Abstract
Neural crest cell (NCC) invasion is a complex sculpting of individual cells into organized migratory streams that lead to organ development along the vertebrate axis. Key to our understanding of how molecular mechanisms modulate the NCC migratory pattern is information about cell behaviors, yet it has been challenging to selectively mark and analyze migratory NCCs in a living embryo. Here, we apply an innovative in vivo strategy to investigate chick NCC behaviors within the rhombomere 4 (r4) migratory stream by combining photoactivation of KikGR and confocal time-lapse analysis of H2B-mRFP1 transfected NCCs. We find that the spatial order of r4 NCC emergence translates into a distal-to-proximal invasion of the 2nd branchial arch. Lead and trailing NCCs display similar average cell speeds and directionalities. Surprisingly, we find that lead NCCs proliferate along the migratory route and grow to outnumber trailing NCCs by nearly 3 to 1. A simple, cell-based computational model reproduces the r4 NCC migratory pattern and predicts the invasion order can be disrupted by slower, less directional lead cells or by environmental noise. Our results suggest a model in which NCC behaviors maintain a spatially-ordered invasion of the branchial arches.
Keywords: neural crest, chick, cranial, photoactivation, KikGR, computational model
Introduction
Cell migration in invasion is a central theme in diverse events including embryogenesis, wound healing and cancer metastasis. Studies of embryonic cell movements have revealed a wide variety of individual and collective cell behaviors, highlighting the complexity to decipher the molecular choreography of the migratory pattern (Keller, 2002; Lecaudey and Gilmour, 2006). Cell migration in the embryo may range from the organization of small clusters of motile cells (~10) as in border cell migration during development of the Drosophila ovary (Montell et al., 1992; Bianco et al., 2007), to the formation of linear arrays or chains of neuronal progenitors in the CNS (Lois et al., 1996; Murase and Horwitz, 2004), and the organization of a tissue that deposits cells in the lateral line of the zebrafish to form mechanosensory organs (Gilmour et al., 2004). Insights into the mechanisms that underlie coordinated cell movements often emerge from elegant in vivo time-lapse imaging strategies (Haas and Gilmour, 2006; Yang et al., 2002; Prasad and Montell, 2007). For example, emerging evidence suggests that extrinsic environmental cues that direct cell movements may only act on a single or lead cluster of cells that in turn instruct trailing cells to follow, through contact or secreted signals (Fulga and Rorth, 2002), or control both lead and trailing cells through the activation of different receptors (Valentin et al., 2007). These signals may change depending on the migratory phase of the cells (Bianco et al., 2007). Thus, insights into the molecular mechanisms that coordinate complex cell movements may come from detailed examinations of cell behaviors in these model systems.
One of the most striking displays of complex coordination of individual cells into organized migratory streams involves neural crest cell (NCC) migration in the vertebrate embryo. The neural crest are a highly migratory population of cells that emerge from the neural tube and accurately invade the surrounding tissue to contribute to a diverse set of peripheral targets, including the face, heart, gut, and peripheral nervous system (Knect and Bronner-Fraser, 2002; Dupin et al., 2006; Anderson et al., 2006; Hutson and Kirby, 2007). NCCs emerge at all axial levels, yet sort into discrete migratory streams that form in a precise spatio-temporal manner (Lumsden et al., 1991; Rickmann et al., 1985; Bronner-Fraser, 1986; Sechrist et al., 1993; Birgbauer et al., 1995; Serbedzija et al., 1989; Raible et al., 1992; Krull et al., 1995; Kulesa and Fraser, 1998; Trainor et al., 2002; Epperlein et al., 2007). In the head, cranial NCCs emerge from rhombomere (r) segments and migrate along a dorsolateral migratory route in discrete streams and target peripheral sites appropriate to the axial level from which they originate (Lumsden et al., 1991; Schilling and Kimmel, 1994; Kontges and Lumsden, 1996). Advances in intravital timelapse imaging in chick, zebrafish, and mouse gut culture have revealed that there are complex NCC migratory behaviors including directed cell movement, wandering, reversal of direction, and collective follow-the-leader movements (Schilling and Kimmel, 1994; Kulesa and Fraser, 1998; Young et al., 2004, Druckenbrod and Epstein, 2005). Despite our increasing knowledge of NCC migratory behaviors, there is relatively little known about the cellular details within NCC migratory streams, such as the extent to which migratory NCCs exchange neighbors, proliferate, and coordinate cell speed and directionality to precisely invade peripheral targets.
In the absence of more detailed information about the migratory properties of NCCs, it is possible to speculate on several models of invasion, including both competitive and cooperative cell-based models where the spatial order of emergence is either impaired or preserved along the migratory route, respectively (Suppl. Fig. 2A). Insights have come from analysis of both trunk and enteric neural crest derived cell (ENCC) migration. In the trunk, cell tracing experiments in chick have shown that NCCs fill positions along distinct migratory pathways in the order in which cells disperse (Serbedzija et al., 1989; Raible and Eisen, 1994). The initial emerging trunk NCCs follow a ventromedial pathway to form the sympathetic ganglia and later emerging NCCs follow the same pathway, but stop near the neural tube to form the dorsal root ganglia (Serbedzija et al., 1989; Raible et al., 1992). Thus, NCCs may follow the same migratory pathway but reach different target sites along the way. At the level of subgroups of NCCs within the same migratory stream, there is evidence from tissue transplantations and time-lapse studies that reveal subgroups of ENCCs within the invading migratory front display distinct cell behaviors (Young et al., 2004; Druckenbrod and Epstein, 2007). Detailed analyses of the ENCCs show increased cell proliferation and unique invasive capabilities of cells at the migratory front suggesting proliferation and cell-cell interactions rather than population pressure or contact inhibition of movement (Bard and Hay, 1975) from trailing cells drive the ENCC invasion of the gut (Simpson et al., 2007). Thus, there appears to be clear differences in cell behaviors between the migratory front and trailing ENCCs during gut invasion.
A major challenge to our understanding of how molecular mechanisms coordinate NCC migratory patterns is detailed in vivo information about cell behaviors and cell proliferation, yet it is difficult to non-invasively mark and follow subgroups of migratory NCCs within a particular stream. The exciting emergence of novel photoactivatable fluorescent proteins (reviewed in Lukyanov et al., 2005; Lippincott-Schwartz and Patterson, 2003) has recently been applied to label single and subgroups of cells within embryos (Stark and Kulesa, 2005; Wacker et al., 2007; Stark and Kulesa, 2007). Here, we take advantage of a new photoconvertible fluorescent protein, KikGR (Tsutsui et al., 2005), and time-lapse confocal imaging to fluorescently mark and follow targeted subpopulations of migratory cranial NCCs within living chick embryos. Photoactivation of a fluorescent protein using laser scanning excitation circumvents the problem of marking subgroups of migratory NCCs at distinct spatial locations along the migratory route without perturbation of local tissue or inadvertent marking of later emerging NCCs that travel along similar pathways. Focusing on the r4 NCC migratory stream, we analyze whether the spatial order of the early migratory stream translates into a distal-to-proximal distribution within the 2nd branchial arch. We measure cell proliferation, cell migratory properties, and cell neighbor relationships of NCCs within front and trailing subgroups of the r4 NCC migratory stream. We formulate a cell-based computational model that reproduces the r4 NCC migratory pattern and use the model to predict mechanisms that modulate the programmed order of invasion.
Materials and Methods
Embryo Preparation
Fertilized white leghorn chicken eggs (supplied by Ozark Hatchery, Oeosho, MO) were incubated at 38°C in a humidified incubator until the appropriate stages. Eggs were rinsed with 70% ethanol and 3 ml of albumin was removed before windowing the eggshell. A solution of 10% India ink (Pelikan Fount; PLK 51822A143, www.mrart.com, Houston, TX) in Howard Ringer’s solution was injected below the area opaca to visualize each embryo. Embryos were staged according to the criteria of Hamburger and Hamilton (HH) (Hamburger and Hamilton, 1951).
Constructs
KikGR (kind gift from Prof. A. Miyawaki Laboratory, Japan, and now commercially available as Kikume Green-Red, AM-V0082, MBL Int’l Corp, Woburn, MA) was originally cloned from stony coral (Favia favus) and modified (Tsutsui et al., 2005). A monomeric red fluorescent protein (mRFP1) (obtained originally from R. Tsien, San Diego, CA, USA) was made to be nuclear-specific (H2B-mRFP1) (kind gift from S. Megason and R. Lansford, Caltech) and used to label the nuclei of individual neural crest cells (NCCs). All constructs were used at a concentration of 5ug/ul.
Cell Labeling
Embryos were injected with expression constructs at HH stages 8–9 (5–7 somites) to label premigratory r3-r5 NCCs. After windowing the eggshell, a sharpened tungsten needle was used to open a small hole in the vitelline membrane above the cranial neural tube and a construct was microinjected directly into the lumen of the cranial neural tube, using a pulled borosilicate glass needle (Sutter; BF100-50-10, Novato, CA) attached to a Picospritzer III (Parker Hannifin Corporation, Fairfield, NJ). The constructs were electroporated into the right side of the dorsal neural tube (using platinum electrodes and an Electro Square Porator ECM 830 (BTX, a division of Genetronics, San Diego, CA) with five 45 millisecond pulses at one second intervals of 20 volts current, to deliver the constructs into premigratory NCCs in the hindbrain. A few drops of sterile Ringer’s solution was applied to the embryo prior to the eggs being sealed with adhesive tape and re-incubated at 38°C in a humidified incubator (Model 1550, G.Q.F. Manufacturing Co., Savannah, GA) until removal at HH 11–13 somites for photoactivation.
Photoactivation of KikGR
The KikGR photoactivatable protein is sensitive to light, as it will cause inadvertent photobleaching and/or photoconversion. It is important to consider the amount of laser and Hg light exposed to the sample. With this in mind, we did not use Hg lamp light on avian embryos. We performed photoconversion of KikGR-labeled cells, including cell target acquisition and selection for photoconversion, following our previously developed protocol for photoactivation of PAGFP (Stark and Kulesa, 2005). Briefly, all photoconversions were performed in ovo, 10–14hrs after electroporation of KikGR, using an upright confocal microscope (LSM 5 Pascal; Carl Zeiss, Thornwood, NY, USA). We located KikGR-expressing cells using normal 488nm laser excitation, 1.5% laser power with a 10X/0.3 (magnification/numerical aperture) objective (Zeiss) at 512x512 pixels. We applied increased detector gain, pinhole diameter, and m=4 frame averaging. If the KikGR expression was too dim to visualize, we gently increased the 488nm laser power above 2%. When we located a region of the r4 NCC migratory stream of interest where cells were expressing KikGR, we used the zoom feature to select a region of interest (AIM software, Zeiss).
