Abstract
We have characterized the homologs of an actin, a troponin I, and a tropomyosin gene in the acoel Symsagittifera roscoffensis. These genes are expressed in muscles and most likely coexpressed in at least a subset of them. In addition, and for the first time for Acoela, we have produced a species-specific muscular marker, an antibody against the tropomyosin protein. We have followed tropomyosin gene and protein expression during postembryonic development and during the posterior regeneration of amputated adults, showing that preexisting muscle fibers contribute to the wound closure. The three genes characterized in this study interact in the striated muscles of vertebrates and invertebrates, where troponin I and tropomyosin are key regulators of the contraction of the sarcomere. S. roscoffensis and all other acoels so far described have only smooth muscles, but the molecular architecture of these is the same as that of striated fibers of other bilaterians. Given the proposed basal position of acoels within the Bilateria, we suggest that sarcomeric muscles arose from a smooth muscle type, which had the molecular repertoire of striated musculature already in place. We discuss this model in a broad comparative perspective.
Muscles exist exclusively in the Eumetazoa, namely the Cnidaria, the Ctenophora, and the Bilateria, and as such they are pivotal to reconstruct and understand the evolution of animals (Burton, 2008).
In the bilateral animals, the muscles are morphologically distinguished in two basic types: the smooth and the striated muscles. In the smooth muscular cells, the myofilaments (i.e. thin actin and thick myosin filaments, which interaction produces the contraction) are poorly arranged. Conversely, in the striated muscles, the myofilaments are highly organized in units called sarcomeres (Ruppert et al., 2004).
The relation of the two different muscle types, to each other and among different taxa, is still not settled and molecular data has just accumulated enough to allow for the first speculations (Seipel and Schmid, 2005). However, one generally accepted scenario suggests that myocytes, i.e. true muscular fibers lacking any epithelial component, are derived from epitheliomuscular cells, which are the most ancestral type of contractile cells (Rieger and Ladurner, 2003). Myoepithelial cells are abundant within the Cnidaria (sea anemones, corals, jellyfish), which are the sister group of the Bilateria. However, in the swimming life stage of some cnidarians, the medusa, there are myocytes too. These are generally suggested to have evolved convergently to the bilaterian muscles as an adaptation to swimming. If these premises are accepted, the origin of true muscles in the Bilateria dates back to its own origin. Recent molecular phylogenies place acoels as the earliest offshoot of all bilateral animals (Hejnol et al., 2009) and, albeit this position is still controversial (Dunn et al., 2008; Egger et al., 2009; Philippe et al., 2011), a full set of evidences, such as morphological characters and the Hox gene complement, support their basal position (Haszprunar, ’96; Hejnol and Martindale, 2008, 2009; Moreno et al., 2009). Accordingly, in order to understand the evolutionary origin of muscular cells and the relationship between the cnidarian and bilaterian muscles, data from these simple worms are crucial.
It is generally accepted that the mesoderm has evolved from the endoderm; however, in most of the Bilateria two mesoderm sources exist: the so-called endomesoderm and the ectomesoderm, which usually develops from ectodermal tissues (Martindale and Henry, ’99; Technau and Scholz, 2003; Martindale et al., 2004). In acoels, muscles and all other mesodermal tissues develop from endomesoderm because they have no ectomesoderm (Henry et al., 2000). Morphogenesis and embryonic development of the musculature have been investigated in two acoel species: Isodiametra pulchra and Symsagittifera roscoffensis (Ladurner and Rieger, 2000; Semmler et al., 2008). Accordingly, in both species, the differentiation of muscles proceeds from the anterior to the posterior pole of the embryo, the circular muscles arise before the longitudinal muscles, and the juvenile and adult musculature originate by adding more fibers to an initial grid. Morphological investigations on muscles, either using electron microscopy (Rieger et al., ’91) or fluorophore-tagged phalloidin and confocal microscopy (Hooge, 2001; Hooge and Tyler, 2005) are more numerous, cover a much greater number of species, and show that the smooth type is the only type of muscle occurring in these animals.
Investigations on the adult body-wall structure and its development are informative for deciphering the interrelationships of taxa and eventually tracing the evolution of new body plans (Wanninger, 2009), though they don’t tell much about the evolution of the muscular tissue itself. Dissecting the molecular fingerprint of muscles in the Acoela could offer important insights into the topic (Arendt, 2008).
