Abstract
Transcription factor II H (TFIIH) is composed of core TFIIH and Cdk-activating kinase (CAK) complexes. Besides transcription, TFIIH also participates in nucleotide excision repair (NER), verifying DNA lesions through its helicase components XPB and XPD. The assembly state of TFIIH is known to be affected by truncation mutations in Xeroderma pigmentosum group G/Cockayne syndrome (XP-G/CS). Here, we showed that CAK component MAT1 was rapidly recruited to UV-induced DNA damage sites, co-localizing with core TFIIH component p62, and dispersed from the damage sites upon completion of DNA repair. While the core TFIIH-CAK association remained intact, MAT1 failed to accumulate at DNA damage sites in fibroblasts harboring XP-B or XP-B/CS mutations. Nevertheless, MAT1, XPD and XPC as well as XPG were able to accumulate at damage sites in XP-D fibroblasts, in which the core TFIIH-CAK association also remained intact. Interestingly, XPG recruitment was impaired in XP-B/CS fibroblasts derived from patients with mild phenotype, but persisted in XP-B/CS fibroblasts from severely affected patients resulting in a nonfunctional preincision complex. An examination of steady-state levels of RNA polymerase II (RNAPII) indicated that UV-induced RNAPII phosphorylation was dramatically reduced in XP-B/CS fibroblasts. These results demonstrated that the CAK rapidly disassociates from the core TFIIH upon assembly of nonfunctional preincision complex in XP-B and XP-B/CS cells. The persistency of nonfunctional preincision complex correlates with the severity exhibited by XP-B patients. The results suggest that XPB and XPD helicases differentially regulate the anchoring of CAK to core TFIIH during damage verification step of NER.
Keywords: Nucleotide excision repair, Xeroderma pigmentosum, Cockayne syndrome, Transcription factor II H, Cdk-activating kinase complex
1. Introduction
Because of its pivotal importance, DNA molecules are often assumed to be stable. In reality, the genome of eukaryotic cells is constantly subjected to challenges by numerous physical and chemical agents from endogenous metabolism and in our environment. Cells utilize several repair pathways to overcome the deleterious effects of DNA damage and maintain their genomic integrity. Nucleotide excision repair (NER) removes a broad variety of double-helix-distorting DNA lesions, including UV-induced cyclobutane pyridine dimers (CPDs) and 6-4 pyrimidine-pyrimidone photoproducts (6-4PPs) [1]. Defects in NER are associated with several rare autosomal recessive genetic disorders including Xeroderma pigmentosum (XP) and Cockayne syndrome (CS) [2]. Seven XP proteins, corresponding to XP complementary group A to G, have been identified, whereas two CS related proteins, CSA and CSB, have been discovered. Selective participation of these components in NER distinguishes two NER sub-pathways, global genomic repair (GGR) which removes DNA lesions from the entire genome, and transcription-coupled repair (TCR) which eliminates DNA lesions from actively transcribed genes [3].
A generally accepted biochemical NER model includes damage recognition, dual incision, and gap-filling DNA synthesis steps [4, 5]. In GGR, the damage-induced DNA distortion is recognized by XPC-hHR23B protein complex [6, 7], and then TFIIH is recruited to open the DNA helix around damage sites and verify the lesions [8-10]. In TCR, lesions are detected by RNA polymerase II (RNAPII) in coordination with the recognition of stalled RNAPII, by CSA, CSB and TFIIH [11-13]. Other NER factors, such as XPA, XPF, XPG and RPA are believed to join the TFIIH-containing complex to form the final preincision complex [14]. The endonuclease XPG and XPF-ERCC1 are responsible for dual incision and the removal of ~24-32 nt oligonucleotide containing the culprit lesion. Subsequent gap-filling DNA synthesis is performed by a concerted action of pol δ or ε, and the cofactors PCNA, RF-C and RPA.
Mammalian TFIIH (referred as holo TFIIH) is organized into core TFIIH, containing the seven subunits XPB, XPD, p62, p52, p44, p34 and p8/TTD-A [15-18], coupled to a Cdk-activating kinase (CAK) complex composed of three subunits Cdk7, cyclin H and MAT1 [19]. TFIIH is a multifunctional protein complex, participating in transcription, NER and cell cycle regulation [17, 20, 21]. In NER, XPB and XPD helicases of TFIIH are involved in unwinding the DNA duplex around the lesion, providing an open DNA structure for subsequent XPG and XPF-ERCC1 cleavage [10]. During transcription, TFIIH functions in harmony with other basal transcription factors, e.g., TFIIB, TFIID, TFIIE and TFIIF. By virtue of its XPB helicase, TFIIH is essential for transcription initiation and promoter escape [22]. In the latter processes, Cdk7 of CAK mediates, at least partially, the phosphorylation of the carboxyl terminal domain (CTD) domain of largest Rpb1 subunit of RNAPII [23]. Therefore, TFIIH must face the challenge of switching its role to cope with diverse functional tasks. Such a challenge is obvious when a transcribing RNAPII is stalled by a DNA lesion, and TFIIH, in collaboration with CSA, CSB and XPG, funnels transcription into the dual incision processing by TCR [12].
TCR defect is related to CS, which is characterized by a wide range of symptoms including severe neurological abnormality [24-26], while defect in GGR is associated with XP symptoms which have characteristics such as extreme sensitivity to sunlight and increased risk of developing sunlight-induced skin cancers. CS patients suffer from skin photosensitivity without cancer predisposition. The cells derived from CS suffer a global impairment in transcription and exhibit a reduced recovery of RNA synthesis following UV exposure [27-29]. CS features can co-exist with XP symptoms (XP/CS) as found in XP-B/CS, XP-D/CS and XP-G/CS [25]. Repair defective XPA and XPF mutations results in XP but not CS symptoms. Thus, NER deficiency cannot explain the CS features of XP-B/CS, XP-D/CS and XP-G/CS. We have previously investigated the molecular and cellular manifestation of TFIIH compositional changes in human XP-G/CS cells and reported that CAK complex was not recruited to DNA damage in XP-G/CS cells [30]. Moreover, kinase activity of CAK is required neither for the assembly of preincision complex nor for GGR. Instead, CAK is involved in regulating phosphorylation and UV-induced degradation of RNAPII. However, it is not clear whether the CAK dissociation is a distinct molecular feature of XP/CS.
In the present study, we have explored whether TFIIH compositional change also occurs in XP-B/CS and XP-D fibroblasts. We found that MAT1 fails to accumulate at DNA damage sites in fibroblasts harboring XP-B or XP-B/CS mutations but not in fibroblasts with XP-D mutations. We confirmed that the core TFIIH and CAK remain associated in XP-B/CS and XP-D fibroblasts despite these cells having reduced XPB and XPD protein levels. Thus, unlike in human XP-G/CS cells, TFIIH undergoes a dramatic compositional change in releasing CAK immediately after TFIIH is recruited to DNA damage in XP-B and XP-B/CS. We also provided the evidence that nonfunctional NER preincision complexes are assembled and persist in severe XP-B/CS fibroblasts. We further examined the steady-state levels of RNAPII and found that the UV-induced RNAPII phosphorylation is reduced and RNAPII degradation is delayed in XP-B/CS fibroblasts. These results suggested that XPB and XPD differentially regulate TFIIH compositional changes during NER and provide meaningful insights on the role of CAK in XP/CS.
2. Materials and Methods
2.1. Cell lines and reagents
Normal human fibroblasts (NHF, OSU2) were established in our laboratory [31]. Primary XP-B fibroblasts from XP patients with mild diseases [GM113025 (/XPCS1BA), GM113026 (/XPCS2BA) and GM21071 (/XP33BR)], or with severe XP/CS complex [GM21072 (/XP183MA) and GM21153 (/XP181MA)] were obtained from the Coriell Cell Repositories (Camden, New Jersey 08103, USA). So were the primary XP-D fibroblasts GM03615 (/XP1BR), GM10428 (/XP17BE), GM10430 (/XP6BE), GM03248 (/XP-CS2) and GM08207/SV40-transformed XP6BE. The clinical and genetic features of mutations in XP-B, XP-B/CS and XP-D cells were previously described [32-36] and are summarized in Supplemental Table 1. The fibroblasts and HeLa cells were grown in DMEM (for NHF and HeLa) or MEM (for primary XP or XP/CS cells) supplemented with 40 mM glutamine, 10% FCS, antibiotics at 37 °C in a humidified atmosphere of 5% CO2.
Rabbit anti-XPC and anti-CPD antibodies were raised in our laboratory as previously described [37-39]. Polycolonal anti-XPB (S-19), anti-XPD (H-150), anti-MAT1 (FL-309), monoclonal anti-MAT1 (F-6), anti-p62 (G-10) and anti-Cdk7 (C4) antibodies were obtained from Santa Cruz biotechnology (Santa Cruz, CA). Monoclonal anti-XPA (12F5) and anti-XPG (8H7) antibodies were purchased from NeoMarkers (Fremont, CA). Monoclonal Ser5-phospho-RNAPII antibody (4H8), fluorescent conjugated Alexa Fluor 488 (goat anti-mouse) and Texas Red (goat anti-rabbit) were acquired from Affinity BioReagents (Golden, CO), Invitrogen (Carlsbad, CA) and Santa Cruz biotechnology, respectively.
