Abstract
C. elegans vulval development is one of the best-characterized systems to study cell fate specification during organogenesis. The detailed knowledge of the signaling pathways determining vulval precursor cell (VPC) fates permitted us to create a computational model based on the antagonistic interactions between the epidermal growth factor receptor (EGFR)/RAS/MAPK and the NOTCH pathways that specify the primary and secondary fates, respectively. A key notion of our model is called bounded asynchrony, which predicts that a limited degree of asynchrony in the progression of the VPCs is necessary to break their equivalence. While searching for a molecular mechanism underlying bounded asynchrony, we discovered that the termination of NOTCH signaling is tightly linked to cell-cycle progression. When single VPCs were arrested in the G1 phase, intracellular NOTCH failed to be degraded, resulting in a mixed primary/secondary cell fate. Moreover, the G1 cyclins CYD-1 and CYE-1 stabilize NOTCH, while the G2 cyclin CYB-3 promotes NOTCH degradation. Our findings reveal a synchronization mechanism that coordinates NOTCH signaling with cell-cycle progression and thus permits the formation of a stable cell fate pattern.
Keywords: Caenorhabditis elegans , cell cycle, modeling, NOTCH, signal transduction
Introduction
During metazoan development, NOTCH signaling is involved in a multitude of cell fate decisions, especially when single cells are selected from a group of equivalent precursor cells (Fanto and Mlodzik, 1999; Baron et al, 2002). Well-studied examples include Drosophila wing formation, mammalian angiogenesis (Liu et al, 2003), or neuronal fate decisions (Hitoshi et al, 2002; Aguirre et al, 2010). Moreover, the NOTCH pathway is deregulated in different types of human cancer (Stylianou et al, 2006; Sharma et al, 2007). The binding of a DSL (Delta/Serrate/LAG-2) family NOTCH ligand activates two specific proteolytic cleavage events that result in the release of the NOTCH intracellular domain (NICD) from the plasma membrane (Baron, 2003). NICD then enters the nucleus, where it interacts with CSL (CBF1, Suppressor of Hairless, LAG-1) family transcription factors to induce the expression of target genes. Therefore, NOTCH signaling is often used as a cell fate switch that can be rapidly turned on. However, since NOTCH signaling also needs to be turned off at specific time points, the turnover of NICD in the nucleus is a critical aspect of NOTCH signaling. Experiments with mammalian cells have shown that NICD phosphorylation and/or deacetylation targets it for degradation (Fryer et al, 2004; Guarani et al, 2011). Furthermore, the F-box and WD repeat containing protein SEL-10/Fbw7 inhibits NOTCH signaling by inducing ubiquitination leading to proteasomal degradation of NICD (Hubbard et al, 1997; Gupta-Rossi et al, 2001). However, the specific mechanisms regulating NICD stability are largely unknown.
The development of the Caenorhabditis elegans hermaphrodite vulva is one of the best-studied systems to investigate the molecular mechanisms governing cell fate determination during organogenesis (Sternberg, 2005). NOTCH signaling has a prominent role in this process (Greenwald, 2005; Sundaram, 2005). During vulval induction, an organizer cell in the somatic gonad, called anchor cell (AC), induces the adjacent VPC P6.p the primary (1°) vulval cell fate by activating the epidermal growth factor receptor (EGFR) signaling pathway (Figure 1A). As a consequence of adopting the 1° fate, P6.p expresses the DSL ligands that activate lateral LIN-12 NOTCH signaling in the neighboring VPCs P5.p and P7.p. NOTCH signaling in P5.p and P7.p prevents these cells from adopting the 1° fate and induces the alternate secondary (2°) fate (blue arrows in Figure 1A). On the other hand, EGFR/RAS/MAPK signaling in P6.p inhibits NOTCH signaling by promoting the endocytosis and subsequent lysosomal degradation of LIN-12 NOTCH, thus allowing P6.p to irreversibly adopt the 1° cell fate (Shaye and Greenwald, 2002). The precise timing of the activation and inactivation of the EGFR/RAS/MAPK and NOTCH signaling pathways is essential to achieve a robust cell fate pattern during vulval development (Euling and Ambros, 1996; Ambros, 1999; Shaye and Greenwald, 2002; Fisher et al, 2007). One important question is therefore to identify the molecular mechanisms that link these signaling pathways to the temporal regulation of vulval development.
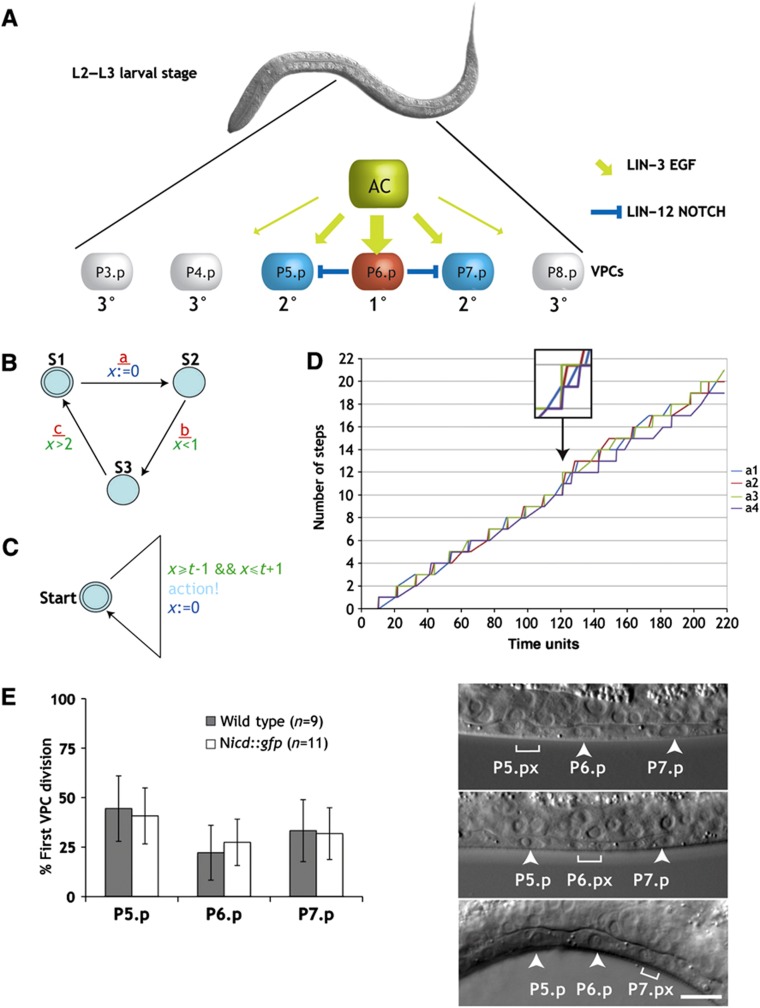
Figure 1.
A model for bounded asynchrony during VPC differentiation. (A) Intercellular signals determining the VPC fates. The AC signal induces the 1° fate in P6.p. P6.p then inhibits its neighbors P5.p and P7.p via lateral LIN-12 NOTCH signaling from adopting the 1° fate and induces the 2° fate. (B) An example illustrating the concept of timed automata. This automaton outputs possible sequences of a, b, and c events. The automaton starts in state S1 from where it can transit to state S2, signaling this with event a. After this transition, the automaton starts measuring time by setting clock x to 0. Then, the transition from S2 to S3 (with output event b) has to occur <1 time unit after a, and the transition from S3 back to S1 (with output event c) has to occur at least 2 time units after a. Overall, the observable communication events are rounds of a, b, and c events, where b is <1 time unit after a and c is at least 2 time units after a. There is no restriction on the time of the next a. (C) A timed automaton modeling an independent scheduler. A run starts in the state start and sets the clock x to 0. The automaton can perform an action between time t−1 and time t+1, and once it performs the action starts counting time again to the next action. Possible behaviors include sequences of actions where the time between every two actions is between t−1 and t+1. Running an independent copy of this automaton in each VPC gives rise to bounded asynchrony. (D) Random simulation of four copies of the timed automaton shown in (C) running in parallel. Every automaton moves between 9 and 11 time units. The first point where one process makes two actions more than another process is after 120 time units where a3 action was performed 12 times and a4 actions only 10 times (arrow and magnification). The earliest time point where such a difference is theoretically possible is after 99 time units, when a process moving very fast could perform 11 actions and a process moving very slowly could perform 9 actions. (E) The 1° and 2° VPCs divide asynchronously without a significant bias for one VPC entering M phase before the others. VPC divisions were observed in wild-type (gray bars) and nicd::gfp (white bars, see below) animals. The panels to the right show three examples of asynchronous VPC divisions where the brackets indicate the dividing VPC and the arrowheads the undivided VPCs. Error bars indicate the standard error as described in Materials and methods. The scale bar represents 10 μm. Source data is available for this figure in the Supplementary Information.
Computational models are excellent tools to describe and systematically analyze the dynamic behavior of a biological system and generate new hypotheses that can be tested experimentally (Kitano, 2002; Fisher and Henzinger, 2007). To this aim, we have previously developed a state-based computational model incorporating the current mechanistic understanding of gene interactions during C. elegans vulval development (Fisher et al, 2005). State-based models are particularly suitable for building mechanistic models of such well-studied biological systems, as they do not require quantitative data relating to the number of molecules or reaction rates. One characteristic feature of our model was the inclusion of multiple modes of crosstalk between the EGFR/RAS/MAPK and LIN-12 NOTCH signaling pathways (Berset et al, 2001; Fisher et al, 2007). Our computational model defines the behavior of biological objects (i.e., the VPCs) over time, based on the various states that an object can enter over its lifetime. Interacting state machines specify causal relationships between state changes in different objects to describe how objects communicate and collaborate. Usually, the state of an object is determined by the states of its parts such as the genes or proteins regulating the process of interest. Each part has its own reaction to the states of other parts. Changes in the state of an object are thus determined by the interdependent state changes of all parts. A hierarchical structure allows one to view a system at different levels of detail. State-based models have previously been used to model a variety of processes such as T-cell activation and differentiation in the thymus (Kam et al, 2001; Efroni et al, 2003, 2005), or pancreatic development (Setty et al, 2008).
