SUMMARY
Crosstalk and complexity within signaling pathways, and their perturbation by oncogenes, limits component-by-component approaches to understanding human disease. Network analysis of how normal and oncogenic signaling can be re-wired by drugs may provide opportunities to target tumors with high specificity and efficacy. Using targeted inhibition of oncogenic signaling pathways combined with DNA damaging chemotherapy, we report that time-staggered EGFR inhibition, but not simultaneous co-administration, dramatically sensitizes a subset of triple-negative breast cancer cells to genotoxic drugs. Systems-level analysis—using high-density time-dependent measurements of signaling networks, gene expression profiles, and cell phenotypic responses in combination with mathematical modeling— revealed an approach for altering the intrinsic state of the cell through dynamic re-wiring of oncogenic signaling pathways. This process converts these cells to a less tumorigenic state that is more susceptible to DNA damage-induced cell death by re-activation of an extrinsic apoptotic pathway whose function is suppressed in the oncogene-addicted state.
INTRODUCTION
Standard therapies for the treatment of human malignancies typically involve the use of chemotherapy or radiation therapy, which function by damaging DNA in both normal and cancerous cells (Lichter and Lawrence, 1995). Our growing understanding of this process suggests that the DNA damage response (DDR) functions as part of a complex network controlling many cellular functions, including cell cycle, DNA repair, and various forms of cell death (Harper and Elledge, 2007). The DDR is highly interconnected with other pro-growth and pro-death signaling networks, which function together to control cell fate in a nonlinear fashion due to multiple levels of feedback and crosstalk. Thus, it is difficult to predict a priori how multiple, often conflicting signals will be processed by the cell, particularly by malignant cells, where regulatory networks often exist in atypical forms. Predicting the efficacy of treatment, and the optimal design of combination therapy, will require a detailed understanding of how the DDR and other molecular signals are integrated and processed, how processing is altered by genetic perturbations commonly found in tumors, and how networks can be ‘rewired’ using drugs individually and in combination (Sachs et al., 2005).
In many forms of breast cancer aberrant hormonal and/or growth factor signaling play key roles in both tumor induction and resistance to treatment (Hanahan and Weinberg, 2000). Moreover, the identification of molecular drivers in specific breast cancer subtypes has led to the development of more efficacious forms of targeted therapy (Schechter et al., 1984; Slamon et al., 1987). In spite of these advances, there are currently no targeted therapies and no established molecular etiologies for triplenegative breast cancers (TNBC)—a heterogeneous mix of breast cancers defined only by the absence of estrogen receptor (ER) or progesterone receptor (PR) expression, and lack of amplification of the HER2 oncogene (Perou et al., 2000). Patients with TNBCs have shorter relapse-free survival and a worse overall prognosis than other breast cancer patients, however, they tend to respond, at least initially, to genotoxic chemotherapy (Dent et al., 2007). Triple-negative patients generally do well if pathologic complete response is achieved following chemotherapy. When residual disease exists, however, the prognosis is typically worse than for other breast cancer subtypes (Abeloff et al., 2008). Thus, identifying new strategies to enhance the initial chemosensitivity of TNBC cells may have substantial therapeutic benefit. We wondered whether a systems biology approach, focused on examining and manipulating the interface between growth factor signaling pathways and DNA damage signaling pathways in tumor cells, could modulate the therapeutic response of this recalcitrant tumor type. We report here that pre-treatment, but not co-treatment or post-treatment of a subset of TNBCs with EGFR inhibitors can markedly synergize their apoptotic response to DNA damaging chemotherapy through dynamic re-wiring of oncogenic signaling networks and unmasking of suppressed pro-apoptotic pathways. These results may have broader implications for the testing, design, and utilization of combination therapies in the treatment of malignant disease.
RESULTS
A critical order and time-dependency for enhanced EGFR inhibition/DNA damage-mediated cell death
Signaling networks can respond to, and can be functionally re-wired by, exposure to specific ligands or drugs (Janes et al., 2005; Janes et al., 2008). It is increasingly clear that these responses are time-dependent. We reasoned that it should, in principle, be possible to dynamically re-wire the DDR network in an insensitive cell through prior exposure to a drug that modulates the network, thereby rendering the cell sensitive to DNA damaging agents. To test this hypothesis, we systematically investigated a series of drug combinations for synergism or antagonism in breast cancer cells using protocols that changed both the order and timing of drug addition.
We combined genotoxic agents with small molecule inhibitors targeting common oncogenic signaling pathways (Figure 1A). We included drugs that are known to be clinically useful in other cancers but to lack efficacy in TNBC individually or in combination (Bosch et al., 2010; Winer and Mayer, 2007). Previous studies using cell culture models of TNBC, for example, reported that EGFR inhibitors in combination with genotoxic compounds such as cisplatin resulted in less than a 10% survival benefit (Corkery et al., 2009); while a randomized phase II trial in TNBC patients reported that addition of cetuximab to carboplatin did not improve outcome (Carey et al., 2008). However, emerging understanding of the complex non-linear and time-dependent interplay between signaling networks argues that a more systematic assessment exploring not only dose but also the order of drug presentation, scheduling and dose duration might uncover cross-pathway effects and efficacious interactions that were missed previously (Fitzgerald et al., 2006). An initial combination screen was therefore performed in a panel of canonical breast cancer cell lines representing those that are hormone-sensitive (MCF7), HER2 over-expressing (MDA-MB-453), or triple-negative (BT-20) (Neve et al., 2006). A first pass of the screen was performed in BT-20 cells scoring for viability and a subset of combinations were then explored more thoroughly, scoring viability, proliferation and apoptotic responses in the panel of three cell lines (Figure 1B–E and S1).
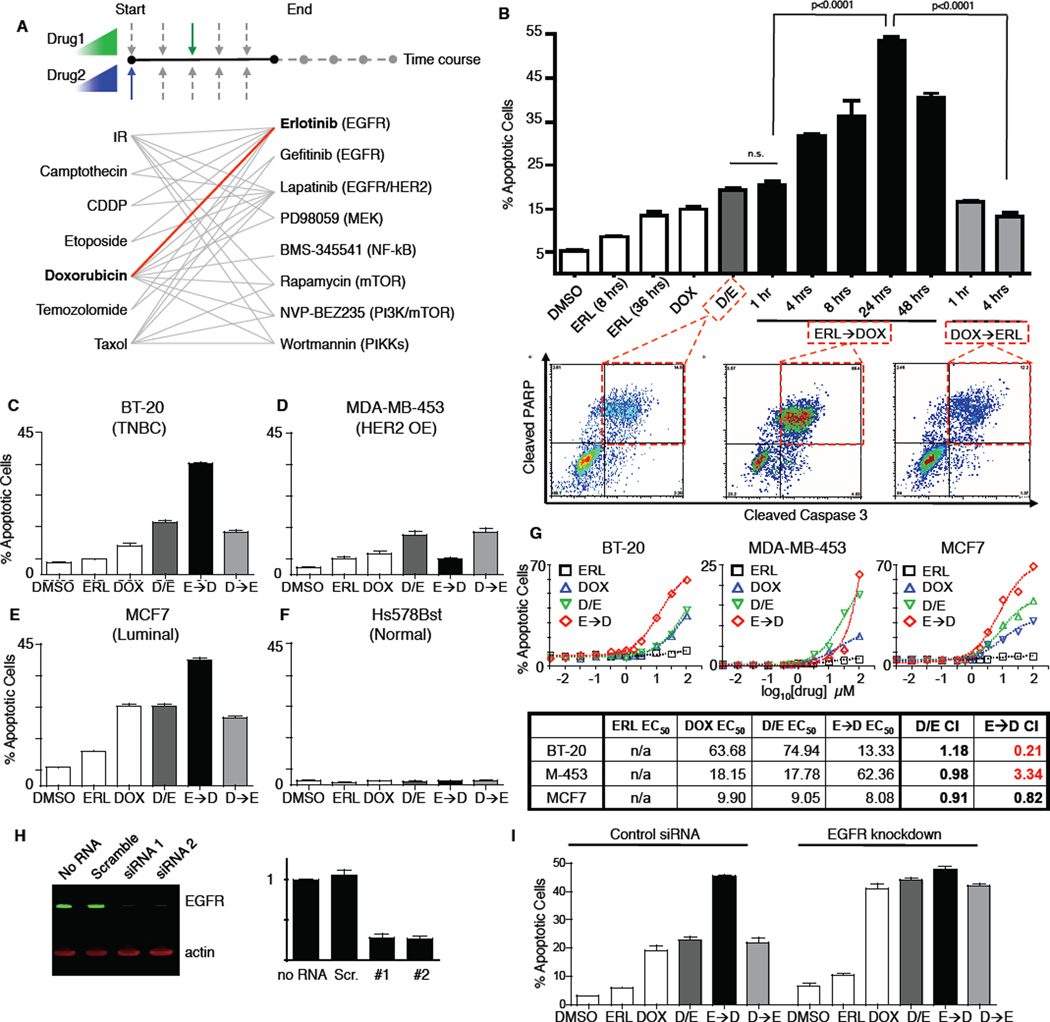
Figure 1. A screen for novel combination treatment reveals dosing schedule-dependent efficacy for killing TNBC cells.