For photoconversion of KikGR with 405nm laser excitation, we recommend using 2–3% laser power at 35X zoom or greater and 4–5% below 35X zoom. If immediate photobleaching occurred, the 405nm power was lowered accordingly for the next cell(s). Immediately after each 405nm excitation scan, we surveyed the loss of green fluorescence signal and gain of red fluorescence signal with the AIM software (Zeiss), using 488nm and 543nm excitation, respectively. We confirmed the photoconversion of KikGR-labeled cells in two ways. First, we measured the red-to-green fluorescence intensity ratio and confirmed the ratio to be greater than 1 (AIM software, Zeiss). Second, we visually confirmed the photoconversion process in 3D using orthogonal section views in xy, xz, and yz (AIM software, Zeiss).
Confocal Imaging
For static imaging, KikGR injected embryos were collected at 24hrs post-photoactivation. Embryos showing expression, surveyed using a fluorescence dissecting microscope (Leica MZFLIII), were mounted dorsal side up on 22×75 mm microslides (48312-024, VWR, West Chester, PA) within PBS contained by silicone grease (79810-99, Dow Corning, Midland, MI) then covered with a 22x22mm glass coverslip (48366 067, VWR). All imaging was performed on an inverted LSM 5 PASCAL microscope (Zeiss), using either a Plan-NeoFluor 10X/0.3 or a 40X/1.2W, C-Apochromat objective (Zeiss). 488nm and 543nm laser lines were used to excite the photoactivated KikGR. Collected z-stacks were projected onto single fields using the AIM Software (Zeiss). Multiple fields were merged using Adobe Photoshop 7.0 (Adobe Systems, Inc., San Jose, CA) to produce a montage of the r4 NCC migratory pattern.
For confocal time-lapse imaging, the microscope (LSM 5 Pascal, Zeiss) was surrounded by an incubator box fashioned from cardboard (4mm thick) and covered with thermal insulation (Kulesa and Kasemeier-Kulesa, 2007). An enclosed heater (115-20, Lyon Electric Company, Inc., Chula Vista, CA) maintained the temperature at 38°C for the duration of the time-lapse acquisition, with only mild temperature fluctuations. Whole embryo explant cultures were prepared according to the method described in Krull and Kulesa (1998). Briefly, embryos were removed from the egg by placing an oval ring of filter paper (1001-185, Whatman International Ltd, Maidstone, England) around the circumference of the embryo and blastoderm and then cutting around the outer edges of the ring. The ring with the embryo attached was placed in sterile Ringer’s solution and gently rinsed. This method left the entire embryo, as well as the surrounding blastoderm intact. The 543nm laser was used to excite H2B-mRFP1 labeled NCC cells. Confocal sections were collected every five mins for up to 12hrs. Collected images were analyzed by concatenating the consecutive time points using the AIM software (Zeiss).
Analysis of KikGR-labeled embryos
After image acquisition, a low pass filter was applied to an entire data set (AIM software, Zeiss). Fluorescence intensities within individual NCCs were measured in 3D using volume z-stack data. The quality of the data set was checked in the following manner. We used the profile setting (AIM software, Zeiss) to construct a straight line through as many cells as possible in the entire r4 NCC migratory stream. The green and red fluorescence intensities were calculated within individual NCCs and the quality of the data set was judged as ‘good’ if a majority of the cells could be easily separated and with a ratio of red/green fluorescence intensity >1.3 (photoactivated). If throughout a majority of the data set the red/green fluorescence ratios were outside of this category, we judged the quality of the labeled cells as not distinct enough to accurately determine a photoactivated from non-photoactivated cell and the data set was not used.
Each NCC migratory stream data set was divided into 4 equal parts starting at the neural tube and extended to the distal most NCC present in the stream. The cells were visually counted as red (photoactivated) or green (non-photoactivated) for each portion of the stream; the profile setting was used (AIM software, Zeiss) to determine the fluorescence intensity ratio. All of the cell counts were recorded into a Microsoft Excel spreadsheet. This was repeated for each embryo. To confirm the distribution of photoactivated NCCs, the data were imported into Metamorph (Molecular Devices, Sunnyvale, CA) and a line scan through the entire NCC migratory stream was obtained. Using the bent line tool, the entire length of the r4 stream (from the neural tube to the distal most NCC) was selected, and the thickness of the line set to cover the majority of the stream. The intensities of the red and green channels and the spatial position (length) along a line profile were automatically recorded into Excel. Origin (OriginLab Corp, Northampton, MA) was used to average the line profiles from each embryo to obtain a global value for the red/green intensities within rectangular regions enclosing the r4 NCC migratory stream.
Time-Lapse Analysis of KikGR-Labeled Cell Divisions
Images of cell division of photoactivated KikGR-labeled NCCs were selected from short (<4hrs) time-lapse imaging sessions of r4 NCC migratory stream dynamics. The spatial locations and timeline of the cell divisions were recorded. In a typical analysis, an individual pre-dividing NCC was circled using the circle selection tool (AIM software, Zeiss) and the average fluorescence intensity in the circled region was calculated, as well as the 2D area. The cell was followed throughout the time-lapse until the frame when it became visually apparent that a cell division had occurred (cell rounding and appearance of a daughter cell). The two progeny were individually circled and the red and green fluorescence intensities, together with the 2D area, were recorded for each of the daughter cells. This process was repeated for each dividing cell. The data were recorded into Excel and plotted.
Cell Tracking Analysis
Premigratory NCCs were labeled in the manner described above and followed over time with confocal imaging (LSM 5 Pascal, Zeiss). NCCs were transfected with H2B-mRFP1 to localize the fluorescence to the nucleus. This allowed for more accurate identification and cell tracking of individual cells than with cytosolic GFP or DiI. Confocal time-lapse imaging data from a typical session was collected in 4D (3D+time), with z-stacks generated at 5 min intervals for up to 11hrs. For each data set, AIM software (Zeiss) generated a 2D projection of the z-stack at each time point to reduce the data to 3D (2D+time). Translational drift, due to the natural motion of the embryo, was corrected by alignment of the images using Image-Pro Plus (Media Cybernetics, Inc., Bethesda, MD). Imaris (Bitplane Inc., Saint Paul, MN) was used to track the H2BmRFP1 transfected cells. Specifically, at the start of each time-lapse tracking session, target NCCs were identified by ‘spot identification’ based on a threshold detection of nuclei size and confirmed by visual inspection. Spots were added or deleted to correspond to any into- or out-of-plane cell movements, and tracked accordingly. Cells were tracked throughout a typical time-lapse session and several quantities were calculated including average cell speed, directionality, and cell trajectory. Directionality was calculated as the straight line distance from the start to final cell position divided by the total distance traveled. To monitor cell divisions, progeny were identified and tracked as new spots and scored as related to the original cell by a ‘branch’. The number of calculated branches corresponded to the number of cell division and care was taken to confirm a cell division (by analyzing prior frames of the time-lapse). However, we could not rule out that close, neighboring cells might appear as a result from some cells rapidly coming into focus.
Computational model of the r4 NCC migratory pattern
The dorsolateral migratory pathway of the r4 NCC stream was treated as a two-dimensional domain; the dorsal neural tube midline represented as the y-axis and the downrange proximal-to-distal distance represented as the x-axis. The y-axis included the region from mid-r3 to mid-r5 (total length of 400um) based on biological data that the r4 NCC migratory stream is comprised of NCCs from r3, r4 and r5. The x-axis was approximated to be 1000um in length, corresponding to our unpublished measurements of the distance from the chick neural tube midline into the 2nd branchial arch at the corresponding HH stage. In the model, NCCs emerge at random locations along the y-axis and are distributed normally around the mid-axial level of r4 at a rate of 2 cells every 15min for a total of 192 cells (24hr). That is, the density of emerging cells remains constant throughout the time course of the experiment. Each simulated NCC decides which direction to move every 15min, based on 3 choices sampled from the local microenvironment around the cell (0<theta<360degrees). The average distance a cell moved was 10um at each time step, which was derived from published in vivo measurements of chick NCC speeds (Kulesa and Fraser, 1998). The environmental signal was modeled as a function (attraction gradient), linear in×(proximo-distal) and Gaussian in y, centered around the middle of r4. Each simulation was run for approximately 96 time intervals (each interval represented 15min; total of 24hrs) to represent the average time for a typical r4 NCC to invade the 2nd branchial arch. The initial emerging 30% of the NCCs in the model migratory stream were pseudo-colored (red) to correspond to the photoactivation cell labeling experiments. Environmental noise was modeled as a percentage (0, 0.10, 0.25) multiplied by the function f(x,y) = sin (100*x*y)/100. The simulated environmental noise was considered to represent that there may be variations in the environmental signal in the form of complexity of the extracellular matrix structure, other neighboring factors, or interactions with other cells. Cell interaction in the model represents simple physical interaction of the cells; no two cells can occupy the same space at the same time. The cells are otherwise independent. An individual NCC’s ability to interpret the environmental signal was perturbed by modulating the number of choices (m) an individual cell made to sample the local microenvironment. This value was set at m=3, and doubled or held constant for either the migratory front or trailing subpopulation of NCCs. The spatial positions of each cell at each time point were plotted in Matlab (MathWorks, Natick, MA, mathworks.com).
Results
Targeted photoactivation of subpopulations of KikGR-labeled neural crest cells within cranial migratory streams
Our first goal was to develop an in vivo technique to selectively and less-invasively fluorescently mark subpopulations of migratory NCCs within an emerging stream. We selected an optical highlighter, KikGR, capable of changing the chromophore emission wavelength from a green-to-red signal, upon UV excitation (Tsutsui et al., 2005). By injecting and electroporating KikGR into the early chick neural tube, we delivered and photoactivated KikGR in selected subpopulations of NCCs, including cells at the lead (distal) or trailing (proximal) portions of the r4 migratory stream. The in ovo photoactivation of KikGR-labeled NCCs was visible as a green-to-red chromophore conversion (Suppl. Fig. 1A), represented as a spectral shift of photoactivated chromophores excited with 488nm and 543nm laser light, respectively. This color change allowed for the visualization of photoactivated KikGR (red fluorescence) in cells within subpopulations of non-photoactivated NCCs (green fluorescence) (Suppl. Fig. 1A-F). We focused our investigation to the cellular details of the r4 NCC migratory stream as a model for other NCC migration since this discrete migratory stream is less than 100um below the surface tissue and more accessible to perturbation and confocal time-lapse imaging. In a typical embryo, photoactivation of KikGR within subgroups of NCCs revealed a clear behavior of the photoactivation as a decrease in green fluorescence within a cell and an increase in red fluorescence (Suppl. Fig. 1C-F).
