Germline mutations in CARD11 that result in constitutive NF-κB activation and selective B cell expansion underlie congenital B cell lymphocytosis.
Abstract
Nuclear factor-κB (NF-κB) controls genes involved in normal lymphocyte functions, but constitutive NF-κB activation is often associated with B cell malignancy. Using high-throughput whole transcriptome sequencing, we investigated a unique family with hereditary polyclonal B cell lymphocytosis. We found a novel germline heterozygous missense mutation (E127G) in affected patients in the gene encoding CARD11, a scaffolding protein required for antigen receptor (AgR)–induced NF-κB activation in both B and T lymphocytes. We subsequently identified a second germline mutation (G116S) in an unrelated, phenotypically similar patient, confirming mutations in CARD11 drive disease. Like somatic, gain-of-function CARD11 mutations described in B cell lymphoma, these germline CARD11 mutants spontaneously aggregate and drive constitutive NF-κB activation. However, these CARD11 mutants rendered patient T cells less responsive to AgR-induced activation. By reexamining this rare genetic disorder first reported four decades ago, our findings provide new insight into why activating CARD11 mutations may induce B cell expansion and preferentially predispose to B cell malignancy without dramatically perturbing T cell homeostasis.
NF-κB represents a family of transcription factors that governs key proliferation, antiapoptosis, and immune function genes in lymphocytes (Lenardo and Baltimore, 1989; Vallabhapurapu and Karin, 2009). Overactive NF-κB is often associated with B cell malignancy (Staudt, 2010). For example, constitutive NF-κB activity in the activated B cell–like (ABC) subtype of diffuse large B cell lymphoma (DLBCL) can result from somatic mutations in various genes including CD79B, MYD88, TNFAIP3, and CARD11 (Shaffer et al., 2012). For B and T lymphocytes, CARD11 acts as a critical signal transducer from the cell surface antigen (Ag) receptor (AgR) to the cytoplasmic IκB kinase (IKK), which in turn activates the canonical NF-κB pathway (Thome et al., 2010). Gain-of-function, somatic CARD11 mutations described in DLBCL specifically localize to the coiled-coil (CC) region of the protein and mimic a chronic BCR signal for NF-κB activation, without requiring Ag stimulation (Lenz et al., 2008a; Davis et al., 2010). Such somatic activating mutations are relatively common in a variety of DLBCL tumors, suggesting CARD11 may be a particularly important target gene in the progression of B cell oncogenesis (Lenz et al., 2008a; Montesinos-Rongen et al., 2010; Dong et al., 2011). However, neither germline-encoded CARD11 mutations nor similar somatic mutations restricted to T cell malignancies have been described to date. How activating mutations in genes involved in NF-κB signaling like CARD11 might selectively predispose to B cell malignancy remains poorly understood.
In this study, we describe the first known germline mutations in CARD11 in four patients with congenital lymphoid hyperplasia driven by excessive, polyclonal accumulation of B lymphocytes but not T lymphocytes. Approximately half of circulating B cells maintain a transitional (T2/T3) phenotype, suggesting that the output of immature B cells from the bone marrow is elevated. We demonstrate that these dominant missense mutations (E127G and G116S) drive constitutive activation of NF-κB in both B and T cells. In contrast to their B cells, however, patient T cells are relatively hyporesponsive upon in vitro stimulation. Our findings identify the underlying genetic cause of a novel hereditary B cell disorder and reveal a potential molecular explanation for why CARD11 mutations may predispose to B but not T lymphoid malignancy.
RESULTS
Novel disease in patients with splenomegaly and polyclonal B cell lymphocytosis
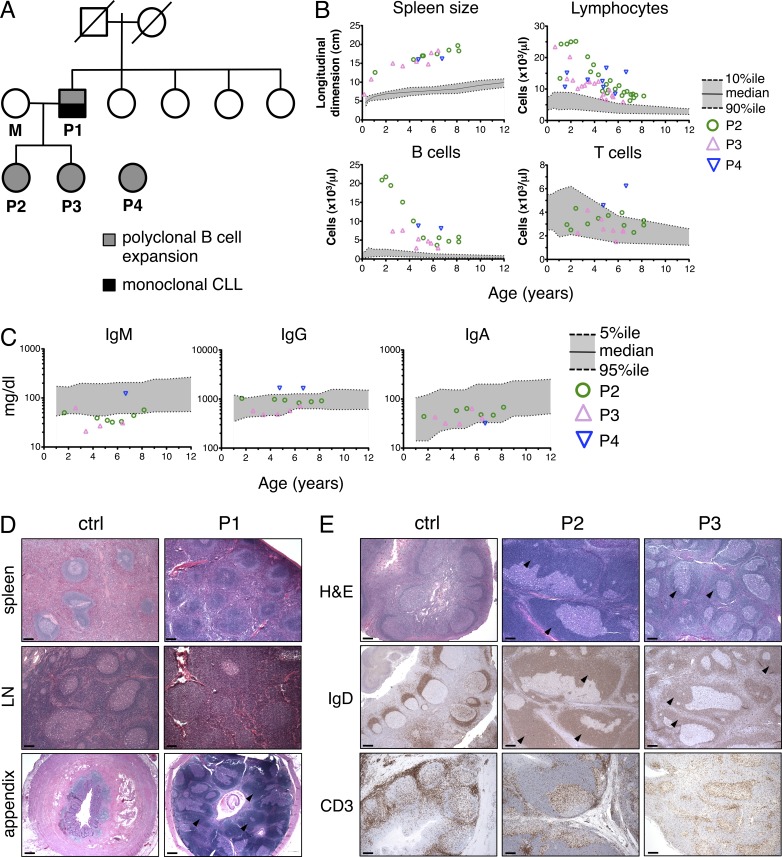
We first investigated a family with a unique form of hereditary polyclonal B cell lymphocytosis (Fig. 1 A; see Supplemental text for detailed case reports). Patient 1 (P1) first presented with splenomegaly and B cell lymphocytosis in infancy (Darte et al., 1971), which persisted into adulthood. When he first entered our study in 2006, we detected a monoclonal expansion of CD20dim, IgM/IgDdim, CD5hi B cells (absolute count >105 × 103/µl), consistent with a diagnosis of B cell chronic lymphocytic leukemia (B-CLL). P1 received hematopoietic stem cell transplantation and has remained healthy since 2007. The parents and four sisters of P1 have no history of similar disease (Fig. 1 A).
Figure 1.
Family pedigree and analysis of B cell lymphocytosis. (A) Family pedigree of P1–3. Circles represent females, squares represent males, and slashes through symbols represent deceased individuals. Solid gray symbols indicate those affected with polyclonal B cell lymphocytosis. Solid black symbols denote progression to monoclonal B-CLL in P1. (B) Total lymphocytes, spleen size, and absolute counts of B cells and T cells in peripheral blood from patients were measured over time by flow cytometry. The gray zone delineates the median and normal range (between 10th and 90th percentile) in age-matched individuals. (C) Serum Ig titers in patients were measured over time by ELISA; gray zone delineates median and normal range (between 5th and 95th percentile). (D) H&E staining of spleen, lymph node, and appendix tissue sections from P1, taken at 34 mo of age (right). Control specimens (ctrl) are shown for comparison (left), including an age-matched lymph node biopsy showing reactive follicular hyperplasia. (E) Histochemical analysis of tonsil sections from control reactive tonsil, P2, and P3 stained with H&E, anti-IgD, and anti-CD3. Black arrowheads mark expanded mantle zones in D and E. Representative photographs are shown. Bars, 500 µm.
P2 and P3 both exhibit the same childhood disease as their father (P1; Fig. 1 A). Despite some recurrent sinus and middle ear infections, both patients are healthy and have no evidence of autoimmunity (see Supplemental text). Notably, both patients failed to generate robust responses to polysaccharide-based vaccines (Table S1). Flow cytometric analysis revealed dramatic elevations of IgM+IgD+CD19+, CD20+ B cells in P2 and P3 peripheral blood, whereas T cell numbers were generally within normal pediatric ranges (Fig. 1 B and Table 1; Rosenberg et al., 1991; Shearer et al., 2003; Piątosa et al., 2010). We did note a slightly elevated proportion of CD4−CD8− double-negative (DN) T cells in these patients, as well as a substantially reduced number of CD45RO+ memory T cells in P3 (Table 1). Although the percentage of memory (CD27+) and class-switched B cell numbers was decreased, only serum IgM titers were slightly below normal ranges (Fig. 1 C). Strikingly, CD10+ immature/transitional B cells in P2 and P3 were significantly elevated in the blood (Table 1). Histological examination of archival P1 spleen, lymph node, and appendix tissue showed florid follicular hyperplasia, containing numerous primary follicles with prominent mantles and marginal zones but few germinal centers (GCs; Fig. 1 D). P2 and P3 tonsillectomy tissue revealed similar follicular hyperplasia, atrophic GCs, and expanded mantles containing an excess of IgD+ naive B cells. However, the number and distribution of CD3+ T cells was normal (Fig. 1 E). Molecular analyses of IgH rearrangements confirmed that B cells from P1 (from childhood splenectomy), P2, and P3 were polyclonal (not depicted).
Table 1.
