Endogenous thymidine plays a critical role in the induction of replication stress in thymocytes.
Abstract
Nucleotide deficiency causes replication stress (RS) and DNA damage in dividing cells. How nucleotide metabolism is regulated in vivo to prevent these deleterious effects remains unknown. In this study, we investigate a functional link between nucleotide deficiency, RS, and the nucleoside salvage pathway (NSP) enzymes deoxycytidine kinase (dCK) and thymidine kinase (TK1). We show that inactivation of dCK in mice depletes deoxycytidine triphosphate (dCTP) pools and induces RS, early S-phase arrest, and DNA damage in erythroid, B lymphoid, and T lymphoid lineages. TK1−/− erythroid and B lymphoid lineages also experience nucleotide deficiency but, unlike their dCK−/− counterparts, they still sustain DNA replication. Intriguingly, dCTP pool depletion, RS, and hematopoietic defects induced by dCK inactivation are almost completely reversed in a newly generated dCK/TK1 double-knockout (DKO) mouse model. Using NSP-deficient DKO hematopoietic cells, we identify a previously unrecognized biological activity of endogenous thymidine as a strong inducer of RS in vivo through TK1-mediated dCTP pool depletion. We propose a model that explains how TK1 and dCK “tune” dCTP pools to both trigger and resolve RS in vivo. This new model may be exploited therapeutically to induce synthetic sickness/lethality in hematological malignancies, and possibly in other cancers.
Replication stress (RS), a common source of DNA damage and genomic instability (Halazonetis et al., 2008), can be caused by deoxyribonucleotide triphosphate (dNTP) deficiency. For example, pharmacological modulators of dNTP synthesis such as hydroxyurea and 5-fluorouracil induce RS (Gagou et al., 2010; Arlt et al., 2011). RS is also triggered by overexpression of ras and cyclin E, which promote cell division in the absence of sufficient dNTP pools to complete genome replication (Bester et al., 2011). How nucleotide metabolism is regulated in rapidly dividing cells to maintain balanced dNTP pools and to prevent RS is not well understood.
Mammalian cells synthesize dNTPs either de novo or via the nucleoside salvage pathway (NSP; Fig. 1 A; Reichard, 1988). The de novo pathway utilizes glucose and amino acids to generate ribonucleotides that are required for RNA synthesis, energy storage, and signal transduction (Evans and Guy, 2004). A fraction of the cellular pool of ribonucleotides is converted into deoxyribonucleotides by ribonucleotide reductase (RNR; Fairman et al., 2011). Nucleoside transporters and deoxyribonucleoside (dN) kinases enable recycling of extracellular dNs originating from DNA degradation in apoptotic cells (Arnér and Eriksson, 1995), liver biosynthetic processes (Fustin et al., 2012), and food intake. The metabolic flux through the NSP is regulated by rate-limiting kinases. Deoxycytidine kinase (dCK) phosphorylates deoxycytidine to produce deoxycytidine monophosphate (dCMP), a precursor of both dCTP and dTTP pools (Sabini et al., 2008). Deoxyadenosine and deoxyguanosine can also be phosphorylated by dCK, albeit with significantly lower efficacy than deoxycytidine (Sabini et al., 2008). Thymidine kinase 1 (TK1) phosphorylates thymidine to generate dTMP, a precursor of thymidine triphosphate (dTTP) pools (Arnér and Eriksson, 1995).
Figure 1.
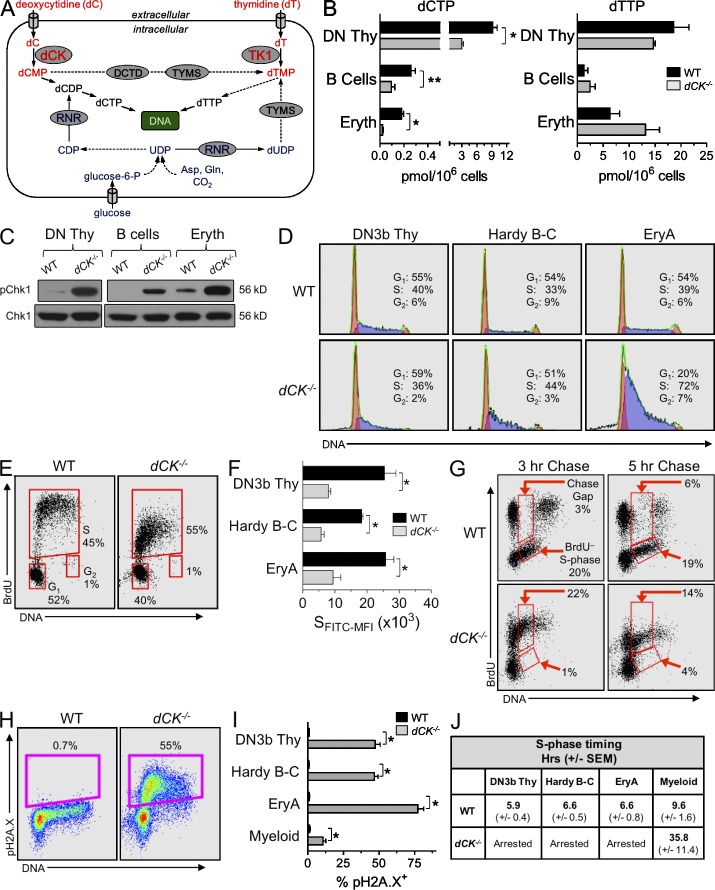
dCK inactivation causes severe RS in lymphoid and erythroid lineages. (A) Schematic of the de novo pathway (blue) and of the nucleoside salvage pathway (NSP, red) inputs into pyrimidine dNTP pools for DNA synthesis. Solid arrows indicate single-step processes; dashed arrows indicate multistep processes with intermediates not named/depicted in the schematic. Gln, glutamine; Asp, aspartate; UDP, uridine diphosphate; CDP, cytidine disphosphate; TYMS, thymidylate synthase; DCTD, dCMP deaminase; dCK, deoxycytidine kinase; TK1, thymidine kinase 1. (B) dCTP and dTTP pools in WT (black bars) and dCK−/− (gray bars) cells; DN Thy, CD4/CD8 double-negative thymocytes; B cells, BM-resident B cell progenitors; Eryth, BM-resident erythroid progenitors. Data are mean values ±SEM for three independent measurements generated from four pooled mice per genotype during each independent measurement. *, P < 0.003; **, P < 0.02. (C) Western blot detection of pChk1 (phosphorylated on Ser345) in lysates from lymphoid and erythroid progenitors. Total CHK1 protein was used as a loading control. (D) Representative examples of total DNA content staining and percentage of cells in the G1, S, and G2/M phases in WT and dCK−/− DN3b thymocytes, Hardy fraction B-C cells, and EryA cells. (E) Representative BrdU incorporation into WT and dCK−/− DN3b thymocytes 1 h after injection of BrdU. Percent of cells in G1, S, and G2 phases of the cell cycle are indicated. (F) Anti-BrdU FITC mean fluorescent intensities of S-phase cells (SFITC-MFI) from WT and dCK−/− DN3b thymocytes, Hardy fraction B-C cells, and EryA cells. Data are mean values ±SEM. n = 4 mice/genotype; *, P < 0.01. (G) Representative BrdU detection in WT and dCK−/− DN3b thymocytes after 3 and 5 h of BrdU chase (n = 4 mice/genotype). Percentages of cells present in chase gap and BrdU− S-phase gates are indicated. (H) Detection of H2A.X phosphorylated on Ser139 (pH2A.X) in DN3b thymocytes by flow cytometry. Percentages of pH2A.X-positive (pH2A.X+) cells are indicated. (I) Quantification of pH2A.X-positive staining in DN3b thymocytes, Hardy B-C cells, and EryA cells from WT (black bars) and dCK−/− (gray bars) mice. n = 7 mice/genotype; *, P < 0.0001. (J) S-phase durations, in hours, calculated using 5-h BrdU chase conditions in DN3b thymocytes, Hardy B-C cells, EryA cells, and BM-resident myeloid cells (CD11b+) from WT and dCK−/− mice. Data are mean values in hours ±SEM. n = 4 mice/genotype.
Although de novo dNTP synthesis is critically important for normal DNA replication and repair (Niida et al., 2010; D’Angiolella et al., 2012; Poli et al., 2012; Pontarin et al., 2012), the role played by the NSP in maintaining the proliferating capacity and genomic integrity of dividing cells is not well understood. In vitro studies using transformed cells and exogenous genotoxic agents demonstrated that both dCK and TK1 are involved in RS and DNA damage responses (DDR; Matsuoka et al., 2007; Chen et al., 2010). However, the precise mechanisms connecting dCK and TK1 to RS and DDR pathways are yet to be characterized. It is also unknown whether dCK and TK1 function in vivo to prevent endogenous RS and DNA damage induction in proliferating primary cells. Analyses of dCK−/− (Toy et al., 2010) and TK1−/− (Dobrovolsky et al., 2003) mice support the existence of a functional link between the NSP, RS, and hematopoietic development. Studies from our group (Toy et al., 2010), and confirmed independently by Choi et al. (2012), document severe developmental abnormalities affecting dCK−/− T cell, B cell, and erythroid lineages. TK1−/− mice also display hematopoietic defects characterized by slightly abnormal secondary lymphoid structures and by elevated levels of micronucleated erythrocytes (Dobrovolsky et al., 2003, 2005).