In this study, we tested through an iterative process of computational modeling, prediction, and experimentation whether cell-cycle progression could provide a mechanism that coordinates the temporal activities of the different signaling pathways involved in VPC fate specification. Although the VPCs progress through the cell cycle in a largely cell-autonomous manner, global synchronization mechanisms imposed by the heterochronic genes that regulate cell-cycle checkpoints may prevent the VPCs from progressing in an uncoordinated manner (Euling and Ambros, 1996; van den Heuvel, 2005). To test this idea, we arrested cell-cycle progression in individual VPCs to ‘de-synchronize’ them. As predicted by our computational model based on bounded asynchrony (Fisher et al, 2008), this perturbation prevented VPCs from adopting a stable cell fate. Moreover, we found a synchronization mechanism acting on the NOTCH signaling pathway. In particular, the termination of the NOTCH signal is tightly coupled to cell-cycle progression, as the degradation of NICD occurs only after entry into the G2 phase. Thus, the formation of a stable cell fate pattern during vulval development involves a strict temporal control of NOTCH signaling during cell-cycle progression.
Results
A state-based model for VPC differentiation based on bounded asynchrony
Our previous models of C. elegans vulval development used high-level abstractions and described the biological process with an interacting state machine (Fisher et al, 2005, 2007). These models were constructed by defining the behavior of sub-components and then letting them work in parallel. In our initial model, there were six identical sub-components, each representing one VPC. There are two standard notions of such parallel composition: synchronous composition and asynchronous composition. Using synchronous composition, all sub-components perform actions together exactly at the same time. When we tried to model the VPC interactions, we found that a synchronous composition was too rigid, making it impossible to break the symmetry within the VPC equivalence group without introducing additional mechanisms for breaking symmetry. Indeed, as the VPCs are equipotent, they all proceeded in exactly the same way and it was only possible to differentiate them if they experienced differences in their environment. For example, in the case of mutants displaying an alternating 1° and 2° fate pattern, a model with a perfectly synchronous progression resulted in all VPCs adopting the 1° cell fate. On the other hand, using an asynchronous composition, where every sub-component performs actions independently, it was possible to differentiate the VPCs. However, some VPCs adopted a 1° instead of a 2° fate because they did not sense the progress of their neighbors and proceeded much faster than others. A completely asynchronous model of VPC differentiation therefore resulted in a variable and unpredictable pattern of cell fates due to excess ‘noise’ in the temporal progression of the VPCs. To limit the degree of asynchrony between the VPCs, we introduced the concept of bounded asynchrony, a notion of concurrency tailored to the modeling of biological cell–cell interactions (Fisher et al, 2008). Bounded asynchrony is achieved via a scheduler that limits the number of steps that one VPC is allowed to get ahead of the others. This allows the components of a system to move independently, while keeping them coupled to a certain degree. The constrained non-deterministic nature of a model incorporating bounded asynchrony captures the variability observed in cells that, although equipotent, assume distinct fates. Although a scheduler may be an artificial mechanism, it does highlight the need to precisely coordinate the timing of different events between the VPCs. Hence, we searched for a distributed computational mechanism that can keep the VPCs together for a long time and requires infrequent global synchronizations. Such a mechanism seems plausible since global synchronization mechanisms do exist in many biological systems including VPC differentiation (Euling and Ambros, 1996). Here, we used timed automata to build such a mechanism (Alur and Dill, 1994). The general principle of timed automata is illustrated in Figure 1B. Abstractly, each timed automaton represents a VPC that takes approximately t time units to perform an action (Figure 1C). We therefore allowed the timed automaton to perform the action between times t−1 and t+1. Depending on the value of t, the system can perform approximately t rounds (i.e., every cell t actions), allowing each cell to perform at the maximum one more action than the other cells. A simulation composed of four such timed automata, where each automaton moves after ∼10 time units, is shown in Figure 1D. In this exemplary simulation, up to time point 120 the difference in number of steps between cells (Δx) does not exceed 1. At time point 120, ‘cell’ a3 (green line) has taken 12 steps, while ‘cell’ a4 (violet line) has only taken 10 steps (Δx=2). The mechanism ensures that Δx=1 up to time point 99, after which the potential difference increases gradually. If an additional mechanism synchronizes these automata every 100 time units, then the difference Δx will continue to be restricted to 1 indefinitely. Thus, a model that combines approximate time keeping together with global synchronization might be an appropriate way to represent the variability that is inherent to all cellular systems.
VPCs enter M phase in a random order
One feature of bounded asynchrony is the fact that the sub-components may enter the next state in an arbitrary order. If VPC differentiation was to be governed by a mechanism resembling bounded asynchrony, then the VPCs should indeed progress through the cell cycle in a slightly asynchronous manner. To address this question, we examined the order in which the VPCs enter the M phase of the cell cycle. For this purpose, we observed the divisions of the proximal VPCs P5.p, P6.p, and P7.p in wild-type larvae using Nomarski optics and scored the time of entry into prometaphase based on nuclear envelope breakdown (Figure 1E). Among the three proximal VPCs, we observed no bias in the order at which they entered M phase, as each of the three VPCs was equally likely to divide first. Using time-lapse imaging, we measured the temporal differences in VPC divisions. The average time span between the first and last proximal VPC divisions was 33.4 min with a maximum divergence of 71 min (n=6). These results indicate that the VPCs adopting 1° and 2° cell fates do indeed enter the M phase in a random order, suggesting that the cell cycle of the VPCs progresses in a slightly asynchronous and unbiased manner.
Cell-cycle arrest interferes with VPC fate specification
We next tested the biological significance of the computational concept of bounded asynchrony by manipulating the cell cycle in single VPCs. We hypothesized that, although the VPCs progress with slightly variable speed through the cell cycle, external mechanisms may keep them ‘more or less’ synchronized and thereby align the relative timing of the signal transduction events between the VPCs. Under this assumption, the repeated use of cell-cycle checkpoints may represent such a synchronization mechanism. The VPCs are born during the first larval stage and remain in the G1 phase until the transition from the second to the third larval stage (Euling and Ambros, 1996). During the second larval stage, 1° cell fate markers can be detected in P6.p, but the other five VPCs remain uncommitted (Ambros, 1999). After the G1/S-phase arrest has been relieved, P5.p and P7.p adopt the 2° fate during the G2 phase, while the 3°, uninduced fate of the distal VPCs (P3.p, P4.p, and P8.p) is only sealed after these cells have divided and fused with the surrounding hypodermis (hyp7).
We therefore performed an experiment to specifically arrest the cell cycle in P6.p without affecting the progression of the other VPCs. For this purpose, we expressed the cyclin-dependent kinase (CDK) inhibitor cki-1 under control of the 1° fate-specific egl-17 promoter (egl-17p::cki-1) to arrest P6.p in the G1 phase without affecting cell-cycle progression in the other VPCs (Hong et al, 1998). We then observed the effect of this intervention on the VPC fate specification using 1° and 2° cell fate markers. As a readout for the 1° cell fate, we used a transcriptional gfp reporter of the RAS/MAP kinase target egl-17 (Burdine et al, 1998), and for the 2° fate, we observed a reporter of the LIN-12 NOTCH target lip-1 (Berset et al, 2001).
In egl-17p::cki-1 animals, in which P6.p was arrested and P5.p and P7.p had progressed to the Pn.px or Pn.pxx stage, expression of the 2° fate marker lip-1p::gfp persisted in the undivided P6.p cells at levels that were significantly higher than those detected in control animals (Figure 2A). To rule out the possibility that the differences in lip-1p::gfp levels between arrested and non-arrested cells are due to a dilution of the GFP signal during the cell divisions, we corrected the signal intensities for the total nuclear areas. Even with this correction, we detected a significant difference in total lip-1p::gfp signal intensity between arrested P6.p and non-arrested P6.p descendants (Supplementary Figure S1). Furthermore, expression of the 1° fate marker egl-17p::gfp in arrested P6.p cells was not affected (Supplementary Figure S2), confirming the previous observation that 1° fate specification begins already during the G1 phase (Ambros, 1999). However, our results also indicate that G1-arrested P6.p cells could not establish a fate-specific gene expression pattern, as they simultaneously expressed 1° and 2° cell fate markers. We conclude that a coordinated cell-cycle progression of the VPCs is necessary for the specification of a stable cell fate as judged by the expression of cell-fate markers.
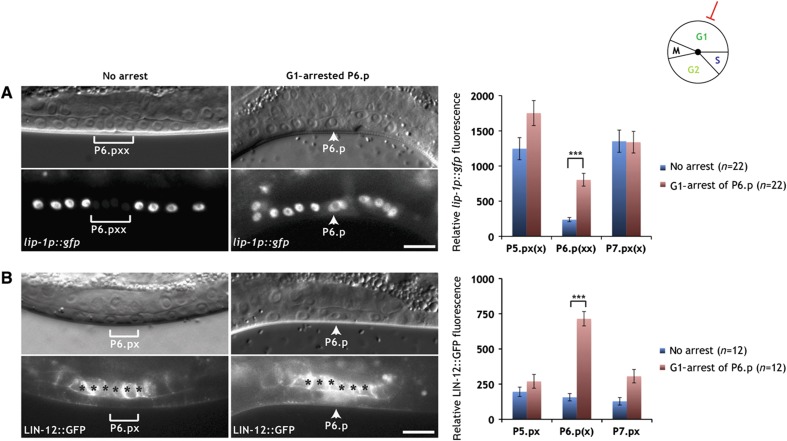
Figure 2.
