Abstract
The genome is folded into domains located in either transcriptionally inert or permissive compartments. Here we used genome-wide strategies to characterize domains during B cell development. Structured Interaction Matrix Analysis revealed that CTCF occupancy was primarily associated with intra-domain interactions, whereas p300, E2A and PU.1 bound sites were associated with intra- and inter-domain interactions that are developmentally regulated. We identified a spectrum of genes that switched nuclear location during early B cell development. In progenitors the transcriptionally inactive Ebf1 locus was sequestered at the nuclear lamina, thereby preserving multipotency. Upon development into the pro-B cell stage Ebf1 and other genes switched compartments to establish de novo intra- and inter-domain interactions that are associated with a B lineage specific transcription signature.
The genome is not merely organized as a linear structure but possesses a distinct three-dimensional configuration. It must fold into an elaborate and coherent pattern to allow genomic elements to find each other with the appropriate frequencies. How the chromatin fiber is organized into higher-order topologies remains to be determined. In the late 20th century, distinct folding arrangements for chromosome topologies were proposed. These involved helical, radial or combined loop-helical folding of the genome1,2. Imaging studies using electron microscopy have suggested that chromosomes consist of clusters of loops separated by linkers3,4. Spatial distance measurements and formaldehyde-cross linking approaches in conjunction with modeling strategies agree well with the organization of the genome into chromatin globules consisting of bundles of loops, related to that originally proposed by the Multi-Loop-Subcompartment (MLS) model5–8.
During developmental progression genes frequently change their nuclear neighborhoods9,10. For example, upon commitment to the B cell lineage the Igh locus moves from the nuclear periphery to more centrally located domains11–14. Following productive Igh locus rearrangement, the non-productive Igh allele moves to the transcriptionally repressive compartment to interact with Igk loci15. Similarly, the Igk locus relocates during B cell development. In small pre-B cells, one of the Igk alleles becomes associated with the repressive compartment to favor rearrangement of the Igk allele in the transcriptionally permissive compartment16.
Recent chromosome capture studies have provided insight into the folding patterns of genomes at a global scale17–23. These studies revealed that genomes are organized into transcriptionally repressive and permissive compartments17,19. The transcriptionally permissive compartments, in turn, are folded into distinct domains organized by intra-domain interactions that involve CTCF18,21,. In the present study we have used genome-wide strategies to characterize chromatin compartments and domains during B cell development. We have identified distinct classes of anchors acting at different length scales. Whereas CTCF occupancy was primarily associated with intra-domain interactions, p300, E2A, Pax5 and PU.1 were involved with both intra- and inter-domain interactions. Intra-domain and inter-domain interactions, involving p300, E2A, Pax5 and PU.1, but not CTCF were developmentally regulated. We found that a large ensemble of genes relocated during developmental progression from the multipotent to the committed pro-B cell stage. Prominent among these was Ebf1, a key developmental regulator that orchestrates the B cell fate. The switching of genes between compartments was closely linked with changes in transcription signatures. Specifically, we found that the Ebf1 locus was tightly associated with the nuclear lamina in multipotent progenitors but relocated away from the lamina in committed pro-B cells. We suggest that the sequestration of key developmental regulators, underpins the mechanism by which the multipotent progenitor cell stage is enforced. Furthermore, we propose that the relocation of chromatin globules between the transcriptionally permissive and repressive compartments and global changes in intra- as well as inter-domain interactions mediated by active enhancer repertoires, underlie the mechanism by which a B lineage specific program of gene expression is established.
RESULTS
Highly conserved folding patterns of B cell genomes
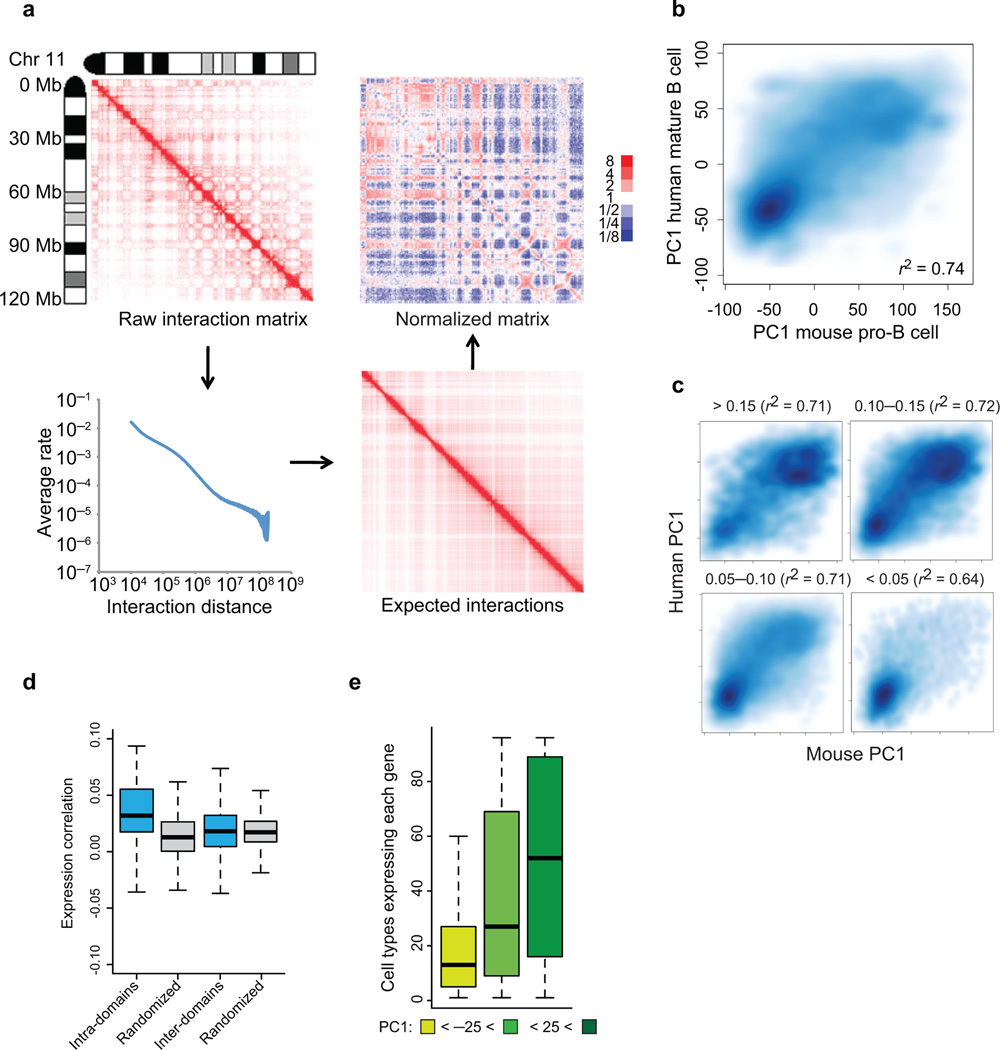
Recent data have demonstrated that spatial distance measurements across the Igh locus agree well with a topology predicted by the MLS model5,6,7. To determine whether at a global scale the pro-B genome architecture is similarly organized we applied Hi-C17,19. Briefly, Rag1- deficient pro-B cells were fixed with ethylene glycol bis(succinimidylsuccinate) (EGS) and formaldehyde or formaldehyde alone, digested with HindIII, filled-in with biotinylated nucleotide, ligated at highly diluted concentrations and analyzed using high-throughput DNA sequencing. From the filtered interaction data, genome-wide interaction matrices of each chromosome were generated (Fig. 1a; Supplementary Table 1; Supplementary Fig. 1a–c) (for a detailed description for filtering and statistical analyses see Supplementary Methods). This analysis revealed clusters of interacting genomic elements across the entire length of pro-B chromosomes (Fig. 1a, Supplementary Fig. 2). The very large majority of genomic interactions were local (<3 Mb) as previously reported (Fig. 1a)17. To identify patterns within the interaction matrix we applied principle component analysis to each chromosome (Supplementary Fig. 1a; top; second row)17. This analysis confirmed that chromosome 11 segregated into transcriptionally permissive and inert compartments (Supplementary Fig. 1a). The coordinates for each region along the first principal component (PC1 values) were highly reproducible across distinct Hi-C replicate experiments with positive values reflecting transcriptionally active compartments and negative values indicating repressive/inert compartments (Supplementary Fig. 1d) and remarkably well conserved between the genomes of murine pro-B cells and syntenic regions derived from human mature B lineage cells (Supplementary Fig. 1a; top; rows 2 and 3; Fig. 1b)17,23. The interactomes of EGS fixed cells, Abelson-transformed pro-B cells, embryonic stem cells and Rag1−/− pro-B cells were also quite similar (Supplementary Fig. 1d–f)22,23. To compare the degree of sequence conservation between transcriptionally active and repressive compartments, as determined by PC1 values, we segregated genomic regions on the basis of PhastCons scores (Fig. 1c)24. PhastCons scores are based on a 38-organism alignment, which assess the probability that DNA sequences are under selective pressure. We found that regions with high PhastCons scores were predominantly present in transcriptionally permissive compartments as compared to the heterochromatic environment (Fig. 1c). To determine domain size across the pro-B cell genome we computed the average size of continuous regions showing positive PC1 values. We found that the pro-B cell genome is predominantly organized as 0.5-3 Mb chromatin globules (Table 1).