(A) Schematic of combinations tested. 7 genotoxic drugs and 8 targeted signaling inhibitors were tested in pair-wise combinations, varying dose, order of presentation, dose duration, and dosing schedule. (B) Apoptosis in BT-20 cells. Cleaved-caspase 3/cleaved-PARP double positive cells were quantified using flow cytometry (bottom panels). In cells treated with DMSO, erlotinib (ERL) or doxorubicin (DOX), apoptosis measurements were performed 8 hrs. after drug exposure or at the indicated times. D/E, ERL→DOX, and DOX→ERL refer to DOX and ERL added at the same time, ERL given at the indicated times before DOX, and DOX given at the indicated times before ERL, respectively. For each, apoptotic measurements were made 8 hrs after the addition of DOX. Erlotinib and doxorubicin were used at 10µM. Mean values ± S.D. of 3 independent experiments, each performed in duplicate, are shown (top panel). (C–F) Apoptosis in different subtypes of breast cancer. Apoptosis was measured as in B. D/E, E→D, and D→E refer to DOX and ERL added at the same time, ERL given 24 hours before DOX, and DOX given 4 hours before ERL, respectively. Data are mean values ± S.D. of 3 independent experiments. (G) Dose-response profiles of erlotinib/doxorubicin drug combinations. Apoptosis was measured as in B. Drugs were added at a 1:1 ratio, and combination index (CI) was calculated according to the Chou-Talalay method. (H) Knockdown of EGFR in BT-20 cells measured 48 hrs. after addition of the indicated siRNA by immunoblotting (left). EGFR expression relative to “no RNA” control is quantified on right. (I) Apoptosis in BT-20 cells +/− EGFR knockdown measured as in B. Scrambled RNAi shown as control. Data shown are the mean ± S.D. of both siRNAs, each performed in biological duplicate.
Consistent with previous reports, we found that inhibition of EGFR using the compound erlotinib was not a potent apoptotic stimulus in TNBC cells when used alone, or when added at the same time as, or shortly before doxorubicin (Figure 1B, left bars 1–6). Surprisingly, however, combinations in which erlotinib was added at least 4 hours prior to doxorubicin showed a markedly enhanced apoptotic response, with cell killing increasing by as much as 500% (Figure 1B, middle bars 7–10). When the order of drug presentation was reversed—doxorubicin given before erlotinib—cell killing was not enhanced relative to treatment with doxorubicin or erlotinib alone (Figure 1B, right bars 11–12). The efficacy of the time-sequenced erlotinib-doxorubicin treatment was analyzed for doxorubicin dose-effect relationships using the Chou-Talalay method (Chou and Talalay, 1984), and found to vary significantly across breast cancer subtypes (Figure 1C–E and G). Whereas chronic EGFR inhibition was synergistic with doxorubicin in killing TNBC BT-20 cells, the same treatment regimen antagonized doxorubicin sensitivity in HER2 over-expressing MDA-MB-453 cells. All temporal erlotinib-doxorubicin combinations tested were merely additive in luminal MCF7 cells. The order and timing of drug addition had little effect in Hs578Bst, a cell line derived from normal peripheral breast tissue, which was generally drug resistant (Figure 1F).
Furthermore this enhanced treatment effect in BT-20 cells was not limited to combinations of doxorubicin and erlotinib. Synergistic killing was also observed following time-staggered pre-treatment of BT-20 cells with either erlotinib, gefitinib, or lapatinib (all EGFR inhibitors) in combination with the DNA damaging agent camptothecin as well as with doxorubicin (Figure S1A–C) (Wood et al., 2004).
Sustained EGFR inhibition suppresses oncogenic signatures and re-wires the intrinsic state of the tumor cells to a more chemosensitive form
Although erlotinib inhibits EGFR and downstream signaling within minutes (Figure S2A and S2B), enhanced cell death in response to DNA damaging agents required pretreatment with erlotinib for several hours. To verify that this was indeed due to on-target inhibition of EGFR, in addition to testing other EGFR inhibitors (above), we knocked down EGFR using two different siRNAs. Like the times-taggered erlotinib-doxorubicin treatment, strong pro-apoptotic responses were observed in BT-20 cells following EGFR knockdown with delayed doxorubicin treatment (Figure 1H–I). Importantly, addition of erlotinib to EGFR knockdown cells had no additional effect, arguing against an off-target effect of the drug. As a further test, we also examined co-administration of higher concentrations of erlotinib instead of time-staggered doses without observing increased apoptosis (Figure S2C). Taken together, these data indicate that enhanced cell death observed using time-staggered erlotinib-doxorubicin combinations is directly mediated by sustained EGFR inhibition.
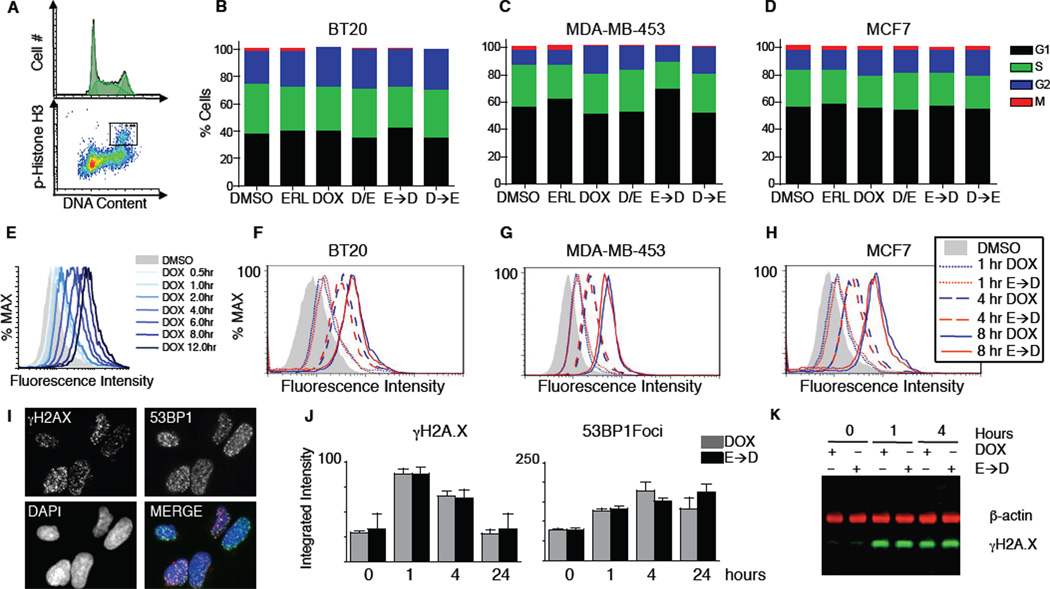
Potential explanations for the increased sensitivity of cells to doxorubicin following sustained EGFR inhibition include modulation of cell cycle progression, altered rates of doxorubicin influx/efflux, or changes in levels of DNA damage itself. To examine these, we monitored cell cycle progression at five time points over 24 hours in our panel of breast cancer cell lines. Although doxorubicin and erlotinib altered cell cycle dynamics depending on the cell type, cells that received both drugs had similar cell cycle profiles regardless of the dosing regimen (Figure 2A–D, S2D); in particular, there is no evidence that cells exposed to the ERL→DOX protocol accumulate in S/G2, the cell cycle stage where doxorubicin may be most effective. Thus, cell cycle modulation cannot explain the unique efficacy of sequential drug exposure. Some membrane pumps can be modulated by EGFR inhibitors (Lopez et al., 2007; Turner et al., 2006) and are responsible for multi-drug resistance in at least some breast cancers (Woehlecke et al., 2003). We therefore measured the intracellular accumulation of doxorubicin by flow cytometry and found that prior treatment with erlotinib did not alter the intracellular doxorubicin concentration (Figure 2E–H). Next as pharmacodynamic markers of doxorubicin action we assayed two indicators of DNA double stranded breaks: phosphorylation of histone H2AX at S139 and formation of 53BP1-containing nuclear foci. Both assays showed similar responses across all treatment conditions (Figure 2I–K). Taken together, these data indicate similar levels of DNA damage and early DNA damage-related signaling in DOX and ERL→DOX treated cells, independent of the efficacy of the combination in cell killing.
Figure 2. Prolonged treatment with erlotinib does not change cell cycle profile, doxorubicin influx/efflux, or the level of DNA damage.
(A–D) Quantitative cell cycle analysis. DNA content and the percentage of mitotic cells were measured by FACS. (A) Example FACS plots from untreated BT-20 cells. (B–D) Cell cycle stage quantified from 3 experiments, each performed in duplicate. Cells were treated as in Figure 1, and data were collected at 6, 8, 12, 24, and 48 hours after DOX treatment. 8 hour data shown for each cell type. (E–H) Doxorubicin retention measured by flow cytometry. (E) Sample time course of BT-20 cells treated with 10µM DOX for the indicated times. (F–H) Cells treated with doxorubicin (DOX) or pre-treated with erlotinib for 24 hrs. prior to DOX (E→D). Cells were collected at 1, 4, or 8 hours after DOX exposure as indicated, and internal doxorubicin fluorescence was measured. (I–J) Quantitative microscopy of the early DNA double stranded break response. (I) Example image of cells treated with DOX for 1 hour and stained for γH2AX, 53BP1, or nuclear content (DAPI). (J) Integrated intensity per nucleus of γH2AX and 53BP1 foci was measured in BT-20 cells after the indicated treatments and times. Mean values +/− S.D. from triplicate experiments shown. (K) Western blot analysis of γH2AX in BT-20 cells. β-actin shown as a loading control.