Control embryos show no aberrant or back photoconversion of KikGR signal
In order to accurately analyze photoactivated KikGR within NCCs, we developed a method using control embryos to find approximate thresholds for photoactivated cells (Suppl. Fig. 1G-N). We selected and prepared a set of control embryos electroporated with KikGR for photoconversion, but not excited with 405nm laser light (Suppl. Fig. 1K,L). After 24hrs of reincubation, we analyzed the embryos for the amounts of red and green fluorescence (Suppl. Fig. 1M,N). Each NCC that contained non-photoactivated KikGR had some amount of both green and red fluorescence when excited with 488nm and 543nm laser light, respectively, which is a feature of KikGR. We determined that there was not any significant change in these fluorescence intensities due to stray room light (Suppl. Fig. L,N). Analysis of the KikGR control embryos provided threshold values for green and red fluorescence. Control embryos in which KikGR-labeled NCCs were entirely photoactivated with 405nm laser light did not appear to display any reverse photoconversion (Suppl. Fig. 1G-J).
Neural crest cells at the front of an emerging r4 migratory stream populate the distal portion of the 2nd branchial arch
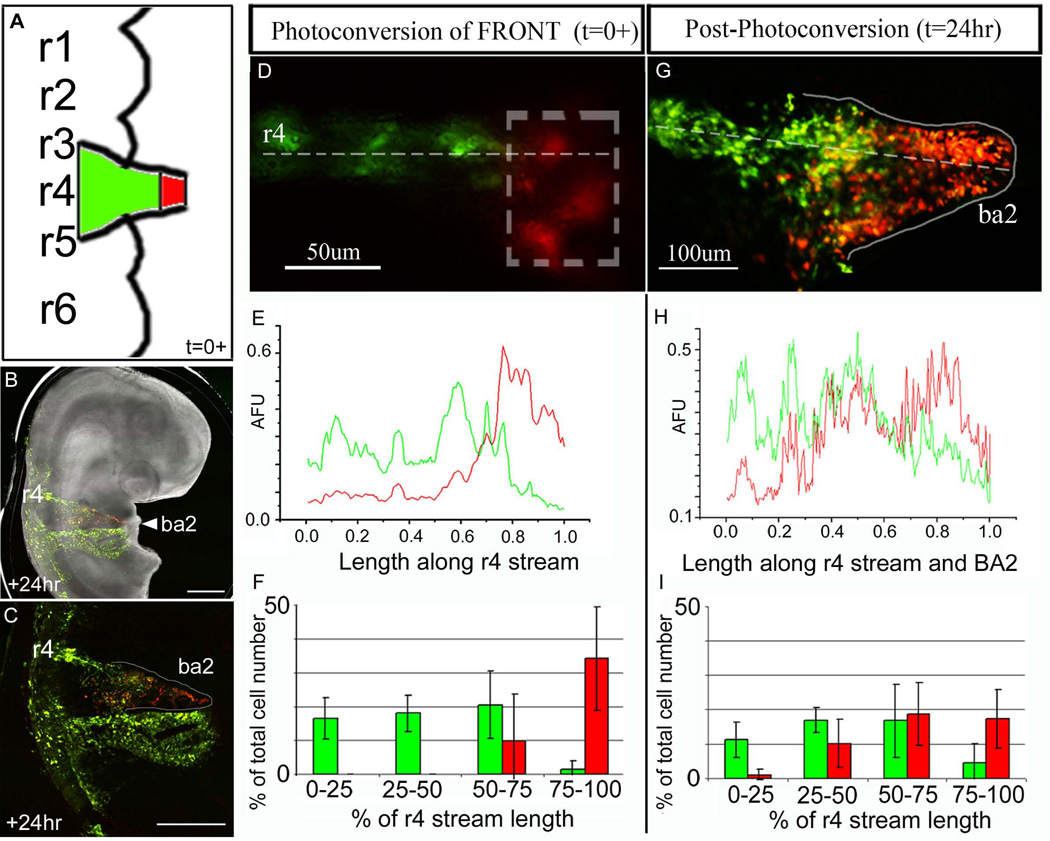
To determine whether the spatial order by which NCCs sort into a migratory stream is translated into a distal-to-proximal invasion order within a branchial arch target, we photoactivated NCCs at the fronts of emerging r4 NCC migratory streams and quantified NCC positions after 24hrs (Fig. 1A-C). With the accuracy of our optical technique, we consistently marked the lead 25% of a typical r4 NCC migratory stream (Fig. 1D-F). The non-photoactivated NCCs comprised the proximal 75% portion of the emerging r4 migratory streams (Fig. 1E,F). The distal portion of the 2nd branchial arch (BA2) was populated by the lead photoactivated NCCs, almost exclusively, when analyzed 24hrs after reincubation (Fig. 1G, H(60–100%), I(75–100% binned range)). We found very few lead photoactivated NCCs along the migratory route and proximal to the BA2 entrance (Fig. 1G, H(0–40%), I(0–25% binned range)). There was some mixing of the photoactivated front and non-photoactivated trailing NCCs within the proximal portion of BA2 (Fig. 1G, I(25–75% binned range), Suppl. Fig. 2B). The fluorescence intensity measurements (AFU) showed the mixing was confined primarily to the region between 40–60% of the migratory route (Fig. 1H). Thus, the photoactivated lead NCCs comprised approximately 60% of the total r4 migratory route, including BA2, 24hrs post-photoactivation.
Figure 1. Targeted fluorescent marking of subpopulations of neural crest cells within the fronts of r4 migratory streams.
(A-D) A typical chick embryo was electroporated with KikGR, the lead 30% of the r4 NCC migratory stream (within the box) was photoactivated 10–14hrs after electroporation, and analyzed at +24hrs after egg re-incubation. (E) A line scan of the intensities (red and green) of multiple embryos (n=7) were averaged together to give a global representation of the line profile throughout the r4 stream at t=0+ (post-photoactivation). (F) The r4 NCC migratory stream was divided into 4 equal parts from the lateral edge of the neural tube to the most distal NCC of the emerging migratory front and the distributed number of photoactivated versus non-photoactivated NCCs. (G) The same embryo in (D) was re-incubated for 24hrs and the NCCs have invaded the 2nd branchial arch. The photoactivated NCCs fill the distal portion of the 2nd branchial arch. (H) A line scan of the intensities (red and green) were averaged together (n=7 embryos) to give a global representation of the line profile throughout the r4 NCC migratory stream at t=24hrs post-photoactivation. (I) The r4 NCC migratory stream at 24hrs post-photoactivation was divided into 4 equal parts from the lateral edge of the neural tube to the most distal NCC within the 2nd branchial arch and shows the distributed number of photoactivated versus non-photoactivated NCCs. BA2=branchial arch 2, r=rhombomere, AFU=arbitrary fluorescence units. The scalebars are as marked and 500um in (B) and (C).
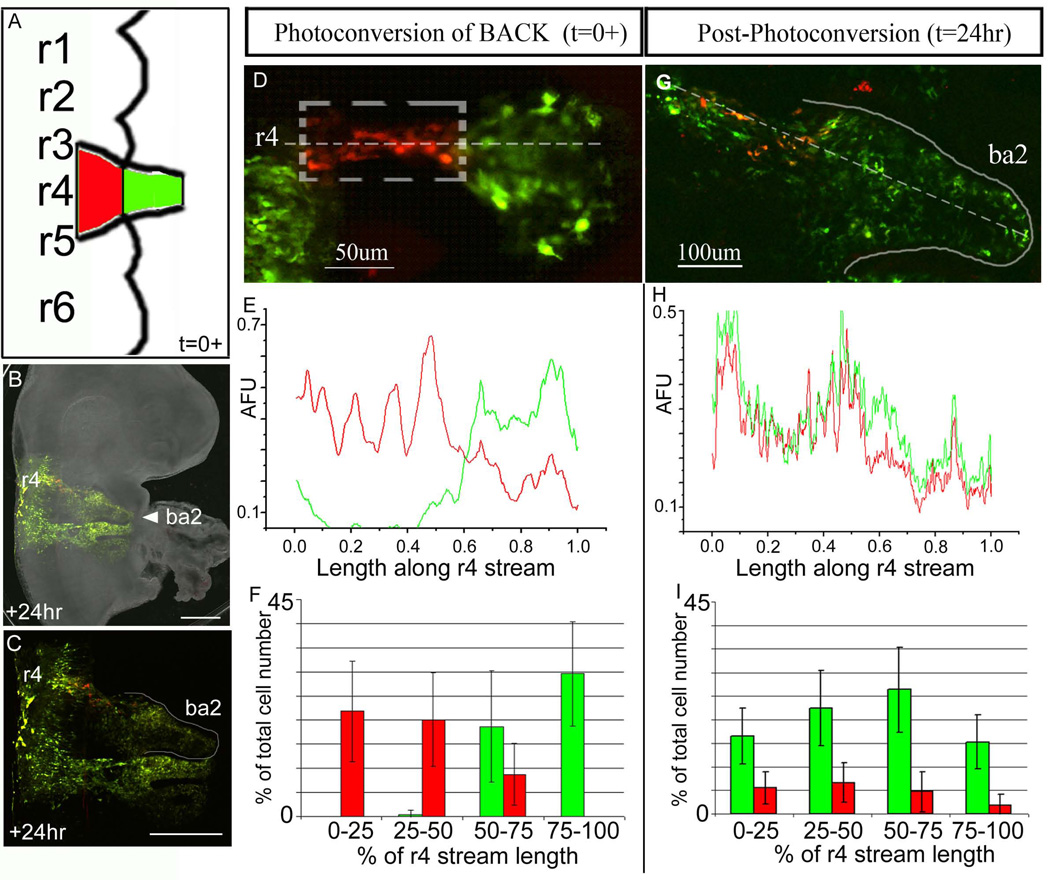
Trailing r4 neural crest cells invade only the proximal portion of the 2nd branchial arch
To determine whether later emerging NCCs within the r4 NCC migratory stream invade the distal portion of the 2nd branchial arch or maintain their spatial order of emergence, we photoactivated NCCs trailing the stream fronts (Fig. 2). With the accuracy of our optical technique, we were consistently able to mark later emerging NCCs at the trailing 60% of a typical r4 NCC migratory stream (Fig. 2D-F). Photoactivated trailing NCCs typically distributed into the proximal portion of the migratory route to the entrance to BA2 (Fig. 2G, I(0–75% binned range)), after embryo reincubation of 24hrs. Analysis of the fluorescence intensities showed that photoactivated trailing NCCs and non-photoactivated NCCs were distributed along the migratory route from near the neural tube to the proximal entrance to BA2 (Fig. 2H (~18–52%), Suppl. Fig. 2B). Very few of these later emerging NCCs migrated into the distal branchial arch (compare the green and red fluorescence intensities and cell counts in Fig. 2G, I(75–100% binned range), H(52–100%)). The addition of further non-photoactivated (green) NCCs in proximal positions was due to newly emerging NCCs (Fig. 2G, I(0–50% binned range)).
