Distinct nuclear subsets of cyclin D3 differ in their subcompartmentalization, function, and regulation.
Abstract
Ubiquitously expressed D-type cyclins are required for hematopoiesis but are dispensable in other cell lineages. Furthermore, within different hematopoietic progenitor populations the D-type cyclins play nonredundant roles. The basis of this lineage and developmental specificity is unknown. In pro–B cells we demonstrate four distinct nuclear D-type cyclin compartments, including one cyclin D3 fraction associated with CDK4 and another phosphoinositide 3-kinase–regulated fraction not required for proliferation. A third fraction of cyclin D3 was associated with the nuclear matrix and repression of >200 genes including the variable (V) gene segments Igkv1-117, Iglv1, and Igh-VJ558. Consistent with different subnuclear compartments and functions, distinct domains of cyclin D3 mediated proliferation and Igk V gene segment repression. None of the cyclin D3 nuclear compartments overlapped with cyclin D2, which was distributed, unbound to CDK4, throughout the nucleus. Furthermore, compartmentalization of the cyclins appeared to be lineage restricted because in fibroblasts, cyclin D2 and cyclin D3 occupied a single nuclear compartment and neither bound CDK4 efficiently. These data suggest that subnuclear compartmentalization enables cyclin D3 to drive cell cycle progression and repress V gene accessibility, thereby ensuring coordination of proliferation with immunoglobulin recombination.
Proliferation in response to mitogenic stimuli is associated with induction of a cascade of regulatory events that facilitates transit past the G1/S checkpoint into cell cycle. At the apex of this cascade are the D-type cyclins, including cyclin D1, cyclin D2, and cyclin D3, which bind and activate cyclin-dependent kinases 4 and 6 (CDK4/6). Cyclin D–CDK4/6 complexes initiate phosphorylation of the retinoblastoma protein (Rb) family members (Rb, p107, and p130), resulting in derepression of E2f transcription factors, the induction of additional cell cycle genes and transition through the G1 checkpoint into S phase (Ciemerych and Sicinski, 2005). Although this model was initially established through analyses of cell lines, gene targeting revealed that the ubiquitous D-type cyclins are required only in the hematopoietic lineages and a few specialized peripheral tissues (Kozar et al., 2004; Malumbres et al., 2004).
Within the hematopoietic lineages, the D-type cyclins provide unique functions. For example, Ccnd2 and Ccnd3, but not Ccnd1, are transcribed and the corresponding proteins are readily detected in B cell progenitors; however, only cyclin D3 is required for early B cell development and for the proliferation of pro–B and pre–B cells (Cooper et al., 2006). Cyclin D3 transcription is dependent on IL-7 receptor (IL-7R) signaling and the downstream activation of STAT5 (Mandal et al., 2009). Pre–B cell receptor (pre-BCR) signaling enhances detectable cyclin D3 protein and this is associated with a cyclin D3–dependent proliferative burst, whereas the detectable levels of cyclin D2 do not change between pro–B and pre–B cells (Cooper et al., 2006). Because both cyclin D2 and cyclin D3 are expressed in proliferating B cell progenitors, it is likely that differences in the D-type cyclin proteins, how they are regulated, and the context in which they are expressed, may determine their contribution to proliferation.
During B lymphopoiesis, proliferation must be tightly coordinated with Ig gene recombination. For light chain recombination, the mutual exclusivity of gene duplication and recombination is ensured by STAT5, which both promotes Ccnd3 transcription (Mandal et al., 2009) and represses Igk recombination (Mandal et al., 2011). However, it is not known if other determiners of cell cycle progression also regulate mechanisms of Ig gene accessibility to reinforce rearrangement after cell cycle exit.
Herein, we demonstrate that there are at least three distinct subnuclear compartments of cyclin D3 in pro–B cells that differ in their regulation and/or function. There are two soluble fractions of cyclin D3, one bound to CDK4 and involved in cell cycle and another independent fraction, regulated by phosphoinositide 3-kinase (PI3K), which is not in functional or biochemical equilibrium with that bound to CDK4. Surprisingly, a third fraction was bound to the nuclear matrix and was associated with repression of several genes including Igk, Igl, and Igh variable (V) gene segments. None of these cyclin D3 compartments overlapped with nuclear cyclin D2, which is dispensable for B lymphopoiesis. These data suggest that cell lineages using D-type cyclins have evolved specialized nuclear mechanisms to enhance CDK4 binding and to enable other lineage-restricted functions including gene repression.
RESULTS
Differential subcellular distribution of cyclin D2 and cyclin D3 in B cell progenitors
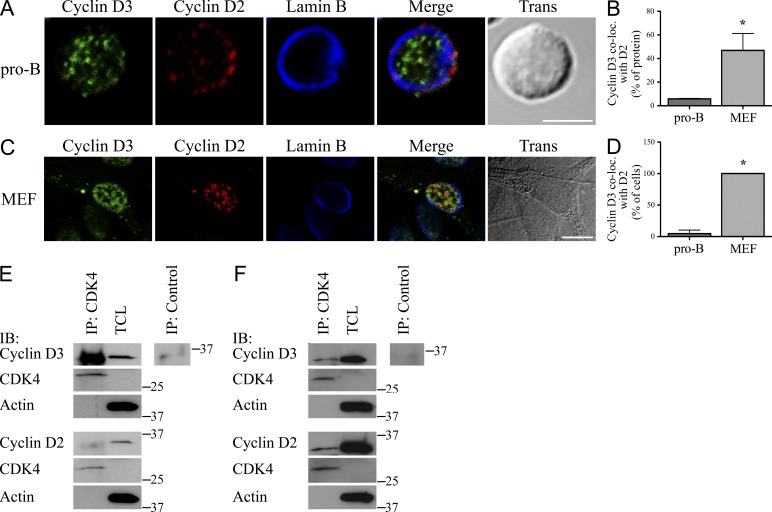
To investigate why cyclin D3 but not cyclin D2 was required for B lymphocyte progenitor proliferation, we used confocal microscopy to examine their relative cellular distribution in IL-7 cultured, proliferating Rag2−/− pro–B cells (Fleming and Paige, 2002). As can be seen in Fig. 1 A, cyclin D2 and cyclin D3 occupy largely nonoverlapping puncta in both the nucleus and cytosol. Per cell, a mean of only 5.8% (±0.004) of expressed cyclin D3 overlapped with cyclin D2 (Fig. 1 B). In contrast, in a representative proliferating nonhematopoietic cell lineage (mouse embryonic fibroblasts [MEFs]) a mean of 46.9 ± 0.1% cyclin D3 colocalized with cyclin D2 (Fig. 1, B and C). When we analyzed the fraction of cells demonstrating significant overlap between cyclin D2 and cyclin D3, 100 ± 0% of MEFs had >15% overlap of cyclin D3 with cyclin D2 compared with only 4.7 ± 5.7% of pro–B cells (Fig. 1 D).
Figure 1.
Differential distribution of nuclear cyclin D2 and cyclin D3 in Rag2−/− pro–B cells but not MEFs. (A and C) Proliferating Rag2−/− pro–B cells (A) or MEFs (C) were fixed, stained, and imaged for cyclin D3, cyclin D2, and Lamin B. Representative single plane confocal images are shown, with merged channel and transmitted light images provided. Bars, 5 µm. (B and D) Quantification of experiments in A and C shown as the mean ± SD derived from three independent experiments. (B) Percentage of detectable cyclin D3 colocalized with cyclin D2 (Manders’ coefficient). *, P < 0.01. (D) Percentage of cells scored positive for colocalization (at least 15% of cyclin D3 colocalized with cyclin D2). *, P < 0.001. (E and F) CDK4 immunoprecipitates and corresponding NP-40 TCLs from Rag2−/− pro–B cells (E) or MEFs (F) were resolved by SDS-PAGE and membranes probed with the indicated antibodies. IP lanes: 8 × 106 cells/sample; TCL lanes: 5 × 105 cells/sample. Relative molecular mass (kD) is as indicated. Representative of three independent experiments.
We next evaluated whether differential localization of cyclin D2 and cyclin D3 in pro–B cells correlated with preferential association of these proteins with CDK4. Immunoprecipitation (IP) of CDK4 from total cell lysates (TCLs) prepared in NP-40 lysis buffer followed by immunoblotting demonstrated appreciable coprecipitation of cyclin D3 but negligible association of cyclin D2 (Fig. 1 E). Both proteins were readily detected in TCLs. In contrast, analysis of CDK4 IPs from MEF TCLs revealed that both cyclin D2 and cyclin D3 formed complexes with the kinase (Fig. 1 F). However, the fraction of either cyclin coprecipitating with CDK4 from MEF TCLs was small compared with total soluble protein. Together, these data indicate that cyclin D2 and cyclin D3 occupy different niches in Rag2−/− pro–B nuclei and this correlates with differential association of the D-type cyclins with CDK4. In a nonhematopoietic cell line, however, cyclin D2 and cyclin D3 largely occupied a similar nuclear niche and, relative to pro–B cells, neither protein efficiently associated with CDK4.
PI3K regulates a labile fraction of cyclin D3 that does not bind CDK4
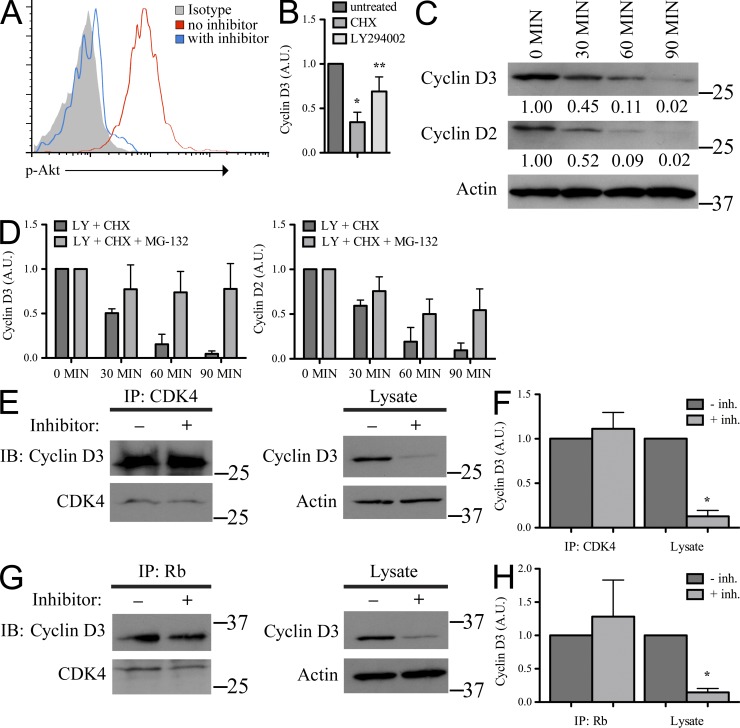
Previously, we reported that PI3K activation enhanced the amount of detectable cyclin D3 protein in pre–B cell lines (Cooper et al., 2006). To explore this potential mechanism of regulation further, Rag2−/− pro–B cells were cultured in IL-7 with or without the PI3K inhibitor LY294002 at sufficient concentrations to inhibit Akt phosphorylation (Fig. 2 A), and NP-40 TCLs of cell aliquots were assayed for cyclin D2 and cyclin D3 expression over time. These experiments were done in the presence of cycloheximide (CHX) to focus our studies on the regulation of existing proteins in the absence of new synthesis. Both CHX and LY294002 independently decreased the amount of cyclin D3 detected in TCLs (Fig. 2 B). When cells were treated with both inhibitors, the half-life of cyclin D2 and cyclin D3 protein was ∼30 min (Fig. 2, C and D) with a nearly complete loss of both proteins by 90 min.
