Abstract
Olfaction in rodents provides an excellent modality for the study of cellular mechanisms of information processing and storage, since a single occurrence of precisely timed stimuli has high survival value. We have followed up preliminary evidence of cytokine and proteinase involvement in normal (as opposed to pathologically-induced) brain plasticity by surveying for the presence of these factors in the olfactory circuitry of the rat. Genes for 25–30 common cytokines and their receptors, and over 30 cell matrix and adhesion molecules were found to be expressed across the olfactory bulb, insular cortex, amygdala, and dorsal hippocampus. We then measured by real-time PCR the transcriptional expression of seven of these genes following a one-time exposure to the novel odor of blueberry bars or cornnuts, in contrast to presentation of the familiar odor of lab chow. In the amygdala significant up-regulation of interleukin-1 receptor 1 (IL1r1), interleukin-4 receptor (IL4r), fibroblast growth factor 13 (FGF13), and cathepsin-H (CtsH) was observed in males in response to the odor of cornnuts only. Changes were less consistent and widespread in the hippocampus, but were again sex specific for three genes: cathepsin-L (CtsL), matrix metalloproteinase-14 (MMP-14) and MMP-16. Our results show that transcription for several specific cytokines, growth factors, and proteinases responds to a one-time exposure to a novel odor, in a manner that tends to be region- and sex-specific. This suggests considerable variation in the way that olfactory information is processed at the cellular level in different brain regions and by the two sexes.
Keywords: amygdala, hippocampus, interleukin, cathepsin, matrix metalloproteinase
1. Introduction
We have sought to identify events that can be associated with, and ultimately explain, the basis for lasting memory storage. Our strategy is to search for candidate genes and gene products at multiple relay points in the information processing circuit for robust and essentially one-trial learning experiences, where exposure to a novel stimulus can be tightly correlated temporally with changes at the cellular level.
Olfaction in rodents is an excellent modality for testing cellular responses to novel stimuli. Odors are discriminated with high specificity by rodents. The information is processed sequentially through well-known neural pathways, and has high survival value, making one-trial learning robust and highly adaptive.
Elements of the immune response are of particular interest to us because of their involvement in the detection and storage of novel information, and their ability to interact in many combinations to account for context and consequence of specific chemical information (Dustin and Colman, 2002; Larson et al., 2002). Molecules that affect the functional association between neurons, including adhesion and the intercellular matrix, are frequently the signaling targets of cytokines, so they are of interest as well. If lasting memory depends on alteration of the functional association between brain cells, these molecules are likely to be involved (Milward et al., 2007).
The objectives of this study, therefore, were to (1) survey the expression of genes for a large number of (a) common cytokines and (b) cell matrix and adhesion molecules, in the amygdala, hippocampus, insular cortex and olfactory bulb, where critical processing of information for olfactory learning occurs; (2) determine whether a one-trial exposure to a novel odor affects the expression of any of those genes; and (3) discuss the implications of our finding for the plausibility that immune-like reactivity plays a role in the processing of new information by brain cells.
2. Results
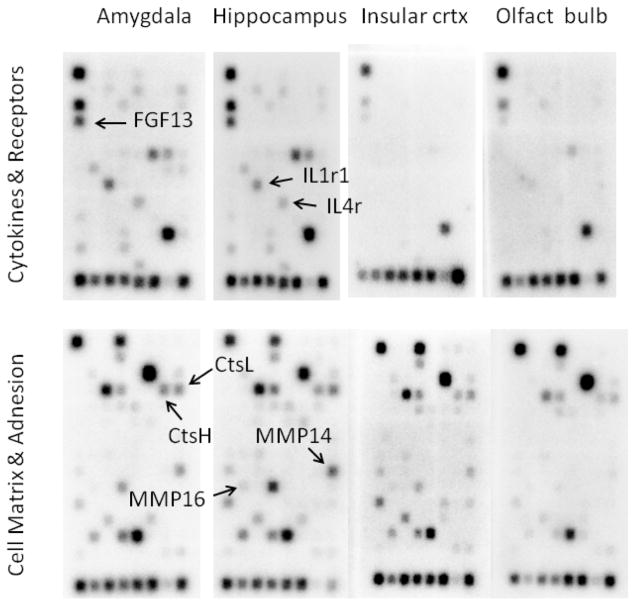
Genes for 23–28 common cytokines and their receptors were expressed in the rat amygdala and hippocampus, with far fewer expressed in the insular cortex and olfactory bulb. Up to 34 cell matrix and adhesion molecules were expressed in the hippocampus, with slightly fewer across the other three brain regions studied (Fig. 1). A complete list of gene expression tested for and detected is given in Supplementary Tables 1 and 2.
