Abstract
Sea ice is believed to be a major factor shaping gene flow for polar marine organisms, but it remains unclear to what extent it represents a true barrier to dispersal for arctic cetaceans. Bowhead whales are highly adapted to polar sea ice and were targeted by commercial whalers throughout Arctic and subarctic seas for at least four centuries, resulting in severe reductions in most areas. Both changing ice conditions and reductions due to whaling may have affected geographic distribution and genetic diversity throughout their range, but little is known about range-wide genetic structure or whether it differed in the past. This study represents the first examination of genetic diversity and differentiation across all five putative stocks, including Baffin Bay-Davis Strait, Hudson Bay-Foxe Basin, Bering-Beaufort-Chukchi, Okhotsk, and Spitsbergen. We also utilized ancient specimens from Prince Regent Inlet (PRI) in the Canadian Arctic and compared them with modern stocks. Results from analysis of molecular variance and demographic simulations are consistent with recent and high gene flow between Atlantic and Pacific stocks in the recent past. Significant genetic differences between ancient and modern populations suggest PRI harbored unique maternal lineages in the past that have been recently lost, possibly due to loss of habitat during the Little Ice Age and/or whaling. Unexpectedly, samples from this location show a closer genetic relationship with modern Pacific stocks than Atlantic, supporting high gene flow between the central Canadian Arctic and Beaufort Sea over the past millennium despite extremely heavy ice cover over much of this period.
Keywords: Ancient DNA, arctic, cetacean, marine mammal, mitochondrial DNA, whaling
Introduction
Sea ice is a dominant feature of the polar environment and is thought to shape patterns of genetic isolation in both marine and terrestrial island species (e.g., Geffen et al. 2007). However, for commercially hunted species such as arctic marine mammals, population genetic structure and diversity also reflect the legacy of whaling and sealing (Roman and Palumbi 2003; Alter et al. 2007; Jackson et al. 2008). Large-scale removals over the last three centuries may have altered pre-whaling genetic differences between populations by disrupting patterns of migration to breeding areas (e.g., Alter et al. 2009) or by eliminating distinct populations from areas and allowing colonization by another stock. Despite these uncertainties, genetic differentiation and stock identity remain important issues for managers and policymakers. Understanding the factors that govern stock structure and gene flow, including the interplay between changing sea-ice conditions and the legacy of whaling, is particularly important for Arctic species that are likely to be affected by climate change, increasing oil and gas development, and shipping, such as the bowhead whale (Balaena mysticetus). Ongoing dramatic declines in sea-ice extent will likely affect genetic exchange rates in bowhead whales as well as other arctic marine mammals such as beluga and walruses (Laidre et al. 2008; O'Corry-Crowe 2008), but evaluating these changes requires the characterization of genetic patterns prior to significant ice loss.
Bowhead whales are the only large baleen whale to occur in the Arctic year-round and are highly adapted to the arctic environment, with the thickest blubber layer of any mammal and the ability to break ice 30–60-cm thick (Marquette 1986). All bowhead whale populations spend summers in the Arctic, but overwinter in subarctic seas, inhabiting polynyas and the marginal ice zone, following seasonally advancing and retreating ice edges (Moore and Reeves 1993). Climatic variations during the Holocene were dramatic across some parts of the species' range, and changes in sea-ice cover over the past several millennia may have shaped gene flow between stocks. In addition, genetic patterns may have been affected by whaling. This species was targeted heavily by commercial whalers throughout Arctic and subarctic seas beginning in Labrador around 1540 and continuing into the early 20th century (Ross 1993), resulting in moderate to severe reductions in population abundance across its range (Woodby and Botkin 1993). Although these reductions likely affected both the amount and geographic distribution of genetic diversity in bowhead whales, relatively little is known about range-wide genetic structure today or how it may have differed before large-scale commercial whaling.
Bowhead whales have been divided into management stocks largely based on geographic discontinuities, including sea ice perceived as a barrier to movement (Moore and Reeves 1993). Until recently, five stocks of bowhead whale have been recognized by the IWC for management purposes: (1) Hudson Bay-Foxe Basin (“HBFB”); (2) _Baffin Bay-Davis Strait (“BBDS”); (3) Beaufort, Chukchi, and Bering Seas (“BCB”); (4) the Okhotsk Sea (“Okhotsk”); and (5) the area of Spitsbergen and the Barents Sea (“Spitsbergen”) (Fig. 1a). Two separate stocks in Canada and Greenland (HBFB and BBDS) were hypothesized based on the assumption that Fury and Hecla Strait represents a geographic barrier to bowhead whales. Persistent ice plugs throughout the Northwest Passages, which are believed to have been stable from roughly 3 kya until the last several years (Vare et al. 2009), are also thought to prevent migration between BCB and Atlantic stocks. However, recent satellite tracking data show that whales occupying Foxe Basin move through Fury and Hecla Strait into Prince Regent Inlet (PRI), an area that has been traditionally classified as belonging to BBDS (Heide-Jørgensen et al. 2006). This evidence, in combination with abundance data on calves and adults in various areas, suggests that bowhead whales in eastern Canada and Greenland may represent one population (“Canada-Greenland”), rather than two (COSEWIC 2009). Likewise, in 2010, satellite telemetry data demonstrated overlap in movement between a BCB individual and a Canada-Greenland individual in Viscount Melville Sound, which was attributed to the recent and dramatic loss of sea ice in the Canadian Arctic (Heide-Jørgensen et al. 2011).
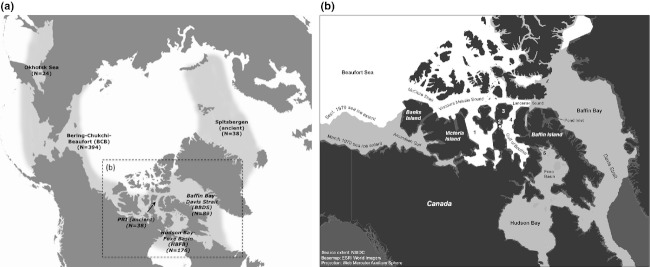
Figure 1.
(a) The full range of bowhead whales across the Arctic (lightly shaded area; Laidre et al. 2008) with sample sizes from region. Italicized names represent data collected in this study. Area inside dotted line shows the location of (b) detailed map of the Canadian Arctic. 1 = McClintock Sound, 2 = Qariaraqyak (PaJS-2) (archeological site from which “PRI” samples were excavated), 3 = Somerset Island, 4 = Prince Regent Inlet, 5 = Fury and Hecla Strait. Also shown are maximum (March – lightest gray) and minimum (September – white) sea-ice extent for 1979 (the earliest year data are available from NSIDC).
Despite these observations, questions remain about range-wide population structure among bowhead whales. In particular, the degree and timing of genetic exchange between Atlantic (HBFB/BBDS, Spitsbergen) and Pacific (BCB, Okhotsk Sea) populations has yet to be fully elucidated. Interchange between the Atlantic and Pacific Oceans through the central Canadian Arctic was likely possible during the warmer conditions of the early and mid-Holocene, but climatic reconstructions indicate a rapid increase in ice cover around 3 kya that excluded bowhead whales from the central channels of the archipelago (Dyke et al. 1996; Vare et al. 2009). A short period of lower ice cover may have occurred just before the start of the Little Ice Age in the early 15th century, followed by an increase in ice cover in the last 400 years (Vare et al. 2009). Although this climatic history indicates the last significant connection through the Canadian Arctic occurred >3 kya, the ability of bowhead whales to navigate cracks and leads in extremely dense pack ice (>90% cover) suggests the possibility of more recent exchange. A recent genetic study compared microsatellite data from western Arctic whales with whales from a location in the eastern Canadian Arctic and found low but significant differentiation, suggesting a small degree of genetic mixing (Givens et al. 2010). Another study compared mitochondrial haplotypes from late Pleistocene to late Holocene Spitsbergen samples with those from the modern BCB population and found a similar result (low but significant differentiation), although the difference disappeared when only the most recent Spitsbergen samples were used (Borge et al. 2007). While both these analyses suggest that there has been some mixing between the Atlantic and Pacific, no previous study has attempted to estimate the magnitude or timing of the most recent exchange, or has incorporated whaling history into genetic data analysis. Population bottlenecks due to whaling can reduce haplotype diversity and can affect haplotype frequency distributions, leading to apparent spatial structure between bottlenecked populations (Alter et al. 2012). Thus, accounting for whaling history is critical for accurate analysis of population structure in heavily exploited species.
Historical and ancient samples represent a valuable but underutilized source of information about marine mammal responses to both climate change and whaling over long periods. Genetic data from such samples have been used to test hypotheses about population response to climate shifts and hunting in many terrestrial species (e.g., Shapiro et al. 2004; Chan et al. 2005; Dalen et al. 2007), but have been less frequently utilized in marine systems (but see, e.g., de Bruyn et al. 2009). For many cetacean species depleted by whaling and now recovering, data from historical and ancient DNA (herein both referred to as “ancient DNA”) can provide an important point of comparison for determining how stock identity and genetic diversity differed before large-scale commercial whaling began.
