Abstract
Tetrahymena thermophila is a free-living ciliate with no exogenous sterol requirement. However, it can perform several modifications on externally added sterols including desaturation at C5(6), C7(8), and C22(23). Sterol desaturases in Tetrahymena are microsomal enzymes that require Cyt b5, Cyt b5 reductase, oxygen, and reduced NAD(P)H for their activity, and some of the genes encoding these functions have recently been identified. The DES5A gene encodes a C-5(6) sterol desaturase, as shown by gene knockout in Tetrahymena. To confirm and extend that result, and to develop new approaches to gene characterization in Tetrahymena, we have now, expressed DES5A in Saccharomyces cerevisiae. The DES5A gene was codon optimized and expressed in a yeast mutant, erg3Δ, which is disrupted for the gene encoding the S. cerevisiae C-5(6) sterol desaturase ERG3. The complemented strain was able to accumulate 74% of the wild type level of ergosterol, and also lost the hypersensitivity to cycloheximide associated with the lack of ERG3 function. C-5(6) sterol desaturases are expected to function at the endoplasmic reticulum. Consistent with this, a GFP-tagged copy of Des5Ap was localized to the endoplasmic reticulum in both Tetrahymena and yeast. This work shows for the first time that both function and localization are conserved for a microsomal enzyme between ciliates and fungi, notwithstanding the enormous evolutionary distance between these lineages. The results suggest that heterologous expression of ciliate genes in S. cerevisiae provides a useful tool for the characterization of genes in Tetrahymena, including genes encoding membrane protein complexes.
Keywords: C-5(6) sterol desaturase, Tetrahymena, endoplasmic reticulum, complementation, Saccharomyces cerevisiae, ergosterol
INTRODUCTION
Sterol composition in eukaryotic organisms is very diverse; while cholesterol is the predominant sterol in vertebrates and ergosterol in fungi, stigmasterol, β-sitosterol and campesterol are the most abundant in plants. More complex and diverse sterol profiles can be found in protozoans and microalgae, with additional sterol ring and side chain modifications such as (de)methylation, (de)saturation and oxidation [1]. In addition, some invertebrates, alveolates (a group to which ciliates belong) and a few flagellated parasites cannot synthesize sterols at all, and are therefore strictly auxotrophic [2]. Interestingly, many of them are nonetheless able to perform structural modifications or rearrangements on the sterol moiety [3, 4].
The ciliate Tetrahymena is an interesting case. This fresh-water protozoan has no sterol requirement and no detectable sterols in its membranes. Instead, it makes and uses tetrahymanol, a compound closely related to hopanoids which function as sterol surrogates in prokaryotes [5]. However, in response to the addition of selected sterols to the growth medium, Tetrahymena suppresses the formation of tetrahymanol and replaces this compound by the added sterol, either with or without prior modification(s). Four sterol-modifying activities have been detected in the ciliate: desaturation in positions C-5(6), C-7(8) and C-22(23) and the removal of the ethyl group on position C24 in C29 sterols [6, 7]. As a result of these transformations, the ciliate accumulates a Δ5,7,22 trien (C27) sterol in its membrane, as the major product.
We have recently identified DES5A (GenBank ID: FJ940725.1) (TTHERM_01194720), the ene encoding a C-5(6) sterol desaturase in the ciliate, by a knockout approach [8]. The gene is divergent in sequence from known C-5(6) sterol desaturases,, raising the question of whether it has also diverged in fundamental mechanistic aspects. One way to explore this question is to ask whether the Tetrahymena gene can rescue sterol desaturation mutants in other organisms. In S. cerevisiae, the C-5(6) sterol desaturase is an endoplasmic reticulum enzyme encoded by the ERG3 gene [9,10], whose disruption blocks ergosterol biosynthesis. In previous studies the yeast erg3 null mutant has been successfully complemented with C-5(6)sterol desaturase homologs from plants, mammals and algae[11, 12], providing a powerful approach to confirming functional homologs. Erg3p, as well as the corresponding enzyme in plants and vertebrates, is a membrane-bound enzyme that requires cytochrome b5 and cytochrome b5 reductase for its activity [13, 14]. Cyt b5 is a small tail anchored protein of the endoplastic reticulum membrane with an N-terminal globular cytosolic haem-binding domain, and a hydrophobic transmembrane domain followed by a carboxyl ER-targeting luminal domain [15]. Interestingly, structural studies of the T. thermophila Cyt b5 suggest that the ciliate protein may differ slightly, both in molecular mass and spectrophotometric absorption peaks from the ones found in mammals and yeast [16]. This apparent divergence may reflect relatively fast evolution of Tetrahymena Cytb5, a phenomenon that has been noted for other ciliate genes, and raises the question of whether fast-evolving Tetrahymena sterol desaturases are still compatible with the microsomal electron transport machinery in distantly-related organisms like S. cerevisiae. This question is underscored by the unusual sterol metabolism in Tetrahymena. To address these questions, we expressed a codon-optimized variant of DES5A in S. cerevisiae, and tested its function as well as its localization. We also expressed the GFP-tagged protein in Tetrahymena, in order to compare localizations in the two distantly-related lineages.