Figure 2.
Soluble cyclin D3, but not that associated with CDK4 and Rb, is regulated by PI3K. (A) Rag2−/− pro–B cells were cultured in IL-7 ± LY294002 for 60 min, fixed, permeabilized, stained for intracellular p-Akt, and analyzed by flow cytometry. (B) Rag2−/− pro–B cells were cultured in IL-7 alone (untreated) or with CHX or LY294002 for 60 min. Samples were prepared in NP-40 lysis buffer, resolved by SDS-PAGE, and membranes were probed for cyclin D3 and Actin. Expression of cyclin D3 was normalized to Actin, standardized as a ratio of the treated sample relative to the expression level in untreated samples (arbitrary units, A.U.), and reported here graphically as the mean ± SD, n = 3. Compared with the untreated sample: *, P < 0.001; **, P < 0.05. (C) Rag2−/− pro–B cells were cultured in IL-7 with LY294002 and CHX for the times indicated. TCLs were prepared and resolved as in B and membranes were probed for cyclin D3, cyclin D2, and Actin (representative experiment, n = 3). Relative molecular mass (kD) is as indicated. Cyclin expression reported below immunoblot in arbitrary units (A.U.) as described in B. (D) Rag2−/− pro–B cells were cultured in IL-7 with LY294002 (LY) and CHX or LY, CHX, and proteasome inhibitor MG-132 for the times indicated, analyzed as in C, and reported here graphically as mean ± SD, n = 3. (E–H) Rag2−/− pro–B cells were cultured in IL-7 with or without inhibitors LY294002 and CHX for 60 min. (E and G) CDK4 (E) or Rb (G) immunoprecipitates and corresponding TCLs were resolved by SDS-PAGE and membranes probed with indicated antibodies. IP lanes: 8 × 106 cells; lysate lanes: 7 × 105 cells. Relative molecular mass (kD) is as indicated. (F and H) Quantification of cyclin D3 from blots normalized to CDK4 (IP lanes) or Actin (TCL lanes), standardized as a ratio of the treated sample relative to the expression level in untreated samples (arbitrary units, A.U.) and shown as mean ± SD, n = 3. *, P < 0.05.
Targeting of the D-type cyclins for nuclear export is thought to result in ubiquitin-mediated proteolysis (Lin et al., 2006). Therefore, we next examined if PI3K activity enhanced cyclin D3 by preventing proteasomal-mediated degradation. Evaluating the preexisting pool of protein in the presence of the proteasome inhibitor MG-132 (Fig. 2 D), inhibiting PI3K did not significantly alter detectable cyclin D3 levels over 90 min (100 vs. 77.7 ± 28.5%, P = 0.2). Cyclin D2 levels were also largely preserved over time. Thus, these data suggest PI3K activity enhances free cyclin D3 by preventing proteolysis.
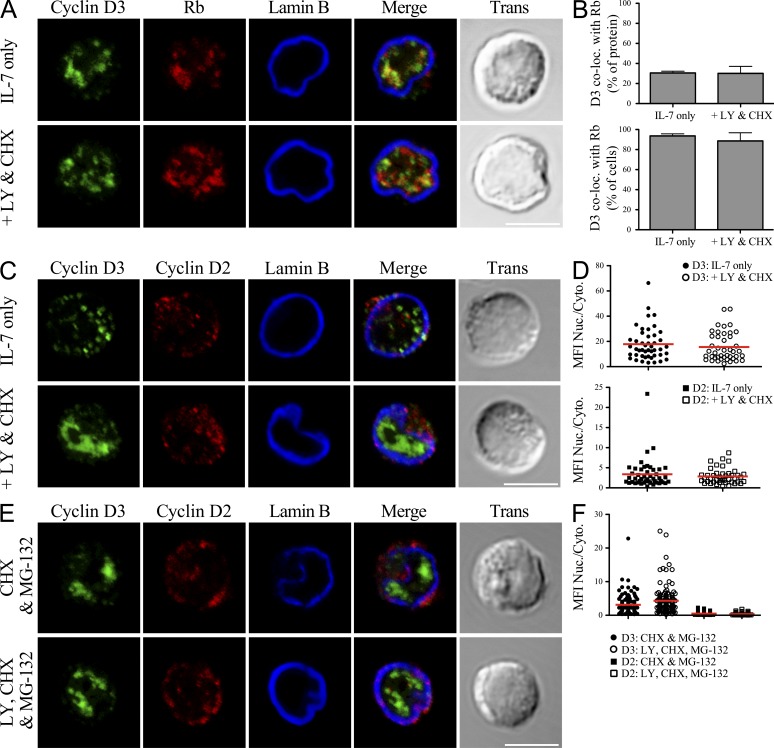
We next examined if PI3K signaling regulated cyclin D3 association with CDK4. Rag2−/− pro–B cells were cultured with or without LY294002 and CHX for 60 min. Cell aliquots were then solubilized in NP-40 lysis buffer and CDK4 IPs were probed in immunoblots with anti-cyclin D3 and anti-CDK4 antibodies. As observed previously (Fig. 2, C and D; Cooper et al., 2006), inhibiting PI3K reduced the amount of cyclin D3 detected in TCLs to <20% of the pretreatment value (Fig. 2, E and F). However, it did not alter the amount of cyclin D3 associated with CDK4. Similarly, IP of Rb demonstrated that inhibiting PI3K did not change the association of Rb with either cyclin D3 or CDK4 (Fig. 2, G and H). Furthermore, as evaluated by confocal microscopy, we observed that the colocalization of cyclin D3 with Rb was not significantly altered upon PI3K inhibition (Fig. 3, A and B). These data indicate that there are at least two soluble fractions of cyclin D3: one unbound labile fraction regulated by PI3K, and a relatively PI3K-insensitive fraction of cyclin D3 bound to cell cycle machinery components.
Figure 3.
Nuclear localization of cyclin D3 is not regulated by PI3K. (A–D) Rag2−/− pro–B cells were cultured in IL-7 alone or with LY294002 (LY) and CHX for 60 min. Cells were fixed, stained, and imaged for cyclin D3, Lamin B, and Rb (A) or cyclin D2 (C). (A and C) Representative single plane confocal images are shown, with merged channel and transmitted light images provided. Bars, 5 µm. (B) Quantification from images of cells treated as in A shown as percentage of detectable cyclin D3 colocalized with Rb (Manders’ coefficient, top) or percentage of cells scored positive for colocalization (at least 15% of cyclin D3 colocalized with Rb, bottom) with mean ± SD derived from three independent experiments. (D) Analysis of single cells treated as in C for the MFI of each protein presented as a ratio of signal detected in the nucleus versus the cytoplasm. Points represent individual cells analyzed across three experiments, with the red bar depicting the mean value for each population. Distribution of cyclin D2 compared with cyclin D3 was statistically different both before and after inhibitor treatment, P < 0.0001. (E and F) Rag2−/− pro–B cells were cultured in IL-7, CHX, and MG-132 ± LY for 60 min. (E) Cells were fixed, stained, and imaged for cyclin D3 and cyclin D2 as in C and F analyzed as in D. Bar, 5 µm. Comparison of cyclin D2 distribution ± LY, P < 0.001. Distribution of cyclin D2 and cyclin D3 was statistically different under both culture conditions, P < 0.0001.
Focusing on the PI3K-sensitive fraction of cyclin D3, we next evaluated whether we could detect nuclear export of cyclin D3 in the absence of PI3K signaling. Rag2−/− pro–B cells were cultured in IL-7 alone or with LY294002 and CHX for 60 min, fixed, and then analyzed by confocal microscopy to evaluate the subcellular distribution of cyclin D2 and cyclin D3 under PI3K-sufficient and -deficient conditions (Fig. 3 C). In untreated cells, cyclin D3 protein was enriched in the nucleus (MFI nucleus/cytoplasm 17.8 ± 1.9; Fig. 3 D). In contrast, cyclin D2 was more evenly distributed between the nucleus and cytosol (3.4 ± 0.5, P < 0.0001). Unexpectedly, the distribution of cyclin D3 and cyclin D2 was not significantly altered upon PI3K inhibition (15.6 ± 1.7, P = 0.4, and 2.8 ± 0.3, P = 0.4, respectively). CHX treatment alone did not appreciably alter the subnuclear distribution of cyclin D3 (unpublished data). Interestingly, we noted that in a fraction of the cells analyzed (33%), the pattern of cyclin D3 staining within the nucleus changed from a punctate to diffuse pattern, as represented in Fig. 3 C.
We were concerned that our experimental system failed to adequately detect exported protein. To circumvent this caveat, we repeated this imaging experiment in the presence of the proteasome inhibitor MG-132 (Fig. 3, E and F). Inhibiting the proteasome enhanced the relative amount of cytosolic cyclin D2 in both the absence (MFI nucleus/cytoplasm 0.5 ± 0.03) and presence (0.3 ± 0.02, P < 0.001) of PI3K inhibition. After inhibiting the proteasome most cyclin D3 appeared to remain localized to the nucleus both in the presence (3.1 ± 0.2) and absence (4.3 ± 0.4) of PI3K signaling.