In this work, we sought to elucidate the biochemical and molecular mechanisms responsible for the hematopoietic phenotypes induced by dCK and TK1 inactivation. We measured dNTP pools, cell cycle kinetics, and RS levels in lymphoid and erythroid lineages from mice lacking dCK or TK1. dNTP deficiency and RS induction were detected in both dCK and TK1 KO models, prompting us to generate dCK/TK1 double KO (DKO) mice to analyze RS dynamics in highly proliferating NSP-deficient hematopoietic precursors. To our surprise, concomitant inactivation of both dCK and TK1 ameliorated, rather than aggravated, the dCK−/− hematopoietic phenotype. To gain additional mechanistic insight into how TK1 inactivation rescued the dCK−/− RS phenotype, we analyzed dCK−/−, TK1−/−, and dCK−/−;TK1−/− DKO T cell precursors in the OP9-DL1 co-culture model system for T lymphocyte development (Taghon et al., 2005). Collectively, our in vivo and in vitro studies demonstrate an antagonistic functional relationship between dCK and TK1 in regulating dCTP pools in vivo and reveal a previously unappreciated role for endogenous thymidine as a biologically active metabolite that is linked to the induction of RS in major hematopoietic lineages.
RESULTS
dCTP pool depletion, RS induction, early S-phase arrest, and DNA damage in dCK−/− lymphoid and erythroid hematopoietic lineages
To investigate the possibility that nucleotide deficiency causes the hematopoietic abnormalities identified in the dCK−/− mice (Toy et al., 2010), we quantified dNTP pools in CD4/CD8 double-negative (DN) thymocytes, BM-resident B cell progenitors, and in nucleated erythroid progenitors in WT and dCK−/− mice. dCTP pools were significantly reduced in all three dCK−/− hematopoietic lineages (Fig. 1 B; *, P < 0.003; **, P < 0.02); dTTP, dGTP, and dATP pools were largely unaffected (Fig. 1 B and Table 1). We next asked whether dCTP deprivation triggers RS in dCK−/− hematopoietic lineages by probing for the presence of the activated form of the key ataxia telangiectasia and Rad3-related protein (ATR) effector and the RS response (RSR) regulator CHK1 kinase (Branzei and Foiani, 2008). Compared with WT cells, dCK−/− lineages displayed significantly higher levels of CHK1 phosphorylated on Ser345 (pChk1), indicating exposure to endogenous RS in vivo (Fig. 1 C).
Table 1.
dATP and dGTP pools (pmol/106 cells) in WT and dCK−/− cells
| dATP pmol/106 cells (±SEM) | dGTP pmol/106 cells (±SEM) | |||||
| DN Thy | B cells | Erythroid | DN Thy | B cells | Erythroid | |
| WT | 1.08 ( 0.62) | 0.64 (±0.37) | 3.71 (±2.15) | 3.93 (±2.27) | 0.76 (±0.44) | 1.47 (±0.85) |
| dCK−/− | 0.80 (±0.46) | 1.19 (±0.69) | 2.32 (±1.34) | 2.00 (±1.16) | 0.69 (±0.40) | 2.10 (±1.21) |
DN Thy, CD4/CD8 double-negative thymocytes; B cells, BM-resident B-cell progenitors; Eryth, BM-resident erythroid progenitors. Data are mean values ±SEM for three independent measurements generated from four pooled mice per genotype during each independent measurement.
To examine the functional consequences of dCTP deficiency and RS, we analyzed the cell cycle profiles of highly proliferative subpopulations that we found to be selectively depleted (e.g., T and B cell precursors) or overrepresented (erythroblast precursors) among dCK−/− hematopoietic progenitor populations (Fig. S1). These highly proliferative subpopulations included DN3b T cell precursors (CD4−, CD8−, CD44−, CD25med-lo, and CD27hi thymocytes; Taghon et al., 2006), Hardy fraction B-C B cell progenitors (IgM−, B220+, CD43hi, and CD19hi; Hardy et al., 2007), and nucleated erythroblast cells (EryA; Ter119+, CD71+, and FSChi; Liu et al., 2006). dCK−/− cells from all three progenitor populations showed abnormal cell cycle profiles (Fig. 1 D), with dCK−/− EryA cells displaying the most pronounced increase in the percentage of cells in S-phase. Each of the three subpopulations appeared to be arrested in early S-phase (Fig. 1 D).
To unequivocally demonstrate that dCK inactivation triggers early S-phase arrest, we performed kinetic measurements of DNA synthesis in vivo by measuring BrdU incorporation into WT and dCK−/− DN3b, Hardy B-C, and EryA cells 1 h after intraperitoneal injection of the probe. The amount of BrdU incorporated per BrdU+ cell was reduced significantly in all three dCK−/− subpopulations (Fig. 1, E and F), a result consistent with impaired DNA synthesis. To confirm this interpretation, we used a BrdU chase approach to analyze productive DNA synthesis in vivo (Begg et al., 1985; Terry and White, 2006). Mice were pulsed with BrdU, and the free circulating probe was allowed to be eliminated (i.e., chased) for 3 and 5 h before analyzing BrdU incorporation. This approach enabled detection and quantification of BrdU chase gaps, which are indicators of productive DNA synthesis (Terry and White, 2006). Although BrdU+ WT cells synthesized sufficient new DNA during the 3- and 5-h chase points to generate typical BrdU chase gaps, BrdU+ dCK−/− cells did not display chase gaps, thus indicating arrested DNA synthesis (Fig. 1 G). Consequently, BrdU+ dCK−/− cells remained stationary in early S-phase (Fig. 1 G). Moreover, few dCK−/− cells entered the S-phase during the 3- and 5-h chase periods (Fig. 1 G, BrdU− S-phase gates), further confirming that dCK inactivation perturbed cell cycle kinetics. The degree of S-phase arrest triggered by dCK inactivation correlated with strong up-regulation of the RSR/DDR marker histone H2A.X phosphorylated on Ser139 (pH2A.X) in early S-phase cells from all three hematopoietic subpopulations (Fig. 1, H and I; *, P < 0.0001). Although the effects of dCK inactivation were highly penetrant in lymphoid and erythroid progenitor populations that have short S-phases (<7 h), myeloid lineage cells, which have a longer S-phase (>9 h), were significantly less affected (Fig. 1, I and J).
Effects of TK1 inactivation on dTTP pools, hematopoietic development, and endogenous RS
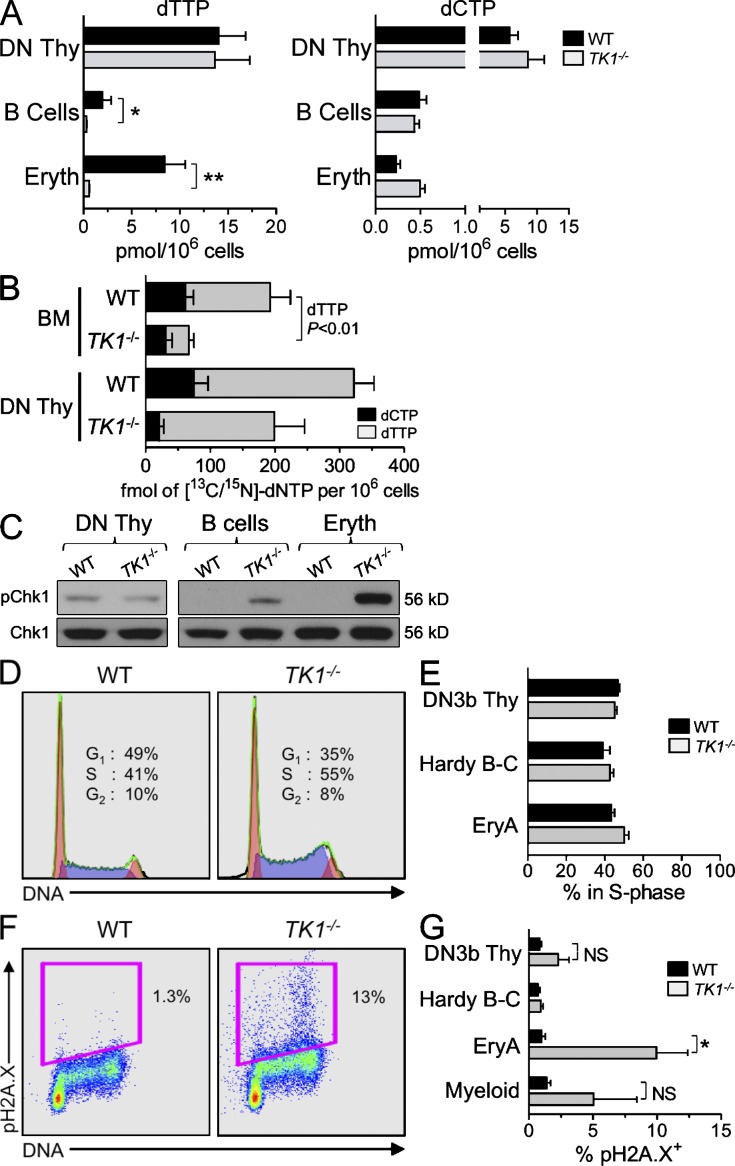
We next determined whether inactivation of TK1, the other cytosolic NSP kinase expressed in mammalian cells (Fig. 1 A), affected hematopoietic development in a manner similar to dCK inactivation. Unlike the severe defects in hematopoiesis induced by dCK inactivation, lymphopoiesis and erythropoiesis appeared to be significantly less affected in the TK1−/− mice (Fig. S1). Nonetheless, dTTP levels were significantly decreased in TK1−/− B cell and erythroid lineages compared with corresponding WT populations (Fig. 2 A; *, P < 0.005; **, P = 0.0001); dCTP pools were largely unaffected (Fig. 2 A).
Figure 2.
