Abstract
Severe reduction in Survival Motor Neuron 1 (SMN1) protein in humans causes Spinal Muscular Atrophy (SMA), a debilitating childhood disease that leads to progressive impairment of the neuro-muscular system. Although previous studies have attempted to identify the tissue(s) in which SMN1 loss most critically leads to disease, tissue-specific functions for this widely expressed protein still remain unclear. Here, we have leveraged RNA interference methods to manipulate SMN function selectively in Drosophila neurons or muscles followed by behavioral and electrophysiological analysis. High resolution measurement of motor performance shows profound alterations in locomotor patterns following pan-neuronal knockdown of SMN. Further, locomotor phenotypes can be elicited by SMN knockdown in motor neurons, supporting previous demonstrations of motor neuron-specific SMN function in mice.
Electrophysiologically, SMN modulation in muscles reveals largely normal synaptic transmission, quantal release and trans-synaptic homeostatic compensation at the larval neuro-muscular junction. Neuronal SMN knockdown does not alter baseline synaptic transmission, the dynamics of synaptic depletion or acute homeostatic compensation. However, chronic glutamate receptor-dependent developmental homeostasis at the neuro-muscular junction is strongly attenuated following reduction of SMN in neurons. Together, these results support a distributed model of SMN function with distinct neuron-specific roles that are likely to be compromised following global loss of SMN in patients. While complementary to, and in broad agreement with, recent mouse studies that suggest a strong necessity for SMN in neurons, our results uncover a hitherto under-appreciated role for SMN in homeostatic regulatory mechanisms at motor synapses.
Keywords: Drosophila, motor behavior, synaptic transmission, homeostasis, Spinal Muscular Atrophy, SMN
1. Introduction
Spinal Muscular Atrophy (SMA) is a severely debilitating childhood disorder that leads to progressive muscle weakness resulting from a loss or degeneration of motor neuron innervation. It is a relatively common autosomal recessive disorder and can affect up to 1 in 6000 babies (Lefebvre et al., 1995; Pearn, 1978; Sleigh et al., 2011). SMA results predominantly from mutations in the Survival Motor Neuron 1 gene (SMN1). Severity is related to the copy number and activity of a near identical homolog in humans, SMN2 (Coovert et al., 1997; Lefebvre et al., 1997). The SMN2 gene carries a C-to-T transition leading to aberrant splicing in 80–90% of the SMN2 transcript and loss of exon 7 (SMNΔ7) resulting in a protein with reduced stability (Lorson et al., 1999; Lorson and Androphy, 2000; Monani et al., 1999). The overall reduction of the SMN protein leads to severe neuromuscular degeneration including the loss of motor neuron cell bodies over time (Balabanian et al., 2007; Jablonka et al., 2000; Monani et al., 2000). SMN is a highly conserved ubiquitously expressed protein that is required critically for normal mRNA splicing since it affects the biogenesis of U snRNP particles across species (small nuclear ribonucleoprotein) (Fischer et al., 1997; Kroiss et al., 2008; Lee et al., 2009; Liu et al., 1997; Meister et al., 2001; Pellizzoni et al., 2002). Complete loss of SMN is cell lethal, therefore, SMA is essentially a disorder that results from reduced SMN availability (Feldkotter et al., 2002; Mailman et al., 2002; McAndrew et al., 1997). Several mouse models of SMA also suggest that full-length SMN is required for cell viability, and that disease severity is closely linked to the dosage of SMN gene product. Thus, total loss of Smn leads to early lethality (Frugier et al., 2000), whereas introduction of two copies of the human SMN2 gene results in normal birth followed by progressive decline in neuromuscular and motor function and subsequent death by post-natal day 7 (PND 7). Introduction of four copies of SMN2, however, leads to complete rescue (Hsieh-Li et al., 2000; Monani et al., 2000). Interestingly, introduction of SMNΔ7 – the mutant form of human SMN missing exon 7 - in an Smn−/−; SMN2+/+ background provides additional rescue, such that animals now die between PND 6 and 13 (Le et al., 2005). Given that SMN is required in all cells, why its reduction leads to specific neuromuscular symptoms or whether SMA symptoms arise from SMN reduction in motor neurons or muscle (or even other tissues), still remain outstanding questions, though recent experiments indicate prominent neuronal functions (Gogliotti et al., 2012; Martinez et al., 2012; Park et al., 2010; Imlach et al., 2012; Lotti et al., 2012).
Since SMN is a highly conserved protein, invertebrate models such as C. elegans and Drosophila have also been used to understand molecular functions for SMN and to more rapidly identify networks of genes within which SMN functions (Briese et al., 2009; Chan et al., 2003; Chang et al., 2008; Dimitriadi et al., 2010; Rajendra et al., 2007; Sen et al., 2011; Sleigh et al., 2011). These studies have revealed remarkable similarities in SMN function in the regulation of U snRNP biogenesis (Cauchi, 2010; Cauchi et al., 2008; Kroiss et al., 2008; Lee et al., 2009; Shpargel et al., 2009) and NMJ development and physiology (Cauchi et al., 2008; Chan et al., 2003; Chang et al., 2008; Praveen et al., 2012; Rajendra et al., 2007; Shpargel et al., 2009). While a number of these studies have used classical loss-of-function alleles in Drosophila Smn, more recent work has made use of gene-targeted RNAi methodologies, that can be used to knock down, but not deplete SMN function in selected tissues in conjunction with the versatile GAL4-UAS system (Brand and Perrimon, 1993; Chang et al., 2008). Thus, RNAi based knockdown is a closer approximation to the situation in SMA patients, and also permits experiments in which tissue-specific reduction in SMN can be achieved in vivo in an otherwise normal animal. If it is true that different tissues have different thresholds for susceptibility to reduction in SMN function, then selective, and relatively mild, knockdown of SMN might uncover fundamentally conserved tissue-specific functions for SMN (Sleigh et al., 2011). Here, we use this strategy to test neuronal versus muscle-specific functions for SMN in Drosophila in the regulation of motor behavior and synaptic physiology (Figure 1). Our results highlight both neuronal and muscle functions for Drosophila Smn, similar to mouse studies, but with stronger neuronal outcomes. But more significantly, they also point to a very precise role for SMN in the regulation of homeostatic mechanisms that maintain parity at the NMJ. These findings are especially relevant in the context of putative developmental roles for SMN in mammals and suggest a unique function for SMN in the homeostatically driven development of mature motor neurons (Foust et al., 2010; Gogliotti et al., 2012; Hammond et al., 2010; Martinez et al., 2012; Sleigh et al., 2011). Behavioral and synaptic phenotypes described in this report should also provide useful platforms for the functional testing of other Smn-interacting genes that have been discovered through genetic and biochemical screening approaches (Dimitriadi et al., 2010; Sen et al., 2011).
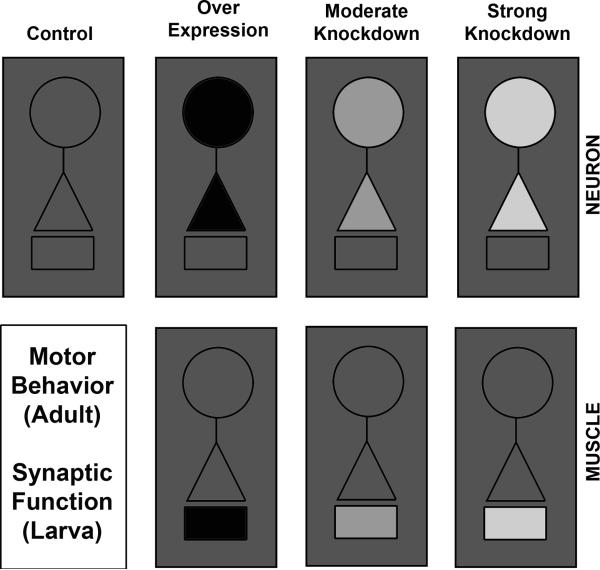
Figure 1. Experimental paradigm to test tissue-specific requirement for SMN in the maintenance of normal motor physiology.
The experimental strategy to test compartment-specific functions for SMN is outlined schematically. SMN is expressed widely in many tissues (grey outer box). In this study, SMN has either been over-expressed (black) in neurons or muscles, or has been reduced through RNAi mediated knockdown (lighter shades of grey reflected moderate to strong knockdown). SMN levels in other tissues are presumed to be intact and comparable to wild type control animals. Experimental animals are then subjected to behavioral assays to measure motor function in adults. Electrophysiological measurements are used to evaluate various aspects of synaptic function at the third instar larval NMJ. In all figures, circles represent the neuronal soma, triangles represent the pre-synaptic compartment and small rectangles represent post-synaptic muscle.
2. Results
2.1 Smn perturbation in neurons or muscle causes behavioral deficits
Gross locomotor defects in Drosophila Smn mutants and in mutants for the fly homolog of Gemin3 – a DEAD-box RNA helicase complexed with SMN, have been reported previously (Cauchi et al., 2008; Chan et al., 2003; Praveen et al., 2012; Shpargel et al., 2009; Imlach et al., 2012). Since Smn mutants are often pupal and adult lethal, these studies revealed abnormal locomotion predominantly at larval stages and showed that systemic or cholinergic neuron-specific add-back of SMN significantly rescued these motor phenotypes. Consequently, a relationship between normal SMN function and motor physiology has been established in Drosophila, similar to human patients and other mammalian model systems (Cauchi et al., 2008; Chan et al., 2003; Shpargel et al., 2009; Imlach et al., 2012). However, it is currently unclear whether locomotor phenotypes in Smn mutants arise from a loss of SMN in neurons or muscle. In this study, we attempted to answer this question using the GAL4-UAS (Brand and Perrimon, 1993) method of tissue-targeted transgene expression and recently available reagents to either over-express the full length wild type SMN gene product or knockdown Smn function using RNA interference (Chang et al., 2008; Sen et al., 2011). We chose to use these RNAi reagents since their efficacy in knocking down endogenous levels of SMN in neurons and muscles, as well as their potency in generating neuro-muscular defects have been well characterized (Chang et al., 2008), and a reduction in SMN, rather than its completely removal, more closely approximates the situation in SMA (western blots in Figure 2A and C). Since RNAi mediated knockdown of Smn does not lead to lethality in most cases, we were also able to measure adult locomotor behavior at a resolution much higher than possible for larvae. Such measurements allow for a thorough analysis of locomotor parameters that will also be useful as screening tools in future genetic interaction experiments. A complementary strategy to rescue strong loss-of-function mutations in SMN using tissue directed rescue has recently been reported (Imlach et al., 2012).