2.2. UV and micropore UV irradiation
The cells in monolayers were washed twice with phosphate-buffered saline (PBS). The 254-nm UV (UV-C) was delivered to cells at a rate of 0.5 J/m2/sec as measured by Model UVX Digital Radiometer. For micropore UV irradiation, cells were grown on glass coverslips to proper density. The cells on coverslips were washed twice with PBS and a 5-μm-isopore polycarbonate filter (Millipore, Bedford, MA) was placed on top of cell monolayer. The coverslips were then irradiated and the cells were processed immediately [0 hour (h)] or maintained in fresh suitable medium for a desired period (0.5, 1, 3 or 24 h) and processed thereafter.
2.3. Immunofluorescence
Immunofluorescence double labeling was performed according to the method established in our laboratory [40, 41]. Briefly, the micropore UV irradiated cells on coverslips were washed twice with cold PBS, permeabilized with 0.5% Triton X-100/PBS on ice for 8 minutes (min) and then fixed with 2% paraformaldehyde in 0.5% Triton X-100/PBS at 4 °C for 30 min. After fixation, the coverslips were rinsed twice with cold PBS and blocked with 20% normal goat serum (NGS) in 0.1% Triton X-100/PBS at 37°C for 2 h. Primary antibody rabbit anti-XPC, anti-XPB, anti-XPD, anti-MAT1, monoclonal anti-p62, anti-XPG or anti-XPA antibody in proper dilution, ranging from 1:50 to 1:1000, were all prepared in 0.1% Tween-20/PBS with 5% NGS and layered on fixed cells for 1 h at room temperature or at 4 °C overnight. Following primary antibody incubation, the cells were washed in 0.1% Tween-20/PBS washing buffer for 4 times, 5 min each. The coverslips were then incubated with fluorescent (Alexa Fluor 488 or Texas Red) secondary antibodies (in 1:200 to 1:1000 dilutions) for another 1 h at room temperature. The coverslips were subsequently washed, mounted and counterstained with DAPI. Immunofluorescent images were captured with a Nikon Fluorescence Microscope E80i (Nikon, Tokyo, Japan) equipped with SPOT analysis software.
2.4. Immunoprecipitation and Western Blot analysis
Fibroblasts were grown to ~70% confluence, left unirradiated or UV-irradiated at a preferred UV dose indicated for individual experiments. The irradiated cells were cultured further for varying repair periods. The cells were then washed twice with PBS and the cell lysates were made in RIPA buffer containing 50 mM Tris-HCl (pH, 7.4), 150 mM NaCl, 5 mM EDTA, 1% NP40, 0.5% Sodium deoxycholate, 0.1% SDS, 0.5 mM PMSF and a complete protease inhibitor cocktail. The cell lysates were cleared by centrifuging at 14, 000 rpm at 4 °C for 15 min. The lysates containing ~0.5 mg proteins were pre-cleared by protein A/G agarose beads (Calbiochem, San Diego, CA) at 4 °C for 3 h and then incubated with 2 μg specific antibodies at 4 °C overnight, followed by immunocapture with ~30 μl of 50% slurry protein A/G agarose beads at 4 °C for 2 h. The immunoprecipitates were collected, washed 4 times with RIPA buffer, resuspended in 40 μl of Laemmli sample buffer, boiled for 10 min and subjected to Western Blot analysis. For Western Blotting, the samples were prepared by immunoprecipitation or by making whole cell extracts in SDS lysis buffer containing 2% SDS, 10% glycerol, 10 mM DDT (freshly added), 62 mM Tris-HCl (pH 6.8) and a complete protease inhibitor cocktail (phosphatase inhibitor phosSTOP was added, as needed). The protein samples were quantitated by DC Bio-Rad Protein Assay, separated by SDS polyacrylamide gel electrophoresis and transferred to a PVDF membrane. The immunoblotting was performed with appropriate primary and secondary antibodies and detected using enhanced chemiluminescence. Autoradiographs were quantitated using NIH ImageJ software.
3. Results
3.1. CAK complex fails to accumulate at DNA damage sites in cells harboring XP-B or XP-B/CS mutations
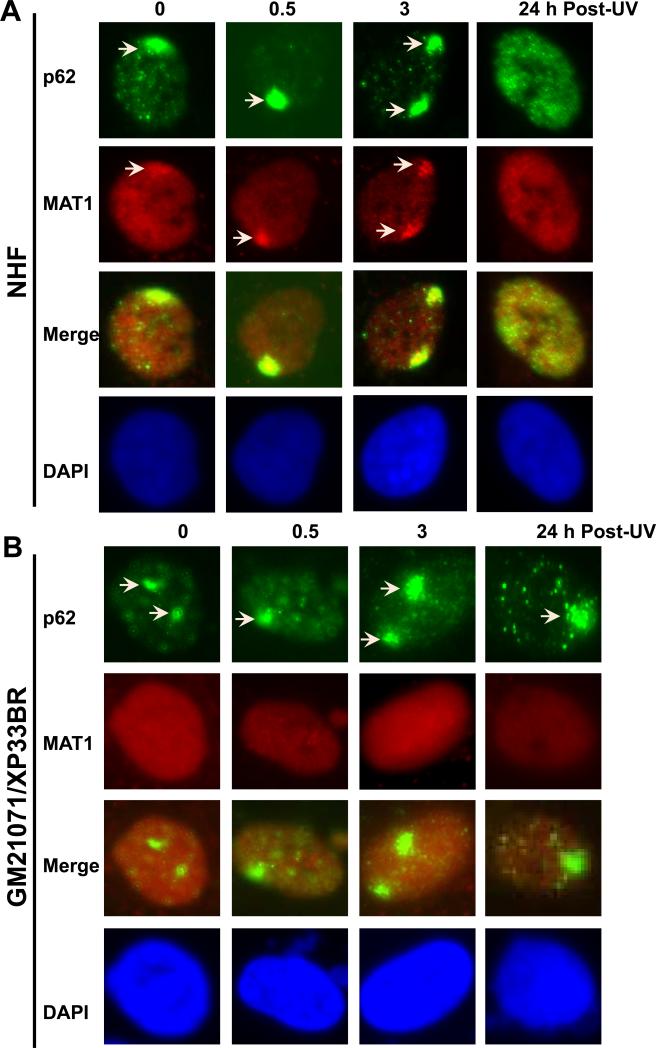
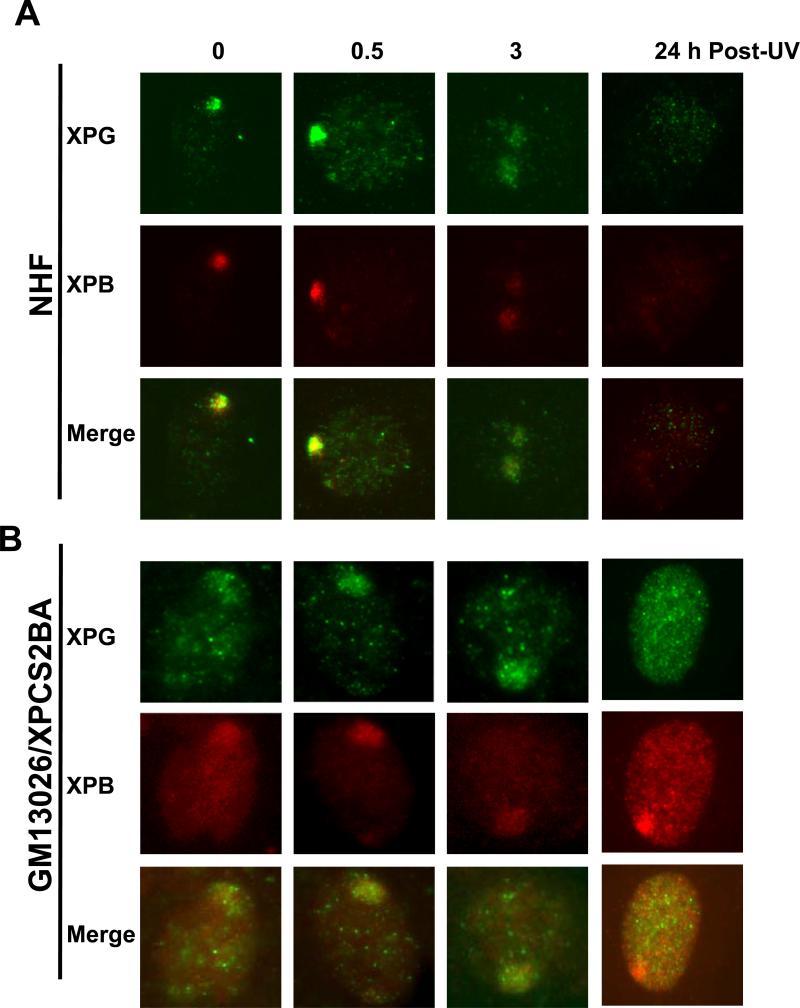
We have previous observed that CAK complex was not recruited to DNA damage in human XP-G/CS cells due to XPG truncation mutation, which resulted in dissociation of CAK from core TFIIH [30]. To investigate whether dissociation of CAK from core TFIIH is a common phenomenon for XP/CS, we first assessed the MAT1 accumulation at DNA damage sites in NHF and in XP fibroblasts harboring XP-B or XP-B/CS mutations. We used micropore UV irradiation to generate localized DNA damage spots and examined the recruitment of TFIIH components p62 and MAT1 to the damage as function of time. Most likely the bulky helix distortion from 6-4PPs in localized spots provokes the accumulation of NER factors such as XPC, XPG and TFIIH [6]. As shown in Fig. 1, p62 accumulated rapidly and formed foci readily visible immediately (0 h) after UV irradiation. These p62 foci co-localized with micropore UV-induced CPD spots but dispersed by 24 h when CPD spots were still distinctly visible (Supplemental Fig. S1A). MAT1 accumulated in the same pattern and the MAT1 foci colocalized with p62 foci in NHF cells. As 6-4PPs are known to have in fast repair kinetics, dispersion of p62 and MAT1 at 24 h would be the result of fully completed 6-4PP repair. The co-localization of p62 with MAT1, XPD, XPB and XPC foci was further confirmed in repair-proficient HeLa cells (Supplemental Fig. S.1B). Additionally, micropore UV induced accumulation of Cdk7, another CAK component, was also detected to co-localize with XPC in HeLa cells. These experiments demonstrated that the holo TFIIH is recruited for NER in repair-proficient cells.
