Abstract
The bacterial ribosome is an important target for many antimicrobial agents. Aminoglycoside antibiotics bind to both 30S and 50S ribosomal subunits, inhibiting translation and subunit formation. During ribosomal subunit biogenesis, ribonucleases (RNases) play an important role in rRNA processing. E. coli cells deficient for specific processing RNases are predicted to have an increased sensitivity to neomycin and paromomycin. Four RNase mutant strains showed an increased growth sensitivity to both aminoglycoside antibiotics. E. coli strains deficient for the rRNA processing enzymes RNase III, RNase E, RNase G, or RNase PH showed significantly reduced subunit amounts after antibiotic treatment. A substantial increase in a 16S RNA precursor molecule was observed as well. Ribosomal RNA turnover was stimulated and an enhancement of 16S and 23S rRNA fragmentation was detected in E. coli cells deficient for these enzymes. This work indicates that bacterial RNases may be novel antimicrobial targets.
Keywords: Escherichia coli, ribonuclease mutants, ribosome assembly, neomycin, paromomycin
Introduction
Bacterial resistance to the most commonly used antimicrobial agents is increasing (Davies and Davies 2010; Rosen 2011). Well examined resistance mechanisms include the acquisition of resistance genes, up-regulation of genes encoding cellular efflux pumps and spontaneous mutations in target genes (Zinner 2007). Development of novel antibacterial agents and the identification of additional bacterial targets have become important research endeavors (Champney 2008).
One significant target of many antibiotics is the bacterial ribosome (Wallis and Schroeder 1997; Champney 2006). The prokaryotic ribosome consists of a large 50S and a small 30S subunit. The 50S subunit is composed of 23S and 5S rRNA and 34 ribosomal proteins. The 30S subunit contains 16S ribosomal RNA and 21 ribosomal proteins. Some antibiotics function by targeting both the 30S and 50S subunits. Neomycin and paromomycin are two aminoglycoside antibiotics that bind to helix 44 of 16S rRNA and stimulate mistranslation of mRNA (Schroeder et al. 2000; Sutcliffe 2005; Arya 2007; Długosz and Trylska 2009). Aminoglycosides also bind to helix 69 of 23S rRNA. Binding to helix 69 diminishes the ability of the ribosome recycling factor to recycle the ribosome (Hirokawa et al. 2005; Borovinskaya et al. 2007; Scheunemann et al. 2010). Both antibiotics have also been shown to inhibit 30S and 50S subunit assembly in various bacteria, including E. coli and Staphylococcus aureus (Mehta and Champney 2002; Mehta and Champney 2003).
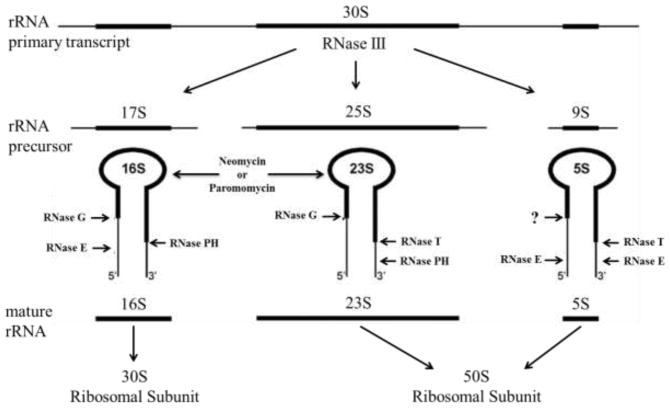
During subunit biogenesis, 16S, 23S, and 5S rRNA transcripts and ribosomal proteins combine to form intermediate precursors. The rRNA in the precursor particles is cleaved and processed by endo- and exoribonucleases to produce the mature subunits (Kaczanowska and Rydén-Aulin 2007; Deutscher 2009). The RNase enzymes involved in the processing of rRNA are indicated in Figure 1. In addition to maturation, ribonucleases are involved in the degradation of rRNA. When an inhibitor, such as an antibiotic, is present in a bacterial cell, specific RNases are utilized to degrade the rRNA to eliminate the stalled precursor (Champney 2006).
Fig. 1.
Bacterial rRNA processing pathways. The major rRNA processing enzymes are indicated. The binding of both aminoglycosides to 16S and 23S rRNAs is also shown (modified from (Davies et al. 2010). John Wiley and Sons used with permission.
It has been previously shown that strains of E. coli deficient for RNase E, RNase II, or polynucleotide phosphorylase (PNPase) displayed an increased sensitivity to erythromycin and azithromycin (Usary and Champney 2001; Silvers and Champney 2005). These mutant E. coli strains were shown to accumulate 23S rRNA and demonstrated a reduced rate of subunit resynthesis after antibiotic removal.