We are currently working to establish the acoel S. roscoffensis as a model system for molecular developmental biology, and we have characterized, for the first time in any acoel species, the expression pattern of three muscular genes, an actin, a tropomyosin, and an inhibitory subunit of the troponin complex. These three proteins interact in the skeletal muscle of vertebrates and have also been identified in several invertebrates, with two of them, actin and tropomyosin, also existing in the cnidarian muscles (Groger et al., ’99; Scholz and Technau, 2003).
Additionally, we have raised a specific antibody against the tropomyosin of S. roscoffensis. In order to understand the dynamic expression of some of the muscular markers in a developmental context, we have followed the expression of tropomyosin, gene and protein, during muscle regeneration.
Regeneration can be easily induced and followed in acoels; although, so far, the process has been poorly studied (Gaerber et al., 2007; De Mulder et al., 2009a; Bery and Martinez, 2011). Although the development of muscles in adults has only been studied during the asexual reproduction of four different Convolutriloba species (Sikes and Bely, 2008), there is no published data available in animals after experimental excision. For the first time in acoels, we describe the regeneration of muscles using a species-specific muscular marker.
MATERIALS AND METHODS
Animal Collection, Rearing, and Fixation
Adult specimens of S. roscoffensis were collected in Carantec (Brittany, France) in 2007 and 2008. The specimens were kept in aquaria with continuous seawater cycling at 15°C. After approximately 1 week, gravid animals released cocoons. These were collected and kept in glass Petri dishes at 15°C as well. Filtered seawater was changed once a day.
Hatchlings were collected and immediately processed for fixation or left to grow for 1–7 days, with filtered seawater replaced once a day. For regeneration studies, adults were sectioned transversally in the mid-body region. The anterior and posterior halves were kept in different dishes. Animals were left to regenerate at 15°C and were subsequently fixed at different time intervals. Within the first 24 hr of regeneration, they were usually fixed at intervals of 4–5 hr.
For immunostaining, specimens were treated with 1% cysteine chloride (pH 7–7.5) in seawater for about 20 min at room temperature, and then rinsed twice in filtered seawater to remove mucous secretions. Subsequently, specimens were relaxed by addition of drops of 7.14% magnesium chloride. Then, the animals were fixed in 4% PFA (dissolved in 0.1 M PBS; pH = 7.5), for either 2 hr at room temperature or overnight at 4°C. The animals were subsequently washed three to five times in PBS and stored at 4°C in PBS+0.1% sodium azide.
For in situ hybridization, the specimens were fixed in a mixture of 0.2% glutaraldehyde+3.7% formaldehyde in PBS for 5 min at room temperature, and then left for 1 hr in 3.7% formaldehyde in PBS at the same temperature. Fixed animals were subsequently washed three to five times in PBS and progressively dehydrated in a methanol series. Specimens were stored in 100% methanol at −20°C.
Gene Isolation and Gene Orthology Analyses
The cDNA clones of this study were identified from an arrayed (and fully sequenced) cDNA library from S. roscoffensis aposymbiotic hatchlings. All inserts were cloned into pBluescript SK- and oriented as to get an antisense riboprobe when using the T7 RNA polymerase (Roche, Hoffman-La Roche Inc., Nutley, NJ), and a sense probe when using the T3 RNA polymerase (Roche).
After regrowing the transformed bacteria from a glycerol stock, the SrAct, SrTnI, and SrTrp were resequenced using the BigDye Terminator v3.1 Cycle sequencing kit (Applied Biosystems, Life Technologies, Carlsbad, CA).
Phylogenetic analyses were performed using protein sequences related to SrAct, SrTnI, and SrTrp, which were downloaded from GenBank. For the actin family we chose muscular and nonmuscular actins, for troponin sequences we used members of all three subunits known to interact in the troponin complex (TnC, TnI, TnT), and for tropomyosin sequences we selected “long-muscular” and “short-nonmuscular” tropomyosin isoforms. Alignments of the protein sequences were done using the MAFFT program, included in the Geneious package (Drummond et al., 2010). Considering the level of sequence identity, the matrices used for generating the alignments were BLOSUM80 for SrAct, BLOSUM30 for SrTnI, and BLOSUM45 for SrTrp. To calculate the trees, a maximum likelihood methodology was performed with RAxML 7.0.3 (Stamatakis et al., 2008) on the Vital-IT server (http://phylobench.vital-it.ch/raxml-bb/), using the model of evolution suggested by ProtTest (Abascal et al., 2005). Accession numbers of the sequences (S1, S2, an S3) and alignments used (S8, S9, and S10) are provided in the supplementary material. The original alignments, in nexus or phylip format, are available on demand.