Figure 1.
The folding pattern of the pro-B cell genome. (a) Strategy for Hi-C data normalization from expected interaction frequencies. Normalized genome-wide contact matrix revealing intrachromosomal interactions involving chromosome 11. Indicated are the ratios of observed versus expected reads. Blue pixels represent lower than expected whereas red pixels reflect higher than expected interaction frequencies. (b) Conservation of PC1 values of murine pro-B cells versus syntenic regions derived from human mature-B lineage cells. Scatter plot of a genome-wide comparison of PC1 values using 50 kb windows derived from mouse chromosome 11 of pro-B cells and syntenic regions derived from human mature-B lineage cells. Intensities of blue pixels correspond to density of indicated regions. (c) Regions with high average PhastCons scores are more closely associated with transcriptionally permissive compartments to regions exhibiting low average PhastCons scores. (d) Coordinate patterns of gene expression and gene localization correlate well within chromatin domains but not with genes dispersed across domains. Distribution of average pair-wise correlations of gene expression values across 96 mouse tissues. Average pair-wise correlations were computed for each active domain for a minimum of ten genes. Values were calculated by comparing genes within the same domain (intra-domain) or by comparing genes in different domains (inter-domain) (P < 1 x10−7) (e) Genes located in the transcriptionally repressive compartment show a more lineage restricted pattern of gene expression as compared to loci positioned in a transcriptionally permissive compartment. For each annotated gene we calculated the number of tissues with detectable expression. The whisker-box plot shows the distribution of these values for genes located in the transcriptionally repressive compartment (PC1 < −25), genes located in regions that show intermediate PC1 values (−25 < PC1< 25), and genes located in the transcriptionally permissive compartment (PC1 > 25). Single Hi-C analyses were performed using either EGS or formaldehyde fixed cells. Gene expression was obtained from BIOGPS that were derived from two independent experiments (GSE10246).
Table 1.
Domain size of the pro-B cell genome compares well with the MLS model. Distribution of domain size across the pro-B cell genome as defined by continuous regions of positive PC1 values across the genome. Upper panel shows distribution of domain size in the transcriptionally permissive compartment. Lower panel indicates distribution of domain size in the repressive compartment.
| Domain size | Domains | Active region |
|---|---|---|
| 5 Mb+ | 37 | 25.0% |
| 3–5 Mb | 51 | 18.4% |
| 1–3 Mb | 138 | 22.5% |
| 500 kb–1 Mb | 177 | 11.7% |
| < 500 kb | 22.4% | |
| Domain size | Domains | Inert region |
|---|---|---|
| 5 Mb+ | 38 | 17.7% |
| 3–5 Mb | 76 | 19.2% |
| 1–3 Mb | 311 | 36.5% |
| 500 kb–1 Mb | 256 | 12.2% |
| < 500 kb | 14.3% | |
The organization of the genome into distinct chromatin globules/domains raises the question as to whether domains are associated with lineage-specific programs of gene expression. To explore this possibility we computed the average pair-wise correlation coefficient of genes in each active domain comprised of a minimum of ten genes using gene expression values derived from 96 mouse tissues, derived from BioGPS (http://biogps.org). These values were then compared to expression patterns of genes dispersed across different chromatin domains. We found that genes that were co-localized within a domain showed a greater tendency to be coordinately expressed as compared to genes located in different domains (P < 1x 10−7) (Fig. 1d). To determine how lineage-specific patterns of gene expression relate to the location of genes in a transcriptionally repressive or permissive compartment, we computed the number of distinct cell types that expressed the ensemble of annotated genes as a function of PC1 values. We found that genes located in inert compartments exhibited a more cell-type restricted pattern of expression as compared to genes showing a more ubiquitous pattern of gene expression (Fig. 1e). These as well as previous observations indicate that the folding patterns of genomes of different cell types and species are highly conserved, predominantly organized as 0.5-3 Mb domains, comprised of clusters of genes that show a tendency to be coordinately transcribed.
Recruitment of promoters to enhancers modulate eRNA abundance
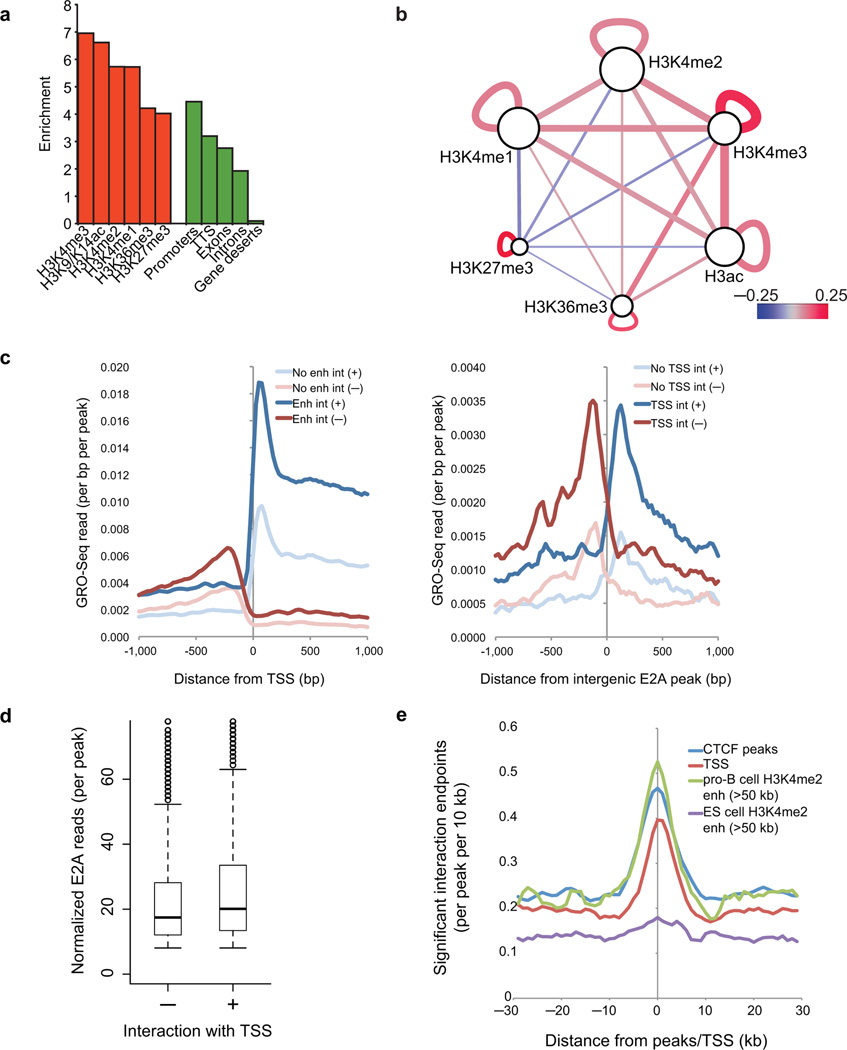
To determine whether and how epigenetic marks pair at genomic features, we computed significant interactions P<0.05) between loci using a window of 10 kb. We identified a total of 64,101 high-confidence interactions in pro-B cells (False Discovery Rate < 10%). Interacting DNA elements were examined for enrichment of promoters (H3K4me3, H3K4me2, H3K9/K14ac), enhancers (H3K4me2/H3K4me1), elongation/splicing (H3K36me3) and repression (H3K27me3) (Fig. 2a)25. We found that the entire spectrum of epigenetic marks was highly enriched across loop attachment regions (Fig. 2b). Notably, loop attachment regions showing H3K27me3 deposition were severely depleted for interactions involving active promoter and enhancer elements, indicating that islands of H3K27me3 (silencing) and H3K4me2 (activation) deposition are spatially segregated across the pro-B cell genome25.
Figure 2.