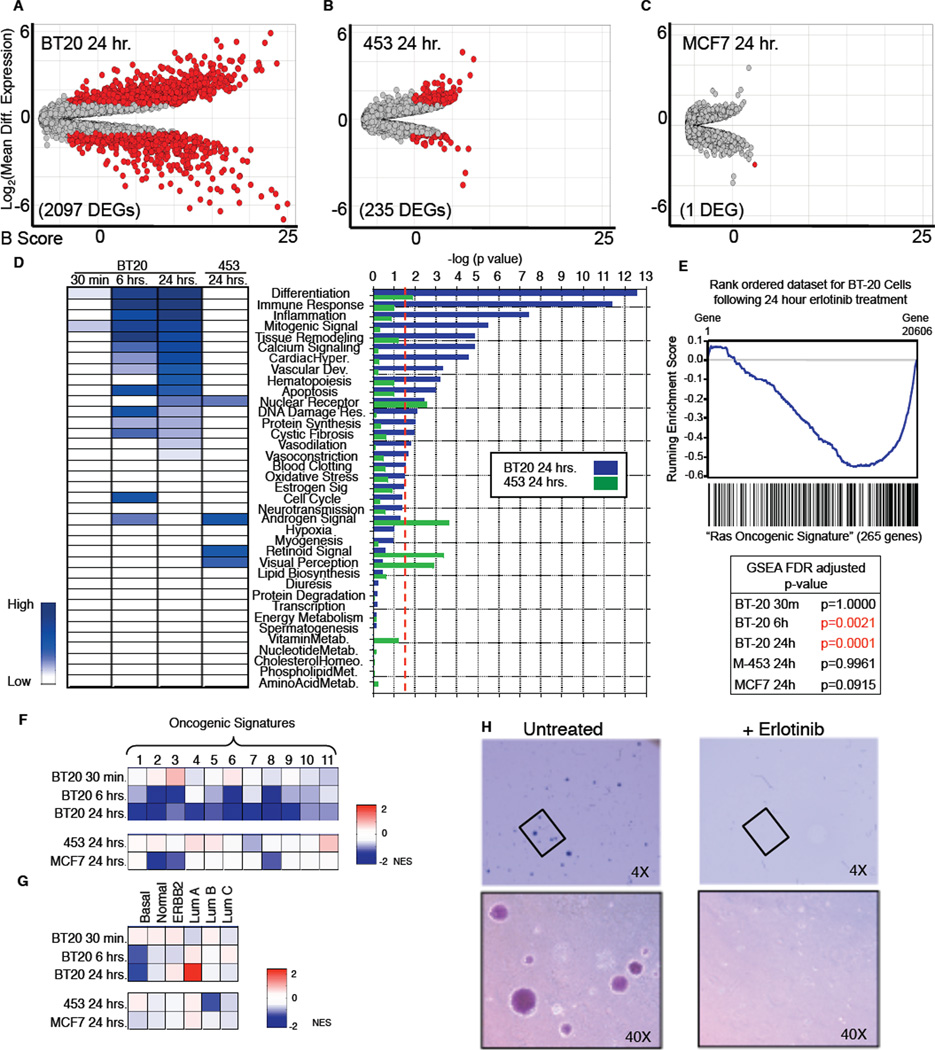
The absence of demonstrable changes in cell cycle states, intracellular doxorubicin concentrations, or doxorubicin-induced DNA damage, suggested that prolonged EGFR inhibition necessary for effective tumor cell killing might result from re-wiring of the signaling networks controlling responses to genotoxic stress. To investigate this idea, we measured changes in gene expression in cells treated with erlotinib alone. In triple-negative BT-20 cells, EGFR inhibition for 30 minutes resulted in few differentially expressed genes (DEGs) (Figure S3A). Following 6 hours of erlotinib treatment, however, we observed over 1200 DEGs, and following 24 hours of treatment, when doxorubicin sensitivity was maximally enhanced, we observed over 2000 DEGs (Figure 3A and S3B). By comparison, in the HER2+ MDA-MB-453 cells, which were desensitized to doxorubicin by erlotinib exposure, we observed only 235 DEGs following 24 hour exposure to erlotinib, and in hormonesensitive MCF7 cells only one gene was significantly altered (Figure 3B–C). Thus, the triple-negative BT-20 cells exhibited progressive and large-scale changes in gene expression following EGFR inhibition that were not observed in cell lines insensitive to the time-staggered ERL→DOX combination.
Figure 3. Triple-negative BT-20 cells are driven by oncogenic EGFR signaling.
(A–C) Differentially expressed genes (DEGs) following erlotinib treatment for 24 hrs vs. untreated cells. Cut-off for DEG was ≥2-fold change and a p-value ≤0.05 (genes that meet both criteria are colored red). B score is the log of the odds of differential expression. (D) DEGs classified using GeneGO “pathway maps”. Heatmap (left) colored according to –log (p-value); (right) p-value cut-off was 0.05 (dotted red line). (E and F) Microarray analysis using GSEA reveals loss of oncogene signatures in BT-20 cells after sustained EGFR inhibition. RAS Oncogenic Signature and false discovery rate (FDR) adjusted p-values shown in E. 11 oncogenic signatures from msigdb shown in F. Boxes are colored according to normalized enrichment score (NES). (G) GSEA reveals a switch from Basal to Luminal A genetic signature in BT-20 cells following sustained EGFR inhibition. Expression analyzed as in F using breast cancer subtype-specific genetic signatures as defined by Sorlie et al. (H) BT-20 cells lose the ability to form colonies in soft agar upon EGFR inhibition. Cells were untreated or treated with ERL, grown in soft agar, and monitored for colony formation 21 days later.
To examine which cellular processes were altered by long-term erlotinib treatment, DEGs in BT-20 cells were categorized by cellular process according to the GeneGO pathway annotation software (Ekins et al., 2007). Significant changes were observed in 16 of 34 GeneGO cellular networks, including those that mediate the DDR, apoptosis, and inflammation (Figure 3D). In contrast, DEGs in MDA-MB- 453 were not only fewer in number, but also lay in networks that did not overlap with those altered in BT-20 cells (Figure 3D). We further analyzed gene expression data using Gene Set Enrichment Analysis (GSEA), a tool for identification of enrichment or depletion of defined gene expression signatures within a rank ordered gene list (Subramanian et al., 2005). The most statistically significant changes in BT-20 cells upon sustained erlotinib exposure were loss of the Ras and MYC oncogenic signatures (Figure 3E). These signatures were not significantly altered in MDA-MB-453 or MCF7 cells treated with erlotinib for 24 hours, or in BT-20 cells exposed to erlotinib for 30 minutes (Figure 3E). Within the GSEA molecular signatures database, there exist 11 oncogenic signatures (Subramanian et al., 2005). GSEA of EGFR-inhibited BT-20 cells showed a similar depletion pattern for all 11 oncogenic signatures (Figure 3F). These changes were not consistently observed in either MDA-MB-453 cells or MCF7 cells following exposure to erlotinib.
Distinct gene expression patterns have been used to define breast cancer subtypes. BT-20 cells, like most triple-negative cells, display a “Basal-like” gene expression signature (Neve et al., 2006). Strikingly, analysis of our expression dataset revealed that chronic erlotinib treatment of BT-20 cells caused progressive time-dependent loss of Basal-like gene expression, with concomitant gain in Luminal A-like gene expression, a breast cancer subtype with the least aggressiveness and best overall prognosis (Figure 3G). In contrast, no such switch in breast cancer subtype patterns of gene expression was observed in HER2 over-expressing MDA-MB-453 cells or hormone sensitive MCF7 cells following erlotinib exposure.
These expression data suggest that the oncogenic potential of BT-20 cells is maintained by chronic EGFR-driven patterns of gene expression, and that this cell state could be remodeled through sustained inhibition of EGFR. To directly test this, we examined the ability of BT-20 cells to form colonies in soft agar, a classic test of transformation that typically shows good correlation with tumorigenic potential in vivo (Montesano et al., 1977). Consistent with the predictions derived from our GSEA, sustained EGFR inhibition with erlotinib potently inhibited soft agar colony formation (Figure 3H).