Fig. 1.
XP-B and XP-B/CS fibroblasts are defective in accumulation of CAK component MAT1 at DNA damage sites in vivo. Normal human fibroblasts (NHF) (A), XP-B fibroblasts XP33BR (B), XP-B/CS fibroblasts XPCS1BA (C) and XP183MA (D) were grown on coverslips, locally irradiated with 100 J/m2 UV through a 5 μm-isopore polycarbonate filter, fixed immediately (0 h) or cultured for 0.5, 3, or 24 h before fixing in 2% paraformaldehyde. The TFIIH component p62 and CAK component MAT1 were visualized by immunofluorescence double labeling using specific antibodies. Nuclei shown in blue are from counterstaining with DAPI. Arrows indicate immunofluorescent foci due to accumulation of THIIH proteins at localized damage sites.
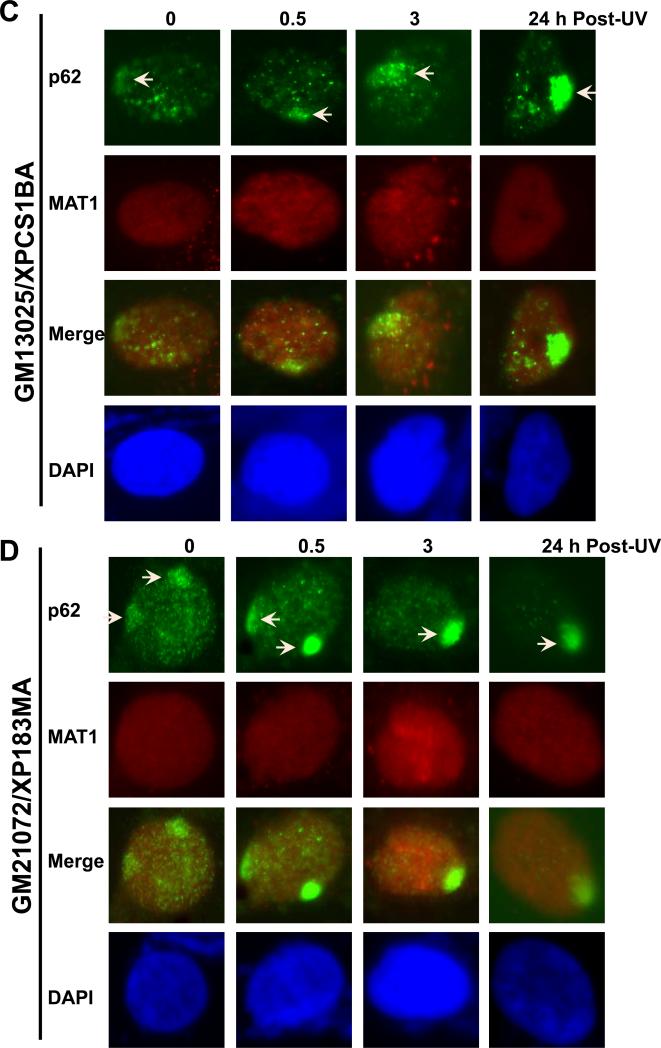
Examination of MAT1 and p62 in human XP-B/CS cells showed a distinctly different pattern as compared with that in NHF. Accumulation of p62 was seen in 0, 0.5, 3 and 24 h time points in XPCS1BA fibroblasts. Failure of p62 dispersion at 24 h is consistent with the defective repair of 6-4PPs in these cells (Fig. 1C). On the other hand, no accumulation of MAT1 was observed at any time point in XPCS1BA fibroblasts. We further tested XPB33BR, XP183MA (Fig. 1B and D), XPBCS2BA and XP181MA (Supplemental Fig. S1C and D) fibroblasts. Not surprisingly, none of these fibroblasts showed MAT1 recruitment, while p62 accumulation followed a clearly identical pattern in all the fibroblasts. To unambiguously confirm the disparate responses, we co-cultured NHF and XP fibroblasts on same coverslips and pre-labeled the cells with latex beads of two different sizes so that immunofluorescence labeling and factor recruitments can be concomitantly performed under identical conditions. Here again, double labeling for γH2AX and MAT1 did not show any accumulation of MAT1 in XPCS2BA, XP183MA and XP181MA fibroblasts, while γH2AX foci were distinctly seen (Supplemental Fig. S2B). Moreover, γH2AX exhibited a clear co-localization with MAT1 in NHF cells (Supplemental Fig. S2). Thus, we concluded that MAT1-containing CAK failed to accumulate at damage sites in XP-B and XP-B/CS cells. Despite this, the accumulation of nonfunctional XPB proteins was still seen in XPCS1BA, XPCS2BA and XP33BR cells, whereas mutant XPB is faintly detectable by anti-XPB S19 antibody in XP183MA and XP181MA fibroblasts. Moreover, the accumulation of XPD exhibited the recruitment kinetics similar to p62 in all tested cells (table 1). Combined results led us to conclude that the nonfunctional core TFIIH is capable of being recruited to damage sites in XP-B and XP-B/CS cells.
Table 1.
Assembly of NER factors in NER deficient XP-B and XP-B/CS fibroblasts
| Factor/Cells/Time | GM13025/XPCS1BA | GM13026/XPCS2BA | GM21071/XP33BR | GM21072/XP183MA | GM21153/XP181MA |
|---|---|---|---|---|---|
| 0 0.5 1 3 24h | 0 0.5 1 3 24h | 0 0.5 1 3 24h | 0 0.5 1 3 24h | 0 0.5 1 3 24h | |
| XPC | 3+ 3+ 3+ 3+ 2+ | 3+ 3+ 3+ 3+ 2+ | 3+ 3+ 3+ 3+ 2+ | 3+ 3+ 3+ 3+ 2+ | 3+ 3+ 3+ 3+ 2+ |
| XPB | 2+ 3+ 3+ 3+ 2+ | 2+ 3+ 3+ 3+ 2+ | 2+ 3+ 3+ 3+ 2+ | - - - - - | - - - - - |
| XPD | 2+ 3+ 3+ 3+ 2+ | 2+ 3+ 3+ 3+ 2+ | 2+ 3+ 3+ 3+ 2+ | 2+ 3+ 3+ 3+ 2+ | 2+ 3+ 3+ 3+ 2+ |
| p62 | 2+ 3+ 3+ 3+ 2+ | 2+ 3+ 3+ 3+ 2+ | 2+ 3+ 3+ 3+ 2+ | 2+ 3+ 3+ 3+ 2+ | 2+ 3+ 3+ 3+ 2+ |
| MAT1 | - - - - - | - - - - - | - - - - - | - - - - - | - - - - - |
| XPG | nd nd nd nd nd | - 1+ 2+ 2+ - | - 1+ 2+ 2+ 1+ | - 2+ 3+ 3+ 2+ | - 2+ 3+ 3+ 2+ |
*Relative post-irradiation recruitment and dispersal kinetics of various NER and TFIIH factors at localized DNA damage sites were examined in XP-B (XP33BR), XP-B/CS fibroblasts from mild symptom patients (XPCS1BA, XPCS2BA), and in fibroblasts from severe symptom patients (XP181MA, XP183MA). Scores of -, 1+, 2+ and 3+ respectively represent < 20, 20-40, 40-60 and 60-80% of the cognate factor-positive foci in the cell population. The percentage are calculated based on foci and cell counts from more than three fields. “nd” indicates “not done”.