Figure 2. Targeted fluorescent marking of subpopulations of trailing neural crest cells within the r4 migratory streams.
(A-D) A typical chick embryo was electroporated with KikGR, the back 60% of the r4 NCC migratory stream was photoactivated (within the dotted box) 10–14hrs after injection, and analyzed at +24hrs after egg re-incubation. (E) A line scan of the intensities (red and green) of multiple embryos (n=8) were averaged together to give a global representation of the line profile along the r4 NCC migratory stream. This profile shows that the trailing 60% of the migratory stream on average was photoactivated at t=0+. (F) The emerging r4 NCC migratory stream was divided into 4 equal parts from the lateral edge of the neural tube to the most distal NCC. The NCCs were counted red or green for each embryo (n=8) and averaged as a percentage of the total cell number versus percentage of the r4 stream length. (G) The same embryo in (D) was re-incubated for 24hrs and the photoactivated NCCs (red) are proximal to the 2nd branchial arch entrance. (H) A line scan of the intensities (red and green) of multiple embryos (n=8) were averaged together to give a global representation of the line profile throughout the r4 stream at 24hrs. (I) The r4 NCC migratory stream at 24hrs was divided into 4 equal parts from the lateral edge of the neural tube to the most distal cell in the 2nd branchial arch. The NCCs were counted red or green for each embryo (n=8). BA2=branchial arch 2, r=rhombomere, AFU=arbitrary fluorescence units. The scalebars are as marked and 500um in (B) and (C).
Cell division does not significantly alter the red-to-green fluorescence ratio in KikGR photoactivated NCCs
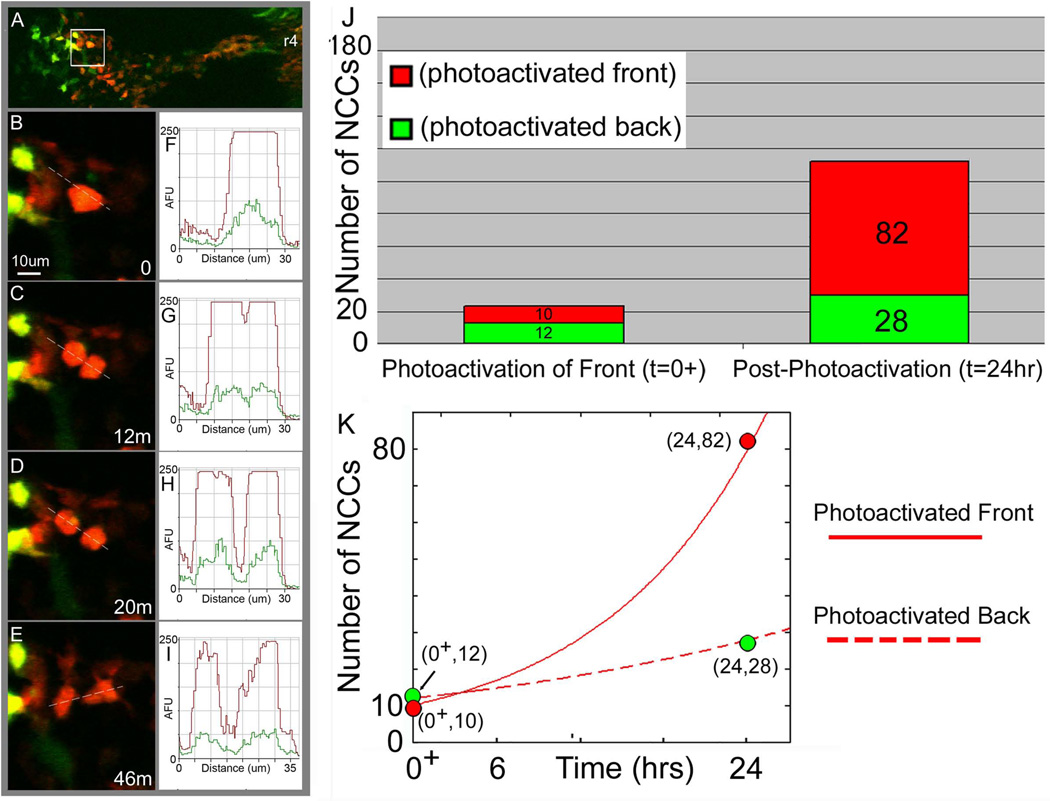
To determine whether there was a loss of photoactivated red fluorescence in KikGR-labeled NCCs after division, we measured fluorescence intensity changes in dividing NCCs using confocal time-lapse imaging (Fig. 3; see also Movie 1). In a typical embryo, migratory NCCs actively divided. Time-lapse analysis revealed NCCs slowed down, rounded up then divided within approximately a 1hr window (Fig. 3B-E). Time-lapse imaging also revealed that prior to cell division, rounded-up NCCs showed a clear photoactivated ratio of red-to-green fluorescence intensities (Fig. 3B,F). Shortly after the visual identification of a distinct NCC progeny (Fig. 3C), there was a clear spatial separation in the red-to-green fluorescence intensities that corresponded to the separation of two distinct NCCs (Fig. 3C,D,G,H). Measurements after cell division (n=7) showed that there was only a minor loss (~5%) in the red-to-green fluorescence intensity ratio in the daughter cells (data not shown; measurements similar to example shown in Fig. 3AI). As the NCCs moved apart and re-established a migratory phenotype (cell protrusions and movement), the NCC progeny continued to display a high level of photoactivated red fluorescence (Fig. 3E,I).
Figure 3. Cell division does not significantly alter the red-to-green fluorescence ratio in KikGR photoactivated NCCs and NCCs at the migratory stream front show higher cell proliferation than trailing NCCs.
KikGR-photoactivated embryos were monitored for short term time-lapse confocal imaging sessions of approximately 4hrs (n=8; multiple cell divisions per time-lapse session). During this time, several (>3) photoactivated NCCs divided and were analyzed for fluorescent intensity measurements before and after the cell division. (A) In a typical time-lapse imaging session, a subpopulation of r4 KikGR photoactivated NCCs were followed over time (at 2min intervals). (B,F) Prior to dividing, NCCs appeared to collapse projections and round-up. An intensity line profile through the NCC shows the red and green fluorescence intensities as a function of the distance along the line (dotted line through the cell). (C,G) NCC division was apparent when a neighboring daughter cell appeared. An intensity line profile through the NCC progeny shows only a minor decrease in the red and green fluorescence intensities as a function of the distance along the line (dotted line through the cell). (D,H) As the daughter cells move apart from each other, the NCCs begin to segregate and (E,I) extend protrusions to move along the migratory route. The scale bar is 10um and the time is in mins. (J) The average number of NCCs are plotted at t=0+ (immediately after photoactivation) for KikGR-expressing NCCs photoactivated at the front of the r4 migratory stream (red) and photoactivated trailing NCCs (green) and at t=24hr later in re-incubated embryos (n=7). (K) The data from the photoactivated front, and photoactivated back are plotted (green and red dots correspond to green and red bars in (J), respectively) and fit to the general equation for cell growth; N(t) = N(0)*2^(kt), N(0)=the number of NCCs that were originally photoactivated and (1/k)=the cell cycle rate. r4=rhombomere 4, AFU=arbitrary fluorescence units, um=microns. The number of NCCs versus time are plotted for the photoactivated front (solid red line, red data points) and photoactivated back (dotted red line, green data points).
Photoactivated NCCs at the r4 migratory stream front show an increase in cell number compared to trailing NCCs after 24hrs
With our observations that lead photoactivated NCCs filled the distal portion of the 2nd branchial arch, we sought to determine whether there were differences in cell proliferation between lead and trailing subgroups of NCCs within the r4 migratory stream. Measurements of the number of photoactivated NCCs at t=0+ and 24hrs later showed an intriguing result. When NCCs at the r4 migratory stream front (~30% of emerging population) were photoactivated, there was approximately an 8-fold increase in the number of photoactivated NCCs within the 2nd branchial arch after 24hrs (Fig. 3J; red region). In contrast, when the trailing portion of the r4 NCC migratory stream was photoactivated, only a small, 2-fold increase in the number of photoactivated NCCs were found in the r4 migratory stream and 2nd branchial arch after 24hrs (Fig. 3J; green region). Fitting of these data to standard cell growth curves revealed a potential difference in NCC doubling times, approximately 8hrs and 20hrs for the lead and trailing subgroups of NCCs, respectively (Fig. 3K; compare solid and dotted lines).