Fig. 1.
Neuroanatomical distribution of genes for cytokines, growth factors, and proteases in the rat brain. The density of each chemiluminescent spot is proportional to the amount of mRNA produced for that gene. The position of seven genes analyzed by qPCR in this study is indicated by arrows.
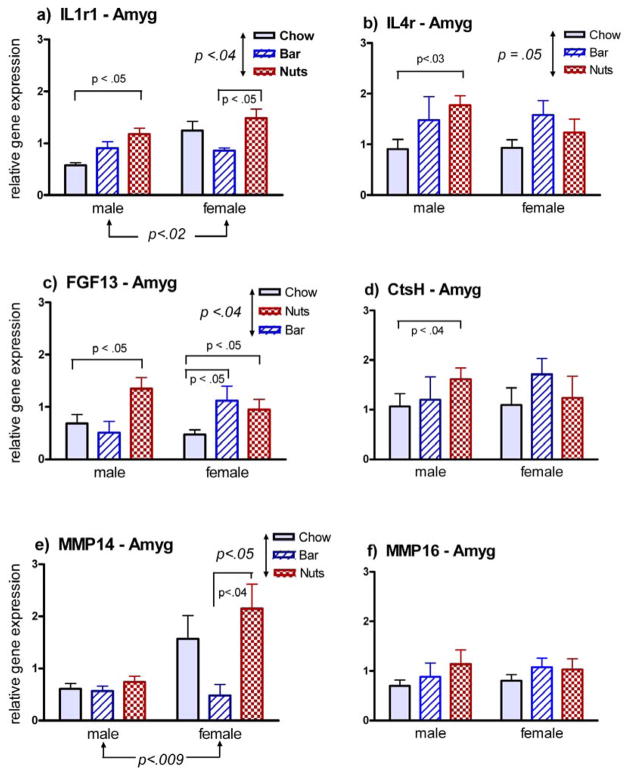
In the amygdala, differential expression in response to novel odors occurred in 5 of the 7 genes whose expression was quantified by qPCR (Fig. 2). In 4 of the 5 differentially expressed genes, the change was restricted to males only, and was characterized by up-regulation in response to the odor of cornnuts but not blueberry bar. In the one case of a differentially expressed gene in females (FGF13, Fig. 2c), the change was up-regulation in response to both blueberry bar and cornnuts. Overall differences in expression attributable to sex were observed for two genes, IL1r1 (Fig. 2a) and MMP-14 (Fig. 2e
Fig. 2.
Gene expression in the amygdala of male and female rats 2 h following exposure to the familiar odor of lab chow, or the novel odor of a blueberry bar (bar) or cornnuts (nuts). Arrows show differences attributable to either sex or odorant by 2-way ANOVA, at the indicated level of significance. Pair-wise comparisons are shown for 2-tailed U-tests at the indicated level of significance. Results for six of the seven genes tested are shown; no changes were seen in this brain region for CtsL.
FGF13.
