Abstract
An NADPH-dependent NO2−-reducing system was reconstituted in vitro using ferredoxin (Fd) NADP+ oxidoreductase (FNR), Fd, and nitrite reductase (NiR) from the green alga Chlamydomonas reinhardtii. NO2− reduction was dependent on all protein components and was operated under either aerobic or anaerobic conditions. NO2− reduction by this in vitro pathway was inhibited up to 63% by 1 mm NADP+. NADP+ did not affect either methyl viologen-NiR or Fd-NiR activity, indicating that inhibition was mediated through FNR. When NADPH was replaced with a glucose-6-phosphate dehydrogenase (G6PDH)-dependent NADPH-generating system, rates of NO2− reduction reached approximately 10 times that of the NADPH-dependent system. G6PDH could be replaced by either 6-phosphogluconate dehydrogenase or isocitrate dehydrogenase, indicating that G6PDH functioned to: (a) regenerate NADPH to support NO2− reduction and (b) consume NADP+, releasing FNR from NADP+ inhibition. These results demonstrate the ability of FNR to facilitate the transfer of reducing power from NADPH to Fd in the direction opposite to that which occurs in photosynthesis. The rate of G6PDH-dependent NO2− reduction observed in vitro is capable of accounting for the observed rates of dark NO3− assimilation by C. reinhardtii.
NO3− assimilation in plants consists of three processes. Initially NO3− is reduced to NO2− by NO3− reductase. This is followed by the subsequent reduction of NO2− to NH4+ by NiR. Finally, the resulting NH4+ is assimilated into amino acids by glutamine synthatase/glutamate synthase (Beevers and Hageman, 1980; Sivarsankar and Oaks, 1996). The Chlamydomonas reinhardtii NO3− assimilatory system is similar to that of higher plants (Barea and Cárdenas, 1975). When grown under NO3−-sufficient conditions, green algae depend on light for NO3− assimilation, whereas cells grown under NO3−-limited conditions are capable of NO3− assimilation in the light and in the dark (Syrett, 1981). The onset of dark NO3− assimilation in N-limited cells stimulates the respiration of starch, thereby providing carbon skeletons for amino acid synthesis and the reductant required for NO3− reduction (Turpin et al., 1997). The onset of NO3− assimilation coincides with the Fd-thioredoxin-dependent activation of G6PDH, the key regulatory enzyme of the OPP pathway (Huppe et al., 1992, 1994; Farr et al., 1994). Physiological and biochemical studies indicated that the source of reductant for NO3− reduction in the dark is the OPP pathway (Vanlerberghe et al., 1992; Huppe et al., 1994). If this is the case, electrons from NADPH must be able to reduce Fd, which is the electron donor to NiR. To date, however, no direct evidence exists to support this hypothesis.
The relationship between the OPP pathway and NO2− reduction has also been investigated in nonphotosynthetic tissues of higher plants (Oaks and Hirel, 1985), where it has also been reported that carbohydrate oxidation via the OPP pathway provides reducing power for NO2− reduction (Emes and Fowler, 1983; Oji et al., 1985; Bowsher et al., 1989; Borchert et al., 1993). However, NiR in roots is also a Fd-dependent enzyme, which cannot utilize directly the NADPH generated by the OPP pathway. This implied the presence of FNR-like proteins that mediate the electron transfer from NADPH to Fd (Oji et al., 1985; Suzuki et al., 1985). Recently, several FNRs and Fds have been purified from nonphotosynthetic plant tissues (Wada et al., 1989; Hirasawa et al., 1990; Morigasaki et al., 1990a, 1990b; Bowsher et al., 1993), but there is still no direct evidence to show FNR and Fd mediating electron transfer from NADPH to NO2− reduction.
The purpose of this study was to test the hypothesis that electrons from NADPH may support NO2− reduction via a FNR- and Fd-mediated electron transfer pathway from NADPH to NO2−. We report the purification of NiR, FNR, and Fd from the green alga C. reinhardtii and the reconstitution of an in vitro electron-transfer system from NADPH to NiR for NO2− reduction via FNR, Fd, and NiR. Furthermore, we have isolated G6PDH from the same source and coupled G6PDH-dependent NADPH generation to NO2− reduction, providing direct evidence for the potential of G6PDH to support NO2− reduction in the dark.
MATERIALS AND METHODS
Chlamydomonas reinhardtii cc-1183 was grown in NO3−-sufficient chemostat cultures as previously described (Huppe and Turpin, 1996). Cells were harvested daily, frozen in liquid N2, and then stored at −80°C until use.