The extensive archeological and stranded remains of bowhead whales across the Arctic provide an opportunity to better understand the factors shaping genetic connectivity in bowhead populations. In this study, we used ancient and modern bowhead control region sequences to compare genetic diversity and population differentiation between: (1) all putative modern management stocks, including sequences from Spitsbergen samples aged 30–3,000 years old (Borge et al. 2007); and (2) modern stocks and ancient samples from PRI, located in the central Canadian Arctic. For the latter comparison, we collected data from the mitochondrial D-loop from bowhead specimens from 500 to 800 years old Thule Inuit house ruins at the east coast of Somerset Island (western side of PRI), and compared them with sequences from the five putative stocks (HBFB, BBDS, BCB, Okhotsk, and Spitsbergen). PRI is situated in the modern-day range of BBDS (Fig. 1b), and ancient samples from this locale are ideal for exploring gene flow between the Pacific and Atlantic populations over the last millennium. We used modern and ancient samples to test the following hypotheses, based on the expectation that persistent ice cover is a barrier to genetic exchange: (1) significant differentiation between Atlantic (HBFB, BBDS, Spitsbergen) and Pacific (BCB, Okhotsk) populations; (2) ancient PRI whales are most closely related to the modern BBDS population; and (3) the last genetic exchange between the Pacific and Atlantic sides of the Canadian Arctic occurred during the mid-Holocene (roughly 3 kya). This approach builds upon previous studies of bowhead whale genetic in two respects: first, we utilized samples from across the entire circumpolar range of bowhead whales and include samples from a late Holocene time period; and second, we used demographic modeling in addition to traditional population structure analyses to test the hypotheses above and to incorporate the impacts of whaling on genetic structure.
Materials and Methods
Ancient sample collection
Samples of preserved baleen and bone were collected from archeological sites on Somerset Island (western side of PRI) as described in Whitridge (2002). Qariaraqyuk (with the Canadian archeological site designation PaJs-2) is a Classic Thule winter village located on the southeastern tip of Somerset Island, (Savelle and McCartney 1994; Whitridge 1999), and was occupied from about AD 1200–1500. It was likely a major winter residential locus for groups who whaled from nearby PaJs-4 in late summer/early fall (Savelle and Wenzel 2003). The site consists of a row of at least 57 sod winter houses, making it the largest precontact winter village in the Canadian Arctic (Whitridge 2002). Six of the houses were excavated in 1993–1994. The samples included in the present analysis consists of specimens of artifactual baleen, including artifacts (vessels, cordage, toys, etc.), refuse from artifact manufacture, and knotted strands that likely represent the structural lashing from whale bone house frameworks. Calibrated radiocarbon dates on heather (Cassiope tetragona), caribou bone (Rangifer tarandus), and willow (Salix sp.) from the house assemblages of which these samples are a part bracket the occupation of the features between 500 and 800 ybp.
The location of this site on the western side of PRI/Gulf of Boothia is on the summering ground of what would today be considered part of the BBDS stock. However, satellite tracking data also indicate that the area is also used by animals from Foxe Basin (Greenland Institute of Natural Resources, unpublished data). Bowhead whales only visit PRI, which is characterized by heavy ice cover, for about 2 months per year. Solid fast ice coverage in the Canadian Arctic Archipelago during fall, winter, and spring forces all cetaceans to move out into open water or to areas with mobile pack ice (Moore and Reeves 1993). This forces animals into relatively small pockets of inhabitable areas in eastern Hudson Strait, West Greenland and recurrent polynias on the east coast of Baffin Island and in Lancaster Sound.
Ancient DNA methods and authentication
All extractions were performed in dedicated ancient DNA facilities at the American Museum of Natural History. No modern whale DNA had been extracted and no amplifications had taken place within this facility. All samples were stored in separate airtight plastic bags until use to prevent cross-contamination. Samples were pretreated to remove potential surface contaminants as described in Rosenbaum et al. (1997). Briefly, all materials used were UV-treated prior to use and bone surfaces were cleaned with kimwipes soaked in ethanol, 10% Clorox, and finally RNAase free H20. Bone surfaces were removed using a clean drill bit treated with HCl and UV light.
Subsamples of bone were obtained using a sterilized drill bit to drill a small hole (<0.5-cm diameter, 3–4-mm deep) to generate ∼0.1g of bone powder. Bone powder was then treated to remove any remaining contaminants by soaking in 10% Clorox for 20 min followed by a rinse in sterile H2O. Baleen was subsampled following the protocol of Rosenbaum et al. (1997). Samples were incubated at 37°C for several hours to overnight with 1.5-mL 0.5M EDTA pH 8.5 in order to decalcify bone and remove inhibitors from humic acid. Following incubation with EDTA, samples were centrifuged at 10,000 rpm for 5 min and supernatant was removed. We performed an additional rinse with 1-mL H20 in order to reduce the EDTA concentration. To extract DNA from the bone pellet, samples were incubated with 0.5-mL Lifton's buffer and 35 uL of 20 mg/mL proteinase K at 56°C for 50 h. Extraction was completed using standard phenol/chloroform purification and ethanol precipitation procedures (Sambrook et al. 1989).
Amplification conditions are given in Rosenbaum et al. (1997). All amplifications were set up in the ancient DNA facility, but thermal cycling was carried out in a separate post-extraction lab. A series of primer pairs that generate overlapping fragments of the mitochondrial D-loop were used, including primers Dlp 1.5 and Dlp 5 that amplify the majority of the variable sites in the cetacean D-loop (Arnason et al. 1993; Baker et al. 1993) and six additional primers detailed in Rosenbaum et al. (1997) that amplify 100–200-bp regions. Three bowhead-specific primers were developed for sequencing (Myst3.3A, Bm96f, and Bm218f; available from authors upon request).
Successful amplification products were sequenced in both directions using fluorescence-labeled dideoxy terminators on an ABI 3700 High-throughput Capillary DNA Sequencer (Applied Biosystems). To authenticate sequences, a subset (∼15%) of samples with unique haplotypes were re-extracted, amplified, and sequenced in both directions in an entirely separate facility (a dedicated ancient DNA facility at Yale University, Department of Ecology and Evolutionary Biology), and scored blind relative to the original sequences.
Modern sequences
Modern D-loop data were generated from biopsy samples collected from areas of Northern Canada and West Greenland including Pelly Bay, Repulse Bay, and Igloolik (previously included in the HBFB stock, N = 176) and Disko Bay, West Greenland and Pangnirtung, Canada (previously included in the BBDS stock, N = 89) collected between 1997 and 2006. Total cellular DNA was extracted from bowhead skin samples using different techniques. Earlier samples (before 2000) were extracted using the methods described in Maiers et al. (1996) with some modifications. The skin tissue was incubated at 37°C for an extended period and had several additions of proteinase K (20 mg/mL) to digest the tissue to the point where it was suitable for extraction. Once this process was complete, in most samples, sufficient quantities of DNA were recovered for analyses. More recent samples (after 2000) were extracted using commercial DNA tissue extraction kits (DNeasy, Qiagen).
A portion of the mitochondrial DNA D-loop was amplified using primers Dlp 1.5 and Dlp 5 that amplify the majority of variable sites in cetaceans (Arnason et al. 1993; Rosenbaum et al. 2002). Automated DNA sequencing of the PCR products was performed using ABI genetic analyzers (Prism 377, 3100, 3130XL) and the related fluorescent dye terminator chemistry. Samples were also genetically profiled at 21 microsatellite loci (using methods described in Givens et al. 2010). These data were used to detect the occurrence of individuals represented multiple times in the dataset due to recapture of animals. Probability of Identity was assessed using the program GeneCap (Wilberg and Dreher 2004). GeneCap is designed to identify matches, but will also flag samples that match at all alleles, but one or two (which may represent true replicates that were undetected due to genotyping errors).
In addition to generating ancient and modern sequences from the Canadian Arctic, we also utilized previously collected D-loop data from the following populations of bowhead whales: (1) BCB Seas (N = 394) (LeDuc et al. 2009); (2) Okhotsk Sea (N = 24) (LeDuc et al. 2009); and (3) Spitsbergen sequences from 30 to 3,000 years in age (N = 38) (Borge et al. 2007).
Genetic analysis
Sequences were cleaned and edited using Sequencher v. 4.0 (GeneCodes), and species identity was determined using the NCBI database (BLAST), as well as a diagnostic character approach for species delimitation (Rosenbaum et al. 2000). Haplotypes from ancient samples were compared with sequences from independent extractions/amplifications to assure sequence authenticity. We examined haplotype frequency distributions within and among ancient and modern populations. Genetic diversity was compared among populations using several measures including haplotype diversity (Hd), nucleotide diversity (π), and sequence diversity (θ[S]), generated using DnaSP v.4.0 (Rozas et al. 2003). Partitioning of genetic variation among sample sets was assessed using analysis of molecular variance (AMOVA [analysis of molecular variance]; Excoffier et al. 1992) generated in ARLEQUIN v2.0 (Schneider et al. 2000). Because of the high substitution rate of mtDNA and incorporation of ancient samples, we expect that both genetic drift and mutation are potentially influencing genetic differentiation, and therefore the ΦST statistic is a more appropriate measure of differentiation than frequency-based FST (which does not take into account molecular distances between haplotypes) (Excoffier 2003; Holsinger and Weir 2009). We assessed a priori geographic stratifications based on four (combining HBFB and BBDS into “Canada-Greenland”) or five putative stocks, and also assessed all Pacific (BCB, Okhotsk) versus all Atlantic (HBFB, BBDS, Spitsbergen) populations. Statistical significance of ΦST values in pairwise population comparisons was determined using 10,000 random permutations of the data matrix variables and a Jukes-Cantor evolutionary model (Jukes and Cantor 1969). In addition, we generated an optimal minimum spanning network using Arlequin v2.0 in order to assess the geographic distribution of haplotypes. Optimal minimum spanning networks utilize haplotype frequency data to determine the most parsimonious relationships between haplotypes.