MATERIAL AND METHODS
Strains, growth conditions and plasmids
The following yeast strains were used in this study: Saccharomyces cerevisiae W303 (MATa/MATα {leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15} [phi+] ) denoted as wild-type strain and the erg3 mutant strain (MATα ade2-1 his3-11,15 leu2-3,112, trp1-1, ura3-1, can1, erg3::TRP1 upc2::HIS3) [17]. These strains were grown aerobically at 30° C either in YPD containing 2% glucose (Merck), 1% Yeast Extract (Oxoid) and 2% peptone (Oxoid) or in synthetic minimal media containing 0.67% Yeast Nitrogen Base (US Biological) supplemented with the appropriate amino acids and 2% glucose as carbon source. The DES5A gene (TTHERM_01194720) was synthesized with codons optimized for expression in S. cerevisiae (Genscript, USA). The ORF was re-amplified with primers containing HindIII/XhoI restriction sites and subcloned into p425GPD, the 2-micron yeast expression vector containing a glyceraldehyde-3-phosphate dehydrogenase promoter [18]. The plasmid was designed pC-5T. For expression of GFP, the enhanced green fluorescent protein was cloned into HindIII/XhoI sites of p425GPD, obtaining the pGFP plasmid, used as a control. For the GFP fused to DES5A construct, the enhanced green fluorescent protein was fused at the C-terminal of DES5A gene by overlapping PCR and cloned into the HindIII/XhoI sites of p425GP, obtaining the pC-5T. GFP plasmid. S. cerevisiae strain were transformed by electroporation and selected on minimal agar plates without leucine [19].
T. thermophila strain CU428 (mpr1- 1/mpr1-1; mp-s, VII), is designated as wild type (WT) in this work. Cells were grown at 30°C in 125 ml Erlenmeyer flasks containing 20 ml SPP medium with the following composition (wt/vol): 1% proteose-peptone (Oxoid, U.K), 0.1% yeast extract (Merck, Germany), 0.2% glucose (Merck, Germany), and 0.003% iron citrate (Sigma-Aldrich).
Plasmid pmEGFP [20] contained a monomeric variant (A206K) of GFP together with the neomycin/paromomycin resistance gene under a cadmium-inducible metallothionein (MTT) promoter (neo 4 cassette) [21].
Construction of DES5A-GFP fusion in Tetrahymena
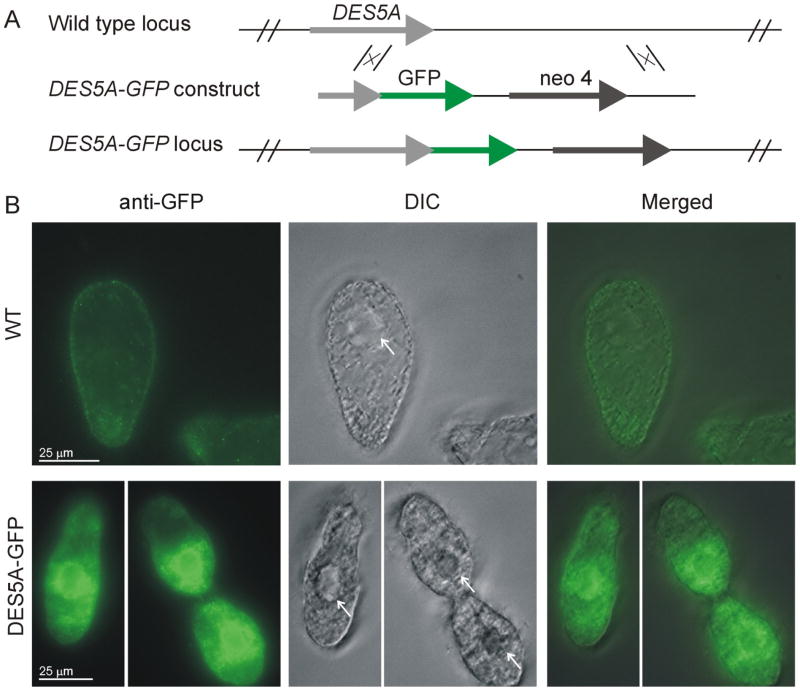
A 791 base pair C-terminal fragment of the DES5A gene, lacking the stop codon, and a downstream flanking region of 910 base pairs were amplified by PCR and cloned into the BamHI and HindIII sites respectively of pmEGFP using directional In-Fusion Cloning techniques (Clontech). The DES5A-GFP construct was then liberated with NheI and XhoI restriction enzymes from the vector and was introduced into the endogenous DES5A locus by homologous integration as diagrammed in Fig. 4A. After biolistic treatment [22], the somatic transformants were selected with 120 μg/ml of paromomycin in the presence of 1.0 ug/ml CdCl2, which induces the MTT1 promoter of the neo 4 expression cassette, and were transferred daily in increasing concentrations of paromomycin to allow phenotypic assortment. Single cells were then isolated and the clones expanded with daily transfers for 2 weeks in 40 mg/ml paromomycin.
Fig. 4.
Localization of GFP tagged Des5Ap in Tetrahymena thermophila (A) Schematic representation of gene replacement in the WT locus of the DES5A gene by targeting the DES5A-GFP construct by homologous recombination, using a cassette in which neo4 confers paramomycin resistance. (B) Immunofluorescence localization of GFP-tagged DES5A. Wild-type (WT) and DES5A-GFP strains were grown to Log phase and fixed for indirect immunofluorescence staining. The GFP was localized by anti-GFP primary antibody followed by labeling with anti-rabbit–Alexa Fluor 488 secondary antibody. DIC: Differential interference contrast microscopy. White arrows indicate the nucleus of the ciliate.