Reagents and Enzymes
Chemical reagents were obtained from commercial sources and were of the highest quality available. Q-Sepharose (fast flow) and phenyl-Sepharose CL-4B were from Pharmacia; Blue-Cellulofine was from Seikagaku-kogyo Company, Ltd. (Tokyo, Japan); Butyl-Toyopearl was from Tosoh Company (Tokyo); DEAE-Fractogel was from EM Science (NJ); Glc oxidase (Aspergillus niger), catalase (bovine liver), and G6P (yeast) were purchased from Sigma; and 6PGDH (yeast) and ICDH (porcine heart) were from Boehringer Mannheim. Fd-Sepharose 4B was prepared by purified C. reinhardtii Fd and CNBr-Sepharose 4B (Pharmacia) according to the protocol provided by the manufacturer.
Isolation of Enzymes
Twenty milliliters of frozen cells (approximately 20 g in fresh weight) was thawed at room temperature. One hundred and eighty milliliters of buffer A containing 50 mm Tris-HCl (pH 7.8), 5 mm EDTA, 5 mm benzamidine, 5 mm 6-aminocaproic acid, 1 mm PMSF, 0.04% (v/v) chemostatin, 2 mm 2-mercaptoethanol, and 3 g of insoluble PVP was added with stirring during sample thawing. The thawed extract homogenate was centrifuged (39,000g for 30 min) and 20 mL of 2% (w/v) protamine sulfate was added to the supernatant (180 mL) to precipitate DNA. The sample was clarified by centrifugation as above and applied to a Q-Sepharose column (1.7 × 20 cm) preequilibrated with buffer A. The column was washed until the A280 returned to baseline and then eluted with a linear gradient of NaCl (0–1.0 m) in buffer A. Enzymes were assayed and the fractions containing NiR, Fd, FNR, and G6PDH activities were pooled separately.
NiR Purification
(NH4)2SO4 was added to the NiR fraction from the Q-Sepharose to a final concentration of 15%. The sample was centrifuged (39,000g for 15 min) and the supernatant was applied to a phenyl-Sepharose column (1.7 × 20 cm) as described by Romero et al. (1987). The column was developed by a 135-mL linear gradient of (NH4)2SO4 (15–0% saturation) in buffer B containing Tris-HCl (pH 7.8), 1 mm EDTA, and 2 mm 2-mercaptoethanol. NiR activity was pooled and brought to 20% saturation with (NH4)2SO4 and centrifuged as above. The supernatant was applied to a butyl-Toyopearl column (1.7 × 20 cm) preequilibrated with buffer B containing 20% saturated (NH4)2SO4. The column was eluted with the same buffer (buffer B containing 20% saturated [NH4]2SO4). The active fractions were pooled and dialyzed overnight against 50 mm Tris-HCl (pH 7.8) and 1 mm EDTA (buffer C). The dialyzed sample was applied to a DEAE-Fractogel column (1 mL) and eluted with a 0 to 0.3 m linear gradient of NaCl (12.5 mL). Fractions with NiR activity were pooled, concentrated by ultrafiltration with a Centricon-50 concentrators (Amicon, Beverly, MA), frozen in liquid N2, and stored at −80°C.
FNR Purification
FNR eluted from the Q-Sepharose column as a broad peak that also contained G6PDH activity. FNR was purified as in Jin et al. (1994) using a Blue-Cellulofine and Fd-Sepharose chromatography. Purified FNR was desalted and concentrated by ultrafiltration with a Centricon-30 concentrators (Amicon) and stored at −80°C. Mung bean leaf FNR was purified as previously described in Jin et al. (1994).
G6PDH Isolation
G6PDH and FNR coeluted from a Blue-Cellulofine column (1.7 × 20 cm) by a linear gradient of NaCl (0–1.0 m) in buffer B. After dialyzing against buffer B overnight, the sample was applied to a Fd-Sepharose column (1.4 × 6 cm) to separate the activities. FNR was bound on the column and G6PDH was passed through. The G6PDH sample was brought to 20% saturation with (NH4)2SO4 and centrifuged (39,000g for 15 min). The supernatant was applied to and eluted from a phenyl-Sepharose column (1.7 × 20 cm) as previously described for NiR except buffer B containing 20% (NH4)2SO4. The active fractions were pooled, concentrated, and desalted on Centricon-10 concentrators (Amicon); frozen in liquid N2; and stored at −80°C.
Fd Purification
Fd eluted from the Q-Sepharose column was further purified by butyl-Toyopearl chromatography according to the method of Jin et al. (1994), except that the concentration of (NH4)2SO4 in the linear gradient was 50 to 0%. Purified Fd was concentrated and desalted on Centricon-10 concentrators and stored at 4°C. Spinach leaf Fd was purified according the method described in Jin et al. (1994).
Rapid Isolation of NiR
One liter of freshly harvested cells (about 1 g in fresh weight) was frozen in liquid N2. The cells were thawed in 2 mL of homogenizing buffer A and centrifuged (18,200g, 3 min). The supernatant was desalted on a PD-10 column (Pharmacia) and the desalted sample was applied to a Blue-Cellulofine column (1.4 × 10 cm) equilibrated with buffer A. The column was washed with buffer A and the fractions showing NiR activity were pooled, concentrated on a Centricon-50 unit (Amicon), frozen in liquid N2, and stored at −80°C.