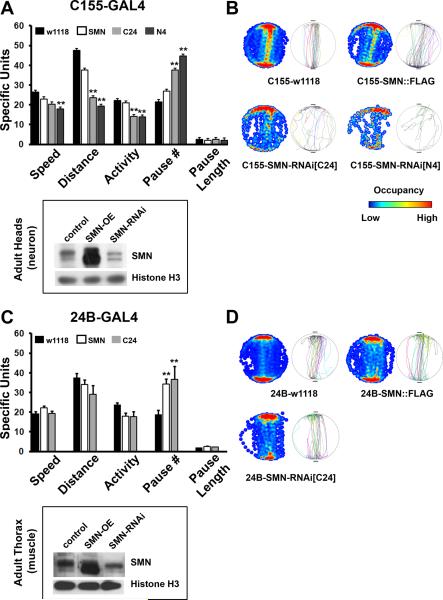
Figure 2. Perturbation of SMN in the nervous system disrupts normal motor behavior.
(A) Various parameters of locomotion as measured by the Buridan's assay are shown for neuronal perturbations of SMN using the elavC155-GAL4 driver line. SMN knockdown using two RNAi lines results in decreased speed, distance covered and overall activity, while the number of pauses is increased significantly without affecting mean pause length. Western blot from adult heads shows strong over-expression (SMN-OE) and significant but not complete knock down (SMN-RNAi) of SMN protein levels in adult heads (a predominantly neuronal population). (B) Single representative fly traces (left circles) and cumulative occupancy plots (right circles with heat maps) show disrupted locomotor behavior following pan-neuronal knockdown of SMN. (C) Muscle perturbation of SMN using the how24B-GAL4 driver line. The number of pauses is significantly increased following either over-expression or knockdown of SMN in the mesoderm. Western blot of protein extract from adult thorax shows strong over-expression (SMN-OE) and significant but not complete knock down of SMN in muscle tissue (SMN-RNAi). (D) Single fly tracks and cumulative occupancy plots show mild impairment of locomotion. Note that muscle expression of SMN-RNAi-N4 is pupal lethal as reported previously. 10 individual animals that are 5–6 days old are tested for each genotype. Error bars are SEM, double asterisks represent p<0.01 as tested by ANOVA.
Pan-neuronal reduction of SMN using the elavC155-GAL4 driver results in significant changes to the normal locomotor patterns of flies in the Buridan's assay (Gotz, 1980). In this assay, 5–6 day old flies are allowed to walk between two diametrically placed landmarks on a circular arena while their movement is recorded in real time followed by off-line analysis (Freeman et al., 2012; Neuser et al., 2008). In general, our results suggest that neuronal loss of SMN leads to slower walking speed, less overall activity and a larger number of pauses between bouts of walking as compared to genetic controls containing the GAL4 line (Figure 2A). These phenotypes are seen when Smn is knocked down using either of the two RNAi lines directed to the C- or N-terminal portion of the Drosophila Smn mRNA (Chang et al., 2008) confirming the specificity of these phenotypes. Neuronal over-expression of SMN, however, does not lead to any discernible defects in locomotion. Single fly tracks and cumulative occupancy plots (heat map coded to show regions of highest occupancy during the performance of the behavior in red) also demonstrate the extent of disruption of locomotor activity in neuronal Smn knockdown animals (Figure 2B).
Changes in SMN levels in muscle do not produce strong locomotor phenotypes in adults (Figure 2C and D). Although muscle-specific roles for SMN have been recognized in both mammalian and Drosophila models (Chan et al., 2003; Chang et al., 2008; Dachs et al., 2011; Lee et al., 2012; Rajendra et al., 2007; Sen et al., 2011), RNAi mediated knockdown or over-expression of wild type SMN does not result in appreciable locomotor defects. Mild changes are seen in the number of pauses when SMN is either more or less than control levels, suggesting that these are likely to result from non-specific effects. Single representative fly tracks as well as occupancy plots further confirm that changes of SMN in muscle tissue do not affect locomotor performance in this assay. Overall, results presented in Figure 2 show the presence of profound locomotor deficits in adult animals following selective knockdown of Smn in neurons. These findings suggest that in Drosophila motor phenotypes previously observed in Smn mutants are likely to be due to loss of SMN in neurons but do not clarify whether SMN is required in specific neuronal subsets for normal locomotor behavior.
2.2. Knockdown of Smn in glutamatergic neurons, but not cholinergic neurons, leads to behavioral deficits
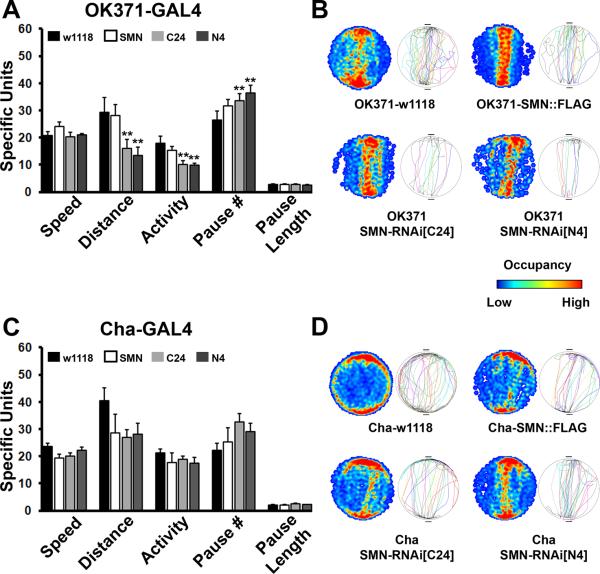
Since pan-neuronal reduction in SMN resulted in locomotor deficits, we asked whether SMN is preferentially required in a subset of neurons for normal motor behavior. Given that motor neurons are strongly implicated in SMA, we first tested if Smn knockdown in adult motor neurons produces behavioral phenotypes. Drosophila motor neurons are glutamatergic. Therefore, we used the VGlut-GAL4 line (OK371) to either over-express or knockdown Smn in all glutamatergic neurons (note that there are many non-motor glutamatergic neurons in the Drosophila nervous system that are also targeted by OK371-GAL4) (Mahr and Aberle, 2006). OK371-GAL4 driven Smn RNAi resulted in phenotypes very reminiscent of pan-neuronal Smn knockdown (Figure 3A and B). While the average speed of walking was not significantly altered, total distance moved and overall activity were significantly reduced, and number of pauses elevated, following Smn knockdown in the glutamatergic nervous system. These phenotypes were not observed following over-expression of Smn in glutamatergic neurons. Single fly tracks and occupancy plots also confirmed aberrant locomotor behavior in the knockdown animals, though these were not as strong as those following pan-neuronal knockdown of Smn. By contrast, Smn perturbation in cholinergic neurons did not have any effect on locomotion at all (Figure 3C and D). Thus, when Smn was over-expressed or knocked down with the Cha-GAL4 driver (Salvaterra and Kitamoto, 2001), none of the parameters were altered significantly and single fly tracks and occupancy plots reiterated normal locomotor behavior in all genotypes tested. Since by all accounts, Cha-GAL4 is a strong driver and has elicited phenotypes in many previous studies including those that have used RNAi mediated knockdown (for examples see Ghosh and Feany, 2004; Kitamoto, 2001; Lima and Miesenbock, 2005; Sakai et al., 2009), we do not favor the idea that the absence of phenotypes is a consequence of weak expression of the SMN-RNAi construct. Instead, taken together with results described in the previous section, these experiments suggest to us that glutamatergic motor neurons are likely to be a prominent locus for Smn-dependent locomotor phenotypes in Drosophila. We next tested whether altered synaptic transmission at motor synapses might underlie behavioral deficits following tissue-specific knockdown of SMN.
Figure 3. SMN reduction in motor neurons is sufficient to impair motor function.
(A, B) Parameters of locomotion, single fly tracks and occupancy plots in animals with glutamatergic neuron specific perturbations of SMN using the VGlut-GAL4 driver (OK371). RNAi mediated knockdown of SMN in glutamatergic neurons recapitulates in large part the effect of pan-neuronal knockdown. Distance and activity are reduced, while the total number of pauses is increased. SMN over-expression does not have any discernible influence on locomotion. (C, D) Results from the Buridan's assay for cholinergic neuron specific perturbations in SMN. None of the parameters for locomotion are significantly affected by altering SMN function in cholinergic neurons. 10 animals are tested for each genotype, error bars are SEM and double asterisks represent p<0.01 as determined by ANOVA.