Time-lapse imaging confirms an ordered progression of the r4 NCC migratory stream and reveals higher cell proliferation at the migratory front
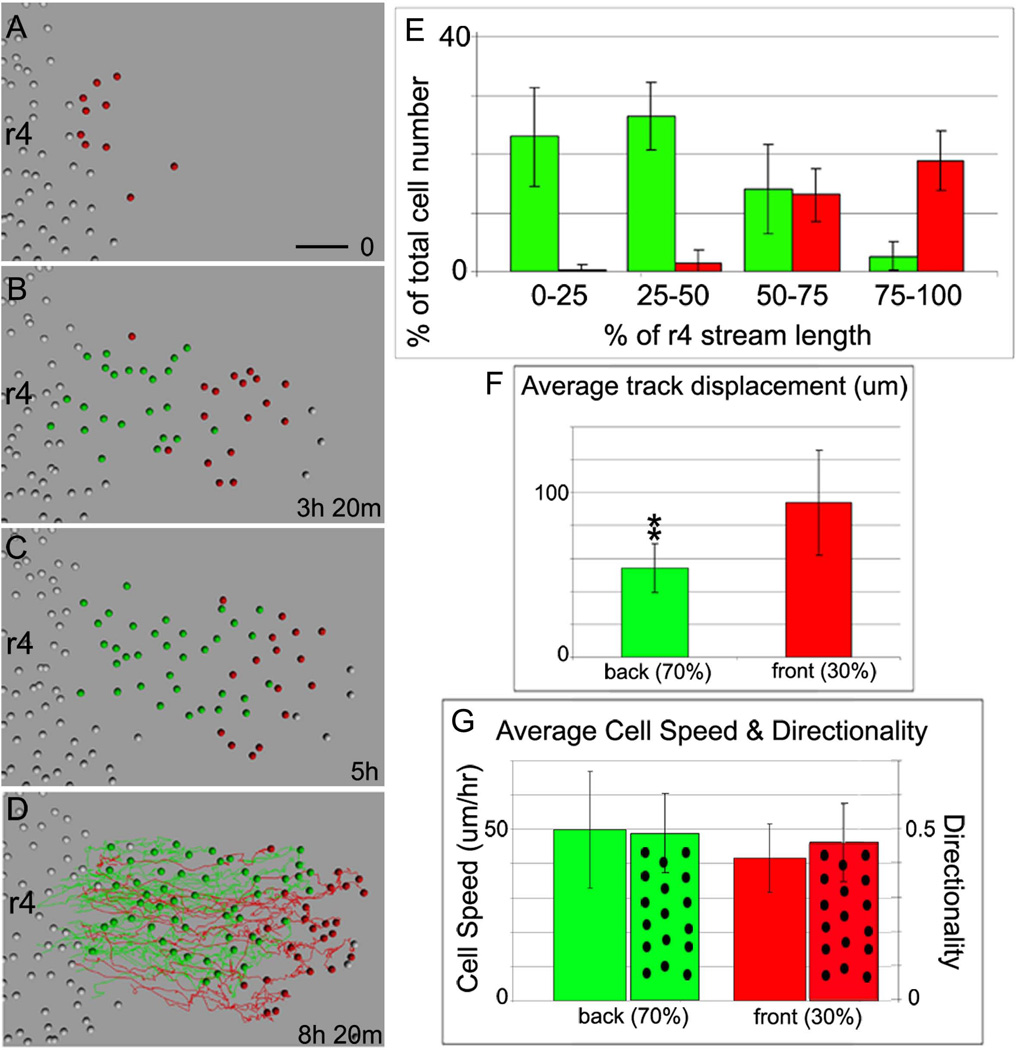
To analyze the in vivo migratory behaviors of subgroups of lead and trailing NCCs within the early emerging r4 migratory stream, we followed individual H2B-mRFP1 transfected NCCs using confocal intravital time-lapse microscopy (Fig. 4A-D; see also Movie 2). For the analysis of the lead and trailing NCCs, we designated the first 30% of emerging NCCs as the front of the r4 migratory stream (Fig. 4A-D; red colored, see also Movie 3). Analysis of the r4 NCC migratory stream over approximately 8hrs revealed that the first 30% of emerging NCCs remained at the front of the migratory stream, while later emerging NCCs trailed more proximally (Fig. 4A-E). Over 50% of the red-colored NCCs remained in the lead 25% and over 90% remained near the lead 50% of the r4 NCC migratory stream (Fig. 4E). There was limited mixing of trailing NCCs with the lead subgroup, as only approximately 5% of the trailing NCCs were found in the lead 25% of the r4 NCC migratory stream (Fig. 4E). Cell tracking analyses revealed that the lead migrating NCC trajectories were displaced further (total lateral distance migrated) on average from r4 (93.9±32um) (Fig. 4F) when compared to the trailing NCC trajectories. Furthermore, since lead NCCs emerged first, those cells were tracked longer over the course of the time-lapse session when compared to later emerging trailing cells (6.8±3hrs compared to 3.4±1.7hrs). The average NCC speed and directionality of the lead 30% NCCs (41.6±10um/hr and 0.46±0.1 respectively) were not significantly different when compared to trailing NCCs (49.9±17um/hr and 0.49±0.1) (Fig. 4G), but there were large variations , as shown by the standard deviations (Fig. 4G).
Figure 4. Time-lapse imaging of H2B-mRFP1 labeled NCCs confirms a spatial order is maintained within the r4 migratory stream and reveals differences in cell proliferation.
(A-D) Selected images from a typical time-lapse imaging session (n=7) show H2B-mRFP1 transfected NCCs (represented by colored spheres) migrating in the r4 NCC stream after cell tracking analysis of raw data. The first 30% of emerging NCCs are pseudo-colored red, while later emerging NCCs are marked in green. (D) Cell trajectories are marked by red or green colored lines over the course of the entire time-lapse. (E) The distribution of the lead (red) and trailing (green) NCCs at the end of the time-lapse sessions, graphed as the percentage of NCCs in 4 different quadrants of the r4 NCC migratory stream. (F) The average total distance an individual NCC traveled along the migratory route is plotted for the lead and trailing subgroups of NCCs as the average track displacement. (G) The calculated average NCC speed and directionality (spotted bars) for each subgroup (lead-red and trailing-green colored bars). r4=rhombomere 4. The scalebar is 50um.
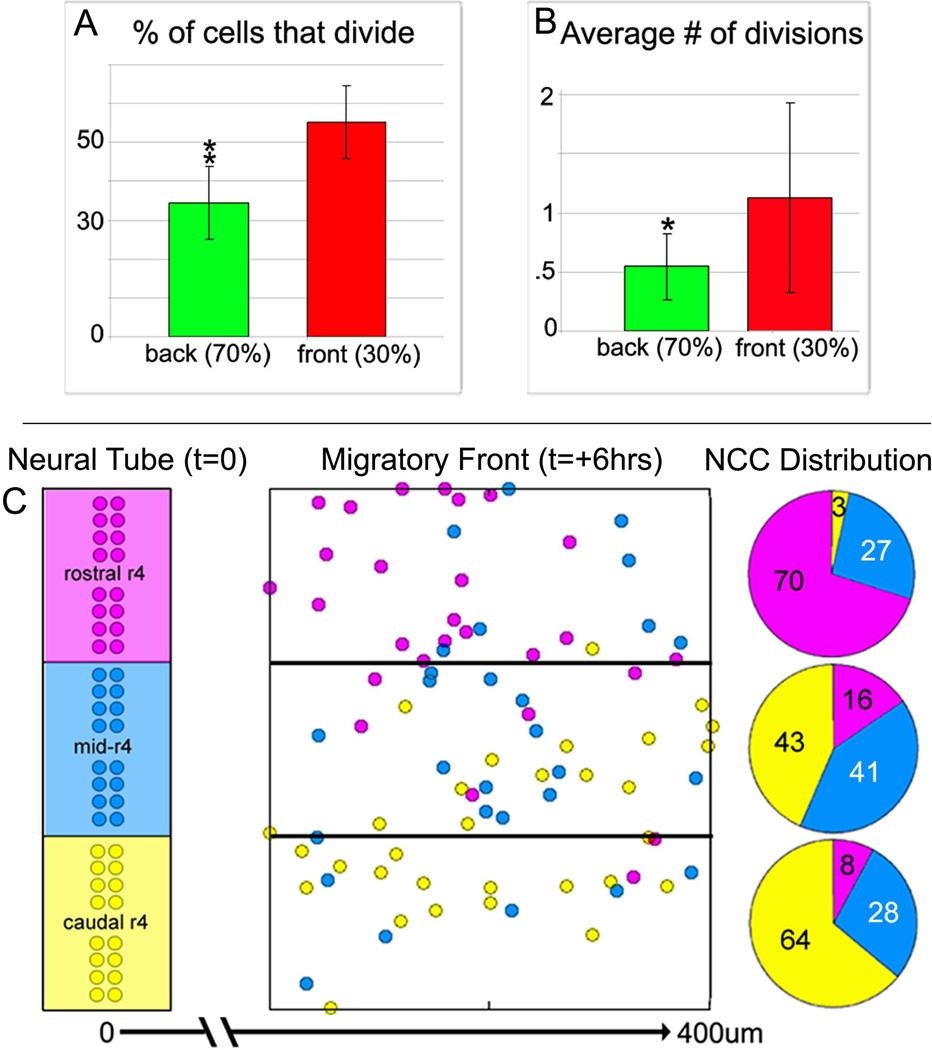
During the cell tracking analysis, we noticed a significant number of NCC divisions along the migratory route. To determine the number and location of NCC divisions within a typical r4 NCC migratory stream, we counted cell divisions from time-lapse data. An average of 52.2% (±9) of the lead NCCs underwent division while 34.4% (±10) of trailing cells performed at least one division (Fig. 5A) within the first 7hrs after emerging from the neural tube (Fig. 5A). NCCs in the front of the r4 migratory stream had a measured cell division rate of 1.13 (±0.8) divisions per cell whereas trailing cells had a cell division rate of 0.54 (±0.3) divisions per cell over the span of approximately 7hrs (Fig. 5B).
Figure 5. Time-lapse analysis reveals differences in cell proliferation between the migratory front and trailing NCCs.
(A) Quantitative measurements of the percentage of dividing NCCs in the lead 30% (red) and the trailing NCCs (green) (from cell tracking data in n=7 time-lapse sessions). (B) Analysis of the number of cell divisions occurring in the subgroup of cells that do undergo division in both lead (red) and trailing cells (green). (C) A plot of the start and final positions of the lead NCCs emerging lateral to r4 and their distribution within the migratory front at the end of the time-lapse sessions, as determined from tracing the backward trajectories of lead NCCs. NCCs are pseudo-colored based on their start position at t=0, as purple (rostral r4), blue (mid-r4), and yellow (caudal r4). The migratory front is segregated into 3 subdomains of rostral, mid, and caudal, and the NCCs distribution as a percentage of the number of NCCs in that particular region is shown as a pie chart. *, significantly different, p<0.1, **, significantly different, p<0.05, (n=7 time-lapse sessions analyzed). r4=rhombomere 4.
The axial level at which an r4 NCC emerged from the neural tube did not predict its rostral-to-caudal position within the migratory front
Further analysis of the lead NCC trajectories showed that some r4 NCCs did not have unidirectional movements, but distributed within different regions of the migratory front (Fig. 5C). Rostral emerging r4 NCCs (rostral 1/3 of r4) tended to be found in the rostral 1/3 of the migratory front at 8hrs, but a small number of NCCs were located in the caudal 1/3 of the migratory front (Fig. 5C). This migratory pattern was similar for NCCs emerging from caudal r4 (Fig. 5C). NCCs emerging from mid-r4 had almost an equal probability of being located in the rostral, mid, or caudal portions of the migratory front at 8hrs (Fig. 5C).