The transcriptionally permissive compartment is spatially segregated into islands of H3K27me3 and H3K4me2. (a) Enrichment of epigenetic marks and genomic annotations such as promoters, transcription start sites, exons, introns and gene deserts (>100 kb from a Refseq gene) at the end points of significant interactions relative to random genomic positions (10 kb window). (b) The transcriptionally permissive compartment is spatially segregated into islands of H3K27me3 and H3K4me2. The relative frequency by which epigenetic marks were associated across loop attachment points was computed and then visualized in Cytoscape. The relative frequency by which epigenetic marks were associated within loop attachment points is indicated by node size. The significance/reproducibility of tethering, in terms of P-values, was visualized by the edge width of connecting paired marked elements. The color (red-blue) is representative of the log ratio of observed frequencies relative to expected frequencies (i.e. the strength of the association). (c) Genome-wide repertoires of enhancers act by looping to promoter regions across large genomic distances. Left, the density of nascent RNAs as determined by GRO-Seq as a function of genomic distance from the transcription start site for promoters that showed looping towards putative enhancers defined by promoter-distal H3K4me2 peaks. Both sense (bold blue) and anti-sense transcription (bold red) initiated from the transcription start site are shown. Nascent transcripts initiated from promoters in the absence of enhancer interactions are indicated for sense (light blue) and anti-sense (light red) transcription. Right, the density of nascent RNAs as determined by GRO-Seq as a function of genomic distance from E2A binding sites located in intergenic H3K4me2 peaks (putative enhancer regions) for enhancers. Both sense (bold blue) and anti-sense transcription (bold red) initiated from the putative enhancer regions are shown. Nascent transcripts initiated from the enhancer regions in the absence of interactions with transcription start site are indicated for sense (light blue) and anti-sense (light red) transcription. (d) Distribution of the normalized number of E2A ChIP-Seq reads per peak at peaks either interacting or lacking an interaction with nearby TSS. (e) Promoter and enhancer elements frequently function as anchors across the pro-B cell genome. Distribution of loop attachment points as a function of genomic distance from transcription start sites (TSS) and transcription factor binding sites (referred to as peaks) are indicated. ChIP-Seq and GRO-Seq experiments were performed one time.
To determine how genomic interactions between regulatory elements relate to nascent transcription at promoter and enhancer regions, we plotted the density of nascent RNAs as determined by GRO (Global nuclear Run On)-Seq as a function of genomic distance from transcription start sites and sites of promoter-distal E2A occupancy (Fig. 2c). Both sense and anti-sense transcription were substantially enhanced for those transcription start sites that showed tethering with putative enhancer elements (Fig. 2c; left). To determine how nascent transcription initiated at enhancer elements (eRNAs) relates to looping to transcription start sites, we plotted the relative abundance of nascent transcripts at intergenic putative enhancer regions bound by E2A (H3K4me2+) (Fig. 2c; right). eRNA abundance was greatly elevated at E2A-bound regions that interact with promoters relative to those that do not (Fig. 2c). However, the degree of E2A occupancy only modestly affects nascent transcript abundance (Fig. 2c,d). Consistent with these observations we found that whereas enhancer elements in the pro-B cell genome are closely associated with B cell-specific interaction end-points, enhancer regions identified in embryonic stem cells were not (Fig. 2e). These data imply that eRNA abundance is elevated at enhancers that interact with promoter regions, suggesting that eRNA transcription is initiated, at least in part, by looping of polymerase II from promoter to enhancer elements.
Putative anchors acting at different length scales
To identify potential anchors associated with the pro-B cell interactome, we focused our analysis on active enhancers and factors known to be involved in early B cell development. Active enhancers marked by p300, as well as factors such as E2A, PU.1, EBF1, CTCF and Rad21 were significantly enriched at loop attachment regions (Fig. 3a). However, we found that anchored regions associated with CTCF and lineage-specific transcription factors (E2A, PU.1, EBF1, FOXO1 were significantly depleted as compared to interactions among themselves (Fig. 3b). Hence, two distinct classes of factors are closely associated with loop attachment regions. On the one hand, lineage-restricted factors pair at regulatory elements with themselves or with other lineage-specific transcription factors, whereas either CTCF-bound sites primarily associate with themselves.
Figure 3.
Two distinct classes of anchors establish the pro-B cell interactome. (a) Potential anchors associated with the pro-B cell interactome are indicated. The enrichment for E2A, PU.1, CTCF, Rad21, Ebf1, c-Myc and p300 occupancy at loop attachment regions are shown. (b) Distinct putative anchors across the pro-B cell interactome are visualized in Cytoscape. The relative frequency by which putative anchors were associated with loop attachment points is indicated by node size. The significance/reproducibility of tethering, in terms of P-values, is visualized by the edge width of connecting paired marked elements. The color (red-blue) is representative of the ratio of observed frequencies relative to expected frequencies (i.e. the strength of the association). (c) Structured Interaction Matrix Analysis (SIMA) identifies distinct classes of anchors acting at different length scales across the pro-B cell interactome. Binding sites within domains were pooled and examined for interactions across the domains to binding sites in other domains (left and middle). These values were compared to peak positions that were randomized 10,000 times and P-values associated with enrichment for interactions were calculated (middle and right). (d) Distinct classes of anchors act at the different length scales. Compartmental interactions across the transcriptionally permissive compartment were examined for enrichment of interactions at sites exhibiting CTCF and E2A occupancy. One independent Hi-C experiment was performed to generate the data presented here. ChIP-Seq experiments were performed in two independent experiments for CTCF and one experiment for p300 and c-myc.
The range of detected significant interactions (FDR<10%), using endpoints spanning windows of 10 kb, was limited to approximately 3 Mb given the sequencing depth (Fig. 1a). Significant interactions were primarily observed within chromatin domains rather than between domains due to the increased Hi-C read coverage for shorter genomic distances. To characterize inter-domain interactions, where Hi-C read pairs were sparse, we developed a novel strategy, named Structured Interaction Matrix Analysis (SIMA). Specifically, we boosted sensitivity by pooling sites of factor occupancy within domains and analyzed their collective Hi-C read counts (Fig. 3c). These values were next compared to peak positions that were randomized within the same domains and P-values associated with enrichments were calculated (Fig. 3c). For domain comparisons with a P-value less than 0.05, we computed the ratio of the value from observed peak positions relative to the average of randomized peak positions plotted across the interaction matrix (Fig. 3d). Using SIMA we found intriguing differences between interactions involving either CTCF or E2A (Fig. 3d; Supplementary Fig. 3). Whereas CTCF interactions were primarily enriched across regions involved in intra-domain interactions, enrichment for E2A occupancy was not restricted to looping within domains but rather appears to be associated with both intra- and inter-domain interactions (Fig. 3d). Thus, SIMA revealed the presence of distinct classes of anchors associated with either intra- and/or inter-domain interactions.