Bacterial RNases have been recently described as potential new antimicrobial target (Eidem et al. 2012). The RNase inhibitor VRC has been shown to impair ribosomal subunit formation in both S. aureus and E. coli (Frazier and Champney 2012a; Frazier and Champney 2012b). The present work was conducted to see if elimination of rRNA processing RNases enhanced the sensitivity of cells to aminoglycoside antibiotics. This work shows that E. coli strains deficient for RNase III, RNase E, RNase G, or RNase PH have an increased sensitivity to neomycin and paromomycin, reflected in reduced subunit synthesis and enhanced rRNA turnover.
Materials and methods
Analysis of cellular growth and viability
Escherichia coli strains used are listed in Table 1. Cultures were grown at 37°C (or 32°C for ts mutants) in tryptic soy broth (TSB). Strains SK5665, SK5729, SK6639 and SK7622 were supplemented with 4 μg/mL thymidine during growth. Growth rates were measured as an increase in cellular density over time using a Klett-Summerson colorimeter. Neomycin or paromomycin were added to cultures at 10μg/mL based on their IC50 values (Mehta and Champney 2002). All cultures were grown for two cellular doublings to approximately 4×108 colony forming units (CFU)/mL. Cellular viability was determined by TSB agar colony counting after serial dilution (Jett et al. 1997).
Table 1.
E. coli strains used in this study
| Strain | Phenotype | Genotype | Reference or source |
|---|---|---|---|
| SK901 | None | F- malA thi- | (Kushner et al. 1977) |
| D10-1 | I | HfrH met- rna-1 relA | (Gesteland 1966) |
| SK7622 | III | F-thyA715 rncD38::kanR | (Babitzke et al. 1993) |
| SK5665 | E | F-thyA715 rne1 | (Arraiano et al. 1988) |
| SK4803 | II | F-gal thi ton sup hasdR4 endAsbcB15 rnb296 | (Donovan and Kushner 1986) |
| N7060 | I/II/PNPase | F- metB1 tryA451 rpsl478 rna919rnb464 pnp13 | (Weatherford et al. 1972) |
| MG1655 I− R− | I/PH/R | Δ rna Δrnr::cam rph-1 | (Chen and Deutscher 2010) |
| GW11 | G | F- zce-726::Tn10, TetR, rng::cat CmR | (Li et al. 1999) |
| SK6639 | PH/PNPase | F-thyA715, CmR, pnp-200, rph-1, λ- | (Cheng and Deutscher 2003) |
| SK5729 | I/II/PNPase/PH | F- thyA715, rna-19, rnb-500, pnp-7, λ-, rph-1 | Sidney Kushner |
Ribosomal subunit assembly assay
Cell cultures were grown in TSB and at a Klett reading of 20, neomycin or paromomycin were added to the cells. After 15 minutes of growth with the antibiotics, 3H uridine (30 Ci/mmol, American Radiochemicals) at a concentration of 1μCi/mL and uridine at 2μg/mL were added. The cells were allowed to grow for two cellular doublings. At that time, uridine was added to 50μg/mL and the cells were incubated for an additional 15 minutes. Cells were collected by centrifugation and stored frozen at −70°C.
Cellular lysates were prepared with lysozyme and DNaseI as previously described (Silvers and Champney 2005). The samples were centrifuged through 5–20% sucrose gradients in S buffer (10 mM Tris-HCl, pH 8.0, 0.5 mM Mg acetate, 50 mM NH4Cl) in an SW41 rotor at 187813 × g for 3.5 hours. Following centrifugation, fractions were collected by pumping the gradient through an ISCO Model UA-5 absorbance monitor set at 254nm. The fractions were collected into vials and mixed with 3mL Scintisafe gel before measuring the 3H uridine radioactivity by liquid scintillation counting.
Agilent Bioanalyzer analysis of total RNA
Bacterial cells were grown as described above with neomycin or paromomycin at 10μg/mL. At a density of 4×108 cells/mL, the cells were collected by centrifugation and RNA was prepared from the cell pellet. Total RNA from cell samples was isolated by phenol/chloroform extraction and ethanol precipitation (Rio et al. 2011). Typically 0.5 to 1μg of RNA was examined using an Agilent Bioanalyzer 2100 and the RNA 6000 nano chip. Integrated gel peak areas were determined with Agilent software.
Analysis of 16S rRNA and 23S rRNA fragmentation by Northern blot hybridization
Biotinylated 16S and 23S specific probes were constructed as previously described (Silvers and Champney 2005). The 16S (241 bp) and 23S (146 bp) DNA probes were amplified from plasmid pKK3535 DNA (Brosius et al. 1980) using the polymerase chain reaction. The 16S primers used were 16S F: GGA GGA AGG TGG GGA TGA CG and 16S R: ATG GTG ACG GGC GGT GTG (nt. nos. 1173–1414). The 23S primers used were 23S F: TAG GGG AGC GTT CTG TAA G and 23S R: CCC ATT AAC GTT GGA C (nt. nos. 1188–1334). The primers were from Life Technologies. PCR products were purified by extraction with equal amounts of phenol and chloroform before precipitating with 2 volumes of ethanol. The pellets were resuspended in 30μL of sterile water. The purified DNA probes were labeled with biotin using the Label-IT biotin labeling kit (Mirus) per the manufacturer’s instructions (Silvers and Champney 2005).