Assay Methods: NiR
Four methods were developed to assay NiR activity.
MV-NiR Assay
The reaction mixture (1 mL) contained 50 mm Tris-HCl (pH 7.8), 2 mm NaNO2, 0.4 mm MV, and 23 mm Na2S2O4 freshly prepared in 0.5 mm NaHCO3. The enzyme sample was added and the reaction was carried out at 30°C for 10 min.
Fd-NiR Assay
The reaction was carried out as described above, except that MV in the reaction mixture was replaced by 20 μm purified C. reinhardtii Fd.
NADPH-Dependent NO2− Reduction Assay
The reaction mixture (0.5 mL), containing 50 mm Tris-HCl (pH 7.8), 0.4 mm NaNO2, 20 nm purified C. reinhardtii FNR, 20 μm purified C. reinhardtii Fd, 10 mm Glc, 100 μg/mL Glc oxidise, 50 μg/mL catalase, and 0.1 unit of NiR (activity calculated by MV-NiR assay), was mixed in a rubber-capped glass chamber (3 mL). To ensure anaerobicity, the mixture was bubbled with N2 for 1 min, and then the reaction was started by injecting NADPH to a final concentration of 1 mm. The assay continued for 15 min at 30°C. NADPH-dependent NO2− reduction was assayed aerobically in the same reaction mixture except that no Glc, Glc oxidase, or catalase were included.
G6PDH-Coupled NO2− Reduction Assay
Reactions were carried out under aerobic conditions and the reaction mixture (0.5 mL) contained 50 mm Tris-HCl (pH 7.8), 2 mm NaNO2, 0.35 unit of G6PDH, 6 mm G6P, 0.1 mm NADP+ (or NADPH), 20 nm purified C. reinhardtii FNR, 20 μm C. reinhardtii Fd, and 0.1 unit of NiR (calculated by MV-NiR activity). The reaction was performed at 30°C for 10 min.
For all of the assays, the disappearance of NO2− was used to calculated enzyme activity and 1 unit of NiR activity catalyzed 1 μmol NO2− reduction per min. NO2− concentration was measured as described by Hirasawa et al. (1989), and a mean of at least three independent measurements was used to calculate NiR activity. The se associated with the reported values was always less than 3%.
Other Enzyme Assays
Fd and FNR activity were measured by the reduction of Cyt c in the presence of NADPH, essentially as described by Morigasaki et al. (1990b). The assay mixture contained 50 mm Tris-HCl (pH 7.8), 40 μm Cyt c, 100 μm NADPH, and 5 μm purified C. reinhardtii Fd for the FNR assay or 20 nm purified C. reinhardtii FNR for the Fd assay. G6PDH activity was measured by monitoring the formation of NADPH in the presence of G6P as described by Farr et al. (1994), except that no 2-mercaptoethanol was added in the reaction mixture.
Other Methods
Gels (10%) were prepared according to the method of Laemmli (1970) and electrophoresed at 100 V for 2 h on a Bio-Rad mini protein gel system. The gel was stained with Coomassie brilliant blue. Protein concentrations were determined by the method of Bradford (1976) using BSA as a standard. All enzyme assays and absorbances were measured with a spectronic array (model 3000, Milton Roy, Spokane, WA).
RESULTS
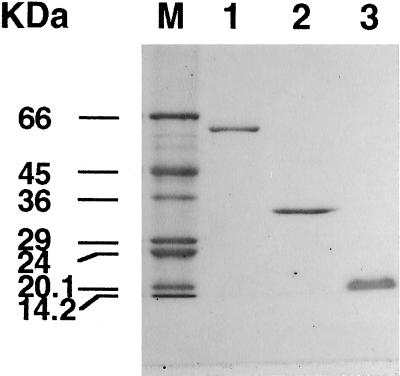
NiR, Fd, and FNR were separated from the same cell extract using Q-Sepharose. Fd and FNR were further purified by standard methods to yield homogeneous proteins (Fig. 1). G6PDH activity copurified with FNR on Q-Sepharose and the affinity column, but was separated by chromatography on Fd-Sepharose. Following this step G6PDH contained no interfering FNR or Fd activity (data not shown). NiR eluted from the Q-Sepharose column as a sharp peak near the beginning of the NaCl gradient. NiR bound to phenyl-Sepharose at 15% (NH4)2SO4, whereas the enzyme did not bind to a different hydrophobic matrix, butyl-Toyopearl, at 20% (NH4)2SO4. The specific activity of NiR increased significantly after chromatography on these two hydrophobic columns (data not shown). A final chromatography step using DEAE-Fractogel yielded NiR, which appeared as a single band on SDS-PAGE (Fig. 1). Purified NiR has absorption maxima at 281, 385, and 544 nm and the ratio of A281/A385 was 1.6 (data not shown). The specific activities of MV-NiR and Fd-NiR were 39 and 14 units mg−1 protein, respectively, which yields a Fd-NiR/MV-NiR ratio of 0.36 (Table I).