2.3. Neuronal perturbation of Smn does not alter baseline and high-frequency synaptic transmission
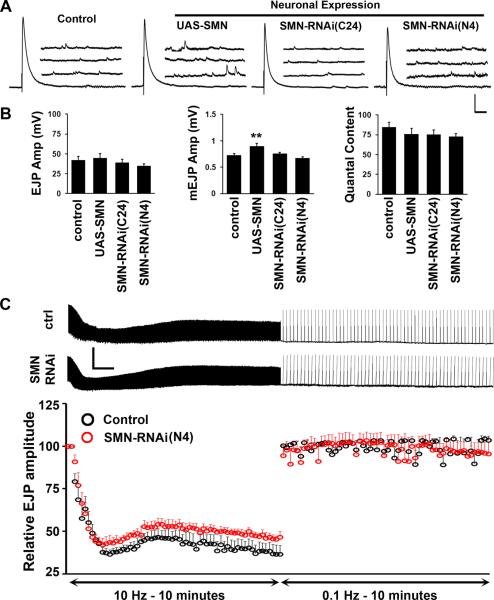
Analysis of adult behavior shows that reduced SMN in motor neurons results in behavioral deficits in a visuo-motor assay. We wondered whether we could detect a possible basis for these motor phenotypes rooted in synaptic physiology. Since the larval neuro-muscular system is simpler and experimentally more accessible than in the adult, we used this model synapse to test our idea. Baseline synaptic transmission was measured at the muscle 6/7 motor synapse in abdominal segment A2 in 1 mM external calcium as reported previously (Freeman et al., 2011; Sen et al., 2011). elavC155-GAL4 was used to drive expression of full-length Smn or RNAi constructs as described in previous sections. Interestingly, we did not observe any meaningful change in synaptic transmission under these conditions (Figure 4A and B). Mean excitatory junction potential (EJP) amplitude, mini evoked junction potential (mEJP) amplitude and the quantal content of release remained unaltered in all genotypes tested, barring a modest increase in mEJP amplitude following over-expression of Smn. These data indicate that normal parameters of synaptic transmission are not affected by Smn perturbation in neurons, although a 50% reduction in the growth of this synapse and the number of synaptic boutons under these conditions of Smn knockdown have been reported previously (Chang et al., 2008).
Figure 4. Baseline synaptic transmission is largely normal under conditions of neuronal SMN modulation.
(A, B) Representative traces (A) and quantification of baseline synaptic transmission at the larval NMJ (muscle 6/7 synapse at abdominal segment A2) following pan-neuronal SMN perturbation (B). mEJP amplitude (quantal size) is significantly different in SMN over-expressing synapses. However, EJP amplitudes or quantal content remain unchanged. 9–10 animals are tested for each genotype. Vertical scale bar = 10 mV for EJP and 5 mV for mEJP; Horizontal scale bar = 100 ms for EJP and 200 ms for mEJP. Mean resting membrane potentials are: Control = 73mV, UAS-SMN = 74mV, SMN-RNAi-C24 = 69mV, SMN-RNAi-N4 = 68.5mV. Mean input resistance values are: Control = 12.5MΩ, UAS-SMN = 12MΩ, SMN-RNAi-C24 = 13.3MΩ, SMN-RNAi-N4 = 12.4MΩ. (C) High-frequency (10 Hz) stimulation in high-calcium (10 mM) containing Ringer's solution leads to gradual synaptic depression to approximately 50% of the initial EJP amplitude, followed by almost instantaneous recovery under low-frequency (0.1 Hz) stimulation. This profile is unchanged when SMN is knocked down pan-neuronally using the N4 RNAi transgene (red spheres) as compared to control animals (black spheres). Errors bars are SEM. 5 animals were tested for each genotype. Horizontal scale bar = 1min, vertical scale bar = 50mV.
We next tested whether Smn knockdown impairs synaptic transmission when the system is stressed during high-frequency stimulation under conditions that maximize quantal content of release. We drove the N4 RNAi line with elavC155-GAL4 since this should result in the strongest pre-synaptic knockdown of Smn. Motor neurons innervating muscle 6/7 were stimulated at 10 Hz for a duration of 10 minutes in an external calcium concentration of 10 mM. Under these conditions more synaptic vesicles are released than can be recovered, leading to a depletion of the readily releasable pool of synaptic vesicles and a rapid decline in the size of the EJP that reflects fewer vesicles being released per action potential. Following this rapid decline, the synapse recovers partially and reaches a stable transmission rate due to vesicle recruitment from the reserve pool of vesicles. After a transition from 10 Hz stimulation to 0.1 Hz stimulation, the synapse recovers almost immediately to pre-stimulus levels since the balance between vesicle fusion and vesicle retrieval is restored. This assay, therefore, examines mechanisms underlying rapid recruitment of distinct vesicle pools and synaptic vesicle fusion and recycling (Dickman et al., 2005; Seabrooke and Stewart, 2011). When challenged with high-frequency stimulation Smn knockdown synapses performed as well as control ones (Figure 4C). The rate and extent of decline, reserve pool recruitment and recovery post-stimulation are not significantly different between knockdown and control animals suggesting normal vesicle dynamics at these synapses. Taken together, results in this section imply that behavioral deficits following neuronal knockdown might not result from obvious changes in baseline synaptic transmission at the neuro-muscular junction. This is in apparent conflict with earlier reports in Drosophila (Chan et al., 2003) and several studies in rodents (Kong et al., 2009; Martinez et al., 2012; Ruiz et al., 2010). However, these studies did not selectively remove Smn in neurons, and synaptic transmission phenotypes in these situations could have arisen from developmental changes triggered by reduced SMN in both neurons and muscles and indeed in other tissues. In a situation where Smn was selectively reduced in rodent motor neurons, synaptic and behavioral defects were also mild and improved dramatically postnatally (Park et al., 2010).
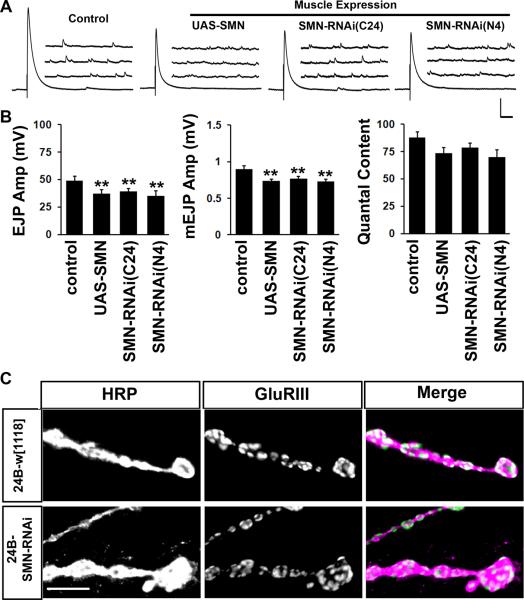
2.4. Smn perturbation in muscle affects quantal size
Increasingly, a role for normal SMN function in muscle is being appreciated (Dachs et al., 2011; Lee et al.; Martinez et al., 2012). These studies suggest that SMN in muscle might contribute to the regulation of normal muscle development and synaptic physiology. In Drosophila, previous experiments have clearly documented post-synaptic enrichment of SMN protein, synaptic growth phenotypes at the larval neuro-muscular junction that have resulted from Smn knockdown (Chang et al., 2008), strong muscle atrophy in Smn mutants (Rajendra et al., 2007) and genetic interactions between Smn and FGF signaling pathways in muscle (Sen et al., 2011). Based on this information, we asked whether Smn perturbations in muscle might result in synaptic phenotypes at the larval NMJ.
how24B-GAL4 driven expression of full-length Smn or Smn-RNAi resulted in reduced mean EJP and mEJP amplitude (Figure 5A and B) resulting in indistinguishably similar quantal content of release across genotypes. Although it seems surprising that both over-expression and knockdown of Smn lead to equivalent synaptic phenotypes, this is in line with the observation that both over-expression and knockdown of Smn in muscle leads to pupal/adult lethality (Chang et al., 2008) and abnormally long pupal cases (our unpublished observations). While the reasons behind the similarity of these phenotypes are unclear, it seems likely that these are indeed due to perturbations in muscle SMN function. Quantal size phenotypes prompted us to examine the staining intensity and distribution of post-synaptic glutamate receptors. A severe disorganization of post-synaptic Acetylcholine receptors at the NMJ has been widely observed in rodent models of SMA (Dachs et al., 2011; Kariya et al., 2008; Kong et al., 2009; Lee et al., 2008; Ling et al.; Murray et al., 2008; Ruiz et al., 2010) and a loss of glutamate receptor staining has also been reported in Drosophila Smn mutants (Chan et al., 2003). However, we did not observe any changes in overall glutamate receptor staining intensity or distribution when Smn was knocked down in muscles using the stronger N4 RNAi transgene (Figure 5C). Similarly, no change in glutamate receptor distribution is observed following neuronal knockdown of Smn either (data not shown). These results suggest that a reduction in SMN exclusively in the muscle (or in neurons) is not sufficient to cause strong synaptic transmission defects or receptor misaggregation, although it is sufficient to cause adult lethality (Chang et al., 2008). Changes in quantal size might also be simply due to small alterations in muscle size (Imlach et al., 2012) that we were not able to detect readily, and therefore, input resistance. It is possible that neuro-muscular symptoms in SMA arise from a progressive distortion in NMJ development or maintenance precipitated by a chronic loss of SMN in both compartments (Martinez et al., 2012). If this were true, compartment-specific knockdown of SMN might reveal the very initial events that trigger this collapse.
Figure 5. SMN knockdown in muscle affects the quantal size of pre-synaptic transmitter release.
(A, B) Representative traces and quantification for baseline synaptic transmission at the third instar larval NMJ following muscle-specific perturbation in SMN. Both over-expression and knockdown of SMN result in smaller EJP and mEJP amplitudes. As a result quantal content is not significantly altered in experimental animals. 9–10 animals are tested for each genotype. Vertical scale bar = 10 mV for EJP and 5 mV for mEJP; Horizontal scale bar = 100 ms for EJP and 200 ms for mEJP. Mean resting membrane potentials are: Control = 74mV, UAS-SMN = 66mV, SMN-RNAi-C24 = 63mV, SMN-RNAi-N4 = 62mV. Mean muscle input resistances are: Control = 13.2MΩ, UAS-SMN = 12.4MΩ, SMN-RNAi-C24 = 12.5MΩ, SMN-RNAi-N4 = 11.8MΩ. (C) Staining of third instar synapses with anti-HRP and anti-GluRIII shows indistinguishable staining intensities and pattern of glutamate receptor distribution following SMN knockdown in muscles. Scale bar = 5 μm.