Sequestration of the Ebf1 locus at the nuclear lamina
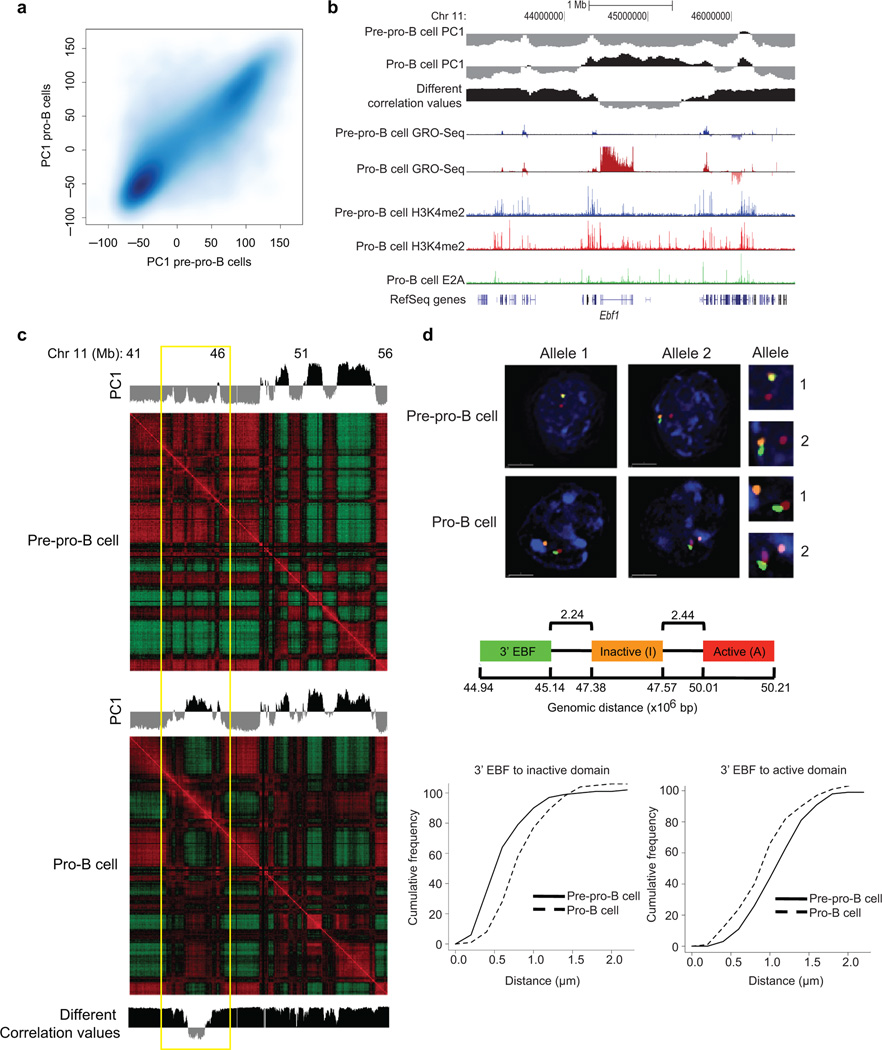
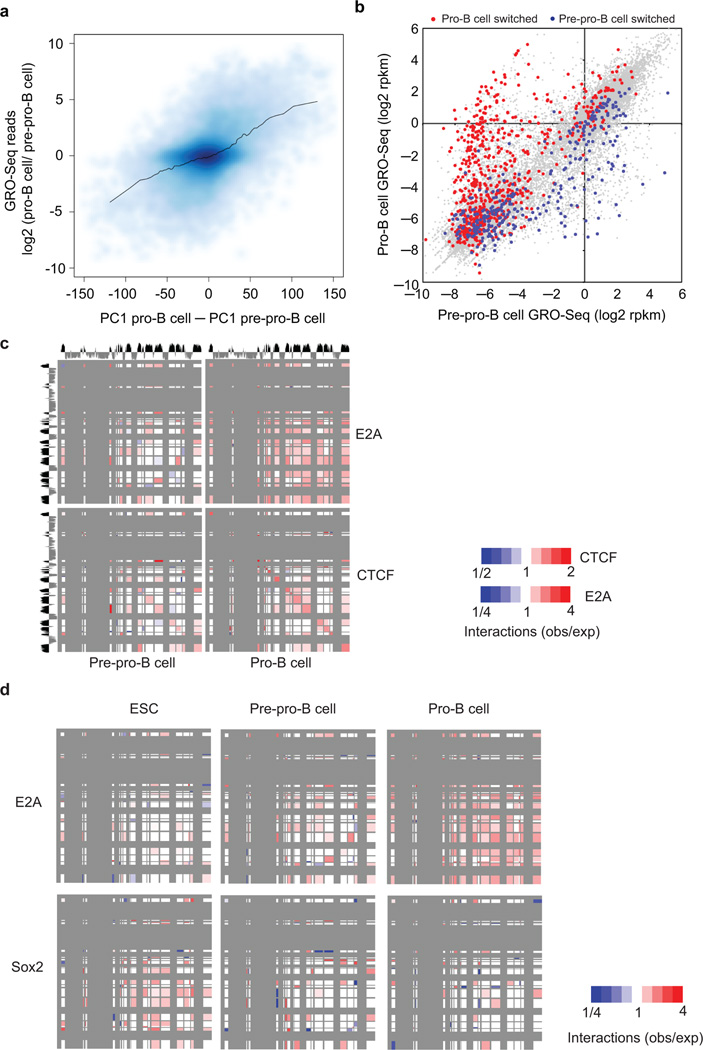
To determine whether developmental progression from the pre-pro-B to the pro-B cell stage is associated with changes in long-range genomic interactions, the interactomes of pre-pro-B cells were generated using Hi-C and compared to those derived from pro-B cells. The pre-pro-B and pro-B interaction matrices were quite similar (Fig. 4a). However, a fraction of loci differed in PC1 values (Fig. 4a and Supplementary Fig. 4a). To further evaluate these differences, Pearson correlation matrices were generated and directly compared (Fig. 4b,c). This analysis revealed a subset of loci that showed PC1 values that differed between pre-pro-B and pro-B cells (Fig. 4b, Supplementary Fig. 4b,c; Supplementary Table 2). Conspicuous among these loci were the Ebf1, Foxo1, Igk and Igl light chain loci that repositioned from the repressive to permissive compartment (Supplementary Fig. 4 and Supplementary Table 2).
Figure 4.
Switching compartments between transcriptionally repressive and permissive nuclear compartments during B lineage specification. (a) B cell development incurs changes in compartment structure. Scatter density plot between pre-pro-B and pro-B defined PC1 values at 25 kb intervals. Genomic regions that switch nuclear environments during the transition from the pre-pro-B to the pro-B cell stage are shown by signals that are located away from the diagonal. (b) Ebf1 locus depicting PC1 values of chromosome 11 of pre-pro-B and pro-B cells in a region surrounding the Ebf1 locus. Inter-experimental correlation is also shown. Nascent RNA (GRO-Seq) and H3K4me2 and E2A ChIP-Seq read densities are indicated. (c) Correlation matrices were derived from chromosome 11 for pre-pro-B and pro-B cells by correlating the normalized interaction ratios for each 25 kb interval against all other regions. The differential correlation values were calculated by comparing the two correlation matrices at each loci. Yellow box indicates the genomic region encoding Ebf1. (d) 3D-FISH in nuclei derived from pre-pro-B and pro-B cells using fluorescently labeled BACs that span three distinct regions across chromosome 11 including the domain containing Ebf1 locus. Digitally magnified pictures of the domains adjacent to the Ebf1 locus are shown. BACs (shown in red, green and orange) were directly labeled with dUTP conjugated to Alexa Fluor dyes. Nuclei were visualized by DAPI staining (blue). Genomic organization of the Ebf1 locus and adjacent regions are indicated. BAC probes specific to the Ebf1 domain (green) and the adjacent constitutively inert domain (orange) as well as a distally located constitutively permissive domain (red) are shown. Lower panel shows the distribution of spatial distances between the probes measured for 102 and 106 alleles in pre-pro-B and pro-B cells, respectively. Data obtained from two 3D-FISH experiments were added and presented.
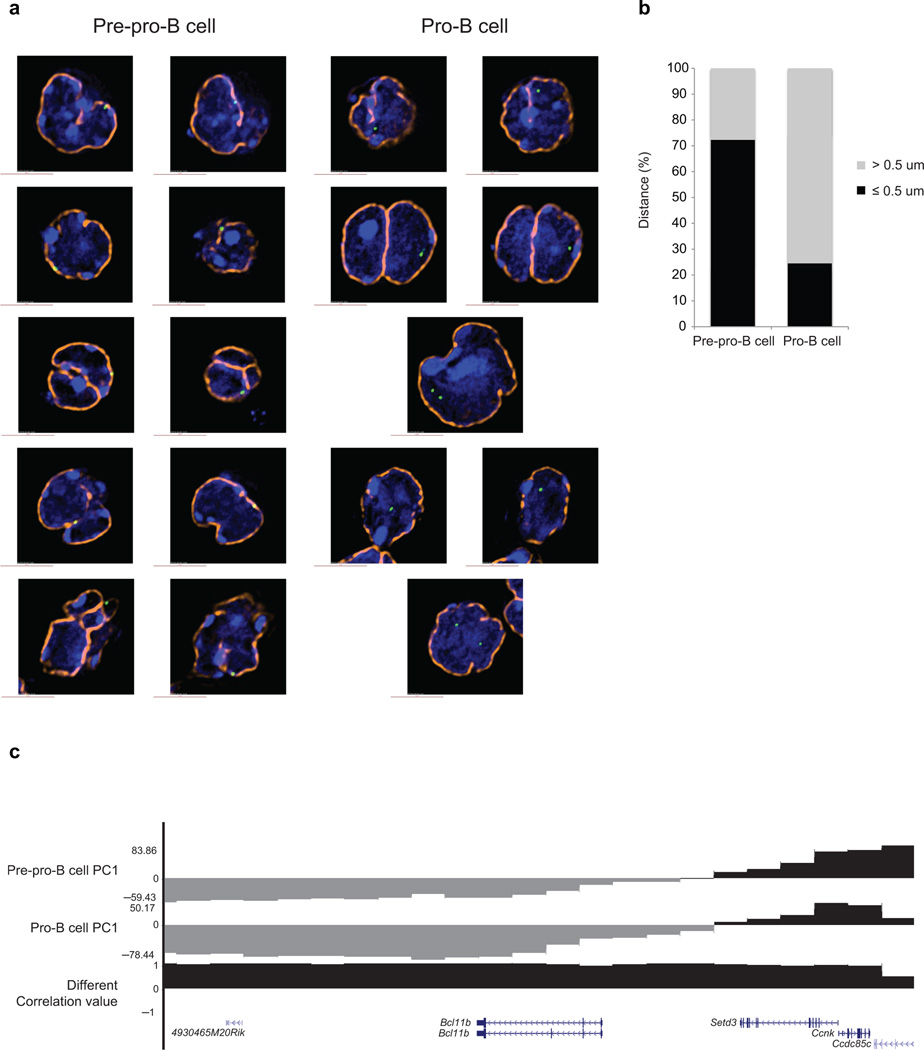
To verify the changes in compartmentalization at the single-cell level we performed three-dimensional fluorescence in situ hybridization (3D-FISH) analysis in conjunction with epifluorescence microscopy. Fluorescently labeled probes were generated to detect Ebf1 localization as well as genomic regions adjacent to the Ebf1 locus (Fig. 4d). Consistent with the Hi-C analysis, the Ebf1 domain in pre-pro-B cells was closely associated with the neighboring region located in the repressive compartment whereas in pro-B cells the Ebf1 locus interacted more frequently with genomic elements localized in the transcriptionally permissive compartment (Fig. 4d). To examine in greater detail how the Ebf1 locus was associated with the transcriptionally repressive environment, we examined the position of the Ebf1 locus with respect to the nuclear lamina. To this end, pre-pro-B and pro-B cells were fixed with formaldehyde, stained for lamina and analyzed. Notably, in pre-pro-B cells the Ebf1 locus was closely associated with the nuclear lamina, whereas in committed Rag1-deficient pro-B cells it was primarily located away from the nuclear lamina (Fig. 5a, 5b).