Genes for numerous cytokines, cell matrix proteinases, and adhesion molecules are expressed in the olfactory pathway
Specific interleukins, cathepsins, and matrix metalloproteinases were up- or down-regulated in response to 1-time exposure to odor of cornnuts or blueberries
Sex-specific changes were observed in transcription of interleukin-1 and -4 receptors, FGF13, and in matrix metalloproteinase-14 in the amygdala
Sex-specific changes occurred in gene expression for the interleukin-1 receptor, cathepsin-L, and for matrix metalloproteinases-14 and -16 in the hippocampus
These results demonstrate cytokine and proteinase involvement in normal information processing functions in the brain, often in a sex-specific manner
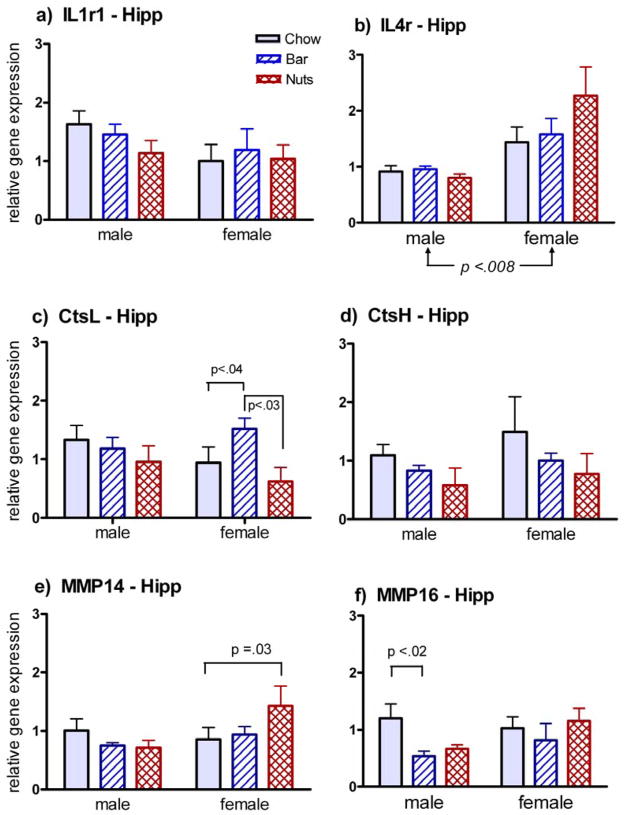
Fewer changes in gene expression were seen in the hippocampus, and those followed no consistent pattern (Fig. 3). Systematic sex differences were observed in only one case, an overall increase in IL4r expression in females (Fig. 3b), though pair-wise differences were also seen for females in up-regulation of CtsL with blueberry bars (Fig. 3c) and MMP-14 with cornnuts (Fig. 3e). Down-regulation of MMP-16 with exposure to blueberry bars (Fig. 3f) was the only change seen in the hippocampus of males.
Fig. 3.
Gene expression in the hippocampus of male and female rats 2 h following exposure to the familiar odor of lab chow, or the novel odor of a blueberry bar (bar) or cornnuts (nuts). Arrows show differences attributable to either sex or odorant by 2-way ANOVA, at the indicated level of significance. Pair-wise comparisons are shown for 2-tailed U-tests at the indicated level of significance. Results for six of the seven genes tested are shown; no changes were seen in this brain region for FGF13.
3. Discussion
The first objective of this study was to survey the presence of gene expression for cytokines and cell matrix and adhesion modulators in different components of the olfactory processing circuitry of the rat brain. We found that genes for up to 28 common cytokines are expressed in the rat amygdala and hippocampus, and from 25 to 34 cell matrix and adhesion molecules are expressed across all four brain regions studied. Interestingly, cytokines were minimally expressed in insular cortex and olfactory bulb (Fig. 1). This observation alone suggests an information processing role for cytokines in the amygdala and hippocampus
Real-time PCR verified changes in the expression of a number of these genes following a one-time presentation of an odor that the rat had never previously experienced. Changes were seen more frequently in the amygdala, and more often in males than females. However, in all three cases of a systematic sex difference, females showed a greater magnitude of up-regulation in response to novel odors than did males (Fig. 2a, 2e, 3b).
Changes in gene expression also tended to be stimulus specific. When changes did occur in males, they more often were responsive to cornnuts only (Figs.2a–d), while females were more likely to respond to both blueberry bar and cornnuts (Figs. 2c, 3c). While the prevalent theoretical assumption is that receptor specificity and precise circuitry provide the basis for conveying odorant identity to central processing areas of the brain (Belluscio et al., 2002; Mori et al., 1999)., the influence of hormones on the maturation and function of those circuits in a sexually dimorphic manner is also well recognized (Wood, 1997; Bakker et al., 2002). The amygdala is a critical integration center for processing olfactory information related to sexual recognition and behavior (Been and Petrulis, 2011; DiBenedictis et al., 2012).