3.2. CAK is associated with core TFIIH in unirradiated XP-B and XP-B/CS fibroblasts
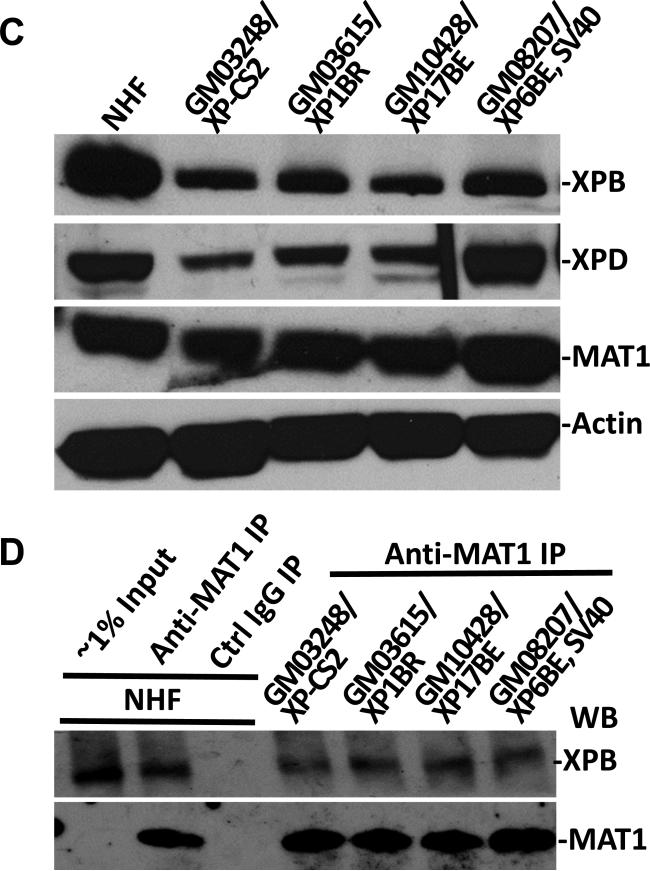
Our observation that CAK does not accumulate at DNA damage site in XP-B and XP-B/CS cells was unexpected, because CAK and core TFIIH association in XPCS1BA cells has previously been reported [42]. To ensure the basic integrity of TFIIH in XP-B and XP-B/CS fibroblasts, we first assessed the steady-state levels of XPB, XPD and MAT1 in these cells (Fig. 2A). The results showed that XPB protein was indeed present in TFIIH albeit at lower levels in XPCS1BA, XPCS2BA and XP33BR cells. Oddly, XPB was virtually undetectable in XP183MA and XP181MA fibroblasts. Perhaps the substitution of 42 amino acids in C-terminal of XPB protein in these cells compromised the detectability of mutant protein by anti-XPB S19 antibody. Examination of XPD and MAT1 also revealed that all XP-B fibroblasts contained comparatively reduced levels of XPD and MAT1 protein, as compared with wild-type XPB containing NHF cells. These results suggested that level of TFIIH is constitutively low in XP-B and XP-B/CS fibroblasts.
Fig. 2.
Physical association of MAT1 with core TFIIH in XP-B and XP-B/CS fibroblasts. (A) Western blotting of XPB, XPD and MAT1 proteins in NHF, XP-B and XP-B/CS fibroblasts. Protein extracts were made in SDS lysis buffer from indicated normal and XP fibroblasts. The proteins were quantitated and resolved by polyacrylamide gel. The proteins transferred on blots were probed with antibodies to XPB, XPD, MAT1 or β-actin. (B and C) Whole cell extracts were made in RIPA buffer from NHF, XP-B and XP-B/CS fibroblasts. Immunoprecipitation was performed by using control (Ctrl) or MAT1 (B) or XPD (C) antibodies. The immunoprecipitates were analyzed by Western blotting with specific XPB, p62 or XPD antibodies.
We subsequently conducted immunoprecipitation experiments to examine the native association of CAK and core TFIIH components in mutant XP cells. The results showed that XPB, p62 and XPD were all present in anti-MAT1 precipitates in these fibroblasts (Fig. 2B). Finally, the anti-XPD immunoprecipitation further confirmed that mutant XPB was present in TFIIH (Fig. 2C). Perhaps, the enrichment of mutant XPB protein from XP183MA and XP181MA cells by anti-MAT1 and Anti-XPD immunoprecipitation rendered the mutant XPB protein detectable by anti-XPB S19 antibody. Taken together, we concluded that CAK is integrally associated with core TFIIH in XP-B and XP-B/CS cells.
3.3. CAK complex accumulates at DNA damage sites in fibroblasts harboring XP-D mutations
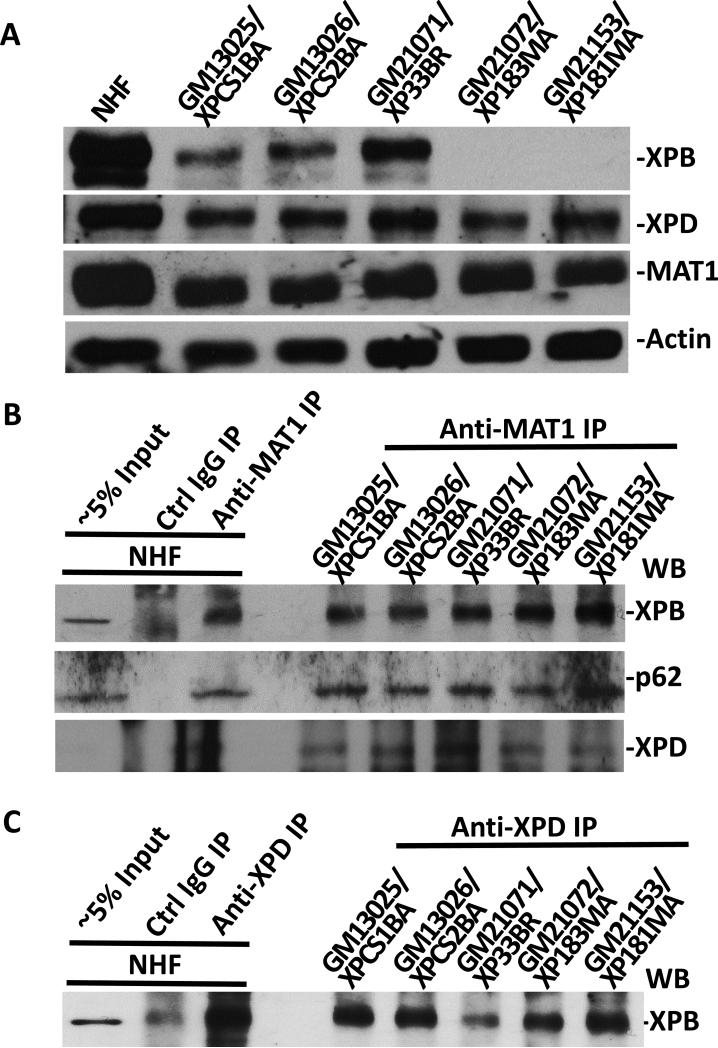
It has been demonstrated that XPD bridges CAK and core TFIIH by interacting with MAT1 and p44 of core TFIIH [43, 44]. We, therefore, examined the assembly of NER preincision complex and any impact on the accumulation of MAT1 in fibroblasts harboring various XPD mutations. As shown in Fig. 3 and Supplemental Fig. 3S, XPC, MAT1, XPD and XPB were all found to prominently accumulate at the damage sites and distinctly co-localize with core TFIIH component p62 in XP1BR, XP17BE, XP6BE and SV40-immortalized XP6BE fibroblasts. Also, and as expected, XPB co-localize with XPG, indicating that holo TFIIH was assembled into the NER preincision complex. Thus, in contrast to XPB mutations, XPD mutations did not affect the accumulation of CAK at DNA damage sites.
Fig. 3.
XP-D fibroblasts accumulate various pre-incision NER factors, including MAT1, at DNA damage sites in vivo. XP-D fibroblasts XP1BR (A) and XP17BE (B) were grown on coverslips, locally UV-irradiated and cultured for 1 h. The cells were then fixed with 2% paraformaldehyde and decorated by immunofluorescence double labeling with indicated specific antibodies. Nuclei were counterstained with DAPI. (C) Western blotting of XPB, XPD and MAT1 proteins in NHF and XP-D fibroblasts. Protein extracts were made in SDS lysis buffer, quantitated and resolved by polyacrylamide gel. The blots were probed with antibodies to XPB, XPD, MAT1and β-actin. (D) Physical association of MAT1 with core TFIIH in NHF and XP-D fibroblasts. Whole cell extracts were made in RIPA buffer from NHF and XP-D fibroblasts. Immunoprecipitation was performed by using control (Ctrl) or MAT1 antibodies and the immunoprecipitates were Western blotting analyzed for presence of XPB.