Computational Model of Stream Formation
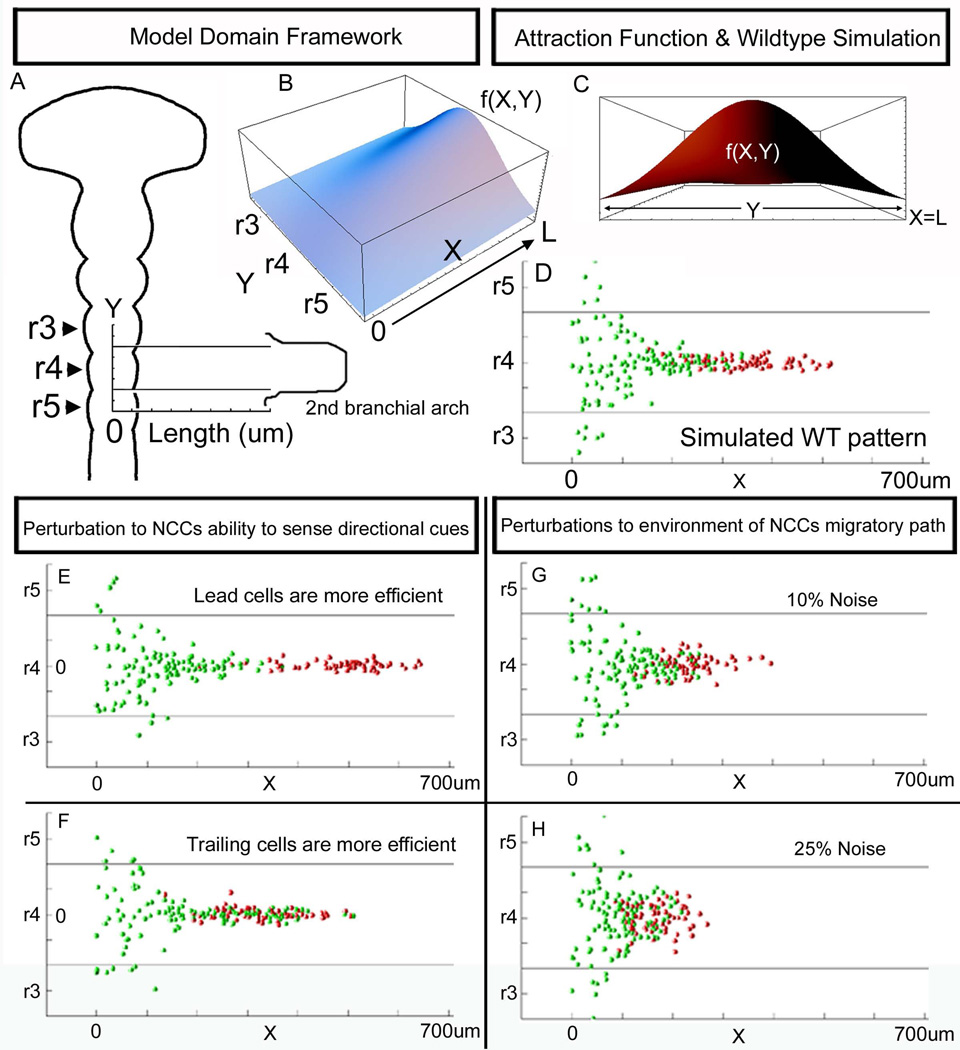
In order to integrate experimental data into a model and simulate hypothetical experiments, we constructed a quantitative framework of the r4 NCC migratory stream (Fig. 6). We constructed an individual cell-based model using assumptions derived from our previous biological data of r3-r5 NCC migratory behaviors and data from other laboratories in normal and perturbed avian embryos. First, we assumed that NCCs from mid-r3 to mid-r5 contribute to the r4 migratory stream (Sechrist et al., 1993; Birgbauer et al., 1995; Kontges and Lumsden, 1996; Kulesa and Fraser, 1998). Second, the length and width of the model simulation 2D domain was based on calculations from time-lapse and static transverse section data in DiI-labeled chick embryos (PMK, RM; unpublished observations) that estimated the length of the r4 migratory pathway from the neural tube to the entrance to the 2nd branchial arch (~700 um) and width of the emerging stream as ~300um (Fig. 6A). We modeled the cell-environmental interactions in the following manner. We assumed a single environmental signal in the form of an attraction gradient; a monotonic increasing function that extended lateral to r4 and peaked at the entrance to BA2 (Fig. 6B). A Gaussian function described this function in the antero-posterior directions (Fig. 6C). This was approximately equivalent to constructing an inhibitory function (repulsion gradient) with inhibition lateral to r3 and r5 and peaked at the neural tube midline, based on evidence of neural crest free exclusion zones adjacent to r3 and r5 (Farlie et al., 1999; Golding et al., 2002; Golding et al., 2004). In the model, each NCC surveyed the local microenvironmental signal in a stochastic manner and decided which direction to migrate based on a choice from 3 selections of the local gradient function. The model simulations were run for a time interval that corresponded to the estimated biological time for r4 NCCs to reach the branchial arches. The number of NCCs in the model corresponded to the number of mid-r3 to mid-r5 NCCs counted from typical r4 NCC migratory streams and simulated to emerge at a specific rate. The model did not include cell proliferation or cell death along the migratory route.
Figure 6. Computational model simulations of r4 NCC migratory stream patterning.
(A) The 2D domain of the computational model was derived from the layout of the NCC migratory stream that emerges lateral to r4 with contribution from r3 and r5. The Y-axis was labeled to correspond to the antero-posterior axis along the neural tube midline, in 50um increments, with Y=0 designated as the axial level of midr4. The X-axis was labeled to correspond to the length along the migratory route from the neural tube to the entrance to the 2nd branchial arch. (B) The environmental signal modeled as an attraction function had the shape of an increasing function with a peak at (X=L,Y=0), (C) that was a normal distribution, f(X,Y). The computational model simulation results for the (D) normal wildtype (WT) pattern, (E) when lead NCCs are more efficient (sample the environment twice as many times as normal), and (F) when trailing cells are more efficient. (G-H) Simulation results of perturbation to the environment (in the form of adding noise to the attraction function f(X,Y) values, (G) 10% noise, and (H) 25% noise. r=rhombomere.
Our first set of computer model simulations revealed that the r4 NCC migratory stream pattern could be reproduced (Fig. 6D). In the model, lead NCCs, pseudo-colored red to correspond to our photoactivation experiments, retained their positions at the migratory front and invaded the distal portion of the migratory route (Fig. 6D). We simulated perturbations to the r4 NCC migratory pattern in two distinct ways. First, we simulated environmental noise, modeled by random noise in the attraction function at 10% and 25% (Fig. 6G, H). In both cases, the spatial order of the r4 NCC migratory stream was disrupted, with increasing mixing of lead and trailing NCCs (Fig. 6G, H, Suppl. Fig. 2B). In contrast, model simulations that perturbed an individual NCCs intrinsic ability to interpret an environmental cue showed the migratory pattern can be similarly disrupted (Fig. 6E, F, Suppl. Fig. 2B). Simulations revealed that by modulating the ability of lead or trailing NCCs to be more (6 choices) or less (3 choices) efficient to sample the environment led to separation of the two subpopulations or mixing between lead and trailing cell subpopulations, respectively (Fig. 6E, F, Suppl. Fig. 2B).
Discussion
Insights into how cellular and molecular mechanisms modulate the neural crest cell (NCC) invasion have been limited by difficulties in targeted cell labeling of migratory NCCs and analysis of migratory stream dynamics. In this paper, photoactivated cell labeling using a newly emerged photoactivatable fluorescent protein, KikGR, has allowed us to analyze the behaviors of subgroups of cranial NCCs within the same migratory stream (Figs. 1,2). Photoactivation using 405nm confocal laser excitation distinctly marked small subgroups of migratory NCCs within the migratory front or trailing NCCs in living embryos, yet involved no grafting or invasive penetration by a glass needle to impale and deliver dye to an individual cell. Thus, we circumvented many of the difficulties inherent in previous cell tracing methods (Schilling and Kimmel, 1994; Kontges and Lumsden, 1996; Baker et al., 1997). We performed controls to ensure that non-photoactivated NCCs subjected to room light and transfer from egg to microscope did not induce photoactivation (Suppl. Fig. 1G,I). We confirmed that KikGR-labeled NCCs subjected to 405nm laser light remained photoactivated for at least 24hrs (Suppl. Fig. 1B) and did not significantly lose photoactivated fluorescence signal after cell division (Fig. 3) and after 24hrs (Stark and Kulesa, 2007), such that we could accurately score photoactivated NCCs after invasion of the branchial arches. Counting of KikGR-photoactivated NCCs and tracking of H2B-mRFP1 transfected NCCs was more accurate than using DiI or cytosolic GFP, since in the latter case the fluorescent label typically becomes punctated into intracellular subregions or throughout the entire cell, making it difficult to distinguish an individual cell in a dense population.
Photoactivation cell labeling revealed that the lead NCCs of the r4 migratory stream invaded the distal region of the 2nd branchial arch (BA2) and trailing NCCs followed to fill in more proximal positions (Suppl. Fig. 2A). This suggests that there is an ordered progression of NCCs along the migratory route that is translated into a distal-to-proximal invasion of BA2, or that NCCs extensively mix along the migratory route then sort into discrete subregions within BA2 shortly after invasion. NCCs within the r4 migratory front entirely filled BA2 with only minimal mixing with trailing NCCs (Fig. 1G-I). Trailing photoactivated NCCs maintained spatial order within the migratory stream, but mixed with trailing and lead non-photoactivated NCCs (Fig. 2G-I). However, when either lead or trailing NCCs were photoactivated, the lead NCC subpopulation almost exclusively filled BA2 (Fig. 1G, 2G). The AFU was a more accurate representation of the photoactivated NCC subpopulations along the migratory route, since the binning of the cell counts into only 4 subregions (Fig. 1I) was not precise to reveal accurate boundaries of the extent of photoactivated NCC distribution. Both measurement techniques clearly showed regions of distinct lead or trailing subpopulations of NCCs. Time-lapse confocal imaging of H2B-mRFP1 transfected NCCs revealed that early emerging NCCs remained at the r4 migratory stream front and later emerging r4 NCCs remained more proximal along the migratory route (Fig. 4A-D). Measurements of the NCC trajectories showed that the lead 30% of NCCs were displaced further downrange from the neural tube when compared to later emerging NCCs (Fig. 4F). These data confirmed the photoactivation cell labeling data (Figs. 1,2) and supported the hypothesis that migratory NCCs maintain a spatial order between the migratory front and trailing NCCs that is translated along the migratory route and into the 2nd branchial arch (Suppl. Fig. 2A).