TK1 inactivation causes only minor RS in hematopoietic cells. (A) dTTP and dCTP pools from WT (black bars) and TK1−/− (gray bars) DN Thymocytes, BM-resident B cell progenitors, and BM-resident erythroid progenitors. Data are mean values ±SEM for three independent measurements. n = 4 mice/genotype/replicate; *, P < 0.005; **, P = 0.0001. (B) Concentrations (fmol per 106 cells) of [13C/15N]-dCTP (black bars) and [13C/15N]-dTTP pools (gray bars) from WT and TK1−/−-nucleated BM cells and DN thymocytes. Data are means ±SEM from n = 5 (WT) and n = 4 (TK1−/−) mice from two independent experiments. (C) Western blot detection of pChk1 in lysates from lymphoid and erythroid progenitors. Total CHK1 protein was used as a loading control. (D) Representative example of total DNA content staining in EryA cells from WT and TK1−/− cells, and (E) quantification of percentage of DN3b thymocytes, Hardy B-C cell, and EryA cells in S-phase as determined by total DNA content staining. Data are mean values ±SEM. n = 3 mice/genotype. (F) Representative example of pH2A.X detection in WT and TK1−/− EryA cells. (G) Quantification of pH2A.X-positive staining in DN3b thymocytes, Hardy B-C cell, EryA cells from WT (black bars), and TK1−/− (gray bars) mice. n = 4 mice/genotype. NS, P > 0.05; *, P < 0.03.
In contrast to dTTP pool depletions in TK1−/− B cell and erythroid lineages, TK1−/− DN thymocytes had normal dTTP pools, suggesting the existence of a compensatory mechanism for dTTP production. dCMP produced by dCK can contribute to the dTTP pool through the sequential actions of deoxycytidine monophosphate deaminase (DCTD) and thymidylate synthase (TYMS; Fig. 1 A; Staub et al., 1988). To determine whether such dCK-dependent contributions occur in vivo to potentially help maintain dTTP pools in the absence of TK1, we measured the relative efficiencies of deoxycytidine-to-thymidine conversion in WT and TK1−/− mice by using [13C/15N]-labeled deoxycytidine ([13C/15N]-dC). Both genotypes were pulsed with [13C/15N]-dC for 30 min, and the conversion of [13C/15N]-dC to [13C/15N]-dCTP and to [13C/15N]-dTTP was determined in target hematopoietic populations using liquid chromatography-tandem mass spectrometry (LC-MS/MS). The [13C/15N]-dTTP to [13C/15N]-dCTP ratio was more than twofold higher in TK1−/− DN thymocytes than in the corresponding WT subpopulation (7.3:1 vs. 3.5:1; Fig. 2 B). Therefore, DN thymocytes may compensate for the loss of the TK1-dependent input in their dTTP pools by converting a larger fraction of dCMP to dTTP (Fig. 2 A). In contrast to DN thymocytes, TK1−/− BM cells displayed a lower [13C/15N]-dTTP to [13C/15N]-dCTP ratio than WT BM cells (1.3:1 vs. 2.1:1; Fig. 2 B). There is an approximate fourfold decrease in the amount of [13C/15N]-dTTP in TK1−/− BM cells relative to WT controls (Fig. 2 B). Collectively, these findings suggest that, compared with TK1−/− DN thymocytes, TK1−/− BM cells are less able to rely upon dCK to maintain dTTP pools.
The dTTP pool depletion observed in TK1−/− BM cells suggested that, similar to their dCK−/− counterparts, these cells would also be subjected to RS induction. BM-resident TK1−/− hematopoietic populations displayed elevated pChk1 levels, whereas TK1−/− DN thymocytes were not affected (Fig. 2 C). These data are consistent with the dTTP pool depletion found in BM TK1−/− and with the normal dTTP pools in TK1−/− thymocytes, respectively (Fig. 2 A). Regardless of their dTTP pool status, proliferating TK1−/− hematopoietic progenitors did not display significant changes in the cell cycle profiles (Fig. 2, D and E). Moreover, in contrast to the highly elevated levels of pH2A.X found in dCK−/− DN3b thymocytes and Hardy B-C B cell progenitors (Fig. 1, H and I), corresponding TK1−/− lymphoid cells did not up-regulate pH2A.X; only TK1−/− EryA cells significantly up-regulated the expression of this DDR marker (Fig. 2, F and G; *, P < 0.03), albeit at levels that were eightfold lower than those observed in dCK−/− EryA cells (Fig. 1 I).
Blocking thymidine salvage alleviates the RS triggered by dCK inactivation in BM-resident hematopoietic progenitors
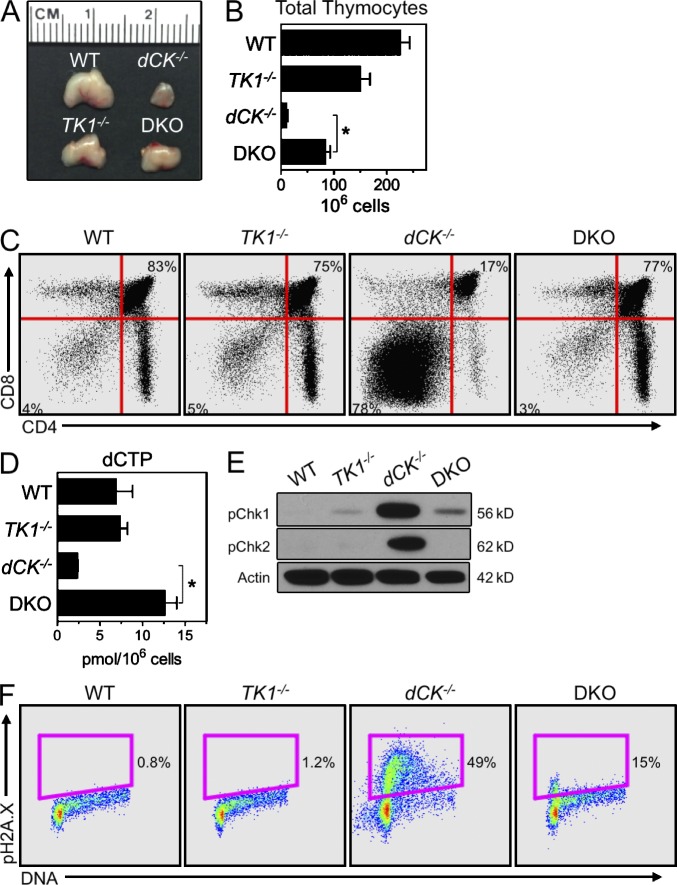
To further investigate the mechanistic basis of dNTP pool imbalances and RS phenotypes observed in the dCK−/− and TK1−/− mice, we examined the consequences of concomitant inactivation of these two NSP kinases. dCK−/−;TK1−/− DKO hematopoietic cells should completely lack the ability to salvage dNs from their extracellular environment and must rely exclusively on the de novo pathway to produce dNTPs for DNA replication and repair. DKO mice were born at sub-Mendelian frequencies (hazard ratio 1.85) and weighed 30% less than WT, TK1−/−, and dCK−/− littermates at 5 wk of age (n = 7/genotype; P < 0.001), indicating that the NSP is important to support animal growth. However, combined inactivation of dCK and TK1 did not further aggravate the defects observed in the dCK single-KO mice. Instead inactivation of TK1 significantly rescued the hematopoietic development in the dCK−/−;TK1−/− DKO mice. Thus, relative to dCK−/− cells, DKO B cell progenitors showed greatly improved differentiation after the Hardy B-C stage (Fig. 3, A-C). Moreover, the cell cycle profile and pH2A.X expression of DKO Hardy B-C cells appeared normal (Fig. 3 A). In the erythroid lineage, dual inactivation of dCK and TK1, partially reduced the relative abundance of EryA cells in the BM (Fig. S1), but DKO EryA cells still displayed an abnormal cell cycle profile with up-regulated pH2A.X expression relative to WT and TK1−/− cells (Fig. 3 D). Nonetheless, these cells were in fact relieved of the early S-phase arrest phenotype characteristic of their dCK−/− counterparts (Fig. 3 D). Intriguingly, pH2A.X up-regulation in DKO EryA cells occurred in mid/late S-phase rather than at the G1/S border corresponding to early stages of DNA synthesis that resulted from dCK inactivation alone (Fig. 3 D). Altogether, erythropoiesis itself in DKO mice appeared significantly normalized, as further indicated by the alleviation of the abnormal extramedullary erythropoiesis manifested as splenomegaly in the dCK−/− mice (Fig. 3 E).
Figure 3.
TK1 inactivation relieves the early S-phase RS in dCK−/− developing B and erythroid cells. (A) Representative examples of B cell development staining of BM samples from WT, dCK−/−, and dCK−/−;TK1−/− DKO mice. IgM and B220 staining of whole BM cells identify Hardy fraction A-D (B220+, IgM−) and Hardy fraction E-F (B220+, IgM+) populations. Hardy fraction A-D cells are sub-gated using CD43 and CD19 expression to identify Hardy Fraction A (CD43hi, CD19−), B-C (CD43hi, CD19+), and D (CD43lo, CD19+). Hardy fraction B-C cells are then analyzed for cell cycle position and pH2A.X expression. Plots are representative of n = 3 mice/genotype. (B and C) Quantification of percentage of total BM cells that are phenotypically Hardy fraction A-D (B), and Hardy fraction B-C (C) populations from WT, dCK−/−, and DKO mice. Data are mean values ±SEM for n = 3/genotype; *, P < 0.04; **, P < 0.01. (D) Representative examples of pH2A.X detection in EryA cells from WT, dCK−/−, TK1−/−, and DKO mice. n = 4 mice/genotype. (E) Representative images of spleens from WT, dCK−/−, TK1−/−, and DKO mice. (F) dCTP and dTTP pool measurements from nucleated BM cells from WT, dCK−/−, TK1−/−, DKO mice. Data are mean values ±SEM. n = 4 mice/genotype. *, P < 0.001.