PI3K does not regulate proliferation during B lymphopoiesis
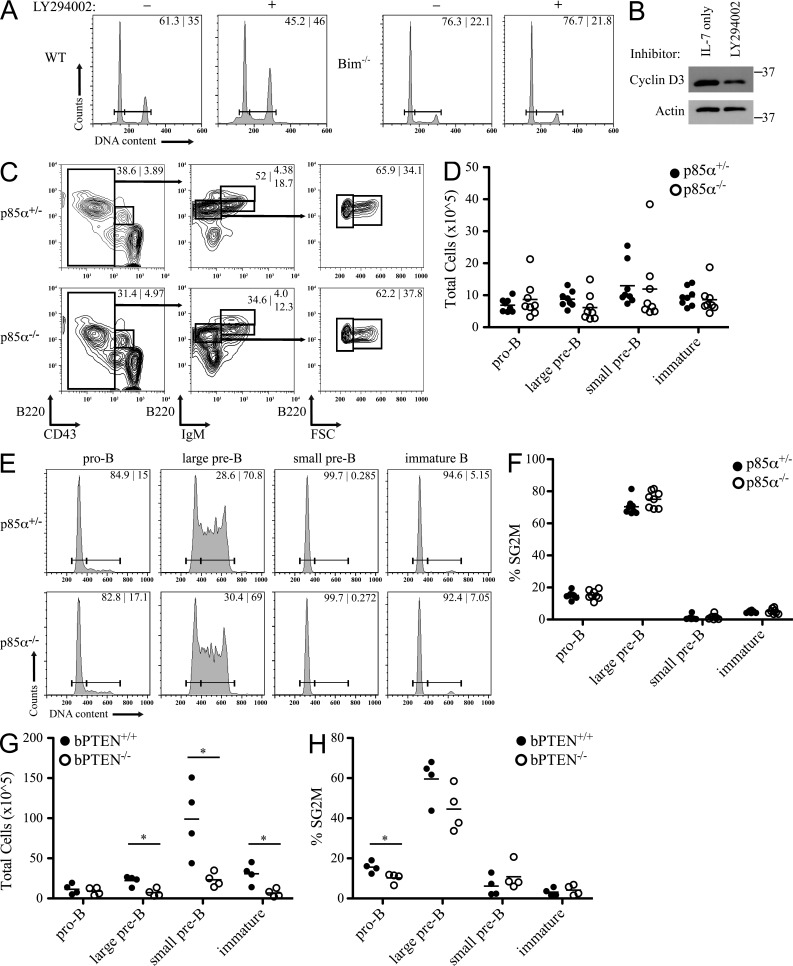
Our data indicate that PI3K regulates detectable levels of cyclin D3 in NP-40 lysates, but that it does not regulate those subfractions involved in cell cycle progression. Therefore, we next examined if PI3K signaling regulates B cell progenitor proliferation. We first isolated pro–B/pre–B cells from WT bone marrow and cultured them with or without LY294002 for 24 h, fixed, and stained with propidium iodide to label DNA content. Analysis by flow cytometry demonstrated that there was no substantial difference in the percentage of cycling cells (SG2M) between the control and LY294002-treated cells (Fig. 4 A). In fact, it appeared that LY294002-treated cells had a greater frequency of proliferating cells; however, these samples also demonstrated a substantial sub-G0/G1 peak, suggestive of enhanced cell death. To abrogate the impact of PI3K inhibition on cell viability, these experiments were repeated in progenitors isolated from the bone marrow of Bim−/− mice. Inhibiting PI3K in Bim−/− B cell progenitors did not alter cell proliferation even though detectable soluble cyclin D3 was reduced (Fig. 4, A and B). Furthermore, treatment did not induce an accumulation of sub-G0/G1 cells. Together, these ex vivo results suggest that PI3K does not regulate progenitor B cell proliferation.
Figure 4.
Alteration of PI3K signaling does not affect B cell progenitor proliferation. (A) B cell progenitors from WT and Bim−/− bone marrow were cultured in the presence of IL-7 with or without LY294002 for 24 h. Cells were then fixed and analyzed for DNA content and the percentage of cells in G1 versus SG2M phase of cell cycle are provided. Data are representative of three independent experiments. (B) B cell progenitors from Bim−/− bone marrow were cultured in the presence of IL-7 with or without LY294002 for 24 h. TCLs were prepared in NP-40 lysis buffer, resolved by SDS-PAGE, and membranes probed for cyclin D3 and Actin. Relative molecular mass (kD) is as indicated. Data are representative of three independent experiments. (C) Flow cytometry for expression of B220, CD43, and IgM by bone marrow cells from p85α+/− and p85α−/− mice. Numbers in contour plots indicate the percentage of cells in each gate. Representative data, n = 8. (D) Relative number of cells in each population determined using total number of bone marrow cells after red blood cell lysis and percentages from flow cytometry. Each point represents a single mouse and bars represent the mean for each population. (E) The corresponding populations from C were analyzed for DNA content. Numbers in histogram plots indicate the percentage of cells in G1 and SG2M phases of cell cycle. (F) Summary of the percentage of cells in SG2M from each population. Each point represents a single mouse and bars represent the mean for each population. (G and H) bPTEN+/+ and bPTEN−/− bone marrow was analyzed as in C and D (G), and the percentage of cells in SG2M from each population was summarized (H), n = 4. Each point represents a single mouse and bars represent the mean for each population. *, P < 0.05.
We next assessed the in vivo role of PI3K in proliferation of B cell progenitors by examining bone marrow from mice lacking the p85α regulatory subunit of PI3K (Suzuki et al., 1999, 2003). p85α−/− mice demonstrate impaired B cell development, with B220+IgM+ mature cells in the spleen, bone marrow, and lymph nodes reduced to less than half of littermate controls, and the remaining B220+ B cells have largely ablated PI3K kinase activity (Suzuki et al., 1999). Single cell bone marrow suspensions were counted, stained for surface markers to resolve pro–B, large pre–B, small pre–B, and immature B cell populations, and evaluated for DNA content by flow cytometry. These analyses revealed that, compared with p85α+/− littermate controls, these B cell progenitor cell populations were conserved in p85α−/− mice (Fig. 4, C and D). We next analyzed cell cycle progression in each progenitor stage with representative histograms provided in Fig. 4 E and a graph summarizing the eight animal pairs analyzed provided in Fig. 4 F. These data demonstrate no statistical difference in proliferation between B cell progenitors in p85α-deficient and -sufficient populations. In complementary experiments, we examined proliferation in B cell progenitors lacking the phosphatase PTEN (PtenloxP/loxPCD19Cre+/−, subsequently referred to as bPTEN−/−) which negatively regulates PI3K activity (Anzelon et al., 2003). Division of the B cell compartment into developmental fractions revealed that although the pro–B cell compartment was intact in bPTEN−/− mice (P = 0.6), there was a reduction in subsequent developmental stages (Fig. 4 G). We next analyzed the fraction of each progenitor population in cell cycle. As summarized in Fig. 4 H, although the fraction of cycling pro–B cells was reduced in bPTEN−/− mice (P < 0.05), the percentage of SG2M cycling pre–B cells from bPTEN−/− and bPTEN+/+ bone marrow was not significantly different. Together these in vivo data demonstrate that PI3K signaling is not essential for inducing proliferation in B cell progenitors.
A PI3K-insensitive fraction of cyclin D3 associates with the nuclear matrix
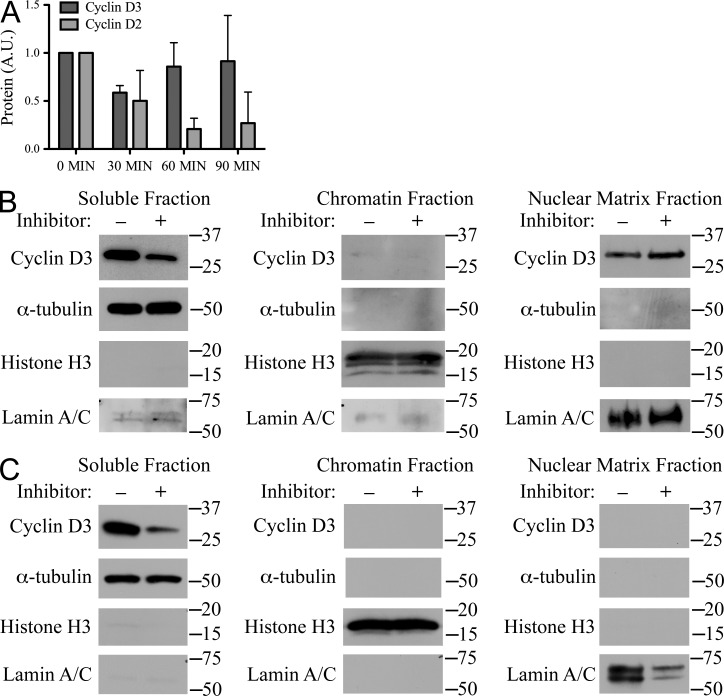
Inhibiting PI3K activity in the absence of new protein synthesis led to a rapid loss of detectable cyclin D3 from TCLs; however, inhibition did not alter the relative abundance of cyclin D3 in the nucleus as detected by confocal microscopy (Fig. 3 C). We postulated that this apparent discrepancy reflected one or more relatively insoluble pools of cyclin D3 in the nucleus. Therefore, we examined the kinetics of cyclin D2 and cyclin D3 disappearance from Rag2−/− pro–B cell samples extracted by boiling in the presence of SDS. As demonstrated in Fig. 2 (C and D), the half-life of cyclin D3 protein detected in NP-40 TCLs was ∼30 min. In contrast, the amount of cyclin D3 detected in SDS-extracted samples did not significantly change over 90 min (P = 0.4; Fig. 5 A). Cyclin D2 decay as detected in SDS lysates paralleled that seen in NP-40 TCLs. These experiments reveal a large insoluble fraction of cyclin D3 present even after loss of PI3K signaling.
Figure 5.
A large fraction of cyclin D3 bound the nuclear matrix. (A) Rag2−/− pro–B cells were cultured in IL-7 with LY294002 and CHX for the times indicated. TCLs were SDS-extracted (5 × 105 cells per sample), resolved by SDS-PAGE, and membranes were probed for cyclin D3, cyclin D2, and Actin. Summary of three experiments, where cyclin expression was normalized to Actin, standardized as a ratio of the treated sample relative to the expression level in untreated samples (arbitrary units, A.U.) and reported graphically as the mean ± SD. (B and C) Rag2−/− pro–B cells (B) or MEFs (C) were cultured with or without the inhibitors LY294002 and CHX for 75 min. Cell aliquots were subjected to subcellular fractionation. Fractions were resolved by SDS-PAGE and membranes probed for cyclin D3 and fraction-specific controls α-tubulin (soluble), Histone H3 (chromatin), and Lamin A/C (nuclear matrix). Relative molecular mass (kD) is as indicated. Data are representative of three independent experiments.