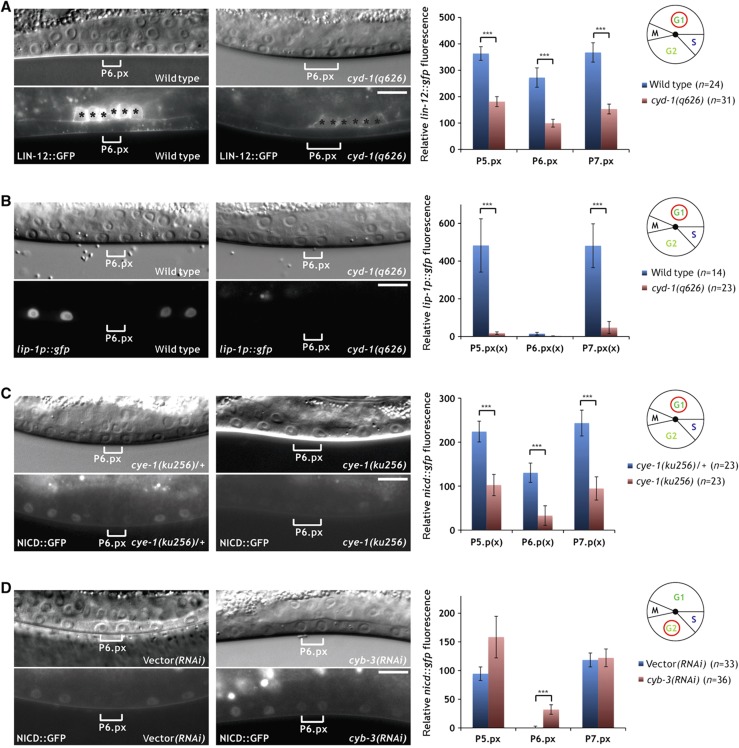
Degradation of LIN-12 NOTCH is blocked in G1-arrested P6.p. (A) Expression of the 2° fate marker lip-1p::gfp persists in the G1-arrested P6.p cell of an egl-17p::cki-1 transgenic animal (arrowheads in the right panels), while lip-1p::gfp expression is downregulated in the P6.p descendants of a sibling that had lost the egl-17p::cki-1 array (brackets in the left panels). (B) LIN-12::GFP persists in the G1-arrested P6.p cell of an egl-17p::cki-1 transgenic animal (arrowheads in the right panels), while LIN-12::GFP is efficiently degraded in the P6.p descendants of a sibling that had lost the egl-17p::cki-1 array (brackets in the left panels). Note the accumulation of the GFP signal in the nucleus and cytoplasm of the arrested P6.p cell. Black asterisks denote uterine LIN-12::GFP expression. In all panels, the corresponding Nomarski images are shown on top. The scale bars represent 10 μm. Quantifications of lip-1p::gfp and intracellular LIN-12::GFP expression are shown to the right. Error bars indicate the standard error and asterisks indicate the significance as described in Materials and methods. Source data is available for this figure in the Supplementary Information.
Termination of NOTCH signaling is linked to cell-cycle progression
The finding that G1-arrested P6.p cells continued to express the 2° cell fate marker suggested that G1 cell-cycle arrest in P6.p might block the proper termination of LIN-12 NOTCH signaling. To investigate this possibility, we examined the expression of a functional, translational LIN-12::GFP reporter that reflects the expression pattern and sub-cellular localization of the endogenous LIN-12 protein (Levitan and Greenwald, 1998; Shaye and Greenwald, 2002). Before vulval induction, LIN-12::GFP is expressed at low levels in all VPCs. Most of the LIN-12 protein is localized to the apical plasma membrane of the VPCs. After vulval induction, LIN-12 is upregulated in the 2° P5.p and P7.p lineages and downregulated in the 1° P6.p lineage.
In control animals lacking the egl-17p::cki-1 transgene, LIN-12::GFP was absent from the 1° P6.p descendants at the Pn.px stage, because LIN-12 NOTCH undergoes rapid endocytosis and degradation in the 1° lineage (Shaye and Greenwald, 2002). In contrast, in egl-17p::cki-1 transgenic animals at the Pn.px stage, in which P6.p had remained undivided, we detected elevated levels of LIN-12::GFP in the cytoplasm and nucleus of P6.p (Figure 2B). However, the LIN-12::GFP signal was lost from the apical plasma membrane, indicating that the endocytosis of the full-length LIN-12 protein in P6.p is not affected by G1 arrest. Since the GFP tag in LIN-12::GFP is inserted in the intracellular domain (NICD) that is cleaved off the transmembrane domain during LIN-12 NOTCH activation, the persisting cytoplasmic and nuclear GFP signal in G1-arrested P6.p cells suggested that the downregulation of the cleaved, intracellular LIN-12 NICD occurs only after exit from the G1 phase. In this way, the termination of the LIN-12 NOTCH signal in the 1° lineage might be coupled to cell-cycle progression.
Degradation of NICD is blocked in G1-arrested VPCs
To directly test the connection between lateral signaling, NICD degradation, and the cell cycle, we integrated via the MosSCI technique (Frokjaer-Jensen et al, 2008) a single copy of an nicd::gfp transgene expressed under control of a bar-1 promoter fragment into the genome at a specific site on chromosome II (zhIs39[nicd::gfp]). Importantly, the bar-1 promoter fragment used drives uniform expression in the six VPCs and their descendants until the Pn.pxx stage (Natarajan et al, 2004; Figure 6A). Thanks to the single copy integration, we achieved a stable and reproducible NICD::GFP expression in the VPCs. The increased dosage of NOTCH signaling caused by nicd::gfp resulted in the ectopic induction of the 2° fate in P3.p, P4.p and P8.p and a Multivulva phenotype in all of the animals examined (Table I, row 1; Supplementary Figure S3). However, P6.p in nicd::gfp animals still adopted the 1° fate as determined by its cell lineage (see insets in Supplementary Figure S3). Moreover, the proximal VPCs in nicd::gfp animals entered M phase in a random order similar to the wild type (Figure 1E).
Table 1. Positive regulation of NOTCH signaling by G1 cyclins.
| Row | Genotype | Induction±s.e. | n |
|---|---|---|---|
| 1 | zhIs039[nicd::gfp] | 5.85±0.04 | 36 |
| 2 | zhEx500[nicd::gfp] | 5.27±0.12]*** | 34 |
| 3 | cyd-1(q626); zhEx500[nicd::gfp] | 4.63±0.14 | 36 |
| 4 | cye-1(ku256)/+; zhIs39[nicd::gfp] | 5.96±0.03]*** | 56 |
| 5 | cye-1(ku256); zhIs39[nicd::gfp] | 5.52±0.09 | 30 |
Induction indicates the average number of induced VPCs (1° or 2° fate) per animal and s.e. the standard error determined by bootstrapping (see Materials and methods). ***Indicates a significance of P<0.001 and n the number of animals scored for each genotype. For cye-1(ku256), the homozygous mutants were compared with their heterozygous siblings on the same plates. Source data is available for this table in the Supplementary Information.
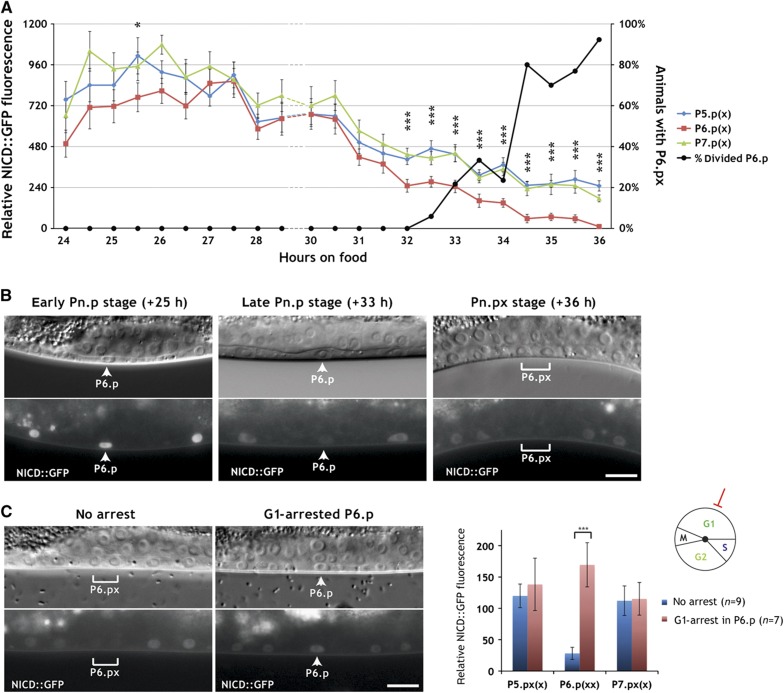
We next performed a time-course analysis to follow NICD::GFP degradation in the induced VPCs and their descendants (Figure 3A and B). NICD::GFP was uniformly expressed in the VPCs of early L2 larvae (+24 h relative to hatching, see Materials and methods). Interestingly, NICD::GFP expression first increased until the mid L2 stage (+27 h) and then started to decline in all VPCs. A significant difference between the 1° and 2° VPCs could first be detected at +32 h, shortly before the first VPCs started dividing (Figure 3A). After this time point, NICD::GFP expression rapidly faded in P6.p. At the Pn.px stage, NICD::GFP was barely detectable in the 1° P6.p descendants, while expression persisted in the 2° P5.p and P7.p descendants, though at lower levels when compared with Pn.p stage animals (right panel in Figure 3B). By the Pn.pxx stage, NICD::GFP was undetectable in P6.p descendants and further reduced in P5.p and P.7.p descendants. Similarly to full-length LIN-12::GFP, NICD::GFP in egl-17p::cki-1 animals persisted in the nuclei of the arrested P6.p cells (Figure 3C). Thus, NICD::GFP is progressively degraded in the induced VPCs and their descendants. Though, the rate of NICD degradation is significantly higher in the 1° than in the 2° lineage and depends on cell-cycle progression.