Figure 1.
SDS-PAGE of NiR, FNR, and Fd purified from C. reinhardtii. The gel was stained with Coomassie brilliant blue. M, Molecular mass standard. Lane 1, One microgram of NiR; lane 2, 1 μg of FNR; and lane 3, 1 μg of Fd.
Table I.
Specific activity of purified and rapidly isolated NiR
| NiR Assays | MV-NiR Activity | Fd-NiR Activity | Ratio (Fd/MV) |
|---|---|---|---|
| units mg−1 protein | |||
| Purified NiR | 39 | 14 | 0.36 |
| Rapidly isolated NiR | |||
| Fresh protein | 3.1 | 3.2 | 1.0 |
| Frozen protein | 2.8 | 1.2 | 0.43 |
Chemically reduced MV and Fd were used in respective NiR assays. For fresh protein, the assay was carried out immediately after isolation; for frozen protein, the assay was carried out after storage at −80°C.
A rapid method was developed to isolate NiR, which exhibited higher relative Fd-NiR activity. Desalted extracts from freshly harvested cells were chromatographed on Blue-Cellulofine, an adenosine-affinity matrix. NiR eluted from the column slightly after the major nonadherent protein peak passed off the column. To improve purification in this step, only fractions containing NiR activity that eluted after the major protein peak were pooled (data not shown). This sample was separated from FNR, G6PDH, or Fd because FNR and G6PDH bound to the Blue-Cellulofine column, and Fd eluted in the main, nonadherent protein peak. The ratio of Fd-NiR/MV-NiR was approximately 1 immediately after isolation; however, it decreased to 0.43 after storage at −80°C (Table I).
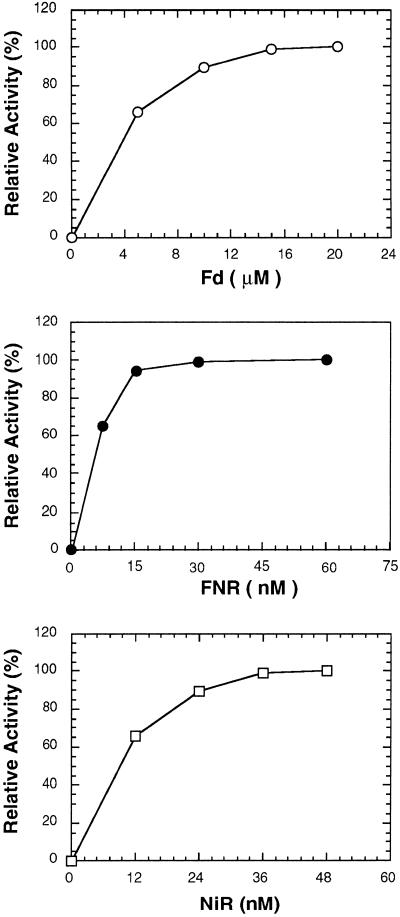
The ability of NADPH to reduce NO2− to NH4+ was examined by reconstituting a NO2−-reducing pathway using the algal FNR, Fd, and the rapidly isolated NiR. NO2− was reduced only in the presence of all components of this reaction (Table II). NO2− reduction was observed under both aerobic and anaerobic conditions, but the reaction was somewhat more effective under anaerobic conditions (Table II). Kinetic studies of the dependency of activity on each of the protein components of the electron-transfer chain under anaerobic conditions revealed that any one could be a limiting factor for the reaction (Fig. 2).
Table II.
Examination of NADPH-dependent NO2− reduction reactions
| Reaction | Specific Activity of NiR |
|---|---|
| units mg−1 protein | |
| Aerobic | |
| Complete reaction | 0.052 |
| −FNR | 0.000 |
| −Fd | 0.000 |
| −NADPH | 0.000 |
| Anaerobic | |
| Complete reaction | 0.079 |
| −FNR | 0.000 |
| −Fd | 0.000 |
| −NADPH | 0.000 |
The complete reaction mixture contained NADPH, FNR, Fd, rapidly isolated NiR, NaNO2, and all other components of the NADPH-dependent NO2− reduction reaction as described in Methods. The assays were carried out under aerobic and anaerobic conditions in the absence (−) of the component indicated.
Figure 2.
Dependency of NADPH-dependent NO2− reduction on Fd (top), FNR (middle), and NiR (bottom). The reactions were carried out under anaerobic conditions as described in Methods.