Six micrograms of total RNA was denatured by heating at 55°C for 10 minutes and separated on a 5% TAE PAGE gel as previously described (Rio et al 2011). A biotinylated RNA standard was run in each gel to provide an estimate of RNA fragment sizes. The intact 16S and 23S rRNAs were evident by ethidium bromide staining of the gel. This region of the gel was removed prior to Northern hybridization to permit a more sensitive detection of the rRNA fragments. After destaining overnight in sterile water, the RNA was transferred onto Nytran nylon membranes using a Turbo blot apparatus (Schleicher & Schuell). The membranes were pre-hybridized in 15mL of 1X pre-hybridization solution (MRC, Inc.) at 42°C for 30 minutes. The membranes were then hybridized overnight at 42°C with 10mL hybridization buffer (50% formamide, 5X SSC, 0.1% sarkosyl, 0.02% SDS and 200μg/mL BSA), 1X background quencher (MRC, Inc.), and 4pmol of the denatured 16S or 23S specific probe (Silvers and Champney 2005). After hybridization, the membranes were washed and the probe detected with horseradish peroxidase using the North2South chemiluminescent hybridization kit (Pierce Chemical Co.). Analysis of the rRNA fragmentation was conducted by imaging membranes in the G Box Imager (SynGene).
Results
RNases play a key role in the processing, maturation and turnover of rRNA (Deutscher 2009; Davies et al. 2010). Figure 1 shows the important rRNA processing enzymes in E. coli. This figure also indicates that the aminoglycoside antibiotics neomycin and paromomycin bind to both 16S and 23S rRNAs (Sutcliffe 2005; Foster and Champney 2008; Scheunemann et al. 2010). It was hypothesized that E. coli cells lacking essential processing RNases would show an increased sensitivity to these compounds. Cell viability assays were performed to determine whether E. coli mutants deficient for any of eight RNases showed an enhanced sensitivity to these antibiotics. As Table 2 shows, E. coli mutants deficient for the rRNA processing RNases all showed enhanced growth sensitivity to neomycin compared with wild type cells. Most of the mutants were also reduced in their relative sensitivity to paromomycin as well. Strains deficient for the degradative RNases I, II and PNPase showed results similar to that of the wild type E. coli cells and were not examined further. Strains with mutations affecting the rRNA processing enzymes RNase III, E, G and PH were particularly sensitive to both aminoglycosides.
Table 2.
Effect of aminoglycosides on viability of wild type and mutant E. coli cells
| CFU (×107/mL) | ||||
|---|---|---|---|---|
|
| ||||
| Strain | RNase mutation | Control | Neomycin | Paromomycin |
| SK901 | None | 133±28.8 | 35±12 (26.3) | 4±0.7 (3.0) |
| SK7622 | III | 157±41.5 | 9±6.7 (5.7) | 4±1.9 (2.5) |
| SK5665 | E | 112±8.5 | 4±0.5 (3.6) | 1±0.01 (0.9) |
| MG1655 I−R− | I/PH/R | 762±19.7 | 88±56.0 (11.5) | 32±3.3 (4.2) |
| GW11 | G | 505±77.7 | 117±44.2 (23.2) | 12±4.4 (2.4) |
| SK5729 | I/II/PNPase/PH | 100±28.4 | 3±1.9 (3.0) | 2±0.6 (2.0) |
Results are the means ± standard error (n=2). The percentage of the control cell number for each strain is listed in parenthesis.