Our finding of sex- and stimulus-specific changes in gene expression, especially in the amygdala, likely reflects a complex interaction between hormones, sexually dimorphic neuronal circuits, and different subcellular signaling pathways unique to specific circuits. The genes demonstrated in this study to be differentially expressed in response to novel odors make strong candidates for future studies using hormonal manipulation to explore the influence of sex and hormones on the processing of olfactory information.
The influence of cytokines on behavior and affect is well established (Larson and Dunn, 2001). The robust response in this study of genes for both IL1r1 and IL4r in the amygdala provides additional evidence that these cytokines play a role in the normal processing of information by brain cells (Vitkovic et al., 2000).
Cathepsins H and L are endopeptidases that generate peptide neurotransmitters and neuromodulators (Brguljan et al., 2003; Hook et al., 2008; Yasothornsrikul et al., 2003). The possibility that cathepsins play a specific role in neuronal plasticity has gained recent traction (Stahl et al., 2007; Wendt et al., 2008; Zaidi and Kalbacher, 2008). This study shows that cathepsins H and L should be considered as candidates for involvement in the neuroplastic basis of olfactory learning.
Matrix metalloproteinases play a key role in normal development as well as pathological conditions (Candelario-Jalil et al., 2009, DeRosa et al., 2008, Bar-Or et al., 2003) Wright and co-workers ((Meighan et al., 2007; Meighan et al., 2006; Olson et al., 2008; Wright and Harding, 2004; Wright and Harding, 2009) have shown that matrix metalloproteinases contribute to synaptic remodeling that could underlie neural plasticity. Our results demonstrate that mere exposure to a novel odor alters gene expression for these enzymes, consistent with those suggestions.
Of particular relevance to this study is that gene expression for some metalloproteinases appears to be regulated by certain cytokines (Contasta et al., 1999; Gottschall and Deb, 1996). For instance, IL-1 plus oncostatin up-regulates the expression of MMP-14 but down-regulates MMP-16 (Milner et al., 2006). In this study, MMP-14 and MMP-16 likewise responded differently to novel odors, in both the amygdala and hippocampus. Thus these two enzymes may likewise play a role in modifying the neural circuitry underlying olfactory learning. Mechanistic studies aimed at unraveling the regulatory interactions among the cytokines would be a next logical step in this research.
Our results do not show conclusively that the genes we studied are involved in olfactory learning. Carefully controlled studies using learning paradigms, such as social transmission of olfactory information (Wrenn, 2004), or conditioned odor aversion (Grigson et al., 1997) should be carried out to reveal which aspects of the behavioral experience (e.g., short-term or long-term consolidation, retrieval, or modulation by affective state) are linked to specific cellular and molecular events. Our data provide a number of candidate molecular targets for focusing those investigations. In conjunction with good anatomical localization, such studies should lead to better insights into neural plasticity in the olfactory system at the cellular level
4. Experimental Procedures
4.1 Animals
This study reports the results of several independent experiments with different groups of young adult rats (Rattus norvegicus, Sprague-Dawley) of both sexes, ranging from 8 to 12 weeks of age. The research was approved by the Institutional Animal Care and Use Committee of the University of Texas at El Paso, in accordance with the United States Public Health Service Guide to the Care and Use of Laboratory Animals.
In preliminary experiments to assess the influence of social transmission, only males were used, to eliminate variation due to sex (Irwin and Byers, 2005). It became apparent, however, that odor by itself could affect gene expression, so new experiments were designed to focus only on exposure to novel odors, and to include both females and males. Sample sizes varied somewhat due to the availability of animals bred in house within a consistent age range. In all experiments rats were housed in group cages in a constant-temperature vivarium on a 12L:12D photoperiod, and fed Purina Lab Chow® with water available ad lib until the last four days of the experiment. The stage of estrus for each female was assessed by cytological examination of vaginal lavage samples obtained immediately after decapitation.
