendations from an International Consensus Workshop. Reprod Sci. 2009;16:335–346. doi: 10.1177/1933719108330568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JL, Bischoff F. Heritability and candidate genes for endometriosis. Reprod Biomed Online. 2003;7:162–169. doi: 10.1016/s1472-6483(10)61746-4. [DOI] [PubMed] [Google Scholar]
- Stefanson H, Geirsson RT, Steinthorsdottir V, Jonsson H, Manolescu A, Kong A, Ingadottir G, Gulcher J, Stefansson K. Genetic factors contribute to the risk of developing endometriosis. Hum Reprod. 2002;17:555–559. doi: 10.1093/humrep/17.3.555. [DOI] [PubMed] [Google Scholar]
- Treloar SA, O'Connor DT, O'Connor VM, Martin NG. Genetic influences on endometriosis in an Australian twin sample. Fertil Steril. 1999;17:701–710. doi: 10.1016/s0015-0282(98)00540-8. [DOI] [PubMed] [Google Scholar]
- Treloar S, Hadfield R, Montgomery G, Lambert A, Wicks J, Barlow DH, O'Connor DT, Kennedy S Int Endogene Study Grp. The International Endogene Study: a collection of families for genetic research in endometriosis. Fertil Steril. 2002;78:679–685. doi: 10.1016/s0015-0282(02)03341-1. [DOI] [PubMed] [Google Scholar]
- Treloar SA, Wicks J, Nyholt DR, Montgomery GW, Bahlo M, Smith V, Dawson G, Mackay IJ, Weeks DE, Bennett ST, et al. Genomewide linkage study in 1,176 affected sister pair families identifies a significant susceptibility locus for endometriosis on chromosome 10q26. Am J Hum Genet. 2005;77:365–376. doi: 10.1086/432960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno S, Zembutsu H, Hirasawa A, Takahashi A, Kubo M, Akahane T, Aoki D, Kamatani N, Hirata K, Nakamura YA. Genome-wide association study identifies genetic variants in the CDKN2BAS locus associated with endometriosis in Japanese. Nat Genet. 2010;42:707–U788. doi: 10.1038/ng.612. [DOI] [PubMed] [Google Scholar]
- Zhao ZZ, Nyholt DR, Le L, Martin NG, James MR, Treloar SA, Montgomery GW. KRAS variation and risk of endometriosis. Mol Hum Reprod. 2006;12:671–676. doi: 10.1093/molehr/gal078. [DOI] [PubMed] [Google Scholar]
- Zhao ZZ, Nyholt DR, Le L, Martin NG, Thomas S, Engwerda C, Randall L, Treloar SA, Montgomery GW. Genetic variation in tumour necrosis factor and lymphotoxin is not associated with endometriosis in an Australian sample. Hum Reprod. 2007;22:2389–2397. doi: 10.1093/humrep/dem182. [DOI] [PubMed] [Google Scholar]
- Zhao ZZ, Pollock PM, Thomas S, Treloar SA, Nyholt DR, Montgomery GW. Common variation in the fibroblast growth factor receptor 2 gene is not associated with endometriosis risk. Hum Reprod. 2008;23:1661–1668. doi: 10.1093/humrep/den035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zondervan K, Cardon L, Desrosiers R, Hyde D, Kemnitz J, Mansfield K, Roberts J, Scheffler J, Weeks DE, Kennedy S. The genetic epidemiology of spontaneous endometriosis in the rhesus monkey. In: Yoshinaga K, Parrott EC, editors. Endometriosis: Emerging Research and Intervention Strategies. vol.955. NY, USA: New York Academy of Sciences; 2002. pp. 233–238. [DOI] [PubMed] [Google Scholar]