We next used subcellular fractionation to further characterize the insoluble compartment of cyclin D3. Aliquots of Rag2−/− pro–B cells cultured with or without LY294002 and CHX for 75 min were fractionated into total soluble, chromatin, and nuclear matrix fractions. These fractions were then immunoblotted with anti-cyclin D3, anti–α-tubulin, anti–Histone H3, and anti–Lamin A/C antibodies (Fig. 5 B). Consistent with our previous results, we observed a reduction in detectable cyclin D3 in the soluble fraction upon inhibitor treatment but we observed negligible cyclin D3 in the chromatin fractions with and without inhibitor treatment. Immunoblotting of the nuclear matrix fractions with anti-cyclin D3 antibodies revealed protein in both samples. Upon inhibition of PI3K, in the experiment shown, the relative intensity of the cyclin D3 band was slightly augmented. However, over several experiments, inhibiting PI3K did not significantly change the mean relative intensities of the bands (unpublished data). In contrast, subcellular fractionation of MEFs (Fig. 5 C) demonstrated PI3K-sensitive cyclin D3 in the soluble fraction without detectable protein in the chromatin or nuclear matrix fractions. Together these observations indicate that cyclin D3 associates with the nuclear matrix in B cell progenitors, but not a nonhematopoietic cell line, and that this fraction of cyclin D3 is not regulated by PI3K.
These findings were surprising, as cyclin D1 has been recently demonstrated to bind chromatin and regulate gene transcription (Bienvenu et al., 2010). However, using chromatin IP (ChIP) directed at known E2f target sites repressed by Rb (Dirlam et al., 2007) and by ChIP followed by deep sequencing (Mandal et al., 2011), we could not detect specific association of endogenous cyclin D3 with chromatin (unpublished data). These observations are consistent with cyclin D3 associating with the nuclear matrix and not chromatin.
Nuclear matrix-associated cyclin D3 represses Ig V genes
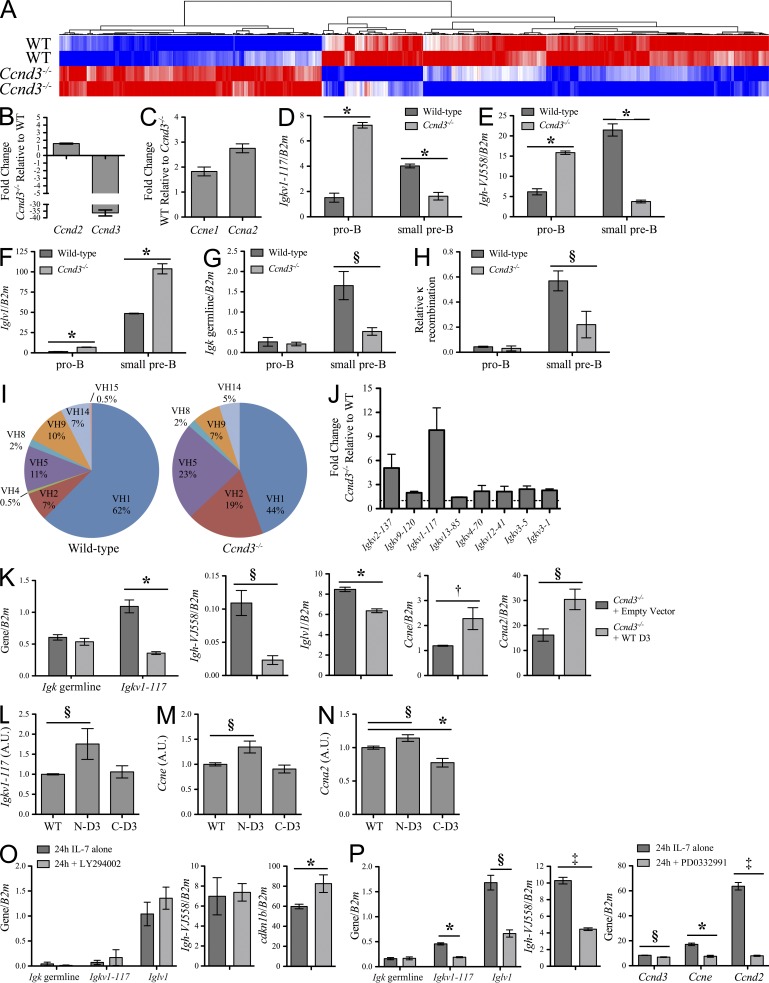
The nuclear matrix has been implicated in several processes important for gene transcription (Andrés and González, 2009). Therefore, to determine if cyclin D3 could be playing a role in regulation of transcription, we compared gene expression profiles of pro–B cells isolated from WT and Ccnd3−/− bone marrow using mRNA microarrays. Duplicate WT and Ccnd3−/− array signals were then averaged to determine targets demonstrating at least a twofold difference in expression. Of these, 472 genes were expressed higher in WT cells and 277 genes were expressed higher in Ccnd3−/− cells (Fig. 6 A and Table S1). As expected (Cooper et al., 2006) and confirmed by quantitative PCR (qPCR) of matched populations, Ccnd3−/− pro–B cells expressed less detectable Ccnd3 and more Ccnd2 than WT cells (Fig. 6 B). Genes expressed higher in WT pro–B cells included factors associated with cell cycle, cell division, and mitosis, including Ccne1 and Ccna2 (Fig. 6 C). Thus, pro–B cells lacking cyclin D3 demonstrate a reduction in cell cycle mediators, agreeing with our previous observation that the fraction of cycling B cell progenitors is reduced in Ccnd3−/− bone marrow (Cooper et al., 2006).
Figure 6.
Dysregulated gene expression in Ccnd3−/− progenitors includes Ig V genes. (A) Heat map representing differential expression of genes in pro–B cells from WT and Ccnd3−/− bone marrow, grouped based on hierarchical clustering, n = 2. (B) qPCR analysis of the expression of Ccnd2 and Ccnd3 in pro–B cells, shown as the fold change in Ccnd3−/− relative to WT. (C) qPCR analysis of the expression of Ccne1 and Ccna2 in pro–B cells, shown as the fold change in WT relative to Ccnd3−/−. Both B and C represent three independent experiments, shown as mean ± SD. (D–G) qPCR analysis of the expression of Igkv1-117 (D), Igh-VJ558 (E), Iglv1 (F), and Igk germline (G) in WT and Ccnd3−/− pro–B and small pre–B cells, normalized to β2 microglobulin (B2m) expression and shown as mean ± SD. Data are representative of three independent experiments. (H) qPCR analysis of Igk rearrangements involving Igkj1, normalized to amplification of Eκi, in WT and Ccnd3−/− pro–B and small pre–B cells and shown as mean ± SD. Data are representative of three independent experiments. (I) Distribution of VH family usage in WT and Ccnd3−/− large pre–B cells. Results represent sequences derived from three independent experiments (WT = 173 and Ccnd3−/− = 162 sequences). (J) qPCR analysis of the expression of Igkv2-137, Igkv9-120, Igkv1-117, Igkv13-85, Igkv4-70, Igkv12-41, Igkv3-5, and Igkv3-1 (organized as most distal to most proximal) in pro–B cells, presented as the fold change (mean ± SD) in Ccnd3−/− relative to WT, n = 3. (K) qPCR analysis of Igk germline, Igkv1-117, Igh-VJ558, Iglv1, Ccne, and Ccna2 in Ccnd3−/− pro–B cells expressing empty vector or WT cyclin D3 (mean ± SD), presented as in D–G. Data are representative of at least three independent experiments. (L–N) qPCR analysis of Igkv1-117 (L), Ccne (M), and Ccna2 (N) in Ccnd3−/− pro–B cells expressing WT cyclin D3 or chimeric proteins containing the N-terminal or C-terminal region of cyclin D3 (N-D3 and C-D3, respectively). Expression of targets was normalized to corresponding WT, N-D3, or C-D3 expression, standardized to values for WT to facilitate comparisons and presented here as mean ± SD in arbitrary units (A.U.). Data are representative of three independent experiments. (O) Rag2−/− pro–B cells were cultured in IL-7 with or without LY294002 for 24 h and assayed by qPCR for the targets indicated (mean ± SD), presented as in D–G. Data are representative of three independent experiments. (P) Rag2−/− pro–B cells were cultured in IL-7 with or without PD0332991 for 24 h and assayed by qPCR for the targets indicated (mean ± SD) as in O. Data are representative of three independent experiments. †, P < 0.05; §, P < 0.01; *, P < 0.001; ‡, P < 0.0001.
Surprisingly, of the genes up-regulated in Ccnd3−/− pro–B cells, we noted multiple Ig V segments including Igkv1-117, Igh-VJ558, and Iglv1. Interestingly, the corresponding constant regions (Igkc, Ighm, and Iglc2 and Iglc3) were not up-regulated in Ccnd3−/− pro–B cells. To confirm these observations, we sorted pro–B and small pre–B cells from WT and Ccnd3−/− bone marrow and evaluated transcript expression by qPCR. In Ccnd3−/− pro–B cells, Igkv1-117 transcript levels were higher than that seen at the WT small pre–B cell stage where cells are actively rearranging Igk (Mandal et al., 2009; Fig. 6 D). Transcription of Igh-VJ558 and Iglv1 was also increased in Ccnd3−/− pro–B cells (Fig. 6, E and F). At the small pre–B cell stage, Ccnd3−/− cells demonstrate reduced levels of Igkv1-117 and Igh-VJ558 transcripts relative to WT, whereas Iglv1 transcripts are further increased in Ccnd3−/− small pre–B cells. However, these differences compared with WT may reflect abnormal development and the severe loss of pre–B cells observed in Ccnd3−/− mice (Cooper et al., 2006). In contrast, Igk germline transcription (Jκ-Cκ; Johnson et al., 2008) was normally repressed in Ccnd3−/− pro–B cells, indicating that there is not a global dysregulation of Igk transcription (Fig. 6 G). Predictably, there was no significant increase in V to Igkj1 recombination in Ccnd3−/− pro–B cells as compared with WT (Fig. 6 H). These data indicate that cyclin D3 selectively represses the transcription of Igk, Igl, and Igh V segments.