Demographic simulations
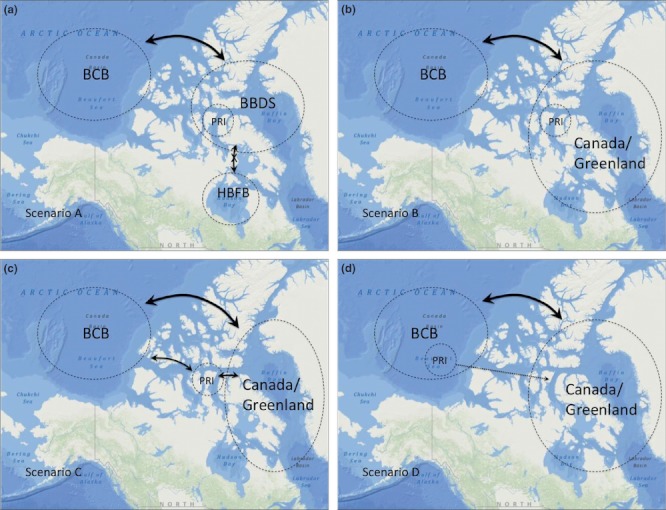
We used a demographic simulation approach to explore whether geographic barriers and whaling may have influenced observed patterns of differentiation. Specifically, we used simulations to determine the expected genetic differentiation between sample sets under scenarios of particular climatic (ice cover) and whaling histories. We used the program BayesSSC (Bayesian Serial SIMCOAL, Anderson et al. 2005) to model demographic scenarios focused on the four sample sets collected in or near the Canadian Arctic (both Atlantic and Pacific): PRI, BBDS, HBFB, and BCB. Other approaches such as IMa (Hey and Nielsen 2007) were considered, but not deemed appropriate for this analysis because of the need to specify samples collected at different time points. The Okhotsk and Spitsbergen populations were not included because these populations are geographically removed from the Canadian Arctic and considerably less is known regarding demographic and whaling histories in these locations. We modeled a total of 36 demographic scenarios between the Atlantic and Pacific. Within each of three basic scenarios (described below), we modeled four subscenarios describing different possibilities for between-stock structure, and three migration rates (m = 0.1, m = 0.01, m = 0.001). In brief, we modeled the following scenarios (see Appendix, Figure A1, Table 3) and residual size as given by Woodby and Botkin (1993), HBFB is reduced to 1–68% of its initial size (Woodby and Botkin 1993), and the BBDS population is reduced to 1–29% of its initial size (Woodby and Botkin 1993). Generation time was assumed to be 52 years in the simulations (Taylor et al. 2007). Simulations were performed using female effective population sizes, which was calculated from census population sizes assuming a 1:1 male:female ratio, 1.5:1 ratio of all individuals to all adults, and an Ne/N ratio of 0.5 (Roman and Palumbi 2003). We used a uniform prior on mutation rate ranging from 2% per my, the fossil-calibrated phylogenetic rate (Roman and Palumbi 2003), to 6.3% per my, which represents the highest rate for baleen whale control region calculated from calibrations using cytochrome-b (Alter and Palumbi 2009), and a mutation model (HKY+G) based on results from MODELTEST (Posada and Crandall 1998). Using these parameters, 10,000 independent genetic datasets were simulated per scenario. We determined whether the simulation results were compatible with the observed genetic difference between ancient samples from PRI and modern samples from BBDS and BCB by sampling the simulated datasets to obtain samples of the same size and age as our empirical datasets. Observed ΦST values between BCB versus BBDS, BCB versus Canada-Greenland, BCB versus PRI, PRI versus BBDS, and PRI versus Canada-Greenland were compared to the distribution of ΦST values between the corresponding simulated datasets. If any of the observed pairwise ΦST values fell outside of the 95% highest posterior density interval of the distribution from simulated datasets, the corresponding demographic scenario was rejected.
Results
Genetic diversity
We obtained sequence data for 38 ancient samples from PRI and 265 modern samples from HBFB and BBDS (Genbank Accession numbers are provided in the Appendix, Table A2). Once aligned with sequences from BCB, Okhotsk, and Spitsbergen, the complete dataset comprised 370 bp of mitochondrial D-loop for a total of 759 samples (Table 1). Probability of Identity (Wilberg and Dreher 2004) was tested for all HBFB and BBDS samples and was found to be sufficient to permit discrimination of individuals (PID HW = 8.1 × 10−31; PID SIB = 1.9 × 10−10). Six duplicated sequences were found (4 in HBFB and 2 in BBDS), and removed from the mtDNA sequence set. For ancient samples, no sequence differences were found between original sequences and samples that were re-extracted and sequenced in an independent facility. Haplotype, nucleotide, and sequence diversity were high for all sample sets examined, with the exception of Okhotsk Sea. Haplotype diversity was significantly higher in PRI (95% confidence intervals: 0.860−0.977) and BCB (0.892−0.935) compared with Canada-Greenland (0.785−0.848) and Okhotsk (0.484−0.775), based on coalescent analyses performed in DNAsp (Rozas et al. 2003).
Table 1.
Number of samples for each sample set (N) and diversity values across populations
| N | S | H | Hd | U | π | θ(S) | |
|---|---|---|---|---|---|---|---|
| PRI (Ancient) | 38 | 26 | 20 | 0.92 | 7 | 0.014 | 7.62 |
| Canada-Greenland | 265 | 26 | 23 | 0.8 | 6 | 0.007 | 5.2 |
| HBFB | 176 | 25 | 20 | 0.81 | 4 | 0.007 | 5.4 |
| BBDS | 89 | 22 | 15 | 0.79 | 2 | 0.008 | 5.34 |
| BCB | 394 | 36 | 54 | 0.9 | 15 | 0.01 | 6.41 |
| Okhotsk | 24 | 24 | 4 | 0.63 | 0 | 0.007 | 3.21 |
| Spitsbergen (Ancient) | 38 | 19 | 17 | 0.85 | 8 | 0.008 | 5.24 |
S, number of segregating sites; H, number of haplotypes; Hd, haplotype diversity; U, unique haplotypes; π, nucleotide diversity; θ(S) based on segregating sites. Canada-Greenland, HBFB and BBDS combined.
Across all samples, a total of 76 haplotypes were observed for an overall haplotype diversity of 0.87. Private haplotypes were observed in all populations with the exception of Okhotsk, and the majority (67%) of these private haplotypes were singletons in the dataset. The most frequently observed haplotype was the same for all sample sets, and the second most frequently observed haplotype was the same for five of the six sample sets (with Okhotsk being the exception).
Population differentiation
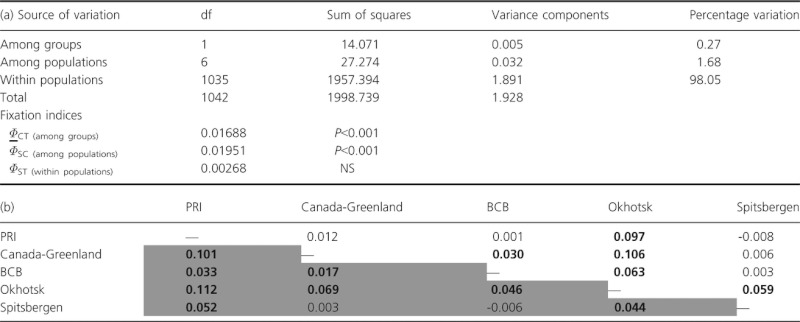
Among-population variation accounted for 1.68% of total variation, compared with 0.27% for the between-group (Atlantic and Pacific) comparison (Table 2a). No differentiation was observed between HBFB and BBDS (ΦST = −0.002, P > 0.1) (See Appendix, Table A3), and these two populations were subsequently grouped together as Canada-Greenland. However, we observed significant ΦST values for most other pairwise population comparisons (Table 2b). The analysis revealed a small but significant amount of differentiation between BCB and Canada-Greenland, and between BCB and PRI. Larger ΦST values were estimated between Okhotsk and all other sample sets, and between PRI and Canada-Greenland. All significant ΦST values remained significant after a False Discovery Rate correction for multiple comparisons (Benjamini and Hochberg 1995) was applied.
Table 2.
(a) Hierarchical AMOVA results, including variation between two groups of putative populations (Pacific and Atlantic), among putative populations, and within populations. (b) Pairwise genetic distances between sample sets: pairwise ΦST values are given below the diagonal and frequency-based FST values are given above. All bold values are significant after a False Discovery Rate correction
 |
The optimal minimum spanning network (Fig. 2) shows the geographic distribution of haplotypes as well as their frequencies, demonstrating a weak geographic signal overall. The most frequently observed haplotypes are distributed widely throughout the Arctic. A large number of singletons are observed for BCB and the two ancient sample sets (PRI and Spitsbergen).
Figure 2.
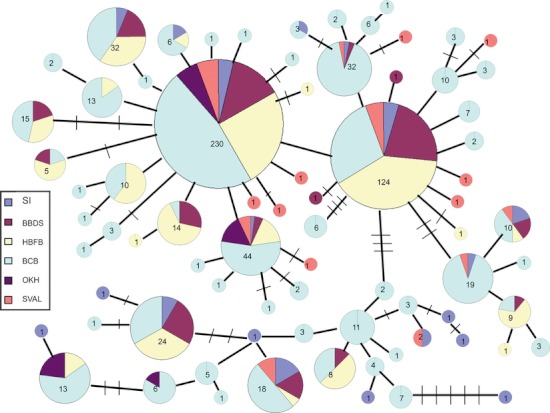
Minimum spanning network for bowhead whale haplotypes created using TCS. Size of circle is proportional to number of samples; hatchmarks represent additional segregating sites.