Flow cytometric phenotype analysis of patient PBMCs
| Marker | P2 | P3 | P4 | Normal Ctrl | ||||
| % | Abs # | % | Abs # | % | Abs # | % | Abs # | |
| B cell lineages | ||||||||
| CD19+ B | 64.9 | 4,703 | 62.8 | 4,710 | 52.7 | 8,105 | 14–33 | 390–1,400 |
| CD20+IgD+IgM+ | 64.2 | 4,658 | 62.3 | 4,673 | 51.7 | 7,951 | 7.4–12.1 | 50–276 |
| CD20+CD10+ | 31.5 | 2,286 | 34.1 | 2,558 | 26.7 | 4,100 | (0.1–3.4) | (2–76) |
| CD20+CD27+ | 1 | 76 | 0.9 | 67 | 0.4 | 62 | 2.9–7.3 | 12–68 |
| CD20+IgG+ | 0.1 | 9 | 0.1 | 9 | 0.1 | 15 | (0.2–2.2) | (4–49) |
| CD20+IgA+ | 0.1 | 5 | 0 | 3 | ND | ND | (0.1–1.3) | (3–9) |
| CD20+CD5+ | 43.6 | 3,164 | 43.4 | 3,255 | 35.7 | 5,491 | (0.7–9.7) | (14–159) |
| T and NK cell lineages | ||||||||
| CD3+ T | 31.5 | 2,287 | 32.9 | 2,464 | 40.5 | 6,229 | 56–75 | 1,400–3,700 |
| CD4+ T | 16.6 | 1,202 | 17.1 | 1,282 | 9.0 | 1,384 | 28–47 | 700–2,200 |
| CD4+CD45RA+ | 10.5 | 760 | 16.6 | 1,245 | 2.5 | 385 | 20–33 | 430–1,500 |
| CD4+CD45RO+ | 5.6 | 405 | 0 | 2 | 1.4 | 215 | 6–17 | 220–660 |
| CD8+ T | 9.6 | 697 | 11.8 | 885 | 27.5 | 4,230 | 16–30 | 490–1,300 |
| CD8+CD45RA+ | 8.4 | 612 | 11.2 | 837 | 7.1 | 1,092 | 16–22 | 380–1,100 |
| CD8+CD45RO+ | 0.5 | 39 | 0.1 | 8 | 5.0 | 769 | 3–11 | 90–440 |
| NK cells | 3.7 | 268 | 4.3 | 325 | 6.9 | 1,061 | 4–17 | 130–720 |
| TCR-α/β+CD4−CD8− | 5.0 | 364 | 3.6 | 272 | 3.9 | 600 | (0.3–1.3) | (6–23) |
| CD4+CD25+Foxp3+ | 1.1 | 83 | 0.6 | 33 | 0.3 | 40 | (1.6–4.3) | (25–89) |
Percentages and absolute cell counts (per microliter of whole blood) of each cell population. Ranges in age-matched pediatric controls are listed on the right, based on published reports (Shearer et al., 2003; Piątosa et al., 2010). Parentheses indicate normal adult controls for which pediatric data were not readily available. Patient ages (years) at time of phenotyping were 8 (P2), 6 (P3), and 6 (P4).
We subsequently identified an unrelated patient (P4) with similar B cell lymphocytosis, splenomegaly, and lymphadenopathy. Like P2 and P3, we noted a significantly reduced percentage of CD27+ memory B cells and an excess of immature CD10+ B cells in the blood (Table 1). P4 was adopted from China at age 4; family history is unavailable. P4 has experienced more frequent infections including bronchitis, pneumonia, otitis media, and Molluscum contagiosum (see Supplemental text), suggesting that immunodeficiency may be more pronounced in P4 compared with P2 and P3. Similar to P2 and P3, P4 did not mount effective antibody (Ab) responses to pneumococcal infection or meningococcal vaccine (Table S1). We also observed an elevated number of CD8+ T cells and NK cells in P4 (Table 1), likely resulting from a chronic active EBV infection. Unlike the other patients, P4 exhibits some evidence of autoimmunity with auto-Abs (antinuclear Abs, anti-C3, and antineutrophil Abs; see Supplemental text) and autoimmune neutropenia. IgM and IgA titers were within normal ranges, whereas IgG titers were slightly increased (Fig. 1 C). No tissue samples were available for further analysis.
Elevated output of transitional B cells from the bone marrow partially accounts for peripheral B cell accumulation
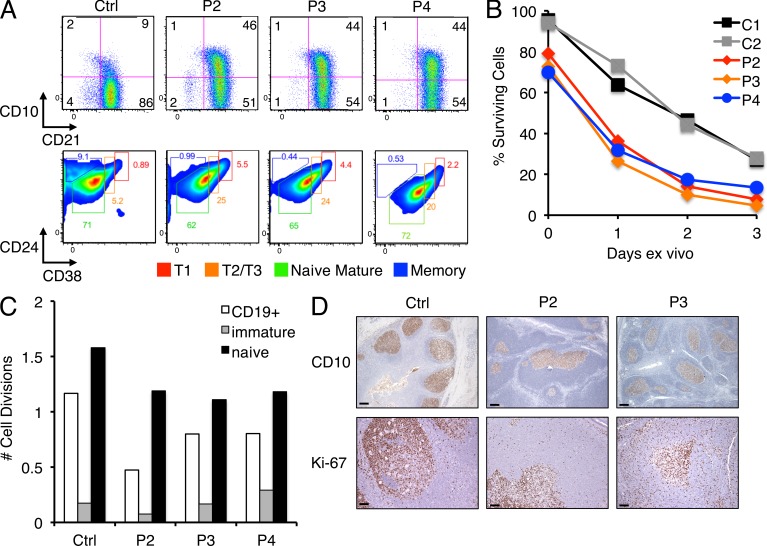
Elevated numbers of peripheral blood B cells expressing markers such as CD5 and CD10 suggested an expansion of immature/transitional B cells in these patients. Further phenotyping revealed that CD10+ B cells uniformly expressed CD21 and intermediate/high levels of CD24 and CD38 (Fig. 2 A), indicating the majority of these cells resemble a transitional T2/T3-like phenotype (Palanichamy et al., 2009). These data suggest that increased output of immature B cells from the bone marrow may partially account for their B cell lymphocytosis, similar to patients undergoing hematopoietic stem cell transplant or B cell depletion therapy (Marie-Cardine et al., 2008; Palanichamy et al., 2009). We next asked whether peripheral B cell accumulation in these patients could result from increased survival or cell turnover. We found that B cells purified from patients’ peripheral blood actually died faster relative to controls ex vivo, consistent with poorer survival noted in transitional B cells (Fig. 2 B). Although stimulation with anti-IgM alone or in combination with the B cell survival factor BAFF (B cell–activating factor) induced blastogenesis and improved B cell survival overall, the faster decline in percentage of viable patient cells remained comparable (not depicted). Quantification of κ-deletion excision circles (KRECs) in sorted mature, naive, and transitional B cells revealed no significant differences in the in vivo replication history of control and patient B cells (Fig. 2 C; van Zelm et al., 2007). Ki-67 staining in P2 and P3 tonsil sections confirmed that proliferating cells were primarily restricted to GCs as noted in the control, with a low, comparable number of dividing cells seen in the follicular mantles (Fig. 2 D). Importantly, CD10 expression was also restricted to the GC in P2 and P3 tonsillar sections (Fig. 2 D), suggesting IgD+ patient B cells that reach peripheral lymphoid compartments down-regulate CD10 as expected as maturation is completed in the follicle. Our findings collectively suggest that B cell lymphocytosis in these patients is driven in part by abnormally high production of B cells in the bone marrow and not by increased survival or turnover of circulating B cells.
Figure 2.
Elevated transitional B cell output in patients without increased cell survival or replication in the peripheral blood. (A) Flow cytometric identification of CD10+CD21+ transitional B cells (top) in patient versus control (Ctrl) PBMCs, including subsets defined by CD24 and CD38 expression (bottom). Gates delineate T1, T2/T3, mature naive, and memory B cells. Gates were adjusted for P4 based on separate matched controls, analyzed on a different day. (B) Ex vivo survival assay for purified naive B cells from patients and controls, with cell death quantitated daily using Annexin V and propidium iodide staining. Data are representative of four independent experiments. (C) Comparison of in vivo replication history by KREC analysis (displayed as number of cell divisions) using sorted transitional and mature naive B cells from controls and patients. The control shown is distinct from that shown in A. (D) CD10 and Ki-67 staining of tonsil sections from control, P2, and P3, with positive staining restricted to GCs. Representative photographs are shown. Bars: (top) 500 µm; (bottom) 200 µm.
Discovery of germline CARD11 mutations in affected patients
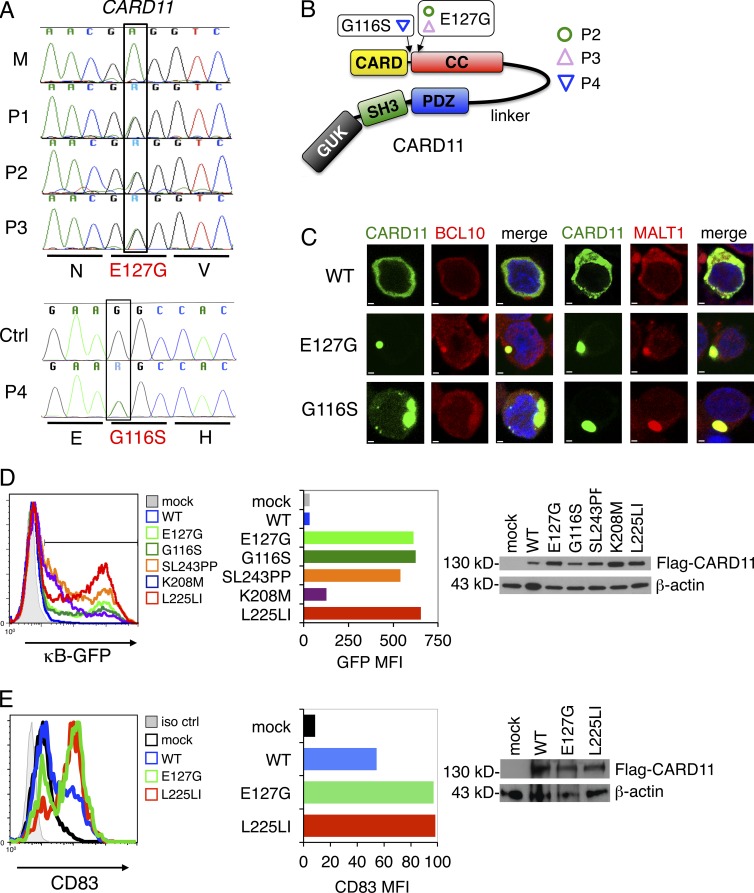
Based on the inheritance pattern observed in patients 1–3, we hypothesized that a single autosomal dominant gene accounted for polyclonal B cell lymphocytosis in this family. Using massively parallel mRNA sequencing (RNA-Seq), we searched for shared mutations in the B cell transcriptome from P2 and P3. Bioinformatics analyses identified a novel heterozygous missense mutation (735A→G) in exon 5 of CARD11 present in all three affected patients (Fig. 3 A). This mutation substitutes a glycine for a phylogenetically conserved glutamic acid residue (E127G) in the N-terminal portion of the CC domain of the CARD11 protein (Fig. 3 B). This mutation was confirmed in genomic DNA from lymphocytes (P1/P2/P3), buccal swabs (P2/P3), and fibroblasts (P1), indicating germline transmission. The mutation was absent in the mother of P2/P3, the 4 sisters of P1, and 100 normal unrelated individuals (not depicted).