Under anaerobic conditions the presence of NADP+ in the reaction mixture decreased the rate of NO2− reduction when the electron donor was NADPH (Table III). NO2− reduction was inhibited by 27% when the NADPH:NADP+ ratio was 10 (1 mm NADPH–0.1 mm NADP+). At a ratio of 1 (1 mm NADPH and 1 mm NADP+), inhibition rose to 63%. The inhibitory effect of NADP+ was not pronounced when chemically reduced MV or Fd was the electron donor (Table III).
Table III.
Effect of NADPH/NADP+ on NADPH-, Fd-, and MV-dependent NO2− reduction
| Ratio of NADPH/NADP+ | Relative
Catalytic Ability
|
||
|---|---|---|---|
| NADPH-NiR | Fd-NiR | MV-NiR | |
| mm | % | ||
| 1.0/0.0 | 100 | 100 | 100 |
| 1.0/0.1 | 73 | 96 | 105 |
| 1.0/1.0 | 37 | 95 | 94 |
The effects of the indicated NADPH/NADP+ ratios on NADPH-, Fd-, and MV-dependent NO2− reduction under anaerobic conditions. Conditions were as described in “Materials and Methods,” except that 0.1 unit (as MV-NiR activity) of rapidly isolated NiR was included in each reaction mixture.
In plant cell chloroplasts, NADPH can be generated either by photosynthetic electron transport or via chloroplastic G6PDH, the key regulatory protein of the OPP pathway. In the presence of partially purified G6PDH from C. reinhardtii, G6P could support in vitro NO2− reduction at a rate of 0.49 unit mg−1 protein (Table IV), approximately 10 times the NADPH-dependent rate (Tables II and IV). G6PDH-coupled NO2− reduction did not occur in the absence of G6P or NADP+, the substrates of G6PDH, or if any other components of the electron-transfer pathway (FNR and Fd) were absent (Table IV). High rates of NO2− reduction were also observed when NADP+ was replaced by NADPH, but only in the presence of G6PDH and G6P (Table IV).
Table IV.
Dependence of G6PDH-coupled NO2− reduction on reaction components
| Reaction | Specific Activity of NiR |
|---|---|
| units mg−1 protein | |
| Complete reaction | 0.49 |
| −FNR | 0.00 |
| −Fd | 0.00 |
| −G6PDH | 0.00 |
| −G6P | 0.00 |
| −NADP+ | 0.00 |
| −NADP+, +NADPH | 0.57 |
| −NADP+, +NADPH, −G6P | 0.076 |
The dependence of G6PDH-dependent NO2− reduction was examined using a complete reaction mixture containing NADP+, G6P, G6PDH, FNR, Fd, rapidly isolated NiR, NaNO2, and all components of the G6PDH-coupled NO2− reduction reaction, as described in Methods. Subsequent experiments were carried out with (+) or without (−) the indicated components in the reaction mixture.
G6PDH-dependent NO2− reduction also showed inhibition with increasing concentrations of NADP+, even though this compound is a substrate of G6PDH. The reaction rate of 16.6 nmol NO2− min−1 as supported by 0.1 mm NADP+ declined to 2.6 nmol NO2− min−1 when 1.0 mm NADP+ was used (Table V). NO2− reduction was observed when the components of this G6PDH coupled reaction were replaced with enzymes from different sources (Table VI). Yeast G6PDH was as effective in this reaction as the C. reinhardtii enzyme; however, replacement of either FNR or Fd with enzymes from leaf sources decreased the reaction rate. 6PGDH and ICDH could replace G6PDH and mediate NO2− reduction in this reaction system (Table VI).
Table V.
Effect of NADPH/NADP+ on G6PDH-coupled NO2− reduction
| Ratio of NADPH/NADP+ | NO2− Reduced |
|---|---|
| mm | nmol min−1 |
| 0.0 /0.0 | 0.00 |
| 0.0 /0.1 | 16.6 |
| 0.0 /1.0 | 2.6 |
| 0.1 /0.0 | 19.3 |
| 0.1 /0.1 | 15.6 |
| 0.1 /1.0 | 1.7 |
| 1.0 /0.0 | 17.8 |
| 1.0 /0.1 | 14.1 |
| 1.0 /1.0 | 0.89 |
The effects of the indicated NADPH/NADP+ ratios on G6PDH- coupled NO2− reduction under aerobic conditions, as described in Methods. Rapidly isolated NiR (0.1 unit of MV-NiR activity) was included in the reaction mixture.
Table VI.