Lead NCC trajectories were not simply unidirectional, but average NCC speeds and directionalities were similar suggesting coordination between lead and trailing NCC subpopulations, but not cell-cell interactions in a follow-the-leader manner. Analysis of lead NCC positions within the migratory front showed that the axial level of emergence did not predict the rostral-to-caudal position within the migratory front (Fig. 5C). Lead NCCs averaged 41.6±10um/hr compared to trailing NCCs at 49.9±17um/hr, with comparable cell directionalities (Fig. 4G). The ordered progression and presumptive coordination between lead and trailing cranial NCCs is exciting and different from other NCC migratory streams. In the gut, there is emerging evidence from elegant time-lapse imaging studies that there are subgroups of NCCs within the invading front that display distinct migratory behaviors and proliferative rates (Young et al., 2004; Druckenbrod and Epstein, 2007; Simpson et al., 2007). The migratory front of ENCCs invades the gut primarily as single strands of cells, with a subgroup of NCCs that sprint forward then stop as trailing NCCs catch up to advance the migratory front (Young et al., 2004; Druckenbrod and Epstein, 2007). Whether cell communication exists between neighboring cranial NCCs and the extent to which this modulates the spatial order of migratory stream progression will be an exciting avenue to explore. In other systems, cell communication has been shown to play a critical role in modulating the collective movement of cohesive groups in Drosophila border cell migration (Bianco et al., 2007) and zebrafish lateral line formation (Valentin et al., 2007). Important observations of cell communication at the individual cell level have been noted in isolated deep cells of Fundulus (Tickle and Trinkaus, 1976). When a blastula cell is stimulated by nudging with a micropipette, a bleb forms on the diametrically opposite side of the cell (Tickle and Trinkaus, 1976). The stimulus for blebbing could be transmitted to a neighboring cell, raising the possibility of cell surface coordination within a migratory stream. We have previously characterized distinct cell morphologies between the lead and trailing r4 NCCs, and observed long and short filopodial extensions within the r4 NCC migratory stream (Teddy and Kulesa, 2004), that may be involved in organizing NCC movements. Comparison of the NCC invasion processes in different regions of the vertebrate embryo and to other systems highlight the complexity of cell behaviors and the mechanisms that modulate these processes.
Lead NCCs dramatically increased in cell number versus trailing NCCs 24hrs after photoactivation or at the end of time-lapse sessions (Figs. 3,5) suggesting that either lack of local confining neighbors or intrinsic cues lead to differences in cell proliferation within subgroups of migrating r4 NCCs. Lead photoactivated NCCs increased in number by approximately 8-fold over 24hrs, while the number of trailing photoactivated NCCs only increased by 2-fold (Fig. 3). Time-lapse data revealed that higher cell proliferation within the r4 NCC migratory front occurs along the migratory route (Fig. 5A,B), rather than a dramatic proliferation of NCCs after branchial arch invasion. Our data support the recent exciting finding that cell proliferation, within the ENCC migratory front, is higher than in the trailing NCC subpopulation and this drives NCC invasion of the gut (Simpson et al., 2007). These data suggest that a subset of the r4 NCCs at the migratory front give rise to the majority of NCC progeny along the migratory route.
Our computational model simulations reproduced the migratory pattern of r4 NCCs and predicted the spatial order of NCCs can be disrupted with alterations to either the environmental signal or intrinsic ability of NCCs to interpret environmental signals (Fig. 6 and Suppl. Fig. 2B). Simulations of the wildtype scenario matched the spatiotemporal pattern of the r4 NCC migratory stream from emergence of NCCs to along the migratory route. This suggested that a single attraction cue lateral to r4 is sufficient to reproduce the r4 NCC migratory pattern (compare Fig. 4D with Fig. 6D) and comparable to the pattern produced if inhibitory signals were substituted lateral to r3 and r5 in the model. Thus, the model simulations point out that the migration pattern is equivalent for cells preferentially moving toward a higher concentration of attractant and away from an inverse concentration of inhibitor. When we perturbed the ability of subgroups of NCCs to sense an environmental signal, we could control the extent of mixing between the lead and trailing cells (Fig. 6E, F, Suppl. Fig. 2B). When we introduced noise to the environmental signal, the NCC invasion pattern was disrupted in a similar manner, but the simulated migratory pattern with 10% noise showed similarity to the actual NCCs migratory pattern (compare Fig. 6G with Fig. 4D). Specifically, the hypothetical experiment that simulated more efficient navigation of lead NCCs produced a migratory pattern very similar to our actual photoactivation experiment of lead NCCs (compare Fig. 1G, 6E). Thus, in the absence of cell proliferation in the computational model, in order to produce the experimental results, the simulations predicted that lead NCCs had to be more efficient navigators (compare Fig. 6E with Fig.1G,H; Suppl. Fig. 2B). Model predictions also suggest two new experiments; first, if we decrease the navigational ability of lead NCCs, the trailing NCCs should mix in with the migratory front and not match speeds to maintain a proper spatial invasion order. Second, if we perturb an environmental signal(s) by manually stirring the paraxial mesodermal tissue adjacent to mid-r3 to mid-r5, do we visualize mixing of the early emerging and trailing NCCs?
Our data support the hypothesis that cranial NCCs maintain a spatial order between early- and late-migrating NCCs along the migratory route and that increased cell proliferation at the migratory front leads to an extensive, ordered invasion into the branchial arches. Our photoactivation experiments support the earlier results that late-migrating chick cranial NCCs are confined to more dorsal positions and that early-migrating NCCs populate distal portions of peripheral targets (Baker et al., 1997; Fig. 1F). However, we did not observe their result that some early-migrating NCCs populate dorsal positions. This may have been due to the difference between the percentage of early-migrating NCCs labeled. We photoactivated the lead 25–30% of the emerging r4 migratory stream (Fig. 1D,E) whereas Baker and colleagues transplanted quail NCCs to approximately 50–60% of the early-migrating NCC stream (Baker et al., 1997; Fig. 1C). Our results also support the prediction in zebrafish that laterally positioned premigratory cranial NCCs retain a lead position and invade distal target sites (Schilling and Kimmel, 1994). Since our goal was to address cranial NCC migratory order and individual cell behaviors within a stream, we did not address cell fate in relation to position within a migratory stream, however future studies using photoactivation cell labeling combined with lineage tracing should help gain insight into this important question. Our proliferation results that showed lead NCCs divided significantly more than trailing NCCs during migration support the data in chick that late-migrating NCCs contribute extensively to jaw cartilage and bone when substituted heterochronically for the early-migrating subpopulation (Baker et al., 1997). This leads to the hypothesis that if both early- and late-migrating chick NCCs give rise to most NCC derivatives, are only a subset of the NCCs within the migratory front dividing more often than other neighboring NCCs?
In summary, our detailed analyses of the lead and trailing cranial NCC migratory behaviors and spatial invasion order into the branchial arches offer a framework for future molecular studies in the head and provide for a wider comparative approach of NCC migratory stream dynamics throughout the vertebrate embryo. Our findings lead us to suggest a model that cranial NCCs subpopulations maintain an ordered progression along the migratory route, but do not merely follow-the-leader and the migratory front proliferates significantly to populate the branchial arches. The application of in vivo photoactivation NCC labeling offers a novel approach for future short term lineage studies to follow up the differences in cell proliferation between lead and trailing subgroups of NCCs. The computational model provided a framework for integration of experimental data and simulations suggested new experiments. When combined with the new data on lead cell proliferation, the model should yield insights into the mechanisms that coordinate cell proliferation and cell migration to maintain migratory order within the stream. By focusing attention on subgroups of cranial NCCs within the same migratory stream, such studies will help to elucidate how molecular mechanisms modulate cell behaviors to reproduce the fascinating pattern of programmed NCC invasion in the head.
Supplementary Material
Acknowledgements
We are grateful to Atsushi Miyawaki for the KikGR photoactivatable protein and helpful comments on the early photoactivation experiments. We appreciate helpful comments on the manuscript from Mary Dickinson. We thank Joel Schwartz and Winfried Wiegraebe for helpful comments on the photoactivation analysis and Joel Schwartz for help with the Origin software. We very much appreciate the kind generosity of Mr. and Mrs. Stowers for support of this research.