Figure 3.
E127G and G116S CARD11 spontaneously aggregate and drive constitutive NF-κB activation. (A) Sanger dideoxy DNA sequencing confirmation of heterozygous missense mutations (A to G substitution or vice versa, designated as purine or “R”) in exon 5 of the CARD11 gene in P1, P2, and P3 (not in mother [M]) and exon 4 of CARD11 in P4 (not in control [Ctrl]). Amino acids are shown in single letter code. (B) Schematic representation of the CARD11 protein (Lenz et al., 2008a). The E127G and G116S amino acid changes within the CC region of CARD11 are denoted. (C) Ectopic expression of WT, E127G, or G116S CARD11 as N-terminal fluorescent fusion proteins (myc/Venus-CARD11) in BJAB B cells. BCL10 or MALT1 were visualized by immunofluorescence staining using Alexa Fluor 594–conjugated secondary Abs. Bars, 1 µm. (D) Plasmids encoding Flag-tagged WT, E127G, G116S, or other DLBCL-derived somatic, active mutant CARD11 were transfected into CARD11-deficient JPM50.6 cells containing a κB-driven GFP reporter gene, and GFP expression was analyzed by flow cytometry. Histograms (left) and MFI of GFP+ cells (middle) are shown, indicating the relative amount of NF-κB activity. Immunoblots confirming expression of Flag-CARD11 proteins are shown at right; β-actin served as a loading control. (E) BJAB B cells were transfected with a GFP expression plasmid plus Flag-tagged CARD11 constructs as listed in D. Flow cytometry of BJAB B cells transfected with a GFP expression plasmid plus Flag-tagged CARD11 constructs as listed in D and then stained with anti-CD83 was performed. Quantitated MFI of CD83 expression in GFP+ cells is shown (middle). Immunoblots confirming expression of CARD11 proteins are shown at right with a β-actin loading control. (C–E) Data are representative of three (C and E) or four (D) independent experiments.
This novel E127G mutation in CARD11 was implicated as the leading disease-causing candidate for several reasons. First, E127G was the only unique mutation shared by P1, P2, and P3 found in a gene expressed exclusively in lymphocytes. We found no other dominant, gain-of-function mutations in these patients that were not reported in the database of common single nucleotide polymorphisms (dbSNP). Second, E127G CARD11 is highly likely to be pathological based on mutation prediction algorithms (PMUT score = 0.838, MutationTaster probability score = 0.997; Ferrer-Costa et al., 2005; Flagella et al., 2006). Finally, this amino acid substitution lies in proximity to several somatic gain-of-function, CC region mutations known to drive constitutive NF-κB activation in DLBCL (Lenz et al., 2008a). In mice, constitutive activation of NF-κB at the level of IKK-β causes mature B cell hyperplasia and independence from the normally limiting B cell survival factor, BAFF (Sasaki et al., 2006). We therefore hypothesized that the germline CARD11 E127G mutation could cause excessive accumulation of mature B cells in our patients via constant NF-κB hyperactivation.
We subsequently subjected purified P4 B cells to RNA-Seq analysis, which revealed a different heterozygous missense mutation in CARD11 (Fig. 3 A). This mutation (G116S) was previously described as a gain-of-function somatic mutant (referred to as mutant 3) in a DLBCL tumor (Lenz et al., 2008a), also located at the N-terminal end of the CARD11 CC domain (Fig. 3 B). This mutation was detected in B cells, T cells, and buccal swabs from P4, suggesting it is also present in the germline. Thus, our identification of a separate germline mutation in P4 strongly suggests that gain-of-function mutations in CARD11 are the primary driver of disease in these patients.
Germline CARD11 mutants spontaneously aggregate and drive constitutive NF-κB activation
Constitutive NF-κB activity induced by somatic CARD11 CC mutations is related to spontaneous multimerization of CARD11 and assembly of a large signalosome, without requiring BCR engagement or CARD11 phosphorylation (Lenz et al., 2008a; Lamason et al., 2010). We also found that ectopically expressed E127G CARD11 dramatically localized into large protein aggregates in BJAB B cells, in contrast to dispersed cytoplasmic distribution of WT (Fig. 3 C). Furthermore, we noted significant colocalization of BCL10 and MALT1, two signaling partners required for CARD11-dependent NF-κB activation, within E127G CARD11 clusters (Fig. 3 C). As reported previously, G116S CARD11 also demonstrated spontaneous aggregation and BCL10/MALT1 colocalization (Fig. 3 C; Lenz et al., 2008a). Similar results were obtained with both mutants when expressed in WT Jurkat T cells (not depicted). These data suggest that both CARD11 mutants aggregate spontaneously in active signaling clusters with BCL10 and MALT1 in lymphocytes.
To definitively test whether E127G CARD11 induces NF-κB activation, we expressed it in CARD11-deficient JPM50.6 T cells containing an NF-κB-driven GFP reporter (Wang et al., 2002). Transfection of E127G CARD11 resulted in an increased proportion and mean fluorescence intensity (MFI) of GFP+ cells relative to WT, reflecting constitutive NF-κB activation comparable with several somatic DLBCL-derived CARD11 mutants described previously, including G116S (Fig. 3 D, left; Lenz et al., 2008a). Similar results were obtained in the BJAB B cell line, using the endogenous NF-κB–dependent marker gene CD83 (Fig. 3 E; Lenz et al., 2008a). Western blots showed comparable expression of the various WT and mutant CARD11 proteins (Fig. 3, D and E, right). Collectively, these data show that like G116S, E127G CARD11 is a dominant, gain-of-function mutant capable of inducing constitutive, AgR-independent NF-κB activation in both B and T cell lines.
Mutant CARD11 and NF-κB function in patient B cells
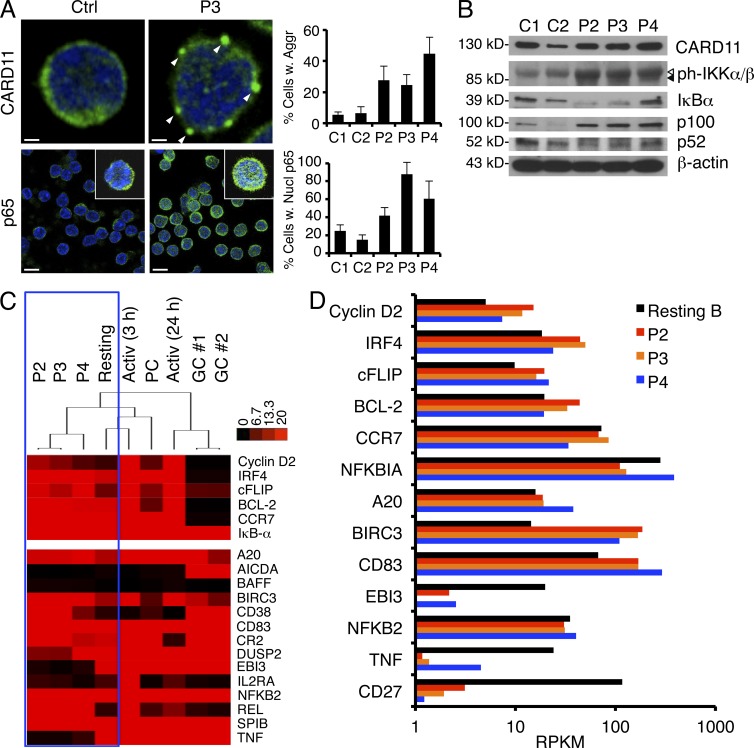
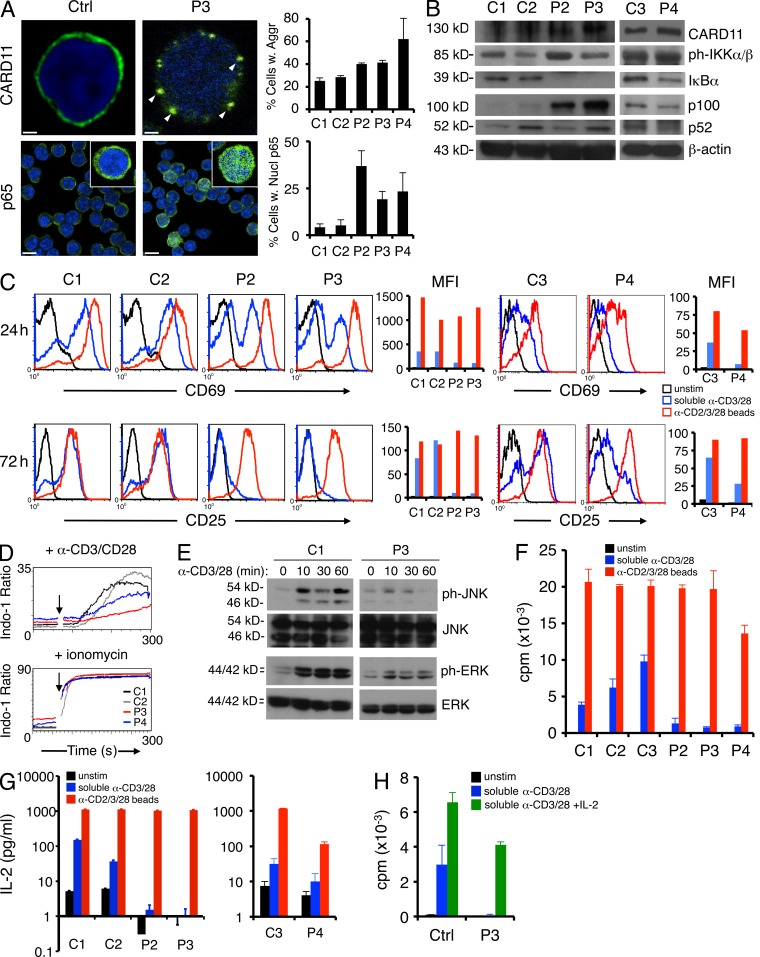
We next examined CARD11 expression and localization in the patients’ primary naive B cells. Quantitative transcript analysis of the RNA-Seq data revealed that ∼46%, 48%, and 54% of CARD11 mRNA transcripts represented the mutant allele in P2, P3, and P4 B cells, respectively, reflecting heterozygous expression (not depicted). By immunofluorescence microscopy, we also observed CARD11 aggregates in patient naive B cells relative to controls, suggesting CARD11 E127G protein is expressed and capable of aggregating spontaneously in the patients’ unstimulated B cells (Fig. 4 A, top). We also noted significantly increased nuclear translocation of the Rel protein p65, a subunit of the classical NF-κB complex, in patients’ B cells compared with controls (Fig. 4 A, bottom). Increased IKK phosphorylation, low steady-state IκB expression, and elevated expression of p65-induced p100 (NFKB2) were also indicative of constitutive, canonical IKK signaling and increased canonical NF-κB activity (Fig. 4 B). Interestingly, an increased ratio of the p100 precursor to p52 in patient cells (Fig. 4 B) suggests alternative NF-κB activation is unaffected or perhaps limited in the presence of active CARD11.