Sterol extraction and GC-MS analysis
To analyze sterols in whole cells, 30 ml of a 48h (stationary) culture were harvested, resuspended in 1.5 ml distilled water and then saponified in 1.5ml 2M NaOH in methanol-water (1:1 v/v) at 80°C for 1 h. Sterols were extracted 3 times with 3ml hexane each and dried under nitrogen flow. The residue was resuspended in 100 μl N-methyl-N-(trimethylsilyl) trifluoroacetamide (MSTFA), and then incubated for 30 min at 70°C. The composition of the steryl trimethylsilyl ester derivatives was analyzed by running samples through an HP-5MS (30 m. × 0.25 mm × 0.25 um, Agilent Technologies) in a Hewlett Packard HP 6890 gas chromatograph. The column was temperature programmed at 10°C/min from 100 to 310°C and subsequently held for 10 min at 310°C. MS was carried out using a HP mass selective detector (model MSD 5973) operated at an ionization voltage of 70 eV with a scan range of 50 to 600 amu. The retention time and mass spectrum of all new peaks obtained were compared to those of standards (Steraloids, USA) and those available in the NIST library.
The sterol content in samples are expressed as percentage of the total amount of sterols present in each sample (% w/w of total sterols).
Localization of GFP tagged proteins
Tetrahymena expressing DES5A-GFP were fixed with 4.2 % paraformaldehyde in 50 mM HEPES pH 7.0 for 10 min at room temperature, and permeabilized with ice-cold 0.1% Triton X-100 in 50 mM HEPES pH 7.0 for 8 min. After being washed with ice-cold HEPES three times, the fixed cells were treated with blocking solution (1% bovine serum albumin in TBS buffer) for 15 min at room temperature, and incubated for 30 min in blocking solution with anti-GFP primary antibody (A-11122; Invitrogen) at a 1:400 dilution. The cells were washed three times for 5 min each with TBS buffer containing 0.1% BSA and subsequently incubated for 30 min at room temperature with donkey anti-rabbit–Alexa Fluor 488 secondary antibody (A21206; Invitrogen) at a 1:200 dilution in blocking solution. After one wash in TBS buffer containing 0.1% BSA and two washes in 50 mM HEPES pH 7.0 the cells were mounted with Trolox antibleaching solution. Digital images were collected using a Carl Zeiss Axio imager M2 fluorescence microscope.
To localize Des5Ap-GFP in yeast, cells were collected by centrifugation for 5 minutes at 600 g and resuspended in 3 ml of freshly prepared 4% paraformaldehyde. After incubation for 20 minutes at room temperature, the suspension was centrifuged for 5 minutes at 350 g and cells were washed 3 times with 2 ml 0.1 M potassium phosphate buffer, pH 7.4 (KPB). Cells were resuspended in 2 ml KPB supplemented with 1.2 M sorbitol and incubated at 4°C overnight. Then cells were collected by centrifugation for 5 minutes at 600 g, resuspended in 0.1% Triton X-100 in PBS and incubated 10 min at room temperature. After washing three times with 1 ml PBS, cells were incubated for 10 minutes with 5 μM Hoechst 33258 stain (prepared in PBS also containing 3% BSA). The cells were washed three times with PBS and resuspended in DakoCytomation Fluorescent Mounting Medium (Dako). Cells were visualized using a Olympus FV300 confocal microscope (model BX61) with acquisition software FluoView version 3.3.
RESULTS
Divergence in Tetrahymena thermophila Cyt b5 enzymes
Our previous analysis indicated that the T. thermophila DES5A gene is a member of the fatty acid hydroxylase superfamily (FAHS) [8]. This enzyme requires the cytochrome b5 microsomal electron transfer for its activity. The family of cytochrome b5-like proteins includes, besides cytochrome b5 itself which is denominated as free Cyt b5, hemoprotein domains covalently associated with other redox domains in diverse fusion proteins.
In querying the Tetrahymena thermophila genome, we identified a large number of putative Cyt b5 genes. By searching for domains corresponding to Cyt b5 (PFAM: PF00173) we identified 13 sequences, six of which possess a sequence length of 109-227 aa and include a hydrophobic transmembrane domain, and which therefore resemble free Cyt b5 proteins in other organisms. The seven remaining sequences are similar but longer and possess other domains such as dopamine beta-monooxygenase (DOMON), catecholamine-binding domain (DOH) and fatty acid desaturase, and which therefore are not free Cyt b5 genes.
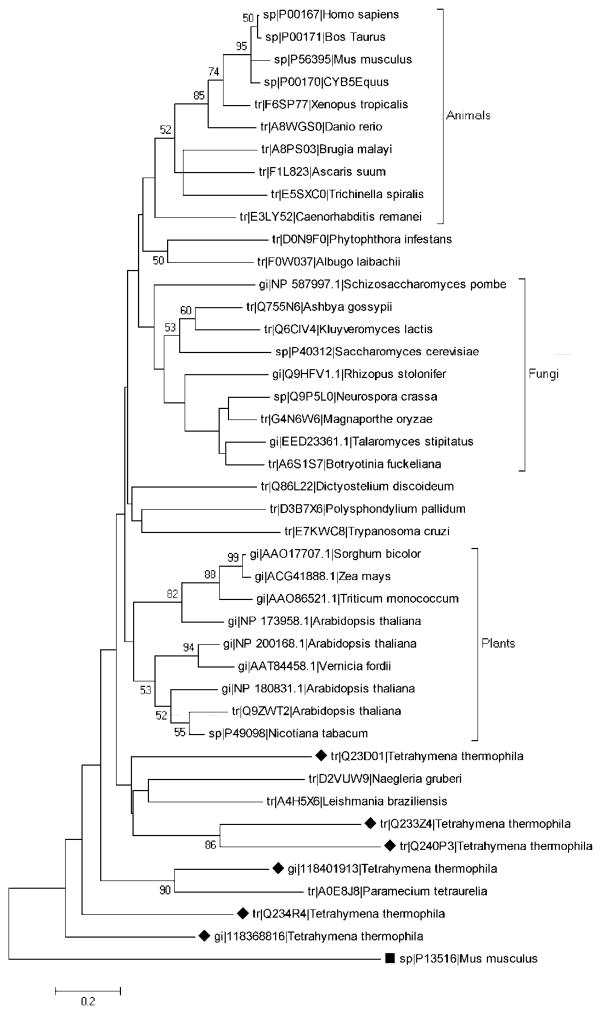
A phylogenetic tree generated by the Maximum likelihood method, based on 43 amino acid sequences of cytochrome b5 proteins including the six putative homologs in Tetrahymena, clearly shows that animal, fungal, and plant proteins cluster according to lineage (Fig. 1). The Tetrahymena sequences do not group in a specific cluster, and thus appear to have undergone the extensive divergence noted previously for protist cytochrome b5 proteins [23].