Demographic simulations
As no genetic differentiation was observed between BBDS and HBFB (see above), we grouped these two stocks into a single population (Canada-Greenland) for the majority of scenarios. The distribution of ΦST values for three pairwise comparisons across 10,000 simulations (m = 0.1) is shown in Figure 3. We compared these simulated distributions with empirically observed ΦST values (Table 2). For the comparison between BCB and either BBDS alone (Scenario A) or Canada-Greenland, the observed value of ΦST falls within the 95% highest posterior density (HPD) interval for 3A, 3B, 3C, and 3D at m = 0.1. For the BCB-PRI comparison, the observed value of ΦST falls within the 95% HPD interval for all scenarios that use m = 0.1 with the exception of 2C. For the comparison between PRI and BBDS alone or Canada-Greenland, the observed ΦST falls within the 95% HPD interval for 1A, 2A, 2B, 1C, 3C, and 3D at m = 0.1. Based on these results, the following scenarios can be excluded (e.g., the range of ΦST values does not include the observed value): 1A, 2A, 3A, 1B, 2B, 3B, 1C, 2C, 1D, and 2D. In other words, for m = 0.1, the only scenarios that are not excluded are 3C (contemporary gene flow with PRI as a separate population) and 3D (contemporary gene flow with PRI ancestral to BCB). For lower migration rates (m = 0.01 and m = 0.001), all scenarios are excluded (see Appendix, Table A4).
Figure 3.
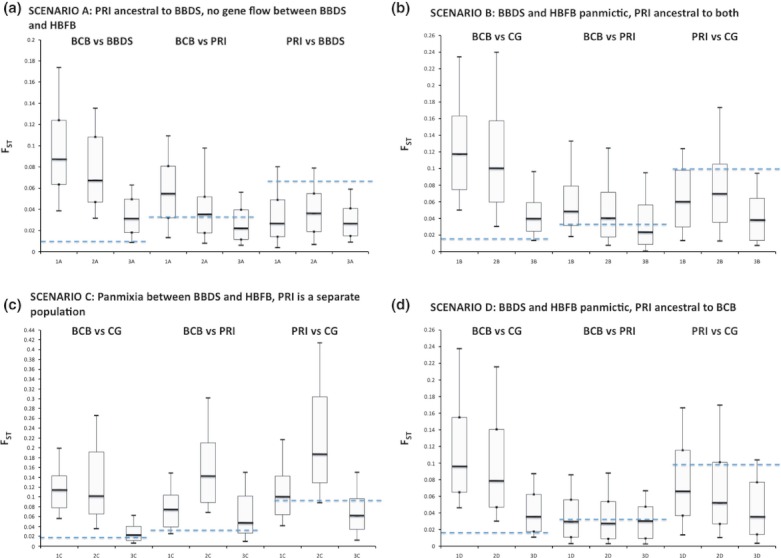
Simulated and empirical pairwise ΦST values between putative populations for m = 0.1. Vertical lines show 95% highest posterior density (HPD) intervals for simulated ΦST values; top and bottom of box indicate 75% HPD intervals and thick horizontal lines show median values for simulations. Dotted horizontal lines show empirically observed pairwise ΦST values. CG = Canada-Greenland (BBDS and HBFB combined). (a) Scenario A; (b) Scenario B; (c) Scenario C; (d) Scenario D (see text for details).
Discussion
Our analysis of the five putative populations of bowhead whales, including data from ancient samples from two locations (PRI and Spitsbergen), represents the first genetic comparison across the entire range of the species and illustrates the utility of ancient specimens in reconstructing the history of genetic exchange in exploited marine mammals. The results indicate that Arctic sea ice has not acted as a strong barrier to migration between the Atlantic and Pacific over the late Holocene as previously assumed, and that genetic diversity has been lost from eastern Canada in the period between ∼500 ybp and the present.
Genetic diversity
Measures of modern diversity are similar to those reported in earlier studies of population-level diversity in the control region of bowhead whales (e.g., LeDuc et al. 2005, 2009; Borge et al. 2007). However, in contrast to Borge et al. (2007)'s finding that genetic diversity was similar between ancient whales from Spitsbergen and modern BCB population, genetic diversity (including Hd, π and θ(S)) in ancient samples from PRI is significantly higher compared with modern Atlantic populations, providing a measure of lost haplotypic diversity over the last millennium (Table 1). Diversity in modern sample sets is broadly consistent with relative sizes of populations: BCB shows the greatest amount of diversity, and Okhotsk the least. These results agree with earlier studies that observed low diversity (7 haplotypes across 67 individuals) in the Sea of Okhotsk (MacLean 2002; LeDuc et al. 2009).
The high number of mitochondrial haplotypes observed across bowhead whale populations may not be atypical for baleen whales. Jackson et al. (2008) observed 38 haplotypes in the southern right whale (Eubalaena australis) population, and estimated pre-exploitation richness at 100–150 haplotypes. We observed 62 haplotypes in the modern dataset alone, without any correction for undersampling or sequence length, which would increase the number of estimated haplotypes.
The relatively high number of unique or population-specific haplotypes among PRI samples (roughly one-fifth of PRI haplotypes are unique to that sample set), as well as the high divergence of several of these haplotypes compared with modern samples (Fig. 2), suggests that lineage diversity in the Canadian Arctic was greater as recently as 500 ybp. This observation represents the first empirical demonstration of lost haplotype diversity in bowhead whales over the last several centuries. Potential causes for this loss, including whaling, are explored below.
Genetic differentiation and timing of exchange between Atlantic and Pacific
In contrast with previous hypotheses about connectivity between BCB and Atlantic populations based on subfossils and sea-ice reconstructions (e.g., Moore and Reeves 1993; Dyke et al. 1996; Vare et al. 2009), AMOVA and simulation results are most consistent with contemporary and high gene flow between the two ocean basins. The degree of differentiation estimated between BCB and Atlantic populations is in agreement with the slight but significant allelic differentiation (Fst = 0.009) between Barrow, Alaska (n = 231) and Igloolik, Canada (n = 37) estimated using 21 microsatellite loci (Givens et al. 2010). These results contrast markedly with estimated divergence times between North Atlantic and North Pacific populations in other whale species (fin whales, 1.05–2.70 Mya (Berube et al. 1998); common minke whales, ∼1.5 Mya (Pastene et al. 2007); humpback whales ∼2–3 Mya (Baker et al. 1993; right whales, >3.5 Mya (Rosenbaum et al. 2000)), and underscore the adaptation of bowhead whales to arctic habitat relative to other baleen whales.
Within ocean basins, we did not observe any consistent relationship between ice barriers and population differentiation. Although BBDS and HBFB have previously been considered separate management stocks based on the assumption that persistent ice in Fury and Hecla Strait would prevent movement between them, we found no significant differentiation between them. These results are consistent with observed movement patterns from satellite telemetry data, but need to be tested with additional markers, which was not possible in the scope of this study.
Ancient samples: relationship of PRI and Spitsbergen to modern populations
Severe population reductions due to whaling may have altered signals of genetic diversity and connectivity between bowhead populations, making it difficult to reconstruct pre-whaling patterns. Data from ancient PRI samples can provide insights into how population structure differed in the past. PRI is located in the summering ground of the modern Canada-Greenland population, but has been connected to the Beaufort Sea (BCB population) via the Northwest Passages intermittently over the Holocene. The data presented here show that the genetic difference between PRI samples from 500 to 800 ybp and modern Canada-Greenland samples is unexpectedly larger than the difference between these ancient samples and modern BCB samples.
If PRI whales were the forbearers of modern Canada-Greenland, as geography would suggest, what could cause such strong genetic differentiation between them? One possibility is that the population bottleneck during the height of whaling caused a dramatic reshuffling of haplotype frequencies between the two sampling time points. However, simulations show that even an extreme (∼71–99%) reduction in the Canada-Greenland population does not explain the degree of genetic differentiation between the two. An alternative explanation, which is supported by simulation results (Scenario C) is that PRI samples represent a genetically unique population or set of maternal lineages based around site fidelity to a summering ground on the western side of the Inlet. Recent observations suggest that PRI is a summering area for female whales with calves and juveniles that move from Foxe Basin in the early summer (Dueck and Ferguson 2008), as well as many subadult and adult whales that spend the spring in Baffin Bay and Davis Strait. Intergenerational fidelity to important habitat has been demonstrated in gray whales (Alter et al. 2009), humpback whales (Baker et al. 1994; Palsbøll et al. 1995), right whales (Schaeff et al. 1993), beluga whales (Brown Gladden et al. 1997), and sperm whales (Lyrholm et al. 1999), and other authors have speculated that fidelity to calving grounds in bowhead whales may also be “behaviorally rigid” (Dueck and Ferguson 2008). McCartney (1979) estimated that whaling sites on the western side of PRI contain on the order of 40% of all archeological whale bone across the Canadian Arctic, suggesting that this region was also important as a summering ground in the past, and whalers referred to PRI as a “nursery ground” (Finley 1990).
While median simulated ΦST values are consistent with PRI as an independent population (Scenario 3C), simulation results support high exchange between PRI and BCB, and did not exclude full panmixia between them (Scenario 3D). These findings support the idea that exchange across the Canadian Arctic was occurring during the late Holocene, prior to the recent decrease in sea ice that has permitted overlap in range between Atlantic and Pacific populations (Heide-Jørgensen et al. 2011).