We next examined the steady-state levels of XPB, XPD and MAT1 and the CAK-core TFIIH association in XP-D cells. Results of Fig. 3C and D showed that XPB and XPD protein levels were reduced in XP-CS2, XP1BR and XP17BE cells but not in SV40-transformed XP6BE fibroblasts. Moreover, no significant differences were noticed in MAT1 protein levels. In immunoprecipitation experiments, XPB were present in all anti-MAT1 precipitates from XP-D fibroblasts. These results indicated that the association of CAK and core TFIIH is not perturbed in XP-D background, and CAK remains wholly attached to core TFIIH upon its recruitment to DNA damage sites in these cells.
3.4. XPG recruitment is impaired in mild XP-B/CS fibroblasts but persists in XP-B/CS fibroblasts from severely affected patients
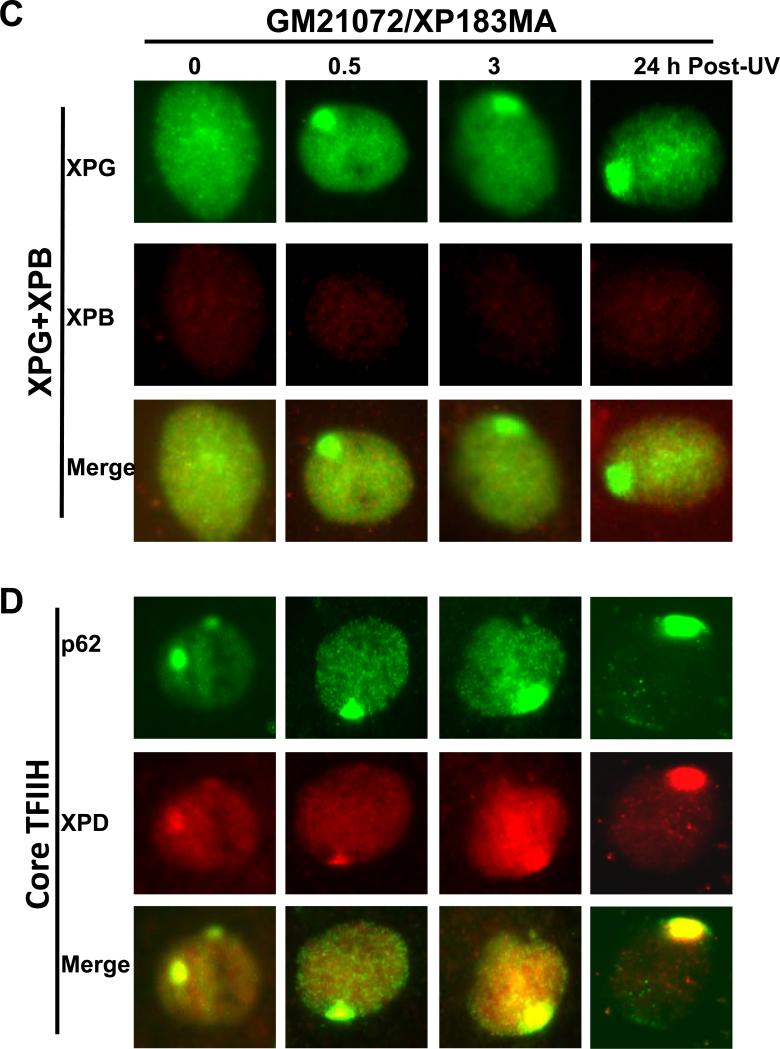
To determine whether diverse XPB mutants have different effects on translocation of XPG and other NER factors, e.g., XPC, XPB, XPD and p62, we examined the accumulation of a variety of factors at localized DNA damage sites (table 1). Immediately after UV irradiation (0 h), the early and upstream damage recognition factor, XPC, was seen to localize in ~60-80% of normal as well as XP fibroblasts. In comparison, XPB, XPD and p62 localization was lower, albeit only slightly as ~40-60% of cell still exhibited a robust recruitment of these factors. On the other hand and as expected, XPB translocation to damage sites could not be detected by the S19 antibody in XP183MA and XP181MA fibroblasts. The translocation of XPG to damage sites in NHF was also rapid as it can be clearly seen immediately (at 0 h) after irradiation. After 24 h, XPG and XPB were not detected in repair-proficient NHF, as these proteins dispersed away due to efficient 6-4PP removal (Fig. 4A). Interestingly, in XPCS1BA fibroblasts, prompt XPG recruitment was not observed at 0 h. A delayed XPG accumulation, however, was seen at 0.5, 1 and 3 h and in a lower percentage of cell population than in NHF (table 1 and Fig. 4B). At 24 h, XPG accumulation was no longer traceable in most (>80%) cells, while XPB, XPC, XPD and p62 were still distinctly present and in a highly significant percentage (~40-60%) of XPCS1BA cells. Similar observation was made in XP33BR fibroblasts. Cumulatively, these factor recruitment and dispersal patterns demonstrated that the XPG recruitment is in particular impaired in mild XP-B and XP-B/CS fibroblasts, both of which harbor a phenylalanine-99 to serine-99 (F99S) missense mutation within their XPB protein.
Fig. 4.
XPG recruitment at DNA damage sites is impaired in XP-B/CS fibroblasts from patients with mild symptoms while persists in fibroblasts from patients with severe symptoms. NHF (A), XP-B/CS fibroblast XPCS2BA (B), XP183MA (C) were grown on coverslips and irradiated with 100 J/m2 UV through a 5 μm-isopore polycarbonate filter. The cells were fixed immediately (0 h) or cultured for indicated repair period and then fixed in 2% paraformaldehyde. The XPG and XPB recruitment at DNA damage sites were examined by immunofluorescence double labeling as described for Fig. 1 to 3. The presence of p62, XPD and their co-localization at DNA damage sites were monitored in XP183MA fibroblasts (D).
In XP183MA fibroblasts derived from severely affected patients, the XPG accumulation was detected at 1 and 3 h in a similar spatio-temporal pattern as XPC, XPD and p62 (table 1). Moreover, the delay of XPG accumulation was also apparent at 0 and 0.5 h (Fig. 4C and D). Surprisingly, a significant number (40-60%) of cells still remained XPG positive at 24 h. This indicated that recruitment of XPG to damage sites was not significantly affected by the alteration of 42 C-terminal amino acids of XPB protein in the cells. Reaffirming this differential response, experiments with XP181MA fibroblasts also exhibited the similar results (Supplemental Fig. S4). Overall, it was concluded that F99S missense mutation rather than C-terminal alteration in XPB protein negatively influences the recruitment of XPG to damage sites.