Figure 4.
CARD11 aggregation and elevated NF-κB activity in patient primary B cells. (A) Expression of CARD11 and p65 visualized by immunofluorescence and confocal microscopy in fixed, purified naive B cells. The percentage of cells with visible CARD11 aggregates (marked by white arrowheads) or >50% p65 in the nucleus was quantified for each sample in by scoring >200 cells from multiple fields. Insets in bottom row show individual cells from the same field. Scoring data represent the mean ± standard deviation from two independent scorers. Bars: (top) 1 µm; (bottom) 10 µm. (B) Representative immunoblots of total naive B cell lysates prepared from normal donor controls (C1 and C2) and patients for proteins listed on the right of each blot. Arrowheads denote phospho–IKK-β (open) and phospho–IKK-α (closed). (A and B) Data are representative of three independent experiments. (C) Hierarchical cluster analysis of RNA-Seq data derived from resting B cells (pooled from six normal donors), activated B cells (3 h, 24 h), GC B cells (two donors), and plasma cells (PC) relative to patients, measured as reads per kilobase of exon model per million mapped reads (RPKM) from RNA-Seq analysis. Heat map projections of selected NF-κB target genes (listed at right) are shown, including NF-κB signature genes (top; Davis et al., 2001). (D) Comparison of digital gene expression (RPKM) for selected NF-κB target genes listed at left.
To compare gene expression in purified patient B cells relative to normal control B cell subsets, we performed hierarchical cluster analysis of RNA-Seq expression data (expressed as reads per kilobase per million mapped reads [RPKM]). As shown in the dendrogram in Fig. 4 C, all three patients clustered together closely (Pearson correlation = 0.94), indicating a highly similar transcriptome expression profile. Patient expression profiles clustered more closely with resting or recently activated (3 h) B cells versus GC B cells. Compared with unstimulated resting B cells, we noted ∼1.5–2-fold higher expression of several signature NF-κB target gene transcripts in patient B cells (Fig. 4, C and D), known to be similarly up-regulated in DLBCL tumors harboring somatic CARD11 mutations, including cyclin D2, IRF4, and cFLIP (Davis et al., 2001; Lenz et al., 2008a). Additional NF-κB responsive genes were even more up-regulated in patient B cells (e.g., BIRC3, CD83, and REL), with certain NF-κB responsive genes up-regulated to greater extent in P2/P3 (e.g., BCL-2) versus P4 (e.g., A20). However, the expression of other genes was comparable or lower than normal resting B cells (Fig. 3 D). Although we did not have transcriptome data from normal, sorted transitional B cell subsets for comparison, the substantial proportion of CD10+ T2/T3-like B cells in patient samples likely contributes to some of these expression differences. For other genes with much lower expression in patient cells (e.g., TNF and CD27), this pattern likely reflects a substantially lower proportion of memory B cells in the original patient sample. Nonetheless, these data collectively suggest that mutant CARD11 expression in B cells drives constitutive, canonical NF-κB signaling with a similar, selective up-regulation of certain NF-κB–dependent genes in all three patients.
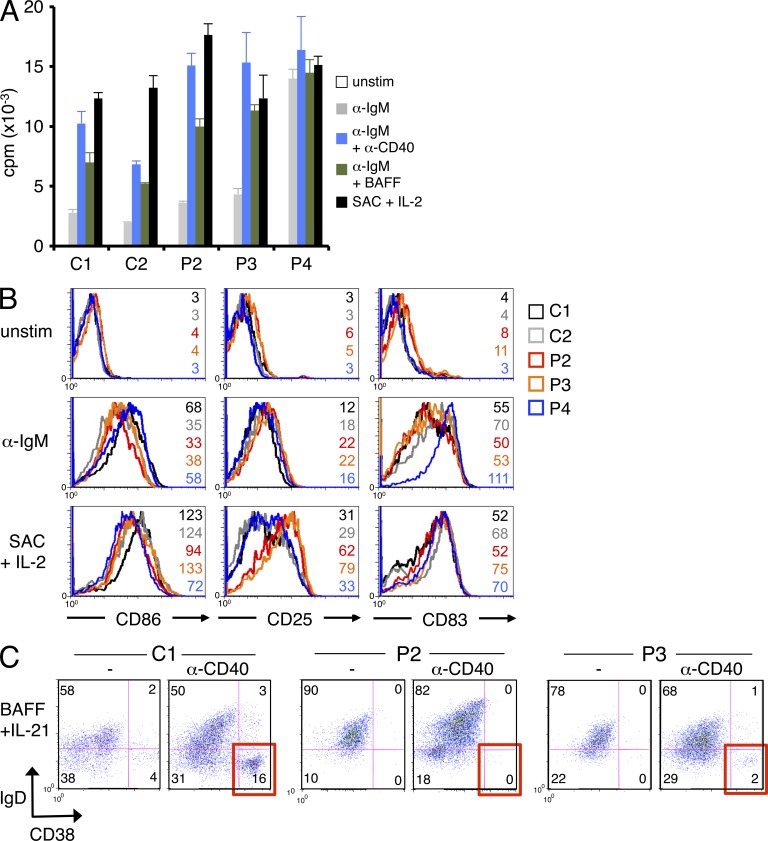
We next assayed activation of purified naive B cells from patients and healthy controls in response to a variety of polyclonal stimuli. Patient B cell proliferation was generally increased relative to controls in response to IgM ligation ± anti-CD40 or BAFF (Fig. 5 A). Mitogenic stimulation with IL-2 and Staphylococcus aureus Cowan strain 1 (SAC), which activates B cells via the BCR and Toll-like receptors, induced equivalent robust proliferation in control and patient B cells (Fig. 5 A). Expression of the BCR and BAFF-R was comparable on control and patient cells (not depicted). Up-regulation of cell surface activation markers was generally comparable with controls, with slightly higher expression of CD25 (P2 and P3) or CD83 (P4) noted for certain stimuli (Fig. 5 B). CD10 and CD38 expression did not remain stable on stimulated B cells (not depicted), making gating on transitional B cells over the course of the experiment unreliable. Considering that transitional B cells, which comprised a greater proportion of the patient B cell pool, are generally less responsive to BCR activation, these data suggest that active CARD11 signaling may enhance patient B cell activation. In contrast, we found that P2 and P3 B cells failed to differentiate into IgD−CD38hi plasmablasts in vitro (Fig. 5 C). Although a larger proportion of T2/T3-like B cells in patient samples may account for some of this difference, these data may also reflect intrinsic defects in patient B cell differentiation that may contribute to impaired generation of GCs or memory B cells. Indeed, Ig production was also decreased after P2 and P3 B cell stimulation (not depicted). Collectively, these findings demonstrate that constitutive NF-κB activity driven by active, mutant CARD11 contributes to abnormal B cell homeostasis.
Figure 5.
Increased proliferation and altered function of patient B cells after stimulation. (A) Proliferation of naive B cells purified from patients and controls was measured by [3H]thymidine incorporation in response to various B cell–activating stimuli. Patient responses were significantly higher than controls for all stimuli (P < 0.03) except SAC. Data show mean ± standard deviation of triplicate wells. (B) Representative protein expression of three NF-κB–responsive surface markers (CD86, CD25, and CD83) was quantified in naive B cells by flow cytometry, before or after polyclonal stimulation with anti-IgM or SAC + 200 U/ml IL-2. MFI values are listed for each histogram shown. With the exception of SAC, patient responses were significantly higher than controls. (C) Purified naive B cells from a normal control donor (C1), P2, and P3 were stimulated with BAFF and IL-21 ± 1 µg/ml anti-CD40 Ab for 10 d. The percentage of IgD−CD38hi plasma cells generated is highlighted in red for the lower right quadrant. (A–C) Data are representative of two (C) or three (A and B) independent experiments.
Mutant CARD11 and NF-κB function in patient T cells
Similar to B cells, resting T cells from all three patients also exhibited evidence of CARD11 aggregation and elevated nuclear translocation of p65 (Fig. 6 A). Altered expression of IκB and p100 were also reflective of constitutive CARD11-driven NF-κB signaling in P2 and P3 (Fig. 6 B), whereas levels of phosphorylated IKK were variable. In P4, phosphorylated IKK was slightly elevated and IκB expression was only slightly decreased, whereas p100 expression was lower than the matched control (Fig. 6 B). Variation in these markers for P4 may signify lower levels of NF-κB activation in the presence of G116S CARD11 or skewed expression caused by the patient’s active EBV infection.
Figure 6.