Fig. 1.
Phylogenetic analysis of cytochrome b5 proteins. The phylogenetic tree of 43 amino acid sequences (UniProtKB) was generated using a Maximum likelihood method with 500 bootstrap replicates with MEGA5 software, as described in Materials and Methods. The black rhombus indicate Tetrahymena thermophila putative cytb5 enzymes. The black box indicates a Mus musculus Acyl-CoA desaturase protein, containing a Cytb5 domain, which is used as an outgroup. The bar indicates percentages of substitution.
Complementation by heterologous expression of DES5A in the Saccharomyces cerevisiae erg3 mutant strain
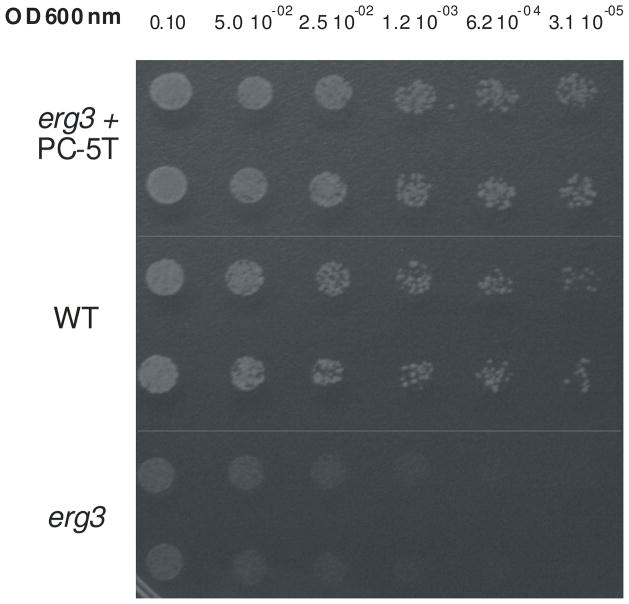
Given the extensive sequence divergence, it is possible that the Tetrahymena Des5Ap enzyme can only function with the Tetrahymena cytochrome b5 providing microsomal electron transfer. Alternatively, the Tetrahymena desaturase may be able to use budding yeast (S. cerevisiae) Cyt b5, , that only shares between 12.1% and 30.1% identify, or 17.6% and 53.4% similarity, with the Tetrahymena homologs. If the latter is true, it would allow us to characterize ciliate genes involved in sterol metabolism by taking advantage of the many tools available in S. cerevisiae. To explore this, we expressed DES5A in a haploid yeast strain deficient in ERG3. Since Tetrahymena utilizes an alternative nuclear genetic code [24], we first optimized codon assignment and frequency bias for expression in yeast. The optimized gene was cloned into a GPD expression shuttle vector (p425-GPD, with LEU2 as selectable marker), obtaining the pC-5T plasmid that was electroporated into the erg3 mutant strain. The Leu+ isolates were checked by PCR for the presence of the DES5A gene and then screened for resistance to low levels of cycloheximide. The erg3 mutant displays a marked hypersensitivity to cycloheximide, compared to wild type Saccharomyces strains [25]. As shown in Fig. 2, the erg3 mutant expressing DES5A (erg3 + pC-5T) shows increased resistance to cycloheximide.
Fig. 2.
Hypersensitivity to cycloheximide in the erg3 null strain is resuced by expression of the DES5A gene. Serial dilutions of the wild type, erg3 null and erg3+pC-5T complemented strains were spotted onto plates containing minimum medium plus 0.01 μg/ml of cycloheximide and grown for 4 days at 28 °C.