An important consideration in evaluating these results is that we were unable to confirm that each ancient sample represents a unique individual because of difficulties genotyping ancient samples using microsatellites. The high number of unique haplotypes among PRI individuals (Hd = 0.92) and the large number of bowhead individuals recovered from the PaJs-2 locality (a minimum of 261 animals site-wide, not including buried bone, which comprised a significant proportion of remains at the site, minimum number of individuals based on same-side proximal mandible counts (Whitridge 2002)) suggest that most, if not all, of the samples represent different individuals. However, any individuals represented twice or more could spuriously reduce the genetic diversity estimate for PRI, and could result in different estimates of genetic differentiation between PRI and other populations. We tested the impact of 1–5 repeated individuals by serially removing repeated sequences from the dataset and recalculating ΦST values (Appendix, Table A5). Values changed by only −0.014% on average for one duplicated individual and by + 0.036% for five duplicated individuals, suggesting that the impact of duplication on the analysis at this level should be low.
The strong differentiation between PRI and modern populations based on AMOVA results contrasts with overall lack of differentiation between the other set of ancient samples, Spitsbergen, with modern populations (with the exception of the comparison with Okhotsk). No differentiation was observed between BCB and Spitsbergen, corroborating earlier findings by Borge et al. (2007) based on fewer BCB samples compared with only the 25 youngest Spitsbergen samples. This, in combination with significant differentiation between BCB versus PRI and BCB versus Canada-Greenland, presents the possibility that gene flow between the Atlantic and Pacific may have occurred clockwise or westward through the East Siberian and Laptev Seas, rather than (or in addition to) through the Canadian Arctic. These results also suggest a contrast between Spitsbergen as a relatively large and demographically open population spread across a wide geographic area (perhaps similar to BCB today), versus PRI, which is located in a geographic cul-de-sac and which does not reflect range-wide genetic diversity to the same degree as Spitsbergen. However, all comparisons using Spitsbergen samples must be interpreted with caution, as noted by Borge et al. (2007), as the temporal spread of sample ages could potentially introduce spurious genetic diversity in the context of a spatial analysis.
The role of climate changes, whaling, and ice entrapment in the extirpation of ancient PRI lineages
What might have caused the disappearance of PRI haplotypes from the modern populations of bowhead whales? Both changing climate at the end of the Holocene and whaling (and perhaps an interaction between the two) may have played a role. The most obvious and significant source of mortality for bowhead whales between 500 ybp and the present was commercial whaling, which eliminated a large part of the population (Woodby and Botkin 1993). By far, the largest proportion of the bowhead catches were taken in Davis Strait and Baffin Bay, and whalers were only capable of entering the dense sea ice of northern PRI late in the whaling period (Ross 1993). No whaling records exist to suggest that commercial whalers visited southern PRI. However, commercial whaling in the Central Canadian Arctic coincided with the end of the Little Ice Age (100–400 ybp), a period of much cooler temperatures that marked the sudden disappearance of Thule Inuit settlements from PRI (Savelle and McCartney 1994). This period of climatic cooling likely resulted in an increase in summer sea ice in PRI and the partial or complete loss of this habitat as an important summering ground for bowheads. Animals that used southern PRI would have been forced to use other, potentially suboptimal or already inhabited, summering areas. These new summering areas, which were likely farther east, may have brought more whales into contact with commercial whaling. Moore and Reeves (1993) note the possibility that during the Little Ice Age, “the bowhead population probably experienced restricted access to the summer feeding range while at the same time being intensively exploited by commercial whalers.” The “west water” fishery targeting the areas around Pond Inlet, Lancaster Sound, PRI and northern Gulf of Boothia began in 1827 (Ross 1993), and the next few decades represent the highest removal rates by far across the entire history of bowhead whaling in eastern Canada/western Greenland (Higdon 2010). Whalers documented how in heavy ice years, land-floe ice blocked the entrances to Pond Inlet and Lancaster Sound and whales were unable to migrate further west (summarized in Higdon 2008). Whalers took large numbers of whales during these “closed seasons” when whales would concentrate along the land floe. Thus, the interplay of climate fluctuations and whaling may have played a role in the loss of genetic diversity in the Canadian Arctic.
Two other explanations for the disappearance of PRI haplotypes are possible (and are not mutually exclusive with each other or the hypothesis of climate change/commercial whaling): (1) Thule Inuit whaling; and (2) ice entrapment. Extensive archeological surveys of Thule sites resulted in minimum estimates of 1,830–2,745 whales from Somerset Island alone (McCartney 1979; summarized in Stoker and Krupnik 1993). These numbers represent catches over a period of 300–400 years, so annual takes would likely have been on the order of 10 or fewer whales. Stoker and Krupnik (1993) estimated eight whales taken per year for Somerset Island. However, Higdon (2010) notes that these numbers may be severe underestimates, as buried bones were not included in these original analyses. Sea-ice entrapments represent another potential source of mass mortality for bowhead whales. The inner Canadian Arctic Archipelago including PRI has severe ice conditions in late autumn and whales that depart late can become entrapped and die. There was a report of a bowhead whale entrapped in Lancaster Sound in 1999 and of narwhals entrapped in the southern part of PRI in 1979 (Heide-Jorgensen et al. 2002). Of all areas where bowhead whales concentrate in large numbers, PRI is undoubtedly the most dangerous area where large-scale entrapments could occur. None of the other areas are so far from open water, mobile pack ice, or recurrent polynias necessary for survival in extreme ice. One very large event, or perhaps several large-scale entrapments over a period of time, could potentially explain the historical loss of genetic diversity in PRI. These possibilities demonstrate the broad range of ecological and anthropogenic forces that could have impacted the distribution of genetic diversity in bowhead whales in the Canadian Arctic and beyond.
Conclusions
These results highlight the complex interplay of factors – including climate history, behavioral ecology, and past exploitation – that shape population genetic patterns in polar marine mammals. In contrast with our initial hypothesis that Pacific and Atlantic populations were last connected during the mid-Holocene (e.g., Dyke et al. 1996), the genetic data presented here instead indicate recent and high gene flow between these areas. At the Holarctic scale, these results suggest that the presence of persistent sea ice does not appear to be a good predictor of genetic exchange in bowhead whales. This finding underscores earlier observations that apparent geographic barriers are not always accurate indicators of population structure in cetaceans (Hoelzel 1994, 1998).
We have attempted to infer a complex demographic history from limited genetic data, and additional genetic information and ancient samples from throughout the region will be needed to further test these hypotheses. Nevertheless, the unique set of maternal lineages found in the central Canadian Arctic (PRI) and the unexpected relationships between this area and other modern populations demonstrate the value of ancient samples in better understanding the role of climatic history and human hunting in shaping genetic diversity and structure in arctic species. Additional ancient samples from across the range of the bowhead whale and integrated data from SNPs and microsatellites would advance analyses beyond the rough estimates possible using mitochondrial data alone. Such studies would allow an unprecedented evaluation of both natural and anthropogenic impacts on genetic variability in a key indicator species in the rapidly changing Arctic.
Acknowledgments
We gratefully acknowledge the assistance of Susan E. Cosens, Denise Tenkula, and Blair Dunn (DFO); HTOs of Pelly Bay, Igloolik, Repulse Bay, Pangnirtung; SWFSC. The sampling of whales in Greenland was funded by the Greenland Institute of Natural Resources, the Commission for Scientific Research in Greenland, the Danish Cooperation for the Environment in the Arctic under the Danish Ministry of Environment and the Department of Fisheries and Oceans. We thank the hunters in Greenland and Canada for assisting with collecting the samples and the University of Copenhagen for providing access to its Arctic Station in Disko Bay. The collection of the bowhead whale samples from the Okhotsk Sea project was conducted as part of the Marine Mammal Project under Area V: Protection of Nature and the Organization of Reserves within the U.S.-Russia Agreement on Cooperation in the Field of Environmental Protection. We thank Jeff Higdon and Rick Leduc, and two internal reviewers at NOAA for helpful comments that greatly improved the manuscript. We thank Danielle LaBruna for assistance with graphics and Robert Suydam for providing helpful information about bowhead ecology.
Appendix
Figure A1.
Cartoon representation of demographic scenarios used in simulations (see text). [Boundaries shown below are intended to convey the general geographic location for putative populations, and are not intended in any way to represent the shape or extent of the range of those populations.]
Table A1.
Parameters used in demographic simulations
| Parameter | Range of values | Distribution | Citations |
|---|---|---|---|
| Mutation rate | 2–6.3% per my | Uniform | Roman and Palumbi 2003; Alter and Palumbi 2009; |
| Generation time | 52 years | NA | Taylor et al. 2007; |
| Length of sequence | 370 bp | NA | NA |
| Initial size (BCB) | 13854 (9466–28475) | Normal | Brandon and Wade 2006; |
| Initial size (BBDS) | 10369 (10333–11,000) | Normal | Woodby and Botkin 1993; Table 10.3 and 10.4 |
| Initial size (HBFB) | 452 (445–680) | Normal | Woodby and Botkin 1993; Table 10.3 and 10.5 |
| Residual population (BCB) | 1000 (100–3000) | Normal | Woodby and Botkin 1993; Table 10.3 |
| Residual population (BBDS) | 1000 (100–3000) | Normal | Woodby and Botkin 1993; Table 10.3 |
| Residual population (HBFB) | 100 (10–300) | Normal | Woodby and Botkin 1993; Table 10.3 |
| Current size (BCB) | 10,470 (8100–13,500) | Normal | George et al. 2004; |
| Current size (BBDS) | 6344 (3119–12916) | Normal | COSEWIC 2009; |
| Current size (HBFB) | 1525 (333–6990) | Normal | COSEWIC 2009 |
Table A2.