4.2 Olfactory stimulation
On each of the final four days of the experiment, food was removed at about 0900 each morning. At about 1700, rats were individually moved to a single cage where they were exposed for 10 min to one of three odors from food just beyond the reach of their nose through a hole at the back of the cage. Control rats (Group C, n = 6 males and 11 females) were exposed to the familiar odor of lab chow. Group B rats (n = 6 males and 4 females) were exposed to the novel odor of blueberry snack bars (Kellogg’s NutriGrain®). Group N rats (n = 6 males and 10 females) were exposed to the novel odor of cornnuts (Kraft Foods, original flavor). Test foods were selected based on preliminary experiments to determine the rats’ preference for a sweet reward and an alternative that was non-sweet and of different texture. The lights went off at 1800. Rats were left in their individual cages for two hours following olfactory stimulation, then weighed and returned to their group home cages on the first three nights, or taken to a dark room and decapitated under red light on the final night.
4.3 Gene Array Analysis
To survey overall gene expression for cytokines and proteinase in the circuitry for olfactory information processing, brains were quickly removed, placed immediately in chilled phosphate-buffered saline, and taken to the dissection lab. Transverse slabs ~ 3 mm thick centered on the infundibulum were made from each brain. Bilateral samples of the amygdala, hippocampus, insular cortex, and olfactory bulb were later dissected from the frozen sections. Total RNA was isolated using the RNAqueous kit (Ambion®) following the manufacturer’s protocol. RNA was reverse transcribed to generate 16-UDP-labeled cRNA probes using TrueLabeling-AMP™ linear RNA Amplification Kit (SABioscience). Probes were hybridized to Oligo GEArrays (SABioscience) ORN-011 for Rat Inflammatory Cytokines and Receptors, and to ORN-013 for Rat Extracellular Matrix and Adhesion Molecules, containing 96 and 111 gene sequences, respectively. After hybridization, arrays were washed and prepared for detection using the GEArray™ Chemiluminescent Detection Kit D-01 (SABioscience). Chemiluminescent images were visualized and photographed with the FX Imager (Bio-Rad), and scanning densitometry software (Molecular One™, Bio-Rad) was used to quantify the intensity of individual spots. The results shown in Fig. 1 were obtained from male brain samples only, but subsequent analyses from females yielded images qualitatively indistiguishable from those for males. A complete list of genes available for hybridization on the arrays, and the ones for which hybridization was visibly detected, are provided in Supplementary Tables 1 and 2.
4.4 Gene selection
Seven genes were selected for quantification of mRNA levels as a measure of transcriptional gene expression on the basis of preliminary studies indicating the possibility that they could be involved in the process of olfactory learning (Byers and Irwin, 2008; Irwin and Byers, 2005). Originally, 13 genes were selected for further study based on an apparent change in level of expression due to social transmission at a confidence level of p<0.05. This was subsequently reduced to eight genes, based on insufficient levels of expression in some samples, or poorly performing primers. One of the eight -- FGF receptor activating protein 1 (Frag1) -- was not affected by novel odors in either brain region for either sex. The remaining seven genes reported here include the interleukin-1 receptor 1 (IL1r1), interleukin-4 receptor (IL4r), fibroblast growth factor 13 (FGF13), cathepsins H and L. (CtsH and CtsL), and matrix metalloproteinases -14 and -16 (MMP-14 and MMP-16)
4.5 Tissue collection and RNA extraction
Follow-up experiments were performed on additional cohorts of animals to analyze changes in the expression of individual candidate genes. Brains were quickly removed, cut into coronal sections 2–3 mm thick, then frozen quickly on dry ice. Micropunched samples of the amygdala and dorsal hippocampus were collected bilaterally with a blunted 16 gauge needle and transferred to an enzyme digestion solution. Samples were not taken from the insular cortex or olfactory bulb in this phase of the study because of the much lower level of expression of the target genes in these two regions (Fig. 1). Total RNA was then isolated from tissue samples using the MELT™ Total RNA Isolation kit (Ambion), following the manufacturer’s protocol for very small tissue sections.
4.6 Verification of differential gene expression
To obtain a realistic measure of inter-animal variation, total RNA was extracted from each regional sample from individual rats. mRNA was then reverse transcribed into unlabeled cDNA templates using the Verso™ cDNA Kit (ABgene, Thermo Scientific). Specific forward and reverse primers for candidate genes (Table 1) were designed using Primer Designer™, Version 2.0 (Scientific & Educational Software), and synthesized by IDT, Inc. (Coralville, IA). The FastStart SYBR Green Master® (Roche Diagnostics) fluorescent labeling kit and custom primers (0.4 mM) were used to perform qPCR using the iCycler™ (Bio-Rad) and the following protocol: 40 cycles, each consisting of 30 sec for melting, 30 sec for annealing, and 20 sec for elongation. Melt curve analysis confirmed a single amplification product for each target gene assayed.