Figure 3.
Time-course analysis of NICD degradation. (A) NICD::GFP levels in P5.p, P6.p, and P7.p were measured every half an hour in around 20 synchronized animals at each time point (left scale). The time is indicated in hours after hatching (see Materials and methods). Error bars indicate the standard errors and asterisks significant differences between 1° and 2° lineages as described in Materials and methods. The percentage of animals with at least one proximal VPC division is indicated by the black line (right scale). (B) Representative pictures of NICD::GFP expression in an early L2 (+25 h, Pn.p stage), a late L2 (+33 h, Pn.p stage), and an early L3 (+36 h, Pn.px stage) larva. Note the uniform NICD::GFP expression in the VPCs at +25 h and the downregulation in P6.p at +33 h (arrowheads) and in P6.px at +36 h (bracket). (C) NICD::GFP persists in the G1-arrested P6.p cell of an egl-17p::cki-1 transgenic animal (arrowheads in the right panels), whereas NICD::GFP is downregulated in the P6.p descendants of a sibling that had lost the egl-17p::cki-1 array (brackets in the left panels). In all panels, the corresponding Nomarski images are shown on top. The scale bars represent 10 μm. A quantification of NICD::GFP expression is shown to the right. Error bars indicate the standard error and asterisks indicate the significance as described in Materials and methods. Source data is available for this figure in the Supplementary Information.
A computational model of VPC fate specification based on the cell cycle
Based on the experimental evidence, which indicated a linkage of VPC fate specification to cell-cycle progression, we incorporated the cell cycle as a key timing factor in our revised computational model. To implement bounded asynchrony in our refined computational model, we used an independent scheduler that recreates the same interactions between cells by limiting the number of steps one process gets ahead of the other as exhibited by the timed automata model described earlier. In our previous VPC models, an artificial delay in LIN-12 NOTCH downregulation was necessary to reproduce the proper behavior of lin-12 and predict the correct fate patterns in lateral signaling mutants (Fisher et al, 2007). With the introduction of the cell cycle, we removed this artificial delay and coupled the inhibition of LIN-12 to the state of the cell-cycle phases G1, S, or G2 (see Supplementary information for a detailed description of the computational model). In a first in silico experiment, we allowed for the inhibition of lateral LIN-12 signaling to happen during the G1 phase. However, this configuration of the model failed to reproduce published experimental data. We further tested a model where inhibition of LIN-12 Notch was forbidden in the G1 phase but allowed during the S or G2 phase. This model was consistent with published experimental results (checked using model checking, see Supplementary Table 2). Furthermore, the coupling between cell-cycle progression and LIN-12 NOTCH degradation was implemented in all VPCs in an identical manner (see Supplementary Figure S17). However, when executing the model we found that LIN-12 levels in VPCs progressing toward a 1° fate stabilized at lower levels than in VPCs adopting a 2° fate. Taken together, the notion of bounded asynchrony introduced into our computational model through the cell cycle predicted that a temporal delay in the termination of the LIN-12 NOTCH signal after G1 is critical for the formation of a stable cell fate pattern.
NICD degradation occurs during the G2 phase
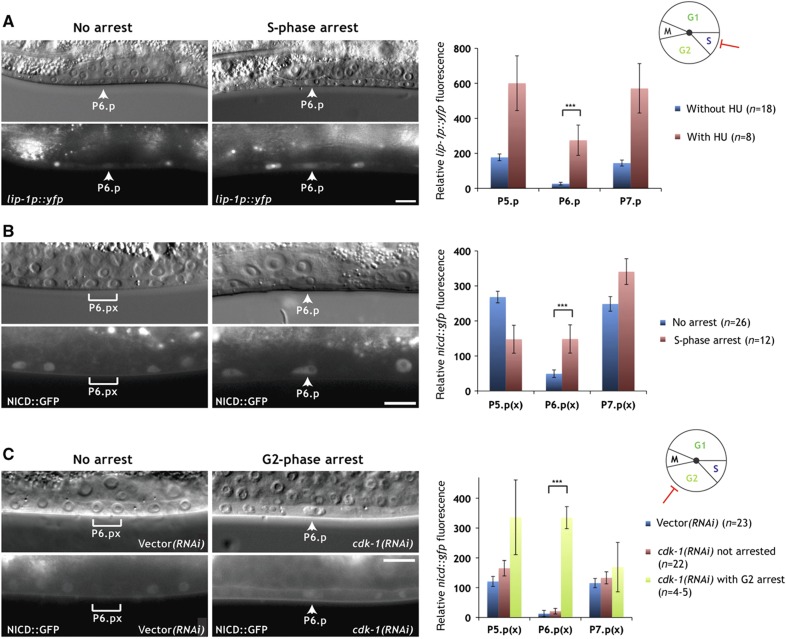
To further narrow down the time window, during which NICD is degraded, we arrested the VPCs in the S phase by hydroxyurea treatment (Ambros, 1999). Expression of the 2° cell fate reporter lip-1p::yfp persisted in the arrested P6.p cells and increased in the adjacent P5.p and P7.p cells (Figure 4A). Overall, lip-1p::yfp reached significantly higher levels in S phase-arrested VPCs compared with the VPCs of untreated control animals. Accordingly, the degradation of NICD::GFP in P6.p was blocked in hydroxyurea-treated animals (Figure 4B).
Figure 4.
NICD is degraded during the G2 phase. (A) Expression of the 2° fate marker lip-1p::yfp persists in P6.p of a hydroxyurea (HU)-treated larva (arrowheads in the right panels), while expression is downregulated in an untreated control animal at the Pn.p stage (arrowheads in the left panels). (B) NICD::GFP persists in the undivided P6.p cell of a hydroxyurea-treated larva (arrowheads in the right panels), but NICD::GFP is downregulated in the P6.p descendants of an untreated control larva at the Pn.px stage (brackets in the left panels). (C) NICD::GFP persists in the G2-arrested P6.p cell of a cdk-1 RNAi-treated animal at the Pn.px stage (arrowheads in the right panels), while NICD::GFP is downregulated in the P6.p descendants of an empty vector RNAi-treated control animal (brackets in the left panels). Note that due to the incomplete penetrance of the cdk-1 RNAi effect, only P6.p had arrested in the example shown in the right panels. In all panels, the corresponding Nomarski images are shown on top. The scale bars represent 10 μm. Quantifications of lip-1p::yfp and NICD::GFP expression are shown to the right. Error bars indicate the standard error and asterisks indicate the significance as described in Materials and methods. Source data is available for this figure in the Supplementary Information.
We then arrested the VPCs in the late G2 phase by RNAi-mediated knockdown of cdk-1, which is essential for the G2-to-M phase progression (Mori et al, 1994; Boxem et al, 1999). Since the cdk-1 RNAi phenotype is incompletely penetrant, this procedure resulted in the random arrest of single or multiple VPCs per affected animal. Similarly to hydroxyurea-treated animals, the undivided VPCs in cdk-1 RNAi animals expressed elevated levels of NICD::GFP, and the bias in P6.p versus P5.p and P7.p-specific expression was lost in the G2-arrested VPCs (Figure 4C). Taken together, the cell-cycle arrest and time-course experiments indicate that NICD is degraded in the VPCs either during the late G2 phase or at the time of entry into the M phase. The persisting expression of NICD::GFP in VPCs arrested by cdk-1 RNAi may also indicate a direct involvement of a CDK-1/Cyclin complex in NICD degradation (see below).
The G1 cyclins CYE-1 and CYD-1 promote while the G2 cyclin CYB-3 inhibits NOTCH activity
To investigate which CDK/Cyclin complexes are responsible for the changes in NOTCH stability and signaling activity during cell-cycle progression, we examined the expression of the full-length LIN-12::GFP and the NICD::GFP reporters as well as the expression of the target gene lip-1 in different cyclin mutants.
We first examined the influence of the G1-specific D-type cyclin gene cyd-1 on NOTCH signaling using the reduction-of-function allele cyd-1(q626) (Tilmann and Kimble, 2005). Although no VPC cell-cycle arrest was observed, cyd-1(q626) mutants displayed reduced levels of LIN-12::GFP in P5.p and P7.p (Figure 5A). Moreover, the expression of the LIN-12 target gene lip-1 was strongly decreased in the P5.p and P7.p descendants of cyd-1(q626) mutants (Figure 5B). Due to the proximity of the nicd::gfp integration site to the cyd-1 locus on LGII, it was not possible to examine NICD::GFP expression in cyd-1(q626) animals. However, cyd-1 RNAi did not cause an obvious reduction in NICD::GFP expression in the VPCs. Rather, in the six cases, in which P6.p was arrested, P6.p continued to express NICD::GFP at high levels similar to the expression pattern observed in egl-17p::cki-1 animals (Supplementary Figure S4; Figure 3C).
Figure 5.
Differential regulation of NOTCH signaling by G1 and G2 cyclins. (A) Localization of LIN-12::GFP to the apical plasma membrane is diminished in the VPC descendants in cyd-1(q626) mutants (right panels) when compared with wild-type controls (left panels). Black asterisks denote uterine LIN-12::GFP expression. (B) Expression of the 2° fate marker lip-1p::gfp is reduced in the P5.p and P7.p descendants in cyd-1(q626) mutants (right panels) compared with wild-type controls (left panels). (C) NICD::GFP expression is reduced in the VPC descendants in homozygous cye-1(ku256) mutants (right panels) when compared with heterozygous cye-1(ku256)/+ controls (left panels). (D) NICD::GFP expression persists in the P6.p descendants of a cyb-3 RNAi-treated animal at the Pn.px stage (brackets in the right panels). Note that in contrast to cdk-1 (Figure 4C), cyb-3 RNAi did not induce a cell-cycle arrest. An empty vector RNAi-treated control animal is shown in the left panels. In all panels, the corresponding Nomarski images are shown on top. The scale bars represent 10 μm. Quantifications of LIN-12::GFP, lip-1p::gfp and NICD::GFP expression are shown to the right. Error bars indicate the standard error and asterisks indicate the significance as described in Materials and methods. Source data is available for this figure in the Supplementary Information
Furthermore, we tested if the cyd-1(q626) mutation modifies the vulval phenotype caused by NICD::GFP expression. For this purpose, we used an extrachromosomal nicd::gfp array (zhEx500) to score vulval induction in the cyd-1(q626) background. The cyd-1(q626) mutation significantly decreased vulval induction in animals carrying the zhEx500[nicd::gfp] array when compared with cyd-1(+); zhEx500[nicd::gfp] controls (Table I, rows 2, 3). Taken together, we conclude that cyd-1 controls NOTCH signaling at the level of the full-length LIN-12 receptor and possibly also downstream of NICD at the level of target gene expression. Since NOTCH signaling prevents the degradation of full-length LIN-12 (Levitan and Greenwald, 1998; Shaye and Greenwald, 2002), it is possible that the reduced expression of the LIN-12::GFP reporter in cyd-1(q626) mutants is caused by a disruption of this positive feedback loop.