To determine if the observation that TK1 inactivation rescued the early S-phase arrest in B cell and erythroid precursors in dCK−/− mice were reflected in biochemical restoration, we investigated whether dNTP pools were also restored to normal levels. This was, indeed, the case (Fig. 3 F; *, P < 0.001); the dCTP pool in DKO BM cells was statistically indistinguishable from that of WT BM cells. In contrast to the dCTP pools, dTTP pools from DKO BM cells were not restored to WT levels and remained at levels comparable to those of TK1−/− BM cells.
TK1 inactivation rescues the development of dCK−/− T cells
Similar to its effects on the development of dCK−/− B cells, TK1 inactivation also rescued dCK−/− T cell development. DKO thymi were significantly larger than those from the dCK−/− mice because of increased overall cellularity (Fig. 4, A and B; *, P < 0.001). In DKO mice, the eightfold increase in thymic cellularity over dCK−/− thymi was accompanied by a striking normalization of a key event in thymic T cell development, specifically the transition from the DN stage to the CD4/CD8 double-positive (DP) stage (Fig. 4 C). Thus, in contrast to the dCK−/− thymocytes, which experience a severe block at the DN to DP transition, DKO thymocytes displayed a DN to DP distribution that was nearly indistinguishable from that of their WT and TK1−/− counterparts (Fig. 4 C). The phenotypic rescue likely reflected the restoration of dCTP pools in the DKO DN thymocytes (Fig. 4 D; *, P < 0.01). The DKO DN thymocytes also expressed significantly reduced levels of the RSR activation markers pChk1 and CHK2 kinase phosphorylated on Thr68 (pChk2) compared with the corresponding dCK−/− population (Fig. 4 E). In addition, both the early S-phase arrest and pH2.AX induction were significantly reduced in DKO cells relative to dCK−/− DN3b thymocytes (Fig. 4 F). Collectively, these findings indicated that elimination of TK1 activity drastically lowered the levels of RS in DKO versus dCK−/− thymocytes, an effect explained by normalization of dCTP pools and reflected by the dramatic differences between T cell development in the DKO versus dCK−/− mice (Fig. 4 A and Fig. S1).
Figure 4.
TK1 inactivation normalizes the development of dCK−/− T cells. (A and B) Gross thymus size (A), and total viable thymocytes recovered (B), from WT, dCK−/−, TK1−/−, and DKO mice. Data are mean values ±SEM. n = 5 mice/genotype; *, P < 0.001. (C) Representative example of CD4 and CD8 staining of total thymocytes from WT, dCK−/−, TK1−/−, and DKO mice. Percentages of DN thymocytes (bottom left gate) and DP thymocytes (top right gate) are indicated. (D) Measurements of dCTP pools in DN thymocytes isolated from WT, dCK−/−, TK1−/−, and DKO mice. Data are mean values ±SEM. n = 2 mice/genotype; *, P < 0.01. (E) Western blot detection of pChk1 and Chk2 phosphorylated on Thr68 (pChk2) in DN thymocytes. Actin was used as a loading control. (F) Representative examples of pH2A.X detection in DN3b thymocytes from WT, dCK−/−, TK1−/−, and DKO mice.
Endogenous thymidine plays a critical role in the induction of a RS phenotype in dCK−/− thymocytes
In in vitro studies, excessive levels of intracellular dTTP have been shown to negatively regulate dCTP pools through allosteric inhibition of RNR-mediated synthesis of deoxycytidine diphosphate (dCDP), the direct precursor of dCTP (Reichard, 1988). It is conceivable that the RS experienced by dCK−/− hematopoietic cells in vivo can be attributed to concomitant inhibition of both salvage and de novo pathways for dCTP production, therefore creating conditions of synthetic sickness/lethality (SSL). As shown in Fig. 5 A, hematopoietic progenitors in BM and thymus are exposed to endogenous concentrations of thymidine that are significantly higher than thymidine levels in other tissues and in plasma. Such high levels of endogenous thymidine may increase dTTP pools via TK1-mediated salvage. Decreased dTTP production caused by TK1 inactivation would relieve the inhibition of RNR’s ability to reduce CDP to dCDP in proliferating dCK−/− cells residing in BM and thymus. The corresponding increase in the output of the de novo pathway would therefore help restore dCTP pools depleted by dCK inactivation.
Figure 5.
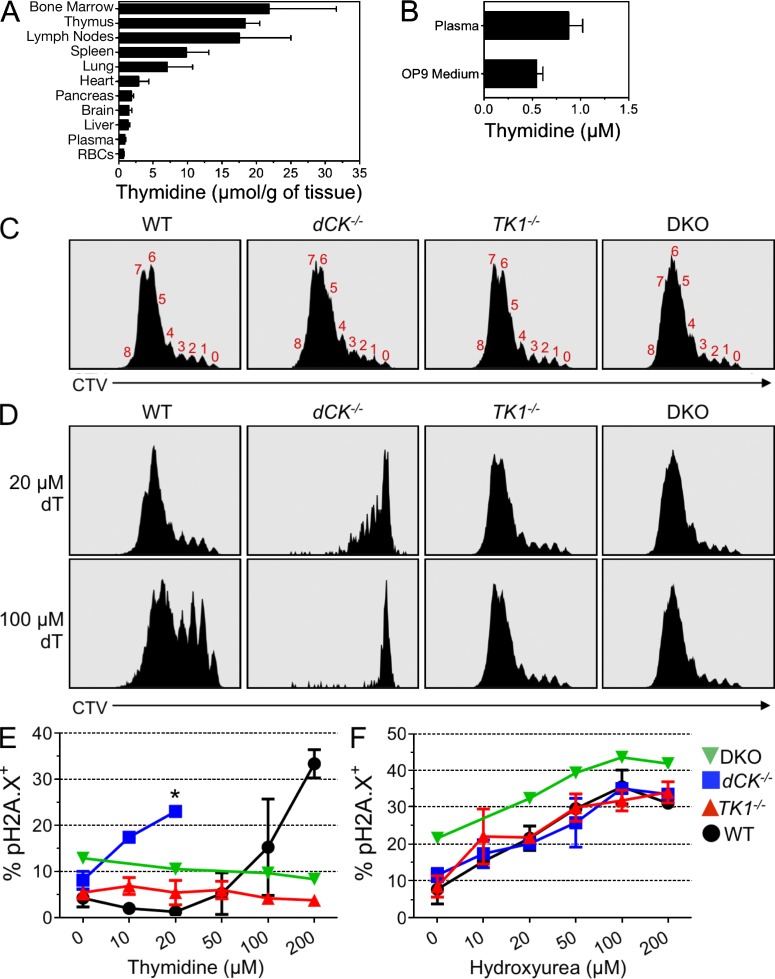
Thymidine induces RS in cultured dCK−/− thymocytes. (A) Relative thymidine abundance in various tissues from C57BL/6 mice as determined by LC-MS/MS. Concentrations given in μmol/g of whole tissue. Data are means ±SEM; n = 5/tissue type. (B) Concentrations of thymidine (in μM) from C57BL/6 plasma and from standard OP9-DL1 culture medium, as determined by LC-MS/MS. Data are means ±SEM; Plasma, n = 7; Media, n = 3. (C) Representative CellTrace Violet (CTV) dye dilution curves from WT, dCK−/−, TK1−/−, and DKO DN3a thymocytes cultured on OP9-DL1 stroma for 4 d without thymidine supplementation in the medium. Red numbers above the distinct CTV peaks reflect the total number of completed cell divisions executed by the thymocyte populations. WT, dCK−/−, and TK1−/−, n = 4; DKO, n = 1. (D) CTV dye dilution curves after 4 d of culturing in the presence of 20 and 100 µM thymidine added to the culture medium. WT, dCK−/−, and TK1−/−, n = 4; DKO, n = 1. (E) Percent of live cells that are pH2A.X positive in WT (black circles), dCK−/− (blue squares), TK1−/− (red up triangles), and DKO (green down triangles) DN3b thymocytes after 48 h of stimulation in increasing concentrations of thymidine. WT, dCK−/−, and TK1−/−, n = 2; DKO, n = 1. *, Cessation of measurements of pH2A.X levels caused by massive cell death (>80% sub-G1 staining; not depicted) induced by exposure of dCK−/− cells to concentrations of thymidine equal to or greater than 50 µM. (F) pH2A.X expression in WT, dCK−/−, TK1−/−, and DKO DN3b thymocytes after 12 h of exposure to increasing concentrations of hydroxyurea 36 h after plating on OP9-DL1 stroma. WT, dCK−/−, and TK1−/−, n = 2; DKO, n = 1.
To test the validity of this model we reasoned that if excessive thymidine salvage in thymus and BM was indeed responsible for the dCK−/− phenotype, then removal of dCK−/− hematopoietic cells from their thymidine-rich in vivo environment should relieve their RS phenotype. To test this prediction, we modeled T cell development in vitro using the OP9-DL1 co-culture system (Taghon et al., 2005). Importantly, LC-MS/MS measurements showed that the OP9-DL1 culture media used in these experiments contained submicromolar amounts of thymidine that are comparable to plasma levels (Fig. 5 B), and are much lower than those measured in thymus and BM (Fig. 5 A). DN3a thymocytes (CD4−, CD8−, CD44−, CD25hi, and CD27lo) from WT, dCK−/−, TK1−/−, and DKO mice were labeled with a fluorescent proliferation dye (CellTrace Violet; CTV), cultured on the OP9-DL1 stroma, and then assessed for cell division four days later by flow cytometric analysis of CTV dye dilution (Quah and Parish, 2012). In contrast to their in vivo phenotype, dCK−/− DN3a thymocytes proliferated robustly in the in vitro co-culture system that contained submicromolar levels of thymidine; dCK−/− cells divided up to 8 times, equivalent to the rate of cell division observed for WT, TK1−/−, and DKO thymocytes over the course of the in vitro study (Fig. 5 C). These findings indicated that salvage of endogenous thymidine in vivo contributes to the induction of S-phase arrest and RS in dCK−/− hematopoietic progenitors.