Antibody Production and Immunostaining
The anti-SrTrp antibody was made against the peptide LDKTNHQLDDANKE of the deduced protein sequence of SrTrp. For predicting the antigenicity of the peptide, based on local average hydrophylicity, the Hopp and Woods’ method (Hopp and Woods, ’81) was applied to the whole aminoacidic sequence of the protein. The highest hydrophilicity is suggested for the peptide containing the residues 64–77.
The peptide was synthesized, purified by semi-preparative reverse-phase HPLC, and its purity was verified by analytical HPLC and amino acid analysis. The corresponding immunogens were prepared by coupling the peptides synthesized to KLH and then injected into rabbits for developing antibodies. The animals were boosted at first after 6 weeks and then every 4 weeks. Blood was collected 2 weeks after each booster injection. The antisera obtained were evaluated for their titer and cross-reactivity using an enzyme-linked immunoabsorbent assay.
Immunostaining of whole mount juveniles or adult specimens was performed following the protocol published by Semmler et al. (2010). The primary antibody working concentration was determined empirically. A 1/50 dilution of anti-SrTrp in 6% Normal Goat Serum (Sigma, St. Louis, MO) in PBS yielded the best signal-to-background ratio. Controls with preimmune serum, at the same working concentration of anti-SrTrp, yielded no signal.
For phalloidin staining, adults’ specimens were first permeabilized in PBS+3% triton for 2 hr at room temperature. Subsequently, they were incubated in a solution of Alexa633-phalloidin (Molecular Probes, Eugene, OR) in PBS diluted 1/40, for 3 hr at room temperature. This step was performed in the dark, as all the following washing steps in PBS (6×10 min). Finally, the specimens were processed for analysis.
All specimens were mounted in either Vectashield (Vector Laboratories, Burlingame, CA) or FluoromountG (Southern Biotech, Birmingham, AL). The preparations were observed and analyzed on either a Leica TCS SP2 or a TCS SPE confocal laser scanning microscope (Leica Microsystems, Wetzlar, Germany).
In Situ Hybridization
Digoxigenin-labeled sense and antisense riboprobes were synthesized using the Dig-RNA labeling kit from Roche (Hoffman-La Roche Ltd., Basel, Switzerland), following the manufacturer’s instructions. The clones recovered in the EST library were used as templates for riboprobe synthesis. All clones contained the complete ORFs and both the 5′ and 3′ untranslated regions. The respective antisense probe lengths for the SrAct, SrTnI, and SrTrp genes were 1,478 bp, 1,480 bp, and 2, 7 kb, respectively.
In situ hybridization on whole mount specimens was performed following the protocol published by Semmler et al. (2010), with the only exception of skipping the proteinase K step plus the following glycine washes and the refixation step. Hybridization was done as follows: SrAct, 0,1 ng/μL of sense or antisense probe concentration and 60°C of hybridization temperature; SrTnI 1ng/μL of sense or antisense probe concentration and 60°C of hybridization temperature; SrTrp 1ng/μL of sense or antisense probe concentration and 50°C of hybridization temperature.
All specimens were mounted in 70% glycerol (Sigma) in PBS and analyzed on a Zeiss Axiophot (Carl Zeiss MicroImaging, GmbH) or on a Leica BMLB (Leica Microsystems, Wetzlar Germany) microscope, both equipped with a ProgRES C3 camera (Jenoptik, Germany).
RESULTS
Molecular Characterization of Actin, Tropomyosin, and Troponin I Orthologs From S. roscoffensis
In order to understand the phylogenetic affinities of some molecular components of the acoel muscles, we isolated a few clones that represent well-known components of the muscular architecture. These clones are the potential homologs of actin, troponin I, and tropomyosin (SrAct, SrTnI, and SrTrp).