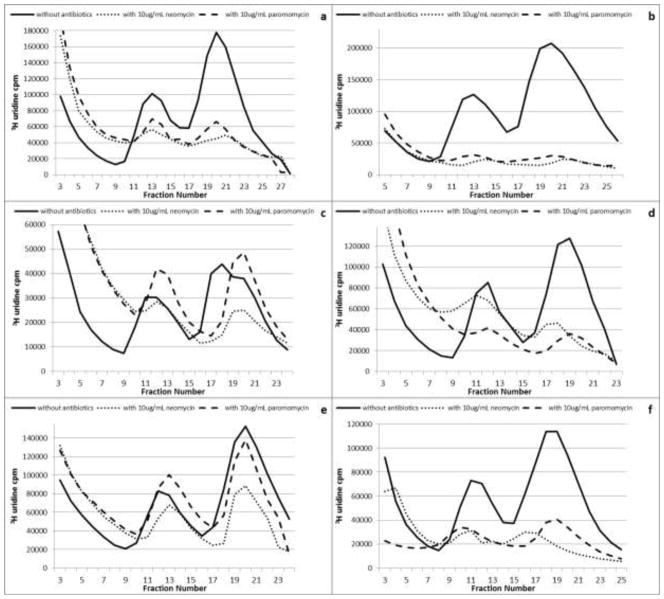
Aminoglycoside antibiotics can bind to both 30S and 50S ribosomal subunits (Sutcliffe 2005; Borovinskaya et al. 2007; Foster and Champney 2008; Długosz and Trylska 2009; Scheunemann et al. 2010). We therefore tested to see if growth with the aminoglycosides impaired the assembly of either subunit compared with the untreated cells. A reduction in the net synthesis of both the 30S and 50S ribosomal subunits was observed for the wild type and each mutant strain examined. Figure 2 shows the sucrose gradient profiles for six of the strains. For untreated control cells the ratio of 3H-rRNA in the 50S and 30S subunits was 2:1 as expected. Both aminoglycosides promoted a reduction in 30S and 50S subunit amounts in all of the RNase mutant strains. 50S subunit amounts were reduced to between 10 and 60% of the untreated control cells while 30S subunit amounts were reduced to between 35 and 75% of the control levels (Fig. 2). The aminoglycosides were most effective against an E. coli strain lacking RNase III and one lacking the four RNases I, II, PNPase and PH (Fig. 2b and 2f). Growth with the aminoglycosides also stimulated an increase in radiolabeled RNA in the top gradient fractions. After treatment with the antibiotics, there was a 30 to 200% increase in the RNA in the top gradient fractions (Fig. 2). This increase is indicative of rRNA degradation (Silvers and Champney 2005). In each case, the labeled RNA lost from the subunit regions was accounted for by an equivalent amount present in the top gradient fractions.
Fig. 2.
Sucrose gradient profiles of 3H uridine labeled ribosomal subunits isolated from E. coli cells grown with or without 10μg/mL antibiotics. Gradient profiles for wild type (a), RNase III deficient (b), RNase E deficient (c), RNase I/PH/R deficient (d), RNase G deficient (e), and RNase I/II/PNPase/PH deficient (f). Results are the means of 2 gradient profiles.
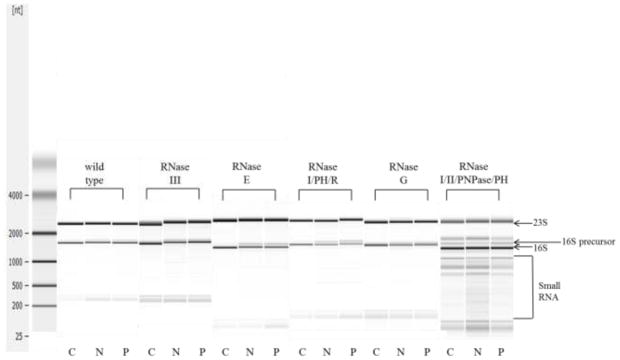
The loss of rRNA processing RNases is predicted to lead to the accumulation of 16S and 23S rRNA precursor species. Total RNA was isolated from each strain after growth without and with the aminoglycosides and the status of 16S precursor rRNA in the cells was examined by Agilent chip analysis (Fig. 3). The precursor 16S rRNA (17S) is 148 nucleotides larger than the mature 16S (1542 nucleotides) and is easily distinguished in the Agilent gel. The precursor to 23S rRNA is only 10 to 16 bases larger than the mature molecule and cannot be distinguished by this procedure (Kaczanowska and Rydén-Aulin 2007). Most of the mutant strains showed an enhanced accumulation of 16S precursor after growth with neomycin or paromomycin (Table 3). The largest increase in 16S precursor rRNA was seen in E. coli mutants deficient for RNase E, PH or R where more than 20% of the total 16S RNA was present in the precursor form (Table 3). The Agilent gels also indicated enhanced rRNA degradation in some mutants after aminoglycoside treatment (Fig. 3, small RNA).
Fig. 3.
Agilent gel analysis of total RNA from wild type and mutant cells grown with and without antibiotics. Control (C), neomycin (N), paromomycin (P)
Table 3.
Distribution of precursor 16S rRNA by Agilent gel analysis
| % 16S rRNA Precursor | ||||
|---|---|---|---|---|
|
| ||||
| Strain | RNase mutation | Control | Neomycin | Paromomycin |
| SK901 | None | 0.0 | 0.3 | 1.0 |
| SK7622 | III | 0.3 | 8.9 | 3.6 |
| SK5665 | E | 0.0 | 7.4 | 12.0 |
| MG1655 I−R− | I/PH/R | 0.8 | 21.8 | 33.1 |
| GW11 | G | 0.0 | 0.0 | 0.0 |
| SK5729 | I/II/PNPase/PH | 32.7 | 33.9 | 28.4 |
The percent of 16S precursor RNA relative to total 16S RNA was computed by analysis of the gel lanes in Fig. 3 using Agilent software. Results are the means of 2 gel analyses.