We next analyzed the effect of the G1-specific E-type cyclin cye-1 using the strong reduction-of-function or null allele ku256 (Fay and Han, 2000). Expression of the NICD::GFP reporter in homozygous cye-1(ku256) mutants was strongly reduced, in both the 1° and the 2° VPCs and their descendants (Figure 5C). Moreover, vulval induction was slightly but significantly suppressed in homozygous cye-1(ku256); nicd::gfp mutants when compared with heterozygous cye-1(ku256)/+; nicd::gfp controls (Table I, rows 4, 5).
Finally, we investigated the influence of the G2-specific B-type cyclins cyb-1, cyb-2, and cyb-3 on NICD stability. Since the available deletion mutations in the cyb genes cause embryonic or larval lethality, we reduced cyb gene activities by RNAi-mediated knockdown. cyb-2 represents two highly similar and functionally redundant genes, cyb-2.1 and cyb-2.2, that were simultaneously affected by the same RNAi treatment. Only cyb-3 RNAi caused an increase in NICD::GFP expression (Figure 5D). Even though RNAi against cyb-3 did not cause a cell-cycle arrest in the VPCs, cyb-3 RNAi animals exhibited significantly higher NICD::GFP levels in the P6.p descendants at the Pn.px stage compared with control RNAi animals (Figure 5D). We thus conclude that both the G1 cyclins CYD-1 and CYE-1 positively regulate LIN-12 NOTCH signaling. CYD-1 appears to act primarily by regulating full-length LIN-12, while CYE-1 seems to exert a stabilizing effect on the intracellular pool of NICD. On the other hand, CYB-3 may act together with CDK-1 during the late G2 phase to directly or indirectly target NICD for degradation before M-phase entry.
The N-terminal ankyrin and the C-terminal PEST domain regulate NICD stability
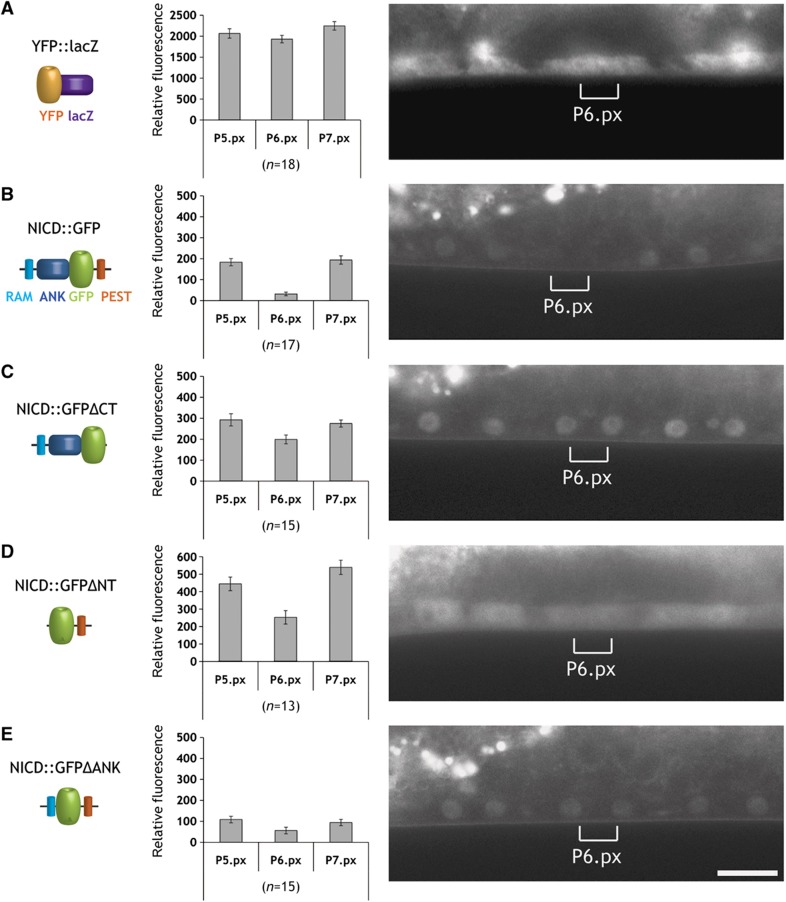
To identify the domains necessary for NICD degradation, we performed a structure function analysis. The LIN-12 NICD is composed of a nuclear localization signal (NLS) at the N-terminus followed by the RBP-Jkappa-associated module (RAM) containing the downregulation targeting signal (DTS) motif, which promotes NOTCH endocytosis (Shaye and Greenwald, 2002, 2005; Kopan and Ilagan, 2009). The central part contains seven ankyrin repeats (ANK) that are essential for the interaction with CSL and other transcription factors and for NICD dimerization. Near its C-terminus, NICD contains a proline/glutamic acid/serine/threonine (PEST) motif that binds to the F-Box protein SEL-10 and regulates protein degradation (Gupta-Rossi et al, 2001). We integrated different NICD::GFP deletion constructs as single copy transgenes into the genome via MosSCI, allowing us to compare expression levels between different transgenes (Frokjaer-Jensen et al, 2008). A construct lacking the C-terminus with the PEST domain (NICD::GFPΔCT) showed persistent expression in the P6.p descendants and elevated levels in the P5.p and P7.p descendants (Figure 6C). However, the reciprocal construct harboring just the C-terminal PEST domain (NICD::GFPΔNT) was only partially downregulated in the 1° lineage (Figure 6D). Since the NICD::GFPΔNT signal was detected mainly in the cytoplasm, we examined if nuclear localization was necessary for proper NICD degradation by deleting the seven ANK repeats but leaving the N-terminal 67 amino acids containing the NLS, RAM, and DTS domains in place (NICD::GFPΔANK). Even though the NICD::GFPΔANK protein localized to the nuclei of the VPCs and their descendants, it was not efficiently downregulated in the 1° lineage. Since the deletion of the N-terminal RAM and DTS resulted in the formation of a very unstable protein that was only weakly expressed at the early or mid L2 stage, before NICD is normally degraded, the combination of ANK and PEST domains alone could not be analyzed. Taken together, our structure function analysis indicates that the PEST domain and the ANK repeats together regulate the NICD degradation.
Figure 6.
Identification of the domains regulating NICD protein stability. (A) Expression pattern of the YFP::LacZ as control, (B) the complete LIN-12 intracellular domain (NICD::GFP), (C) a C-terminal truncation of 87 amino acids containing the PEST domain (NICD::GFPΔCT), (D) an N-terminal deletion of 384 amino acids removing the RAM domain and ANK repeats (NICD::GFPΔNT), and (E) an internal deletion of 324 amino acids removing the ANK repeats (NICD::GFPΔANK). All constructs were expressed using the VPC-specific bar-1 promoter fragment (Natarajan et al, 2004) and inserted via MosSCI as single copy transgenes at the same location on LGII. Expression was scored in the descendants of P5.p, P6.p, and P7.p at the Pn.px stage. For each construct, a quantification of the GFP levels is shown in the bar graphs in the center. Error bars indicate the standard error as described in Materials and methods. Representative animals for each construct are shown in the right panels. The brackets indicate the position of the P6.p descendants. The scale bar represents 10 μm. Source data is available for this figure in the Supplementary Information.
Discussion
We have developed a new state-based computational model for cell fate specification during C. elegans vulval development and experimentally tested the predictions made by the model. When using interacting state-machine models to describe a biological behavior, we are facing the question of how to compose the components comprising the model. We previously found that the two standard notions of concurrency, in this context, synchrony and asynchrony, are not appropriate to model the VPC interactions (Fisher et al, 2008). Synchronous composition is too rigid, making it impossible to break the symmetry between processes without the introduction of additional timing mechanisms. On the other hand, asynchronous composition introduces a difficulty in deciding how long to wait for a signal that may never arrive, again requiring artificial timing mechanisms. We have therefore implemented bounded asynchrony in our previous model of VPC specification (Fisher et al, 2007). This allows the components of a system to move independently with a certain degree of randomness, while keeping them coupled within certain boundaries.
The concept of bounded asynchrony may apply particularly well to the case of VPC fate specification in C. elegans. First, the progression of VPCs through the cell cycle does indeed occur with a certain degree of variability, as the VPCs enter the M phase in a random order. After a long G1-phase arrest during the L2 larval stage, the VPCs enter S phase at the L2-to-L3 transition (Euling and Ambros, 1996) and progress through the remainder of the cell cycle in a more or less synchronized way. Second, cell-cycle progression and signaling events are coupled, as a cell cycle-dependent sequence of 1° and 2° cell fate specification has previously been observed (Ambros, 1999). Our data suggest that the degradation of NICD during the late G2 phase is an additional mechanism used to establish this temporal order in 1° and 2° fate specification.