The clone SrAct is 1,478 bp long and it contains an open reading frame (ORF) of 1,129 nucleotides, which conceptual translation is a 377 amino acids–long protein. Phylogenetic analysis was performed using full-length sequences of cytoplasmic and muscular isoforms from several taxa (supplementary material, S8). In agreement with previous results, we have recovered the muscular actins of the vertebrates and those of the insects as independent monophyletic groups (Vandekerckhove and Weber, ’84; Mounier et al., ’92) (supplementary material, S4). SrAct clusters with the deuterostomes’ muscular actins; however, the bootstrap values for the nodes in this topology are very low with RAxML (supplementary material, S4).
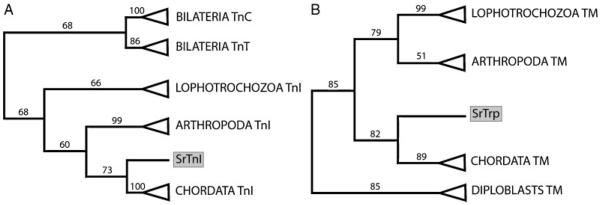
The clone SrTnI is 1,480 bp long and it contains a 529 bp ORF. The deduced translation of the ORF results in a protein of 175 amino acids. Phylogenetic analyses using sequences of all three subunits of the troponin complex from distantly related taxa show that SrTnI is a true ortholog of the inhibitory subunit of the troponin complex (Fig. 1A and supplementary material S5).
Figure 1.
Schematic representation of phylogenetic trees calculated by RAxML. All nodes with support values below 50 have been collapsed. (A) Tree calculated using the WAG model. The tree includes all the three subunits of the Troponin complex. TnC, calcium-binding subunit. TnI, inhibitory subunit. TnT, tropomyosin-binding subunit. The grey box highlights the acoel sequence SrTnI. (B) Tree calculated using the RETREV model. The tree includes long and short isoforms of the tropomyosin proteins which are not specifically shown for easy representation purposes. The acoel sequence, SrTrp, is grey boxed.
The clone SrTrp is 2.7 Kb long and has an 858 bp ORF. The encoded protein is 285 amino acids long and is a “long-type” tropomyosin (supplementary material, S3 and S10). The long forms of the tropomyosin include 38 amino acids long N-terminal motif, which is highly conserved even among distantly related organisms, but is never recovered in nonbilateral animals (green box in S7). Given the complexity of the gene and the lack of information on paralogous genes and different isoforms in acoels, we have included in the analysis both long and short isoforms from as many taxa across the Eumetazoa as possible (supplementary material, S10). The phylogenetic trees show that tropomyosins of the diploblasts (Cnidaria and Ctenophora) cluster separately from the tropomyosins of Bilateria. It is noteworthy that SrTrp clusters with the chordates’ muscular isoforms in the RAxML analyses (Fig. 1B and supplementary material, S6).
Remarkably, all our phylogenies would support a position of the acoels within the deuterostomes, as recently suggested by Philippe et al. (2011), and contradict the so far accepted basal postion of the acoels within the Bilateria. However, our phylogenies are based on single gene analysis (one single gene per tree) and the support values for the “deteurostome” topologies are low (all below 85).