To confirm the hypothesis that TK1-dependent thymidine salvage mediates S-phase arrest and RS in dCK−/− hematopoietic progenitors, we titrated thymidine into the DN3a/OP9-DL1 co-cultures and determined how this affected the rate of cell proliferation. The addition of 20 µM thymidine to the co-culture medium significantly blocked the proliferation of dCK−/− cells without affecting WT cell division (Fig. 5 D, top row). Further increasing the thymidine concentration from 20 to 100 µM completely and specifically blocked the proliferation of dCK−/− cells (Fig. 5 D, bottom row). As expected, TK1−/− and DKO cells were unaffected by thymidine at either concentration (Fig. 5 D), because of their inability to salvage this deoxyribonucleoside.
To verify that the thymidine-induced proliferation block was caused by RS induction, we assayed pH2A.X activation in DN3b/OP9-DL1 co-cultures in response to thymidine concentrations varying from 10 to 200 µM. dCK−/− thymocytes strongly induced pH2A.X after exposure to thymidine concentrations as low as 10 µM. pH2A.X expression by dCK−/− cells peaked at 20 µM thymidine (Fig. 5 E); higher thymidine concentrations triggered massive cell death in the dCK−/− DN3b/OP9-DL1 co-cultures (percent sub-G1 cells >80% at 50 µM or greater thymidine concentrations; n = 3 independent experiments; unpublished data). WT cells were 10-fold more resistant to thymidine than dCK−/− cells; TK1−/− and DKO cells were completely resistant to thymidine at all tested concentrations (Fig. 5 E).
Lastly, to determine whether dCK−/− cells were intrinsically more susceptible to RS induced by nucleotide deprivation via a TK1-independent mechanism, we titrated the RNR inhibitor hydroxyurea into the DN3b/OP9-DL1 co-cultures, and then measured RS induction 12 h later. WT, dCK−/−, TK1−/−, and DKO cells were equally susceptible to hydroxyurea concentrations ranging from 10–200 µM (Fig. 5 F), suggesting that dCK plays a specific role in preventing RS induced by thymidine. In conclusion, data from the OP9-DL1 co-culture in vitro T cell development system showed that dCK-deficient cells were hypersensitive to RS induced by low amounts of thymidine through a TK1-dependent mechanism.
DISCUSSION
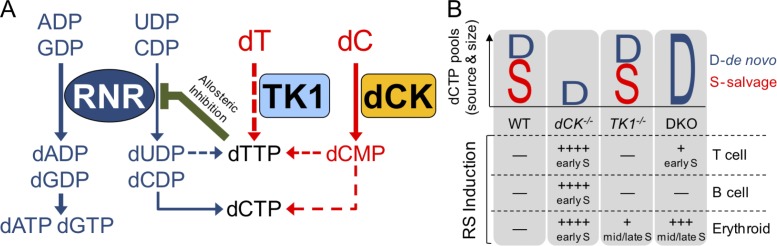
Using genetic models of individual and combined deficiencies in the cytosolic dN kinases dCK and TK1, we demonstrate that the dN salvage pathway contributes both to induction and to prevention of RS during hematopoietic development. We also show that endogenous thymidine itself is a highly active metabolite that induces RS in vivo, an observation reminiscent of a widely used in vitro experimental approach for synchronizing cells in cell cycle known as the “thymidine block” (Xeros, 1962). In the course of a typical thymidine block experiment, cultured cells exposed to high concentrations of thymidine undergo S-phase arrest through a TK1-dependent mechanism (Gagou et al., 2010). It is through the kinase action of TK1 that exogenously added thymidine is trapped in cells, thus enabling its conversion to dTTP in the cytosol. dTTP is not only a precursor of DNA but also a strong allosteric inhibitor of RNR’s affinity for its pyrimidine ribonucleotide substrates CDP and uridine diphosphate (UDP; Fairman et al., 2011). dTTP-mediated inhibition of CDP conversion to dCDP depletes dCTP pools, thus causing S-phase arrest (Bjursell and Reichard, 1973; Larsson et al., 2004). The thymidine block approach has been thought of traditionally as an in vitro “laboratory tool” (Gentry, 1992) with little, if any, relevance to in vivo conditions. Our data challenge this assumption by establishing thymidine block as a metabolic phenomenon that normally occurs in BM and thymus during hematopoiesis. We show that, in the absence of dCK activity, physiological concentrations of thymidine in these tissues are sufficient to induce dCTP deprivation and severe RS in proliferating T cell, B cell, and erythroid precursors. By enabling these cells to salvage deoxycytidine, dCK replenishes dCTP pools depleted by dTTP produced from thymidine via TK1, thereby exerting an important role in hematopoiesis. The two cytosolic dN salvage kinases thus play paradoxically opposing roles; TK1 in the absence of dCK induces RS, whereas dCK functions to prevent RS in TK1-expressing cells exposed to elevated levels of endogenous thymidine (Fig. 6, A and B). Importantly, the degree of dependence on the NSP kinases to avoid endogenous RS varied across different hematopoietic lineages, with the erythroid lineage being most affected, followed by T and B cell progenitors and then by myeloid precursors (Fig. 1 J and Fig. 6 B).
Figure 6.
Deoxyribonucleoside salvage kinases induce and resolve RS during hematopoiesis. (A) Under normal conditions, the RNR complex reduces purine ribonucleotide diphosphates (ADP and GDP) and pyrimidine ribonucleotide diphosphates (CDP and UDP) to contribute to dNTP pools (dATP, dGTP, dTTP, and dCTP). Although in hematopoietic cells RNR appears to be solely responsible for producing purine dNTP pools, the majority of dTTP pools are produced from salvaged thymidine, which is present in abundant amounts in thymus and BM. dCK may also contribute to dTTP pools as shown in Fig. 1 A and Fig. 2 C. Elevated dTTP levels prevent RNR from reducing CDP to dCDP and possibly UDP to dUDP via allosteric inhibition. To maintain dCTP pools, rapidly dividing hematopoietic cells rely on deoxycytidine salvage via dCK. (B) Graphical representation of the source (D-de novo, S-salvage) and size (height of D or S) of dCTP pools in various hematopoietic lineages. In the absence of dCK activity (dCK−/− column), dCTP pools become insufficient, leading to severe RS (++++) and DNA synthesis arrest in early S-phase in T cell, B cell, and erythroid cell precursors. In the absence of TK1 activity (TK1−/− column), dCTP pools are unaffected and only mild RS (+) occurs in late S-phase in erythroid precursors. The mild RS may be caused by an imbalanced dUTP/dTTP ratio in the absence of TK1. When both dCK and TK1 are inactivated (DKO column), de novo dCDP production is de-repressed and, subsequently, dCTP pools are restored to WT levels. DKO thymocytes have measurable, but overall mild, levels of RS (+) in early S-phase. The absence of NSP is well tolerated by B cell precursors, but it results in severe late S-phase RS (+++) in erythroid precursors.
The causal relationship between TK1 activity, inhibition of de novo dCTP production, and dependency on dCK to avoid thymidine-induced endogenous RS inevitably leads to the question of why rapidly dividing hematopoietic cells engage in what at first sight would appear to be excessive salvage of thymidine. Further adding to the puzzle of why hematopoietic cells express high levels of TK1 is that our data show that hematopoiesis appears to be much less severely affected by TK1 inactivation than by the loss of dCK activity. Although additional studies are needed to fully elucidate the biological significance of TK1 by further analyses of the consequences of its inactivation on hematopoiesis, our preliminary data indicate that TK1 may play an important role in regulating the dUTP/dTTP balance; we observe an approximately fourfold increase in dUTP/dTTP ratio in TK1−/− BM cells relative to WT BM cells (unpublished data). A proper dUTP/dTTP ratio is needed to reduce misincorporation of dUTP into DNA (Melnyk et al., 1999), thereby preventing the formation of U:A pairs. Excessive occurrences of such pairs may have cytotoxic effects (Hagen et al., 2006) and would require the intervention of uracil-DNA glycosylases and of base excision repair mechanisms to prevent induction of RS. TK1 activity could promote the maintenance of a low dUTP/dTTP ratio in three ways. First, through preferential phosphorylation of thymidine versus deoxyuridine caused by a 20-fold difference in affinity in favor of thymidine (Munch-Petersen et al., 1991), TK1 is likely to generate significantly more dTMP than dUMP. In turn, this would increase the substrate availability for thymidylate kinase, an enzyme recently shown to play an important role in preventing dUTP incorporation into the DNA during DNA repair (Hu et al., 2012). Second, by opposing the activity of cytosolic nucleotidases, which dephosphorylate dTMP, TK1 activity will prevent the loss of thymidine containing deoxyribonucleotides from dividing cells (He and Skog, 2002). Third, by increasing cytosolic dTTP pools, TK1 would promote the allosteric inhibition of RNR’s ability to reduce not only CDP but also UDP (Fairman et al., 2011; Fig. 6 B), therefore preventing excessive accumulation of dUTP caused by high RNR activity in proliferating cells. If confirmed by future studies, this hypothesis may explain why the dTTP-mediated allosteric regulation of RNR’s substrate specificity is mostly conserved among members of the three classes of RNRs, despite wide differences in their primary and quaternary structures (Reichard, 1993).