Figure 5.
The Ebf1 locus is closely associated with the nuclear lamina in pre-pro-B cells. (a) Immuno-FISH in nuclei derived from pre-pro-B and pro-B cells using a fluorescently labeled BAC encoding the Ebf1 domain as in previous figure and lamin antibodies. Digitally magnified pictures of nuclear location of the Ebf1 locus in pre-pro-B and pro-B cells are shown. BACs (shown in green) were directly labeled with dUTP conjugated to Alexa dyes. Nuclei were visualized by DAPI staining (blue). Nuclear lamina are shown in orange. (b) The graph shows the fraction of spatial distances (<500 nm) separating the Ebf1 domain from the nuclear lamina in pre-pro-B and pro-B cells, respectively. 112 and 102 measurements were done in pre-pro-B and pro-B cells, respectively. (c) PC1 values for the Bcl11b locus in pre-pro-B and pro-B cells are indicated. Data obtained from two experiments were added and presented.
To determine whether developmental regulators associated with alternative cell lineages were also sequestered in the transcriptional repressive compartment, PC1 values were determined for the GATA1, Gfi1, Tcf7, Cebpa, Cebpb, Bcl11b and Id2 loci. The majority of these loci were located in the transcriptional permissive compartment in both multipotent progenitors and pro-B cells. Notably, however, we found that in both cell types the Bcl11b locus was associated with the transcriptionally repressive compartment (Fig. 5c). Thus, these observations indicate that in multipotent progenitors, the Ebf1 and Bcl11b loci, encoding for key regulators that promote the B and T cell fates, respectively, are positioned in the transcriptionally repressive compartment.
Nuclear repositioning, epigenetics and gene expression
To examine whether spatial repositioning of loci relates to differences in programs of gene expression, we plotted the ratio of nascent RNA abundance in pro-B versus pre-pro-B cells against PC1 values (Fig. 6a). Indeed, loci that switched from a transcriptionally repressive to permissive compartment showed an increase in nascent RNA abundance whereas loci that relocated from a transcriptionally permissive to a repressive compartment exhibited a decrease in nascent transcripts (Fig. 6a). Globally, in pro-B cells nascent transcript abundance of 1534 was elevated whereas 1623 annotated genes showed decreased amounts as compared to prepro-B cells (Fig. 6b). Of the genes that showed increased transcript abundance in pro-B cells, 240 (16%) were found in regions that switched from a repressive to a permissive environment, while 87 (5%) of the genes whose expression declined in pro-B cells were found in compartments that changed from transcriptionally permissive to repressive compartments (Fig. 6b and Supplementary Tables 2,3). Regions associated with transcription of non-coding RNAs, micro-RNAs as well as eRNAs showed similar patterns (data not shown). As predicted, we found that E2A, EBF1 and Foxo1 occupancy were significantly enriched across domains that were actively transcribed in pro-B cells (Supplementary Fig. 5a).
Figure 6.
Repositioning of loci during developmental progression from a transcriptionally repressive to a permissive compartment is accompanied either with activation of gene expression or silencing by deposition of H3K27me3. (a) Switching nuclear environments is tightly linked with changes in nascent transcription as measured by GRO-Seq. For each 25 kb interval of the genome, the difference in nascent RNA read density (log2) was plotted versus the differential PC1 values derived from pre-pro-B and pro-B cells. Black line represents moving average values for 100 regions. (b) Relocating chromatin domains from an inert to a transcriptionally permissive compartment during developmental progression of B-lineage cells is associated with changes in gene expression. Pre-pro-B and pro-B RPKM (log2) values for RefSeq genes were plotted against each other. Red dots represent genes located within regions that switched from a transcriptionally repressive to a transcriptionally permissive compartment. Blue dots represent genes in regions that switched from a transcriptionally permissive compartment in pre-pro-B to an inert compartment in pro-B cells. (c) Loop-attachment points associated with E2A occupancy in pro-B cells change during developmental progression. E2A and CTCF binding sites found in pre-pro-B and pro-B cell domains were examined using SIMA. Specifically, reads were pooled and examined for interactions across the transcriptionally permissive environment to binding sites in other compartments. These values were compared to peak positions that were randomized and P-values associated with enrichment for paired interactions were calculated and visualized with red boxes representing higher than expected frequencies and blue boxes representing lower than expected frequencies. Matrices on the left use pre-pro-B Hi-C interactions for SIMA calculations and matrices on the right use pro-B Hi-C interactions. (d) Loop-attachment points enriched for E2A and Sox2 were examined in embryonic stem cells (ESC), pre-pro-B and pro-B cells using SIMA. E2A binding sites were identified in the pro-B cell genome whereas Sox2 occupancy was derived from embryonic stem cells. The data presented were derived from a single Hi-C experiment.
However, not the entire spectrum of genes that switched from the repressive to permissive compartment showed an increase in nascent transcription (Fig. 6b). Rather a substantial fraction of genes showed no or minimal changes in nascent RNA abundance (Fig. 6b). To examine this group of genes in more detail, we compared deposition of H3K27me3 to nascent transcript abundance for genes that switched from the repressive to the permissive compartment. Notably, we found that H3K27me3 abundance was enriched across genes that switched from the transcriptionally repressive to the permissive compartment but whose nascent transcript amounts were equivalent in pre-pro-B versus pro-B cells (Supplementary Fig. 5b). Thus, repositioning of loci during developmental progression from the transcriptionally repressive to the permissive compartment is accompanied either by activation of gene expression or silencing by deposition of H3K27me3.
Repositioning of genes during ontogeny
The findings described above indicate large-scale changes in compartmentalization of genes during B cell development. Do such changes also occur during ontogeny? To address this question we compared PC1 values derived from the pro-B and embryonic stem cell interactomes (Supplementary Fig. 6a). Indeed, we found a subset of loci that appeared to have relocated from a transcriptionally permissive chromatin environment in embryonic stem cells to a repressive environment in pro-B cells (Supplementary Fig. 6a).
Previous studies have indicated that transcriptionally active domains showed high densities for Short INterspersed Elements (SINE), representing short DNA sequences originating from reverse transcribed small nuclear RNAs. The enrichment of SINE elements across transcriptionally active regions prompted us to examined for the presence of SINE elements in both embryonic stem cells as well as pro-B cells23,26. Consistent with these observations, we found that transcriptionally active regions showed relatively high densities of SINE elements (Supplemental Fig. 6b). In contrast, Long INterspersed Elemens (LINE), a cluster of actively transcribed genomic repeat elements, showed a reverse pattern and were significantly enriched across the repressive compartment (Supplementary Fig. 6b).
Why is a subset of the pro-B cell genome containing high SINE density located in the transcriptionally repressive compartment? To address this question, we considered the possibility that SINE-rich regions localized in the transcriptionally repressive chromatin compartment were in fact localized in transcriptionally permissive compartments during earlier stages of ontogeny. Consistent with this prediction, we found that SINE-enriched regions localized in a repressive chromatin compartment in pro-B cells tend to be highly enriched for H3K4me2 in embryonic stem cells (Supplementary Fig. 6c,d). These data imply that during embryogenesis a subset of chromatin domains switch from a transcriptionally permissive to a repressive compartment. Thus, in pro-B cells the majority of SINE-rich domains are located in a transcriptionally permissive compartment but a fraction of SINE-rich domains located in the transcriptionally repressive compartment represents domains that switched from a transcriptionally permissive to a repressive nuclear compartment during an earlier stage in ontogeny.