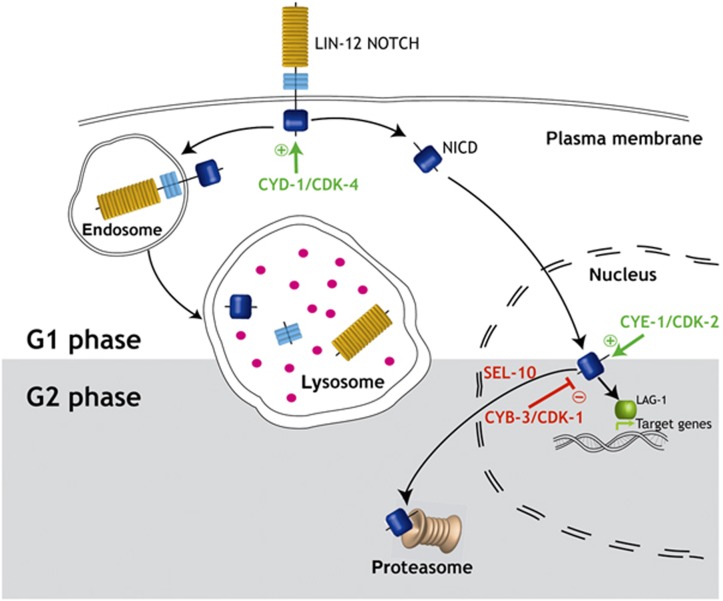
By arresting the cell cycle of the VPC that normally adopts the 1° cell fate in the G1 phase without affecting the progression of the other VPCs, we could experimentally test the concept of bounded asynchrony. Even though 1° fate specification in P6.p occurs already toward the end of the G1 phase (Ambros, 1999), G1-arrested P6.p cells could not adopt a stable 1° cell fate, resulting in the simultaneous expression of 1° and 2° cell fate markers. We traced this instability in fate specification back to a persisting expression of LIN-12 NOTCH in the arrested cells. It has previously been reported that 1° cell fate specification results in the rapid endocytosis of full-length LIN-12 from the apical plasma membrane, a process that depends on Ser/Thr phosphorylation of a cis-acting downregulation (DTS) element in the cytoplasmic portion of LIN-12 (Shaye and Greenwald, 2005). Endocytosis of LIN-12 is followed by rapid degradation in the lysosomes. Interestingly, the LIN-12::GFP protein in G1-arrested VPCs did not remain on the apical cell membrane, where it is normally concentrated in VPCs adopting the 2° cell lineage. Rather, the intracellular LIN-12 cleavage product (NICD) accumulated in the cytoplasm and nucleus. We conclude that the endocytosis and lysosomal degradation of full-length LIN-12 is already occurring during the G1 phase of the cell cycle (Ambros, 1999), or it is not linked to cell-cycle progression at all. We thus postulate the existence of a second LIN-12 NOTCH degradation pathway that is tightly coupled to cell-cycle progression and dedicated to the destruction of NICD in the nucleus (Figure 7). This nuclear NICD degradation pathway is only effective during the G2 phase of the cell cycle.
Figure 7.
Model for cell-cycle regulation of LIN-12 NOTCH signaling. During the G1 phase of the cell cycle, LIN-12 NOTCH is cleaved upon binding to a DSL ligand, thereby releasing the NICD fragment that activates transcription of target genes. Alternatively, NOTCH can undergo endocytosis followed by lysosomal degradation. The activity of the G1-specific CDK-4/CYD-1 and CDK-2/CYE-1 complexes positively regulate LIN-12 NOTCH signaling in P5.p, P6.p, and P7.p by stabilizing full-length NOTCH at the apical plasma membrane and NICD in the nucleus, respectively. Degradation of NICD in P6.p occurs during the G2 phase, when activation of the G2-specific CDK-1/CYB-3 complex terminates NOTCH signaling by inducing ubiquitin-mediated proteasomal degradation of NICD.
On the basis of our findings, we propose a model, in which NICD is stabilized and accumulates in the VPCs during the G1 phase thanks to the activity of the CDK-4/CYD-1 and CDK-2/CYE-1 complexes. In analogy to Drosophila dacapo, constitutive expression of CKI-1 blocks entry into S phase most likely by inhibiting CDK-2/CYE-1 activity (de Nooij et al, 1996; Boxem and van den Heuvel, 2001; van den Heuvel, 2005). Similarly to its mammalian homologs, CKI-1 and CDK-4/CYD-1 may form a complex, which sequesters and inhibits CKI-1 and at the same time stabilizes CDK-4/CYD-1 (LaBaer et al, 1997; Cheng et al, 1999; Boxem and van den Heuvel, 2001). Therefore, arresting a 1° VPC in the G1 phase by CKI-1 overexpression may increase LIN-12 NOTCH activity due to the persisting activity of CDK-4/CYD-1 and the absence of G2 CDK/Cyclin activity. After the VPCs have completed the S phase, activation of the CDK-1/CYB-3 complex during the late G2 phase may directly or indirectly target NICD for degradation, resulting in the termination of the NOTCH signal in the 1° cell lineage before M-phase entry. The nature of the signals targeting NICD for destruction and causing a higher rate of NICD decay in the 1° than in the 2° lineage remains to be identified. In vertebrate cells, the stability of NICD is negatively regulated by phosphorylation of the PEST domain and deacetylation of the ANK repeats (Cornell et al, 1999; Gupta-Rossi et al, 2001; Oberg et al, 2001; Wu et al, 2001; Guarani et al, 2011). Accordingly, our in vivo structure function analysis indicated that both the ANK repeats and the PEST domain are required for efficient degradation of NICD in the VPCs.
Taken together, these findings point at a temporal sequence of signaling events and a mechanism, by which the coordinated progression of VPCs through the cell cycle allows the formation of a stable cell fate pattern. First, activation of the EGFR/RAS/MAPK pathway by the AC signal during the G1 phase induces the 1° cell fate in P6.p. Still during the G1 phase, LIN-12 NOTCH signaling is activated in the 2° cells as a consequence of high EGFR/RAS/MAPK activity in the 1° cell, which leads to the induction of DSL ligand expression by P6.p (Tax et al, 1994; Chen and Greenwald, 2004). The 1° versus 2° cell fate decision, however, can only be sealed after the G1 arrest has been relieved and NICD has been degraded in the 1° cell lineage during the late G2 phase. Thus, by linking the inactivation of the key cell fate determinant NOTCH to cell-cycle progression, a boundary is created that prevents VPCs from becoming de-synchronized. The heterochronic gene pathway that regulates the G1-to-S checkpoint in the VPCs may represent a biological equivalent of the global ‘scheduler’ in the computational model (Euling and Ambros, 1996; van den Heuvel, 2005). The early heterochronic genes maintain the expression of the CDK inhibitor CKI-1 during the L2 larval stage. Expression of cki-1 ceases due to a switch in heterochronic gene expression at the L2-to-L3 transition, thus lifting the boundary and allowing VPCs to enter S phase (Hong et al, 1998).
In many cases of cell fate specification, the coordination of different intercellular signaling pathways is critically dependent on spatial and temporal control mechanisms linked to the cell cycle, which determines a cell’s competence to respond to extrinsic signals (Gomer and Firtel, 1987; McConnell and Kaznowski, 1991; Weigmann and Lehner, 1995). Cell-cycle control of developmental decisions may be particularly important in situations where a cell is sensitive to multiple signals that specify distinct outcomes. Therefore, specific cell fate choices need to be executed with a certain priority or temporal sequence. To link different steps of cell fate determination to specific cell-cycle phases could be a general strategy used to temporally coordinate cell fate choices among equivalent cells. This strategy allows the sequencing and prioritizing of different developmental programs within a single cell lineage. In complex multicellular environments, coupling cell-cycle progression to signal transduction could provide a means to limit the number of cells in a population that can respond to extrinsic signals at a given time point. Thus, the deregulation of the cell cycle observed in many tumor cells may not only affect cell proliferation but also the responsiveness of the cells to growth factors, causing cell fate transformations or de-differentiation. While numerous studies have demonstrated a strong influence of intercellular signaling on cell-cycle progression (Boxem and van den Heuvel, 2002; Clayton et al, 2008; Yamaguchi et al, 2010), fewer studies focused on the impact of the cell cycle on signaling (Moore et al, 2007; Davidson et al, 2009; Ali et al, 2011). Bounded asynchrony achieved through cell-cycle control of signal transduction could be a global principle utilized during the development of multicellular organisms. Finally, by exploring the various mechanisms linking cell-cycle progression to signaling we will better understand how deregulation of the cell cycle promotes tumor development.
Materials and methods
Timed automata
A timed automaton is a mathematical model for describing systems that have discrete states that interact with continuous time. Its behavior consists of discrete changes of states and continuous evolution of time. A given automaton describes the possible sequence of changes in discrete states and their timing. Technically, the automaton has one or more clocks that measure time. These clocks can be reset when the automaton changes its discrete states, and in turn, the automaton uses the clocks to determine how long to stay in states and which discrete state changes are possible. This leads to state sequences (with their timing) to be possible (accepted) in the automaton or impossible (rejected). Events attached to transitions of the automaton can serve as a communication mechanism between multiple timed automata. A system is modeled by creating a timed automaton that produces all the possible behaviors of this system and not more. An example of a timed automaton and an explanation of its possible behaviors are given in Figure 1B–D. We used the tool Uppaal for modeling and analysis of timed automata (Larsen et al, 1997).