The role of the NSP in nonhematopoietic tissues
Our findings indicate that the de novo synthesis pathway can maintain dNTP pools at levels that are sufficient to support most hematopoietic proliferation. However, the observation that DKO mice are born at sub-Mendelian frequencies and that surviving animals display growth retardation indicates that the NSP is important in supporting overall growth. It is possible that the growth defects affecting the DKO mice may result from increased stress forced upon other biosynthetic pathways in the absence of the NSP. The utilization of dN salvage for dNTP synthesis requires significantly less ATP and NADPH than does de novo synthesis (Lunt and Vander Heiden, 2011). For example, de novo synthesis of 1 mol of either dCTP or dTTP starting from glucose, glutamine, aspartate, and bicarbonate requires 6 moles of ATP each, while the same amount of dCTP or dTTP can be generated via salvage at a cost of 3 moles of ATP (Voet and Voet, 2004). ATP and NADPH savings afforded by using pyrimidine salvage instead of de novo synthesis could then be spent by cells to generate other essential biomass, such as proteins and phospholipids. An alternative explanation for the growth defects in the DKO mice involves mitochondrial dysfunction due to imbalances in the dNTP pools in this organelle. Defects in mitochondrial dNTP synthesis have been shown to cause severe developmental abnormalities (Eriksson and Wang, 2008; Zhou et al., 2008; Zhou et al., 2010; González-Vioque et al., 2011). Since in cycling cells mitochondrial dNTP pools correlate linearly with the cytoplasmic pools (Gandhi and Samuels, 2011), imbalances in cytosolic dNTP pools resulting from defects in the NSP may impair mitochondrial DNA synthesis and thus trigger mitochondrial stress manifested as embryonic lethality and neonatal growth retardation.
Nucleotide deficiency, cellular transformation, and the relationship between the NSP and the RSR pathway
Nucleotide deficiency has recently been shown to promote cellular transformation (Bester et al., 2011). The study by Bester et al. described how overexpression either of viral E-proteins or of mammalian Cyclin E in cultured human keratinocytes overrides normal cell cycle regulation and triggers premature S-phase entry, which in turn causes RSR pathway activation and subsequent mutation induction. Presumably, this chain of events is initiated by the inability of E-protein/Cyclin E–overexpressing keratinocytes to generate sufficient dNTP pools to support DNA replication and repair. The nucleotide deficiency observed here in our in vivo genetic systems resembles that induced by Bester et al. (2011) in their cell culture system. However, despite the severe nucleotide deficiency and strong activation of the RSR pathway observed in the dCK−/− hematopoietic lineages, we did not detect increased rates of cancer in the dCK−/− mice when these animals were followed for 12 mo after birth. It is possible that onset of nucleotide deficiency-induced cancer in our model will occur at an even older age. It is also conceivable that the protective effects of the RSR pathway in dCK−/− mice are sufficient to eliminate potentially oncogenic cells, whereas a yet to be defined aspect of the keratinocyte system permits the propagation of cells carrying fixed DNA mutations. Future studies will be required to determine the consequences of concomitant inactivation of the NSP and of key components of the RSR pathway. In this context, it is important to note the similarities between the defects in erythropoiesis observed in the NSP-deficient mice and defects in this hematopoietic lineage documented in mouse models of ATR-Seckel (Murga et al., 2009) and of Chk1 haploinsufficiency (Boles et al., 2010).
Therapeutic implications
The demonstration of a functional link between endogenous thymidine, dN kinases, nucleotide deficiency, and RS induction highlights a potential therapeutic strategy to induce RS overload, and ultimately cell death, in hyperproliferative diseases such as cancer. Thymidine therapy has been attempted in human malignancies, based on the ability of this nucleoside to induce S-phase arrest of cultured cancer cells by inhibiting their de novo dCTP synthesis. Although well tolerated in patients, thymidine had limited efficacy when used as single-agent therapy in acute lymphoid and myeloid leukemias (Kufe et al., 1980). By demonstrating that dCK, which is frequently overexpressed in hematological malignancies, significantly contributes to dCTP pools in vivo, the current study may have revealed an important mechanism of resistance to thymidine and thus explain the failure of thymidine in initial clinical trials. Combining thymidine with a small molecule dCK inhibitor (dCKi) would induce SSL by blocking both dCTP-producing pathways. Whether an adequate therapeutic window exists for thymidine/dCKi SSL will be determined in subsequent studies.
MATERIALS AND METHODS
Animals.
Mice were bred and housed under specific pathogen–free conditions and were treated in accordance with the UCLA Animal Research Committee protocol guidelines. The dCK−/− mice were generated and bred as previously described and backcrossed to C57BL/6 for n = 7 generations (Toy et al., 2010). TK1−/− mice were rederived from cryopreserved embryonic stem cells stored at the UC Davis Repository (stock VG18248) and genotyped as previously described (Dobrovolsky et al., 2003). dCK/TK1 DKO mice were generated by first intercrossing dCK+/− mice to TK1+/− mice to generate dCK+/− TK1+/− progeny. dCK+/− TK1+/− mice were then intercrossed together to generate dCK/TK1 DKO mice at potential 1:16 frequency. Age-matched (5–12-wk-old) WT, dCK−/−, TK1−/−, and DKO littermates were used for all experiments except for the endogenous thymidine determinations from whole tissue. Tissues for endogenous thymidine measurements were extracted from 6-wk-old C57BL/6 mice purchased from the UCLA Radiation Oncology breeding colony.
Tissue preparation.
Single-cell suspensions were prepared from BM by flushing femur, tibia, and humerus bones with DMEM supplemented with 2% FBS using 25-G needles, followed by 70-µm filtration. Whole BM was depleted of mature red blood cells by overlaying cell suspensions on a solution of 16% iodixanol (D1556; Sigma-Aldrich), 0.63% NaCl, 10 mM Hepes, pH 7.4, and 0.1% NaN3, followed by centrifugation at 900 g for 15 min. The mononuclear cell–containing supernatant was transferred and washed twice before antibody staining or dNTP extraction. Single-cell suspensions of thymus were prepared by mechanical dissociation using frosted glass slides in DMEM supplemented with 2% FBS and 50 µg/ml DNaseI (Roche) and passed through 70-µm sterile filters. CD4−/CD8− thymocytes were purified from whole thymocyte suspensions using combined CD4 and CD8 negative selection kits (Invitrogen).
Immunophenotyping antibodies.
The following antibodies from eBioscience were used for B cell, erythroid cell, and myeloid cell phenotyping from whole BM: B220 (clone RA3-6B2) PE-Cy7 and APC-eFluor780; IgM (clone II/41) FITC, PerCP-eFluor710; CD43 (clone eBioR2/60) PE; CD19 (clone 1D3) APC and PE-Cy7; Ter119 (clone TER-119) PerCP-Cy5.5 and PE-Cy7; CD71 (clone R17217) APC and PE; and CD11b (clone M1/70) APC-eFluor780. The following antibodies from eBioscience were used for thymocyte phenotyping: CD4 (clone L3T4) FITC, PE, Alexa Fluor 700, and PE-Cy7; CD8a (clone 53–6.7) PE, PE-Cy7, and PerCP-eFluor710; CD25 (clone PC61.5) APC and PE-Cy7; CD44 (clone IM7) APC-eFluor780; CD27 (clone LG.7F9) PE; and CD45 (clone 30-F11) PE-Cy7. Hematopoietic stem cell phenotyping from whole BM: Lineage cocktail PE, CD150 PE-Cy5 (clone TC15-12F12.2), (BioLegend); CD127 APC-eFluor780 (clone A7R34), Flk2/Flt3 Biotin (clone A2F10), Sca-1 PerCP-Cy5.5 (clone D7), c-Kit PE-Cy7 (clone 2B8), CD34 eFluor660 (clone RAM34), CD16/32 Alexa Fluor 700 (clone 93; eBioscience); streptavidin PE-Alexa Fluor 610 (Invitrogen).
Flow cytometry analyses.
All flow cytometry data were acquired on four- and five-laser LSRII cytometers (BD) for analysis, and FACS purification of cells was performed on four-laser BD FACSAriaII cell sorters running BD FACSDiva6 software (BD). All cytometry data were analyzed using FlowJo software (Tree Star).
DNA content staining and intracellular detection of BrdU and pH2A.X.
BrdU (1 mg/mouse) was administered by intraperitoneal injection. Cells were collected, antibody-stained for surface antigens, and then processed for intracellular detection of BrdU and pH2A.X using a BrdU-FITC kit staining protocol and reagents (BD). Cells were then stained with BrdU-FITC and/or pH2A.X antibodies conjugated to FITC (Millipore; clone JBW301) or Alexa Fluor 647 (BD; clone N1-431). Total DNA content was assessed by staining with DAPI (Roche) at 1 µg/ml final concentration in PBS containing 2% FBS.
CTV labeling of thymocytes.
Single-cell suspensions of thymocytes were resuspended in PBS containing 0.5% FBS at a cell density of 50 × 106 cells/ml. 5 mM stocks of CTV dye (Invitrogen) dissolved in DMSO were diluted to 50 µM in PBS/0.5% FBS and then added at a dilution of 1:10 to the cell suspension (5 µM final concentration). Cells were mixed well, incubated at 37°C for 20 min, and then washed twice with 40 ml of PBS/5% FBS. Cells were then stained with fluorescent antibodies to identify DN3a thymocytes and purified via FACS.
OP9-DL1 co-cultures.
OP9 cells were purchased from American Type Culture Collection (CRL-2749) and transduced by retroviral infection with delta-like Ligand 1 vector (DL1), provided by J. Carlos Zúñiga-Pflücker (University of Toronto, Toronto, Ontario, Canada), and FACS-sorted based on their GFP fluorescence. OP9-DL1 cells were maintained in Minimum Essential Medium, Alpha Modification (Sigma-Aldrich) supplemented with 20% FBS, 100 U/ml penicillin, 100 µg/ml streptomycin. Thymocyte/OP9-DL1 co-cultures were initiated by plating 50 × 103 OP9-DL1 cells in 250 µl of media/well in 48-well tissue culture dishes at day −1. On day 0, DN3a thymocytes were FACS purified and resuspended in OP9-DL1 media supplemented with 10 ng/ml Flt3-ligand (PeproTech) and 10 ng/ml IL-7 (PeproTech) at a cell density of 105 cells/ml. The single-cell suspensions (250 µl) were then plated atop OP9-DL1 monolayers (25 × 103 cells; 5 ng/ml final concentrations of Flt3-ligand and IL-7 in a 500 µl total volume). Co-cultures were incubated for 2 or 4 d. Thymocytes were harvested by forceful pipetting, stained for developmental antigens and intracellular markers, and analyzed by flow cytometry.