3.5. UV-induced RNAPII phosphorylation was reduced and the RNAPII degradation was delayed in severe XP-B/CS fibroblasts
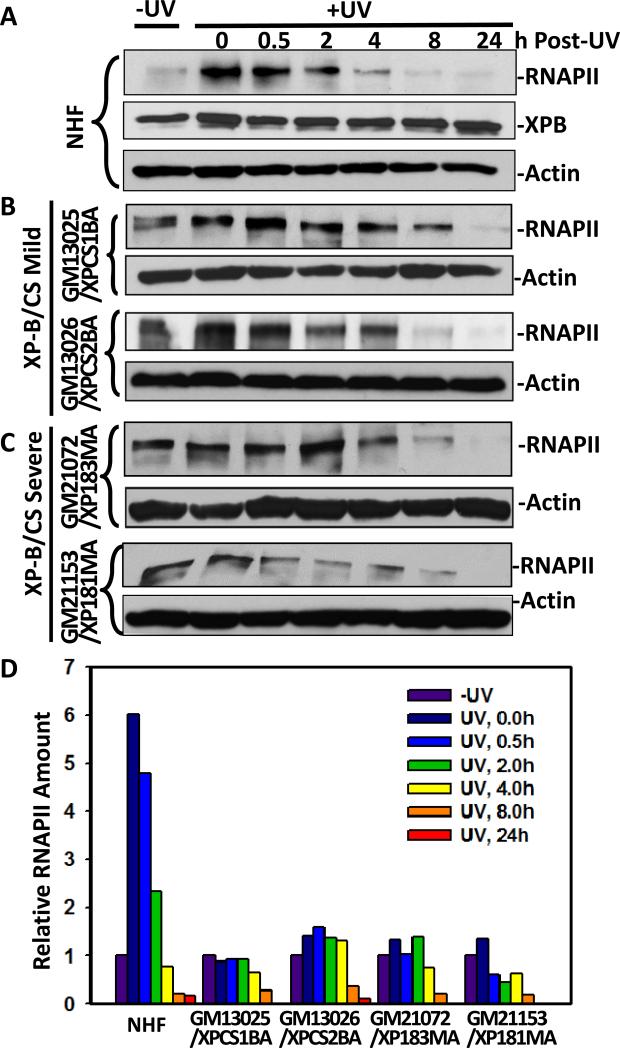
During transcription, XPB and XPD helicases are essential for the open complex formation. CAK in turn phosphorylates Serine 5 (Ser5) of the C-terminal domain of large subunit of RNAPII. This phosphorylation enables RNAPII to initiate mRNA elongation [45]. When transcribing RNAPII is stalled by an UV-induced photolesion, TFIIH funnels transcription into damage processing by the TCR machinery. The RNAPII is eventually degraded by ubiquitin-mediated proteolysis and Ser5 phosphorylation of RNAPII plays an important role in this degradation [46]. We have previously demonstrated that pre-existing RNAPII phosphorylation relies on kinase activity of CAK as inhibition of CAK abolished pre-existing RNAPII phosphorylation and delayed RNAPII degradation [30]. Thus, the absence of CAK accumulation at DNA damage sites observed in XP-B/CS mutant cells impelled us to examine the impact on UV-induced RNAPII phosphorylation in these cells. As shown in Fig. 5, the level of Ser5-phosphoryled RNAPII in NHF increased up to 6-fold immediately (0 h) after UV irradiation and then decreased gradually over a 24-h period due presumably to UV-induced and ubiquitin-mediated proteolysis [47-49]. No significant changes in XPB and the control β-actin levels were seen in NHF. However, the RNAPII Ser5-phosphorylation was not significantly induced in XPCS1BA, XPCS2BA, XP183MA or XP181MA fibroblasts. Semi-quantitative estimates of protein levels in autoradiographs indicated that relative amount of Ser5-phosphorylated RNAPII decreased slower in XP-B/CS cells as compared with that in NHF, where the Ser5-phosphorylated RNAPII level was elevated immediately UV irradiation (Fig. 5D). Thus, both RNAPII phosphorylation and degradation after UV irradiation was affected in XP-B/CS cells.
Fig. 5.
UV-induced RNAPII Ser5-phosphorylation and degradation in XP-B/CS fibroblasts. The exponentially growing NHF (A), XPCS1BA, XPCS2BA (B), XP181MA and XP183MA (C) fibroblasts were UV-irradiated at a dose of 20 J/m2 and maintained for indicated repair period in fresh medium. The protein extracts were then made in SDS lysis buffer, and the proteins were quantitated and examined by Western blotting using specific anti-XPB and anti-phospho-RNAPII or anti-β-actin antibodies. (D) Representative Western blot images were quantitated using ImageJ software and arbitrary amount of RNAPII were estimated relative to protein levels found without UV irradiation.
4. Discussion
The relationship between genotype and phenotype in TFIIH mutations is complicated by its dual role in transcription and NER. It is well accepted that XP is phenotypically related to the deficiency of DNA repair. By contrast, CS is considered to be related to the problems of both transcription and TCR. In this study, we demonstrated a defect in CAK accumulation at damage sites in XP-B and XP-B/CS fibroblasts. Moreover, we found that XPG recruitment persists in XP-B/CS fibroblasts derived from severely affected XP-B patients but not from mildly affected XP-B patients. We further described a reduction in UV-induced RNAPII phosphorylation and a slower RNAPII degradation in XP-B/CS fibroblasts as possible consequences of CAK dissociation during TCR.
4.2. Defect in CAK accumulation at damage sites and change in TFIIH subunit composition in response to DNA damage in XP-B and XP-B/CS fibroblasts
Recent discovery of the role of XPG in stabilizing TFIIH [42] provokes a model explaining XP-B/CS and XP-G/CS phenotypes. According to the model, defect in specific enzymatic functions of XPB, XPD and XPG proteins results in XP and the specific XP/CS features could result from the dissociation of CAK from TFIIH [25]. In present study, our experiments showed a defect in CAK accumulation at damage sites in XP-B/CS fibroblasts. We also detected a decrease in levels of cellular XPB, XPD and MAT1, which may consequently results in a reduced level of TFIIH. Nevertheless, lower levels of TFIIH could not be the cause of our inability of detecting CAK accumulation. First, TFIIH components, e.g., p62 and XPD, were clearly detected in our immunofluorescence dual-labeling experiments; second, in primary XP-D fibroblasts we observed a similar reduction in XPB and XPD. Yet, CAK accumulation depicted by robust MAT1 recruitment was constantly detected. When integrity of TFIIH was examined, we found that the XPB, p62 and XPD proteins were present in anti-MAT1 and the XPB in anti-XPD precipitates, indicating that the CAK remains associated with core TFIIH in XPB mutant fibroblasts. Thus, it is possible that the defect in CAK accumulation at damage sites in XP-B/CS fibroblasts reflects a rapid change in TFIIH subunit composition upon its assembly into preincision complex. It has been recently reported that CAK is released from core TFIIH in repair-proficient cells during NER [50]. Further, the release of the CAK promotes the incision of the damaged oligonucleotide and thereby the repair of the DNA. In our experiments, MAT1 in NHF disappeared from damage sites 24 h after UV irradiation and it happened together with the disappearance of p62 ( Fig 1A) and other NER factors such as XPC, XPB, XPD and XPG [30]. Therefore, in our experiments, the CAK dispersal from damage sites in NHF might not reflect the release of CAK from TFIIH. It would rather signify the completion of 6-4PP repair. By contrast, XP-B and XP-B/CS fibroblasts are not able to repair 6-4PPs. The dispersion of CAK in these cells could not be the result of 6-4PP repair. It is very likely that the nonfunctional mutant XPB protein accelerates the release of CAK when TFIIH is recruited to DNA damage. In XP-D fibroblasts, CAK accumulation appears to be normal and comparable to that in NHF. Therefore, mutant XPD does not accelerate the release of CAK but retains TFIIH at DNA damage sites. Based on these results, we hypothesized that XPB and XPD differentially regulate TFIIH compositional change during NER.
4.3. XPG recruitment is differentially affected by F99S and C-terminal alteration in XPB proteins
Our examination on XPG recruitment revealed a differential effect of XPB mutant alterations. In XP33BR and XPCS2BA fibroblasts, XPG recruitments were not seen initially at 0 h and were also significantly reduced at later time points. This is in agreement with the previous report in which XPCS1BA was examined [51]. XPCS1BA and XPCS2BA are mild XP-B/CS fibroblasts derived from siblings, and both are homozygous for F99S missense mutation. Interestingly, the same F99S mutation was present in the paternal allele in XP33BR cells, where the maternal allele has a substitution, which results in an arginine at position 425 being converted to a stop codon. Therefore, F99S missense mutation appears to be specifically related to the failure of XPG's retention at damage sites. By contrast, a delay of XPG recruitment was seen in XP183MA and XP181MA severe XP-B/CS fibroblasts at 0 h. Whereas, the XPG accumulation was not affected at 0.5, 1 or 3 h and, consistent with a previous report, persisted up to 24 h [52]. As the delay occurred in all XP-B/CS cells, it is likely related to lower level of TFIIH due to the decrease in defective XPB protein level. The persistence of XPG recruitment, in essence, would specifically relate to the alteration of C-terminal 42 amino acids of XPB protein, and not simply to the loss of XPB function. It is worthy of note that persistence of XPF recruitment has also been reported in XP183MA and XP181MA fibroblasts [52]. Thus, the persistence of nonfunctional preincision complex is correlated with severity of XP-B patients. It would be interesting to learn how the nonfunctional preincision complexes residing at damaged chromatin affects other DNA templated processes, .e.g., DNA replication, transcription and translesion DNA synthesis.
4.4. UV-induced RNAPII phosphorylation and degradation in severe XP-B/CS fibroblasts
The NER factor recruitment/accumulation, examined by immunofluorescence double labeling, only reflect the cellular events of GGR [6]. The failure of CAK accumulation at damage sites in XP-B, XP-B/CS and XP-G/CS (Fig. 1 and [30]) would therefore be an evidence of the TFIIH compositional change in GGR but not in TCR. To probe the regulatory role of CAK in TCR, we previously approached the question using chemical genetic testing system, where the kinase activity of a genetically modified Cdk7 is sensitive to unnatural ATP analog 1-NMPP1. We demonstrated that inhibition of Cdk7 diminished the pre-existing and the immediate RNAPII Ser5-phosphorylation after UV irradiation. In the present study, we observed an immediate dramatic increase in RNAPII Ser5-phosphorylation in NHF, but not in XP-B/CS (XPCS1BA, XPCS2BA, XP183MA or XP181MA) fibroblasts. Moreover, a slower decrease in Ser5-phosphorylated RNAPII over 24-h post-UV period was also observed in XP-B/CS. It is suggested that the TFIIH compositional change may also occur in XP-B/CS fibroblasts during TCR.