Identifying developmentally regulated anchors
The data described above identified distinct classes of factors that were associated with intra-and/or inter-domain interactions. To determine whether these interactions are developmentally regulated, we used SIMA to examine for enrichment of putative anchors across loop attachment regions in multipotent pre-pro-B and pro-B cells. We found that the pattern of genomic interactions associated with CTCF occupancy did not differ significantly between pre-pro-B versus pro-B cells (Fig. 6c). However, the pattern of intra- and inter-domain interactions, associated with E2A occupancy, differed significantly in pre-pro-B as compared to pro-B cells. As expected, intra- and interdomain interactions that were associated with E2A occupancy in pro-B cells were absent in E2A-deficient multipotent progenitors (Fig. 6c). Thus, the developmental progression from the pre-pro-B to the pro-B cell stage is associated with large-scale changes in loop attachment regions associated with E2A but not CTCF occupancy.
To identify additional potential anchors associated with the pro-B cell interactome, we applied SIMA to examine PU.1, Pax5 and Foxo1 occupancy (Supplementary Fig. 5c)26–30. We used p300 occupancy to examine for enhancers involved in intra-and inter-domain interactions (Supplementary Fig. 6c). This analysis revealed that overall the spectrum of loop-attachment regions enriched for enhancers as well as Pax5, PU.1 and Foxo1 occupancy were distinct from CTCF but quite similar, albeit not identical, as shown for E2A (Supplementary Fig. 5c). The active enhancer repertoire, as marked by p300 occupancy, also appeared to be associated with both intra- and inter-domain interactions (Supplementary Fig. 6c).
To determine how transcription factor occupancy associated with loop attachment regions change during embryogenesis we used SIMA for enrichment of E2A and Sox2 occupancy28 in embryonic stem cells, pre-pro-B cells and pro-B cells (Fig. 6d). As expected, E2A-associated binding sites identified in the pro-B cell genome were not enriched at loop-attachment regions in embryonic stem cells (Fig. 6d). As predicted the reverse pattern was observed for Sox2 occupancy gated on binding sites detected in embryonic stem cells (Fig. 6d). In sum, these data indicate that loop-attachment regions associated with lineage-specific transcription factors are developmentally regulated.
Functional Vκ regions cluster indicating close spatial proximity
To determine whether changes in nuclear location are associated with changes in chromatin topology, we compared the Ebf1 and Igk interactomes between pre-pro-B and pro-B cells. We found a wide spectrum of “de novo” genomic interactions across the Ebf1 locus in pro-B cells as compared to pre-pro-B cells, indicative of widespread changes in topology (Supplementary Fig. 7). Looping across the Ebf1 locus predominantly involved promoter-promoter, enhancer-enhancer as well as enhancer-promoter interactions (Supplementary Fig. 7).
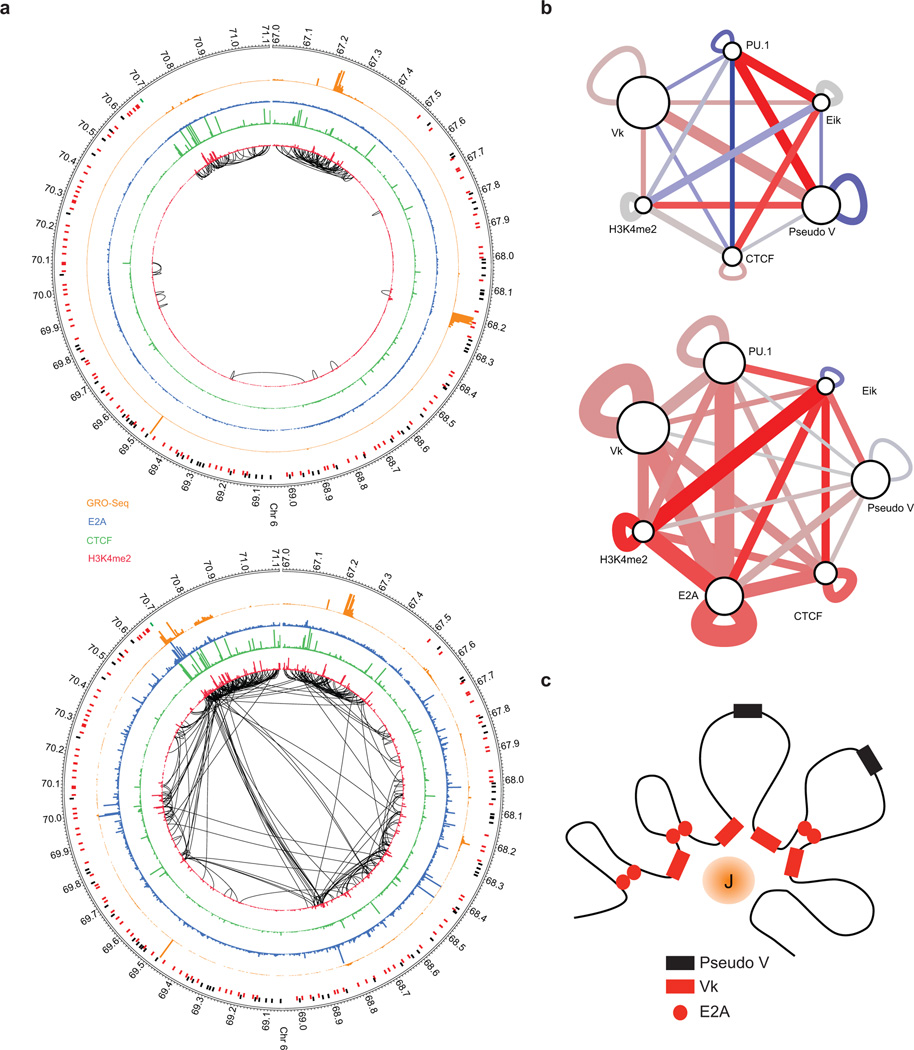
Striking changes in genomic interactions during developmental progression were also observed in genomic interactions across the Igk locus (Fig. 7a). Looping was mainly found adjacent to the Igk locus in pre-pro-B cells whereas in pro-B cells we observed an intricate and elaborate pattern of interactions involving both small and large loops (Fig. 7a). Conspicuous were interactions involving the Igk intronic enhancer (Eiκ) and genomic elements across the Igk locus (Fig. 7a). Many of these interactions were closely associated with E2A occupancy (Fig. 7a). To examine genomic interactions in greater detail, we applied SIMA for transcription factors, epigenetic marks, joining and variable regions (Fig. 7b). Strikingly, Vκ regions that were utilized in the Igk repertoire appeared to cluster in pro-B cells but not in pre-pro-B cells. In contrast, pseudo-V regions did not gather (Fig. 7b). The clustering of Vκ elements indicates close spatial proximity permitting the entire repertoire of functional Vκ regions, separated by large genomic distances, to encounter Jκ elements with similar probabilities (Fig. 7c).
Figure 7.
Switching nuclear compartments is closely associated with remodeling of chromatin architecture. (a) Extensive remodeling of chromatin structure is associated with the transition from the pre-pro-B to the pro-B cell stage. Circos diagram of genomic regions including the Igk locus representing significant intrachromosomal interactions in pre-pro-B (top) versus pro-B cells (bottom) (P < 0.05). H3K4me2 deposition is indicated in red. CTCF occupancy is shown in green. E2A occupancy is indicated in blue. GRO-Seq read density is shown in orange. The intronic enhancer (Eik)-Jκ region is indicated by the green bar. Pseudo V segments are indicated in black bars whereas functional V segments are indicated in red bars. (b) Vκ regions cluster across the pro-B cell interactome. The significance/reproducibility of tethering between various genomic regions was determined by SIMA and then visualized using Cytoscape. The width of edges are proportional to the log P-value of association, and the color (red-blue) is representative of the ratio of observed interaction frequencies relative to expected frequencies (i.e. the strength of the association). (c) Model depicting depicting the Igk locus. In this model a subset of Vκ segments are anchored in close spatial proximity, surrounding an inner cavity containing the Jκ elements. Data presented were obtained from two Hi-C experiments representing pre-pro-B and pro-B cells.