We next evaluated whether cyclin D3 globally regulates Ig V segment usage. At the pro–B cell stage, cells actively rearrange the Igh locus and successful rearrangements are expressed on the cell surface in the context of the pre-BCR, signaling developmental progression to the pre–B cell stage. To determine whether dysregulated Igh transcription at the pro–B cell stage impacts VH usage in recombined heavy chains, we cloned and sequenced V(D)J rearrangements from WT and Ccnd3−/− large pre–B cells (Fuxa et al., 2004). In both WT and Ccnd3−/− cells, ∼30% of sequences analyzed demonstrated out-of-frame or in-frame with stop codon rearrangements. Rearrangements yielding successful in-frame V(D)J recombination (WT = 173, Ccnd3−/− = 162) were then grouped by VH family (Fig. 6 I; Johnston et al., 2006). As expected, WT sequences demonstrated a large proportion of VH1 (J558 family) sequences (ten Boekel et al., 1997). In contrast, Ccnd3−/− sequences showed a more preferential usage of VH2 (Q52 family) and VH5 (7183 family) segments. These data indicate that cyclin D3 shapes the repertoire of VH genes expressed in pro–B cells.
We next examined whether cyclin D3 repressed other Igk V segments besides Igkv1-117. qPCR of mRNA from Ccnd3−/− pro–B cells demonstrated increased transcription of V segments spanning the Igk locus (Aoki-Ota et al., 2012) including Igkv2-137, Igkv9-120, Igkv1-117, Igkv13-85, Igkv4-70, Igkv12-41, Igkv3-5, and Igkv3-1 (Fig. 6 J). Both Igkv1-117 and Igkv2-137 displayed marked derepression in Ccnd3−/− pro–B cells, whereas most others demonstrated approximately a twofold increase in transcripts. These findings suggest that cyclin D3 both globally represses and modulates V segment transcription across the Igk locus.
To directly determine if cyclin D3 expression mediates transcriptional regulation at the Igk locus, we infected cultured Ccnd3−/− progenitors with control retrovirus (MIGR1-IRES-GFP, empty vector) or retrovirus encoding WT cyclin D3 and assessed pro–B cells expressing GFP. As expected, cyclin D3 expression did not alter Igk germline transcription (Jκ-Cκ); however, in the presence of cyclin D3, Igkv1-117, Igh-VJ558, and Iglv1 transcription was significantly reduced (Fig. 6 K). Additionally, ectopic expression of cyclin D3 resulted in enhanced transcription of downstream cell cycle targets Ccne and Ccna2 (Fig. 6 K). Thus, restoring cyclin D3 in Ccnd3−/− pro–B cells enhanced expression of the cell cycle machinery and repressed Ig V transcription.
We speculated that these two cellular functions could be assigned to different regions of the cyclin D3 protein. If so, expressing particular regions of cyclin D3 in the context of an irrelevant molecule (cyclin D2) should either up-regulate the cell cycle machinery or repress Ig V transcription in Ccnd3−/− progenitors. Therefore, we created chimeric constructs containing either the N- or C-terminal 1/3 of cyclin D3 paired with cyclin D2 for the remaining protein. Immunoblotting of 3T3 cells expressing virus encoding hemagglutinin-tagged WT cyclin D3, a chimeric protein containing the N-terminal third of cyclin D3 (N-D3), or a chimeric protein containing the C-terminal third of cyclin D3 (C-D3) demonstrated expression of protein at the expected molecular mass of ∼34 kD (not depicted). We then infected Ccnd3−/− progenitors with empty virus or virus encoding WT cyclin D3, N-D3, or C-D3. After 48 h, GFP+B220+CD43hi cells were assayed for expression of viral transcripts as well as Igkv1-117, Ccne, and Ccna2 expression (Fig. 6, L and N). As compared with WT protein, C-D3, but not N-D3, was as effective at repressing Igkv1-117 transcription. In contrast, ectopic expression of N-D3, but not C-D3, was at least as effective, if not more effective, at augmenting Ccne and Ccna2 transcription as WT cyclin D3.
Although it is evident that different cellular functions of cyclin D3 can be mapped to disparate regions of the protein, it is not clear how these differential functions relate to the discrete pools of cyclin D3 described herein. We have demonstrated that PI3K signaling is necessary for maintaining the free fraction of cyclin D3 but not for that associated with the nuclear matrix. Therefore, to determine the relative contribution of each cyclin D3 fraction to Ig V gene repression, Rag2−/− pro–B cells were cultured in IL-7 with or without LY294002 for 24 h and then assayed for expression of several targets by qPCR. As expected, Igk germline transcription is negligible both in the presence and absence of PI3K signaling, whereas inhibitor treatment did induce cdkn1b (Fig. 6 O; Herzog et al., 2009). However, PI3K inhibition did not induce Igkv1-117, Iglv1, or Igh-VJ558 transcription. Next, to determine whether cyclin D3–mediated repression of Ig V genes is dependent on the kinase activity of CDK binding partners, Rag2−/− pro–B cells were cultured in IL-7 with or without CDK4/6 inhibitor PD0332991 for 24 h. As demonstrated in Fig. 6 P, CDK4 inhibition did not induce Ig V gene expression, whereas it decreased expression of Ccne and Ccna2. Together, these data suggest cyclin D3–mediated repression of Ig V genes is independent of the PI3K-regulated fraction of cyclin D3 and does not require CDK4 kinase activity. Rather, our results implicate the nuclear matrix-bound fraction of cyclin D3 in Ig gene repression.
DISCUSSION
Our data reveal that in pro–B cells, there are at least three distinct subnuclear compartments of cyclin D3 that differ in their spatial organization, function, and regulation. As might be expected, one soluble fraction of cyclin D3 associates with the cell cycle machinery. This cyclin D3 compartment, in complex with CDK4 and Rb, is not regulated by PI3K signaling. Consistent with previous results, a second soluble fraction of cyclin D3 is regulated by PI3K signaling (Cooper et al., 2006); however, this is a minor fraction of total cyclin D3 and does not participate in proliferation. The largest fraction of cyclin D3 bound the nuclear matrix (Mariappan et al., 2007), was insensitive to PI3K signaling, and was associated with the repression of a large subset of genes including all assayed Igk V gene segments, Iglv1, and Igh-VJ558. None of these fractions of cyclin D3 overlap with cyclin D2, which exists free within the nucleus not engaged with determiners of proliferation. These findings suggest that, in addition to the mechanisms controlling cyclin D3 expression, the location of cyclin D3 within the nucleus and the molecules it interacts with are important determiners of cyclin D3 function.
It is not known how cyclin D3 segregates into the different subnuclear niches in pro–B cells. However, these mechanisms are likely cell type specific. Pro–B cells, but not MEFs, have a nuclear matrix-associated fraction of protein. Of the soluble pool of protein, MEFs demonstrate extensive overlap between nuclear cyclin D2 and cyclin D3. Furthermore, both cyclin D2 and cyclin D3 associate with CDK4, yet the fraction of each D-type cyclin associated with CDK4 in MEFs was far less than the soluble fraction of cyclin D3 associated with CDK4 in pro–B cells. This suggests that the spatial organization of the cyclins in pro–B cells may enable preferential and relatively efficient association of cyclin D3 with CDK4. Correspondingly, cyclin D3 is required for proliferation and proper developmental progression of B cell progenitors, whereas the D-type cyclins are dispensable for MEF proliferation. We postulate that the different cyclin D3 compartments within pro–B cells might be necessary to drive productive interactions of cyclin D3 with its effectors. In cells that lack these nuclear compartments, other core cell cycle machinery components, such as cyclin E and CDK2, may drive proliferation (Kozar and Sicinski, 2005). Despite these obvious differences, there are some biochemical similarities in how the D-type cyclins are regulated in these cell types. As is observed in fibroblasts (Diehl et al., 1997), in pro–B cells the soluble free fraction of cyclin D3, along with cyclin D2, was degraded by the proteasome upon PI3K attenuation. However, in pro–B cells this mechanism only regulates a minor fraction of cyclin D3 that was detectable biochemically but not by confocal microscopy. These data suggest that regulatory pathways present in nonhematopoietic cells are not circumvented in B cell progenitors. Rather, we propose that additional mechanisms are present in B cell progenitors that enable the use of cyclin D3 as a cell cycle regulatory molecule.
One striking finding was that cyclin D3 could act as a transcriptional repressor of >200 genes, including the Ig V genes. The effects of cyclin D3 on Igk V gene transcription were particularly profound. Loss of cyclin D3 led to complete derepression of Igkv1-117, whereas adding back cyclin D3 to Ccnd3−/− cells was sufficient to fully repress Igkv1-117 transcription. Additionally, Ccnd3−/− pro–B cells demonstrated dysregulation of transcription of several other V segments across the Igk locus. However, cyclin D3–mediated repression was restricted to V regions, as loss of cyclin D3 did not derepress Igk J or C regions. Correspondingly, cyclin D3 deficiency was not sufficient to allow promiscuous Igk recombination. These results were expected, as Jκ and Cκ are repressed epigenetically by STAT5 binding to the Eκi (Mandal et al., 2011), a regulatory pathway independent of cyclin D3.
Cyclin D3 also potently repressed other Ig V regions, including the distal Ig heavy chain V region VJ558, indicating that, in pro–B cells, cyclin D3 is an important regulator of V gene transcription. However, enhanced transcription of Igh-VJ558 did not correlate with increased representation of VH1 family usage in Ccnd3−/− progenitors. Large pre–B cells lacking cyclin D3 demonstrate skewed VH segment usage as compared with WT, with an overrepresentation of the proximal VH2 (Q52) and VH5 (7183) family variable regions (Johnston et al., 2006). VH segments from these families are typically overrepresented in B cell progenitors before pre-BCR selection, whereas later developmental populations express a VH repertoire that better reflects germline complexity and that pairs more effectively with surrogate light chain (ten Boekel et al., 1997). Therefore, the skewed repertoire seen in Ccnd3−/− pre–B cells might arise from abnormal pre–B cell selection (Cooper et al., 2006). Alternatively, although the accessibility hypothesis proposes that sterile transcripts precede activation of recombination (Yancopoulos and Alt, 1985), the presence of these transcripts is not sufficient to predict recombination. In Pax5−/− pro–B cells, although sterile transcripts from V7183 and VJ558 families are detected at levels similar to WT, VJ558-to-DJ transcripts are strongly reduced in the absence of Pax5, whereas V7183-to-DJ transcripts remain comparable to WT (Hesslein et al., 2003). These results demonstrate that sterile transcription and V(D)J recombination can be discordant. Whether reflecting aberrant pre-BCR selection, and/or other mechanisms, our results demonstrate that cyclin D3 influences which V(D)J rearrangements are expressed in large pre–B cells.