Creation of a data driven model for combined EGFR inhibition/DNA damage
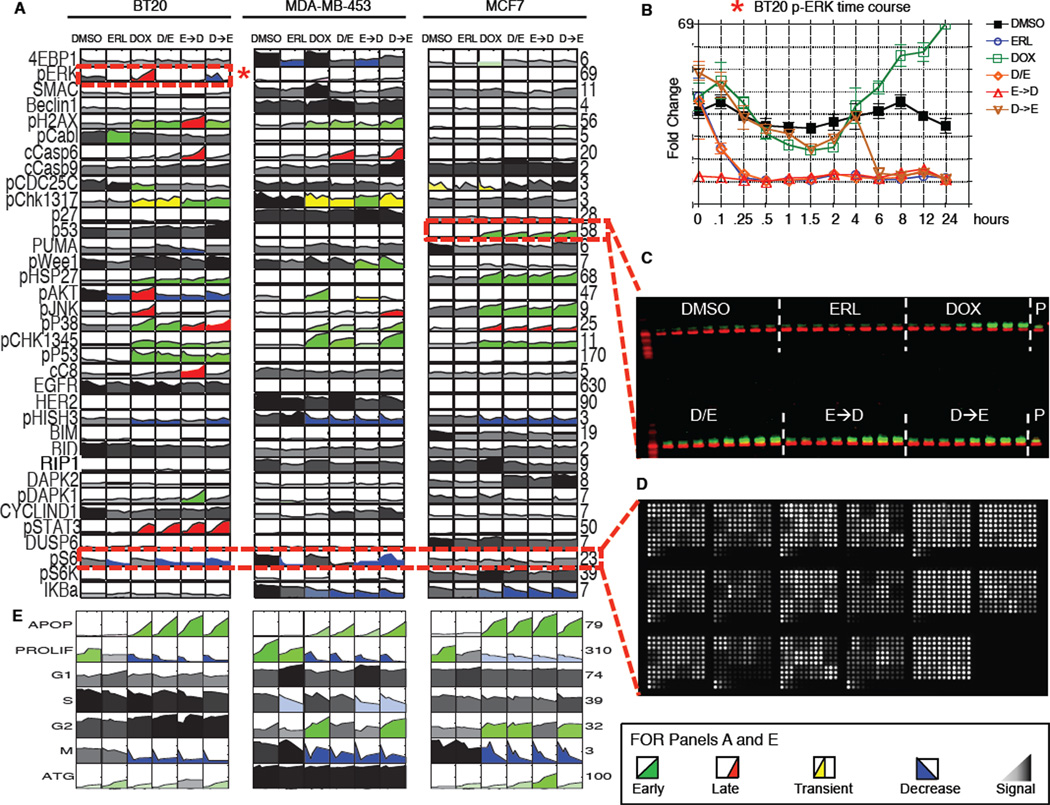
To better understand the biochemical changes in signaling that accompany time-staggered ERL→DOX treatment we used quantitative high-throughput reverse phase protein microarrays and quantitative Western blotting to measure the levels or activation states of 35 signaling proteins within multiple signaling pathways at 12 time points following exposure to erlotinib and doxorubicin individually and in combination (Figure 4A–D and see S4 for description of the selection of 35 proteins for analysis) (MacBeath, 2002). Oncogenic signaling networks typically exhibit multiple levels of feedback and crosstalk with other networks, rendering single protein measurements ineffective in predicting complex cellular responses to drugs such as those leading to DNA damage-induced apoptosis (Fitzgerald et al., 2006). We therefore constructed a multifactorial data-driven mathematical model relating signaling “inputs” to phenotypic “outputs. ” In addition to examining signaling pathways known to contribute to the DDR, we used our list of differentially expressed genes (Figure 3) to identify other proteins that might function as critical signaling nodes. This DEG-expanded list of signaling proteins extends far beyond the canonical components of the DDR, including proteins involved in apoptotic and non-apoptotic death, growth and stress responses, and cytokine/inflammatory signaling (Figure S4A). Specific proteins whose measurement was motivated by gene expression data included BIM, BID, capase-8, 4E-BP1, S6K, Stat3, DUSP6, and IKB. Phenotypic responses including cell cycle arrest and progression, autophagy, and apoptotic and non-apoptotic cell death were scored at 6 time points using luminescent microplate assays, flow cytometry, and automated microscopy (Figure 4E, S4C–F). All signaling and phenotypic response measurements were performed in biological and experimental triplicate in BT-20, MDA-MB-453, and MCF7 cells. In total, this dataset comprised more than 45,000 measurements of molecular signals, and 2,000 measurements of cellular responses (Figure 4A and 4E), revealing many changes in cell state and phenotype associated with drug exposure.
Figure 4. A systems level signal-response dataset collected using a variety of high-throughput techniques.
(A–D) (A) The complete signaling dataset for 3 different breast cancer sub-types following combined EGFR inhibition and genotoxic chemotherapy treatments as in Figure 1. Each box represents an 8- or 12-point time course of biological triplicate experiments. Time course plots are colored by response profile, with early sustained increases in signal colored green, late sustained increases colored red, and transient increases colored yellow. Decreases in signal are colored blue. Signals that are not significantly changed by treatment are shaded grey to black with darkness reflecting signal strength. Numbers to the right of each plot report fold-change across all conditions/cells. (B) Sample detailed signaling time course from panel A, highlighted by dashed box and asterisk, showing p-ERK activation in BT-20cells. Mean values ± S.D. of 3 experiments shown. (C) 48-sample Western blots analyzed using 2-color infrared detection. Each gel contained an antibody-specific positive control (P) for blot-toblot normalization. The example shown is one of three gels for total p53 in MCF7 cells (p53 in green; β-actin in red). (D) Reverse phase protein lysate microarrays were used to analyze targets of interest when array-compatible antibodies were available. The slide shown contains ~2,500 lysate spots (experimental and technical triplicates of all of our experimental samples, and control samples used for antibody calibration), probed for phospho-S6. (E) The complete cellular response dataset, colored as in A.
Several mathematical modeling approaches were employed to relate signaling data to cell phenotypes. Initial modeling efforts used principal component analysis (PCA) to identify co-variation between signals, while partial least squares (PLS) regression was used to identify statistically significant co-variation between molecular signals and corresponding cellular responses (Figure S4B) (Janes and Yaffe, 2006). In both PCA and PLS modeling, vectors were constructed whose elements contained quantitative measures of the level/state/activity of specific signaling proteins. The vectors were then reduced to a set of principal components, calculated so that each additional PCA or PLS dimension maximally captures information not captured by preceding components. This process was iteratively repeated until additional principal components ceased to capture meaningful data, as judged relative to experimental noise.
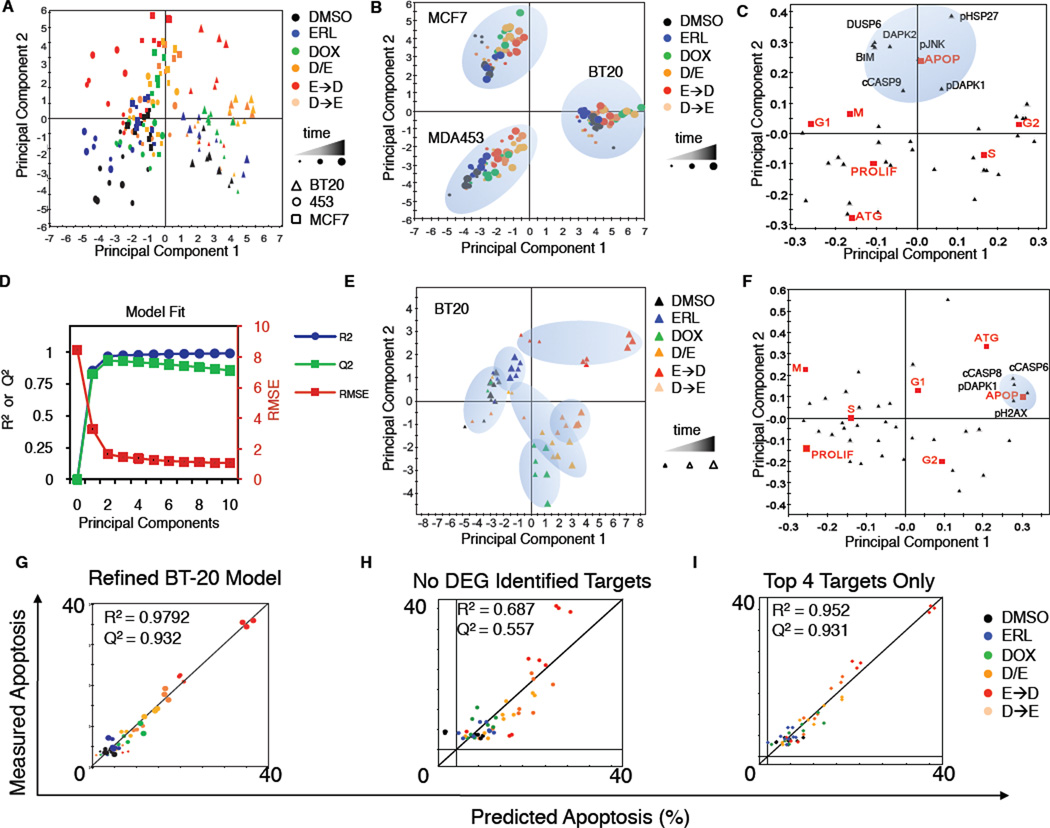
Following PCA, multiplex data from MDA-MB-453 cells projected negatively along principal component one (PC1), data from BT-20 cells projected positively along PC1, and MCF7 data were largely neutral (Figure 5A and S5A). Thus, the first principal component captured cell type-specific variance in the data. In contrast, data from all cell types projected similarly along PC2, but in a manner that was drug-dependent. Data from DMSO or erlotinib-treated cells not exposed to doxorubicin projected negatively along PC2 whereas data from cells co-treated with doxorubicin and erlotinib, or exposed to ERL→DOX sequentially, projected positively along PC2. Finally, data from cells treated with doxorubicin alone or DOX→ERL were largely neutral along PC2. Thus, the second principal component captured signaling variance from treatment-specific modulation of the signaling networks regardless of cell type (Figure 5A). These data suggest that, although significant differences in the state of the networks exist between cell lines, the drugs we applied modulated signaling networks in similar ways across all lines examined. PLS analysis linking signals to responses gave similar results, with differences between the cell lines now captured in both PC1 and PC2, and treatment-specific variance emerging in the third principal component PC3 (Figure 5B and S5B–E). The expected differences that we observed between these cell types captured by both PCA and PLS analyses confirms that the signaling molecules we measured can be used to define both the cell-type specific and drug treatment specific differences between these cells.
Figure 5. A PLS model accurately predicts phenotypic responses from time-resolved molecular signals.