Western blots.
Purified hematopoietic cell populations were lysed in 1X RIPA buffer containing 1X Halt Protease/Phosphatase Inhibitor (78440; Thermo Fisher Scientific); supernatants were isolated after centrifugation at 17,000 g for 15 min. Lysates were mixed with 1X Laemmli-SDS loading buffer, boiled, electrophoresed, and transferred to nitrocellulose membranes for immunoblotting. Monoclonal rabbit anti–phospho-CHK1 (Ser345; clone 133D3); monoclonal mouse anti-CHK1 (clone 2G1D5); and monoclonal rabbit anti-Phospho-CHK2 (Thr48; clone C13C1) were purchased from Cell Signaling Technology. Monoclonal mouse anti-actin (clone MM2/193) was purchased from Sigma-Aldrich.
Intracellular dNTP pool measurements.
Purified hematopoietic cell populations were counted and pelleted. Pellets were then suspended in 1 ml of ice-cold 60% methanol, vortexed for 1 min and stored overnight at -20°C. The following day, the lysates were boiled for 3 min and then centrifuged for 15 min at 17,000 g at 4°C. Supernatants were evaporated overnight in a SVC100H SpeedVac Concentrator (Savant). Dry pellets were resuspended in 100 µl ddH2O, vortexed and centrifuged for 15 min at 17,000 g at 4°C to clear insoluble debris. 5 µl of concentrated lysate was used in a 25-µl reaction volume. Reactions were performed for 2 h according to previously described protocols (Mathews and Wheeler, 2009).
In vivo measurements of dCK-dependent incorporation of [13C/15N]-deoxycytidine into dCTP and dTTP pools.
Mice were injected intraperitoneally with 200 µl of 2.5 mM uniformly labeled [13C/15N]-deoxycytidine (Cambridge Isotopes). Mice were euthanized 30 min after injection to harvest BM cells and DN thymocytes. Cells were washed with ice cold PBS/2% FBS, and then extracted with 60% methanol overnight at −20°C, evaporated, and the dry pellets were resuspended in 100–500 µl of 5 mM hexylamine. dTTP/dCTP concentrations were then determined by LC-MS/MS using an Agilent 6460 Triple Quad LC/MS system. The stationary phase was a Phenomenex Gemini-NX C18 column (2.0 × 100 mM) with 5 mM hexylamine in ddH2O and acetonitrile as the mobile phases.
Whole-tissue nucleoside concentration measurements.
Solid tissues were homogenized at 4°C in acetonitrile/methanol (9:1) using the BulletBlender Tissue Homogenizer (NextAdvance) according to the manufacturer’s specifications. BM cells were flushed using ice-cold PBS, filtered into single-cell suspensions, pelleted, and resuspended in homogenization buffer. Whole blood was collected in heparinized tubes, and then centrifuged to separate plasma and blood cells. Both fractions were extracted directly with the homogenization buffer. Extracted samples were evaporated, and the dry pellets were resuspended in ddH2O. Thymidine concentrations were then determined by LC-MS/MS. The stationary phase was a Thermo Fisher Hypercarb column (2.1 × 100 mM) with 0.1% formic acid in ddH2O and 0.1% formic acid in acetonitrile as the mobile phases.
Statistical analyses.
All statistics presented as means of biological replicates with standard error of the mean (±SEM). P-value significances were calculated using one sample Student’s t test function in GraphPad Prism 5 (GraphPad Software).
Online supplemental material.
Figure S1 shows representative examples of hematopoietic stem cell phenotyping, thymocyte phenotyping, BM B cell developmental phenotyping, BM erythroid phenotyping for WT, dCK−/−, TK1−/−, and DKO mice. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20121061/DC1.
Supplementary Material
Acknowledgments
We thank the flow cytometry cores of the UCLA Jonsson Comprehensive Cancer Center, UCLA Broad Stem Cell Research Center, and the UCLA Institute of Molecular Medicine for use of their equipment. We acknowledge Dr. Kym F. Faull and the UCLA Pasarow Mass Spectrometry laboratory for their assistance with LC-MS/MS measurements. We acknowledge Drs. Steve Bensinger and Mike Teitell for critically reading the manuscript.
This work was funded by In Vivo Cellular and Molecular Imaging Centers Developmental Project Award National Institutes of Health P50 CA86306 (C.G. Radu, H.R. Herschman, and W.R. Austin), and the US National Cancer Institute grant 5U54 CA119347 (C.G. Radu). O.N. Witte is an Investigator of the Howard Hughes Medical Institute.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- ATR
- ataxia telangiectasia and Rad3-related protein
- dCK
- deoxycytidine kinase
- DDR
- DNA damage response
- DKO
- double KO
- LC-MS/MS
- liquid chromatography-tandem mass spectrometry
- NSP
- nucleoside salvage pathway
- RS
- replication stress
- RNR
- ribonucleotide reductase
- RSR
- RS response
- SSL
- synthetic sickness/lethality
- TK1
- thymidine kinase 1
- UDP
- uridine diphosphate
References
- Arlt M.F., Ozdemir A.C., Birkeland S.R., Wilson T.E., Glover T.W. 2011. Hydroxyurea induces de novo copy number variants in human cells. Proc. Natl. Acad. Sci. USA. 108:17360–17365 10.1073/pnas.1109272108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnér E.S., Eriksson S. 1995. Mammalian deoxyribonucleoside kinases. Pharmacol. Ther. 67:155–186 10.1016/0163-7258(95)00015-9 [DOI] [PubMed] [Google Scholar]
- Begg A.C., McNally N.J., Shrieve D.C., Kärcher H. 1985. A method to measure the duration of DNA synthesis and the potential doubling time from a single sample. Cytometry. 6:620–626 10.1002/cyto.990060618 [DOI] [PubMed] [Google Scholar]
- Bester A.C., Roniger M., Oren Y.S., Im M.M., Sarni D., Chaoat M., Bensimon A., Zamir G., Shewach D.S., Kerem B. 2011. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell. 145:435–446 10.1016/j.cell.2011.03.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjursell G., Reichard P. 1973. Effects of thymidine on deoxyribonucleoside triphosphate pools and deoxyribonucleic acid synthesis in Chinese hamster ovary cells. J. Biol. Chem. 248:3904–3909 [PubMed] [Google Scholar]
- Boles N.C., Peddibhotla S., Chen A.J., Goodell M.A., Rosen J.M. 2010. Chk1 haploinsufficiency results in anemia and defective erythropoiesis. PLoS ONE. 5:e8581 10.1371/journal.pone.0008581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D., Foiani M. 2008. Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell Biol. 9:297–308 10.1038/nrm2351 [DOI] [PubMed] [Google Scholar]
- Chen Y.L., Eriksson S., Chang Z.F. 2010. Regulation and functional contribution of thymidine kinase 1 in repair of DNA damage. J. Biol. Chem. 285:27327–27335 10.1074/jbc.M110.137042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi O., Heathcote D.A., Ho K.K., Müller P.J., Ghani H., Lam E.W., Ashton-Rickardt P.G., Rutschmann S. 2012. A deficiency in nucleoside salvage impairs murine lymphocyte development, homeostasis, and survival. J. Immunol. 188:3920–3927 10.4049/jimmunol.1102587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angiolella V., Donato V., Forrester F.M., Jeong Y.T., Pellacani C., Kudo Y., Saraf A., Florens L., Washburn M.P., Pagano M. 2012. Cyclin F-mediated degradation of ribonucleotide reductase M2 controls genome integrity and DNA repair. Cell. 149:1023–1034 10.1016/j.cell.2012.03.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrovolsky V.N., Bucci T., Heflich R.H., Desjardins J., Richardson F.C. 2003. Mice deficient for cytosolic thymidine kinase gene develop fatal kidney disease. Mol. Genet. Metab. 78:1–10 10.1016/S1096-7192(02)00224-X [DOI] [PubMed] [Google Scholar]
- Dobrovolsky V.N., McGarrity L.J., VonTungeln L.S., Mittelstaedt R.A., Morris S.M., Beland F.A., Heflich R.H. 2005. Micronucleated erythrocyte frequency in control and azidothymidine-treated Tk+/+, Tk+/- and Tk−/− mice. Mutat. Res. 570:227–235 10.1016/j.mrfmmm.2004.11.006 [DOI] [PubMed] [Google Scholar]
- Eriksson S., Wang L. 2008. Molecular mechanisms of mitochondrial DNA depletion diseases caused by deficiencies in enzymes in purine and pyrimidine metabolism. Nucleosides Nucleotides Nucleic Acids. 27:800–808 10.1080/15257770802146197 [DOI] [PubMed] [Google Scholar]
- Evans D.R., Guy H.I. 2004. Mammalian pyrimidine biosynthesis: fresh insights into an ancient pathway. J. Biol. Chem. 279:33035–33038 10.1074/jbc.R400007200 [DOI] [PubMed] [Google Scholar]
- Fairman J.W., Wijerathna S.R., Ahmad M.F., Xu H., Nakano R., Jha S., Prendergast J., Welin R.M., Flodin S., Roos A., et al. 2011. Structural basis for allosteric regulation of human ribonucleotide reductase by nucleotide-induced oligomerization. Nat. Struct. Mol. Biol. 18:316–322 10.1038/nsmb.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fustin J.M., Doi M., Yamada H., Komatsu R., Shimba S., Okamura H. 2012. Rhythmic nucleotide synthesis in the liver: temporal segregation of metabolites. Cell Rep. 1:341–349 10.1016/j.celrep.2012.03.001 [DOI] [PubMed] [Google Scholar]