Sterol profile in S. cerevisae WT, erg3 mutant and erg3 + pC-5T-complemented strain
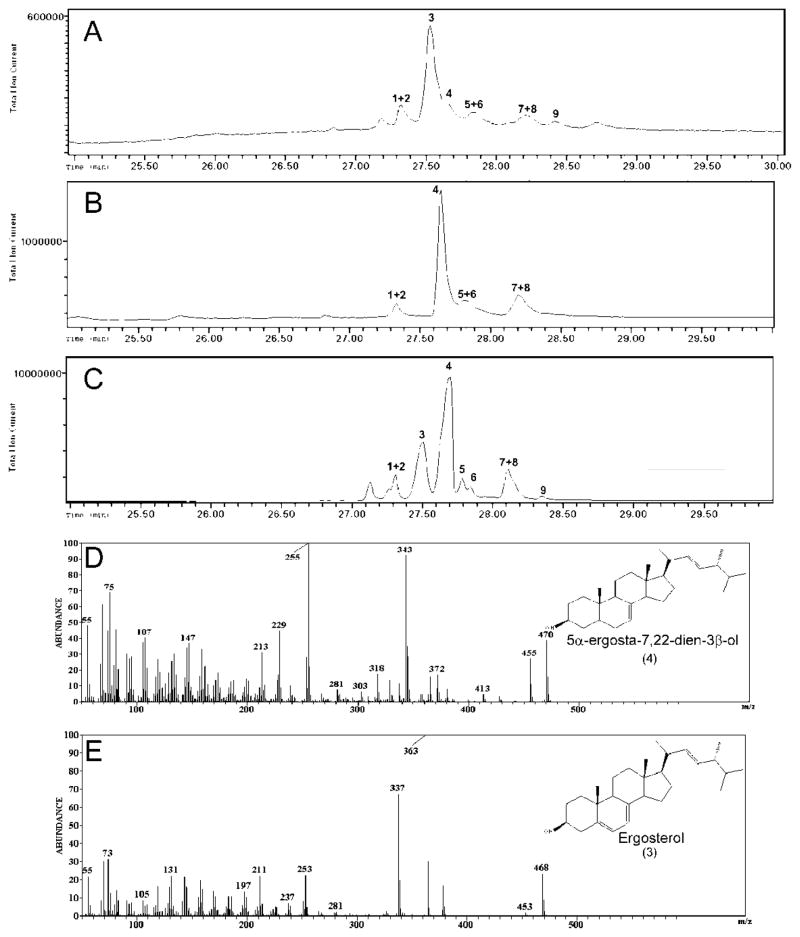
We performed GC-MS analysis to determine the sterol composition of WT, the erg3 mutant and erg3+pC-5T transformed cells (Fig 3 and Table 1). Major differences in sterol composition were noted between WT and erg3Δ strains in ergosterol and 5α-ergosta-7,22-dien-3β-ol content. In particular, erg3Δ cells accumulated less than 1% the level of ergosterol found in WT cells. Importantly, expression of the pC-5T plasmid in erg3Δ cells restored ergosterol synthesis in the mutant to 74% of the WT level. This result suggests that DES5A could significantly but incompletely complement the ERG3 deletion. The conclusion that complementation was incomplete was also supported by analysis of intermediates in ergosterol synthesis. The erg3 mutant accumulated an ergosterol precursor, 5α-ergosta-7,22-dien-3β-ol (47%) as the major sterol. This precursor makes up only 12% of the sterol pool in WT cells, but accumulates to high levels (41%) in the erg3Δ strain expressing DES5A. The profile of other intermediates in the ergosterol biosynthesis pathway, such as fecosterol (5α-ergosta-8-en-3β-ol) and lanosterol were also slightly different between the three strains (Table 1).
Fig. 3.
GC-MS analysis of sterols. A: S. cerevisiae WT 303. B: erg3 null. C: erg3+p C-5T strain. D and E: mass spectra of 5α-ergosta-7,22-dien-3β-ol and ergosterol trimethylsilyl derivatives respectively. The compounds were identified with the NIST library. Peaks: 1 - zymosterol, 2 - 5α-ergosta-8,22-dien-3β-ol, 3 - ergosterol, 4 - 5α-ergosta-7,22-dien-3β-ol, 5 - fecosterol, 6 - 5α-ergosta-8-en-3β-ol, 7 - 5α-ergosta-7,24-dien-3β-ol, 8 - 5α-ergosta-7-en-3β-ol, 9 - lanosterol.
Table 1.
Sterol profile in S. cerevisae WT W303, erg3 null mutant and erg3+ pC-5T complemented strains
| Peak | m/z TMS | Sterol | Sterol content in strains (% w/w)* | ||
|---|---|---|---|---|---|
| Wild type | erg3 | erg3 + pC-5T | |||
| 1 | 456 | Zymosterol | 11 | 6 | 7 |
| 2 | 470 | 5α-ergostan-8,22-dien-3β-ol | |||
| 3 | 468 | Ergosterol | 46 | <1 | 34 |
| 4 | 470 | 5α-ergostan-7,22-dien-3β-ol | 12 | 47 | 41 |
| 5 | 470 | Fecosterol | 14 | 15 | 1 |
| 6 | 472 | 5α-ergostan-8-en-3β-ol | 3 | ||
| 7 | 470 | 5α-ergostan-7,24-dien-3β-ol | 11 | 16 | 12 |
| 8 | 472 | 5α-ergostan-7-en-3β-ol | |||
| 9 | 498 | Lanosterol | 6 | 16 | 2 |
Sterol content is expressed as percentage of total sterols isolated in each strain (% w/w)
Sub-cellular localization of Des5Ap
Based on work in other systems, we predicted that Des5Ap would function at the level of the endoplasmic reticulum. The desaturase contains the C-terminal di-lysine motif, QIKQKKN, which have been demonstrated to be necessary for the retention of ER integral membrane proteins in different organisms[26]. To localize the enzyme in Tetrahymena, and to ask whether it localizes similarly when expressed in yeast, we C-terminally tagged both Des5Ap and the variant optimized for yeast expression, with eGFP. To avoid potential misexpression artifacts in Tetrahymena, we targeted the tagged gene to its endogenous locus [21]. T. thermophila wild type cells (CU428 strain) were transformed with a DES5A-GFP construct that integrated in the somatic (expressed) macronucleus, so that the tagged gene was under the control of the endogenous promoter, as depicted in Fig 4A. Due to high autofluorescent background even in wild type cells, we increased the selectivity of the signal using anti-GFP antibody and indirect immunofluorescence microscopy. As shown in Fig. 4B, Des5Ap-GFP showed a strong signal around the nucleus, in a pattern that is consistent with nuclear envelope localization, and a more diffuse one in the cytoplasm. This pattern is consistent with ER localization in these cells[27, 28].