Effects of heterologous components and different dehydrogenases on related NO2− reduction
| Reaction | NO2− Reduced |
|---|---|
| nmol min−1 | |
| Complete reaction | 16.6 |
| G6PDH (yeast) | 17.4 |
| FNR (mung bean) | 12.6 |
| Fd (spinach) | 9.7 |
| 6PGDH (porcine heart) | 8.0 |
| ICDH (yeast) | 6.3 |
The effects of replacing the protein components of G6PDH-coupled NO2− reduction reaction with heterologous proteins and different dehydrogenases. The complete reaction mixture contained NADP+, G6P, G6PDH, FNR, Fd, rapidly isolated NiR, NaNO2, and all components of the G6PDH-coupled NO2− reduction reaction, as described in Methods. The respective C. reinhardtii component was replaced by an identical concentration of G6PDH from yeast; FNR from mung bean leaf, and Fd from spinach leaf. In 6PGDH- and ICDH-coupled reactions, G6PDH and G6P were replaced by the same amount of 6PGDH (porcine heart) and 6-phosphogluconate, ICDH (yeast), and the isocitrate.
DISCUSSION
NADPH can support in vitro NO2− reduction in the presence of NiR, FNR, and Fd (Table II). Removal of NiR, FNR, or Fd from the reaction mixture prevented NO2− reduction. The rate of NO2− reduction could also be limited by the concentration of each of these proteins (Fig. 2). Analysis showed that in a crude extract of C. reinhardtii, the concentrations of Fd, FNR, and NiR were approximately 4.5, 0.10, and 0.14 μm, respectively (data not shown). These data suggested that the concentration of Fd may be the limiting factor for the rate of NO2− reduction in vivo. The failure of NADPH to support NO2− reduction in the absence of Fd confirms that the algal NiR is a Fd-dependent enzyme and cannot use NADPH directly. The dependency of electron transport on FNR demonstrates for the first time, to our knowledge, that FNR can couple Fd reduction with the oxidation of NADPH. This reaction proceeds only slightly better under anaerobic conditions, indicating that the oxidation of components of this pathway with atmospheric O2 may not be a significant factor in vivo.
During the purification of NiR, comparison of MV- and Fd-dependent activity revealed that Fd became a relatively less-effective electron donor as the enzyme was purified (Table I). To maintain high Fd-NiR activity required the rapid separation of enzyme from fresh cells. It has been suggested that NiR may contain a subunit with a molecular mass of 24 kD that is specifically involved in binding Fd (Romero et al., 1987; Hirasawa et al., 1989); therefore, the drop in the ratio of Fd-NiR/MV-NiR activity could result from the loss of this subunit. However, our purified NiR retained substantial Fd-NiR activity, and no second subunit for this protein appeared on SDS-PAGE even when the gels were overloaded (data not shown). This result is consistent with that reported by Pajuelo et al. (1993), but the ratio of Fd-/MV-NiR activity reported in this study is somewhat lower than the value of 0.82 that they reported for C. reinhardtii NiR.
Several reports of work on heterotrophic tissues have indicated that NADPH must be the electron donor for dark NO2− reduction (Oji et al., 1985; Suzuki et al., 1985; Bowsher et al., 1989); however, it has been suggested that high concentrations of NADPH would be necessary to support the reduction of Fd by FNR (Bowsher et al., 1989). NADPH-dependent NO2− reduction was extremely sensitive to the presence of NADP+, dropping almost 30% even at a NADPH:NADP+ ratio of 10:1 (Table III). As NADP+ did not affect either MV- or Fd-NiR activity (Table III), it can be concluded that NADP+ was not inhibiting the activity of either NiR or Fd. The inhibitory effect of NADP+ must therefore be mediated through FNR directly. Additional support for this conclusion is the observation that the reduction of Cyt c by NADPH via FNR and Fd is also inhibited in the presence of NADP+ (data not shown).
The sensitivity of the reconstituted NADPH-dependent system to NADP+ levels supports the idea that high ratios of NADPH/NADP+ would have to be maintained for NADPH to be able to serve as an electron donor to Fd. Measurements of pyridine nucleotides in N-limited algae have shown that the ratio of NADPH/NADP+ ranges from approximately 2 in the light to approximately 4 in the dark (Huppe et al., 1992, 1994; Vanlerberghe et al., 1992), which is within the range yielding high NiR activity in vitro (Table V).
The onset of NO3− assimilation in N-limited algae has been shown to result in increased activity of the OPP pathway resulting from G6PDH activation (Huppe et al., 1992, 1994; Vanlerberghe et al., 1992). Previous studies demonstrated that the addition of G6P to broken plastids allowed NADPH to serve as an electron donor for NO2− reduction (Oji et al., 1985). These investigators suggested that G6PDH served to regenerate NADPH from NADP+, thereby maintaining a high NADPH/NADP+ ratio, favoring NO2− reduction. In our reconstituted system NADPH generated by G6PDH supported NO2− reduction at rates nearly 10-fold greater than that when NADPH was added alone (Tables II and IV). When G6PDH-dependent NADP+ recycling was prevented by withholding G6P in the presence of NADPH, the rate of NO2− reduction dropped to the NADPH-dependent rate (Tables II and IV). This demonstrated that the key factor in allowing G6PDH-enhanced rates of NO2− reduction was the role of G6PDH in scavenging NADP+. This role for G6PDH could be replaced by G6PDH from other sources or by 6PGDH or ICDH, both of which consume NADP+ and produce NADPH (Table VI). These results indicated that the OPP pathway supports dark NO2− reduction in the chloroplast in at least two ways: (a) it generates the NADPH that is necessary to reduce Fd and subsequently NO2−, and (b) it consumes the NADP+ produced by FNR in the reduction of Fd, thereby releasing FNR from NADP+ inhibition. Although substitution of a nonplant G6PDH did not affect the rate of NO2− reduction, replacement of either Fd or FNR did lower the rate of reaction (Table VI). Fd has also been noted to interact more favorably with another reductase, Fd-thioredoxin, when both proteins were from similar sources (Huppe et al., 1990).