(A) Principal components analysis of co-variation between signals. Scores plot represents an aggregate measure of the signaling response for each cell type under each treatment condition at a specified time, as indicated by the colors and symbols in the legend. (B and C) Scores and loadings for a PLS model. (B) Scores calculated and plotted as in panel A, except the principal components now reflect co-variation between signals and responses. (C) PLS loadings plotted for specific signals and responses projected into principal component space. (D–M) BT-20 cell line-specific model calibration. (D) R2, Q2, and RMSE for BT-20 models built with increasing numbers of principal components. (E, F) Scores and loadings plots, respectively, for a 2 component model of BT-20 cells. (G–I) Apoptosis as measured by flow cytometry, or as predicted by our model using jack-knife cross-validation. R2 reports model fit, and Q2 reports model prediction accuracy. (G) Final refined model of apoptosis in BT-20. (H) BT-20 model minus targets identified as DEGs in microarray analysis. (I) Model using only the top 4 signals: c-caspase-8, c-caspase-6, p-DAPK1, and pH2AX.
Based on these cell-type specific differences in the global PCA/PLS model we next built models for each cell line in isolation, focusing primarily on triple-negative BT-20 cells. To optimize the BT-20 PLS model, we compared fitness measures such as R2 (percent of variance captured by model), Q2 (percent of variance predicted by the model using a leave-one-out-cross-validation approach), and rootmean- squared error (RMSE; the mean deviation between model and data) across models containing increasing numbers of principal components. With BT-20 data alone, greater than 97 percent of the variance linking signals to responses under different conditions of drug treatment was captured by two principal components. Incorporation of additional components actually reduced the predictive ability of the model (Figure 5D), a common finding reflecting the addition of noise when components with little predictive value are added. Similar trends were observed for each of the other cell lines. To derive molecular understanding from the models, we projected the loading vectors (i.e. individual signals and responses) into PLS component space. We observed a strong anti-correlation between the apoptotic and proliferative responses (Figure 5C and 5F)) that was captured by the first principal component in the BT-20 model (Figure 5F) and by the second principal component in the aggregate cell line model (Figure 5C). To further test model quality, we compared each measured cellular response in isolation to that predicted by the model using jack-knife based cross-validation (Figure S5F-L). Our model was particularly accurate at predicting apoptosis following treatment (Figure 5G), and moderately good at predicting proliferation and autophagy (Figure S5K and S5L). Other responses (G1, G2, S) were not predicted as accurately, likely due to the limited dynamic range in our cell cycle response dataset (Figure S5G–J).
PLS modeling reveals chemo-sensitization following network re-wiring is driven by caspase-8 activation
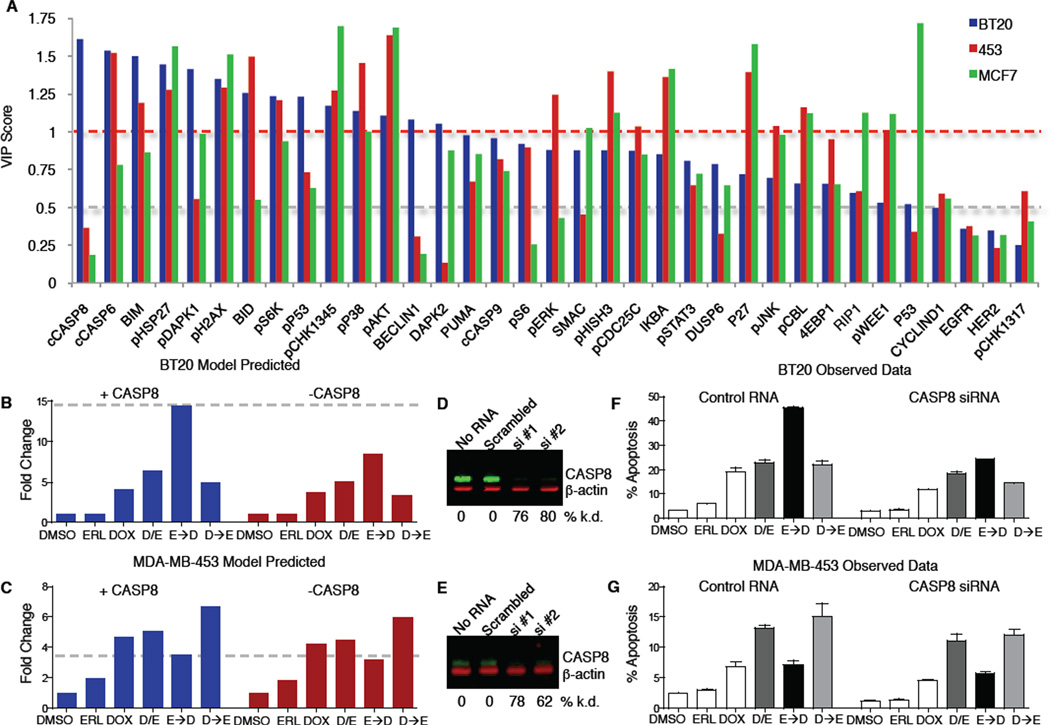
Since PLS models of individual cell lines could accurately predict apoptosis, we analyzed the models to identify specific proteins or signals that might account for the enhanced sensitivity of BT-20 cells to doxorubicin following EGFR inhibition. The BT-20 two-component PLS model identified four signals—cleaved caspase-8, cleaved-caspase-6, phospho-DAPK1, and phospho-H2AX—that were highly covariant with apoptosis (Figure 5F). Remarkably, a model including only these 4 signals was just as accurate at predicting apoptosis as the complete 35-signal model (Figure 5G–I). Notably, of these signals, only pDAPK1 would have been identified using the aggregate cell line PLS model (Figure 5C). We reasoned that the enhanced sensitivity of BT-20 cells to doxorubicin mediated by erlotinib pretreatment likely involved one of these molecular signals. We therefore calculated and plotted the “variable importance in the projection” (VIP) score for each signal (Figure 6A). The VIP score reports the sum (over all model dimensions) of each variable x (molecular signals in this case), weighted by the amount of the cellular response y (apoptosis) explained by variable x. Strikingly, caspase-8—an initiator caspase in death receptor-mediated apoptosis—was the single most important variable for predicting apoptosis in BT-20 cells, and simultaneously among the least important variables in MDA-MB-453 and MCF7 cells. Caspase-8 has previously been implicated in cell death mediated by EGFR inhibition in other contexts (Kang et al., 2010; Morgillo et al., 2011), however erlotinib alone did not cause death in any of our cell types. Instead, apoptosis in these cells, and the potential importance of caspase-8, resulted from their exposure to the genotoxic agent doxorubicin. In most cells, DNA damage activates cell intrinsic apoptosis mediated through caspase-9 (c.f. Figure 5C), not caspase-8 (Kim, 2005). Thus, the strong influence of caspase-8 was unexpected.
Figure 6. Enhanced sensitivity to doxorubicin is mediated by caspase-8 activation.
(A) VIP scores for predicting apoptosis plotted for each cell line specific PLS model. VIP score >1 indicates important x variables that predict y responses, while signals with VIP scores < 0.5 indicate unimportant x variables. (B, C) Model-generated predictions of apoptosis with (blue) or without (red) caspase-8 activation 8 hrs. after the indicated treatments in BT-20 (B) and 453 (C). (D, E) Western blot verifying caspase-8 knockdown in BT-20 (D) and453 (E). (F, G) Measured apoptosis 8 hrs. after the indicated treatment in cells expressing control RNA or caspase-8 siRNA. (F) BT-20. (G) 453. In both (F) and (G) apoptotic values represent mean response ± S.D. from both siRNAs, each in duplicate.
As an in silico test for the importance of caspase-8 in particular erlotinib/doxorubicin protocols, we set caspase-8 activity to zero in the model while leaving all other variables unchanged. The BT-20 model specifically predicted a dramatic decrease in the enhanced sensitivity to doxorubicin following sustained erlotinib treatment (Figure 6B), with much smaller decreases in apoptosis under all other treatments. In contrast, in the apoptosis model for MDA-MB-453 cells predicted no change following loss of caspase-8 activity under any conditions (Figure 6C). To test these predictions experimentally, 2 separate caspase-8 siRNAs were used in both BT-20 cells and MDA-MB-453 cells (Figure 6D and 6E). In excellent agreement with the model, knockdown of caspase-8 mitigated the enhanced cell death following erlotinib treatment in BT-20 cells, while having minimal effect on apoptosis following other treatment combinations (Figure 6F). Furthermore, caspase-8 knockdown had little effect on apoptosis in MDA-MB-453 under any condition (Figure 6G). To further assess model predictions and evaluate the relative importance of caspase-8 in the enhanced doxorubicin-induced apoptosis, we tested several other model-generated predictions, including proteins predicted to contribute strongly (caspase-6), moderately (Beclin-1) or weakly (RIP1) to apoptosis in BT-20 cells. Based on the VIP plot and loadings projections: caspase-6 is predicted to be a strong driver of the apoptotic response in BT-20 and MDA-MB-453 cells, but not MCF7 cells; Beclin-1 is predicted to be moderately anti-apoptotic in BT-20 cells, but have no role in the other cell lines; and RIP1 is predicted to be weakly anti-apoptotic in BT-20 and MDA-MB- 453 cells, but strongly anti-apoptotic in MCF7 cells. As shown in Figure S6, we were able to confirm these cell type dependences using siRNA, and confirm the relative magnitude of the effect of each target on the apoptotic response following various combinations of erlotinib and/or doxorubicin. Importantly, although caspase-6 contributed strongly to cell death in BT-20 cells, caspase 8 remained the strongest predictor. None of the other targets tested modulated the apoptotic response to the same extent as caspase-8, further highlighting its importance. Thus, the increased cell killing by ERL→DOX treatment in BT-20 cells appears to involve rewiring of the DNA damage response allowing activation of both cell intrinsic and extrinsic apoptotic programs to contribute to cell death.