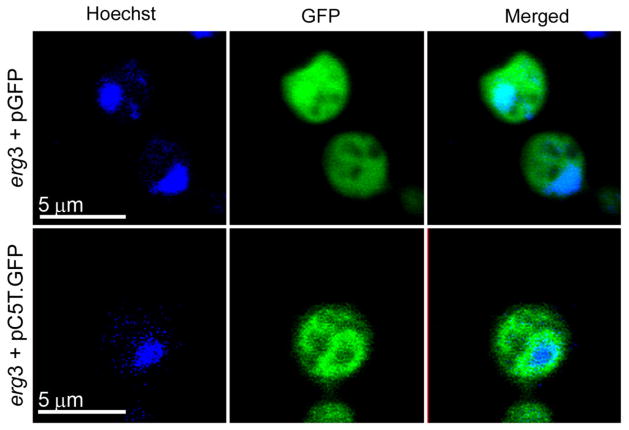
To assay localization of the C-5(6) sterol desaturase from Tetrahymena thermophila in yeast, the DES5A gene fused to eGFP was cloned into a GPD expression shuttle vector obtaining the pC-5T. GFP plasmid, which was introduced into the S. cerevisiae erg3Δ strain. A perinuclear signal, typical of ER anchored proteins [10], was observed in the complemented strain (Fig 5.). Expression of eGFP by itself under the same promoter (pGFP), produced only diffuse cytoplasmic fluorescence (Fig. 5). These results are consistent with those reported for other ER localized proteins in the ciliate [27] and with studies of the microsomal C-5 sterol desaturase in Saccharomyces cerevisiae [10].
Fig. 5.
Localization of GFP-tagged Des5Ap in Saccharomyces cerevisiae. The erg3 strain expressing GFP by itself (top) and GFP-tagged DES5A (bottom) were grown in minimum medium and analyzed at early log phase by direct fluorescence microscopy. DNA was stained with Hoechst.
DISCUSSION
We found that the S. cerevisiae erg3 null mutant could be partially complemented by expression of the T. thermophila DES5A gene, confirming that Des5Ap has C-5(6) sterol desaturase activity. This enzyme is likely to require the activity of a cytochrome b5 -dependent microsomal electron transport system, as previously shown in Tetrahymena for a stearoyl-CoA desaturase [29]. Although the cytochrome b5 sequences are highly divergent between S. cerevisiae and T. thermophila, our results indicate that there is significant functional conservation since the ciliate sterol desaturase can function in the context of fungal cytochrome b5. These results are in concordance with the work of Desmond and Gribaldo [2] which proposed that the last eukaryotic common ancestor already harbored many enzymes for sterol biosynthesis, and that subsequent evolution over the eukaryotic tree occurred by modifications or gene losses, as reported for other enzymes involved in sterol biosynthesis. Overall, our current analysis, based on expression of a ciliate gene in an evolutionarily-distant species, complements our previous studies based on disruption of the gene in T. thermophila via homologous recombination [8].
Sterol C-5(6) desaturases belong to the fatty acid hydroxylase superfamily (pfam: PF04116). These enzymes are integral membrane proteins that localize in the endoplasmic reticulum. An ER retention signal for similar proteins that has been characterized in other systems, consists of a C-terminal di-lysine or di-arginine motif [26]. Examples of sterol C-5(6) desaturasas containing this motif are found in different organisms, such us Saccharomyces cerevisiae (P32353 UniProtKB accession number), Mus musculus (O88822) and Arabidopsis thaliana (Q39208). Tetrahymena Des5Ap contains the QIKQKKN sequence at the C-terminal predicted cytosolic domain, which may be functioning as an ER retention signal. This is in accordance with ER localization in both organisms (Fig. 4 and Fig. 5). Other ciliates proteins with C-terminal di-lysine motif have been also localized in ER like the Paramecium tetraurelia Ca2+-ATPase [30], implying that this type of mechanisms of ER retention for integral proteins are conserved in ciliates.
The ability to express ciliate proteins in heterologous systems with strong experimental tools, like S. cerevisiae, may be strongly advantageous for characterizing their activities. A handful of T. thermophila proteins and one enzymatic RNA have previously been expressed in yeast, namely Histone H2A[31], the translation release factor eRF1[32], the cytosolic methionine salvage pathway protein mtnBD [33] and the ribozyme Group I intron [34]. Our report adds to this list, and moreover shows that a membrane-associated Tetrahymena protein that requires a compatible Cyt b5/Cyt b5reductase electron transport system, can function in a heterologous context. Our results are similar to a recent report in which a Δ5 fatty acid desaturase gene from another ciliate, Paramecium tetraurelia, was functionally expressed in S. cerevisiae [35], but with the significant difference that the Paramecium enzyme, a so-called “front-end” desaturase, contains a fused N-terminal Cyt b5-like domain and therefore does not require the activity of the yeast Cyt b5.
The rescue of erg3Δ by DES5A expression was incomplete, as evidenced by the only partial restoration of ergosterol levels in the complemented strain. Similar results have been reported when mammalian or plant C-5(6) sterol desaturases were expressed in S. cerevisiae. For example, the enzyme from Arabidopsis restored 46% of ergosterol accumulation when expressed in the erg3Δ mutant [36]. The incomplete complementation may be a consequence of aberrant transcriptional regulation of the transgenes, but downstream steps (e.g., translation, or trafficking) may also be non-optimal for the transgenes. In summary, our results suggest that functional complementation of Saccharomyces cerevisiae mutants can be used as an efficient approach for the analysis of genes in Tetrahymena. This includes both the study of membrane protein complexes, and the characterization of genes whose functions have not yet been established.