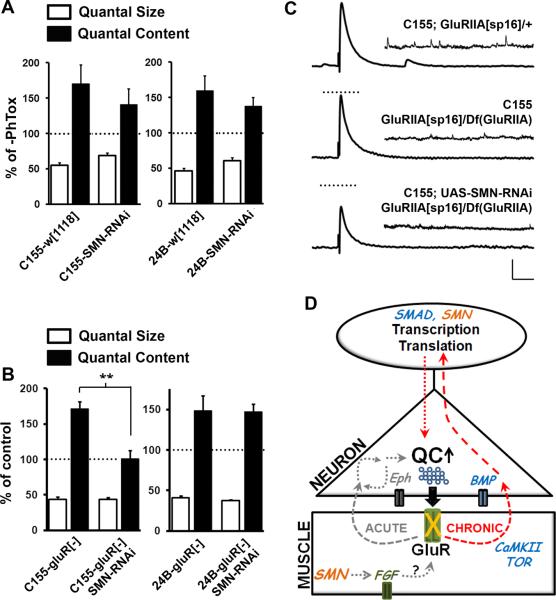
2.5. Pre-synaptic knockdown of Smn abolishes long-term homeostatic compensation at the NMJ
Rodent models of SMA show early synaptic transmission deficits at the NMJ that typically worsen with age (Dachs et al., 2011; Kariya et al., 2008; Kong et al., 2009; Lee et al., 2012; Ling et al., 2011; Murray et al., 2008; Ruiz et al., 2010). Additionally, recent studies suggest that homeostatic feedback mechanisms that maintain parity of synaptic transmission might be compromised when SMN levels are reduced in motor neurons. In one study, selective reduction of SMN in motor neurons resulted in reduced quantal content of transmission at post-natal day 8 (PND8) in mice (Park et al., 2010). Strikingly, these defects were sharply reduced by PND10–12, and by 3 months, NMJs showed overcompensation as they displayed higher mini-end plate potentials (mEPPs), EPPs and quantal content than control animals (Park et al., 2010). These data reveal aberrations in homeostatic mechanisms at the synapse following neuronal knockdown of Smn. Conversely, two other studies have addressed the tissue-specificity of SMN by rescuing SMN expression in either neurons or muscle in an Smn mutant background (Gogliotti et al., 2012; Martinez et al., 2012). While neuronal, but not muscle, add-back of SMN was able to rescue synaptic transmission defects at the NMJ in Smn mutants, surprisingly, neuronal supplementation of SMN also prevented loss of central motor neuron synaptic input. This observation suggests that under normal conditions neuronal SMN might regulate retrograde signaling mechanisms that maintain homeostatic drive at synapses.
To directly evaluate whether SMN plays a role in homeostatic signaling mechanisms in neurons, we used two assays that test either a rapid, gene expression-independent or a long-term, gene expression requiring form of homeostasis at the larval NMJ. An activity-dependent block of post-synaptic glutamate receptors at the larval NMJ using the wasp venom Philanthotoxin (PhTox) triggers a fast homeostatic response from the pre-synapse that results in a significant increase in quantal content, while quantal size remains predictably reduced (Frank et al., 2006; Frank et al., 2009). We tested whether Smn knockdown in neurons or muscles affected this homeostatic response. Figure 6A shows that PhTox-dependent homeostatic responses remained robust in either case. Thus, reduction of SMN using the N4 RNAi transgene did not alter the synapse's ability to engage a fast homeostatic increase in quantal content following glutamate receptor attenuation with PhTox (reflected in the reduced mEJP amplitudes in Figure 6A). These data suggest that local signaling mechanisms that are involved in this form of homeostasis do not require SMN activity (Figure 6D).
Figure 6. Loss of SMN in neurons impairs long-term homeostasis at the neuro-muscular junction.
(A) PhTox induced rapid homeostatic compensation at the NMJ is normal following SMN knockdown in either neurons or muscle. Shown are the percentage reduction in mEJP amplitude (quantal size) and percentage increase in quantal content induced by a 10 minute bath application of PhTox. 10 animals were tested for each genotype. (B) Persistent developmental homeostasis (increased quantal content) induced by a genetic reduction in the GluRIIA subunit of glutamate receptors at the NMJ is lost when SMN is knocked down pre-synaptically in neurons. Reduction of SMN in muscles, however, does not impair this form of homeostatic compensation. (C) Representative traces showing the absence of homeostasis in neuronal SMN knockdown animals. 8–10 animals were tested for each genotype. Vertical scale bar = 10 mV for EJP and 2 mV for mEJP; Horizontal scale bar = 100 ms for EJP and 1 s for mEJP. (D) Model schematic showing a role for SMN in chronic homeostatic plasticity at the NMJ. Interestingly, this form of plasticity also requires normal BMP signaling, a pathway known to genetically interact with SMN. Chronic homeostasis is likely to rely on long-term changes in gene expression that might require normal SMN function in neurons. SMN also functions in muscles to modulate outputs of FGF signaling that control post-synaptic properties.
Since aberrations in homeostasis in rodent models of SMA occur over a developmental time period, we next tested a form of homeostatic compensation at the larval NMJ that is longer term and likely requires changes in gene expression. A strong genetic reduction in one glutamate receptor subunit, GluRIIA, leads to severely attenuated post-synaptic currents and smaller mEJP amplitudes (Davis et al., 1998; Petersen et al., 1997). However, over developmental time, the pre-synapse compensates through an increase in quantal content (Davis et al., 1998 1999; Haghighi et al., 2003; Penney et al., 2012; Petersen et al.,1997). When we reduced SMN in neurons in a GluRIIA mutant background, we saw a virtual absence of homeostatic compensation (Figure 6B and C and Supplementary Table 1). By contrast, SMN reduction in muscles did not affect this form of homeostasis. These observations point to very specific neuronal functions for SMN in the control of long-term homeostatic responses. Such a function would also be consistent with a general role for SMN in the regulation of RNA metabolism and gene expression (Figure 6D).
3. Discussion
Although there are obvious differences between Drosophila and mammalian neuro-muscular organization, modeling SMA in flies is a productive approach given the high degree of conservation in SMN function and the ease and rapidity of genetic analysis in flies (Dimitriadi et al., 2010). Indeed, previous work in Drosophila has suggested that Smn regulates U snRNP biogenesis (Cauchi, 2010; Kroiss et al., 2008; Lee et al., 2009; Shpargel et al., 2009) and loss of SMN results in both synaptic and motor defects that are comparable to those seen in mouse models of SMA (Chan et al., 2003; Chang et al., 2008; Rajendra et al., 2007; Shpargel et al., 2009). Recent work has also made use of RNA interference to knockdown Smn in target tissues (Chang et al., 2008; Sen et al., 2011) to better mimic the situation in SMA patients. Experiments with these validated RNAi reagents have uncovered morphological phenotypes at the NMJ when SMN was knocked down in either neurons or muscle (Chang et al., 2008; Sen et al., 2011). In the current study we have used the same RNAi reagents in conjunction with the GAL4-UAS system to reduce SMN in neurons or muscle tissue followed by an assessment of such perturbation on motor performance and synaptic transmission at the NMJ (Figure 1). Muscle knockdown of SMN did not result in strong behavioral or synaptic transmission defects, though similar manipulations have been shown to affect synaptic morphology in larvae and lead to adult lethality (Chang et al., 2008; Sen et al., 2011). This suggests a situation in Drosophila that is very similar to that in mice, where SMN plays a role in muscles to ultimately impact lifespan, but is not critically required for normal synaptic function (Gogliotti et al., 2012; Martinez et al., 2012). On the other hand, we find distinct deficits in visuo-motor performance when SMN is reduced neuronally, though baseline synaptic transmission under these conditions is largely unaffected. This contrasts with earlier findings in Drosophila (Chan et al., 2003) and is likely due to the compartment-specific knockdown of SMN in our experiments. Interestingly, a recent report suggests that larval locomotor phenotypes in strong hypomorphic mutations in SMN arise from SMN deficiency in cholinergic neurons (Imlach et al., 2012). While our complementary manipulations do not reveal any effect of cholinergic knock down of SMN on locomotor activity in adult flies (perhaps due to insufficient knock down with the RNAi reagent available to us), we were able to identify a very specific loss of homeostatic compensation between the pre- and post-synapse when SMN was reduced neuronally (Figure 6D). We propose that this might reflect an early and subtle event in SMA etiology that predates more dramatic SMA sequelae comprising loss of normal NMJs and neuro-muscular degeneration.
Severe mouse models of SMA (e.g. Smn−/−; SMN2+/+) show clearly discernible loss of neuro-muscular morphology, strong synaptic transmission defects, loss of pre-synaptic inputs to spinal motor neurons, muscle degeneration and eventual death of motor neurons (Balabanian et al., 2007; Jablonka et al., 2000; Monani et al., 2000). Several studies have also used Cre-loxP derived conditional knockout models of SMA in mice to address the question of tissue-specificity of SMN vis-à-vis the incidence of SMA-like symptoms. Complete removal of exon 7 in specific tissues led to cell lethality, a situation, though consistent with a vital role for SMN in all cells, significantly different from that found in SMA patients (Cifuentes-Diaz et al., 2001; Cifuentes-Diaz et al., 2002; Frugier et al., 2000; Nicole et al., 2003; Vitte et al., 2004). Recent studies have more successfully approximated the patient condition, while trying to distinguish tissue-specific functions for SMN in vivo (Gogliotti et al., 2012; Martinez et al., 2012; Park et al., 2010). In one study, Smn exon 7 was removed in neurons in a background where SMN2 was introduced (Park et al., 2010). Surprisingly, these mice showed mild SMA symptoms that improved with age. However, this manipulation did uncover subtle homeostatic regulatory phenotypes at the neuro-muscular junction (NMJ) that may not have been noticed in the presence of the typically severe SMA phenotypes (Kariya et al., 2008; Michaud et al., 2010; Murray et al., 2010; Murray et al., 2008; Ruiz et al., 2010). In another study, full-length SMN protein was added back to either neurons or muscle in a mutant SMN background that otherwise results in viable “SMA” mice (Lutz et al., 2011; Martinez et al., 2012). This study revealed that while both neuronal and muscle add-back of SMN could significantly rescue survival and motor behavior, only neuronal expression rescued synaptic function at the NMJ and also motor neuron somal synapses, through putative homeostatic mechanisms (Martinez et al., 2012). A third study showed, by adding back SMN selectively to motor neurons using an Hb9-Cre, that SMN supplementation in motor neurons could rescue the vast majority of motor defects and restored normal sensory-motor synapses (Gogliotti et al., 2012). Together, these observations hint at neuron-specific roles for SMN in the homeostatic regulation of normal synaptic connectivity and transmission in mammals.