Haplotype names and Genbank Accession numbers. PRI = number of individuals with Hap_1, etc. found in PRI. ECA = number of individuals with Hap_1, etc. found in ECA
| Haplotype number | PRI | ECA | Genbank Accession # |
|---|---|---|---|
| 1 | 6 | 76 | JX507921 |
| 2 | 9 | 87 | JX507922 |
| 3 | 3 | 4 | JX507923 |
| 4 | 2 | 14 | JX507924 |
| 5 | 1 | 9 | JX507925 |
| 6 | 2 | 17 | JX507926 |
| 7 | 0 | 13 | JX507927 |
| 8 | 2 | 3 | JX507928 |
| 9 | 1 | 1 | JX507929 |
| 10 | 0 | 5 | JX507930 |
| 11 | 0 | 8 | JX507931 |
| 12 | 0 | 4 | JX507932 |
| 13 | 0 | 1 | JX507933 |
| 14 | 0 | 1 | JX507934 |
| 15 | 0 | 7 | JX507935 |
| 16 | 0 | 6 | JX507936 |
| 17 | 0 | 2 | JX507937 |
| 18 | 1 | 1 | JX507938 |
| 19 | 0 | 1 | JX507939 |
| 20 | 0 | 2 | JX507940 |
| 21 | 0 | 1 | JX507941 |
| 22 | 0 | 1 | JX507942 |
| 23 | 1 | 0 | JX507943 |
| 24 | 1 | 0 | JX507944 |
| 25 | 1 | 0 | JX507945 |
| 26 | 1 | 0 | JX507946 |
| 27 | 1 | 0 | JX507947 |
| 28 | 1 | 0 | JX507948 |
| 29 | 1 | 0 | JX507949 |
| 30 | 1 | 0 | JX507950 |
| 31 | 1 | 0 | JX507951 |
| 32 | 1 | 0 | JX507952 |
| 33 | 1 | 0 | JX507953 |
| 34 | 1 | 0 | JX507954 |
Table A3.
Pairwise genetic distances between sample sets assuming five stocks. Pairwise ΦST values are given below the diagonal and frequency-based FST are given above. All bold values are significant at P < 0.05 after a False Discovery correction and bold values in italics are significant at P < 0.001
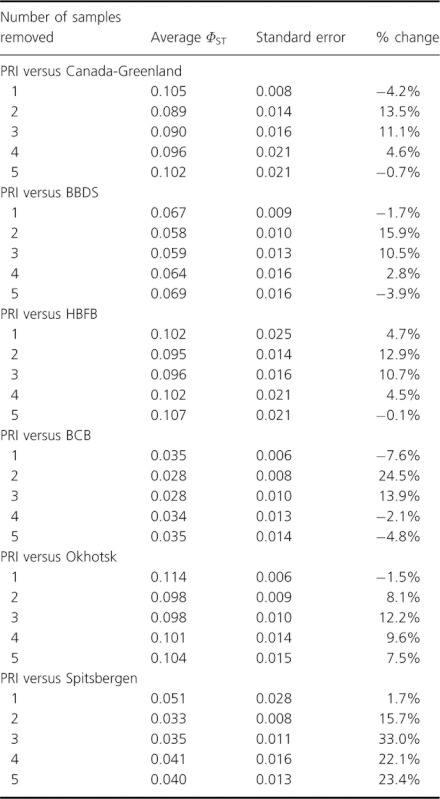 |
Table A4.
(a) Distribution of ΦST values for all scenarios using a migration value of m = 0.01. (b) Distribution of ΦST values for all scenarios using a migration value of m = 0.001
| (a) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| BCB-BBDS | BCB-PRI | PRI-BBDS | |||||||
| SCENARIO | 1A | 2A | 3A | 1A | 2A | 3A | 1A | 2A | 3A |
| 1% HPD | 0.057 | 0.041 | 0.033 | 0.032 | 0.014 | 0.016 | 0.000 | 0.004 | 0.003 |
| MEDIAN | 0.107 | 0.082 | 0.078 | 0.073 | 0.042 | 0.053 | 0.030 | 0.030 | 0.040 |
| 99% HPD | 0.202 | 0.127 | 0.153 | 0.135 | 0.090 | 0.110 | 0.080 | 0.084 | 0.078 |
| BCB-Canada/Greenland | BCB-PRI | PRI-Canada/Greenland | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 1B | 2B | 3B | 1B | 2B | 3B | 1B | 2B | 3B | |
| 1% HPD | 0.069 | 0.054 | 0.048 | 0.012 | 0.011 | 0.011 | 0.039 | 0.032 | 0.031 |
| MEDIAN | 0.127 | 0.102 | 0.095 | 0.031 | 0.029 | 0.042 | 0.085 | 0.067 | 0.072 |
| 99% HPD | 0.246 | 0.209 | 0.207 | 0.072 | 0.060 | 0.079 | 0.227 | 0.141 | 0.185 |
| 1C | 2C | 3C | 1C | 2C | 3C | 1C | 2C | 3C | |
|---|---|---|---|---|---|---|---|---|---|
| 1% HPD | 0.058 | 0.109 | 0.031 | 0.022 | 0.094 | 0.014 | 0.039 | 0.124 | 0.016 |
| MEDIAN | 0.110 | 0.207 | 0.081 | 0.072 | 0.187 | 0.050 | 0.089 | 0.248 | 0.062 |
| 99% HPD | 0.194 | 0.347 | 0.185 | 0.163 | 0.378 | 0.117 | 0.191 | 0.515 | 0.201 |
| 1D | 2D | 3D | 1D | 2D | 3D | 1D | 2D | 3D | |
|---|---|---|---|---|---|---|---|---|---|
| 1% HPD | 0.054 | 0.042 | 0.037 | 0.007 | 0.006 | 0.003 | 0.027 | 0.023 | 0.020 |
| MEDIAN | 0.127 | 0.102 | 0.095 | 0.031 | 0.029 | 0.042 | 0.085 | 0.067 | 0.072 |
| 99% HPD | 0.276 | 0.236 | 0.235 | 0.082 | 0.067 | 0.088 | 0.262 | 0.159 | 0.213 |
| (b) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| BCB-BBDS | BCB-PRI | PRI-BBDS | |||||||
| SCENARIO | 1A | 2A | 3A | 1A | 2A | 3A | 1A | 2A | 3A |
| 1% HPD | 0.061 | 0.065 | 0.056 | 0.040 | 0.048 | 0.043 | 0.002 | 0.000 | 0.000 |
| MEDIAN | 0.122 | 0.116 | 0.135 | 0.095 | 0.087 | 0.103 | 0.034 | 0.027 | 0.031 |
| 99% HPD | 0.268 | 0.216 | 0.245 | 0.166 | 0.165 | 0.186 | 0.081 | 0.081 | 0.091 |
| BCB-Canada/Greenland | BCB-PRI | PRI-Canada/Greenland | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 1B | 2B | 3B | 1B | 2B | 3B | 1B | 2B | 3B | |
| 1% HPD | 0.075 | 0.077 | 0.069 | 0.046 | 0.039 | 0.060 | 0.013 | 0.016 | 0.009 |
| MEDIAN | 0.179 | 0.173 | 0.173 | 0.122 | 0.109 | 0.142 | 0.048 | 0.053 | 0.051 |
| 99% HPD | 0.392 | 0.344 | 0.368 | 0.302 | 0.310 | 0.310 | 0.193 | 0.152 | 0.132 |
| 1C | 2C | 3C | 1C | 2C | 3C | 1C | 2C | 3C | |
|---|---|---|---|---|---|---|---|---|---|
| 1% HPD | 0.071 | 0.100 | 0.077 | 0.048 | 0.066 | 0.048 | 0.077 | 0.117 | 0.085 |
| MEDIAN | 0.159 | 0.208 | 0.172 | 0.117 | 0.173 | 0.128 | 0.176 | 0.249 | 0.218 |
| 99% HPD | 0.348 | 0.386 | 0.331 | 0.255 | 0.379 | 0.274 | 0.412 | 0.455 | 0.385 |
| 1D | 2D | 3D | 1D | 2D | 3D | 1D | 2D | 3D | |
|---|---|---|---|---|---|---|---|---|---|
| 1% HPD | 0.074 | 0.058 | 0.064 | 0.000 | 0.002 | 0.004 | 0.046 | 0.036 | 0.053 |
| MEDIAN | 0.166 | 0.150 | 0.189 | 0.036 | 0.023 | 0.027 | 0.125 | 0.111 | 0.147 |
| 99% HPD | 0.324 | 0.281 | 0.303 | 0.081 | 0.074 | 0.083 | 0.276 | 0.237 | 0.327 |
Table A5.