Patient T cells expressing mutant CARD11 are hyporesponsive to polyclonal stimulation. (A) Expression of CARD11 and p65 visualized by immunofluorescence and confocal microscopy in fixed resting T cells, quantified as described for Fig. 4 A. Insets in bottom row show individual cells from the same field. Bars: (top) 1 µm; (bottom) 10 µm. (B) Representative immunoblots of total T cell lysates prepared from normal donor controls (C1, C2, and C3) and patients for the indicated proteins. A separate matched control (C3) was compared with P4 for subsequent experiments. (C) Representative flow cytometric analysis of CD69 and CD25 measured ± stimulation with either soluble anti-CD3ε + anti-CD28 mAbs (blue lines) or bead-coupled anti-CD2/anti-CD3/anti-CD28 mAbs (red lines). MFI values are plotted at right in the corresponding color. (D) Calcium flux in purified resting T cells from controls (C1 and C2) and patients (P3 and P4) as measured by flow cytometry after stimulation with anti-CD3/anti-CD28 + 1 µg/ml protein A. Addition of ionomycin served as a positive control (bottom). (E) Western blot analysis of ERK and JNK phosphorylation relative to total ERK/JNK levels, after anti-CD3/anti-CD28 Ab stimulation of T cells cycling in IL-2. (F) Proliferation of resting T cells purified from patients and controls was measured by [3H]thymidine incorporation in counts per minute to stimuli described in C. Patient responses to soluble Ab stimulation were significantly lower than controls (P = 3.97 × 10−4). (G) IL-2 accumulation measured by ELISA in cell supernatants 4 d after stimulation as described in C. (H) Proliferation assay for purified control and P3 T cells stimulated with anti-CD3/CD28 Abs ± 100 U/ml of exogenous recombinant IL-2. (A and D–G) Data are representative of two (A, D, and E), three (F and G), or four (C) independent experiments. (A and F–H) Data show mean ± standard deviation of triplicate wells.
In contrast to B cells, blastogenesis and up-regulation of T cell activation markers CD69 and CD25 were markedly decreased in purified patient T cells stimulated with soluble agonistic anti-CD3/CD28 Abs for 24–72 h (Fig. 6 C). This defect in CD69 and CD25 up-regulation was less pronounced in P4 T cells, relative to a matched control. TCR expression was comparable in controls and patients (not depicted). Although some of this difference may be attributed to a higher proportion of naive and/or DN T cells relative to controls (Table 1), patients’ memory T cells also failed to up-regulate TCR-induced activation markers (not depicted). These defects were largely rescued by stimulating with anti-CD2/CD3/CD28 Abs coupled to beads, which provide a stronger co-stimulatory signal (Fig. 6 C; Boussiotis et al., 1994). We also observed decreased calcium flux in purified P3 and P4 T cells compared with controls after stimulation with anti-CD3/CD28 cross-linked with protein A (Fig. 6 D), suggesting TCR signaling is impaired in the presence of active CARD11 mutants. Activated patient T cells expanded normally in the presence of exogenous IL-2 (not depicted), but we observed defective phosphorylation of extracellular signal-regulated kinase (ERK) and JNK upon restimulation (Fig. 6 E), consistent with an early TCR signaling defect. Strikingly, patient T cells failed to proliferate and secreted less IL-2 in response to soluble Ab stimulation (Fig. 6, F and G). Activation with anti-CD2/CD3/CD28–coupled beads restored proliferation and IL-2 production in patient T cells, although P4 T cell responses remained significantly reduced relative to control T cells (Fig. 6, F and G). Addition of exogenous IL-2 with soluble Abs also rescued patient (P3) T cell proliferation (Fig. 6 H). Collectively, these data suggest patient T cells are rendered mildly anergic by chronic mutant CARD11 signaling (Chappert and Schwartz, 2010; Piątosa et al., 2010).
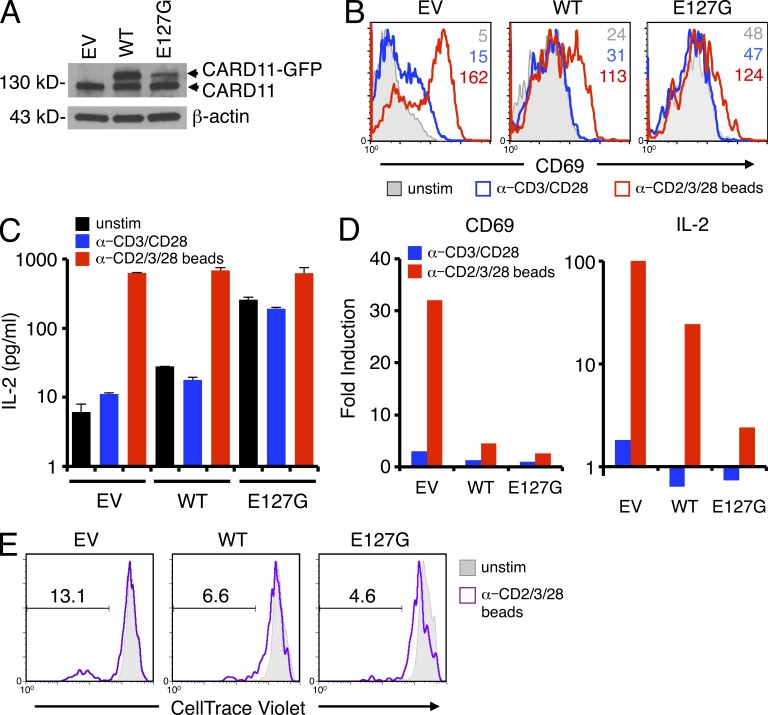
We next tested whether ectopic expression of E127G CARD11 could render normal T cells hyporesponsive to AgR stimulation. Primary human T cells were transfected with WT or E127G CARD11 fused to GFP (Fig. 7 A). Acute overexpression of CARD11 in primary human T cells resulted in elevated CD69 expression and IL-2 production (relative to GFP alone) before stimulation, which was more pronounced in cells expressing E127G versus WT CARD11 (Fig. 7, B and C). Indeed, we observed some aggregation of WT CARD11-GFP (not depicted), suggesting spontaneous signaling can occur simply with overexpression. However, stimulation with either anti-CD3/CD28 Abs or anti-CD2/CD3/CD28–coated beads induced proportionally less CD69 expression and IL-2 secretion relative to background in GFP+ CARD11 transfectants, particularly for E127G (Fig. 7, B and C; summarized in Fig. 7 D). Proliferation of GFP+ T cells in response to Ab-coated beads followed a similar pattern, despite lower T cell responsiveness in general after electroporation. In contrast to GFP+ cells alone, cells overexpressing WT or E127G CARD11-GFP divided one to three times. However, the proportion of cells that underwent robust cell division (five to seven times) after stimulation was decreased in WT CARD11-GFP+ cells and further reduced in E127G CARD11-GFP+ cells (Fig. 7 E). These data suggest that active CARD11 signaling resulting in constitutive NF-κB activation diminishes the maximum T cell response to AgR stimulation, closely resembling the phenotype of mouse T cells expressing constitutively active IKK-β (Krishna et al., 2012). Thus our findings suggest that constitutive NF-κB activity driven by germline-encoded, gain-of-function CARD11 mutants may be stimulatory in B cells but partially inhibitory in T cells, offering a molecular explanation for the abnormal expansion and potential for malignancy of B cells but not T cells in these patients.
Figure 7.
Ectopic overexpression of E127G CARD11 in normal T cells reduces magnitude of AgR-induced response. (A) Representative expression of endogenous or GFP-fused CARD11 in normal T cells transfected with empty vector (EV), WT, or E127G CARD11-GFP. (B) Representative flow cytometric analysis of CD69 expression on transfected cells stimulated as in Fig. 6 C. MFI values are listed for each histogram. (C) IL-2 secretion measured by ELISA in transfected cell supernatants 3 d after stimulation as described in Fig. 6 C. Data show mean ± standard deviation of triplicate wells. (D) Fold induction of CD69 expression and IL-2 secretion was calculated by normalizing CD69 MFI or IL-2 concentration in stimulated cells to unstimulated cells for each transfection. (E) Representative proliferation, as measured by dilution of CellTrace Violet dye, of unstimulated and bead-stimulated transfected T cells. The marker denotes the percentage of T cells that divided five to seven times. (B–E) Data are representative of two independent experiments.
DISCUSSION
In this study, we describe a novel congenital disorder featuring the first known germline mutation in CARD11 in a family affected by polyclonal expansion of naive B cells, splenomegaly, and potential predisposition to B cell malignancy (Darte et al., 1971). A separate patient with highly similar disease manifestations harbors a germline G116S CARD11 mutation, which was previously characterized in DLBCL as a gain-of-function somatic mutation (Lenz et al., 2008a). Although both mutations cause constitutive NF-κB activity in unstimulated lymphocytes, our findings suggest that the germline mutation does not transform a B cell clone outright. Instead, in addition to the oncogenic potential of constitutive NF-κB hyperactivation, germline CARD11 mutants may promote B cell malignancy by dramatically increasing the number of naive B cells that may undergo secondary genomic alterations and become malignant (Lenz et al., 2008b; Lenz and Staudt, 2010), as exemplified in P1 (see Supplemental text). It is well established that genomic instability inherent in the GC reaction is a primary driver of mutagenesis and B cell transformation (Küppers, 2005). Indeed, a monoallelic deletion of tumor suppressor genes located in chromosome 13q14.3, associated with ∼50% of B-CLL cases, was detected in P1 B-CLL cells and may have contributed to oncogenesis (see Supplemental text; Pekarsky et al., 2010). However, preliminary evidence suggests that the P1 tumor does not harbor recurrent mutations recently associated with B-CLL (Fabbri et al., 2011; Puente et al., 2011). Our studies are ongoing to determine the nature of secondary mutations in B-CLL tumor cells from P1 and the frequency of CARD11 mutations specifically in B-CLL.
Our analysis indicated patient B cells did not appear to proliferate excessively in the periphery, but instead may have accumulated from increased B cell output from the bone marrow, as indicated by elevated CD10+ transitional B cells in the blood and consistent with hyperplasia reported in P1 bone marrow (Darte et al., 1971). Patients’ peripheral blood B cells died more rapidly ex vivo, consistent with heightened sensitivity of transitional B cells to cell death (Palanichamy et al., 2009). However, survival of transitional and/or naive B cells that complete maturation in peripheral lymphoid tissues may have been enhanced, explaining the continuous increase in spleen size over time (Fig. 1 B). This phenotype resembles mice expressing constitutively active IKK-β in the B cell lineage, which results in accumulation of longer-lived, resting mature B cells that are no longer dependent on BAFF for survival (Sasaki et al., 2006). BAFF normally acts as a sensitive limiting factor for controlling peripheral B cell numbers; indeed, excess BAFF in circulation is associated with higher B cell production, autoimmune disease, and B cell malignancy (Khan, 2009). However, neither serum BAFF levels nor surface expression of BAFF receptor (BR3) was elevated in our patients, suggesting B cell accumulation occurs independently of increased BAFF signaling through the alternative NF-κB pathway. Increased levels of unprocessed p100 in patient cells further suggest the alternative NF-κB activity is less engaged. Instead, we speculate that constitutive, canonical NF-κB signaling via mutant CARD11 drives greater output of immature/transitional B cells from the bone marrow and may activate prosurvival programs (e.g., BCL2 up-regulation) that permit the accumulation of noncycling and perhaps self-reactive immature B cells in vivo that would otherwise be deleted or anergized in response to chronic BCR engagement (Healy and Goodnow, 1998). It will be interesting to determine the extent of autoreactivity in the patients’ B cell repertoire given that the transitional B cell stage is thought to represent a key negative selection checkpoint for self-reactive B cell clones (Carsetti et al., 1995).