Phenotypic end-points in severe SMA mouse models closely mirror SMA pathology in patients, particularly Type1 SMA. However, the progressive nature of these phenotypes, increased fetal expression of SMN and the general observation that SMA phenotypes can be most effectively rescued by adding back full length SMN perinatally, suggest that defining events in SMA pathology might occur at very early stages of development (Baumer et al., 2009; Burlet et al., 1998; Butchbach et al., 2010; Foust et al., 2010; Gabanella et al., 2005; Hammond et al., 2010; Murray et al., 2010; Narver et al., 2008). Thus, obvious target tissues in SMA such as motor neurons and muscle might be exquisitely susceptible to a reduction in SMN function such that subtle defects in neuro-muscular development or physiology are the first to appear at these loci – consistent with the threshold hypothesis for SMA (Sleigh et al., 2011). Milder models of SMA in mice such as those mentioned in the previous section, seem to support this idea in some ways since they reveal aberrations in the homeostatic regulation of synaptic connectivity that may not be discernible in the presence of more severe phenotypes (Gogliotti et al., 2012; Martinez et al., 2012; Park et al., 2010). These observations point to a fundamental impairment in communication between motor neurons and their pre-synaptic input following loss of SMN. Over time, this might lead to an asynchrony in an otherwise tightly coordinated program of synaptic development leading to eventual synaptic degeneration.
Our experiments in Drosophila probably represent relatively mild manipulations of Smn. Consistent with this idea, animals are largely viable and do not show a dramatic loss of locomotion at larval stages that is seen in Smn loss-of-function mutants (Chan et al., 2003; Shpargel et al., 2009). However, neuronal knockdown of Smn does result in adult locomotor dysfunction that can be discerned with sophisticated visuo-motor assays. Further electrophysiological analysis at the larval NMJ provides a possible underlying mechanism, with the obvious caveat that these recordings are made at larval stages, while our behavioral analysis is in adults. Although most parameters of synaptic transmission are near normal, we detect specific phenotypes when Smn is perturbed in neurons or muscles. Muscle knockdown of Smn results in altered quantal size. Although we did not detect any changes in glutamate receptor staining intensity or distribution, increased mEJP size might still result from altered post-synaptic receptor sensitivity as electrophysiological measures are typically more sensitive than immuno-histochemistry. Alternatively, altered quantal size might result from changes in synaptic vesicle size or neurotransmitter packaging, or from small changes in muscle size and input impedance. Neuronal knockdown of Smn leads to a defect in homeostatic compensation. A severe impairment of synaptic transmission as described previously (Chan et al., 2003), might not have allowed the detection of this phenotype. In general, this phenotype finds resonance with defects in homeostatic regulatory mechanisms that have been highlighted previously in mild SMA models in mice (Gogliotti et al., 2012; Martinez et al., 2012; Park et al., 2010) and might represent a “weak link” that is most susceptible to a loss of SMN in neurons. It is also conceivable that an SMN-dependent loss of calibration in homeostatic drive is one of the earliest events that disrupts developmental coupling between the pre- and post-synapse during a critical period in synaptogenesis. It is interesting to note that this phenotype is only uncovered when Glutamate receptor expression is reduced, suggesting that low neuronal SMN limits the capacity for a homeostatic response when the demand for such a response is high. Further experiments are required to test whether this prediction holds in mouse models of SMA.
Our results might also help in understanding the molecular function of SMN in neurons. While SMN is clearly involved in U snRNP biogenesis, it is not clear whether this function of SMN is the one most compromised in neurons (Buhler et al., 1999; Gabanella et al., 2007; Pellizzoni et al., 1999; Shpargel and Matera, 2005; Wan et al., 2005). For example, in Drosophila, Smn null mutants show normal mRNA splicing (Praveen et al., 2012; Rajendra et al., 2007). In addition, low transgene-mediated expression of wild type SMN rescues SMA-like phenotypes in flies without any improvement in snRNA levels (Praveen et al., 2012). By contrast, a recent report implicates aberrant U12 splicing events in a collection of U12 intron containing genes following SMN perturbations (Lotti et al., 2012). While snRNA levels have not been tested in our manipulations, phenotypic analysis shows clearly that a gene expression-requiring homeostatic compensation is completely abolished through a neuron-specific reduction in SMN (red arrows in Figure 6D). By contrast, a rapid gene expression-independent form of homeostasis at the NMJ is normal (grey arrows in Figure 6D), suggesting perhaps a role for SMN in the regulation of nuclear gene expression. Whether this occurs through control of mRNA metabolism, remains to be investigated. However, it is interesting to note that this form of long-term homeostasis also requires BMP/TGF-β signaling in the pre-synapse, a signaling pathway recently shown to interact with SMN in flies (Chang et al., 2008; Haghighi et al., 2003). In this model, one might envisage SMN playing a role either in the relay of a homeostatic “signal” in the pre-synaptic compartment, or in the execution of a suitably graded response (Figure 6D). Although our studies have previously shown a role for SMN in the regulation of FGF signaling in the muscle, this function does not seem to be necessary for homeostatic regulation of synaptic transmission (Sen et al., 2011). Finally, it is of note that motor neurons in Drosophila are glutamatergic. Whether GluR-dependent homeostatic signaling in flies has a bearing on cholinergic neuro-muscular synapses or glutamatergic sensory-motor synapses in mammals remains to be seen.
4. Experimental Procedure
4.1. Drosophila strains, genetics and husbandry
Drosophila strains were reared in standard corn meal–dextrose–yeast containing food at 25 °C in controlled humidity incubators under a constant 12 h light:dark cycle. elavC155-GAL4 and how24B-GAL4 have been described previously (Brand and Perrimon, 1993; Luo et al., 1994). UAS-SMN, UAS-SMN-RNAi[C24] and UAS-SMN-RNAi[N4] were from Spyros Artavanis-Tsakonas and have been validated and described earlier (Chang et al., 2008). GluRIIA mutants (GluRIIASp16) and deficiency (Df(2l)cl-h4) were from Pejmun Haghighi (Penney et al., 2012).
4.2. Antibodies, immuno-histochemistry and western blotting
Larval dissection, staining and confocal microscopy were performed according to standard protocols (Franciscovich et al., 2008). Briefly, larvae were dissected in Ca2+ free HL3 ringer's solution (in mM): NaCl 70, KCl 5, MgCl2 20, NaHCO3 10, Sucrose 115, Trehalose 5, BES 5 (pH 7.2). Dissections were fixed in 4% paraformaldehyde, stained with primary antibody overnight, and followed by incubation in Alexa Fluor conjugated secondary antibody. Rabbit anti-dGluRIII (Marrus et al., 2004) was used at 1:1000 and Alexa-568 conjugated anti-HRP (Molecular Probes) was used at 1:500. Anti-rabbit Alexa 488 conjugated secondary antibody (Molecular Probes) was used at 1:400. An inverted 510 Zeiss LSM microscope was used for imaging. For quantitative fluorescence care was taken to prepare samples identically. Samples were imaged and analyzed double blind, and all imaging was interleaved such that control and experimental samples were imaged alternately on the same day. Confocal settings including black level (offset/contrast), gain, pixel dwell time and the number of iterative samplings for noise reduction were kept constant. Western blotting was carried out according to standard procedures (Freeman et al., 2012). Rabbit polyclonal anti-SMN antibodies were used at 1:1000 (Chang et al., 2008).
4.3. Electrophysiology
Baseline evoked and spontaneous junction potentials were measured as described previously (Sen et al., 2011). Briefly, wandering third instar larvae were dissected in normal HL3 ringer's solution (Stewart et al., 1994) (in mM): NaCl 70, KCl 5, MgCl2 20, CaCl2 1, NaHCO3 10, Sucrose 115, Trehalose 5, BES 5 (pH 7.2). Intracellular recording electrodes with tip resistances between 25–50 MΩ were filled with 3 M KCl. Only those recordings were used where the resting membrane potential was more polarized than −60 mV and the muscle input resistance was greater than 10 MΩ. Muscle 6 (VL3) in abdominal segment A2 was used for all recordings. A train of 25 supra-threshold stimuli at 0.5 Hz was delivered in each experiment to record compound EJPs from both Is and Ib terminals, from which mean peak EJP values were obtained by averaging the last 20 traces. EJP amplitudes were corrected for non-linear summation using Martin's correction factor, assuming a reversal potential of 0 mV and a membrane capacitance factor of 0.55 (Kim et al., 2009; McLachlan and Martin, 1981). 8–10 separate animals were used for each genotype. A 2 minute continuous recording was used to measure mEJP amplitude. Traces were low-pass filtered at 1 kHz and analyzed using Clampfit or Mini-Analysis programs (Synaptosoft). 8–10 separate animals were analyzed for each genotype. Quantal content was determined by dividing the mean corrected EJP amplitude for a given synapse by the mean mEJP amplitude. Statistical significance was determined using one-way ANOVA. Representative traces were plotted using MS-Excel from episodic recordings exported from Clampfit as axon text files. High-frequency stimulation was carried out in a modified HL3 Ringer's solution containing 10 mM Ca2+ as described previously (Dickman et al., 2005) (in mM): NaCl 70, KCl 5, MgCl2 10, CaCl2 10, NaHCO3 10, Sucrose 115, Trehalose 5, BES 5 (pH 7.2). Rapid PhTox (Sigma Aldrich Inc.) mediated homeostasis was measured as described earlier in 0.3 mM Ca2+ containing HL3.1 Ringer's solution (Feng et al., 2004; Frank et al., 2006) (in mM): NaCl 70, KCl 5, MgCl2 10, CaCl2 0.3, NaHCO3 10, Sucrose 115, Trehalose 5, BES 5 (pH 7.2). Recordings in a loss of GluRIIA background were carried out in the same HL3.1 Ringer's with 0.3 mM Ca2+.