Table 1.
Primers used for q-PCR, in the forward (F) and reverse (R) directions
| Gene | Sequence (5′ → 3′) |
|---|---|
| IL1r1- F | gcc agt cat ctg aag agc |
| IL1r1- R | gcc aag tgg taa gtg tgt c |
| IL4r - F | gct gag gtc tgt gct aag g |
| IL4r – R | cta tgc cag gac tgc tgt g |
| FGF13 – F | cct ctc ctt cct act gtc c |
| FGF13 – R | caa tgc cac tgt tcc ac |
| CtsH – F | ggc tat gga gaa cag aat gg |
| CtsH – R | gta tgg ctg gtg atg tc |
| CtsL – F | ggt ggt tgg cta tgg tta tg |
| CtsL – R | gaa ctc ctt cgg atg tag tg |
| MMP-14 – F | ctg aag gta gag cca gg |
| MMP-14 – R | ctt gtc cag cag tga acg |
| MMP-16 – F | gat gga cca aca gac cga g |
| MMP-16 - R | gga ctg aag aac ggc aga tac |
All samples were run in duplicate along with a standard curve to calculate copy numbers of mRNA for each gene, and normalized to ribosomal 18S RNA as an internal standard.
4.7 Statistical Analysis
To determine if stage of estrus affected gene expression in female rats, results from those in proestrus or estrus were first separated out from results for all females for each gene. Expression profiles were virtually identical for all rats compared to those in proestrus or estrus only in all but two cases, and in those, a two-way analysis of variance (ANOVA) confirmed that the proestrus/estrus females showed no significant differences in the expression of any gene from that shown by all females. Therefore, in all following analyses, stage of estrus was disregarded.
We next determined by two-way ANOVA if results differed systematically according to sex or to the odorants presented. Statistical interactions were frequent, indicating that odorants affected gene expression differently in the two sexes. A Mann-Whitney two-tailed U-test was used to test for individual pair-wise differences. This is a non-parametric test, used because of the substantial differences in variance among many of the samples. All statistical tests were carried out with software embedded in GraphPad® Prism 4 for Windows.
Supplementary Material
Many cytokines and cell matrix proteinases are expressed in the olfactory pathway.
Gene expression was altered in response to a single exposure to novel odors
Olfactory stimulation up-regulated IL1r1, IL4r, FGF13, and MMP-14 in the amygdala
Sex affected changes in IL1r1 and MMP-14 in amygdala, and IL4r in hippocampus
Cytokines and proteinases play a role in normal olfactory information processing
Acknowledgments
This work was supported by a grant (#R15-DC005179) from NIH-NICDC, and by infrastructure support from NIH-NCRR-RCMI (#G12-RR08124) to the University of Texas at El Paso.
Abbreviations
- IL1r1
interleukin-1 receptor 1
- IL4r
interleukin-4 receptor
- FGF13
fibroblast growth factor 13
- CtsH
cathepsin-H
- CtsL
cathepsin-L
- MMP-14
matrix metalloproteinase-14
- MMP-16
matrix metalloproteinase-16
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Louis N. Irwin, Email: lirwin@utep.edu.
Donna M. Byers, Email: dbyers2@utm.edu.