DISCUSSION
Recently global studies using formaldehyde cross-linking approaches have provided insight into the folding patterns of genomes17–23. These studies revealed that genomes are organized into transcriptionally repressive and permissive compartments17. Compartments themselves are organized into domains that are structured by intra-domain interactions, primarily mediated by CTCF21,23. Here we have developed Structured Interaction Matrix Analysis (SIMA) to identify additional factors associated with loop attachment regions. Consistent with previous observations, we found that CTCF occupancy was primarily associated with genomic interactions within chromatin globules. We also identify novel putative anchors such as E2A, PU.1, EBF1 and Pax5 that appear to bind collaboratively across loop-attachment regions. Thus, different classes of putative anchors establish the intra-domain interactome. SIMA also allowed us to detect inter-domain interactions. Inter-domain interactions primarily are mediated by enhancer-enhancer interactions involving transcriptionally active domains such as transcription factories, since they are closely associated with p300 bound sites31,32,33. We note that distinct inter-domain interactions are rather infrequent, reflecting that genomic interactions between domains occur only in a small fraction of the populace.
The data aforementioned indicate that at least two distinct classes of anchors establish the pro-B cell interactome; first, architectural proteins such as CTCF acting inside domains and second, transcriptional regulators such as E2A and PU.1 acting within and between chromatin domains. How do these two classes, CTCF versus enhancer binding proteins, differ? The biochemical activities and regulatory functions of CTCF and E2A, Pu.1 are quite distinct. Whereas CTCF bound sites are not enriched across transcriptionally active regions, E2A and PU.1 occupancy are primarily associated with active enhancer repertoires29,30. Furthermore, tethers associated with CTCF occupancy are not developmentally regulated, whereas the spectrum of loop attachment regions, enriched for E2A and PU.1 bound sites, differs extensively between the pre-pro-B and pro-B cell stage. Across what genomic regions do the latter contacts originate? Since p300 occupancy overlaps with the very large majority of E2A and PU.1 binding sites, we suggest that this spectrum of genomic interactions primarily involve enhancer elements34. Do any of such factors play a direct role in establishing genome topology, or is there a hierarchy of primary and secondary participants? We would like to consider the possibility that the E2A proteins act as primary players. E2A proteins have been particularly well studied for their roles in antigen receptor assembly35,36. Enforced expression of the E2A encoded isoform, E47, readily promotes the assembly of antigen receptor genes in embryonic kidney cells37,38. We previously noted that a significant fraction of the Vκ regions were located within close genomic proximity from E2A binding sites39. Here we observed that in the bulk population functional Vκ but not pseudo Vκ repertoires appeared to cluster. The clustering of functional Vκ elements implies close spatial proximity. Since E2A occupancy is closely associated with a subset of V regions we propose that E2A binding directly promotes the assembly of Vk regions (Fig. 7c). Such a configuration would permit the entire Igk V region repertoire, separated by vast genomic distances, to encounter the Jκ elements with similar probabilities providing an equal playing field for the variable region repertoire. We suggest that similarly the E2A proteins act directly to mediate enhancer-enhancer as well as enhancer-promoter interactions inside chromatin domains as well as between domains. Ultimately, biochemical approaches will be needed to determine whether indeed the E2A proteins directly function as anchors.
The observations described here also show that in multipotent progenitors the Ebf1 locus is sequestered at the nuclear lamina. Is sequestration of key developmental regulators at the nuclear lamina a general principle that governs developmental specific gene expression? Strikingly, upon closer inspection we found that in E2A-deficient multipotent progenitors, Bcl11b, a key regulator that promotes T lineage commitment, is also located in the heterochromatic environment40,41,42. Why are the Ebf1 and Bcl11b loci located in the heterochromatic environment in progenitor cells? We suggest that the nuclear lamina serves to ensure efficient silencing. In other words, being located in a transcriptionally repressive compartment would not readily lead to the aberrant activation of gene expression since the entire neighborhood is predominantly in a silent state. On the other hand, transcriptionally inactive genes that are localized across the transcriptionally permissive compartment might be subject to aberrant activation, for example by aberrant looping of nearby located active enhancers. Furthermore, we suggest that key developmental regulators such as EBF1 and Bcl11b, need to be efficiently silenced in multipotent progenitors since inappropriate activation of such genes may readily lead to premature differentiation. It is now well established that the E2A proteins act indirectly and directly to induce the expression of EBF130,43,44. However, during early hematopoiesis E2A expression is already high in multipotent hematopoietic progenitors and it has remained an open question as to why E2A does not activate EBF1 expression prematurely at the multipotent progenitor cell stage45. The data described here provide a mechanism. The Ebf1 locus in multipotent progenitors is sequestered away from transcription factories and tightly associated with the nuclear lamina, preventing premature activation and developmental progression towards the B cell lineage. Similarly, the Bcl11b locus might be sequestered in the heterochromatic compartment in hematopoietic progenitor cells to suppress premature developmental progression towards the T cell lineage. Hence, we propose that the positioning of key developmental regulators at the nuclear lamina in progenitor cells is a general principle to ensure efficient silencing.
The critical question now to be addressed is how the Ebf1 and Bcl11b loci are sequestered in the heterochromatic environment and how they are released at the appropriate developmental stage. Recent studies have identified ThPok as a factor mediating the sequestration of the genes at the nuclear lamina46. It is conceivable that ThPok might similarly be involved in recruiting the Ebf1 or Bcl11b locus to the nuclear lamina. However, since EBF1 and Bcl11 expression leads to either the induction of a B lineage- or T lineage-specific program of gene expression, it seems more likely that they are sequestered to the lamina by distinct tethers. Identifying such tethers and how they are regulated during hematopoiesis deserves further scrutiny.
METHODS
Mice
Animal studies were approved by the IACUC-UCSD. All mice used were maintained onto a C57BL/6 background.
Cell culture
E2a−/− hematopoietic progenitors were isolated and grown in the presence of IL7, SCF and FLT3L as described previously46. Rag1-deficient pro-B cells were grown in Opti-MEM 10% FCS, 2% glutamine, penicillin, and streptomycin, 50 M -mercaptoethanol in the presence of IL7 and SCF for 7 days as described47.
ChIP-Seq
Chromatin was immunoprecipitated as described30. Details were found in supplementary information. Antibodies used in these experiments were: anti-dimethyl-Histone H3 (Lys4) (07-030, Millipore), anti-trimethyl-Histone H3 (K27) (07-449, Millipore), anti-CTCF (07-729, Millipore), anti-Rad21 (ab992, Abcam), anti-H3K36me3 (ab9050, Abcam), anti-c-Myc (sc-764x, Santa Cruz Biotechnology), and anti-p300 (sc-585, Santa Cruz Biotechnology) antibody.
Imaging
Fluorescent in situ hybridization (FISH) and image acquisition were done as described previously47. The BAC probes used for this study were RP23-118P17, RP23-286D14, and RP24-74E7 and were obtained from the BACPAC Resource Center (BPRC) at Children’s Hospital Oakland Research Institute. 3D fluorescent images were acquired on a deconvolution microscope (Deltavision) using a 100x objective. Optical sections (z stacks) of 0.2 µm apart were obtained in the DAPI, FITC, Red, and Cy5 channels. The distances separating the probes were measured using the center of mass as focal points. The center of mass was determined by SoftWorx software package. Chromatic aberration was corrected using TetraSpeck™ (size of 0.2 µm) measurements as described previously6. Fluorescent in situ hybridization (FISH) and image acquisition were done as described previously47 with the following changes. The BAC probe used for Ebf1 domain was RP23-118P17 as listed above. It was labeled with Alexa Fluor 488-5-dUTP. The nuclear lamina was stained first with the primary anti-Lamin A and B antibodies (sc-6214 and sc-6217) from Santa Cruz Biotechnology, followed by the secondary staining using donkey anti-goat IgG antibody conjugated to Alexa Flour 594 (A11058) (Invitrogen). Fluorescent images were acquired on a deconvolution microscope (Deltavision) using a 100x objective. Optical sections (z stacks) of 0.2 m apart were obtained in the DAPI, FITC, and Red channels. The distances between the nuclear lamina and the Ebf1 locus were measured in 2D using SoftWorx software package.
Hi-C analysis
Computational data analysis was performed with software developed in-house. Hi-C analysis was integrated into HOMER, a general Next-gen sequencing analysis suite. The software is freely available (http://biowhat.ucsd.edu/homer/). Specialized visualization of HOMER analysis files was accomplished using Java Tree View, Circos and Cytoscape software. Details are described in the supplementary information section.
Supplementary Material
ACKNOWLEDGMENTS
We thank G. Hardiman, C. Ludka, L. Edsall, S. Kuan, C. Espinoza, U. Wagner, J. Sprague and Z. Ye for help with Solexa DNA sequencing. We thank J. Dixon for help during the initial phase of Hi-C analysis. We thank B. Wold for suggesting EGS fixation. We thank B. Ren for generously allowing access to the Illumina Hi-Seq instrumentation. Y.C.L. was supported by the American Recovery and Reinvestment Act (ARRA PHS 3RO1AI082850). CBo was supported by the EMBO and Swiss National Science Foundation. We thank the members of the Murre laboratory for comments on the manuscript. The studies were supported by grants from the NIH to C.K.G. and C.M (AI082850A and AI00880).