4.4. Behavioral analysis
2–3 day old flies were collected and their wings cut close to the thorax. These flies were then allowed to recover for an additional 3 days. Flies were individually placed on a circular platform 10 cm in diameter and surrounded by a moat of water 2 cm in width. Flies normally walked back and forth between two diametrically opposite vertical black bars within a brightly illuminated cylinder. A 5 mega-pixel webcam mounted centrally above the platform recorded their movement for a period of 5 minutes from within a Buridan tracking program from Bjorn Brembs. These recordings were then analyzed using custom designed Buridan analysis software (from Bjorn Brembs) written in the statistical package R. Results from this analysis were plotted in MS-Excel. Statistical significance was determined using one-way ANOVA. Single representative fly tracks and occupancy plots were also generated using the Buridan analysis software.
Supplementary Material
Highlights
Knock down of dSmn in the nervous system impairs locomotion
Knock down of dSmn in muscle does not impair locomotion
Synaptic transmission is normal following neuronal or muscle knock down of dSmn
Homeostasis of synaptic transmission is impaired by neuronal dSmn knock down
Acknowledgments
The authors thank Spyros Artavanis-Tsakonas for the UAS-SMN and UAS-SMN-RNAi reagents, Pejmun Haghighi for the GluRIIA stocks, Graeme Davis for help with PhTox experiments and Aaron DiAntonio for anti-GluRIII antibodies. S.S. also thanks Julien Colomb and Bjorn Brembs for help with the Buridan's assay and Anindya Sen for useful discussions.
Abbreviations
- NMJ
Neuro-muscular junction
- SMA
Spinal Muscular Atrophy
- SMN
Survival Motor Neuron
- UAS
Upstream Activating Sequence
- RNAi
RNA interference
- FGF
Fibroblast Growth Factor
- BMP
Bone Morphogenetic Protein
- GluR
Glutamate Receptor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
6. References
- 1.Balabanian S, Gendron NH, MacKenzie AE. Histologic and transcriptional assessment of a mild SMA model. Neurol Res. 2007;29:413–24. doi: 10.1179/016164107X159243. [DOI] [PubMed] [Google Scholar]
- 2.Baumer D, Lee S, Nicholson G, Davies JL, Parkinson NJ, Murray LM, Gillingwater TH, Ansorge O, Davies KE, Talbot K. Alternative splicing events are a late feature of pathology in a mouse model of spinal muscular atrophy. PLoS Genet. 2009;5:e1000773. doi: 10.1371/journal.pgen.1000773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–15. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 4.Briese M, Esmaeili B, Fraboulet S, Burt EC, Christodoulou S, Towers PR, Davies KE, Sattelle DB. Deletion of smn-1, the Caenorhabditis elegans ortholog of the spinal muscular atrophy gene, results in locomotor dysfunction and reduced lifespan. Hum Mol Genet. 2009;18:97–104. doi: 10.1093/hmg/ddn320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buhler D, Raker V, Luhrmann R, Fischer U. Essential role for the tudor domain of SMN in spliceosomal U snRNP assembly: implications for spinal muscular atrophy. Hum Mol Genet. 1999;8:2351–7. doi: 10.1093/hmg/8.13.2351. [DOI] [PubMed] [Google Scholar]
- 6.Burlet P, Huber C, Bertrandy S, Ludosky MA, Zwaenepoel I, Clermont O, Roume J, Delezoide AL, Cartaud J, Munnich A, Lefebvre S. The distribution of SMN protein complex in human fetal tissues and its alteration in spinal muscular atrophy. Hum Mol Genet. 1998;7:1927–33. doi: 10.1093/hmg/7.12.1927. [DOI] [PubMed] [Google Scholar]
- 7.Butchbach ME, Rose FF, Jr., Rhoades S, Marston J, McCrone JT, Sinnott R, Lorson CL. Effect of diet on the survival and phenotype of a mouse model for spinal muscular atrophy. Biochem Biophys Res Commun. 2010;391:835–40. doi: 10.1016/j.bbrc.2009.11.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cauchi RJ. SMN and Gemins: `we are family' … or are we?: insights into the partnership between Gemins and the spinal muscular atrophy disease protein SMN. Bioessays. 2010;32:1077–89. doi: 10.1002/bies.201000088. [DOI] [PubMed] [Google Scholar]
- 9.Cauchi RJ, Davies KE, Liu JL. A motor function for the DEAD-box RNA helicase, Gemin3, in Drosophila. PLoS Genet. 2008;4:e1000265. doi: 10.1371/journal.pgen.1000265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan YB, Miguel-Aliaga I, Franks C, Thomas N, Trulzsch B, Sattelle DB, Davies KE, van den Heuvel M. Neuromuscular defects in a Drosophila survival motor neuron gene mutant. Hum Mol Genet. 2003;12:1367–76. doi: 10.1093/hmg/ddg157. [DOI] [PubMed] [Google Scholar]
- 11.Chang HC, Dimlich DN, Yokokura T, Mukherjee A, Kankel MW, Sen A, Sridhar V, Fulga TA, Hart AC, Van Vactor D, Artavanis-Tsakonas S. Modeling spinal muscular atrophy in Drosophila. PLoS One. 2008;3:e3209. doi: 10.1371/journal.pone.0003209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cifuentes-Diaz C, Frugier T, Tiziano FD, Lacene E, Roblot N, Joshi V, Moreau MH, Melki J. Deletion of murine SMN exon 7 directed to skeletal muscle leads to severe muscular dystrophy. J Cell Biol. 2001;152:1107–14. doi: 10.1083/jcb.152.5.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cifuentes-Diaz C, Nicole S, Velasco ME, Borra-Cebrian C, Panozzo C, Frugier T, Millet G, Roblot N, Joshi V, Melki J. Neurofilament accumulation at the motor endplate and lack of axonal sprouting in a spinal muscular atrophy mouse model. Hum Mol Genet. 2002;11:1439–47. doi: 10.1093/hmg/11.12.1439. [DOI] [PubMed] [Google Scholar]
- 14.Coovert DD, Le TT, McAndrew PE, Strasswimmer J, Crawford TO, Mendell JR, Coulson SE, Androphy EJ, Prior TW, Burghes AH. The survival motor neuron protein in spinal muscular atrophy. Hum Mol Genet. 1997;6:1205–14. doi: 10.1093/hmg/6.8.1205. [DOI] [PubMed] [Google Scholar]
- 15.Dachs E, Hereu M, Piedrafita L, Casanovas A, Caldero J, Esquerda JE. Defective neuromuscular junction organization and postnatal myogenesis in mice with severe spinal muscular atrophy. J Neuropathol Exp Neurol. 2011;70:444–61. doi: 10.1097/NEN.0b013e31821cbd8b. [DOI] [PubMed] [Google Scholar]
- 16.Davis GW, DiAntonio A, Petersen SA, Goodman CS. Postsynaptic PKA controls quantal size and reveals a retrograde signal that regulates presynaptic transmitter release in Drosophila. Neuron. 1998;20:305–15. doi: 10.1016/s0896-6273(00)80458-4. [DOI] [PubMed] [Google Scholar]
- 17.Dickman DK, Horne JA, Meinertzhagen IA, Schwarz TL. A slowed classical pathway rather than kiss-and-run mediates endocytosis at synapses lacking synaptojanin and endophilin. Cell. 2005;123:521–33. doi: 10.1016/j.cell.2005.09.026. [DOI] [PubMed] [Google Scholar]
- 18.Dimitriadi M, Sleigh JN, Walker A, Chang HC, Sen A, Kalloo G, Harris J, Barsby T, Walsh MB, Satterlee JS, Li C, Van Vactor D, Artavanis-Tsakonas S, Hart AC. Conserved genes act as modifiers of invertebrate SMN loss of function defects. PLoS Genet. 2010;6:e1001172. doi: 10.1371/journal.pgen.1001172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feldkotter M, Schwarzer V, Wirth R, Wienker TF, Wirth B. Quantitative analyses of SMN1 and SMN2 based on real-time lightCycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am J Hum Genet. 2002;70:358–68. doi: 10.1086/338627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng Y, Ueda A, Wu CF. A modified minimal hemolymph-like solution, HL3.1, for physiological recordings at the neuromuscular junctions of normal and mutant Drosophila larvae. J Neurogenet. 2004;18:377–402. doi: 10.1080/01677060490894522. [DOI] [PubMed] [Google Scholar]
- 21.Fischer U, Liu Q, Dreyfuss G. The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell. 1997;90:1023–9. doi: 10.1016/s0092-8674(00)80368-2. [DOI] [PubMed] [Google Scholar]
- 22.Foust KD, Wang X, McGovern VL, Braun L, Bevan AK, Haidet AM, Le TT, Morales PR, Rich MM, Burghes AH, Kaspar BK. Rescue of the spinal muscular atrophy phenotype in a mouse model by early postnatal delivery of SMN. Nat Biotechnol. 2010;28:271–4. doi: 10.1038/nbt.1610. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Franciscovich AL, Mortimer AD, Freeman AA, Gu J, Sanyal S. Overexpression screen in Drosophila identifies neuronal roles of GSK-3 beta/shaggy as a regulator of AP-1-dependent developmental plasticity. Genetics. 2008;180:2057–71. doi: 10.1534/genetics.107.085555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frank CA, Kennedy MJ, Goold CP, Marek KW, Davis GW. Mechanisms underlying the rapid induction and sustained expression of synaptic homeostasis. Neuron. 2006;52:663–77. doi: 10.1016/j.neuron.2006.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frank CA, Pielage J, Davis GW. A presynaptic homeostatic signaling system composed of the Eph receptor, ephexin, Cdc42, and CaV2.1 calcium channels. Neuron. 2009;61:556–69. doi: 10.1016/j.neuron.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freeman A, Franciscovich A, Bowers M, Sandstrom DJ, Sanyal S. NFAT regulates pre-synaptic development and activity-dependent plasticity in Drosophila. Mol Cell Neurosci. 2011;46:535–47. doi: 10.1016/j.mcn.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Freeman A, Pranski E, Miller RD, Radmard S, Bernhard D, Jinnah HA, Betarbet R, Rye DB, Sanyal S. Sleep fragmentation and motor restlessness in a Drosophila model of restless legs syndrome. Curr Biol. 2012;22:1142–8. doi: 10.1016/j.cub.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frugier T, Tiziano FD, Cifuentes-Diaz C, Miniou P, Roblot N, Dierich A, Le Meur M, Melki J. Nuclear targeting defect of SMN lacking the C-terminus in a mouse model of spinal muscular atrophy. Hum Mol Genet. 2000;9:849–58. doi: 10.1093/hmg/9.5.849. [DOI] [PubMed] [Google Scholar]