- Gagou M.E., Zuazua-Villar P., Meuth M. 2010. Enhanced H2AX phosphorylation, DNA replication fork arrest, and cell death in the absence of Chk1. Mol. Biol. Cell. 21:739–752 10.1091/mbc.E09-07-0618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi V.V., Samuels D.C. 2011. A review comparing deoxyribonucleoside triphosphate (dNTP) concentrations in the mitochondrial and cytoplasmic compartments of normal and transformed cells. Nucleosides Nucleotides Nucleic Acids. 30:317–339 10.1080/15257770.2011.586955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry G.A. 1992. Viral thymidine kinases and their relatives. Pharmacol. Ther. 54:319–355 10.1016/0163-7258(92)90006-L [DOI] [PubMed] [Google Scholar]
- González-Vioque E., Torres-Torronteras J., Andreu A.L., Martí R. 2011. Limited dCTP availability accounts for mitochondrial DNA depletion in mitochondrial neurogastrointestinal encephalomyopathy (MNGIE). PLoS Genet. 7:e1002035 10.1371/journal.pgen.1002035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen L., Peña-Diaz J., Kavli B., Otterlei M., Slupphaug G., Krokan H.E. 2006. Genomic uracil and human disease. Exp. Cell Res. 312:2666–2672 10.1016/j.yexcr.2006.06.015 [DOI] [PubMed] [Google Scholar]
- Halazonetis T.D., Gorgoulis V.G., Bartek J. 2008. An oncogene-induced DNA damage model for cancer development. Science. 319:1352–1355 10.1126/science.1140735 [DOI] [PubMed] [Google Scholar]
- Hardy R.R., Kincade P.W., Dorshkind K. 2007. The protean nature of cells in the B lymphocyte lineage. Immunity. 26:703–714 10.1016/j.immuni.2007.05.013 [DOI] [PubMed] [Google Scholar]
- He Q., Skog S. 2002. Involvement of a substrate cycle between thymidine and thymidylate in the regulation of DNA precursor pool in ehrlich ascites tumour. Cell. Physiol. Biochem. 12:305–314 10.1159/000067900 [DOI] [PubMed] [Google Scholar]
- Hu C.M., Yeh M.T., Tsao N., Chen C.W., Gao Q.Z., Chang C.Y., Lee M.H., Fang J.M., Sheu S.Y., Lin C.J., et al. 2012. Tumor cells require thymidylate kinase to prevent dUTP incorporation during DNA repair. Cancer Cell. 22:36–50 10.1016/j.ccr.2012.04.038 [DOI] [PubMed] [Google Scholar]
- Kufe D.W., Beardsley P., Karp D., Parker L., Rosowsky A., Canellos G., Frei E., III 1980. High-dose thymidine infusions in patients with leukemia and lymphoma. Blood. 55:580–589 [PubMed] [Google Scholar]
- Larsson K.M., Jordan A., Eliasson R., Reichard P., Logan D.T., Nordlund P. 2004. Structural mechanism of allosteric substrate specificity regulation in a ribonucleotide reductase. Nat. Struct. Mol. Biol. 11:1142–1149 10.1038/nsmb838 [DOI] [PubMed] [Google Scholar]
- Liu Y., Pop R., Sadegh C., Brugnara C., Haase V.H., Socolovsky M. 2006. Suppression of Fas-FasL coexpression by erythropoietin mediates erythroblast expansion during the erythropoietic stress response in vivo. Blood. 108:123–133 10.1182/blood-2005-11-4458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunt S.Y., Vander Heiden M.G. 2011. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell Dev. Biol. 27:441–464 10.1146/annurev-cellbio-092910-154237 [DOI] [PubMed] [Google Scholar]
- Mathews C.K., Wheeler L.J. 2009. Measuring DNA precursor pools in mitochondria. Methods Mol. Biol. 554:371–381 10.1007/978-1-59745-521-3_22 [DOI] [PubMed] [Google Scholar]
- Matsuoka S., Ballif B.A., Smogorzewska A., McDonald E.R., III, Hurov K.E., Luo J., Bakalarski C.E., Zhao Z., Solimini N., Lerenthal Y., et al. 2007. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 316:1160–1166 10.1126/science.1140321 [DOI] [PubMed] [Google Scholar]
- Melnyk S., Pogribna M., Miller B.J., Basnakian A.G., Pogribny I.P., James S.J. 1999. Uracil misincorporation, DNA strand breaks, and gene amplification are associated with tumorigenic cell transformation in folate deficient/repleted Chinese hamster ovary cells. Cancer Lett. 146:35–44 10.1016/S0304-3835(99)00213-X [DOI] [PubMed] [Google Scholar]
- Munch-Petersen B., Cloos L., Tyrsted G., Eriksson S. 1991. Diverging substrate specificity of pure human thymidine kinases 1 and 2 against antiviral dideoxynucleosides. J. Biol. Chem. 266:9032–9038 [PubMed] [Google Scholar]
- Murga M., Bunting S., Montaña M.F., Soria R., Mulero F., Cañamero M., Lee Y., McKinnon P.J., Nussenzweig A., Fernandez-Capetillo O. 2009. A mouse model of ATR-Seckel shows embryonic replicative stress and accelerated aging. Nat. Genet. 41:891–898 10.1038/ng.420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niida H., Shimada M., Murakami H., Nakanishi M. 2010. Mechanisms of dNTP supply that play an essential role in maintaining genome integrity in eukaryotic cells. Cancer Sci. 101:2505–2509 10.1111/j.1349-7006.2010.01719.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poli J., Tsaponina O., Crabbé L., Keszthelyi A., Pantesco V., Chabes A., Lengronne A., Pasero P. 2012. dNTP pools determine fork progression and origin usage under replication stress. EMBO J. 31:883–894 10.1038/emboj.2011.470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontarin G., Ferraro P., Bee L., Reichard P., Bianchi V. 2012. Mammalian ribonucleotide reductase subunit p53R2 is required for mitochondrial DNA replication and DNA repair in quiescent cells. Proc. Natl. Acad. Sci. USA. 109:13302–13307 10.1073/pnas.1211289109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quah B.J., Parish C.R. 2012. New and improved methods for measuring lymphocyte proliferation in vitro and in vivo using CFSE-like fluorescent dyes. J. Immunol. Methods. 379:1–14 10.1016/j.jim.2012.02.012 [DOI] [PubMed] [Google Scholar]
- Reichard P. 1988. Interactions between deoxyribonucleotide and DNA synthesis. Annu. Rev. Biochem. 57:349–374 10.1146/annurev.bi.57.070188.002025 [DOI] [PubMed] [Google Scholar]
- Reichard P. 1993. From RNA to DNA, why so many ribonucleotide reductases? Science. 260:1773–1777 10.1126/science.8511586 [DOI] [PubMed] [Google Scholar]
- Sabini E., Hazra S., Ort S., Konrad M., Lavie A. 2008. Structural basis for substrate promiscuity of dCK. J. Mol. Biol. 378:607–621 10.1016/j.jmb.2008.02.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub M., Spasokukotskaja T., Benczur M., Antoni F. 1988. DNA synthesis and nucleoside metabolism in human tonsillar lymphocyte subpopulations. Acta Otolaryngol. Suppl. 454:118–124 10.3109/00016488809125014 [DOI] [PubMed] [Google Scholar]
- Taghon T.N., David E.S., Zúñiga-Pflücker J.C., Rothenberg E.V. 2005. Delayed, asynchronous, and reversible T-lineage specification induced by Notch/Delta signaling. Genes Dev. 19:965–978 10.1101/gad.1298305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghon T., Yui M.A., Pant R., Diamond R.A., Rothenberg E.V. 2006. Developmental and molecular characterization of emerging beta- and gammadelta-selected pre-T cells in the adult mouse thymus. Immunity. 24:53–64 10.1016/j.immuni.2005.11.012 [DOI] [PubMed] [Google Scholar]
- Terry N.H., White R.A. 2006. Flow cytometry after bromodeoxyuridine labeling to measure S and G2+M phase durations plus doubling times in vitro and in vivo. Nat. Protoc. 1:859–869 10.1038/nprot.2006.113 [DOI] [PubMed] [Google Scholar]
- Toy G., Austin W.R., Liao H.I., Cheng D., Singh A., Campbell D.O., Ishikawa T.O., Lehmann L.W., Satyamurthy N., Phelps M.E., et al. 2010. Requirement for deoxycytidine kinase in T and B lymphocyte development. Proc. Natl. Acad. Sci. USA. 107:5551–5556 10.1073/pnas.0913900107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voet D. Voet J.G. 2004. Biochemistry. J. Wiley & Sons, Hoboken, NJ [Google Scholar]
- Xeros N. 1962. Deoxyriboside control and synchronization of mitosis. Nature. 194:682–683 10.1038/194682a0 [DOI] [PubMed] [Google Scholar]
- Zhou X., Solaroli N., Bjerke M., Stewart J.B., Rozell B., Johansson M., Karlsson A. 2008. Progressive loss of mitochondrial DNA in thymidine kinase 2-deficient mice. Hum. Mol. Genet. 17:2329–2335 10.1093/hmg/ddn133 [DOI] [PubMed] [Google Scholar]
- Zhou X., Johansson M., Solaroli N., Rozell B., Grandien A., Karlsson A. 2010. Hematopoiesis in the thymidine kinase 2 deficient mouse model of mitochondrial DNA depletion syndrome. J. Inherit. Metab. Dis. 33:231–236 10.1007/s10545-010-9102-x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.