Footnotes
Accession codes. GEO: GSE40173
AUTHOR CONTRIBUTIONS
YCL, RM and SH designed and performed the majority of experiments. SH performed Gro-Seq. CBe performed the bioinformatics, analyzed data, suggested novel approaches and developed SIMA. MM, KM and VC performed ChIP-Seq analysis. CBo provided novel insights into Igκ locus topology. YCL, CBe and CM wrote the manuscript with contributions from CKG, RM and SH. CKG and CM supervised the study.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Sedat J, Manuelidis L. A direct approach to the structure of eukaryotic chromosomes. Cold Spring Harbor Symp. Quant. Biol. 1977;42:331–350. doi: 10.1101/sqb.1978.042.01.035. [DOI] [PubMed] [Google Scholar]
- 2.Rattner JB, Lin CC. Radial loops and helixcal coils coexist in methaphase chromosomes. Cell. 1985;42:291–296. doi: 10.1016/s0092-8674(85)80124-0. [DOI] [PubMed] [Google Scholar]
- 3.Paulson JR, Laemmli UK. The structure of histone depleted metaphase chromosomes. Cell. 1977;12:817–828. doi: 10.1016/0092-8674(77)90280-x. [DOI] [PubMed] [Google Scholar]
- 4.Pienta KJ, Coffey DS. A structural analysis of the nuclear matrix and DNA loops in the organization of the nucleus and chromosome. J. Cell Sci. Suppl. 1984;1:123–135. doi: 10.1242/jcs.1984.supplement_1.9. [DOI] [PubMed] [Google Scholar]
- 5.Munkel C, Langowski J. Chromosome structure described by a polymer model. Phys. Rev. 1998;E57(5B):5888–5896. [Google Scholar]
- 6.Jhunjhunwala S, et al. The 3D-structure of the immunoglobulin heavy chain locus: Implications for long-range genomic interactions. Cell. 2008;133:265–279. doi: 10.1016/j.cell.2008.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bau. D, et al. The three-dimensional folding of the a-globin gene domain reveals formation of chromatin globules. Nat. Struct. Mol. Biol. 2011;18:107–114. doi: 10.1038/nsmb.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo C, et al. Two forms of loops generate the chromatin conformation of the immunoglobulin heavy chain locus. Cell. 2011;147:332–343. doi: 10.1016/j.cell.2011.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peric-Hupkes D, et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol. Cell. 2010;38:603–613. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schneider R, Grosschedl R. Dynamics and interplay of nuclear architecture, genome organization and gene expression. Genes Dev. 2007;21:3027–3042. doi: 10.1101/gad.1604607. [DOI] [PubMed] [Google Scholar]
- 11.Brown KE, Baxtger J, Graf D, Merkenschlager M, Fisher AG. Dynamic repositioning of genes in the nucleus of lymphocytes preparing for cell division. Mol. Cell. 1999;3:2017–217. doi: 10.1016/s1097-2765(00)80311-1. [DOI] [PubMed] [Google Scholar]
- 12.Kosak ST, et al. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- 13.Fuxa M, et al. Pax5 induces V-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Devel. 2004;18:411–422. doi: 10.1101/gad.291504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roldan E, et al. Locus 'decontraction' and centromeric recruitment contribute to allelic exclusion of the immunoglobulin heavy-chain gene. Nat. Immunol. 2005;6:31–41. doi: 10.1038/ni1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hewitt SL, Chaumeil J, Skok JA. Chromosome dynamics and the regulation of V(D)J recombination. Immunol. Rev. 2010;237:43–54. doi: 10.1111/j.1600-065X.2010.00931.x. [DOI] [PubMed] [Google Scholar]
- 16.Goldmit M, et al. Epigenetic ontogeny of the IgK locus during B cell development. Nat. Immunol. 2005;6:198–203. doi: 10.1038/ni1154. (2005) [DOI] [PubMed] [Google Scholar]
- 17.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fullwood MJ, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kalhor R, Tjong H, Jayathilaka N, Alber F, Chen L. Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. Nat. Biotechnol. 2011;30:90–98. doi: 10.1038/nbt.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sexton T, et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 21.Li G, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, et al. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell. 2012;148:908–921. doi: 10.1016/j.cell.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 26.Schmidt D, et al. Waves of retrotransposon expansion remodel genome organization and CTCF binding in multiple mammalian lineages. Cell. 2012;148:335–348. doi: 10.1016/j.cell.2011.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ochiai K, et al. A self-reinforcing regulatory network triggered by limiting IL-7 activates pre-BCR signaling and differentiation. Nat. Immunol. 2012;13:300–307. doi: 10.1038/ni.2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen X, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 29.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–89. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin YC, et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat. Immunol. 2010;11:635–643. doi: 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iborra FJ, Pombo A, Jackson DA, Cook PR. Active RNA polymerase are localized within discrete transcription factories in human nuclei. J. Cell Sci. 1996;109:142–2436. doi: 10.1242/jcs.109.6.1427. [DOI] [PubMed] [Google Scholar]
- 32.Carter D, Chakalova L, Osborne CS, Dai YF, Fraser P. Long-range chromatin regulatory interactions in vivo . Nat. Genet. 2010;32:623–626. doi: 10.1038/ng1051. [DOI] [PubMed] [Google Scholar]
- 33.Corcoran AE. The epigenetic role of non-coding RNA transcription and nuclear organization in immunoglobulin repertoire generation. Semin. Immunol. 2010;6:353–361. doi: 10.1016/j.smim.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 34.Simonis M, et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome capture-on-chip (4C) Nat. Genet. 2006;11:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 35.Jhunjhunwala S, van Zelm MC, Peak M, Murre C. Chromatin architecture and the generation of antigen receptor diversity. Cell. 2009;138:435–448. doi: 10.1016/j.cell.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Inlay MA, Tina H, Lin T, Xu Y. Important roles for E protein binding sites within the immunoglobulin kappa chain intronic enhancer in activating Vkappa to Jkappa rearrangement. J. Exp Med. 2004;200:1205–1211. doi: 10.1084/jem.20041135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romanow WR, et al. E2A and EBF Act in Synergy with the V(D)J Recombinase to Generate a Diverse Immunoglobulin Repertoire in Non-Lymphoid Cells. Mol. Cell. 2000;5:343–353. doi: 10.1016/s1097-2765(00)80429-3. [DOI] [PubMed] [Google Scholar]
- 38.Sakamoto S, et al. E2A and CBP/p300 act in synergy to promote chromatin accessibility of the immunoglobulin k locus. J. Immunol. 2012;188:5547–5560. doi: 10.4049/jimmunol.1002346. [DOI] [PubMed] [Google Scholar]
- 39.Bossen C, Mansson R, Murre C. Chromatin topology and the regulation of antigen receptor assembly. Ann. Rev. Immunol. 2012;30:337–356. doi: 10.1146/annurev-immunol-020711-075003. [DOI] [PubMed] [Google Scholar]
- 40.Li L, Leid M, Rothenberg EV. An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science. 2010;329:89–93. doi: 10.1126/science.1188989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ikawa T, et al. An essential developmental checkpoint for production of the T cell lineage. Science. 2010;329:93–96. doi: 10.1126/science.1188995. [DOI] [PubMed] [Google Scholar]
- 42.Li P, et al. Reprogamming of T cells to natural killer-like cells upon Bcl11b deletion. Science. 2010;329:85–89. doi: 10.1126/science.1188063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Welinder E, et al. E2A and HEB act in concert to induce the expression of FOXO1 in the common lymphoid progenitor. Proc. Natl. Acad. Sci. USA. 2011;108:17402–17407. doi: 10.1073/pnas.1111766108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seet CS, Brumbaugh RL, Kee BL. Early B cell factor promotes B lymphopoiesis with reduced interleukin 7 responsiveness in the absence of E2A. J. Exp. Med. 2004;199:1689–1700. doi: 10.1084/jem.20032202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Semerad CL, Mercer EM, Inlay MA, Weissman IL, Murre C. E2A proteins maintain the hematopoietic stem cell pool and promote the maturation of myelolymphoid and myeloerythroid progenitors. Proc. Natl. Acad. Sci. USA. 2009;106:1930–1935. doi: 10.1073/pnas.0808866106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zullo JM, et al. DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell. 2012;149:1474–1487. doi: 10.1016/j.cell.2012.04.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.