- 29.Gabanella F, Carissimi C, Usiello A, Pellizzoni L. The activity of the spinal muscular atrophy protein is regulated during development and cellular differentiation. Hum Mol Genet. 2005;14:3629–42. doi: 10.1093/hmg/ddi390. [DOI] [PubMed] [Google Scholar]
- 30.Gabanella F, Butchbach ME, Saieva L, Carissimi C, Burghes AH, Pellizzoni L. Ribonucleoprotein assembly defects correlate with spinal muscular atrophy severity and preferentially affect a subset of spliceosomal snRNPs. PLoS One. 2007;2:e921. doi: 10.1371/journal.pone.0000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghosh S, Feany MB. Comparison of pathways controlling toxicity in the eye and brain in Drosophila models of human neurodegenerative diseases. Hum Mol Genet. 2004;13:2011–8. doi: 10.1093/hmg/ddh214. [DOI] [PubMed] [Google Scholar]
- 32.Gogliotti RG, Quinlan KA, Barlow CB, Heier CR, Heckman CJ, Didonato CJ. Motor neuron rescue in spinal muscular atrophy mice demonstrates that sensory-motor defects are a consequence, not a cause, of motor neuron dysfunction. J Neurosci. 2012;32:3818–29. doi: 10.1523/JNEUROSCI.5775-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gotz KG. Visual guidance in Drosophila. Basic Life Sci. 1980;16:391–407. doi: 10.1007/978-1-4684-7968-3_28. [DOI] [PubMed] [Google Scholar]
- 34.Haghighi AP, McCabe BD, Fetter RD, Palmer JE, Hom S, Goodman CS. Retrograde control of synaptic transmission by postsynaptic CaMKII at the Drosophila neuromuscular junction. Neuron. 2003;39:255–67. doi: 10.1016/s0896-6273(03)00427-6. [DOI] [PubMed] [Google Scholar]
- 35.Hammond SM, Gogliotti RG, Rao V, Beauvais A, Kothary R, DiDonato CJ. Mouse survival motor neuron alleles that mimic SMN2 splicing and are inducible rescue embryonic lethality early in development but not late. PLoS One. 2010;5:e15887. doi: 10.1371/journal.pone.0015887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hsieh-Li HM, Chang JG, Jong YJ, Wu MH, Wang NM, Tsai CH, Li H. A mouse model for spinal muscular atrophy. Nat Genet. 2000;24:66–70. doi: 10.1038/71709. [DOI] [PubMed] [Google Scholar]
- 37.Imlach WL, Beck ES, Choi BJ, Lotti F, Pellizzoni L, McCabe BD. SMN is required for sensory-motor circuit function in Drosophila. Cell. 2012;151:427–39. doi: 10.1016/j.cell.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jablonka S, Schrank B, Kralewski M, Rossoll W, Sendtner M. Reduced survival motor neuron (Smn) gene dose in mice leads to motor neuron degeneration: an animal model for spinal muscular atrophy type III. Hum Mol Genet. 2000;9:341–6. doi: 10.1093/hmg/9.3.341. [DOI] [PubMed] [Google Scholar]
- 39.Kariya S, Park GH, Maeno-Hikichi Y, Leykekhman O, Lutz C, Arkovitz MS, Landmesser LT, Monani UR. Reduced SMN protein impairs maturation of the neuromuscular junctions in mouse models of spinal muscular atrophy. Hum Mol Genet. 2008;17:2552–69. doi: 10.1093/hmg/ddn156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim SM, Kumar V, Lin YQ, Karunanithi S, Ramaswami M. Fos and Jun potentiate individual release sites and mobilize the reserve synaptic vesicle pool at the Drosophila larval motor synapse. Proc Natl Acad Sci U S A. 2009;106:4000–5. doi: 10.1073/pnas.0806064106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kitamoto T. Conditional modification of behavior in Drosophila by targeted expression of a temperature-sensitive shibire allele in defined neurons. J Neurobiol. 2001;47:81–92. doi: 10.1002/neu.1018. [DOI] [PubMed] [Google Scholar]
- 42.Kong L, Wang X, Choe DW, Polley M, Burnett BG, Bosch-Marce M, Griffin JW, Rich MM, Sumner CJ. Impaired synaptic vesicle release and immaturity of neuromuscular junctions in spinal muscular atrophy mice. J Neurosci. 2009;29:842–51. doi: 10.1523/JNEUROSCI.4434-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kroiss M, Schultz J, Wiesner J, Chari A, Sickmann A, Fischer U. Evolution of an RNP assembly system: a minimal SMN complex facilitates formation of UsnRNPs in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2008;105:10045–50. doi: 10.1073/pnas.0802287105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Le TT, Pham LT, Butchbach ME, Zhang HL, Monani UR, Coovert DD, Gavrilina TO, Xing L, Bassell GJ, Burghes AH. SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum Mol Genet. 2005;14:845–57. doi: 10.1093/hmg/ddi078. [DOI] [PubMed] [Google Scholar]
- 45.Lee JB, Lee KA, Hong JM, Suh GI, Choi YC. Homozygous SMN2 deletion is a major risk factor among twenty-five Korean sporadic amyotrophic lateral sclerosis patients. Yonsei Med J. 2012;53:53–7. doi: 10.3349/ymj.2012.53.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee L, Davies SE, Liu JL. The spinal muscular atrophy protein SMN affects Drosophila germline nuclear organization through the U body-P body pathway. Dev Biol. 2009;332:142–55. doi: 10.1016/j.ydbio.2009.05.553. [DOI] [PubMed] [Google Scholar]
- 47.Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–65. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 48.Lefebvre S, Burlet P, Liu Q, Bertrandy S, Clermont O, Munnich A, Dreyfuss G, Melki J. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat Genet. 1997;16:265–9. doi: 10.1038/ng0797-265. [DOI] [PubMed] [Google Scholar]
- 49.Lima SQ, Miesenbock G. Remote control of behavior through genetically targeted photostimulation of neurons. Cell. 2005;121:141–52. doi: 10.1016/j.cell.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 50.Ling KK, Lin MY, Zingg B, Feng Z, Ko CP. Synaptic defects in the spinal and neuromuscular circuitry in a mouse model of spinal muscular atrophy. PLoS One. 2011;5:e15457. doi: 10.1371/journal.pone.0015457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Q, Fischer U, Wang F, Dreyfuss G. The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell. 1997;90:1013–21. doi: 10.1016/s0092-8674(00)80367-0. [DOI] [PubMed] [Google Scholar]
- 52.Lorson CL, Hahnen E, Androphy EJ, Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci U S A. 1999;96:6307–11. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lorson CL, Androphy EJ. An exonic enhancer is required for inclusion of an essential exon in the SMA-determining gene SMN. Hum Mol Genet. 2000;9:259–65. doi: 10.1093/hmg/9.2.259. [DOI] [PubMed] [Google Scholar]
- 54.Lotti F, Imlach WL, Saieva L, Beck ES, Hao Le T, Li DK, Jiao W, Mentis GZ, Beattie CE, McCabe BD, Pellizzoni L. An SMN-dependent U12 splicing event essential for motor circuit function. Cell. 2012;151:440–54. doi: 10.1016/j.cell.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luo L, Liao YJ, Jan LY, Jan YN. Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes Dev. 1994;8:1787–802. doi: 10.1101/gad.8.15.1787. [DOI] [PubMed] [Google Scholar]
- 56.Lutz CM, Kariya S, Patruni S, Osborne MA, Liu D, Henderson CE, Li DK, Pellizzoni L, Rojas J, Valenzuela DM, Murphy AJ, Winberg ML, Monani UR. Postsymptomatic restoration of SMN rescues the disease phenotype in a mouse model of severe spinal muscular atrophy. J Clin Invest. 2011;121:3029–41. doi: 10.1172/JCI57291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mahr A, Aberle H. The expression pattern of the Drosophila vesicular glutamate transporter: a marker protein for motoneurons and glutamatergic centers in the brain. Gene Expr Patterns. 2006;6:299–309. doi: 10.1016/j.modgep.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 58.Mailman MD, Heinz JW, Papp AC, Snyder PJ, Sedra MS, Wirth B, Burghes AH, Prior TW. Molecular analysis of spinal muscular atrophy and modification of the phenotype by SMN2. Genet Med. 2002;4:20–6. doi: 10.1097/00125817-200201000-00004. [DOI] [PubMed] [Google Scholar]