The fact that prominent lymphocytosis is largely restricted to the B cell compartment is surprising in our patients, because CARD11 mediates AgR-triggered activation of NF-κB in both B and T cells (Thome et al., 2010). Although a recent study reported that adoptive transfer of bone marrow cells expressing constitutively active CARD11 resulted in Th2-mediated inflammation in mice (Blonska et al., 2012), we did not observe a similar phenotype in our patients. In fact, patient T cells were hyporesponsive to AgR stimulation and did not proliferate in vitro unless stronger co-stimulation was provided. A modestly elevated proportion of CD4−CD8− DN T cells in patient samples, which are typically unresponsive and die rapidly in vitro, cannot explain this phenomenon alone. More importantly, our investigation marshals clinical evidence consonant with conclusions from a recent study by Krishna et al. (2012). They showed that chronic NF-κB signaling, triggered by transgenic expression of constitutively active IKK-β (ca–IKK-β) in mice, also renders T cells hyporesponsive to Ag stimulation, including defects in calcium flux, ERK phosphorylation, induction of activation markers, proliferation, and IL-2 secretion. Unlike ca–IKK-β mouse T cells, however, our patients’ T cells did not show evidence of increased apoptosis or exhaustion (e.g., increased PD-1 expression; unpublished data). Consistent with the classical two-signal paradigm of T cell activation, we speculate that T cell hyporesponsiveness in mutant CARD11 patients may reflect a mild form of anergy induced by a chronic AgR-like signal 1 driven by E127G CARD11 in the absence of a strong co-stimulatory signal 2 (Chappert and Schwartz, 2010). Rescued proliferation in the presence of CD2 ligation or exogenous IL-2 lends credence to this model. In contrast, B cell proliferation can be triggered by BCR cross-linking alone, mimicked by mutant CARD11. Impaired T cell responses may be linked to increased infections in mutant CARD11 patients, particularly in P4.
Our findings logically suggest overactive CARD11/NF-κB signaling may skew lymphocyte development and function in favor of selective B cell expansion. However, we also observed defects in Ig secretion and plasma cell differentiation in vitro for P2 and P3 mature B cells. The presence of mutant CARD11 throughout B cell development may paradoxically enlarge the pool of transitional and naive B cells that are nonetheless hampered in their ability to differentiate and function beyond the naive stage, including maintenance of a memory B cell pool. We hypothesize mutant CARD11 signaling during thymocyte development may also select for hyporesponsive T cell clones. Defects in T cell help to B cells may partly explain the paucity of GCs and few autoimmune manifestations in P2 and P3 to date. In contrast, deficiencies in T cell–independent humoral responses to polysaccharide Ags reinforce the idea of intrinsic dysregulation of B cell responsiveness/function with mutant CARD11 present, even if more self-reactive clones escape negative selection in transitional stages. Further studies are required to distinguish the differential influence of E127G versus G116S CARD11 expression on AgR signaling in B and T cells, selective induction of NF-κB–dependent genes, and lymphocyte development. Indeed, dynamic changes in these processes may also reflect a variable equilibrium in the translation and self-induced, ubiquitin-mediated degradation of different CARD11 mutants (Moreno-García et al., 2010), although we did not observe abnormal CARD11 protein expression in our patients’ cells.
In conclusion, our findings reveal the genetic basis of a hereditary disorder of B cell lymphocytosis first described in an unsolved case report four decades ago. Our study of these patients illuminates how AgR signaling must be regulated differently by CARD11 in B and T cells, even though the proximal signaling machinery is nearly identical. Because of the contrasting features in lymphocytes, we designate this condition “B cell expansion with NF-κB and T cell anergy” (BENTA) disease. Although B cells are generally more susceptible to malignant transformation because of AID-induced genomic instability, the novel disease we describe here may help to explain the preponderance of B cell rather than T cell malignancies specifically associated with CARD11 activating mutations.
MATERIALS AND METHODS
Cells, tissues, and treatments.
Patients were enrolled, and blood/tissue samples were obtained with informed consent through a protocol established by the Lymphocyte Molecular Genetics Unit and approved by the Institutional Review Board (IRB) of the National Institutes of Health (NIH). Experiments involving patients were performed at National Institute of Allergy and Infectious Diseases (NIAID), conforming to IRB protocol. Jurkat T cell (E6 and JPM50.6) and BJAB B cell lines were maintained in RPMI 1640 medium (Lonza) supplemented with 10% FBS, 2 mM glutamine, and 100 U/ml each of penicillin and streptomycin (Invitrogen). Tissue samples were obtained as prestained slides (P1) or as formalin-fixed paraffin-embedded specimens (P2 and P3 tonsil). Sections were stained with hematoxylin and eosin (H&E), anti-IgD (Dako), anti-CD3, anti-CD10, or anti-Ki67 (Dako) Abs for immunohistochemistry analysis. PBMCs were isolated from whole blood using Ficoll-Paque PLUS (Roche) density gradient centrifugation and ACK Lysis Buffer (Invitrogen). Naive B cells and total T cells were purified by negative selection using magnetic bead sorting kits (Naive B Cell Isolation kit II and Pan T Cell Isolation kit II; Miltenyi Biotec). Purity of negatively selected populations routinely exceeded 95%. B cell stimulations were performed in complete RPMI medium (see above) using SAC (Sigma-Aldrich) + 200 U/ml rIL-2 (PeproTech), 25 µg/ml anti-IgM F(ab′)2 (Jackson ImmunoResearch Laboratories, Inc.), 1 µg/ml anti-CD40, 200 ng/ml rBAFF, and/or rIL-21 (R&D Systems). T cells were stimulated with 2 µg/ml each of anti-CD3ε and anti-CD28 mAbs (BD) or MACS iBead particles (1:2 bead/cell ratio) loaded with biotinylated anti-CD2, anti-CD3ε, and anti-CD28 Abs (Miltenyi Biotec) in complete RPMI.
Flow cytometry.
Unless otherwise noted, all Abs were obtained from BD. Standard flow cytometry methods were used for cell surface staining and analysis of unstimulated cells. In brief, 0.5–1 × 106 cells were washed in cold FACS buffer (1× PBS, 1% FBS, and 0.1% sodium azide) and stained for 30 min on ice with appropriate Abs. B cells were stained before and after (72 h) stimulation with 5 mg FITC anti-CD86, PE anti-CD25, PE anti-CD83, or APC anti-CD19 Abs. For in vitro plasma cell differentiation, cells were stained with PE anti-IgD and FITC anti-CD38 10 d after stimulation. Purified T cells were stained with 5 mg FITC anti-CD2, PE anti-CD69, and APC anti-CD25 before or after (24, 72 h) stimulation. Cells were subsequently washed in cold FACS buffer and acquired on an LSR-II flow cytometer (BD). Analysis was performed using FlowJo software (Tree Star).
B cell survival was measured ex vivo over 3 d by staining with Annexin V–FITC (BD) and propidium iodide (Sigma-Aldrich) to identify dying cells. Calcium flux was measured in T cells loaded with 5 µM Indo-1-AM calcium-sensitive dye (Invitrogen) in Ringers solution. The change in ratio of Indo-1 fluorescence emission was detected over time on an LSR-II flow cytometer.
IgH PCR analysis.
DNA was extracted from peripheral blood using a Magtration System 8Lx (Precision System Science USA) or from formalin-fixed paraffin-embedded tissue using a DNA FFPE Tissue kit (QIAGEN). PCR was performed using consensus primers to VH framework region (FR) II or III and joining region (JH) of the IgH gene according to the method of Ramasamy et al. (1992). PCR products were separated by standard acrylamide gel electrophoresis or capillary electrophoresis on an ABI 3130xl Genetic Analyzer with electropherograms analyzed using GeneMapper software version 4.0 (Applied Biosystems).
High-throughput RNA sequencing.
RNA was isolated from purified B cells from P2 and P3 using an RNeasy kit (QIAGEN), and 1 mg RNA was used for constructing cDNA libraries. The standard Illumina Pipeline for RNA-Seq was used, using paired-end 108-bp runs with each sample run in one sequencing lane, yielding 20 million reads per sample. Sequences were mapped back to both RefSeq and Ensemble transcript models using the BWA algorithm, yielding a median resequencing coverage of 10×. Single nucleotide variants were reported that deviated from the human reference genome sequence (build 37), were observed in both sequencing directions, represented >20% of the resequencing coverage at a particular base pair position, and were not known single nucleotide polymorphisms in the dbSNP database of NCBI (build #131). A total of 110,004 and 106,444 putative single nucleotide variants were detected in samples from P2 and P3, respectively.
We filtered thousands of unique mutations from both datasets not reported in dbSNP to meet the following criteria: (a) >5× transcript read coverage to eliminate artifactual base changes, (b) nonsynonymous mutations in the coding region, (c) present in both P2 and P3, and (d) evidence of heterozygosity. Applying these criteria reduced the number of possible disease-causing mutations to 214 missense, 8 nonsense, and 54 frameshift alterations. Shared mutations were further prioritized according to their relative expression in B cells, restricted expression in lymphocytes, and relevance to B cell development and function, based on published literature. The presence of several candidate mutations in P1, P2, and P3 (but not in the mother of P2/P3) was checked using conventional Sanger DNA sequencing. Based on this prioritization scheme, top candidate mutations were identified in CARD11, INPP5D (SHIP-1), and TCF3 (E2A). Among these, only CARD11 is exclusively expressed in lymphocytes. Mutations in INPP5D (G244D and E334G) were scored as nonpathogenic polymorphisms (MutationTaster probability scores = 0.9986 and 0.9952). The G362D mutation in TCF3 was recently reported as a rare polymorphism in dbSNP (accession no. rs148928381). Therefore, E127G CARD11 emerged as the top disease-causing candidate mutation in P1–3. RNA-Seq data from P4 was subjected to similar analyses to reveal a germline heterozygous mutation (G116S) in CARD11.