C. elegans strains and constructs
Standard methods were used for maintaining and manipulating C. elegans (Brenner, 1974). The following mutations and transgenes were used: LGI: cye-1(ku256) (Fay and Han, 2000), ayIs4[egl-17::gfp] (Burdine et al, 1998), LGII: zhIs39[bar-1p::nicd::gfp::unc-54 3′utr, unc-119(+)], zhIs49[bar-1p::yfp::unc-54 3′utr, unc-119(+)], zhIs56[bar-1p::nicd::gfpΔCT::unc-54 3′utr, unc-119(+)], zhIs50.1[bar-1p::nicd::gfpΔNT::unc-54 3′utr, unc-119(+)], zhIs55[bar-1p::nicd::gfpΔANK::unc-54 3-1p::nicd::gfp+)], mfIs41[lip-1::yfp, myo-2::rfp] (gift of Marie-Anne Felix), cyd-1(q626) (Tilmann and Kimble, 2005), unc-4(e120), LGIII: unc-119(ed3), zhIs4[lip-1::gfp] (Berset et al, 2001), LGIV: dpy-20(e1282), LGV: arIs92[egl-17::cfp, tax-3::gfp], arIs82[LIN-12::GFP; unc-4(+); egl-17p::LacZ] (Shaye and Greenwald, 2002), Balancer chromosome: +/hT2[dpy-18(h662)]; +/hT2[bli-4(e937)] (I;III), extrachromosomal arrays: zhEx314, zhEx315[egl-17p::cki-1, lin-48::gfp], zhEx334, zhEx367[egl-17p::cki-1, myo-2::mcherry], zhEx500[bar-1p::nicd::gfp::unc-54 3′utr, unc-119(+); myo-2p::mCherry].
Plasmid constructs
GFP reporter constructs had been made by gateway cloning (MultiSite Gateway® Three-Fragment Vector Construction Kit; Invitrogen) using PCFJ150 as final destination vector. The reporter constructs were inserted as single copies into the C. elegans genome using MosSCI (Frokjaer-Jensen et al, 2008). The entry vectors listed in Supplementary Table 2 were sub-cloned using the listed primers and genomic DNA as template, unless otherwise stated.
Additional plasmids used were pVT363(egl-17p::cki-1) (Hong et al, 1998), pTJ1157(lin-48p::gfp) (Johnson et al, 2001), backbone plasmids and plasmids containing co-injection markers for MosSCI (Frokjaer-Jensen et al, 2008).
Germline transformation
Worms were transformed by microinjection as described (Mello et al, 1991).
Plasmids were injected at a concentration of 50 ng/μg together with the co-injection marker pTJ1157 (50 ng/μg) or pCFJ90 (2.5 ng/μg).
To integrate single copies of transgenes, the MosSCI method was used as described (Frokjaer-Jensen et al, 2008).
Time-course experiment
Eggs were obtained by bleaching adult animals for 10 min in 1 ml 400 mM NaOH, 7% sodium hypochlorite followed by three washing steps with M9 or water. The eggs were allowed to hatch on an NGM plates without food. To obtain a highly synchronized population of larvae, all the L1 larvae that hatched during a 30-min interval were collected by mouth pipetting, placed on growth plates with food and cultivated at 20°C. After 24 h about 20 worms were analyzed every 30 min until the VPCs had divided. Due to the limited number of larvae that could be collected over a single 30-min time period, the time-course experiment shown in Figure 3 was done in batches over three different days (first day: 24–27 h; second day: 27.5–28.5 h; third day: 30–36 h).
S-phase arrest
Eggs obtained by bleaching (see above) were allowed to hatch in M9 medium overnight without food. After transferring the L1 larvae to NGM plates with food, they were cultivated at 20°C for 28 h, when early L3 larvae were placed for 14 h on NGM plates containing 40 mM hydroxyurea with food. Control animals remained on NGM plates without hydroxyurea.
RNAi experiments
For RNA interference, the feeding method described by Kamath et al (2001) was used. The worms were synchronized by bleaching (see time-course experiment). L1 larvae were placed on growth media plates containing 3 mM IPTG, 50 μg/ml ampicillin and 50 μg/ml tetracycline and bacteria of specific RNAi strains. Worms were allowed to grow for 36 h at 20°C and analyzed.
C. elegans microscopy and image analysis
Animals of the indicated stages (Pn.p, Pn.px, or Pn.pxx) were placed in 4 μl of 5 mM tetramisole solution in M9 on 4% agarose pads. Fluorescent images were acquired on a Leica DMRA wide-field microscope equipped with a cooled CCD camera (Hamamatsu Orca ER) controlled by the Openlab 5 software package (Improvision). For quantification of GFP intensity, images were acquired with the same microscope, camera and software settings using GFP-specific filter sets. The intensity of GFP expression was measured using the measurement tool in the Openlab software. Each measurement was standardized to the background intensity in the same animal (GFP intensity in arbitrary units). Microsoft Excel (Version 11.5.3) and an R 2.11.1 script were used for data processing and statistical analysis. Data were analyzed by the bootstrap method (10 000 bootstrap samples). The standard errors of the samples were estimated by the bootstrap method (Efron, 1981). The significance levels ***P<0.001, **P<0.01, and *P<0.05 were determined by calculating the P-value for the differences between two samples and corrected for multiple testing according to Bonferroni.
Supplementary Material
Supplementary Figures S1-S22, Supplementary Tables S1-S2
Acknowledgments
We wish to thank all members of the Hajnal laboratory for support and critical input into this work. We are especially grateful to Itay Nakdimon, Matthias Morf, Michael Walser, and Adrian Streit for comments on the manuscript. We thank Victor Ambros for providing the egl-17p::cki-1 plasmid; Eric Jorgensen for providing the MosSCI vectors; Judith Kimble for the cyd-1 allele q626; Min Han for the cye-1 allele ku256; and the C. elegans genetics center for strains. We thank Matthias Morf for writing the R script that was needed for statistical data analysis. This work was funded by the Kanton of Zurich, the FP7 PANACEA project (grant no. 222936) and a grant from the Swiss National Science Foundation to AH.
Author contributions: JF and AH conceived the project and designed the computational model and laboratory experiments; SN-S, IR, MA, JF, and AH performed the laboratory experiments; AB, NP, and JF constructed the computational model; and NP wrote the timed automaton; SN-S, IR, MA, JF, and AH performed statistical analysis and data interpretation of the laboratory experiments; and AB, NP, and JF performed the analysis of the computational model; SN-S, JF, and AH wrote the manuscript.
References
- Aguirre A, Rubio ME, Gallo V (2010) Notch and EGFR pathway interaction regulates neural stem cell number and self-renewal. Nature 467: 323–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali F, Hindley C, McDowell G, Deibler R, Jones A, Kirschner M, Guillemot F, Philpott A (2011) Cell cycle-regulated multi-site phosphorylation of Neurogenin 2 coordinates cell cycling with differentiation during neurogenesis. Development 138: 4267–4277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alur R, Dill D (1994) A theory of timed automata. Theor Comput Sci 126: 183–235 [Google Scholar]
- Ambros V (1999) Cell cycle-dependent sequencing of cell fate decisions in Caenorhabditis elegans vulva precursor cells. Development 126: 1947–1956 [DOI] [PubMed] [Google Scholar]
- Baron M (2003) An overview of the Notch signalling pathway. Semin Cell Dev Biol 14: 113–119 [DOI] [PubMed] [Google Scholar]
- Baron M, Aslam H, Flasza M, Fostier M, Higgs JE, Mazaleyrat SL, Wilkin MB (2002) Multiple levels of Notch signal regulation (review). Mol Membr Biol 19: 27–38 [DOI] [PubMed] [Google Scholar]
- Berset T, Hoier EF, Battu G, Canevascini S, Hajnal A (2001) Notch inhibition of RAS signaling through MAP kinase phosphatase LIP-1 during C. elegans vulval development. Science 291: 1055–1058 [DOI] [PubMed] [Google Scholar]
- Boxem M, Srinivasan DG, van den Heuvel S (1999) The Caenorhabditis elegans gene ncc-1 encodes a cdc2-related kinase required for M phase in meiotic and mitotic cell divisions, but not for S phase. Development 126: 2227–2239 [DOI] [PubMed] [Google Scholar]
- Boxem M, van den Heuvel S (2001) lin-35 Rb and cki-1 Cip/Kip cooperate in developmental regulation of G1 progression in C. elegans. Development 128: 4349–4359 [DOI] [PubMed] [Google Scholar]
- Boxem M, van den Heuvel S (2002) C. elegans class B synthetic multivulva genes act in G(1) regulation. Curr Biol 12: 906–911 [DOI] [PubMed] [Google Scholar]
- Brenner S (1974) The genetics of Caenorhabditis elegans. Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdine RD, Branda CS, Stern MJ (1998) EGL-17(FGF) expression coordinates the attraction of the migrating sex myoblasts with vulval induction in C. elegans. Development 125: 1083–1093 [DOI] [PubMed] [Google Scholar]
- Chen N, Greenwald I (2004) The lateral signal for LIN-12/Notch in C. elegans vulval development comprises redundant secreted and transmembrane DSL proteins. Dev Cell 6: 183–192 [DOI] [PubMed] [Google Scholar]
- Cheng M, Olivier P, Diehl JA, Fero M, Roussel MF, Roberts JM, Sherr CJ (1999) The p21(Cip1) and p27(Kip1) CDK 'inhibitors' are essential activators of cyclin D-dependent kinases in murine fibroblasts. EMBO J 18: 1571–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton JE, van den Heuvel SJ, Saito RM (2008) Transcriptional control of cell-cycle quiescence during C. elegans development. Dev Biol 313: 603–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell M, Evans DA, Mann R, Fostier M, Flasza M, Monthatong M, Artavanis-Tsakonas S, Baron M (1999) The Drosophila melanogaster Suppressor of deltex gene, a regulator of the Notch receptor signaling pathway, is an E3 class ubiquitin ligase. Genetics 152: 567–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson G, Shen J, Huang YL, Su Y, Karaulanov E, Bartscherer K, Hassler C, Stannek P, Boutros M, Niehrs C (2009) Cell cycle control of wnt receptor activation. Dev Cell 17: 788–799 [DOI] [PubMed] [Google Scholar]