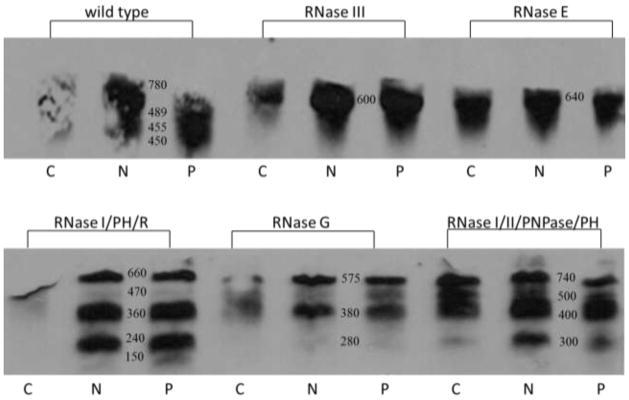
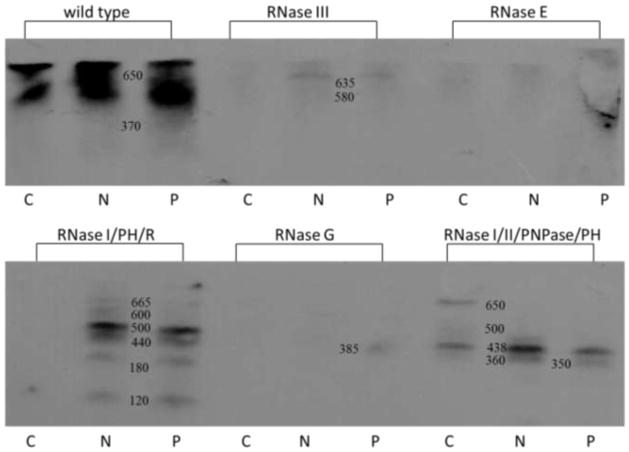
Ribosomal RNA degradation and turnover in the mutant strains was examined further. Fragmentation of 16S and 23S rRNA stimulated by antibiotic treatment was examined by Northern hybridization analysis. Figure 4 shows the result of Northern blots hybridized with a 16S rRNA probe, and Figure 5 shows the same E. coli RNA samples examined for rRNA fragmentation with a 23S rRNA probe. Enhanced 16S rRNA fragmentation was observed in each of the antibiotic treated samples in compared to the untreated control. E. coli deficient for RNase III, R+PH, G and I/II/PNPase/PH displayed the greatest increase in 16S rRNA fragmentation when compared with the mutant strain controls grown without neomycin or paromomycin (Fig. 4). Fragmentation of 23S rRNA was also detected by Northern hybridization analysis for the wild type and RNase mutant strains (Fig. 5). The largest increases in 23S rRNA were observed in strains deficient for RNase III, I/PH/R, and I/II/PNPase/PH after antibiotic treatment These results enhance the observations made by the sucrose gradient analysis indicating rRNA degradation after antibiotic treatment.
Fig. 4.
Northern blot analysis of 16S rRNA fragmentation for wild type and RNase mutant E. coli cells. RNA was isolated from cells grown without and with antibiotics. RNA sequences were identified by hybridization with a 16S DNA probe. Control (C), neomycin (N), paromomycin (P)
Fig. 5.
Northern blot analysis of 23S rRNA fragmentation for wild type and RNase mutant E. coli cells. RNA was isolated from cells grown without and with antibiotics. RNA sequences were identified by hybridization with a 23S DNA probe. Control (C), neomycin (N), paromomycin (P)
Discussion
Ribonucleases function in both rRNA processing and degradation (Deutscher 2009). RNases III, E, G, and PH are involved in the processing of precursor rRNA during subunit synthesis (Davies et al. 2010); Fig. 1). Elimination of specific RNases by mutation was predicted to enhance the effectiveness of antibiotics which stall ribosomal subunit formation. E. coli cells with a mutation in the gene for RNase E showed an enhanced sensitivity to erythromycin and the accumulation of a precursor to the 50S subunit (Usary and Champney 2001). E. coli cells deficient for RNase II or PNPase were increased in their sensitivity to azithromycin and showed an impairment of 23S rRNA function (Silvers and Champney 2005). Turnover of the rRNA was stimulated and 50S ribosomal subunit formation was impaired by azithromycin treatment. In addition, recovery of subunit formation after antibiotic removal was delayed. Based on these findings, it was hypothesized that E. coli cells deficient for specific processing RNases would display an increased sensitivity to aminoglycosides.
The wild type strain with a full complement of processing RNases showed a significant decrease in cell viability and a decrease in the formation of both subunits after antibiotic treatment (Table 2 and Fig. 2). A small amount of 16S precursor rRNA was found and enhanced rRNA fragmentation was apparent in the hybridization analysis.