HIGHLIGHTS.
Ciliate DES5A gene restores ergosterol biosynthesis in erg3 S. cerevisiae strain.
DES5A gene reverses the hypersensitivity to cycloheximide in erg3 strain.
Des5Ap is localized in endoplasmic reticulum of T. thermophila and S. cerevisiae.
Acknowledgments
We wish to thank A. Uttaro and S. Najle (Instituto de Biología Molecular y Celular de Rosario, Argentina) for their gifts of strain erg3 and plasmid p425GPD, which were used in this work. We thank K. Hellingwerf (University of Amsterdam, The Netherlands) for the critical reading of this manuscript, and J. Briguglio and L. Bright (University of Chicago, USA) for their assistance. A.D. Nusblat and C.B. Nudel are members of the Carrera del Investigador Científico, CONICET, Argentina. This work was supported by grants PIP 01937, ANPCYT-PICT 1155-2008 and UBACYT B095, and NSF MCB-1051985 and NIH RO1 GM77607 to APT.
Abbreviation key
- Cyt
Cytochrome
- ER
endoplasmic reticulum
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Volkman JK. Sterols in microorganisms. Appl Microbiol Biotechnol. 200;60:495–506. doi: 10.1007/s00253-002-1172-8. [DOI] [PubMed] [Google Scholar]
- 2.Desmond E, Gribaldo S. Phylogenomics of sterol synthesis: insights into the origin, evolution, and diversity of a key eukaryotic feature. Genome Biol Evol. 2009;1:364–81. doi: 10.1093/gbe/evp036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vinci G, Xia X, Veitia RA. Preservation of genes involved in sterol metabolism in cholesterol auxotrophs: facts and hypotheses. PloS One. 2008;3:e2883. doi: 10.1371/journal.pone.0002883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ikekawa N, Morisaki M, Fujimoto Y. Sterol metabolism in insects: dealkylation of phytosterol to cholesterol. Acc Chem Res. 1993;26:139–46. [Google Scholar]
- 5.Thompson GA, Bamberry RJ, Nozawa Y. Further Studies of the Lipid Coposition and Biochemical Properties of Tetrahymena pyriformis Membrane Systems. Biochemistry. 1971;10:4441–7. doi: 10.1021/bi00800a014. [DOI] [PubMed] [Google Scholar]
- 6.Conner RL, Mallory FB, Landrey JR, Iyengar CW. The conversion of colesterol to delta-5,7,22-cholestatrien-3-beta-ol by Tetrahymena pyriformis. J Biol Chem. 1969;244:2325–33. [PubMed] [Google Scholar]
- 7.Nes WR, Alcaide A. Dealkylation of 24-ethylsterols by Tetrahymena pyriformis. Lipids. 1975;10:140–4. doi: 10.1007/BF02534151. [DOI] [PubMed] [Google Scholar]
- 8.Nusblat AD, Najle SR, Tomazic ML, Uttaro AD, Nudel CB. C-5(6)sterol desaturase from Tetrahymena thermophila: Gene identification and knockout, sequence analysis, and comparison to other C-5(6) sterol desaturases. Eukaryot Cell. 2009;8:1287–97. doi: 10.1128/EC.00057-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arthington BA, Bennett LG, Skatrud PL, Guynn CJ, Barbuch RJ, Ulbright CE, et al. Cloning, disruption and sequence of the gene encoding yeast C-5 sterol desaturase. Gene. 1991;102:39–44. doi: 10.1016/0378-1119(91)90535-j. [DOI] [PubMed] [Google Scholar]
- 10.Natter K, Leitner P, Faschinger A, Wolinski H, McCraith S, Fields S, et al. The spatial organization of lipid synthesis in the yeast Saccharomyces cerevisiae derived from large scale green fluorescent protein tagging and high resolution microscopy. Mol Cell Proteomics. 2005;4:662–72. doi: 10.1074/mcp.M400123-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Husselstein T, Schaller H, Gachotte D, Benveniste P. Delta7-sterol- C-5-desaturase: molecular characterization and functional expression of wild-type and mutant alleles. Plant Mol Biol. 1999;39:891–906. doi: 10.1023/a:1006113919172. [DOI] [PubMed] [Google Scholar]
- 12.Brumfield KM, Moroney JV, Moore TS, Simms TA, Donze D. Functional characterization of the Chlamydomonas reinhardtii ERG3 ortholog, a gene involved in the biosynthesis of ergosterol. PLoS One. 2010;5:e8659. doi: 10.1371/journal.pone.0008659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Honjo K, Ishibashi T, Imai Y. Partial purification and characterization of lathosterol 5-desaturase from rat liver microsomes. J Biochem. 1985;97:955–9. doi: 10.1093/oxfordjournals.jbchem.a135137. [DOI] [PubMed] [Google Scholar]
- 14.Taton M, Rahier A. Plant sterol biosynthesis: identification and characterization of higher plant delta 7-sterol C-5(6)-desaturase. Arch Biochem Biophys. 1996;325:279–88. doi: 10.1006/abbi.1996.0035. [DOI] [PubMed] [Google Scholar]
- 15.Mitoma J, Ito A. The carboxy-terminal 10 amino acid residues of cytochrome b5 are necessary for its targeting to the endoplasmic reticulum. EMBO J. 1992;11:4197–203. doi: 10.1002/j.1460-2075.1992.tb05513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fukushima H, Takeda T, Sasaki N, Watanabe T, Nozawa Y. Purification and characterization of microsomal cytochrome b560ms from a unicellular eukaryote Tetrahymena pyriformis. J Biol Chem. 1983;258:11991–6. [PubMed] [Google Scholar]