Regulation of Ig V segment accessibility remains poorly defined. Upon transition from prepro–B to pro–B cells, Igk V segments move away from the nuclear membrane to more centrally located nuclear domains. However, repositioning is not sufficient for transcriptional activation (Jhunjhunwala et al., 2009). Furthermore, although histone modifications have been described throughout D and J genes, Igh and Igk V segments appear to be relatively devoid of histone marks (Degner-Leisso and Feeney, 2010). These observations suggest that cyclin D3 represses Ig V transcription through mechanisms that do not depend upon epigenetic modification.
Cyclin D3 was diffusely distributed throughout the nucleoplasm and not enriched at the nuclear envelope, indicating an association with nucleoplasmic matrix and not nuclear lamina (Dechat et al., 2010). The nucleoplasmic matrix has been implicated in several important transcriptional functions including assembly of RNA polymerase II transcription centers, binding of transcription factors, and chromatin organization within the nuclear interior. Modulation of any of these activities, or regulating accessibility to them, might be sufficient to explain the effect of cyclin D3 on Ig V region transcription. Our observations are broadly consistent with descriptions of cell cycle–independent roles for cyclin D1 (Bienvenu et al., 2010; Jirawatnotai et al., 2011), which can bind to genomic regulatory regions via transcription factors and recruit CREB binding protein histone acetyltransferase. This chromatin binding was associated with both the induction and repression of gene products (Bienvenu et al., 2010), suggesting that cyclin D1 may play complex roles in gene regulation. As we found no evidence of chromatin binding, our data indicate that cyclin D3 can perform similar functions as cyclin D1 but through fundamentally different mechanisms.
In pro–B cells, cyclin D3 was a much more potent repressor of Ig V gene transcription than inducer of CDK4/6-dependent cell cycle targets. This is consistent with observations in vivo that Ccnd3−/− mice only have a partial proliferative defect at the pro–B cell stage (Cooper et al., 2006). Rather, cyclin D3 appears to play a critical role in pre–B cell proliferation. This might reflect some minor redundancy with cyclin D2 in pro–B cells. However, the pro–B and pre–B cell populations are normal in Ccnd2−/− mice and, as demonstrated here, cyclin D2 does not efficiently form complexes with CDK4. Alternatively, pro–B cells could use cell cycle components downstream of CDK4/6 to proliferate (Kozar and Sicinski, 2005).
It is reasonable that, beyond their shared roles in cell cycle progression, the D-type cyclins function differently. Based on peptide sequence similarity, chicken cyclin D1, cyclin D2, and the putative cyclin D3 are more closely related to the corresponding human homologues than to each other (Withers et al., 1991; Xiong et al., 1991, 1992; Inaba et al., 1992; Li et al., 1995; Lahti et al., 1997; Caldwell et al., 2005). Although all proteins contain the N-terminal LxCxE motif (implicated in Rb binding), a cyclin box spanning the cyclin-N and cyclin-C domains and a cyclin signature CDK binding motif, the remaining sequences are divergent between family members (Musgrove et al., 2011). Perhaps not surprisingly, the N-D3 chimera containing the LxCxE and CDK binding motifs from cyclin D3 promoted up-regulation of cell cycle factors. Conversely, the less well characterized C-terminal region of cyclin D3 was able to repress Igkv1-117 transcription. Thus, although the conserved D-type cyclin motifs are associated with cell cycle regulation, regions of divergence may impart cell cycle–independent and specific roles to each cyclin D protein.
It was unexpected that PI3K activation did not contribute to proliferation. PI3K activation is required for proliferation in peripheral B cells (Fruman et al., 1999; Suzuki et al., 1999, 2003; Clayton et al., 2002; Jou et al., 2002; Okkenhaug et al., 2002) and lies downstream of the IL-7R (Corcoran et al., 1996), which contributes to proliferation in pro–B and pre–B cells (Milne and Paige, 2006). PI3K may contribute to B cell development by regulating Rag1/2 expression (Ramadani et al., 2010). As demonstrated here and elsewhere (Cooper et al., 2006), PI3K activation enhances detectable soluble cyclin D3 levels. However, in vitro studies indicate that PI3K does not regulate that fraction of cyclin D3 involved in determining proliferation, whereas in vivo studies confirm normal proliferation in p85α−/− and bPTEN−/− progenitors. Our findings are consistent with recent findings that Akt1−/−Akt2−/− fetal liver cells can competitively reconstitute bone marrow B lymphopoiesis when transferred with WT fetal liver into irradiated congenic hosts (Calamito et al., 2010). In contrast, all mature B cell populations were underrepresented in Akt1−/−Akt2−/− + WT chimeras, further arguing that PI3K signaling in progenitor and peripheral B cell populations performs different functions.
Based on our data, we propose that compartmentalization of cyclin D3 enables it to effectively mediate very different functions during B lymphopoiesis including cell cycle progression and repression of Ig V gene accessibility. Coupling these two divergent functions through one molecule helps ensure that proliferation and gene recombination remain mutually exclusive events. The mechanisms segregating cyclin D3 into these different nuclear compartments are unclear. However, comparisons between pro–B cells and MEFs suggest that compartmentalization of the D-type cyclins is a feature of those cells dependent on them for proliferation. Therefore, hematopoietic nuclei must contain unique structures or functions that enable D-type cyclin function (Zong et al., 2000; Dobreva et al., 2003). Identifying these cell-specific features should provide further mechanistic insights into why the D-type cyclins are indispensible for hematopoiesis.
MATERIALS AND METHODS
Mice.
Rag2−/− and Bim+/− and C57BL/6 (WT) mice were purchased from Taconic and The Jackson Laboratory, respectively. Ccnd3−/− mice were maintained as described previously (Cooper et al., 2006). Mice were used at 6–10 wk of age, housed in the GCIS barrier animal facility, and experiments were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee of the University of Chicago. Mixed background p85α+/− mice (Suzuki et al., 1999) were backcrossed to BALB/c for 12 generations. Animals were housed in the animal facility at Keio University School of Medicine under specific pathogen-free conditions and analyzed at 7–8 wk of age. Experimental protocols were approved by the Animal Care and Use Committee of Keio University and were performed in accordance with the Institutional Guidelines. bPTEN−/− (PtenloxP/loxPCD19Cre+/−) and control (Pten+/+CD19Cre+/−) mice were analyzed at 7–9 wk of age (Anzelon et al., 2003). Animals were maintained in the animal facility of the Sanford-Burnham Medical Research Institute (SBMRI). All protocols were approved by the Institutional Animal Care and Use Committee at the SBMRI and were performed in accordance with institutional guidelines and regulations.
Isolation and in vitro culture of cells.
Rag2−/− pro–B cells were isolated from bone marrow with a MACS separation column (Miltenyi Biotec) for isolation of CD19+ (1D3; BD) cells or by depleting lineage-positive cells using antibodies against CD3ε (145-2C11), CD4 (RM4-5), CD8α (53–6.7), CD11b (M1/70), CD11c (HL3), Gr-1 (RB6-8C5), NK1.1 (PK136), and TER-119 (all from BD), and IgG + IgM F(ab)2 (Jackson ImmunoResearch Laboratories; Fig. 5, I and J only). Bim−/− (and littermate WT) and Ccnd3−/− pro–/pre–B cell progenitors were also obtained by lineage depletion. Cells were cultured in complete Opti-MEM containing 7.5% (vol/vol) FBS and 10 ng/ml IL-7 (Mandal et al., 2009). MEFs (CF551.2.1; gift from M. Barbacid, Spanish National Cancer Research Centre, Madrid, Spain) were cultured as previously described (Malumbres et al., 2004) and analyzed as asynchronized, proliferating cells. For inhibitor studies, cells were cultured in the presence of 25 µg/ml CHX (Sigma-Aldrich), 5 µM LY294002 (TOCRIS), 0.5 µM PD0332991 (ChemieTek), and/or 5 µM MG-132 (Biomol-Enzol Life Sciences) for the times indicated.
Confocal microscopy and image analysis.
Cells were fixed and stained as described previously (Zhang et al., 2007) with primary antibodies to cyclin D2 (M-20), cyclin D3 (C-16), Lamin B (M-20; all from Santa Cruz Biotechnology, Inc.), or Rb (554136; BD) and secondary antibodies coupled to Alexa Fluor dyes (Invitrogen). Images were collected using a TCS SP2 AOBS or SP5-II-STED-CW confocal microscope (63× objective; Leica) and analyzed as single plane confocal images. To quantify colocalization between two signals in individual cells, we used the ImageJ (National Institutes of Health) JACoP plug-in (Manders’ coefficient) with a threshold of 15 to score positive colocalization (Bolte and Cordelières, 2006). To measure the location of cyclin D staining in cells, Lamin B was used to demarcate the nuclear and cytoplasmic regions of interest (ROI) and the ImageJ ROI Manager was used to measure MFI for each protein in each region. Values are presented as a ratio of signal (nucleus/cytoplasm). For each experiment, at least 30 randomly selected cells were scored.
TCLs, IP, fractionation, and immunoblot.
TCLs obtained with 1% (vol/vol) Nonidet-P40 (Mandal et al., 2009) or SDS extractions (375 mM Tris, 6% SDS, 15% β-mercaptoethanol, 30% glycerol, and 0.15% Bromophenol blue) were prepared. Where indicated, TCLs were subjected to IP using 1 µg of antibody (anti-CDK4 [DCS-31; BioSource], anti-Rb, and mouse IgG1 κ isotype control [P3; eBioscience]) coupled to protein G–Sepharose beads (Thermo Fisher Scientific). Subcellular fractionation protocol was modified from that previously published (Fey et al., 1984) to include digestion of the chromatin with 100 U/ml DNase I (QIAGEN), 10 µg/ml RNase A (Sigma-Aldrich), and a 1:15 dilution Micrococcal Nuclease (Thermo Fisher Scientific) for 90 min. Equal cell equivalents of each TCL or volumes of each fraction were resolved by SDS-PAGE and transferred to Immuno-Blot PVDF membranes (Bio-Rad Laboratories). Blots were probed with antibodies against cyclin D2, cyclin D3, CDK4, Actin (MAB1501; Millipore), α-tubulin (YOL 1/34; Accurate Chemical), Histone H3 (9715; Cell Signaling Technology), and Lamin A/C (H-110; Santa Cruz Biotechnology, Inc.).
Flow cytometry.