RNase III is responsible for the initial cleavage of the precursor rRNA (Deutscher 2009). This study found that the addition of neomycin or paromomycin to E. coli cells deficient for RNase III led to an approximate 90% reduction in cell viability (Table 2). Examination of ribosomal subunit assembly showed that when either antibiotic was added, the amounts of 30S subunits were reduced by more than 60% and the 50S subunit level was reduced by approximately 85% (Fig. 2). RNA analysis by Agilent chip showed that there was a large increase in the 16S precursor, an indication of subunit precursor accumulation (Table 3; (Silvers and Champney 2005). Finally, analysis of 16S and 23S rRNA by Northern blotting hybridization revealed an increase in the 16S rRNA fragmentation and a change in the fragmentation pattern of 23S rRNA (Fig. 4 and Fig. 5). These results are consistent with the important role of RNase III in generating the rRNA precursors from the primary transcript.
RNases G and E function in processing of the 5′ end of 16S rRNA (Li et al. 1999). RNase G is also involved in processing of 23S rRNA (Song et al. 2011; Gutgsell and Jain 2012). We found that the absence of either RNase led to a decrease in cell viability when neomycin or paromomycin were present. The loss of RNase E appeared to have a greater on cell viability reduction than the absence of RNase G (Table 2). In both E. coli strains, there was a decrease in the 30S and 50S ribosomal subunit amounts with neomycin appearing to have a stronger effect in assembly reduction than paromomycin (Fig. 2). Agilent chip analysis revealed a larger increase in 16S precursor for RNase E deficient cells but no apparent 16S precursor for RNase G deficient cells (Table 3). Further analysis of 16S and 23S rRNA by Northern blotting hybridization demonstrated that both RNase E and G deficient cells showed enhanced fragmentation of 16S and 23S rRNAs (Fig. 4 and Fig. 5).
RNase PH is involved in 23S rRNA processing through cleavage of the 3′ end. Without this cleavage additional RNases, such as RNase T, cannot complete the maturation process (Gutgsell and Jain 2012). A double mutant strain lacking the processing RNase PH and the degradative enzyme RNase R showed impairment in the formation of both subunits by both sucrose gradient analysis and by the accumulation of precursor 16S rRNA. Also very large increases in the fragmentation of both RNA species were seen. The loss of RNase PH in a quadruple deficient mutant lacking the degradative enzymes RNase II and PNPase led to a very severe reduction in cell viability when neomycin or paromomycin were present. This strain also showed an approximate 80% reduction in the assembly of both the 30S and 50S ribosomal subunit when the aminoglycosides were used. Lastly, analysis of bacterial RNA by Agilent chip revealed the presence of the 16S precursor with and without antibiotic treatment (Table 3). Significant amounts of 16S and 23S rRNA oligonucleotides were also found under both circumstances (Fig. 4 and Fig. 5). In both of these mutant strains, impairment of rRNA processing and degradation significantly enhanced the antibiotic effectiveness. Ribonuclease I, also missing in both strains, is a periplasmic enzyme and is not normally involved in ribosomal RNA processing (Gesteland 1966).
The clearest results in this study were observed with strains containing single mutations in RNases III, E, and G. The results indicating major roles for the processing enzymes RNase III, E and G in antibiotic sensitivity are apparent. The interpretation of the results with the strain defective for RNases PH, and R and the strain missing four enzymes is less certain. Both strains showed major effects resulting from antibiotic treatment but the combination of nuclease mutations precludes a clear understanding of the role of the missing individual proteins. It is well established that in ribonuclease defective mutants, other ribonucleases can complement the defect in numerous ways. Studies have shown that due to the homology between RNases E and G, the loss of RNase E in E. coli cells can be complemented by an overexpression of RNase G (Lee et al. 2002). Additionally, when RNase E is absent in E. coli cells, there is an increase in the degradative enzyme, RNase R, indicating that the loss of one RNase involved in RNA degradation can lead to the up-regulation of another RNase involved in rRNA turnover in the bacterial cell (Cairrão and Arraiano 2006).
The enzymes identified in this study, which increase sensitivity to aminoglycosides, are different from the RNases shown to affect erythromycin and azithromycin sensitivity. The difference in enzymes in the two cases may result, in part, from the differences in the rRNA transcription and subunit assembly sequence. Aminoglycosides affect both 30S and 50S ribosomal subunit synthesis while macrolides impair only affect 50S ribosomal subunit assembly (Champney and Tober 2000; Usary and Champney 2001; Mehta and Champney 2002). During transcription, 16S rRNA is transcribed first and initiates 30S precursor assembly prior to 23S and 5S rRNA synthesis for 50S precursor formation. Aminoglycoside antibiotic stalling of 30S subunit formation leads to the accumulation of a 21S precursor, whose rRNA will not be further processed in specific RNase deficient mutants. Macrolide antibiotic stalling of 50S assembly generates a 32S precursor to the 50S particle which can be removed from the cell by the rRNA degradative enzymes like RNase II and PNPase as observed (Silvers and Champney 2005).