- 59.Marrus SB, Portman SL, Allen MJ, Moffat KG, DiAntonio A. Differential localization of glutamate receptor subunits at the Drosophila neuromuscular junction. J Neurosci. 2004;24:1406–15. doi: 10.1523/JNEUROSCI.1575-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martinez TL, Kong L, Wang X, Osborne MA, Crowder ME, Van Meerbeke JP, Xu X, Davis C, Wooley J, Goldhamer DJ, Lutz CM, Rich MM, Sumner CJ. Survival Motor Neuron Protein in Motor Neurons Determines Synaptic Integrity in Spinal Muscular Atrophy. J Neurosci. 2012;32:8703–8715. doi: 10.1523/JNEUROSCI.0204-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McAndrew PE, Parsons DW, Simard LR, Rochette C, Ray PN, Mendell JR, Prior TW, Burghes AH. Identification of proximal spinal muscular atrophy carriers and patients by analysis of SMNT and SMNC gene copy number. Am J Hum Genet. 1997;60:1411–22. doi: 10.1086/515465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McLachlan EM, Martin AR. Non-linear summation of end-plate potentials in the frog and mouse. J Physiol. 1981;311:307–24. doi: 10.1113/jphysiol.1981.sp013586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meister G, Hannus S, Plottner O, Baars T, Hartmann E, Fakan S, Laggerbauer B, Fischer U. SMNrp is an essential pre-mRNA splicing factor required for the formation of the mature spliceosome. EMBO J. 2001;20:2304–14. doi: 10.1093/emboj/20.9.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Michaud M, Arnoux T, Bielli S, Durand E, Rotrou Y, Jablonka S, Robert F, Giraudon-Paoli M, Riessland M, Mattei MG, Andriambeloson E, Wirth B, Sendtner M, Gallego J, Pruss RM, Bordet T. Neuromuscular defects and breathing disorders in a new mouse model of spinal muscular atrophy. Neurobiol Dis. 2010;38:125–35. doi: 10.1016/j.nbd.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 65.Monani UR, Lorson CL, Parsons DW, Prior TW, Androphy EJ, Burghes AH, McPherson JD. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum Mol Genet. 1999;8:1177–83. doi: 10.1093/hmg/8.7.1177. [DOI] [PubMed] [Google Scholar]
- 66.Monani UR, Sendtner M, Coovert DD, Parsons DW, Andreassi C, Le TT, Jablonka S, Schrank B, Rossoll W, Prior TW, Morris GE, Burghes AH. The human centromeric survival motor neuron gene (SMN2) rescues embryonic lethality in Smn(−/−) mice and results in a mouse with spinal muscular atrophy. Hum Mol Genet. 2000;9:333–9. doi: 10.1093/hmg/9.3.333. [DOI] [PubMed] [Google Scholar]
- 67.Murray LM, Lee S, Baumer D, Parson SH, Talbot K, Gillingwater TH. Pre-symptomatic development of lower motor neuron connectivity in a mouse model of severe spinal muscular atrophy. Hum Mol Genet. 2010;19:420–33. doi: 10.1093/hmg/ddp506. [DOI] [PubMed] [Google Scholar]
- 68.Murray LM, Comley LH, Thomson D, Parkinson N, Talbot K, Gillingwater TH. Selective vulnerability of motor neurons and dissociation of pre- and post-synaptic pathology at the neuromuscular junction in mouse models of spinal muscular atrophy. Hum Mol Genet. 2008;17:949–62. doi: 10.1093/hmg/ddm367. [DOI] [PubMed] [Google Scholar]
- 69.Narver HL, Kong L, Burnett BG, Choe DW, Bosch-Marce M, Taye AA, Eckhaus MA, Sumner CJ. Sustained improvement of spinal muscular atrophy mice treated with trichostatin A plus nutrition. Ann Neurol. 2008;64:465–70. doi: 10.1002/ana.21449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Neuser K, Triphan T, Mronz M, Poeck B, Strauss R. Analysis of a spatial orientation memory in Drosophila. Nature. 2008;453:1244–7. doi: 10.1038/nature07003. [DOI] [PubMed] [Google Scholar]
- 71.Nicole S, Desforges B, Millet G, Lesbordes J, Cifuentes-Diaz C, Vertes D, Cao ML, De Backer F, Languille L, Roblot N, Joshi V, Gillis JM, Melki J. Intact satellite cells lead to remarkable protection against Smn gene defect in differentiated skeletal muscle. J Cell Biol. 2003;161:571–82. doi: 10.1083/jcb.200210117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Park GH, Maeno-Hikichi Y, Awano T, Landmesser LT, Monani UR. Reduced survival of motor neuron (SMN) protein in motor neuronal progenitors functions cell autonomously to cause spinal muscular atrophy in model mice expressing the human centromeric (SMN2) gene. J Neurosci. 2010;30:12005–19. doi: 10.1523/JNEUROSCI.2208-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pearn J. Incidence, prevalence, and gene frequency studies of chronic childhood spinal muscular atrophy. J Med Genet. 1978;15:409–13. doi: 10.1136/jmg.15.6.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pellizzoni L, Charroux B, Dreyfuss G. SMN mutants of spinal muscular atrophy patients are defective in binding to snRNP proteins. Proc Natl Acad Sci U S A. 1999;96:11167–72. doi: 10.1073/pnas.96.20.11167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pellizzoni L, Yong J, Dreyfuss G. Essential role for the SMN complex in the specificity of snRNP assembly. Science. 2002;298:1775–9. doi: 10.1126/science.1074962. [DOI] [PubMed] [Google Scholar]
- 76.Penney J, Tsurudome K, Liao EH, Elazzouzi F, Livingstone M, Gonzalez M, Sonenberg N, Haghighi AP. TOR is required for the retrograde regulation of synaptic homeostasis at the Drosophila neuromuscular junction. Neuron. 2012;74:166–78. doi: 10.1016/j.neuron.2012.01.030. [DOI] [PubMed] [Google Scholar]
- 77.Petersen SA, Fetter RD, Noordermeer JN, Goodman CS, DiAntonio A. Genetic analysis of glutamate receptors in Drosophila reveals a retrograde signal regulating presynaptic transmitter release. Neuron. 1997;19:1237–48. doi: 10.1016/s0896-6273(00)80415-8. [DOI] [PubMed] [Google Scholar]
- 78.Praveen K, Wen Y, Matera AG. A Drosophila Model of Spinal Muscular Atrophy Uncouples snRNP Biogenesis Functions of Survival Motor Neuron from Locomotion and Viability Defects. Cell Rep. 2012;1:624–31. doi: 10.1016/j.celrep.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rajendra TK, Gonsalvez GB, Walker MP, Shpargel KB, Salz HK, Matera AG. A Drosophila melanogaster model of spinal muscular atrophy reveals a function for SMN in striated muscle. J Cell Biol. 2007;176:831–41. doi: 10.1083/jcb.200610053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ruiz R, Casanas JJ, Torres-Benito L, Cano R, Tabares L. Altered intracellular Ca2+ homeostasis in nerve terminals of severe spinal muscular atrophy mice. J Neurosci. 2010;30:849–57. doi: 10.1523/JNEUROSCI.4496-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sakai T, Kasuya J, Kitamoto T, Aigaki T. The Drosophila TRPA channel, Painless, regulates sexual receptivity in virgin females. Genes Brain Behav. 2009;8:546–57. doi: 10.1111/j.1601-183X.2009.00503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Salvaterra PM, Kitamoto T. Drosophila cholinergic neurons and processes visualized with Gal4/UAS-GFP. Brain Res Gene Expr Patterns. 2001;1:73–82. doi: 10.1016/s1567-133x(01)00011-4. [DOI] [PubMed] [Google Scholar]
- 83.Seabrooke S, Stewart BA. Synaptic transmission and plasticity are modulated by nonmuscle myosin II at the neuromuscular junction of Drosophila. J Neurophysiol. 2011;105:1966–76. doi: 10.1152/jn.00718.2010. [DOI] [PubMed] [Google Scholar]
- 84.Sen A, Yokokura T, Kankel MW, Dimlich DN, Manent J, Sanyal S, Artavanis-Tsakonas S. Modeling spinal muscular atrophy in Drosophila links Smn to FGF signaling. J Cell Biol. 2011;192:481–95. doi: 10.1083/jcb.201004016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shpargel KB, Matera AG. Gemin proteins are required for efficient assembly of Sm-class ribonucleoproteins. Proc Natl Acad Sci U S A. 2005;102:17372–7. doi: 10.1073/pnas.0508947102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shpargel KB, Praveen K, Rajendra TK, Matera AG. Gemin3 is an essential gene required for larval motor function and pupation in Drosophila. Mol Biol Cell. 2009;20:90–101. doi: 10.1091/mbc.E08-01-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sleigh JN, Buckingham SD, Esmaeili B, Viswanathan M, Cuppen E, Westlund BM, Sattelle DB. A novel Caenorhabditis elegans allele, smn-1(cb131), mimicking a mild form of spinal muscular atrophy, provides a convenient drug screening platform highlighting new and pre-approved compounds. Hum Mol Genet. 2011;20:245–60. doi: 10.1093/hmg/ddq459. [DOI] [PubMed] [Google Scholar]
- 88.Sleigh JN, Gillingwater TH, Talbot K. The contribution of mouse models to understanding the pathogenesis of spinal muscular atrophy. Dis Model Mech. 2011;4:457–67. doi: 10.1242/dmm.007245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stewart BA, Atwood HL, Renger JJ, Wang J, Wu CF. Improved stability of Drosophila larval neuromuscular preparations in haemolymph-like physiological solutions. J Comp Physiol A. 1994;175:179–91. doi: 10.1007/BF00215114. [DOI] [PubMed] [Google Scholar]
- 90.Vitte JM, Davoult B, Roblot N, Mayer M, Joshi V, Courageot S, Tronche F, Vadrot J, Moreau MH, Kemeny F, Melki J. Deletion of murine Smn exon 7 directed to liver leads to severe defect of liver development associated with iron overload. Am J Pathol. 2004;165:1731–41. doi: 10.1016/S0002-9440(10)63428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wan L, Battle DJ, Yong J, Gubitz AK, Kolb SJ, Wang J, Dreyfuss G. The survival of motor neurons protein determines the capacity for snRNP assembly: biochemical deficiency in spinal muscular atrophy. Mol Cell Biol. 2005;25:5543–51. doi: 10.1128/MCB.25.13.5543-5551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.