Expression Pattern of SrAct, SrTnI, SrTrp Genes and the SrTrp Protein
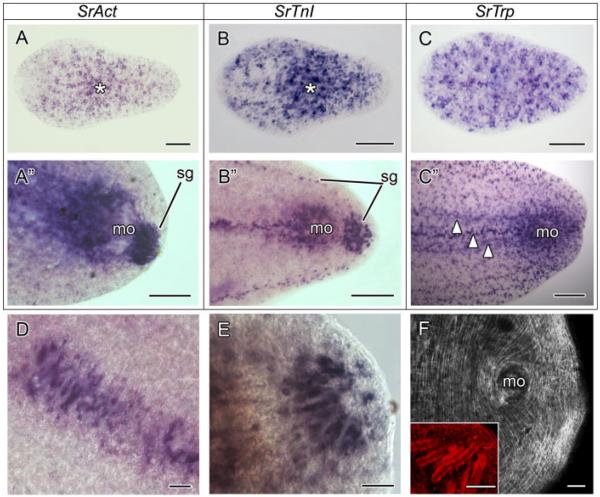
SrAct is broadly expressed in juveniles of S. roscoffensis. This pattern is not surprising because the gene is very likely to be expressed in differentiating myoblasts and myocytes, which are expected to be widely distributed along the whole body axis of a growing juvenile (Ladurner and Rieger, 2000; Semmler et al., 2008). However, the expression pattern of the gene is not uniform along the antero–posterior body axis (Fig. 2A). The number of SrAct positive cells is lower in the anterior-most part of the juvenile, around the frontal organ, whereas the highest density of positive cells is always found in the region of the mouth, where the U-shaped, accessory, and ring muscles are located (Fig. 2A, asterisk; Semmler et al., 2008). In adult worms, SrAct is also widely expressed along the body, but the highest signal is seen around the male genital opening and the region anterior to it (Fig. 2A”). The male genital opening is surrounded by circular and gonopore-specific muscles and is equipped with accessory muscles in the anterior region (Semmler et al., 2008), which is also strongly stained. The highest signal though is found in special muscular cells, the muscle mantles of sagittocysts (Fig. 2A”). These are present only in adults, between the female and male genital openings, at the posterior-most tip of the body, and along the posterior lateral edges (Fig. 2A”, B”, D, E; Semmler et al., 2008).
Figure 2.
SrAct, SrTnI, and SrTrp gene expression patterns in the acoel Symsagittifera roscoffensis. Anterior is toward the left in all aspects. (A) SrAct expression in a juvenile. The positive cells appear scattered along the AP axis of the juvenile. The highest concentration of actin-expressing cells is around the mouth region (asterisk), where the mouth accessory muscles and U-shaped muscles are present. (A”) SrAct expression in the posterior tip of an adult. The expression is restricted to the male genital opening (mo), the region frontal to it, and the muscle mantles of sagittocysts (sg). (B) SrTnI expression in a juvenile specimen. The expression of the troponin I gene appears scattered with the highest concentration of positive cells in the mouth region (asterisk). (B”) SrTnI expression in the posterior part of an adult. The expression of SrTnI is recovered in the region of the male gonopore (mo), in a longitudinal band anterior to it, and in the muscle mantles of sagittocyts at the posterior-most tip and in two lateral longitudinal bands (sg). (C) SrTrp expression in a juvenile. Contrary to the SrAct and the SrTnI genes, SrTrp is more broadly expressed along the body of the juvenile. The frontal region has a greater number of SrTrp-positive cells if compared with SrAct and SrTnI. (C”) SrTrp expression in the posterior part of an adult. As in the juvenile, the SrTrp-positive cells are more scattered than SrAct and SrTnI ones. High expression levels are seen in the region of the male gonopore and in longitudinal bands anterior to it (arrowheads). (D) Detail of the muscle mantles of the sagittocysts in the region between the female and the male genital opening as revealed by the anti-SrTnI riboprobe. (E) Detail of the muscle mantles of the saggitocysts, revealed by the anti-SrTnI riboprobe, at the posterior-most tip of an adult specimen. (F) Anti-SrTrp antibody staining in the caudal end of an adult. The antibody detects all types of muscles, except for the muscle mantles that surround the sagittocysts. The latter, labeled by the phalloidin, are shown in the inset. Scale bars: A-C 50 μm, A”–C” 100 μm, D–F and inset 25 μm. mo, male genital opening; Sg, muscle mantle of the sagittocyst.
The gene SrTnI is also broadly expressed in the juvenile S. roscoffensis. Its expression is reminiscent of that of SrAct. A lower number of SrTnI positive cells is found in the anterior part of the juvenile, whereas the majority of SrTnI-expressing cells congregate around the mouth (Fig. 2B, asterisk). In adults, the male genital opening and the muscle mantles of the sagittocysts are clearly stained by the SrTnI probe (Fig. 2B”).
The expression patterns of SrAct and SrTnI are remarkably similar, especially in the adult stage where the highest expression domains correspond to the male gonopore and the muscle mantles of the sagittocysts.