Time-staggered inhibition of EGFR enhances apoptotic response in a subset of TNBC cells and other oncogene driven cells
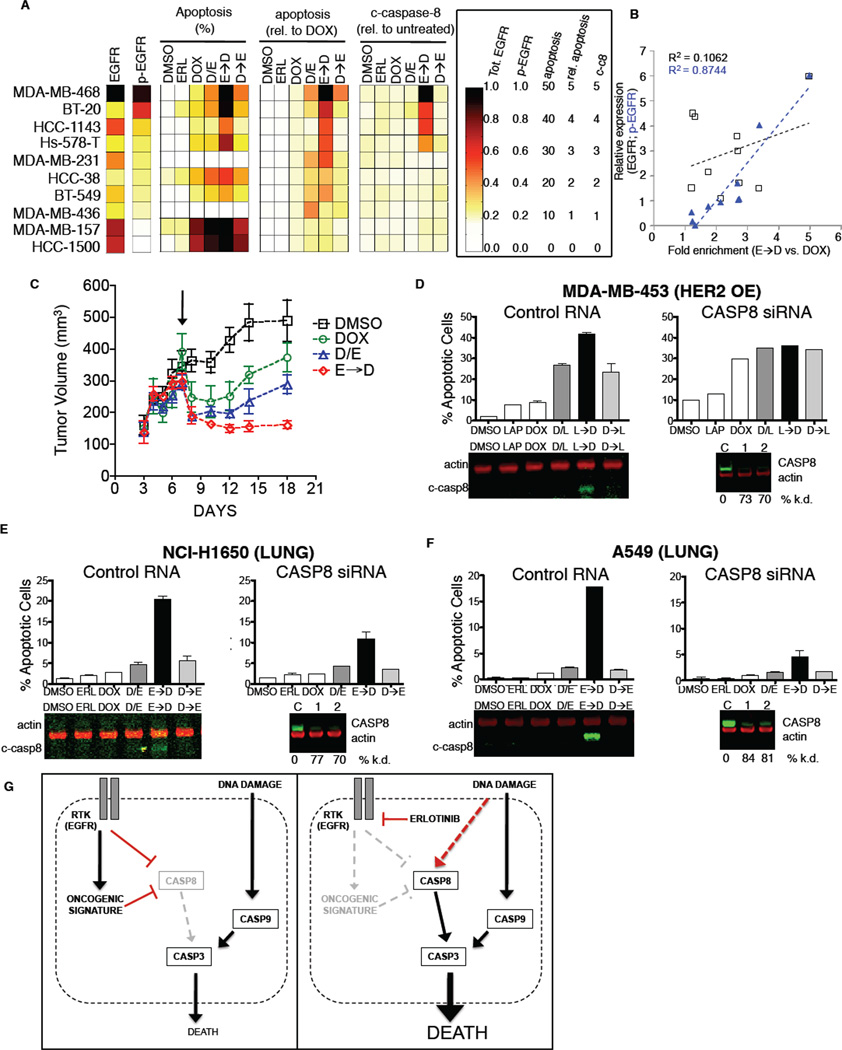
To examine whether the efficacy of time-staggered ERL→DOX treatment was unique to BT-20 cells or potentially a more general phenomenon of TNBC cells, we examined a handful of other triplenegative cell lines (Neve et al., 2006). The selected cell lines have markedly different growth rates, EGFR expression levels, and p53 states (Figure S7A). Despite these differences, sustained EGFR inhibition enhanced sensitivity to doxorubicin in 9 of 10 triple-negative cell lines tested. A synergistic effect, however, was observed in only 4 of the 10 TNBC lines (Figure 7A and S7A–B). To identify potential reasons for this, we measured total EGFR protein levels and basal EGFR activation by immunoblotting. Our quantitative measurement of EGFR protein expression was very similar to previously reported values (Neve et al., 2006), and correlated only very weakly with sensitivity to ERL→DOX treatment (Fig. 7A and 7B). In marked contrast, the levels of basal EGFR activity exhibited a much higher correlation (Figure 7A and 7B). Furthermore, in those TNBC cell lines in which ERL→DOX treatment was synergistic, we consistently observed caspase-8 cleavage following sequential administration, but not other drug treatments, suggesting a similar mechanism of enhanced apoptosis in these cells as that observed in BT-20 cells (Figure 7A and 7B). Taken in context with our observation that EGFR signaling drives expression of an oncogenic gene expression signature in BT-20 cells, these findings suggest that a subset of triple-negative cell lines are similarly driven by aberrant EGFR signaling. Importantly, however, these cells could not be distinguished by measuring EGFR gene amplification or EGFR abundance. Instead, they are unique in displaying high levels of activated (phosphorylated) EGFR as a biomarker of response to time-staggered EGFR inhibition and cytotoxic treatment.
Figure 7. Time-staggered inhibition of EGFR signaling enhances apoptotic response in a subset of TNBC cells and other EGFR driven cells.
(A) Panel of TNBC cell lines with a wide range of EGFR expression levels. Heatmap for total EGFR expression, p-EGFR (Y1173), percent apoptosis, apoptosis relative to DOX alone, and casp-8 cleavage. Apoptosis measured as in Figure 1. EGFR and p-EGFR expression measured by Western blotting of untreated cells. Cleaved casp-8 measured by Western blot 8 hrs. after exposure to DOX. (B) EGFR activity—but not total EGFR expression—is correlated with sensitivity to time-staggered ERL→DOX combination. Fold enrichment of cell death observed in E→D relative to DOX alone regressed against total EGFR or p-EGFR (pY1173) as measured in untreated cells for the 10 TNBC cell lines shown in Figure 7A. R2 reports the linear fit for each trend line. (C) BT-20 cells grown as xenograft tumors in nude mice. Arrow indicates intraperitoneal administration of indicated drugs. Mean tumor volume ± SEM shown from 4 animals for each treatment condition. (D–F) Time-staggered inhibition of HER2 in HER2 driven breast cancer cells (D) or EGFR in lung cancer cells (E–F) causes casp-8 activation and sensitization to DOX. Apoptosis measured as in Figure 1 for cells exposed to a control RNA (left in each panel) or siRNA targeting casp-8 (right in each panel). Caspase-8 activation was monitored 8 hrs. after doxorubicin treatment (c-casp8, shown beneath the Control RNA plots). Validation of caspase-8 knockdown shown below the CASP8 siRNA plots. (D) HER2 overexpressing MDA-MB-453 cells treated with lapatinib. (E–F) Lung cancer cells treated with erlotinib. (E) NCI-H1650. (F) A-549. (G) A model for enhanced cell death after DNA damage by chronic EGFR inhibition in triple-negative breast cancer cells.
We next investigated whether the initial chemo-sensitizing effects of an ERL→DOX protocol could be observed when treating EGFR-driven triple-negative tumors in vivo. BT-20 cells were injected into the flanks of nude mice, and tumors allowed to form for seven days before treatment with either doxorubicin alone, or erlotinib-doxorubicin combinations. Following a single dose of doxorubicin alone, a marked reduction in tumor volume was observed over the first three days after treatment. The residual tumors continued to grow, however, reaching pretreatment volume after approximately 14 days (Figure 7C). A similar trend was observed for tumors co-treated with erlotinib and doxorubicin, although the initial reduction in tumor size was greater. In contrast, when mice were given erlotinib 8 hours prior to doxorubicin, the tumors not only exhibited a similar initial reduction in size, but also failed to re-grow throughout the 14-day monitoring period. Thus, the chemo-sensitizing effect of sequential ERL→DOX treatment seen in culture was also observed in vivo. These results suggest that time-staggered inhibition of EGFR in combination with DNA damaging agents could be a potentially useful therapeutic strategy for treating a subset of triple-negative tumors, particularly those with high basal levels of phosphorylated EGFR.
We next examined whether the principle of time-staggered inhibition would sensitize other breast cancer subtypes to doxorubicin. In contrast to BT-20 cells, MDA-MB-453 cells were not sensitized by sustained EGFR inhibition, but instead were desensitized to DNA-damaging chemotherapy (Figure 1D). However, MDA-MB-453—and other widely used cell lines like BT-474—have a well-established oncogene addiction to HER2 (Neve et al., 2006). We therefore tested time-staggered inhibition of HER2 using the drug lapatinib (a potent inhibitor of both EGFR and HER2), in combination with doxorubicin in these cells. In both MDA-MB-453 and BT-474 cells, in contrast to the desensitization caused by pre-treatment with erlotinib, we observed that lapatinib pre-treatment enhanced sensitivity to doxorubicin to a similar extent as the enhancement observed with erlotinib in BT-20 and other EGFR driven TNBC cells (Figure 7C and S7C). Importantly, while all temporal combinations of lapatinib and doxorubicin were synergistic in HER2 over-expressing cells, pretreatment with lapatinib resulted in the largest increase in apoptosis. Furthermore, caspase-8 cleavage was only observed following LAP→DOX treatment of HER2-driven cells but not by other drug combinations. Knockdown of caspase-8 in these cells eliminated the specific component of enhanced cell death observed only in the pre-treatment condition (Figure 7C and S7C), suggesting that this portion of the overall cell death was driven by caspase-8 activity.