Sequences were serially removed from sample sets. For each population comparison, ΦST was calculated and averaged across all possible combinations of removed samples, and compared with the observed ΦST value
| Number of samples removed | Average ΦST | Standard error | % change |
|---|---|---|---|
| PRI versus Canada-Greenland | |||
| 1 | 0.105 | 0.008 | −4.2% |
| 2 | 0.089 | 0.014 | 13.5% |
| 3 | 0.090 | 0.016 | 11.1% |
| 4 | 0.096 | 0.021 | 4.6% |
| 5 | 0.102 | 0.021 | −0.7% |
| PRI versus BBDS | |||
| 1 | 0.067 | 0.009 | −1.7% |
| 2 | 0.058 | 0.010 | 15.9% |
| 3 | 0.059 | 0.013 | 10.5% |
| 4 | 0.064 | 0.016 | 2.8% |
| 5 | 0.069 | 0.016 | −3.9% |
| PRI versus HBFB | |||
| 1 | 0.102 | 0.025 | 4.7% |
| 2 | 0.095 | 0.014 | 12.9% |
| 3 | 0.096 | 0.016 | 10.7% |
| 4 | 0.102 | 0.021 | 4.5% |
| 5 | 0.107 | 0.021 | −0.1% |
| PRI versus BCB | |||
| 1 | 0.035 | 0.006 | −7.6% |
| 2 | 0.028 | 0.008 | 24.5% |
| 3 | 0.028 | 0.010 | 13.9% |
| 4 | 0.034 | 0.013 | −2.1% |
| 5 | 0.035 | 0.014 | −4.8% |
| PRI versus Okhotsk | |||
| 1 | 0.114 | 0.006 | −1.5% |
| 2 | 0.098 | 0.009 | 8.1% |
| 3 | 0.098 | 0.010 | 12.2% |
| 4 | 0.101 | 0.014 | 9.6% |
| 5 | 0.104 | 0.015 | 7.5% |
| PRI versus Spitsbergen | |||
| 1 | 0.051 | 0.028 | 1.7% |
| 2 | 0.033 | 0.008 | 15.7% |
| 3 | 0.035 | 0.011 | 33.0% |
| 4 | 0.041 | 0.016 | 22.1% |
| 5 | 0.040 | 0.013 | 23.4% |
Conflict of Interest
None declared.
References
- Alter SE, Palumbi SR. Comparing evolutionary patterns and variability in the mitochondrial control region and cytochrome b in three species of Baleen whales. J. Mol. Evol. 2009;68:97–111. doi: 10.1007/s00239-008-9193-2. [DOI] [PubMed] [Google Scholar]
- Alter SE, Rynes E, Palumbi SR. DNA evidence for historic population size and past ecosystem impacts of gray whales. Proc. Natl. Acad. Sci. USA. 2007;104:15162–15167. doi: 10.1073/pnas.0706056104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter SE, Ramirez SF, Nigenda S, Urban Ramirez J, Bracho LR, Palumbi SR. Mitochondrial and nuclear genetic variation across calving lagoons in eastern North Pacific gray whales (Eschrichtius robustus. J. Hered. 2009;100:34–46. doi: 10.1093/jhered/esn090. [DOI] [PubMed] [Google Scholar]
- Alter SE, Newsome S, Palumbi SR. Pre-whaling genetic diversity and population ecology in eastern Pacific gray whales: insights from ancient DNA and stable isotopes. PLoS ONE. 2012;7:e35039. doi: 10.1371/journal.pone.0035039. doi: 10.1371/journal.pone.0035039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson CNK, Ramakrishnan U, Chan YL, Hadly EA. Serial SimCoal: a population genetic model for data from multiple populations and points in time. Bioinformatics. 2005;21:1733–1734. doi: 10.1093/bioinformatics/bti154. [DOI] [PubMed] [Google Scholar]
- Arnason U, Gullberg A, Widegren B. Cetacean mitochondrial DNA control region: sequences of all extant baleen whales and two sperm whale species. Mol. Biol. Evol. 1993;10:960–970. doi: 10.1093/oxfordjournals.molbev.a040061. [DOI] [PubMed] [Google Scholar]
- Baker CS, Perry A, Bannister JL, Abernathy RB, Weinrich MT, Abernaty B, et al. Abundant mitochondrial DNA variation and world-wide population structure in humpback whales. Proc. Natl. Acad. Sci. USA. 1993;90:8239–8243. doi: 10.1073/pnas.90.17.8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CS, Slade RW, Bannister JL, Abernethy RB, Weinrich MG, Lien J, et al. Hierarchical structure of mitochondrial DNA gene flow among humpback whales Megaptera novaeangliae, world-wide. Mol. Ecol. 1994;5:283–287. doi: 10.1111/j.1365-294x.1994.tb00071.x. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
- Berube M, Aguilar A, Dendanto D, Larsen F, Notobartolo di Sciara G, Sears R, et al. Population genetic structure of North Atlantic, Mediterranean Sea and Sea of Cortez fin whales, Balaenoptera physalus (Linneaeus 1758): analysis of mitochondrial and nuclear loci. Mol. Ecol. 1998;7:585–599. doi: 10.1046/j.1365-294x.1998.00359.x. [DOI] [PubMed] [Google Scholar]
- Borge T, Bachmann L, Bjornstad G, Wiig O. Genetic variation in Holocene bowhead whales from Svalbard. Mol. Ecol. 2007;16:2223–2235. doi: 10.1111/j.1365-294X.2007.03287.x. [DOI] [PubMed] [Google Scholar]
- Brandon J, Wade PR. Assessment of the Bering-Chukchi-Beaufort Seas stock of bowhead whales using Bayesian model averaging. J. Cetacean Res. Manage. 2006;8:225–239. [Google Scholar]
- Brown Gladden JG, Ferguson MM, Clayton JW. Matriarchal genetic population structure of North American beluga whales Delphinapterus leucas (Cetacea: Monodontidae) Mol. Ecol. 1997;6:1033–1046. doi: 10.1046/j.1365-294x.1997.00275.x. [DOI] [PubMed] [Google Scholar]
- de Bruyn M, Hall BL, Chauke LF, Baroni C, Koch PL, Hoelzel AR. Rapid response of a marine mammal species to Holocene climate and habitat change. PLoS Genet. 2009;5:e1000554. doi: 10.1371/journal.pgen.1000554. doi: 10.1371/journal.pgen.1000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YL, Lacey EA, Pearson OP, Hadly EA. Ancient DNA reveals Holocene loss of genetic diversity in a South American rodent. Biol. Lett. 2005;1:423–426. doi: 10.1098/rsbl.2005.0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COSEWIC. COSEWIC assessment and update status report on the Bowhead Whale Balaena mysticetus, Bering-Chukchi-Beaufort population and Eastern Canada-West Greenland population, in Canada. Ottawa: Committee on the Status of Endangered Wildlife in Canada; 2009. p. vii + 49. ( http://www.sararegistry.gc.ca/status/status_e.cfm) [Google Scholar]
- Dalen L, Nystrom V, Valdiosera C, Germonpre M, Sablin M, Turner E, et al. Ancient DNA reveals lack of postglacial habitat tracking in the arctic fox. Proc. Natl. Acad. Sci. USA. 2007;104:6726–9. doi: 10.1073/pnas.0701341104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dueck L, Ferguson SH. Habitat use by bowhead whales (Balaena mysticetus) of the Eastern Canadian Arctic. 2008. DFO Science Advisory Secretariat Research Document 2008/082. Available via http://www.dfo-mpo.gc.ca/csas-sccs/publications/resdocs-docrech/2008/2008_082-eng.htm, 27 pp. [Google Scholar]
- Dyke AS, Hooper J, Savelle JM. A history of sea ice in the Canadian Arctic Archipelago based on postglacial remains of the bowhead whale (Balaena mysticetus. Arctic. 1996;49:235–255. [Google Scholar]
- Excoffier L. Analysis of Population Subdivision. In: Balding D, Bishop M, Cannings C, editors. Handbook of statistical genetics. 2nd ed. Vol. 2. New York: John Wiley & Sons, Ltd; 2003. pp. 713–750. [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley KJ. Isabella Bay, Baffin Island: an important historical and present-day concentration area for the endangered bowhead whale (Balaena mysticetus) of the eastern Canadian Arctic. Arctic. 1990;43:137–152. [Google Scholar]
- Geffen E, Waidyaratne S, Dalen L, Angerbjorn A, Vila C, Hersteinsson P, et al. Sea ice occurrence predicts genetic isolation in the Arctic fox. Mol. Ecol. 2007;16:4241–4255. doi: 10.1111/j.1365-294X.2007.03507.x. [DOI] [PubMed] [Google Scholar]
- George JC, Zeh J, Suydam R, Clark C. Abundance and population trend (1978–2001) of Western Arctic bowhead whales surveyed near Barrow, Alaska. Mar. Mamm. Sci. 2004;20:755–773. [Google Scholar]
- Givens GH, Huebinger RM, Patton JC, Postma LD, Lindsay M, Suydam RS, et al. Population genetics of bowhead whales (Balaena mysticetus) in the western Arctic. Arctic. 2010;63:1–12. [Google Scholar]
- Heide-Jorgensen MP, Richard P, Ramsay M, Akeeagok S. Three recent ice entrapments of Arctic cetaceans in West Greenland and the eastern Canadian High Arctic. NAMMCO Sci. Publ. 2002;4:143–148. [Google Scholar]
- Heide-Jørgensen MP, Laidre KL, Jensen MV. Satellite tracking: bowhead whales in Baffin Bay. Mar. Mamm. Sci. 2006;22:34–45. [Google Scholar]
- Heide-Jørgensen MP, Laidre KL, Quakenbush LT, Citta JJ. The Northwest Passage opens for bowhead whales. Biol. Lett. 2011;8:270–273. doi: 10.1098/rsbl.2011.0731. doi: 10.1098/rsbl.2011.0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J, Nielsen R. Integration within the Felsenstein equation for improved Markov chain Monte Carlo methods in population genetics. PNAS. 2007;104:2785–2790. doi: 10.1073/pnas.0611164104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higdon J. Commercial and subsistence harvests of bowhead whales (Balaena mysticetus) in eastern Canada and West Greenland. 2008. DFO Canadian Science Advisory Secretariat Research Document 2008/008. Available via http://www.dfo-mpo.gc.ca/csas-sccs/Publications/ResDocs-DocRech/2008/2008_008-eng.htm, 59 pp. [Google Scholar]