- de Nooij JC, Letendre MA, Hariharan IK (1996) A cyclin-dependent kinase inhibitor, Dacapo, is necessary for timely exit from the cell cycle during Drosophila embryogenesis. Cell 87: 1237–1247 [DOI] [PubMed] [Google Scholar]
- Efron B (1981) Nonparametric estimates of standard error: the jackknife, the bootstrap and other methods. Biometrika 68: 589–599 [Google Scholar]
- Efroni S, Harel D, Cohen IR (2003) Toward rigorous comprehension of biological complexity: modeling, execution, and visualization of thymic T-cell maturation. Genome Res 13: 2485–2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni S, Harel D, Cohen IR (2005) Reactive animation: realistic modeling of complex dynamic systems. Computer 38: 38–47 [Google Scholar]
- Euling S, Ambros V (1996) Heterochronic genes control cell cycle progress and developmental competence of C. elegans vulva precursor cells. Cell 84: 667–676 [DOI] [PubMed] [Google Scholar]
- Fanto M, Mlodzik M (1999) Asymmetric Notch activation specifies photoreceptors R3 and R4 and planar polarity in the Drosophila eye. Nature 397: 523–526 [DOI] [PubMed] [Google Scholar]
- Fay DS, Han M (2000) Mutations in cye-1, a Caenorhabditis elegans cyclin E homolog, reveal coordination between cell-cycle control and vulval development. Development 127: 4049–4060 [DOI] [PubMed] [Google Scholar]
- Fisher J, Henzinger TA, Mateescu M, Piterman N (2008) Bounded asynchrony: concurrency for modeling cell-cell interactions. In Formal Methods in Systems Biology Fisher J (ed) pp 17–32. Springer-Verlag: Cambridge, UK [Google Scholar]
- Fisher J, Henzinger TA (2007) Executable cell biology. Nat Biotechnol 25: 1239–1249 [DOI] [PubMed] [Google Scholar]
- Fisher J, Piterman N, Hajnal A, Henzinger TA (2007) Predictive modeling of signaling crosstalk during C. elegans vulval development. PLoS Comput Biol 3: e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher J, Piterman N, Hubbard EJ, Stern MJ, Harel D (2005) Computational insights into Caenorhabditis elegans vulval development. Proc Natl Acad Sci USA 102: 1951–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frokjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, Grunnet M, Jorgensen EM (2008) Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet 40: 1375–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer CJ, White JB, Jones KA (2004) Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol Cell 16: 509–520 [DOI] [PubMed] [Google Scholar]
- Gomer RH, Firtel RA (1987) Cell-autonomous determination of cell-type choice in Dictyostelium development by cell-cycle phase. Science 237: 758–762 [DOI] [PubMed] [Google Scholar]
- Greenwald I (2005) LIN-12/Notch signaling in C. elegans. WormBook 1–16, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.10.1, http://www.wormbook.org
- Guarani V, Deflorian G, Franco CA, Kruger M, Phng LK, Bentley K, Toussaint L, Dequiedt F, Mostoslavsky R, Schmidt MH, Zimmermann B, Brandes RP, Mione M, Westphal CH, Braun T, Zeiher AM, Gerhardt H, Dimmeler S, Potente M (2011) Acetylation-dependent regulation of endothelial Notch signalling by the SIRT1 deacetylase. Nature 473: 234–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta-Rossi N, Le Bail O, Gonen H, Brou C, Logeat F, Six E, Ciechanover A, Israel A (2001) Functional interaction between SEL-10, an F-box protein, and the nuclear form of activated Notch1 receptor. J Biol Chem 276: 34371–34378 [DOI] [PubMed] [Google Scholar]
- Hitoshi S, Alexson T, Tropepe V, Donoviel D, Elia AJ, Nye JS, Conlon RA, Mak TW, Bernstein A, van der Kooy D (2002) Notch pathway molecules are essential for the maintenance, but not the generation, of mammalian neural stem cells. Genes Dev 16: 846–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y, Roy R, Ambros V (1998) Developmental regulation of a cyclin-dependent kinase inhibitor controls postembryonic cell cycle progression in Caenorhabditis elegans. Development 125: 3585–3597 [DOI] [PubMed] [Google Scholar]
- Hubbard EJ, Wu G, Kitajewski J, Greenwald I (1997) sel-10, a negative regulator of lin-12 activity in Caenorhabditis elegans, encodes a member of the CDC4 family of proteins. Genes Dev 11: 3182–3193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AD, Fitzsimmons D, Hagman J, Chamberlin HM (2001) EGL-38 Pax regulates the ovo-related gene lin-48 during Caenorhabditis elegans organ development. Development 128: 2857–2865 [DOI] [PubMed] [Google Scholar]
- Kam N, Cohen IR, Harel D (2001) The immune system as a reactive system: modeling T cell activation with statecharts. Human-Centric Computing Languages and Environments Proceedings IEEE Symposia, 2001, pp15–22http://ieeexplore.ieee.org/xpl/articleDetails.jsp?arnumber=995228 [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J (2001) Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol 2: RESEARCH0002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano H (2002) Systems biology: a brief overview. Science 295: 1662–1664 [DOI] [PubMed] [Google Scholar]
- Kopan R, Ilagan MX (2009) The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137: 216–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBaer J, Garrett MD, Stevenson LF, Slingerland JM, Sandhu C, Chou HS, Fattaey A, Harlow E (1997) New functional activities for the p21 family of CDK inhibitors. Genes Dev 11: 847–862 [DOI] [PubMed] [Google Scholar]
- Larsen KG, Pettersson P, Yi W (1997) Uppaal in a Nutshell. J Softw Tools Technol Transfer 1: 134–152 [Google Scholar]
- Levitan D, Greenwald I (1998) LIN-12 protein expression and localization during vulval development in C. elegans. Development 125: 3101–3109 [DOI] [PubMed] [Google Scholar]
- Liu ZJ, Shirakawa T, Li Y, Soma A, Oka M, Dotto GP, Fairman RM, Velazquez OC, Herlyn M (2003) Regulation of Notch1 and Dll4 by vascular endothelial growth factor in arterial endothelial cells: implications for modulating arteriogenesis and angiogenesis. Mol Cell Biol 23: 14–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell SK, Kaznowski CE (1991) Cell cycle dependence of laminar determination in developing neocortex. Science 254: 282–285 [DOI] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V (1991) Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J 10: 3959–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore NL, Narayanan R, Weigel NL (2007) Cyclin dependent kinase 2 and the regulation of human progesterone receptor activity. Steroids 72: 202–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori H, Palmer RE, Sternberg PW (1994) The identification of a Caenorhabditis elegans homolog of p34cdc2 kinase. Mol Gen Genet 245: 781–786 [DOI] [PubMed] [Google Scholar]
- Natarajan L, Jackson BM, Szyleyko E, Eisenmann DM (2004) Identification of evolutionarily conserved promoter elements and amino acids required for function of the C. elegans beta-catenin homolog BAR-1. Dev Biol 272: 536–557 [DOI] [PubMed] [Google Scholar]
- Oberg C, Li J, Pauley A, Wolf E, Gurney M, Lendahl U (2001) The Notch intracellular domain is ubiquitinated and negatively regulated by the mammalian Sel-10 homolog. J Biol Chem 276: 35847–35853 [DOI] [PubMed] [Google Scholar]
- Setty Y, Cohen IR, Dor Y, Harel D (2008) Four-dimensional realistic modeling of pancreatic organogenesis. Proc Natl Acad Sci USA 105: 20374–20379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma VM, Draheim KM, Kelliher MA (2007) The Notch1/c-Myc pathway in T cell leukemia. Cell Cycle 6: 927–930 [DOI] [PubMed] [Google Scholar]
- Shaye DD, Greenwald I (2002) Endocytosis-mediated downregulation of LIN-12/Notch upon Ras activation in Caenorhabditis elegans. Nature 420: 686–690 [DOI] [PubMed] [Google Scholar]
- Shaye DD, Greenwald I (2005) LIN-12/Notch trafficking and regulation of DSL ligand activity during vulval induction in Caenorhabditis elegans. Development 132: 5081–5092 [DOI] [PubMed] [Google Scholar]
- Sternberg PW (2005) Vulval development. WormBook 1–28, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.6.1, http://www.wormbook.org [DOI] [PMC free article] [PubMed]
- Stylianou S, Clarke RB, Brennan K (2006) Aberrant activation of notch signaling in human breast cancer. Cancer Res 66: 1517–1525 [DOI] [PubMed] [Google Scholar]
- Sundaram MV (2005) The love-hate relationship between Ras and Notch. Genes Dev 19: 1825–1839 [DOI] [PubMed] [Google Scholar]
- Tax FE, Yeargers JJ, Thomas JH (1994) Sequence of C. elegans lag-2 reveals a cell-signalling domain shared with Delta and Serrate of Drosophila. Nature 368: 150–154 [DOI] [PubMed] [Google Scholar]
- Tilmann C, Kimble J (2005) Cyclin D regulation of a sexually dimorphic asymmetric cell division. Dev Cell 9: 489–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel S (2005) Cell-cycle regulation. WormBook1–16ed. The C. elegans Research Community, WormBook, doi:10.1895/wormbook.1.28.1, http://www.wormbook.org [DOI] [PMC free article] [PubMed]
- Weigmann K, Lehner CF (1995) Cell fate specification by even-skipped expression in the Drosophila nervous system is coupled to cell cycle progression. Development 121: 3713–3721 [DOI] [PubMed] [Google Scholar]
- Wu G, Lyapina S, Das I, Li J, Gurney M, Pauley A, Chui I, Deshaies RJ, Kitajewski J (2001) SEL-10 is an inhibitor of notch signaling that targets notch for ubiquitin-mediated protein degradation. Mol Cell Biol 21: 7403–7415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Cubizolles F, Zhang Y, Reichert N, Kohler H, Seiser C, Matthias P (2010) Histone deacetylases 1 and 2 act in concert to promote the G1-to-S progression. Genes Dev 24: 455–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1-S22, Supplementary Tables S1-S2