Finally, we examined whether the efficacy of time-staggered inhibition of EGFR was limited to breast cancer cells. Many lung cancers, for example, contain either high levels of phosphorylated wild-type EGFR or mutations within EGFR itself. We therefore tested our ERL→DOX treatment protocol on NCI-H1650 cells, a lung cancer cell that contains an in frame deletion that is commonly seen in lung cancers (Sordella et al., 2004), as well as A549 and NCI-H358, cells that have high levels of phosphorylated wild-type EGFR possibly due to HER2 amplification (Balko et al., 2006; Diaz et al., 2010; Helfrich et al., 2006; Rusnak et al., 2007). Remarkably, in all three lung cancer cell lines we found that time-staggered inhibition of EGFR using erlotinib caused a dramatic sensitization to killing by doxorubicin that was associated with caspase-8 cleavage (Figure 7E–F and S7D). Furthermore, knockdown of caspase-8 largely abrogated the enhanced cell death observed in the pre-treatment condition, exactly as seen in the setting of TNBCs. Thus, time-staggered inhibition of EGFR in cells with highly active EGFR signaling may be a generalizable approach to potentiate the effects of DNA damaging chemotherapy.
DISCUSSION
In this study, we describe a systematic time- and dose-dependent approach to identifying drug combinations that are efficacious in killing cancer cells depending on changes in the order and duration of drug exposure. We found that EGFR inhibition dramatically sensitizes a subset of TNBCs to DNA damage if the drugs are given sequentially but not simultaneously. Furthermore, our transcriptional, proteomic, and computational analyses of signaling networks and phenotypes in drug-treated cells revealed that the enhanced treatment efficacy results from dynamic network re-wiring of an oncogenic signature maintained by active EGFR signaling to unmask an apoptotic processes that involves activation of caspase-8. The enhanced sensitivity to damaging agents that we observed required sustained inhibition of EGFR because the phenotype did not result from the rapid direct inhibition of the oncogene, but rather from modulation of an oncogene driven transcriptional network as indicated schematically in the model shown in Figure 7G. Furthermore, our data suggest that it is activity of the EGFR pathway, rather than EGFR expression per se, that determines whether time-staggered inhibition will result in synergistic killing. Since EGFR can be activated through a diverse set of genetic alterations, some of which do not necessarily include EGFR itself (Sun et al., 2011), these findings highlight the need to understand network connectivity and dynamics (Pawson and Linding, 2008). Conversely, these observations suggest that EGFR phosphorylation may constitute a useful biomarker of response to time-staggered inhibition in at least some tumor types that are EGFR driven, including some TNBCs and lung cancers.
A key consequence of the erlotinib-dependent dynamic remodeling of the DDR network is activation of caspase-8 following DNA damage. The mechanism of caspase-8 activation is unclear, since it is generally thought to be specific to receptor-mediated apoptosis triggered by ligands such as TNF and TRAIL. Possibilities include feedback activation by caspase-3, possibly involving caspase-6 (Albeck et al., 2008); direct activation of death receptors by DDR proteins (Yoon et al., 2009); or an autocrine/paracrine mechanism involving an as-yet unidentified death ligand. Distinguishing between these and other possibilities will be a focus for future studies.
Combinatorial drug effects are complex, even for relatively specific drugs like EGFR inhibitors. Our understanding of compensation and network re-wiring is currently not sufficient to allow a priori predictions of the cellular response, particularly in cancer cells where signaling networks often exist in atypical forms. Our work highlights the utility of experimental examination of time-staggered combination treatments for their anti-cancer effects, particularly when combined with an analysis of signaling pathways and responses using mathematical modeling. These types of approaches may facilitate the identification of efficacious drug combinations and new therapeutic targets, and the design of different types of clinical trials, to study the killing of oncogene addicted tumors through druginduced dynamic re-wiring of signaling pathways.
EXPERIMENTAL PROCEDURES
Cellular response assays
Apoptosis
Following the treatment time course, cells were washed, trypsinized, fixed in 4% paraformaldehyde for 15 minutes at room temperature, resuspended in ice cold methanol and incubated overnight at −20°C. Cells were then washed twice in PBS-Tween and stained with antibodies against cleaved caspase-3 and PARP. Secondary Alexa-conjugated antibodies were used for visualization in a BD FacsCaliber flow cytometer.
Cell cycle analysis
Cells were fixed in 70% ethanolovernight at −20°C, permeabilized with 0.25% Triton X-100 for 20 minutes at 4°C, blocked with 1% BSA, and incubated with anti-phospho-Histone H3 antibody. Following washing, cells were incubated with Alexa488-conjugated secondary antibody on ice, washed, and stained with propidium iodide (PI) prior to analysis. Data were analyzed using the Dean-Jett-Fox algorithm.
Cell viability/proliferation
Cells were plated at 5,000 cells/well in 96-well plates. Metabolic viability was determined using CellTiterGlo (Promega) according to the manufacturer’s protocol.
Western blotting and antibodies
Cells were lysed in a manner that would allow samples to be used for both Western blot analysis and reverse phase protein microarray. See Supplemental Information for a detailed description of the cell lysis protocol and antibodies used in this study.
Data generated by quantitative Western blot was pre-processed prior to use in computational modeling. Raw signals for each protein target of interest were quantified and background subtracted using the Li-COR Odyssey software, divided by β-actin signals to normalize for loading differences, then each normalized signal was divided by a reference sample contained on each gel, for gel-to-gel normalization.
Reverse phase protein microarray
Reverse phase protein microarrays were printed on a fee-for-service basis through Aushon Biosystems. Validation of antibodies, staining and analysis of array data were performed as described previously (Sevecka and MacBeath, 2006).
Immunofluorescence Microscopy
Cells were seeded onto coverslips and treated for the indicated times. For autophagy analysis, cells were stably transfected with a mCHERRY-GFP-LC3 reporter construct. Cells were fixed and stained with primary antibody targeting either p-H2AX or 53BP1 and DAPI as above. Data reported are integrated intensity of pH2AX or 53BP1 foci per nucleus. For autophagy measurements, cells were scored positive” if the number of GFP and mCHERRY puncta significantly increased relative to untreated cells. ~100 cells were counted in a double-blind fashion per condition. Each experiment was performed in experimental triplicate.
RNA expression analysis by microarray
RNA was extracted from cells using the RNAeasy Kit (Qiagen). Affymetrix Human U133 Plus 2.0 microarrays were hybridized, labeled and processed on a fee-for-service basis through the MIT BioMicro Center. Microarray data were obtained from 3 independent biological replicates per time point, and analyzed using LIMMA.
Computational modeling and statistics
Unless otherwise noted, all statistical analyses were performed using Graphpad Prism, and graphs were created using Microsoft Excel, Spotfire, Matlab, DataRail, or SIMCA-P. Analysis of flow cytometry data was performed using FloJo. Analysis of RNA expression microarray data was performed either using GSEA or using GeneGO as indicated.
Data Driven Modeling
Data driven modeling and the application of partial-least squares to biological data have been described in detail previously (Janes and Yaffe, 2006). All data were variance scaled to non-dimensionalize the different measurements. Model predictions were made via cross-validation. Model fitness was calculated using R2, Q2, and RMSE, as described previously by Gaudet et al. (2005). Variable importance in projection (VIP) was calculated following Janes et al. (2008).
Xenograft Tumor Model
For in vivo tumor regression assays, 107 BT-20 cells in PBS were mixed 1:1 with matrigel on ice, injected subcutaneously into the hindflanks of nude mice (NCR nu/nu, Taconic). Tumors were allowed to form for 7 days. Mice were then treated i.p. with doxorubicin (4 mg/kg), or a combination of doxorubicin and erlotinib (25 mg/kg) with erlotinib either given at the same time as doxorubicin (D/E), or given 8 hours prior to doxorubicin (E→D). Tumors were monitored for 14 days after the treatment phase, and volume estimated using the ½ L × W2 formula. These experiments were approved by the Massachusetts Institute of Technology Committee on Animal Care (CAC).
Supplementary Material
HIGHLIGHTS.