The present research has identified rRNA processing RNases as important in the effectiveness of aminoglycoside antibiotic inhibitory activity. Inhibition of processing RNases by small molecule inhibitors (Frazier and Champney 2012a; Frazier and Champney 2012b) or RNA interference methods could represent an attractive new antibiotic target which can enhance the effectiveness of other antimicrobial agents.
Acknowledgments
Mutant RNase deficient E. coli strains were gifts from Dr. Murray Deutscher (University of Miami) and Dr. Sidney Kushner (University of Georgia). This research was supported by an NIH AREA grant and an ETSU Graduate School research grant.
References
- Arraiano CM, Yancey SD, Kushner SR. Stabilization of discrete mRNA breakdown products in ams pnp rnb multiple mutants of Escherichia coli K-12. J Bacteriol. 1988;170:4625–4633. doi: 10.1128/jb.170.10.4625-4633.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arya DP, editor. Aminoglycoside Antibiotics. John Wiley & Sons, Inc; Hoboken, New Jersey: 2007. [Google Scholar]
- Babitzke P, Granger L, Olszewski J, Kushner SR. Analysis of mRNA decay and rRNA processing in Escherichia coli multiple mutants carrying a deletion in RNase III. J Bacteriol. 1993;175:229–239. doi: 10.1128/jb.175.1.229-239.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borovinskaya MA, Pai RD, Zhang W, Schuwirth BS, Holton JM, Hirokawa G, Kaji H, Kaji A, Cate JH. Structural basis for aminoglycoside inhibition of bacterial ribosome recycling. Nat Struct Mol Biol. 2007;14:727–732. doi: 10.1038/nsmb1271. [DOI] [PubMed] [Google Scholar]
- Brosius J, Dull TJ, Noller HF. Complete nucleotide sequence of a 23S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci U S A. 1980;77:201–204. doi: 10.1073/pnas.77.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairrão F, Arraiano CM. The role of endoribonucleases in the regulation of RNase R. Biochem Biophys Res Commun. 2006;343:731–737. doi: 10.1016/j.bbrc.2006.03.040. [DOI] [PubMed] [Google Scholar]
- Champney WS. The other target for ribosomal antibiotics: inhibition of bacterial ribosomal subunit formation. Infect Disord Drug Targets. 2006;6:377–390. doi: 10.2174/187152606779025842. [DOI] [PubMed] [Google Scholar]
- Champney WS, editor. New Antibiotic Targets. Humana Press Inc; Totowa, New Jersey: 2008. [Google Scholar]
- Champney WS, Tober CL. Specific inhibition of 50S ribosomal subunit formation in Staphylococcus aureus cells by 16-membered macrolide, lincosamide, and streptogramin B antibiotics. Curr Microbiol. 2000;41:126–135. doi: 10.1007/s002840010106. [DOI] [PubMed] [Google Scholar]
- Chen C, Deutscher MP. RNase R is a highly unstable protein regulated by growth phase and stress. RNA. 2010;16:667–672. doi: 10.1261/rna.1981010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng ZF, Deutscher MP. Quality control of ribosomal RNA mediated by polynucleotide phosphorylase and RNase R. Proc Natl Acad Sci U S A. 2003;100:6388–6393. doi: 10.1073/pnas.1231041100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies BW, Köhrer C, Jacob AI, Simmons LA, Zhu J, Aleman LM, Rajbhandary UL, Walker GC. Role of Escherichia coli YbeY, a highly conserved protein, in rRNA processing. Mol Microbiol. 2010;78:506–518. doi: 10.1111/j.1365-2958.2010.07351.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010;74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher MP. Maturation and degradation of ribosomal RNA in bacteria. Prog Mol Biol Transl Sci. 2009;85:369–391. doi: 10.1016/S0079-6603(08)00809-X. [DOI] [PubMed] [Google Scholar]
- Donovan WP, Kushner SR. Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc Natl Acad Sci U S A. 1986;83:120–124. doi: 10.1073/pnas.83.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Długosz M, Trylska J. Aminoglycoside association pathways with the 30S ribosomal subunit. J Phys Chem B. 2009;113:7322–7330. doi: 10.1021/jp8112914. [DOI] [PubMed] [Google Scholar]
- Eidem TM, Roux CM, Dunman PM. Wiley Interdiscip Rev RNA. 2012. RNA decay: a novel therapeutic target in bacteria. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster C, Champney WS. Characterization of a 30S ribosomal subunit assembly intermediate found in Escherichia coli cells growing with neomycin or paromomycin. Arch Microbiol. 2008;189:441–449. doi: 10.1007/s00203-007-0334-6. [DOI] [PubMed] [Google Scholar]