Digital gene expression was calculated as reads per kilobase of exon model per million mapped reads (RPKM). Quantitative transcript analysis of the RNA-Seq data was performed using the TopHat alignment program, with redundant reads removed. Hierarchical cluster analysis was performed using Cluster 3.0 software, and heat map dendrograms were visualized using TreeView.
Cell sorting and KREC assay.
PBMCs were stained with the anti–human mAbs FITC-conjugated anti-CD21 (Beckman Coulter), PerCP-Cy-5-5–conjugated anti-CD19 and PE-Cy7–conjugated anti-CD27 (eBioscience), and APC-conjugated anti-CD10 (BD) and sorted on a FACSAria instrument (BD). The following B cell fractions were sorted: CD10+CD21+CD27− immature/transitional and CD10−CD21+CD27− naive cells. The replication history of sorted fractions was evaluated using a PCR-based assay for measuring KRECs. The ratio of KREC joints (signal joint) to the Jκ-Cκ recombination genomic joints (coding joint) was determined as previously described (Moir et al., 2008).
Immunoblotting.
Cells were lysed in 1% NP-40 lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40, and 0.5% sodium deoxycholate) plus Complete protease inhibitors (Roche) for 30 min on ice. Insoluble material was cleared by centrifugation. Protein concentration was determined by BCA assay (Thermo Fisher Scientific), and 30 mg total protein was separated by SDS-PAGE and subsequently transferred to nitrocellulose membrane. After blocking in 5% milk, membranes were probed with the following Abs: anti-CARD11 (1D12), anti-phospho–IKK-α/β (Cell Signaling Technology), anti-Flag (M2; Sigma-Aldrich), anti–IκB-α, anti-p100/p52 (Santa Cruz Biotechnology, Inc.), and anti–β-actin (Sigma-Aldrich). Bound Abs were detected using horseradish peroxidase–conjugated secondary Abs (SouthernBiotech) and enhanced chemiluminescence (ECL; Thermo Fisher Scientific).
Confocal microscopy.
Cells adhered to poly-l-lysine–coated slides were fixed for 20 min with 2% paraformaldehyde (Electron Microscopy Sciences) and then permeabilized for 5 min in 0.1% Triton X-100/PBS. After blocking for 30 min in 1% FCS/PBS, cells spots were incubated with the following Abs: anti-CARD11 (Abcam), anti-BCL10, anti-MALT1, and/or anti-p65 (Santa Cruz Biotechnology, Inc.). Cells were washed in PBS and then incubated with goat anti–rabbit secondary Ab conjugated to Alexa Fluor 488 or 568 (Invitrogen) plus a 1:10,000 dilution of Hoechst 33342 (30 min per Ab). After washing in PBS, coverslips were attached with Fluoromount (SouthernBiotech). Fluorescent images were acquired on a confocal microscope (SP5; Leica) using a 63× oil immersion objective.
Statistical analysis.
Two-tailed Student’s t tests were applied to test whether differences in CARD11 aggregation and p65 nuclear translocation measured by confocal microscopy (n ≥ 3) were significant between normal controls and patient samples. P-values were calculated as follows: percentage of T cells with CARD11 aggregates (C1 vs. C2, P = 0.19; C1 vs. P2, P = 0.05; C1 vs. P3, P = 0.017; C1 vs. P4, P = 0.23; C2 vs. P2, P = 0.008; C2 vs. P3, P = 0.03; C2 vs. P4, P = 0.26), percentage of B cells with CARD11 aggregates (C1 vs. C2, P = 0.57; C1 vs. P2, P = 0.04; C1 vs. P3, P = 0.02; C1 vs. P4, P = 0.008; C2 vs. P2, P = 0.04; C2 vs. P3, P = 0.007; C2 vs. P4, P = 0.01), percentage of T cells with >50% nuclear p65 (C1 vs. C2, P = 0.19; C1 vs. P2, P = 9.42 × 10−4; C1 vs. P3, P = 4.76 × 10−4; C1 vs. P4, P = 2.9 × 10−4; C2 vs. P2, P = 2.30 × 10−6; C2 vs. P3, P = 1.23 × 10−4; C2 vs. P4, P = 1.58 × 10−6), and percentage of B cells with >50% nuclear p65 (C1 vs. C2, P = 0.008; C1 vs. P2, P = 0.014; C1 vs. P3, P = 5.75 × 10−5; C1 vs. P4, P = 0.005; C2 vs. P2, P = 9.48 × 10−4; C2 vs. P3, P = 5.49 × 10−5; C2 vs. P4, P = 5.5 × 10−4). For B cell proliferation assays, p-values from Student’s t tests were calculated as follows: anti-IgM (controls vs. patients, P = 0.034), anti-IgM + anti-CD40 (controls vs. patients, P = 1.61 × 10−5), anti-IgM + BAFF (controls vs. patients, P = 3.17 × 10−5), and SAC + IL-2 (controls vs. patients, P = 0.072). For T cell proliferation assays, p-values from Student’s t tests were calculated as follows: soluble anti-CD3/anti-CD28 (controls vs. patients, P = 3.97 × 10−4) and anti-CD2/3/28 beads (controls vs. patients, P = 0.048; controls vs. P4, P = 1.81 × 10−6).
CARD11 transfections.
WT CARD11 was cloned into the p3xFLAG expression vector via EcoRI and BamHI. The original WT Venus-CARD11 fusion construct was provided by N. Bidère (Institut National de la Santé et de la Recherche Médicale, Hôpital Paul-Brousse, Villejuif, France). Site-directed mutagenesis to obtain E127G and L225LI CARD11 was performed using specific primers for linear amplification, followed by DpnI digestion of methylated DNA. Mutations were verified by Sanger sequencing (Rocky Mountain Laboratories, NIAID, NIH). To create fluorescent fusion proteins, WT and E127G CARD11 were subcloned into the pCEFL-myc/Venus expression vector using EcoRI or into pmaxGFP (Lonza) using EcoRI and BamHI. JPM50.6 cells (5 × 106/transfection) were transfected either with 5–10 µg DNA using a BTX Electroporator (BTX Harvard Apparatus, 260 V, 950 mF) or 5 µg DNA using an Amaxa 4D Nucleofector (kit SE, Program CL-120; Lonza). BJAB cells (5 × 106/transfection) were transfected with 5 µg DNA using an Amaxa 4D Nucleofector (kit SF, Program EH-100). Primary human T cells were transfected with 4 mg DNA using an Amaxa 4D Nucleofector (kit P3, HF program for unstimulated T cells). Proliferation of CARD11-GFP+ cells was measured using CellTrace Violet (Invitrogen) according to the manufacturer’s instructions.
Proliferation assays.
Purified B and T cells were plated at 105 cells/well in triplicate (200 µl/well) in 96-well flat-bottom pates and stimulated as described in Cells, tissues, and treatments. 3 d later, wells were pulsed with 1 µCi of tritiated [3H]thymidine (PerkinElmer) and incubated for 24 h before collection on a mash harvester. Incorporation of [3H]thymidine was quantified as counts per million.
ELISAs.
Secreted IL-2 in the cell culture supernatant was quantified by ELISA, using the Ready-Set-Go Human IL-2 ELISA kit (eBioscience) according to the manufacturer’s instructions. Absorbance at 450 nm was detected using a BioTek Synergy H1m microplate reader.
Online supplemental material.
The supplemental text contains detailed clinical case reports. Table S1 shows the humoral response to vaccination in P2, P3, and P4. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20120831/DC1.
Supplementary Material
Acknowledgments
We thank the patients and their family for participating in this study. We also thank Drs. Steve Porcella and Mr. Kent Barbian of the Rocky Mountain Laboratories Genomics Unit for sequencing and microarray support, Drs. Hilaire Meuwissen and Maxine Hetherington for patient referrals, Drs. Alan Wayne, Gerald Marti, Koneti Rao, and Joanne Porter for clinical care, Ms. Margaret Brown and Ms. Shakuntala Gurprasad for flow cytometry support, Drs. Yu Zhang and Nicholas Beckloff for bioinformatics expertise, and Dr. Nicolas Bidère for supplying plasmids. We also thank Drs. Yogesh Jeelall, Keisuke Horikawa, and Chris Goodnow for helpful discussions and input.
This work was supported by the Intramural Research Program of the National Institutes of Health, the National Institute of Allergy and Infectious Diseases, and the National Cancer Institute and an intramural grant from Uniformed Services University of the Health Sciences (to A.L. Snow and J.R. Stinson).
No potential conflict of interest relevant to this article was reported.