- 17.Davies BS, Wang HS, Rine J. Dual activators of the sterol biosynthetic pathway of Saccharomyces cerevisiae: similar activation/regulatory domains but different response mechanisms. Mol Cell Biol. 2005;25:7375–85. doi: 10.1128/MCB.25.16.7375-7385.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mumberg D, Müller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–22. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 19.Ausubel FM, Frederick M. Current Protocols in Molecular Biology. New York: Wiley; 1991. [Google Scholar]
- 20.Bright LJ, Kambesis N, Nelson SB, Jeong B, Turkewitz AP. Comprehensive analysis reveals dynamic and evolutionary plasticity of Rab GTPases and membrane traffic in Tetrahymena thermophila. PLoS Genet. 2010;6:e1001155. doi: 10.1371/journal.pgen.1001155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kataoka K, Schoeberl UE, Mochizuki K. Modules for C-terminal epitope tagging of Tetrahymena genes. J Microbiol Methods. 2010;82:342–6. doi: 10.1016/j.mimet.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cassidy-Hanley D, Bowen J, Lee JH, Cole E, VerPlank LA, Gaertig J, et al. Germline and somatic transformation of mating Tetrahymena thermophila by particle bombardment. Genetics. 1997;146:135–47. doi: 10.1093/genetics/146.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gostincar C, Turk M, Gunde-Cimerman N. The Evolution of Fatty Acid Desaturases and Cytochrome b5 in Eukaryotes. J Membrane Biol. 2010;233:63–72. doi: 10.1007/s00232-010-9225-x. [DOI] [PubMed] [Google Scholar]
- 24.Horowitz S, Gorovsky MA. An unusual genetic code in nuclear genes of Tetrahymena. Proc Natl Acad Sci U S A. 1985;82:2452–5. doi: 10.1073/pnas.82.8.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith SJ, Parks LW. The ERG3 gene in Saccharomyces cerevisiae is required for the utilization of respiratory substrates and in heme-deficient cells. Yeast. 1993;9:1177–87. doi: 10.1002/yea.320091104. Erratum in: Yeast 1994; 10:557. [DOI] [PubMed] [Google Scholar]
- 26.Teasdale RD, Jackson MR. Signal-mediated sorting of membrane proteins between the endoplasmic reticulum and the golgi apparatus. Annu Rev Cell Dev Biol. 1996;12:27–54. doi: 10.1146/annurev.cellbio.12.1.27. [DOI] [PubMed] [Google Scholar]
- 27.Xie R, Clark KM, Gorovsky MA. Endoplasmic reticulum retention signal-dependent glycylation of the Hsp70/Grp170-related Pgp1p in Tetrahymena. Eukaryot Cell. 2007;6:388–97. doi: 10.1128/EC.00366-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rahaman A, Elde NC, Turkewitz AP. A dynamin-related protein required for nuclear remodeling in Tetrahymena. Curr Biol. 2008;18:1227–33. doi: 10.1016/j.cub.2008.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sasaki N, Fukushima H, Watanabe T, Nozawa Y. Studies on Tetrahymena microsomal electron transport systems: solubilization of microsomal electron transport enzymes involved in fatty acid desaturation. Comp Biochem Physiol B. 1984;79:219–23. doi: 10.1016/0305-0491(84)90016-6. [DOI] [PubMed] [Google Scholar]
- 30.Hauser K, Pavlovic N, Klauke N, Geissinger D, Plattner H. Green fluorescent protein-tagged sarco(endo)plasmic reticulum Ca2+-ATPase overexpression in Paramecium cells: isoforms, subcellular localization, biogenesis of cortical calcium stores and functional aspects. Mol Microbiol. 2000;37:773–87. doi: 10.1046/j.1365-2958.2000.02038.x. [DOI] [PubMed] [Google Scholar]
- 31.Liu X, Bowen J, Gorovsky MA. Either of the major H2A genes but not an evolutionarily conserved H2AF/Z variant of Tetrahymena thermophila can function as the sole H2A gene in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2878–87. doi: 10.1128/mcb.16.6.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karamyshev AL, Ito K, Nakamura Y. Polypeptide release factor eRF1 from Tetrahymena thermophila: cDNA cloning, purification and complex formation with yeast eRF3. FEBS Lett. 1999;457:483–8. doi: 10.1016/s0014-5793(99)01089-3. [DOI] [PubMed] [Google Scholar]
- 33.Salim HM, Negritto MC, Cavalcanti AR. 1+1 = 3: a fusion of 2 enzymes in the methionine salvage pathway of Tetrahymena thermophila creates a trifunctional enzyme that catalyzes 3 steps in the pathway. PLoS Genet. 2009;5:e1000701. doi: 10.1371/journal.pgen.1000701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Good L, Elela SA, Nazar RN. Tetrahymena ribozyme disrupts rRNA processing in yeast. J Biol Chem. 1994;269:22169–72. [PubMed] [Google Scholar]
- 35.Tavares S, Grotkjær T, Obsen T, Haslam RP, Napier JA, Gunnarsson N. Metabolic engineering of Saccharomyces cerevisiae for production of Eicosapentaenoic Acid, using a novel {Delta}5-Desaturase from Paramecium tetraurelia. Appl Environ Microbiol. 2011;77:1854–61. doi: 10.1128/AEM.01935-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Husselstein T, Schaller H, Gachotte D, Benveniste P. Delta7-sterol- C-5-desaturase: molecular characterization and functional expression of wild-type and mutant alleles. Plant Mol Biol. 1999;39:891–906. doi: 10.1023/a:1006113919172. [DOI] [PubMed] [Google Scholar]