- Frazier AD, Champney WS. Inhibition of Ribosomal Subunit Synthesis in Escherichia coli by the Vanadyl Ribonucleoside Complex. Archives of Microbiology. 2012a doi: 10.1007/s00284-013-0350-5. In Submission. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier AD, Champney WS. The vanadyl ribonucleoside complex inhibits ribosomal subunit formation in Staphylococcus aureus. J Antimicrob Chemother. 2012b doi: 10.1093/jac/dks182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesteland RF. Isolation and characterization of ribonuclease I mutants of Escherichia coli. J Mol Biol. 1966;16:67–84. doi: 10.1016/s0022-2836(66)80263-2. [DOI] [PubMed] [Google Scholar]
- Gutgsell NS, Jain C. Role of precursor sequences in the ordered maturation of E. coli 23S ribosomal RNA. RNA. 2012;18:345–353. doi: 10.1261/rna.027854.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa G, Nijman RM, Raj VS, Kaji H, Igarashi K, Kaji A. The role of ribosome recycling factor in dissociation of 70S ribosomes into subunits. RNA. 2005;11:1317–1328. doi: 10.1261/rna.2520405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jett BD, Hatter KL, Huycke MM, Gilmore MS. Simplified agar plate method for quantifying viable bacteria. Biotechniques. 1997;23:648–650. doi: 10.2144/97234bm22. [DOI] [PubMed] [Google Scholar]
- Kaczanowska M, Rydén-Aulin M. Ribosome biogenesis and the translation process in Escherichia coli. Microbiol Mol Biol Rev. 2007;71:477–494. doi: 10.1128/MMBR.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushner SR, Maples VF, Champney WS. Conditionally lethal ribosomal protein mutants: characterization of a locus required for modification of 50S subunit proteins. Proc Natl Acad Sci U S A. 1977;74:467–471. doi: 10.1073/pnas.74.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Bernstein JA, Cohen SN. RNase G complementation of rne null mutation identifies functional interrelationships with RNase E in Escherichia coli. Mol Microbiol. 2002;43:1445–1456. doi: 10.1046/j.1365-2958.2002.02848.x. [DOI] [PubMed] [Google Scholar]
- Li Z, Pandit S, Deutscher MP. RNase G (CafA protein) and RNase E are both required for the 5′ maturation of 16S ribosomal RNA. EMBO J. 1999;18:2878–2885. doi: 10.1093/emboj/18.10.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta R, Champney WS. 30S ribosomal subunit assembly is a target for inhibition by aminoglycosides in Escherichia coli. Antimicrob Agents Chemother. 2002;46:1546–1549. doi: 10.1128/AAC.46.5.1546-1549.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta R, Champney WS. Neomycin and paromomycin inhibit 30S ribosomal subunit assembly in Staphylococcus aureus. Curr Microbiol. 2003;47:237–243. doi: 10.1007/s00284-002-3945-9. [DOI] [PubMed] [Google Scholar]
- Rio DC, Ares M, Jr, Hannon GJ, Nilsen TW. Cold Spring Harbor Protocols. 2011. RNA: A Laboratory Manual. [Google Scholar]
- Rosen T. Antibiotic resistance: an editorial review with recommendations. J Drugs Dermatol. 2011;10:724–733. [PubMed] [Google Scholar]
- Scheunemann AE, Graham WD, Vendeix FA, Agris PF. Binding of aminoglycoside antibiotics to helix 69 of 23S rRNA. Nucleic Acids Res. 2010;38:3094–3105. doi: 10.1093/nar/gkp1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder R, Waldsich C, Wank H. Modulation of RNA function by aminoglycoside antibiotics. EMBO J. 2000;19:1–9. doi: 10.1093/emboj/19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvers JA, Champney WS. Accumulation and turnover of 23S ribosomal RNA in azithromycin-inhibited ribonuclease mutant strains of Escherichia coli. Arch Microbiol. 2005;184:66–77. doi: 10.1007/s00203-005-0017-0. [DOI] [PubMed] [Google Scholar]
- Song WS, Lee M, Lee K. RNase G participates in processing of the 5′-end of 23S ribosomal RNA. J Microbiol. 2011;49:508–511. doi: 10.1007/s12275-011-1198-7. [DOI] [PubMed] [Google Scholar]
- Sutcliffe JA. Improving on nature: antibiotics that target the ribosome. Curr Opin Microbiol. 2005;8:534–542. doi: 10.1016/j.mib.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Usary J, Champney WS. Erythromycin inhibition of 50S ribosomal subunit formation in Escherichia coli cells. Mol Microbiol. 2001;40:951–962. doi: 10.1046/j.1365-2958.2001.02438.x. [DOI] [PubMed] [Google Scholar]
- Wallis MG, Schroeder R. The binding of antibiotics to RNA. Prog Biophys Mol Biol. 1997;67:141–154. doi: 10.1016/s0079-6107(97)00011-4. [DOI] [PubMed] [Google Scholar]
- Weatherford SC, Rosen L, Gorelic L, Apirion D. Escherichia coli strains with thermolabile ribonuclease II activity. J Biol Chem. 1972;247:5404–5408. [PubMed] [Google Scholar]
- Zinner SH. Antibiotic use: present and future. New Microbiol. 2007;30:321–325. [PubMed] [Google Scholar]