In summary, we demonstrated a TFIIH compositional change in CAK dissociation after TFIIH is recruited to DNA damage sites in XP-B and XP-B/CS fibroblasts. Such CAK dissociation is not exclusively related to XP-B/CS. We also showed that the persistence of nonfunctional preincision complex is associated with severe XP-B/CS, while the reduction in UV-induced RNAPII Ser5-phosphorylation and degradation occurs in all XP-B/CS. How these changes may contribute to high risk of skin cancer and other phenotypes of XP/CS patients warrants additional exploration.
Supplementary Material
Highlights.
! ! While TFIIH is intact, its CAK component fails to accumulate at DNA damage sites in XP-B and XP-B/CS but not in XP-D cells.
! ! XPG recruitment is impaired in XP-B/CS cells harboring a F99S mutation but persists in XP-B/CS cells with C-terminal alteration in XPB protein.
! ! UV-induced RNAPII phosphorylation is dramatically reduced and the RNAPII degradation is delayed in XP-B/CS cells.
! ! CAK rapidly disassociates from core TFIIH upon assembly of nonfunctional preincision complex in XP-B and XP-B/CS cells.
! ! The overall work suggests a differential role of XPB and XPD helicases in regulating the anchoring of CAK to core TFIIH during NER.
Acknowledgements
The work was supported by Public Health service Grants ES2388, ES12991 & CA93413 to AAW from National Institute of Health. The authors thank Dr. Qi-en Wang for help with immunofluorescence experiments and Dr. Alo Ray for advice on discussion and manuscript preparation.
Abbreviation
- TFIIH
Transcription factor II H
- CAK
Cdk-activating kinase complex
- UV
ultraviolet light
- 6-4PP
6-4 pyrimidine-pyrimidone photoproduct
- CPD
cyclobutane pyridine dimer
- NER
Nucleotide excision repair
- GGR
global genomic repair
- TCR
transcription-coupled repair
- RNAPII
RNA polymerase II
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
None.
REFERENCES
- 1.Lindahl T, Wood RD. Quality control by DNA repair. Science. 1999;286:1897–1905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 2.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 3.Hanawalt PC. Subpathways of nucleotide excision repair and their regulation. Oncogene. 2002;21:8949–8956. doi: 10.1038/sj.onc.1206096. [DOI] [PubMed] [Google Scholar]
- 4.Petit C, Sancar A. Nucleotide excision repair: From E.coli to man. Biochimie. 1999;81:15–25. doi: 10.1016/s0300-9084(99)80034-0. [DOI] [PubMed] [Google Scholar]
- 5.Araujo SJ, Tirode F, Coin F, Pospiech H, Syvaoja JE, Stucki M, Hubscher U, Egly JM, Wood RD. Nucleotide excision repair of DNA with recombinant human proteins: definition of the minimal set of factors, active forms of TFIIH, and modulation by CAK. Genes Dev. 2000;14:349–359. [PMC free article] [PubMed] [Google Scholar]
- 6.Volker M, Mone MJ, Karmakar P, Van Hoffen A, Schul W, Vermeulen W, Hoeijmakers JH, van Driel R, Van Zeeland AA, Mullenders LH. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell. 2001;8:213–224. doi: 10.1016/s1097-2765(01)00281-7. [DOI] [PubMed] [Google Scholar]
- 7.Fitch ME, Nakajima S, Yasui A, Ford JM. In vivo recruitment of XPC to UV-induced cyclobutane pyrimidine dimers by the DDB2 gene product. J. Biol. Chem. 2003;278:46906–46910. doi: 10.1074/jbc.M307254200. [DOI] [PubMed] [Google Scholar]
- 8.Yokoi M, Masutani C, Maekawa T, Sugasawa K, Ohkuma Y, Hanaoka F. The Xeroderma pigmentosum group C protein complex XPC-HR23B plays an important role in the recruitment of transcription factor IIH to damaged DNA. J. Biol. Chem. 2000;275:9870–9875. doi: 10.1074/jbc.275.13.9870. [DOI] [PubMed] [Google Scholar]
- 9.Evans E, Moggs JG, Hwang JR, Egly JM, Wood RD. Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. EMBO, J. 1997;16:6559–6573. doi: 10.1093/emboj/16.21.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans E, Fellows J, Coffer A, Wood RD. Open complex formation around a lesion during nucleotide excision repair provides a structure for cleavage by human XPG protein. EMBO J. 1997;16:625–638. doi: 10.1093/emboj/16.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brueckner F, Hennecke U, Carell T, Cramer P. CPD damage recognition by transcribing RNA polymerase II. Science. 2007;315:859–862. doi: 10.1126/science.1135400. [DOI] [PubMed] [Google Scholar]
- 12.Sarker AH, Tsutakawa SE, Kostek S, Ng C, Shin DS, Peris M, Campeau E, Tainer JA, Nogales E, Cooper PK. Recognition of RNA polymerase II and transcription bubbles by XPG, CSB, and TFIIH: insights for transcription-coupled repair and Cockayne Syndrome. Mol. Cell. 2005;20:187–198. doi: 10.1016/j.molcel.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 13.Fousteri M, Vermeulen W, Van Zeeland AA, Mullenders LH. Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol. Cell. 2006;23:471–482. doi: 10.1016/j.molcel.2006.06.029. [DOI] [PubMed] [Google Scholar]
- 14.Sugasawa K, Okamoto T, Shimizu Y, Masutani C, Iwai S, Hanaoka F. A multistep damage recognition mechanism for global genomic nucleotide excision repair. Genes Dev. 2001;15:507–521. doi: 10.1101/gad.866301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giglia-Mari G, Coin F, Ranish JA, Hoogstraten D, Theil A, Wijgers N, Jaspers NG, Raams A, Argentini M, Van der Spek PJ, Botta E, Stefanini M, Egly JM, Aebersold R, Hoeijmakers JH, Vermeulen W. A new, tenth subunit of TFIIH is responsible for the DNA repair syndrome trichothiodystrophy group A. Nat. Genet. 2004;36:714–719. doi: 10.1038/ng1387. [DOI] [PubMed] [Google Scholar]
- 16.Giglia-Mari G, Miquel C, Theil AF, Mari PO, Hoogstraten D, Ng JM, Dinant C, Hoeijmakers JH, Vermeulen W. Dynamic interaction of TTDA with TFIIH is stabilized by nucleotide excision repair in living cells. PLoS. Biol. 2006;4:e156. doi: 10.1371/journal.pbio.0040156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coin F, Egly JM. Ten years of TFIIH. Cold Spring Harb. Symp. Quant. Biol. 1998;63:105–110. doi: 10.1101/sqb.1998.63.105. [DOI] [PubMed] [Google Scholar]
- 18.Coin F, Bergmann E, Tremeau-Bravard A, Egly JM. Mutations in XPB and XPD helicases found in xeroderma pigmentosum patients impair the transcription function of TFIIH. EMBO J. 1999;18:1357–1366. doi: 10.1093/emboj/18.5.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roy R, Adamczewski JP, Seroz T, Vermeulen W, Tassan J-P, Schaeffer L, Nigg EA, Hoeijmakers JHJ, Egly J-M. The MO15 cell cycle kinase is associated with the TFIIH transcription-DNA repair factor. Cell. 1994;79:1093–1101. doi: 10.1016/0092-8674(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 20.Hoogstraten D, Nigg AL, Heath H, Mullenders LH, van Driel R, Hoeijmakers JH, Vermeulen W, Houtsmuller AB. Rapid switching of TFIIH between RNA polymerase I and II transcription and DNA repair in vivo. Mol. Cell. 2002;10:1163–1174. doi: 10.1016/s1097-2765(02)00709-8. [DOI] [PubMed] [Google Scholar]
- 21.Matsuno M, Kose H, Okabe M, Hiromi Y. TFIIH controls developmentally-regulated cell cycle progression as a holocomplex. Genes Cells. 2007;12:1289–1300. doi: 10.1111/j.1365-2443.2007.01133.x. [DOI] [PubMed] [Google Scholar]
- 22.Bradsher J, Coin F, Egly JM. Distinct roles for the helicases of TFIIH in transcript initiation and promoter escape. J. Biol. Chem. 2000;275:2532–2538. doi: 10.1074/jbc.275.4.2532. [DOI] [PubMed] [Google Scholar]
- 23.Feaver WJ, Svejstrup JQ, Henry NL, Kornberg RD. Relationship of CDK-activating kinase and RNA polymerase II CTD kinase TFIIH/TFIIK. Cell. 1994;79:1103–1109. doi: 10.1016/0092-8674(94)90040-x. [DOI] [PubMed] [Google Scholar]
- 24.Andressoo JO, Hoeijmakers JH, de WH. Nucleotide excision repair and its connection with cancer and ageing. Adv. Exp. Med. Biol. 2005;570:45–83. doi: 10.1007/1-4020-3764-3_3. [DOI] [PubMed] [Google Scholar]