- Higdon JW. Commercial and subsistence harvests of bowhead whales (Balaena mysticetus) in eastern Canada and West Greenland. J. Cetacean Res. Manage. 2010;11:185–216. (accepted January 2010) [Google Scholar]
- Hoelzel AR. Ecology and genetics of whales and dolphins. Annu. Rev. Ecol. Syst. 1994;25:377–399. [Google Scholar]
- Hoelzel AR. Geneti structure of cetacean poulations in sympatry, parapatry and mixed assemblages: implications for conservation policy. J. Hered. 1998;98:451–458. [Google Scholar]
- Holsinger KE, Weir BS. Genetics in geographically structured populations: defining, estimating and interpreting Fst. Nat. Rev. Genet. 2009;10:639–650. doi: 10.1038/nrg2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JA, Patenaude NJ, Carroll L, Baker CS. How few whales were there after whaling? Inference from contemporary mtDNA diversity. Mol. Ecol. 2008;17:236–251. doi: 10.1111/j.1365-294X.2007.03497.x. [DOI] [PubMed] [Google Scholar]
- Jukes TH, Cantor CR. Evolution of protein molecules. In: Munro HN, editor. Mammalian protein metabolism. New York: Academic Press; 1969. pp. 21–132. [Google Scholar]
- Laidre KL, Stirling I, Lowry LF, Wiig O, Heide Jorgensen MP, Ferguson SH. Quantifying the sensitivity of Arctic marine mammals to climate-induced habitat change. Ecol. Appl. 2008;18(Suppl):S97–S125. doi: 10.1890/06-0546.1. [DOI] [PubMed] [Google Scholar]
- LeDuc RG, Dizon AE, Burdin AM, Blockin SA, George JC, Brownell RL., Jr Genetic analyses (mtDNA and microsatellites) of Okhotsk and Bering/Chukchi/Beaufort Seas populations of bowhead whales. J. Cetacean Res. Manage. 2005;7:107–111. [Google Scholar]
- LeDuc RG, Martien KK, Morin PA, Hedrick N, Robertson K, Taylor BL, et al. Mitochondrial genetic variation in bowhead whales in the western Arctic. J. Cetacean Res. Manage. 2009;10:2. [Google Scholar]
- Lyrholm T, Leimar O, Johanneson B, Gyllensten U. Sex-biased dispersal in sperm whales: contrasting mitochondrial and nuclear genetic structure of global populations. Proc. R. Soc. Lond. B. 1999;266:347–354. doi: 10.1098/rspb.1999.0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean SA. Occurrence, behavior, and genetic diversity of bowhead whales in the western Sea of Okhotsk, Russia. College Station, TX: Texas A&M Univ; 2002. p. 114. MS thesis. [Google Scholar]
- Maiers LD, Friesen MK, Wiens AV, Clayton JW. Use of DNA microsatellites in beluga whale (Delphinapterus leucas) population genetics. 1996. Canadian Technical Report of Fisheries and Aquatic Sciences 2115, v + 17pp. [PubMed] [Google Scholar]
- Marquette WM. Bowhead whale. In: Haley D, editor. Marine mammals of the eastern North Pacific and Arctic waters, second edition. Seattle: Pacific Search Press; 1986. pp. 83–93. [Google Scholar]
- McCartney AP. A processual consideration of Thule whale bone houses. In: McCartney AP, editor. Thule Eskimo culture: an anthropological retrospective. Archaeological Survey of Canada, Mercury Series 88. Ottawa: National Museum of Man; 1979. pp. 301–323. [Google Scholar]
- Moore SE, Reeves RR. Distribution and movement. In: Burns JJ, Montague JJ, Cowles CJ, editors. The bowhead whale. Lawrence, Kansas: Society for Marine Mammalogy; 1993. pp. 313–386. [Google Scholar]
- O'Corry-Crowe G. Climate change and the molecular ecology of arctic marine mammals. Ecol. Appl. 2008;18:S56–S76. doi: 10.1890/06-0795.1. [DOI] [PubMed] [Google Scholar]
- Palsbøll PJ, Clapham PJ, Matilla DK, Larsen F, Sears R, Siegismund HR, et al. Distribution of mtDNA haplotypes in North Atlantic humpback whales: the influence of behavior on population structure. Mar. Ecol. Prog. Ser. 1995;116:1–10. [Google Scholar]
- Pastene LA, Goto M, Kanda N, Zerbini AN, Kerem D, Watanabe K, et al. Radiation and speciation of pelagic organisms during periods of global warming: the case of the common minke whale, Balaenoptera acutorostrata. Mol. Ecol. 2007;16:1481–1495. doi: 10.1111/j.1365-294X.2007.03244.x. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Roman J, Palumbi SR. Whales before whaling. Science. 2003;301:508–510. doi: 10.1126/science.1084524. [DOI] [PubMed] [Google Scholar]
- Rosenbaum HC, Egan MG, Clapham PJ, Brownell RL, DeSalle R. An effective method for isolating DNA from historical specimens of baleen. Mol. Ecol. 1997;6:677–681. doi: 10.1046/j.1365-294x.1997.00230.x. [DOI] [PubMed] [Google Scholar]
- Rosenbaum HC, Brownell RL, Jr, Brown MW, Schaeff C, Portway V, White BN, et al. World-wide genetic differentiation of Eubalaena: questioning the number of right whale species. Mol. Ecol. 2000;9:1793–1802. doi: 10.1046/j.1365-294x.2000.01066.x. [DOI] [PubMed] [Google Scholar]
- Rosenbaum HC, Weinrich MT, Stoleson SA, Gibbs JP, Baker CS, DeSalle R. The effect of differential reproductive success on population genetic structure: correlations of life history with matrilines in humpback whales of the Gulf of Maine. J. Hered. 2002;93:389–399. doi: 10.1093/jhered/93.6.389. [DOI] [PubMed] [Google Scholar]
- Ross WG. Commercial whaling in the North Atlantic sector. In: Burns JJ, Montague JJ, Cowles CJ, editors. The bowhead whale. Lawrence, Kansas: Society for Marine Mammalogy; 1993. pp. 511–577. [Google Scholar]
- Rozas J, Sanchez-Delbarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maiatis T. Molecular coloning: a laboratory manual. 2nd ed. Vol. 1. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Savelle JM, McCartney AP. Thule Inuit bowhead whaling: a biometrical analysis. In: Morrison D, Pilon J-L, editors. Threads of Arctic prehistory: papers in honour of William E Taylor Jr. Archaeological Survey of Canada Mercury Series. Ottawa: Canadian Museum of Civilization; 1994. pp. 281–310. 149. [Google Scholar]
- Savelle JM, Wenzel GW. Out of Alaska: reconstructing the social structure of prehistoric Canadian Thule culture. In: Habu J, Savelle JM, Koyama S, Hongo H, editors. Senri ethnological studies no 63. Osaka: National Museum of Ethnology; 2003. pp. 103–121. [Google Scholar]
- Schaeff CM, Kraus SD, Brown MW, White BN. Assessment of the population structure of western North Atlantic right whales (Eubalaena glacialis) based on sighting and mtDNA data. Can. J. Zool. 1993;71:339–345. [Google Scholar]
- Schneider S, Roessli D, Excoffier L. Arlequin: a software for population genetics data analysis. Ver 2.000. Genetics and Biometry Laboratory, Department of Anthropology, University of Geneva: 2000. [Google Scholar]
- Shapiro B, Drummond AJ, Rambaut A, Wilson MC, Matheus PE, Sher AV, et al. Rise and fall of the Beringian steppe bison. Science. 2004;306:1561–1565. doi: 10.1126/science.1101074. [DOI] [PubMed] [Google Scholar]
- Stoker SW, Krupnik II. Subsistence whaling. In: Burns JJ, Montague JJ, Cowles CJ, editors. The bowhead whale. Lawrence, Kansas: Society for Marine Mammalogy; 1993. pp. 579–629. [Google Scholar]
- Taylor BL, Chivers SJ, Larese J, Perrin WF. Generation length and percent mature estimates for IUCN assessments of cetaceans. 2007. National Marine Fisheries Service Administrative Report LJ-07-01. Available via http://swfsc.noaa.gov/uploadedfiles/divisions/prd/publications/, 24 pp. [Google Scholar]
- Vare LL, Masse G, Gregory TR, Smart CW, Belt ST. Sea ice variations in the central Canadian Arctic Archipelago during the Holocene. Quatern. Sci. Rev. 2009;28:1354–1366. [Google Scholar]
- Whitridge PJ. The prehistory of Inuit and Yupik whale use. Revista de Arqueología Americana. 1999;16:99–154. [Google Scholar]
- Whitridge PJ. Social and ritual determinants of whale bone transport at a Classic Thule winter site in the Canadian Arctic. Int. J. Osteoarchaeol. 2002;12:65–75. [Google Scholar]
- Wilberg MJ, Dreher BP. GENECAP: a program for analysis of multilocus genotype data for non-invasive sampling and capture-recapture population estimation. Mol. Ecol. Notes. 2004;4:783–785. [Google Scholar]
- Woodby DA, Botkin DB. Stock sizes prior to commercial whaling. In: Burns JJ, Montague JJ, Cowles CJ, editors. The bowhead whale. Lawrence, Kansas: Society for Marine Mammalogy; 1993. pp. 387–407. [Google Scholar]