The potential for G6PDH-coupled NO2− reduction must be considered in the context of the observed rates of dark NO3− assimilation rates in C. reinhardtii of approximately 30 μmol mg−1 chlorophyll h−1 (L.K. McPhee, personal communication). Given the observed G6PDH-NiR/MV-NiR ratio of 0.175 (0.49 unit mg−1 protein/2.8 units mg−1 protein; Tables I and IV) and a MV-NiR activity in the crude extract of 260 μmol NO2− mg−1 chlorophyll h−1 (T. Jin, unpublished data), the G6PDH-coupled system could support in vitro assimilation of (260 × 0.175) at approximately 45 μmol NO2− mg−1 Chl h−1. As a result, the observed in vitro rate of G6PDH-dependent NO2− reduction is sufficient to account for the observed rates of dark NO3− assimilation.
The results of this study demonstrate for the first time, to our knowledge, that NADPH may reduce Fd via FNR and thereby support NO2− reduction. The reversible character of the reaction catalyzed by FNR is the key point of this reconstituted system. The equilibrium between reduced/oxidized Fd and NADPH/NADP+ drives the FNR reaction in the appropriate direction. Therefore, to have Fd serve as an efficient electron donor to NiR, the ratio of NADPH/NADP+ must be very high. When G6PDH is used to generate NADPH and to consume NADP+, NiR activity increased approximately 10-fold primarily due to the consumption of NADP+, thereby releasing FNR from NADP+ inhibition. The rates of NO2− reduction supported in this way account for the observed rates of whole-cell NO3− assimilation in the dark.
Abbreviations:
- FNR
Fd NADP+ oxidoreductase
- G6P
Glc-6-P
- G6PDH
G6P dehydrogenase
- ICDH
isocitrate dehydrogenase
- MV
methyl viologen
- NiR
nitrite reductase
- OPP
oxidative pentose phosphate
- 6PGDH
6-phosphogluconate dehydrogenase
Footnotes
This work was supported by the Natural Sciences and Engineering Research Council of Canada.
LITERATURE CITED
- Barea JL, Cárdenas J. The nitrate-reducing enzyme system of Chlamydomonas reinhardii. Arch Microbiol. 1975;105:21–25. doi: 10.1007/BF00447107. [DOI] [PubMed] [Google Scholar]
- Beevers L, Hageman RH (1980) Nitrate and nitrite reduction. In BJ Miflin, eds, The Biochemistry of Plants, Vol 5. Academic Press, New York, pp 115–168
- Borchert S, Harborth J, Schünemann D, Hoferichter P, Heldt HW. Studies of the enzymic capacities and transport properties of pea root plastids. Plant Physiol. 1993;101:303–312. doi: 10.1104/pp.101.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowsher CG, Dunber B, Emes MJ. The purification and properties of ferredoxin-NADP+ oxidoreductase from roots of Pisum sativum L. Protein Expr Purif. 1993;4:512–518. doi: 10.1006/prep.1993.1067. [DOI] [PubMed] [Google Scholar]
- Bowsher CG, Hucklesby DP, Emes MJ. Nitrite reduction and carbohydrate metabolism in plastids purified from roots of Pisum sativum L. Planta. 1989;177:359–366. doi: 10.1007/BF00403594. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Emes MJ, Fowler MW. The supply of reducing power for nitrite reduction in plastids of seedling pea roots (Pisum sativum L.) Planta. 1983;158:97–102. doi: 10.1007/BF00397700. [DOI] [PubMed] [Google Scholar]
- Farr TJ, Huppe HC, Turpin DH. Coordination of chloroplastic metabolism in N-limited Chlamydomonas reinhardtii by redox modulation. I. The activation of phosphoribulosekinase and glucose-6-phosphate dehydrogenate is relative to the photosynthetic supply of electrons. Plant Physiol. 1994;105:1037–1042. doi: 10.1104/pp.105.4.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirasawa M, Chang KT, Knaff DB. Characterization of a ferredoxin:NADP+ oxidoreductase from a non-photosynthetic plant issues. Arch Biochem Biophys. 1990;276:251–258. doi: 10.1016/0003-9861(90)90035-w. [DOI] [PubMed] [Google Scholar]
- Hirasawa M, Gray KA, Sung J, Knaff DB. Spinach nitrite reductase: subunit composition and siroheme redix potential. Arch Biochem Biophys. 1989;275:1–10. doi: 10.1016/0003-9861(89)90342-1. [DOI] [PubMed] [Google Scholar]