Bone marrow was collected from p85α−/−, bPTEN−/−, or control mice, erythrocytes were lysed, and cells were stained (Cooper et al., 2006) with anti-B220 (RA3-6B2), anti-CD43 (S7), anti-IgM (R6-60.2; all from BD; or II/41, eBioscience), and 5 µg/ml 7-AAD (Sigma-Aldrich) to resolve pro–B (B220+CD43+IgM−), large pre–B (B220+CD43−IgM−FCShigh), small pre–B (B220+CD43−IgM−FCSlow), and immature B (B220+IgM+CD43−) populations plus DNA content using a FACSAria or FACSCanto (BD) and FlowJo software. Bim−/− and control progenitors were fixed and incubated in a solution containing propidium iodide and analyzed on a C6 cytometer (Accuri) as previously described (Mandal et al., 2009). For intracellular phospho-Akt S473 (4058; Cell Signaling Technology) staining, Rag2−/− pro–B cells were fixed with Cytofix and permeabilized with Phosflow Perm Buffer III as recommended by the manufacturer (BD).
Derivation of WT cyclin D3 and cyclin D2/D3 chimeric constructs and retroviral transduction.
Mouse Ccnd2 and Ccnd3 cDNAs were cloned downstream of a double hemagglutinin tag, mutating the initiating Met to Ala. Nested PCR using these templates was performed to produce cyclin D2/D3 chimeric proteins as follows: N-terminal chimera (N-D3) with cyclin D3 aa 2–94 followed by cyclin D2 93–289; and C-terminal chimera (C-D3) with cyclin D2 aa 2–170, followed by cyclin D3 173–292. WT cyclin D3 and chimeric constructs were then subcloned into the MIGR1 retroviral vector. Retroviruses containing constructs were produced by transient transfection of PLAT-E packaging cell lines, and infection of Ccnd3−/− B cell progenitors or 3T3 cells was done as previously described (Mandal et al., 2009). After 48 h, GFP+B220+CD43hi pro–B or GFP+ 3T3 cells were isolated by cell sorting.
Microarray and qPCR analysis.
Bone marrow was collected from WT and Ccnd3−/− mice, erythrocytes lysed, cells suspended in staining buffer (10% [vol/vol] FBS in PBS), and stained (Cooper et al., 2006) with antibodies against CD3 (17A2), CD8α, CD11b, NK1.1, Ter119, CD43 (BD), CD4 (GK1.5), B220 (RA3-6B2; both from eBioscience), and IgM (115–097-020, Jackson ImmunoResearch Laboratories; or II/41, eBioscience). Pro–B (Lin−B220+CD43hiIgM−) cells were isolated by cell sorting (FACSAria). Total RNA was extracted using TRIzol (Invitrogen) and RNeasy kit (QIAGEN) and samples prepared and hybridized to Affymetrix Mouse 430 2.0 Expression arrays according to Affymetrix protocol at the Functional Genomics Facility (University of Chicago). Duplicate samples for WT and Ccnd3−/− were processed for concurrent RNA extraction and hybridization. Data were annotated according to the Affymetrix database available as of July, 2011.
The resulting dataset was analyzed for statistically significant differential expression using the NIA Array Analysis Tool (Sharov et al., 2005). Of the 45,100 probe sets on the array, 1,900 were retained. Genes were then ranked by fold change and those demonstrating at least a twofold difference were retained. In cases where multiple probe sets mapped to one gene, the probe set with the largest differential expression was retained. Overall, 472 genes were evaluated as higher in WT and 277 as higher in Ccnd3−/− pro–B cells. Heat mapping was performed using HierarchicalCluster on GenePattern (Reich et al., 2006) and grouping by keywords was performed using DAVID (Huang et al., 2009a,b).
Specific gene expression data were validated using qPCR. Pro–B and small pre–B (Lin−B220+CD43lo/−IgM−FSClo) cells were isolated, total RNA extracted, and reverse transcribed with SuperScript III reverse transcription (Invitrogen). Alternatively, Rag2−/− progenitors were isolated by MACS negative selection and cultured with or without LY294002 or PD0332991 for 24 h before RNA isolation. Retrovirally transduced progenitors were cell sorted as GFP+B220+CD43hi followed by RNA extraction and cDNA synthesis. Samples were then analyzed by qPCR in triplicate (primers, Table S2) as previously described (Mandal et al., 2011). For Igk recombination, genomic DNA was isolated from the indicated populations and analyzed by qPCR using a degenerate Vk primer (degVk) and a primer downstream of Igkj1 (k-J1-R). Amplification of the Eκi (Q-iEnhL and Q-iEnhR) was used to control for the amount of DNA (Johnson et al., 2008).
V(D)J recombination sequence analysis.
WT and Ccnd3−/− large pre–B (Lin−B220+CD43lo/−IgM−FSChi) cells were isolated, total RNA extracted, and reverse transcribed. Resulting cDNA was amplified by PCR (primers, Table S2) as previously described (Fuxa et al., 2004). PCR products were cloned into the pCR2.1-TOPO vector (Invitrogen) and plasmid DNA was purified. Bacterial clones were randomly selected for sequencing (100 clones per genotype per experiment, n = 3) and V(D)J segments determined using NCBI’s IgBLAST (KABAT Ig domain system). For sequences that aligned as in-frame rearrangements, VH segments were then grouped into families as described by Johnston et al. (2006).
Statistical analysis.
To analyze statistical significance, we used either paired or unpaired distribution Student’s t test (Prism 5.0a software; GraphPad Software). Significant p-values (<0.05) are reported and in some cases are rounded to facilitate comparisons.
Online supplemental material.
Tables S1 shows genes differentially expressed between WT and Ccnd3−/− Lin−B220+CD43hiIgM− pro–B cells. Table S2 shows primers used in the study. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20120800/DC1.
Supplementary Material
Acknowledgments
We thank Y. Hirata (Keio University) and C. Labno, D. Leclerc, R. Duggin, M. Olson, J. Cao, J. Kim, and S. Karki (University of Chicago) for technical assistance.
This work was supported by a Grant-in-Aid for Scientific Research (B) (#18390155) from the Japan Society for the Promotion of Science (S. Koyasu) and National Institutes of Health grants 5T32GM007183-35 (S.E. Powers) and GM088847 (M.R. Clark).
The authors declare that they have no competing financial interests.
Footnotes
Abbreviations used:
- CHX
- cycloheximide
- IP
- immunoprecipitation
- MEF
- mouse embryonic fibroblast
- PI3K
- phosphoinositide 3-kinase
- pre-BCR
- pre–B cell receptor
- qPCR
- quantitative PCR
- TCL
- total cell lysate
References
- Andrés V., González J.M. 2009. Role of A-type lamins in signaling, transcription, and chromatin organization. J. Cell Biol. 187:945–957 10.1083/jcb.200904124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzelon A.N., Wu H., Rickert R.C. 2003. Pten inactivation alters peripheral B lymphocyte fate and reconstitutes CD19 function. Nat. Immunol. 4:287–294 10.1038/ni892 [DOI] [PubMed] [Google Scholar]
- Aoki-Ota M., Torkamani A., Ota T., Schork N., Nemazee D. 2012. Skewed primary Igκ repertoire and V-J joining in C57BL/6 mice: implications for recombination accessibility and receptor editing. J. Immunol. 188:2305–2315 10.4049/jimmunol.1103484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienvenu F., Jirawatnotai S., Elias J.E., Meyer C.A., Mizeracka K., Marson A., Frampton G.M., Cole M.F., Odom D.T., Odajima J., et al. 2010. Transcriptional role of cyclin D1 in development revealed by a genetic-proteomic screen. Nature. 463:374–378 10.1038/nature08684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S., Cordelières F.P. 2006. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224:213–232 10.1111/j.1365-2818.2006.01706.x [DOI] [PubMed] [Google Scholar]
- Calamito M., Juntilla M.M., Thomas M., Northrup D.L., Rathmell J., Birnbaum M.J., Koretzky G., Allman D. 2010. Akt1 and Akt2 promote peripheral B-cell maturation and survival. Blood. 115:4043–4050 10.1182/blood-2009-09-241638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell R.B., Kierzek A.M., Arakawa H., Bezzubov Y., Zaim J., Fiedler P., Kutter S., Blagodatski A., Kostovska D., Koter M., et al. 2005. Full-length cDNAs from chicken bursal lymphocytes to facilitate gene function analysis. Genome Biol. 6:R6 10.1186/gb-2004-6-1-r6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciemerych M.A., Sicinski P. 2005. Cell cycle in mouse development. Oncogene. 24:2877–2898 10.1038/sj.onc.1208608 [DOI] [PubMed] [Google Scholar]
- Clayton E., Bardi G., Bell S.E., Chantry D., Downes C.P., Gray A., Humphries L.A., Rawlings D., Reynolds H., Vigorito E., Turner M. 2002. A crucial role for the p110Δ subunit of phosphatidylinositol 3-kinase in B cell development and activation. J. Exp. Med. 196:753–763 10.1084/jem.20020805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A.B., Sawai C.M., Sicinska E., Powers S.E., Sicinski P., Clark M.R., Aifantis I. 2006. A unique function for cyclin D3 in early B cell development. Nat. Immunol. 7:489–497 10.1038/ni1324 [DOI] [PubMed] [Google Scholar]
- Corcoran A.E., Smart F.M., Cowling R.J., Crompton T., Owen M.J., Venkitaraman A.R. 1996. The interleukin-7 receptor alpha chain transmits distinct signals for proliferation and differentiation during B lymphopoiesis. EMBO J. 15:1924–1932 [PMC free article] [PubMed] [Google Scholar]
- Dechat T., Adam S.A., Taimen P., Shimi T., Goldman R.D. 2010. Nuclear lamins. Cold Spring Harb. Perspect. Biol. 2:a000547 10.1101/cshperspect.a000547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degner-Leisso S.C., Feeney A.J. 2010. Epigenetic and 3-dimensional regulation of V(D)J rearrangement of immunoglobulin genes. Semin. Immunol. 22:346–352 10.1016/j.smim.2010.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl J.A., Zindy F., Sherr C.J. 1997. Inhibition of cyclin D1 phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin-proteasome pathway. Genes Dev. 11:957–972 10.1101/gad.11.8.957 [DOI] [PubMed] [Google Scholar]