- 25.Scharer OD. Hot topics in DNA repair: the molecular basis for different disease states caused by mutations in TFIIH and XPG. DNA Repair (Amst) 2008;7:339–344. doi: 10.1016/j.dnarep.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nouspikel T. Nucleotide excision repair and neurological diseases. DNA Repair (Amst) 2008;7:1155–1167. doi: 10.1016/j.dnarep.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 27.van Gool AJ, Van der Horst GT, Citterio E, Hoeijmakers JH. Cockayne syndrome: defective repair of transcription? EMBO J. 1997;16:4155–4162. doi: 10.1093/emboj/16.14.4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balajee AS, May A, Dianov GL, Friedberg EC, Bohr VA. Reduced RNA polymerase II transcription in intact and permeabilized Cockayne syndrome group B cells. Proc. Natl. Acad. Sci. U. S. A. 1997;94:4306–4311. doi: 10.1073/pnas.94.9.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dubaele S, Egly JM. Cockayne syndrome, between transcription and DNA repair defects. J. Eur. Acad. Dermatol. Venereol. 2002;16:220–226. doi: 10.1046/j.1468-3083.2002.00453.x. [DOI] [PubMed] [Google Scholar]
- 30.Arab HH, Wani G, Ray A, Shah ZI, Zhu Q, Wani AA. Dissociation of CAK from core TFIIH reveals a functional link between XP-G/CS and the TFIIH disassembly state. PLoS. One. 2010;5:e11007. doi: 10.1371/journal.pone.0011007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Venkatachalam S, Denissenko MF, Wani AA. DNA repair in human cells: quantitative assessment of bulky anti-BPDE DNA adducts by non-competitive immunoassays. Carcinogenesis. 1995;16:2029–2036. doi: 10.1093/carcin/16.9.2029. [DOI] [PubMed] [Google Scholar]
- 32.Vermeulen W. Clinical heterogeneity within xeroderma pigmentosum associated with mutations in the DNA repair and transcription gene ERCC3. 1994. [PMC free article] [PubMed]
- 33.Vermeulen W. Xeroderma pigmentosum complementation group H falls into complementation group D. 1991. [DOI] [PubMed]
- 34.Boyle J, Ueda T, Oh KS, Imoto K, Tamura D, Jagdeo J, Khan SG, Nadem C, Digiovanna JJ, Kraemer KH. Persistence of repair proteins at unrepaired DNA damage distinguishes diseases with ERCC2 (XPD) mutations: cancer-prone xeroderma pigmentosum vs. non-cancer-prone trichothiodystrophy. Hum. Mutat. 2008;29:1194–1208. doi: 10.1002/humu.20768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oh KS, Khan SG, Jaspers NG, Raams A, Ueda T, Lehmann A, Friedmann PS, Emmert S, Gratchev A, Lachlan K, Lucassan A, Baker CC, Kraemer KH. Phenotypic heterogeneity in the XPB DNA helicase gene (ERCC3): xeroderma pigmentosum without and with Cockayne syndrome. Hum. Mutat. 2006;27:1092–1103. doi: 10.1002/humu.20392. [DOI] [PubMed] [Google Scholar]
- 36.Takayama K, Salazar EP, Lehmann A, Stefanini M, Thompson LH, Weber CA. Defects in the DNA repair and transcription gene ERCC2 in the cancer-prone disorder xeroderma pigmentosum group D. Cancer Res. 1995;55:5656–5663. [PubMed] [Google Scholar]
- 37.Wani AA, D'Ambrosio SM, Alvi NK. Quantitation of pyrimidine dimers by immunoslot blot following sublethal UV-irradiation of human cells. Photochem. Photobiol. 1987;46:477–482. doi: 10.1111/j.1751-1097.1987.tb04798.x. [DOI] [PubMed] [Google Scholar]
- 38.El-Mahdy MA, Zhu Q, Wang QE, Wani G, Praetorius-Ibba M, Wani AA. Cullin 4A-mediated proteolysis of DDB2 protein at DNA damage sites regulates in vivo lesion recognition by XPC. J. Biol. Chem. 2006;281:13404–13411. doi: 10.1074/jbc.M511834200. [DOI] [PubMed] [Google Scholar]
- 39.Wang QE, Zhu Q, Wani G, El-Mahdy MA, Li J, Wani AA. DNA repair factor XPC is modified by SUMO-1 and ubiquitin following UV irradiation. Nucleic Acids Res. 2005;33:4023–4034. doi: 10.1093/nar/gki684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang QE, Zhu Q, Wani MA, Wani G, Chen J, Wani AA. Tumor supressor p53 dependent recruitment of nucleotide excision repair ractors XPC and TFIIH to DNA damage. DNA Repair. 2003;2:483–499. doi: 10.1016/s1568-7864(03)00002-8. [DOI] [PubMed] [Google Scholar]
- 41.Zhu Q, Wani G, Arab HH, El-Mahdy MA, Ray A, Wani AA. Chromatin restoration following nucleotide excision repair involves the incorporation of ubiquitinated H2A at damaged genomic sites. DNA Repair (Amst) 2009;8:262–273. doi: 10.1016/j.dnarep.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ito S, Kuraoka I, Chymkowitch P, Compe E, Takedachi A, Ishigami C, Coin F, Egly JM, Tanaka K. XPG stabilizes TFIIH, allowing transactivation of nuclear receptors: implications for Cockayne syndrome in XP-G/CS patients. Mol. Cell. 2007;26:231–243. doi: 10.1016/j.molcel.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 43.Dubaele S, Proietti De SL, Bienstock RJ, Keriel A, Stefanini M, Van HB, Egly JM. Basal transcription defect discriminates between xeroderma pigmentosum and trichothiodystrophy in XPD patients. Mol. Cell. 2003;11:1635–1646. doi: 10.1016/s1097-2765(03)00182-5. [DOI] [PubMed] [Google Scholar]
- 44.Keriel A, Stary A, Sarasin A, Rochette-Egly C, Egly JM. XPD mutations prevent TFIIH-dependent transactivation by nuclear receptors and phosphorylation of RARalpha. Cell. 2002;109:125–135. doi: 10.1016/s0092-8674(02)00692-x. [DOI] [PubMed] [Google Scholar]
- 45.Gerber HP, Hagmann M, Seipel K, Georgiev O, West MA, Litingtung Y, Schaffner W, Corden JL. RNA polymerase II C-terminal domain required for enhancer-driven transcription. Nature. 1995;374:660–662. doi: 10.1038/374660a0. [DOI] [PubMed] [Google Scholar]
- 46.Yasukawa T, Kamura T, Kitajima S, Conaway RC, Conaway JW, Aso T. Mammalian Elongin A complex mediates DNA-damage-induced ubiquitylation and degradation of Rpb1. EMBO J. 2008;27:3256–3266. doi: 10.1038/emboj.2008.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ratner JN, Balasubramanian B, Corden J, Warren SL, Bregman DB. Ultraviolet radiation-induced ubiquitination and proteasomal degradation of the large subunit of RNA polymerase II -Implications for transcription-coupled DNA repair. Journal of Biological Chemistry. 1998;273:5184–5189. doi: 10.1074/jbc.273.9.5184. [DOI] [PubMed] [Google Scholar]
- 48.Luo Z, Zheng J, Lu Y, Bregman DB. Ultraviolet radiation alters the phosphorylation of RNA polymerase II large subunit and accelerates its proteasome-dependent degradation. Mutat. Res. 2001;486:259–274. doi: 10.1016/s0921-8777(01)00097-0. [DOI] [PubMed] [Google Scholar]
- 49.Anindya R, Aygun O, Svejstrup JQ. Damage-induced ubiquitylation of human RNA polymerase II by the ubiquitin ligase Nedd4, but not Cockayne syndrome proteins or BRCA1. Mol. Cell. 2007;28:386–397. doi: 10.1016/j.molcel.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 50.Coin F, Oksenych V, Mocquet V, Groh S, Blattner C, Egly JM. Nucleotide excision repair driven by the dissociation of CAK from TFIIH. Mol. Cell. 2008;31:9–20. doi: 10.1016/j.molcel.2008.04.024. [DOI] [PubMed] [Google Scholar]
- 51.Zotter A, Luijsterburg MS, Warmerdam DO, Ibrahim S, Nigg A, van Cappellen WA, Hoeijmakers JH, van DR, Vermeulen W, Houtsmuller AB. Recruitment of the nucleotide excision repair endonuclease XPG to sites of UV-induced dna damage depends on functional TFIIH. Mol. Cell Biol. 2006;26:8868–8879. doi: 10.1128/MCB.00695-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oh KS, Imoto K, Boyle J, Khan SG, Kraemer KH. Influence of XPB helicase on recruitment and redistribution of nucleotide excision repair proteins at sites of UV-induced DNA damage. DNA Repair (Amst) 2007;6:1359–1370. doi: 10.1016/j.dnarep.2007.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.