References
- Bakker J, Honda S, Harada N, Balthazart J. The aromatase knock-out mouse provides new evidence that estradiol is required during development in the female for the expression of sociosexual behaviors in adulthood. J Neurosci. 2002;22:9104–12. doi: 10.1523/JNEUROSCI.22-20-09104.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Or A, Nuttall RK, Duddy M, Alter A, Kim HJ, Ifergan I, Pennington CJ, Bourgoin P, Edwards DR, Yong VW. Analyses of all matrix metalloproteinase members in leukocytes emphasize monocytes as major inflammatory mediators in multiple sclerosis. Brain. 2003;126:2738–49. doi: 10.1093/brain/awg285. [DOI] [PubMed] [Google Scholar]
- Been LE, Petrulis A. Chemosensory and hormone information are relayed directly between the medial amygdala, posterior bed nucleus of the stria terminalis, and medial preoptic area in male Syrian hamsters. Horm Behav. 2011;59:536–48. doi: 10.1016/j.yhbeh.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belluscio L, Lodovichi C, Feinstein P, Mombaerts P, Katz LC. Odorant receptors instruct functional circuitry in the mouse olfactory bulb. Nature. 2002;419:296–300. doi: 10.1038/nature01001. [DOI] [PubMed] [Google Scholar]
- Brguljan PM, Turk V, Nina C, Brzin J, Krizaj I, Popovic T. Human brain cathepsin H as a neuropeptide and bradykinin metabolizing enzyme. Peptides. 2003;24:1977–84. doi: 10.1016/j.peptides.2003.09.018. [DOI] [PubMed] [Google Scholar]
- Byers D, Irwin L. Gene expression changes in the brain during olfactory learning are region and context dependent. Society for Neuroscience Annual Meeting. Neuroscience Meeting Planner, Program No. 362.18; Washington D.C. 2008. [Google Scholar]
- Candelario-Jalil E, Yang Y, Rosenberg GA. Diverse roles of matrix metalloproteinases and tissue inhibitors of metalloproteinases in neuroinflammation and cerebral ischemia. Neuroscience. 2009;158:983–94. doi: 10.1016/j.neuroscience.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contasta I, Berghella AM, Pellegrini P, Del Beato T, Casciani CA, Adorno D. Relationships between the activity of MMP1/TIMP1 enzymes and the TH1/TH2 cytokine network. Cancer Biother Radiopharm. 1999;14:465–75. doi: 10.1089/cbr.1999.14.465. [DOI] [PubMed] [Google Scholar]
- DeRosa DC, Ryan PJ, Okragly A, Witcher DR, Benschop RJ. Tumor-derived death receptor 6 modulates dendritic cell development. Cancer Immunol Immunother. 2008;57:777–87. doi: 10.1007/s00262-007-0413-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiBenedictis BT, Ingraham KL, Baum MJ, Cherry JA. Disruption of urinary odor preference and lordosis behavior in female mice given lesions of the medial amygdala. Physiol Behav. 2012;105:554–9. doi: 10.1016/j.physbeh.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dustin ML, Colman DR. Neural and immunological synaptic relations. Science. 2002;298:785–9. doi: 10.1126/science.1076386. [DOI] [PubMed] [Google Scholar]
- Gottschall PE, Deb S. Regulation of matrix metalloproteinase expressions in astrocytes, microglia and neurons. Neuroimmunomodulation. 1996;3:69–75. doi: 10.1159/000097229. [DOI] [PubMed] [Google Scholar]
- Grigson PS, Shimura T, Norgren R. Brainstem lesions and gustatory function: II.. The role of the nucleus of the solitary tract in Na+ appetite, conditioned taste aversion, and conditioned odor aversion in rats. Behav Neurosci. 1997;111:169–79. [PubMed] [Google Scholar]
- Hook V, Funkelstein L, Lu D, Bark S, Wegrzyn J, Hwang SR. Proteases for processing proneuropeptides into peptide neurotransmitters and hormones. Annu Rev Pharmacol Toxicol. 2008;48:393–423. doi: 10.1146/annurev.pharmtox.48.113006.094812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin L, Byers D. Cytokines induce changes in the functional association of brain cells during olfactory learning. J Neurochem. 2005;94(Suppl 2):39. [Google Scholar]
- Larson SJ, Dunn AJ. Behavioral effects of cytokines. Brain Behav Immun. 2001;15:371–87. doi: 10.1006/brbi.2001.0643. [DOI] [PubMed] [Google Scholar]