References
- Anderson RB, Newgreen DF, Young HM. Neural crest and the development of the enteric nervous system. Adv Exp Med Biol. 2006;589:181–196. doi: 10.1007/978-0-387-46954-6_11. [DOI] [PubMed] [Google Scholar]
- Baker CVH, Bronner-Fraser MB, LeDouarin N, Teillet M. Early- and late-migrating cranial neural crest cell populations have equivalent developmental potential in vivo. Development. 1997;124:3077–3087. doi: 10.1242/dev.124.16.3077. [DOI] [PubMed] [Google Scholar]
- Bard JBL, Hay ED. The behavior of fibroblasts from the developing avian cornea. J Cell Biol. 1975;67:400–418. doi: 10.1083/jcb.67.2.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco A, Poukkula M, Cliffe A, Mathieu J, Luque CM, Fulga TA, Rorth P. Two distinct modes of guidance signalling during collective migration of border cells. Nature. 2007;448(7151):362–365. doi: 10.1038/nature05965. [DOI] [PubMed] [Google Scholar]
- Birgbauer E, Sechrist J, Bronner-Fraser M, Fraser S. Rhombomeric origin and rostrocaudal reassortment of neural crest cells revealed by intravital microscopy. Development. 1995;121:935–945. doi: 10.1242/dev.121.4.935. [DOI] [PubMed] [Google Scholar]
- Bronner-Fraser M. Analysis of the early stages of trunk neural crest migration in avian embryos using monoclonal antibody HNK-1. Dev Biol. 1986;115(1):44–55. doi: 10.1016/0012-1606(86)90226-5. [DOI] [PubMed] [Google Scholar]
- Dupin E, Creuzet S, LeDouarin NM. The contribution of the neural crest to the vertebrate body. Adv Exp Med Biol. 2006;589:96–119. doi: 10.1007/978-0-387-46954-6_6. [DOI] [PubMed] [Google Scholar]
- Druckenbrod NR, Epstein ML. The pattern of neural crest advance in the cecum and colon. Dev Biol. 2005;287(1):125–133. doi: 10.1016/j.ydbio.2005.08.040. [DOI] [PubMed] [Google Scholar]
- Druckenbrod NR, Epstein ML. Behavior of enteric neural crest-derived cells varies with respect to the migratory wavefront. Dev Dyn. 2007;236(1):84–92. doi: 10.1002/dvdy.20974. [DOI] [PubMed] [Google Scholar]
- Epperlein HH, Selleck MA, Meulemans D, Mchedlishvili L, Cerny R, Sobkow L, Bronner-Fraser M. Migratory patterns and developmental potential of trunk neural crest cells in the axolotl embryo. Dev Dyn. 2007;236(2):389–403. doi: 10.1002/dvdy.21039. [DOI] [PubMed] [Google Scholar]
- Farlie PG, Kerr R, Thomas P, Symes T, Minichiello J, Hearn CJ, Newgreen D. A paraxial exclusion zone creates patterned cranial neural crest cell outgrowth adjacent to rhombomeres 3 and 5. Dev. Biol. 1999;213:70–84. doi: 10.1006/dbio.1999.9332. [DOI] [PubMed] [Google Scholar]
- Fulga TA, Rorth P. Invasive cell migration is initiated by guided growth of long cellular extensions. Nat Cell Biol. 2002;4:715–719. doi: 10.1038/ncb848. [DOI] [PubMed] [Google Scholar]
- Gilmour D, Knaut H, Maischein HM, Nusslein-Volhard C. Towing of sensory axons by their migrating target cells in vivo. Nat Neurosci. 2004;7:491–492. doi: 10.1038/nn1235. [DOI] [PubMed] [Google Scholar]
- Golding JP, Dixon M, Gassmann M. Cues from neuroepithelium and surface ectoderm maintain neural crest-free regions within cranial mesenchyme of the developing chick. Development. 2002;129:1095–1105. doi: 10.1242/dev.129.5.1095. [DOI] [PubMed] [Google Scholar]
- Golding JP, Sobieszczuk D, Dixon M, Coles E, Christiansen J, Wilkinson D, Gassmann M. Roles of erbB4, rhombomere-specific, and rhombomere-independent cues in maintaining neural crest-free zones in the embryonic head. Dev Biol. 2004;266:361–372. doi: 10.1016/j.ydbio.2003.11.003. [DOI] [PubMed] [Google Scholar]
- Haas P, Gilmour D. Chemokine signaling mediates self-organizing tissue migration in the zebrafish lateral line. Dev Cell. 2006;10:673–680. doi: 10.1016/j.devcel.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. J Morph. 1951;88:49–92. doi: 10.1002/aja.1001950404. [DOI] [PubMed] [Google Scholar]
- Hutson MR, Kirby ML. Model systems for the study of heart development and disease. Cardiac neural crest and conotruncal malformations. Semin Cell Dev Biol. 2007;18(1):101–110. doi: 10.1016/j.semcdb.2006.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller R. Shaping the vertebrate body plan by polarized embryonic cell movements. Science. 2002;298:1950–1954. doi: 10.1126/science.1079478. [DOI] [PubMed] [Google Scholar]
- Knect AK, Bronner-Fraser M. Induction of the neural crest: a multigene process. Nat. Rev. Genet. 2002;3:453–461. doi: 10.1038/nrg819. [DOI] [PubMed] [Google Scholar]
- Kontges G, Lumsden A. Rhombencephalic neural crest segmentation is preserved throughout craniofacial ontogeny. Dev. 1996 Oct;122(10):3229–3242. doi: 10.1242/dev.122.10.3229. [DOI] [PubMed] [Google Scholar]
- Krull CE, Collazo A, Fraser SE, Bronner-Fraser M. Segmental migration of trunk neural crest: time-lapse analysis reveals a role for PNA-binding molecules. Development. 1995;121:3733–3743. doi: 10.1242/dev.121.11.3733. [DOI] [PubMed] [Google Scholar]
- Krull CE, Kulesa PM. Embryonic explant and slice preparations for studies of cell migration and axon guidance. Curr Top Dev Biol. 1998;36:145–159. doi: 10.1016/s0070-2153(08)60500-1. [DOI] [PubMed] [Google Scholar]
- Kulesa PM, Fraser SE. Neural crest cell dynamics revealed by time-lapse video microscopy of whole chick explant cultures. Dev. Biol. 1998;204:327–344. doi: 10.1006/dbio.1998.9082. [DOI] [PubMed] [Google Scholar]
- Kulesa, Kasemeier-Kulesa Construction of a heated incubation chamber around a microscope stage for time-lapse imaging. CSH Protocols. 2007 doi: 10.1101/pdb.prot4792. [DOI] [PubMed] [Google Scholar]
- Lecaudey V, Gilmour D. Organizing moving groups during morphogenesis. Curr Opin Cell Biol. 2006;18:102–107. doi: 10.1016/j.ceb.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Patterson GH. Development and use of fluorescent protein markers in living cells. Science. 2003;300(5616):87–91. doi: 10.1126/science.1082520. [DOI] [PubMed] [Google Scholar]
- Lois C, Garcia-Verdugo JM, Alvarez-Buylla A. Chain migration of neuronal precursors. Science. 1996;271:978–981. doi: 10.1126/science.271.5251.978. [DOI] [PubMed] [Google Scholar]
- Lukyanov KA, Chudakov DM, Lukyanov S, Verkhusha VV. Innovation: Photoactivatable fluorescent proteins. Nat Rev Mol Cell Biol. 2005;6(11):885–891. doi: 10.1038/nrm1741. [DOI] [PubMed] [Google Scholar]
- Lumsden A, Sprawson N, Graham A. Segmental origin and migration of neural crest cells in the hindbrain region of the chick embryo. Development. 1991;113:1281–1291. doi: 10.1242/dev.113.4.1281. [DOI] [PubMed] [Google Scholar]
- Montell DJ, Rorth P, Spradling AC. Slow border cells, a locus required for a developmentally regulated cell migration during oogenesis, encodes Drosophila C/EBP. Cell. 1992;71:51–62. doi: 10.1016/0092-8674(92)90265-e. [DOI] [PubMed] [Google Scholar]
- Murase S, Horwitz AF. Directions in cell migration along the rostral migratory stream: the pathway for migration in the brain. Curr Top Dev Biol. 2004;61:135–152. doi: 10.1016/S0070-2153(04)61006-4. [DOI] [PubMed] [Google Scholar]
- Prasad M, Montell DJ. Cellular and molecular mechanisms of border cell migration analyzed using time-lapse live-cell imaging. Dev Cell. 2007;12:997–1005. doi: 10.1016/j.devcel.2007.03.021. [DOI] [PubMed] [Google Scholar]
- Raible DW, Wood A, Hodsdon W, Henion PD, Weston JA, Eisen JS. Segregation and early dispersal of neural crest cells in the embryonic zebrafish. Dev Dyn. 1992;195(1):29–42. doi: 10.1002/aja.1001950104. [DOI] [PubMed] [Google Scholar]
- Raible DW, Eisen JS. Restriction of neural crest cell fate in the trunk of the embryonic zebrafish. Dev. 1994;120(3):495–503. doi: 10.1242/dev.120.3.495. [DOI] [PubMed] [Google Scholar]
- Rickmann M, Fawcett JW, Keynes RJ. The migration of neural crest cells and the growth of motor axons through the rostral half of the chick somite. J Embryol Exp Morphol. 1985;90:437–455. [PubMed] [Google Scholar]
- Sechrist J, Serbedzija GN, Scherson T, Graser SE, Bronner-Fraser M. Segmental migration of the hindbrain neural crest does not arise from its segmental generation. Development. 1993;118:691–703. doi: 10.1242/dev.118.3.691. [DOI] [PubMed] [Google Scholar]
- Schilling TF, Kimmel CB. Segment and cell type lineage restrictions during pharyngeal arch development in the zebrafish embryo. Development. 1994;120:483–494. doi: 10.1242/dev.120.3.483. [DOI] [PubMed] [Google Scholar]
- Serbedzija GN, Bronner-Fraser M, Fraser SE. A vital dye analysis of the timing and pathways of avian trunk neural crest cell migration. Dev. 1989;106(4):809–816. doi: 10.1242/dev.106.4.809. [DOI] [PubMed] [Google Scholar]
- Simpson MJ, Zhang DC, Mariani M, Landman KA, Newgreen DF. Cell proliferation drives neural crest cell invasion of the intestine. Dev Biol. 2007;302(2):553–568. doi: 10.1016/j.ydbio.2006.10.017. [DOI] [PubMed] [Google Scholar]
- Stark DA, Kulesa PM. Photoactivatable green fluorescent protein as a single-cell marker in living embryos. Dev Dyn. 2005;233(3):983–992. doi: 10.1002/dvdy.20385. [DOI] [PubMed] [Google Scholar]
- Stark DA, Kulesa PM. An in vivo comparison of photoactivatable fluorescent proteins in an avian embryo model. Dev Dyn. 2007;236(6):1583–1594. doi: 10.1002/dvdy.21174. [DOI] [PubMed] [Google Scholar]
- Teddy JM, Kulesa PM. In vivo evidence for short- and long-range cell communication in cranial neural crest cells. Dev. 2004;131:6141–6151. doi: 10.1242/dev.01534. [DOI] [PubMed] [Google Scholar]
- Tickle C, Trinkaus JP. Observations on nudging cells in culture. Nature. 1976;261:413. doi: 10.1038/261413a0. [DOI] [PubMed] [Google Scholar]
- Trainor PA, Sobieszczuk D, Wilkinson D, Krumlauf R. Signalling between the hindbrain and paraxial tissues dictates neural crest migration pathways. Dev. 2002 Jan;129(2):433–442. doi: 10.1242/dev.129.2.433. [DOI] [PubMed] [Google Scholar]
- Tsutsui H, Karasawa S, Shimizu H, Nukina N, Miyawaki A. Semi-rational engineering of a coral fluorescent protein into an efficient highlighter. EMBO Rep. 2005;6(3):233–238. doi: 10.1038/sj.embor.7400361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young HM, Bergner AJ, Anderson RB, Enomoto H, Milbrandt J, Newgreen DF, Whitington PM. Dynamics of neural crest-derived cell migration in the embryonic mouse gut. Dev Biol. 2004;270(2):455–473. doi: 10.1016/j.ydbio.2004.03.015. [DOI] [PubMed] [Google Scholar]
- Valentin G, Haas P, Gilmour D. The chemokine SDF1a coordinates tissue migration through the spatial restricted activation of Cxcr7 and Cxcr4b. Curr Biol. 2007;17(12):1026–1031. doi: 10.1016/j.cub.2007.05.020. [DOI] [PubMed] [Google Scholar]
- Wacker SA, Oswald F, Wiedenmann J, Knochel W. A green to red photoactivatable protein as an analyzing tool for early vertebrate development. Dev Dyn. 2007;236(2):473–480. doi: 10.1002/dvdy.20955. [DOI] [PubMed] [Google Scholar]
- Yang X, Dormann D, Munsterberg AE, Weijer CJ. Cell movement patterns during gastrulation in the chick are controlled by positive and negative chemotaxis mediated by FGF4 and FGF8. Dev Cell. 2002;3:425–437. doi: 10.1016/s1534-5807(02)00256-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.