- Huppe HC, de Lamotte-Guéry F, Jacquot JP, Buchanan BB. The ferredoxin/thioredoxin system of a green alga, Chlamydomonas reinhardtii: identification and characterization of thioredoxins and ferredoxin-thioredoxin reductase components. Planta. 1990;180:341–351. [PubMed] [Google Scholar]
- Huppe HC, Farr TJ, Turpin DH. Coordination of chloroplastic metabolism in N-limited Chlamydomonas reinhardtii by redox modulation. II. Redox modulation activates the oxidative pentose phosphate pathway during photosynthetic nitrate assimilation. Plant Physiol. 1994;105:1043–1048. doi: 10.1104/pp.105.4.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppe HC, Turpin DH. Appearance of novel glucose-6-phosphate dehydrogenase isoforms in Chlamydomonas reinhardtii during growth on nitrate. Plant Physiol. 1996;110:1431–1433. doi: 10.1104/pp.110.4.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppe HC, Vanlerberghe GC, Turpin DH. Evidence for activation of the oxidative pentose phosphate pathway during photosynthetic assimilation of NO3− but not NH4+ by a green alga. Plant Physiol. 1992;100:2096–2099. doi: 10.1104/pp.100.4.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin T, Morigasaki S, Wada K. Purification and characterization of two ferredoxin-NADP+ oxidoreductase isoforms from the first foliage leaves of mung bean (Vigna radiata) seedlings. Plant Physiol. 1994;106:697–702. doi: 10.1104/pp.106.2.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Morigasaki S, Takata K, Sanada Y, Wada K, Yee BC, Shin S, Buchanan BB. Novel forms of ferredoxin and ferredoxin-NADP reductase from spinach roots. Arch Biochem Biophys. 1990a;283:75–80. doi: 10.1016/0003-9861(90)90614-5. [DOI] [PubMed] [Google Scholar]
- Morigasaki S, Takata K, Suzuki T, Wada K. Purification and characterization of ferredoxin-NADP+ oxidoreductase-like enzyme from radish root tissues. Plant Physiol. 1990b;93:896–901. doi: 10.1104/pp.93.3.896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oaks A, Hirel B. Nitrogen metabolism in roots. Annu Rev Plant Physiol. 1985;36:345–365. [Google Scholar]
- Oji Y, Watanabe M, Wakiuchi N, Okamoto S. Nitrite reduction in barley-root plastids: dependence on NADPH coupled with glucose-6-phosphate and 6-phosphogluconate dehydrogenases, and possible involvement of an electron carrier and a diaphorase. Planta. 1985;165:85–90. doi: 10.1007/BF00392215. [DOI] [PubMed] [Google Scholar]
- Pajuelo E, Borrero JA, Márquez AJ. Immunological approach to subunit composition of ferredoxin-nitrite reductase from Chlamydomonas reinhardtii. Plant Sci. 1993;95:9–21. [Google Scholar]
- Romero LC, Galván F, Vega JM. Biochem Biophys Acta. 1987;914:55–63. [Google Scholar]
- Sivarsankar S, Oaks A. Nitrate assimilation in higher plants: the effects of metabolites and light. Plant Physiol Biochem. 1996;34:609–620. [Google Scholar]
- Suzuki A, Oaks A, Jacquot J-P, Vidal J, Gadal P. An electron transport system in maize roots for reactions of glutamate synthase and nitrite reductase. Physiological and immunochemical properties of the electron carrier and pyridine nucleotide reductase. Plant Physiol. 1985;78:374–378. doi: 10.1104/pp.78.2.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syrett PJ. Nitrogen metabolism of microalgae. Can Bull Fish Aquat Sci. 1981;219:182–210. [Google Scholar]
- Turpin DH, Weger HG, Huppe HC. Interactions between photosynthesis, respiration and nitrogen assimilation. In: Dennis DT, Turpin DH, Lefebvre DD, Layzell DB, editors. Plant Metabolism, Ed 2. Addison Wesley. Reading, MA: Longman; 1997. pp. 509–524. [Google Scholar]
- Vanlerberghe GC, Huppe HC, Vlossak KDM, Turpin DH. Plant Physiol. 1992;99:495–500. doi: 10.1104/pp.99.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada K, Onda M, Matsubara H. Amino acid sequences of ferredoxin isoproteins from radish roots. J Biochem. 1989;105:619–625. doi: 10.1093/oxfordjournals.jbchem.a122714. [DOI] [PubMed] [Google Scholar]