- Larson SJ, Romanoff RL, Dunn AJ, Glowa JR. Effects of interleukin-1beta on food-maintained behavior in the mouse. Brain Behav Immun. 2002;16:398–410. doi: 10.1006/brbi.2001.0634. [DOI] [PubMed] [Google Scholar]
- Meighan PC, Meighan SE, Davis CJ, Wright JW, Harding JW. Effects of matrix metalloproteinase inhibition on short- and long-term plasticity of schaffer collateral/CA1 synapses. J Neurochem. 2007;102:2085–96. doi: 10.1111/j.1471-4159.2007.04682.x. [DOI] [PubMed] [Google Scholar]
- Meighan SE, Meighan PC, Choudhury P, Davis CJ, Olson ML, Zornes PA, Wright JW, Harding JW. Effects of extracellular matrix-degrading proteases matrix metalloproteinases 3 and 9 on spatial learning and synaptic plasticity. J Neurochem. 2006;96:1227–41. doi: 10.1111/j.1471-4159.2005.03565.x. [DOI] [PubMed] [Google Scholar]
- Milner JM, Rowan AD, Cawston TE, Young DA. Metalloproteinase and inhibitor expression profiling of resorbing cartilage reveals pro-collagenase activation as a critical step for collagenolysis. Arthritis Res Ther. 2006;8:R142. doi: 10.1186/ar2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milward EA, Fitzsimmons C, Szklarczyk A, Conant K. The matrix metalloproteinases and CNS plasticity: an overview. J Neuroimmunol. 2007;187:9–19. doi: 10.1016/j.jneuroim.2007.04.010. [DOI] [PubMed] [Google Scholar]
- Mori K, Nagao H, Yoshihara Y. The olfactory bulb: coding and processing of odor molecule information. Science. 1999;286:711–5. doi: 10.1126/science.286.5440.711. [DOI] [PubMed] [Google Scholar]
- Olson ML, Meighan PC, Brown TE, Asay AL, Benoist CC, Harding JW, Wright JW. Hippocampal MMP-3 elevation is associated with passive avoidance conditioning. Regul Pept. 2008;146:19–25. doi: 10.1016/j.regpep.2007.07.004. [DOI] [PubMed] [Google Scholar]
- Stahl S, Reinders Y, Asan E, Mothes W, Conzelmann E, Sickmann A, Felbor U. Proteomic analysis of cathepsin B- and L-deficient mouse brain lysosomes. Biochim Biophys Acta. 2007;1774:1237–46. doi: 10.1016/j.bbapap.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitkovic L, Bockaert J, Jacque C. “Inflammatory” cytokines: neuromodulators in normal brain? J Neurochem. 2000;74:457–71. doi: 10.1046/j.1471-4159.2000.740457.x. [DOI] [PubMed] [Google Scholar]
- Wendt W, Lubbert H, Stichel CC. Upregulation of cathepsin S in the aging and pathological nervous system of mice. Brain Res. 2008;1232:7–20. doi: 10.1016/j.brainres.2008.07.067. [DOI] [PubMed] [Google Scholar]
- Wood RI. Thinking about networks in the control of male hamster sexual behavior. Horm Behav. 1997;32:40–5. doi: 10.1006/hbeh.1997.1403. [DOI] [PubMed] [Google Scholar]
- Wrenn CC. Social transmission of food preference in mice. Curr Protoc Neurosci. 2004;Chapter 8(Unit 8):5G. doi: 10.1002/0471142301.ns0805gs28. [DOI] [PubMed] [Google Scholar]
- Wright JW, Harding JW. The brain angiotensin system and extracellular matrix molecules in neural plasticity, learning, and memory. Progress in Neurobiology. 2004;72:263–293. doi: 10.1016/j.pneurobio.2004.03.003. [DOI] [PubMed] [Google Scholar]
- Wright JW, Harding JW. Contributions of matrix metalloproteinases to neural plasticity, habituation, associative learning and drug addiction. Neural Plast. 2009:579382. doi: 10.1155/2009/579382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasothornsrikul S, Greenbaum D, Medzihradszky KF, Toneff T, Bundey R, Miller R, Schilling B, Petermann I, Dehnert J, Logvinova A, Goldsmith P, Neveu JM, Lane WS, Gibson B, Reinheckel T, Peters C, Bogyo M, Hook V. Cathepsin L in secretory vesicles functions as a prohormone-processing enzyme for production of the enkephalin peptide neurotransmitter. Proc Natl Acad Sci U S A. 2003;100:9590–5. doi: 10.1073/pnas.1531542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi N, Kalbacher H. Cathepsin E: a mini review. Biochem Biophys Res Commun. 2008;367:517–22. doi: 10.1016/j.bbrc.2007.12.163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.