The efficacy of combination treatments for cancer depends on drug order and timing
Dynamic re-wiring of signaling networks by drugs can provide therapeutic benefit
Sustained but not acute EGFR inhibition enhances TNBC killing by DNA damaging drugs
EGFR activity but not EGFR expression is a biomarker of response to this treatment
ACKNOWLEDGEMENTS
We thank Sandra Morandell, Kylie Huang, Jose McFaline, and Jessica Reddy for technical assistance in early stages of this study; the Koch Institute core facilities for their technical support, including Eliza Vasile (microscopy), Glen Paradis (flow cytometry), and especially Charlie Whittaker (bioinformatics) for his guidance in analyzing microarray experiments. We further thank Doug Lauffenburger and members of the CDP community for fruitful discussions and advice throughout the development and execution of this study. This work was supported by NIH grants CA112967 and GM68762 to P.K.S. and M.B.Y, ES015339 to M.B.Y. and DOD fellowship BC097884 to M.J.L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abeloff M, Wolff A, Weber B, Zaks T, Sacchini V, McCormick B. Cancer of the breast. In: Abeloff M, Armitage J, Niederhuber J, Kastan M, McKenna W, editors. Abeloff's Clinical Oncology. Maryland Heights, MO: Churchill Livingstone; 2008. [Google Scholar]
- Albeck JG, Burke JM, Spencer SL, Lauffenburger DA, Sorger PK. Modeling a snap-action, variable-delay switch controlling extrinsic cell death. PLoS Biol. 2008;6:2831–2852. doi: 10.1371/journal.pbio.0060299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balko JM, Potti A, Saunders C, Stromberg A, Haura EB, Black EP. Gene expression patterns that predict sensitivity to epidermal growth factor receptor tyrosine kinase inhibitors in lung cancer cell lines and human lung tumors. BMC Genomics. 2006;7:289. doi: 10.1186/1471-2164-7-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch A, Eroles P, Zaragoza R, Vina JR, Lluch A. Triple-negative breast cancer: molecular features, pathogenesis, treatment and current lines of research. Cancer Treat Rev. 2010;36:206–215. doi: 10.1016/j.ctrv.2009.12.002. [DOI] [PubMed] [Google Scholar]
- Carey L, Rugo H, Marcom P, Irvin W, Ferraro M, Burrows E, He X, Perou C, Winer E. TBCRC 001: EGFR inhibition with cetuximab added to carboplatin in metastatic triple-negative (basal-like) breast cancer. J Clin Oncol. 2008;26 [Google Scholar]
- Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- Corkery B, Crown J, Clynes M, O'Donovan N. Epidermal growth factor receptor as a potential therapeutic target in triple-negative breast cancer. Ann Oncol. 2009;20:862–867. doi: 10.1093/annonc/mdn710. [DOI] [PubMed] [Google Scholar]
- Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P, Narod SA. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2007;13:4429–4434. doi: 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- Diaz R, Nguewa PA, Parrondo R, Perez-Stable C, Manrique I, Redrado M, Catena R, Collantes M, Penuelas I, Diaz-Gonzalez JA, et al. Antitumor and antiangiogenic effect of the dual EGFR and HER-2 tyrosine kinase inhibitor lapatinib in a lung cancer model. BMC Cancer. 2010;10:188. doi: 10.1186/1471-2407-10-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekins S, Nikolsky Y, Bugrim A, Kirillov E, Nikolskaya T. Pathway mapping tools for analysis of high content data. Methods Mol Biol. 2007;356:319–350. doi: 10.1385/1-59745-217-3:319. [DOI] [PubMed] [Google Scholar]
- Fitzgerald JB, Schoeberl B, Nielsen UB, Sorger PK. Systems biology and combination therapy in the quest for clinical efficacy. Nat Chem Biol. 2006;2:458–466. doi: 10.1038/nchembio817. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Helfrich BA, Raben D, Varella-Garcia M, Gustafson D, Chan DC, Bemis L, Coldren C, Baron A, Zeng C, Franklin WA, et al. Antitumor activity of the epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor gefitinib (ZD1839, Iressa) in non-small cell lung cancer cell lines correlates with gene copy number and EGFR mutations but not EGFR protein levels. Clin Cancer Res. 2006;12:7117–7125. doi: 10.1158/1078-0432.CCR-06-0760. [DOI] [PubMed] [Google Scholar]
- Janes KA, Albeck JG, Gaudet S, Sorger PK, Lauffenburger DA, Yaffe MB. A systems model of signaling identifies a molecular basis set for cytokine-induced apoptosis. Science. 2005;310:1646–1653. doi: 10.1126/science.1116598. [DOI] [PubMed] [Google Scholar]
- Janes KA, Reinhardt HC, Yaffe MB. Cytokine-induced signaling networks prioritize dynamic range over signal strength. Cell. 2008;135:343–354. doi: 10.1016/j.cell.2008.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janes KA, Yaffe MB. Data-driven modelling of signal-transduction networks. Nat Rev Mol Cell Biol. 2006;7:820–828. doi: 10.1038/nrm2041. [DOI] [PubMed] [Google Scholar]
- Kang N, Zhang JH, Qiu F, Tashiro S, Onodera S, Ikejima T. Inhibition of EGFR signaling augments oridonin-induced apoptosis in human laryngeal cancer cells via enhancing oxidative stress coincident with activation of both the intrinsic and extrinsic apoptotic pathways. Cancer Lett. 2010;294:147–158. doi: 10.1016/j.canlet.2010.01.032. [DOI] [PubMed] [Google Scholar]
- Kim R. Recent advances in understanding the cell death pathways activated by anticancer therapy. Cancer. 2005;103:1551–1560. doi: 10.1002/cncr.20947. [DOI] [PubMed] [Google Scholar]
- Lichter AS, Lawrence TS. Recent advances in radiation oncology. N Engl J Med. 1995;332:371–379. doi: 10.1056/NEJM199502093320607. [DOI] [PubMed] [Google Scholar]
- Lopez JP, Wang-Rodriguez J, Chang C, Chen JS, Pardo FS, Aguilera J, Ongkeko WM. Gefitinib inhibition of drug resistance to doxorubicin by inactivating ABCG2 in thyroid cancer cell lines. Arch Otolaryngol Head Neck Surg. 2007;133:1022–1027. doi: 10.1001/archotol.133.10.1022. [DOI] [PubMed] [Google Scholar]
- MacBeath G. Protein microarrays and proteomics. Nat Genet. 2002;32(Suppl):526–532. doi: 10.1038/ng1037. [DOI] [PubMed] [Google Scholar]
- Montesano R, Drevon C, Kuroki T, Saint Vincent L, Handleman S, Sanford KK, DeFeo D, Weinstein IB. Test for malignant transformation of rat liver cells in culture: cytology, growth in soft agar, and production of plasminogen activator. J Natl Cancer Inst. 1977;59:1651–1658. doi: 10.1093/jnci/59.6.1651. [DOI] [PubMed] [Google Scholar]
- Morgillo F, D'Aiuto E, Troiani T, Martinelli E, Cascone T, De Palma R, Orditura M, De Vita F, Ciardiello F. Antitumor activity of bortezomib in human cancer cells with acquired resistance to anti-epidermal growth factor receptor tyrosine kinase inhibitors. Lung Cancer. 2011;71:283–290. doi: 10.1016/j.lungcan.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson T, Linding R. Network medicine. FEBS Lett. 2008;582:1266–1270. doi: 10.1016/j.febslet.2008.02.011. [DOI] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Rusnak DW, Alligood KJ, Mullin RJ, Spehar GM, Arenas-Elliott C, Martin AM, Degenhardt Y, Rudolph SK, Haws TF, Jr, Hudson-Curtis BL, et al. Assessment of epidermal growth factor receptor (EGFR, ErbB1) and HER2 (ErbB2) protein expression levels and response to lapatinib (Tykerb, GW572016) in an expanded panel of human normal and tumour cell lines. Cell Prolif. 2007;40:580–594. doi: 10.1111/j.1365-2184.2007.00455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs K, Perez O, Pe'er D, Lauffenburger DA, Nolan GP. Causal protein-signaling networks derived from multiparameter single-cell data. Science. 2005;308:523–529. doi: 10.1126/science.1105809. [DOI] [PubMed] [Google Scholar]
- Schechter AL, Stern DF, Vaidyanathan L, Decker SJ, Drebin JA, Greene MI, Weinberg RA. The neu oncogene: an erb-B-related gene encoding a 185,000-Mr tumour antigen. Nature. 1984;312:513–516. doi: 10.1038/312513a0. [DOI] [PubMed] [Google Scholar]
- Sevecka M, MacBeath G. State-based discovery: a multidimensional screen for smallmolecule modulators of EGF signaling. Nat Methods. 2006;3:825–831. doi: 10.1038/NMETH931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- Sordella R, Bell DW, Haber DA, Settleman J. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science. 2004;305:1163–1167. doi: 10.1126/science.1101637. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledgebased approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Aceto N, Meerbrey KL, Kessler JD, Zhou C, Migliaccio I, Nguyen DX, Pavlova NN, Botero M, Huang J, et al. Activation of multiple proto-oncogenic tyrosine kinases in breast cancer via loss of the PTPN12 phosphatase. Cell. 2011;144:703–718. doi: 10.1016/j.cell.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JG, Gump JL, Zhang C, Cook JM, Marchion D, Hazlehurst L, Munster P, Schell MJ, Dalton WS, Sullivan DM. ABCG2 expression, function, and promoter methylation in human multiple myeloma. Blood. 2006;108:3881–3889. doi: 10.1182/blood-2005-10-009084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winer EP, Mayer EL. Optimizing Treatment of "Triple-Negative" Breast Cancer. SABCS 2007: Improving Outcomes in Advanced and Metastatic Breast Cancer. 2007 [Google Scholar]
- Woehlecke H, Osada H, Herrmann A, Lage H. Reversal of breast cancer resistance protein-mediated drug resistance by tryprostatin A. Int J Cancer. 2003;107:721–728. doi: 10.1002/ijc.11444. [DOI] [PubMed] [Google Scholar]
- Wood ER, Truesdale AT, McDonald OB, Yuan D, Hassell A, Dickerson SH, Ellis B, Pennisi C, Horne E, Lackey K, et al. A unique structure for epidermal growth factor receptor bound to GW572016 (Lapatinib): relationships among protein conformation, inhibitor off-rate, and receptor activity in tumor cells. Cancer Res. 2004;64:6652–6659. doi: 10.1158/0008-5472.CAN-04-1168. [DOI] [PubMed] [Google Scholar]
- Yoon CH, Kim MJ, Park MT, Byun JY, Choi YH, Yoo HS, Lee YM, Hyun JW, Lee SJ. Activation of p38 mitogen-activated protein kinase is required for death receptor-independent caspase-8 activation and cell death in response to sphingosine. Mol Cancer Res. 2009;7:361–370. doi: 10.1158/1541-7786.MCR-08-0069. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.