- Dirlam A., Spike B.T., Macleod K.F. 2007. Deregulated E2f-2 underlies cell cycle and maturation defects in retinoblastoma null erythroblasts. Mol. Cell. Biol. 27:8713–8728 10.1128/MCB.01118-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobreva G., Dambacher J., Grosschedl R. 2003. SUMO modification of a novel MAR-binding protein, SATB2, modulates immunoglobulin mu gene expression. Genes Dev. 17:3048–3061 10.1101/gad.1153003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fey E.G., Wan K.M., Penman S. 1984. Epithelial cytoskeletal framework and nuclear matrix-intermediate filament scaffold: three-dimensional organization and protein composition. J. Cell Biol. 98:1973–1984 10.1083/jcb.98.6.1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming H.E., Paige C.J. 2002. Cooperation between IL-7 and the pre-B cell receptor: a key to B cell selection. Semin. Immunol. 14:423–430 10.1016/S1044532302000775 [DOI] [PubMed] [Google Scholar]
- Fruman D.A., Snapper S.B., Yballe C.M., Davidson L., Yu J.Y., Alt F.W., Cantley L.C. 1999. Impaired B cell development and proliferation in absence of phosphoinositide 3-kinase p85alpha. Science. 283:393–397 10.1126/science.283.5400.393 [DOI] [PubMed] [Google Scholar]
- Fuxa M., Skok J., Souabni A., Salvagiotto G., Roldan E., Busslinger M. 2004. Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev. 18:411–422 10.1101/gad.291504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog S., Reth M., Jumaa H. 2009. Regulation of B-cell proliferation and differentiation by pre-B-cell receptor signalling. Nat. Rev. Immunol. 9:195–205 10.1038/nri2491 [DOI] [PubMed] [Google Scholar]
- Hesslein D.G.T., Pflugh D.L., Chowdhury D., Bothwell A.L.M., Sen R., Schatz D.G. 2003. Pax5 is required for recombination of transcribed, acetylated, 5′ IgH V gene segments. Genes Dev. 17:37–42 10.1101/gad.1031403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Sherman B.T., Lempicki R.A. 2009a. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37:1–13 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Sherman B.T., Lempicki R.A. 2009b. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Inaba T., Matsushime H., Valentine M., Roussel M.F., Sherr C.J., Look A.T. 1992. Genomic organization, chromosomal localization, and independent expression of human cyclin D genes. Genomics. 13:565–574 10.1016/0888-7543(92)90126-D [DOI] [PubMed] [Google Scholar]
- Jhunjhunwala S., van Zelm M.C., Peak M.M., Murre C. 2009. Chromatin architecture and the generation of antigen receptor diversity. Cell. 138:435–448 10.1016/j.cell.2009.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirawatnotai S., Hu Y., Michowski W., Elias J.E., Becks L., Bienvenu F., Zagozdzon A., Goswami T., Wang Y.E., Clark A.B., et al. 2011. A function for cyclin D1 in DNA repair uncovered by protein interactome analyses in human cancers. Nature. 474:230–234 10.1038/nature10155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K., Hashimshony T., Sawai C.M., Pongubala J.M.R., Skok J.A., Aifantis I., Singh H. 2008. Regulation of immunoglobulin light-chain recombination by the transcription factor IRF-4 and the attenuation of interleukin-7 signaling. Immunity. 28:335–345 10.1016/j.immuni.2007.12.019 [DOI] [PubMed] [Google Scholar]
- Johnston C.M., Wood A.L., Bolland D.J., Corcoran A.E. 2006. Complete sequence assembly and characterization of the C57BL/6 mouse Ig heavy chain V region. J. Immunol. 176:4221–4234 [DOI] [PubMed] [Google Scholar]
- Jou S.-T., Carpino N., Takahashi Y., Piekorz R., Chao J.-R., Carpino N., Wang D., Ihle J.N. 2002. Essential, nonredundant role for the phosphoinositide 3-kinase p110delta in signaling by the B-cell receptor complex. Mol. Cell. Biol. 22:8580–8591 10.1128/MCB.22.24.8580-8591.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozar K., Sicinski P. 2005. Cell cycle progression without cyclin D-CDK4 and cyclin D-CDK6 complexes. Cell Cycle. 4:388–391 10.4161/cc.4.3.1551 [DOI] [PubMed] [Google Scholar]
- Kozar K., Ciemerych M.A., Rebel V.I., Shigematsu H., Zagozdzon A., Sicinska E., Geng Y., Yu Q., Bhattacharya S., Bronson R.T., et al. 2004. Mouse development and cell proliferation in the absence of D-cyclins. Cell. 118:477–491 10.1016/j.cell.2004.07.025 [DOI] [PubMed] [Google Scholar]
- Lahti J.M., Li H., Kidd V.J. 1997. Elimination of cyclin D1 in vertebrate cells leads to an altered cell cycle phenotype, which is rescued by overexpression of murine cyclins D1, D2, or D3 but not by a mutant cyclin D1. J. Biol. Chem. 272:10859–10869 10.1074/jbc.272.16.10859 [DOI] [PubMed] [Google Scholar]
- Li H., Grenet J., Kidd V.J. 1995. Structure and gene expression of avian cyclin D2. Gene. 167:341–342 10.1016/0378-1119(95)00598-6 [DOI] [PubMed] [Google Scholar]
- Lin D.I., Barbash O., Kumar K.G.S., Weber J.D., Harper J.W., Klein-Szanto A.J.P., Rustgi A., Fuchs S.Y., Diehl J.A. 2006. Phosphorylation-dependent ubiquitination of cyclin D1 by the SCF(FBX4-alphaB crystallin) complex. Mol. Cell. 24:355–366 10.1016/j.molcel.2006.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres M., Sotillo R., Santamaría D., Galán J., Cerezo A., Ortega S., Dubus P., Barbacid M. 2004. Mammalian cells cycle without the D-type cyclin-dependent kinases Cdk4 and Cdk6. Cell. 118:493–504 10.1016/j.cell.2004.08.002 [DOI] [PubMed] [Google Scholar]
- Mandal M., Powers S.E., Ochiai K., Georgopoulos K., Kee B.L., Singh H., Clark M.R. 2009. Ras orchestrates exit from the cell cycle and light-chain recombination during early B cell development. Nat. Immunol. 10:1110–1117 10.1038/ni.1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M., Powers S.E., Maienschein-Cline M., Bartom E.T., Hamel K.M., Kee B.L., Dinner A.R., Clark M.R. 2011. Epigenetic repression of the Igk locus by STAT5-mediated recruitment of the histone methyltransferase Ezh2. Nat. Immunol. 12:1212–1220 10.1038/ni.2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariappan I., Gurung R., Thanumalayan S., Parnaik V.K. 2007. Identification of cyclin D3 as a new interaction partner of lamin A/C. Biochem. Biophys. Res. Commun. 355:981–985 10.1016/j.bbrc.2007.02.060 [DOI] [PubMed] [Google Scholar]
- Milne C.D., Paige C.J. 2006. IL-7: a key regulator of B lymphopoiesis. Semin. Immunol. 18:20–30 10.1016/j.smim.2005.10.003 [DOI] [PubMed] [Google Scholar]
- Musgrove E.A., Caldon C.E., Barraclough J., Stone A., Sutherland R.L. 2011. Cyclin D as a therapeutic target in cancer. Nat. Rev. Cancer. 11:558–572 10.1038/nrc3090 [DOI] [PubMed] [Google Scholar]
- Okkenhaug K., Bilancio A., Farjot G., Priddle H., Sancho S., Peskett E., Pearce W., Meek S.E., Salpekar A., Waterfield M.D., et al. 2002. Impaired B and T cell antigen receptor signaling in p110delta PI 3-kinase mutant mice. Science. 297:1031–1034 [DOI] [PubMed] [Google Scholar]
- Ramadani F., Bolland D.J., Garcon F., Emery J.L., Vanhaesebroeck B., Corcoran A.E., Okkenhaug K. 2010. The PI3K isoforms p110alpha and p110delta are essential for pre-B cell receptor signaling and B cell development. Sci. Signal. 3:ra60 10.1126/scisignal.2001104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich M., Liefeld T., Gould J., Lerner J., Tamayo P., Mesirov J.P. 2006. GenePattern 2.0. Nat. Genet. 38:500–501 10.1038/ng0506-500 [DOI] [PubMed] [Google Scholar]
- Sharov A.A., Dudekula D.B., Ko M.S.H. 2005. A web-based tool for principal component and significance analysis of microarray data. Bioinformatics. 21:2548–2549 10.1093/bioinformatics/bti343 [DOI] [PubMed] [Google Scholar]
- Suzuki H., Terauchi Y., Fujiwara M., Aizawa S., Yazaki Y., Kadowaki T., Koyasu S. 1999. Xid-like immunodeficiency in mice with disruption of the p85alpha subunit of phosphoinositide 3-kinase. Science. 283:390–392 10.1126/science.283.5400.390 [DOI] [PubMed] [Google Scholar]
- Suzuki H., Matsuda S., Terauchi Y., Fujiwara M., Ohteki T., Asano T., Behrens T.W., Kouro T., Takatsu K., Kadowaki T., Koyasu S. 2003. PI3K and Btk differentially regulate B cell antigen receptor-mediated signal transduction. Nat. Immunol. 4:280–286 10.1038/ni890 [DOI] [PubMed] [Google Scholar]
- ten Boekel E., Melchers F., Rolink A.G. 1997. Changes in the V(H) gene repertoire of developing precursor B lymphocytes in mouse bone marrow mediated by the pre-B cell receptor. Immunity. 7:357–368 10.1016/S1074-7613(00)80357-X [DOI] [PubMed] [Google Scholar]
- Withers D.A., Harvey R.C., Faust J.B., Melnyk O., Carey K., Meeker T.C. 1991. Characterization of a candidate bcl-1 gene. Mol. Cell. Biol. 11:4846–4853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Connolly T., Futcher B., Beach D. 1991. Human D-type cyclin. Cell. 65:691–699 10.1016/0092-8674(91)90100-D [DOI] [PubMed] [Google Scholar]
- Xiong Y., Menninger J., Beach D., Ward D.C. 1992. Molecular cloning and chromosomal mapping of CCND genes encoding human D-type cyclins. Genomics. 13:575–584 10.1016/0888-7543(92)90127-E [DOI] [PubMed] [Google Scholar]
- Yancopoulos G.D., Alt F.W. 1985. Developmentally controlled and tissue-specific expression of unrearranged VH gene segments. Cell. 40:271–281 10.1016/0092-8674(85)90141-2 [DOI] [PubMed] [Google Scholar]
- Zhang M., Veselits M., O’Neill S., Hou P., Reddi A.L., Berlin I., Ikeda M., Nash P.D., Longnecker R., Band H., Clark M.R. 2007. Ubiquitinylation of Ig beta dictates the endocytic fate of the B cell antigen receptor. J. Immunol. 179:4435–4443 [DOI] [PubMed] [Google Scholar]
- Zong R.-T., Das C., Tucker P.W. 2000. Regulation of matrix attachment region-dependent, lymphocyte-restricted transcription through differential localization within promyelocytic leukemia nuclear bodies. EMBO J. 19:4123–4133 10.1093/emboj/19.15.4123 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.