Footnotes
Abbreviations used:
- Ab
- antibody
- Ag
- antigen
- AgR
- Ag receptor
- BAFF
- B cell–activating factor
- B-CLL
- B cell chronic lymphocytic leukemia
- CC
- coiled coil
- DLBCL
- diffuse large B cell lymphoma
- DN
- double negative
- ERK
- extracellular signal-regulated kinase
- GC
- germinal center
- IKK
- IκB kinase
- KREC
- κ-deletion excision circle
- MFI
- mean fluorescence intensity
- SAC
- Staphylococcus aureus Cowan strain 1
References
- Blonska M., Joo D., Zweidler-McKay P.A., Zhao Q., Lin X. 2012. CARMA1 controls Th2 cell-specific cytokine expression through regulating JunB and GATA3 transcription factors. J. Immunol. 188:3160–3168 10.4049/jimmunol.1102943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussiotis V.A., Freeman G.J., Griffin J.D., Gray G.S., Gribben J.G., Nadler L.M. 1994. CD2 is involved in maintenance and reversal of human alloantigen-specific clonal anergy. J. Exp. Med. 180:1665–1673 10.1084/jem.180.5.1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carsetti R., Köhler G., Lamers M.C. 1995. Transitional B cells are the target of negative selection in the B cell compartment. J. Exp. Med. 181:2129–2140 10.1084/jem.181.6.2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappert P., Schwartz R.H. 2010. Induction of T cell anergy: integration of environmental cues and infectious tolerance. Curr. Opin. Immunol. 22:552–559 10.1016/j.coi.2010.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darte J.M., McClure P.D., Saunders E.F., Weber J.L., Donohue W.L. 1971. Congenital lymphoid hyperplasia with persistent hyperlymphocytosis. N. Engl. J. Med. 284:431–432 10.1056/NEJM197102252840807 [DOI] [PubMed] [Google Scholar]
- Davis R.E., Brown K.D., Siebenlist U., Staudt L.M. 2001. Constitutive nuclear factor κB activity is required for survival of activated B cell–like diffuse large B cell lymphoma cells. J. Exp. Med. 194:1861–1874 10.1084/jem.194.12.1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R.E., Ngo V.N., Lenz G., Tolar P., Young R.M., Romesser P.B., Kohlhammer H., Lamy L., Zhao H., Yang Y., et al. 2010. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 463:88–92 10.1038/nature08638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong G., Chanudet E., Zeng N., Appert A., Chen Y.W., Au W.Y., Hamoudi R.A., Watkins A.J., Ye H., Liu H., et al. 2011. A20, ABIN-1/2, and CARD11 mutations and their prognostic value in gastrointestinal diffuse large B-cell lymphoma. Clin. Cancer Res. 17:1440–1451 10.1158/1078-0432.CCR-10-1859 [DOI] [PubMed] [Google Scholar]
- Fabbri G., Rasi S., Rossi D., Trifonov V., Khiabanian H., Ma J., Grunn A., Fangazio M., Capello D., Monti S., et al. 2011. Analysis of the chronic lymphocytic leukemia coding genome: role of NOTCH1 mutational activation. J. Exp. Med. 208:1389–1401 10.1084/jem.20110921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer-Costa C., Gelpí J.L., Zamakola L., Parraga I., de la Cruz X., Orozco M. 2005. PMUT: a web-based tool for the annotation of pathological mutations on proteins. Bioinformatics. 21:3176–3178 10.1093/bioinformatics/bti486 [DOI] [PubMed] [Google Scholar]
- Flagella M., Bui S., Zheng Z., Nguyen C.T., Zhang A., Pastor L., Ma Y., Yang W., Crawford K.L., McMaster G.K., et al. 2006. A multiplex branched DNA assay for parallel quantitative gene expression profiling. Anal. Biochem. 352:50–60 10.1016/j.ab.2006.02.013 [DOI] [PubMed] [Google Scholar]
- Healy J.I., Goodnow C.C. 1998. Positive versus negative signaling by lymphocyte antigen receptors. Annu. Rev. Immunol. 16:645–670 10.1146/annurev.immunol.16.1.645 [DOI] [PubMed] [Google Scholar]
- Khan W.N. 2009. B cell receptor and BAFF receptor signaling regulation of B cell homeostasis. J. Immunol. 183:3561–3567 10.4049/jimmunol.0800933 [DOI] [PubMed] [Google Scholar]
- Krishna S., Xie D., Gorentla B., Shin J., Gao J., Zhong X.P. 2012. Chronic activation of the kinase IKKβ impairs T cell function and survival. J. Immunol. 189:1209–1219 10.4049/jimmunol.1102429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küppers R. 2005. Mechanisms of B-cell lymphoma pathogenesis. Nat. Rev. Cancer. 5:251–262 10.1038/nrc1589 [DOI] [PubMed] [Google Scholar]
- Lamason R.L., McCully R.R., Lew S.M., Pomerantz J.L. 2010. Oncogenic CARD11 mutations induce hyperactive signaling by disrupting autoinhibition by the PKC-responsive inhibitory domain. Biochemistry. 49:8240–8250 10.1021/bi101052d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenardo M.J., Baltimore D. 1989. NF-kappa B: a pleiotropic mediator of inducible and tissue-specific gene control. Cell. 58:227–229 10.1016/0092-8674(89)90833-7 [DOI] [PubMed] [Google Scholar]
- Lenz G., Staudt L.M. 2010. Aggressive lymphomas. N. Engl. J. Med. 362:1417–1429 10.1056/NEJMra0807082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz G., Davis R.E., Ngo V.N., Lam L., George T.C., Wright G.W., Dave S.S., Zhao H., Xu W., Rosenwald A., et al. 2008a. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 319:1676–1679 10.1126/science.1153629 [DOI] [PubMed] [Google Scholar]
- Lenz G., Wright G.W., Emre N.C., Kohlhammer H., Dave S.S., Davis R.E., Carty S., Lam L.T., Shaffer A.L., Xiao W., et al. 2008b. Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proc. Natl. Acad. Sci. USA. 105:13520–13525 10.1073/pnas.0804295105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie-Cardine A., Divay F., Dutot I., Green A., Perdrix A., Boyer O., Contentin N., Tilly H., Tron F., Vannier J.P., Jacquot S. 2008. Transitional B cells in humans: characterization and insight from B lymphocyte reconstitution after hematopoietic stem cell transplantation. Clin. Immunol. 127:14–25 10.1016/j.clim.2007.11.013 [DOI] [PubMed] [Google Scholar]
- Moir S., Ho J., Malaspina A., Wang W., DiPoto A.C., O’Shea M.A., Roby G., Kottilil S., Arthos J., Proschan M.A., et al. 2008. Evidence for HIV-associated B cell exhaustion in a dysfunctional memory B cell compartment in HIV-infected viremic individuals. J. Exp. Med. 205:1797–1805 10.1084/jem.20072683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montesinos-Rongen M., Schmitz R., Brunn A., Gesk S., Richter J., Hong K., Wiestler O.D., Siebert R., Küppers R., Deckert M. 2010. Mutations of CARD11 but not TNFAIP3 may activate the NF-kappaB pathway in primary CNS lymphoma. Acta Neuropathol. 120:529–535 10.1007/s00401-010-0709-7 [DOI] [PubMed] [Google Scholar]
- Moreno-García M.E., Sommer K., Shinohara H., Bandaranayake A.D., Kurosaki T., Rawlings D.J. 2010. MAGUK-controlled ubiquitination of CARMA1 modulates lymphocyte NF-kappaB activity. Mol. Cell. Biol. 30:922–934 10.1128/MCB.01129-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanichamy A., Barnard J., Zheng B., Owen T., Quach T., Wei C., Looney R.J., Sanz I., Anolik J.H. 2009. Novel human transitional B cell populations revealed by B cell depletion therapy. J. Immunol. 182:5982–5993 10.4049/jimmunol.0801859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekarsky Y., Zanesi N., Croce C.M. 2010. Molecular basis of CLL. Semin. Cancer Biol. 20:370–376 10.1016/j.semcancer.2010.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piątosa B., Wolska-Kuśnierz B., Pac M., Siewiera K., Gałkowska E., Bernatowska E. 2010. B cell subsets in healthy children: reference values for evaluation of B cell maturation process in peripheral blood. Cytometry B Clin. Cytom. 78:372–381 [DOI] [PubMed] [Google Scholar]
- Puente X.S., Pinyol M., Quesada V., Conde L., Ordóñez G.R., Villamor N., Escaramis G., Jares P., Beà S., González-Díaz M., et al. 2011. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 475:101–105 10.1038/nature10113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy I., Brisco M., Morley A. 1992. Improved PCR method for detecting monoclonal immunoglobulin heavy chain rearrangement in B cell neoplasms. J. Clin. Pathol. 45:770–775 10.1136/jcp.45.9.770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg H.K., Markowitz R.I., Kolberg H., Park C., Hubbard A., Bellah R.D. 1991. Normal splenic size in infants and children: sonographic measurements. AJR Am. J. Roentgenol. 157:119–121 [DOI] [PubMed] [Google Scholar]
- Sasaki Y., Derudder E., Hobeika E., Pelanda R., Reth M., Rajewsky K., Schmidt-Supprian M. 2006. Canonical NF-kappaB activity, dispensable for B cell development, replaces BAFF-receptor signals and promotes B cell proliferation upon activation. Immunity. 24:729–739 10.1016/j.immuni.2006.04.005 [DOI] [PubMed] [Google Scholar]
- Shaffer A.L., III, Young R.M., Staudt L.M. 2012. Pathogenesis of human B cell lymphomas. Annu. Rev. Immunol. 30:565–610 10.1146/annurev-immunol-020711-075027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer W.T., Rosenblatt H.M., Gelman R.S., Oyomopito R., Plaeger S., Stiehm E.R., Wara D.W., Douglas S.D., Luzuriaga K., McFarland E.J., et al. ; Pediatric AIDS Clinical Trials Group 2003. Lymphocyte subsets in healthy children from birth through 18 years of age: the Pediatric AIDS Clinical Trials Group P1009 study. J. Allergy Clin. Immunol. 112:973–980 10.1016/j.jaci.2003.07.003 [DOI] [PubMed] [Google Scholar]
- Staudt L.M. 2010. Oncogenic activation of NF-kappaB. Cold Spring Harb. Perspect. Biol. 2:a000109 10.1101/cshperspect.a000109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thome M., Charton J.E., Pelzer C., Hailfinger S. 2010. Antigen receptor signaling to NF-kappaB via CARMA1, BCL10, and MALT1. Cold Spring Harb. Perspect. Biol. 2:a003004 10.1101/cshperspect.a003004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallabhapurapu S., Karin M. 2009. Regulation and function of NF-kappaB transcription factors in the immune system. Annu. Rev. Immunol. 27:693–733 10.1146/annurev.immunol.021908.132641 [DOI] [PubMed] [Google Scholar]
- van Zelm M.C., Szczepanski T., van der Burg M., van Dongen J.J. 2007. Replication history of B lymphocytes reveals homeostatic proliferation and extensive antigen-induced B cell expansion. J. Exp. Med. 204:645–655 10.1084/jem.20060964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., You Y., Case S.M., McAllister-Lucas L.M., Wang L., DiStefano P.S., Nuñez G., Bertin J., Lin X. 2002. A requirement for CARMA1 in TCR-induced NF-kappa B activation. Nat. Immunol. 3:830–835 10.1038/ni824 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.