The tropomyosin gene, SrTrp, is expressed all along the body axis of juveniles (Fig. 2C). However, compared with SrAct and SrTnI, its expression is more broad and uniform. There is no lack of SrTrp-expressing cells in the frontal part of the animal and the higher density of positive cells in the mouth region is less pronouced (Fig. 2C). In the adult, similar to what we detect in the juvenile, the SrTrp-positive cells are more evenly distributed, compared with SrAct- and SrTnI-positive cells (Fig. 2C”). The highest expression of the gene occurs at the male gonopore. The strongly labeled longitudinal bands found in front of the male genital organ might coincide with known strong muscles present between the female and male gonopore, although they could also be longitudinal nerve cords or longitudinal bands of gland cells (Fig. 2C”, arrowheads). This has to be determined using detailed histological analysis. Surprisingly, no tropomyosin expression is found in the muscle mantles of the sagittocysts.
In all cases, control in situ hybridizations run with sense probes of all the three genes yielded no signal (data not shown).
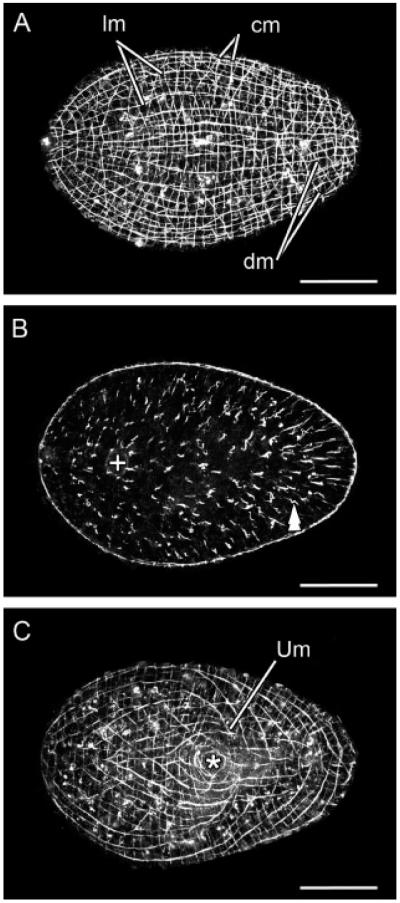
A better understanding of the tropomyosin pattern is uncovered by the use of a complementary tool, a specific antibody. The specific anti-SrTrp antibody recognizes the peptide LDKTNHQLDDANKE, which is located at the N-terminal region of the protein, covering the residues 64–77 of the deduced translation of the SrTrp gene. The anti-SrTrp antibody recognizes specifically the muscles of S. roscoffensis. When applied to juveniles, it reveals the body-wall musculature, the inner thick longitudinal, outer thin circular, and intermediate diagonal crossover muscles on the dorsal side (Fig. 3A), plus the longitudinal, diagonal, circular, and U-shaped muscles, as well as specialized ring muscles and accessory fibers associated with the ventral side of the mouth (Fig. 3C). Besides the body-wall musculature anti-SrTrp stains parenchymal muscles, showing that dorsoventral muscles are denser in the posterior half of the juvenile than in the anterior one (Fig. 3B, double arrowhead). This asymmetry correlates with the presence of prominent organs, such as the nervous system, the statocyst, and the frontal organ, which are all located in the anterior half of the body (Bery et al., 2010).
Figure 3.
SrTrp protein expression detected by a specific anti-SrTrp antibody in a juvenile specimen. Anterior is toward the left in all aspects. (A) Confocal projection of dorsal sections. Muscles of the body wall are evident. (B) Confocal projection of sections in the mid-body plane. Parenchymal muscles are highlighted by the double arrowhead. They run dorsoventrally and their density is higher in the region posterior to the mouth than anterior to it. The statocyst is highlighted by the symbol+. (C) Confocal projection of ventral sections. In addition to the longitudinal, circular, and diagonal muscles, specialized U-shaped muscles are present on the ventral side of the animal. The mouth opening (asterisk) is surrounded by specialized circular muscles. Scale bars: 50 μm in all aspects. hpa, hours post amputation. cm, circular muscles; dm, diagonal muscles; lm, longitudinal muscles; Um, U-shaped muscles.
In adult specimens, the overall organization of the body-wall musculature is maintained. Specialized muscles associated with the copulatory organs are detected at the ventral side. The male genital opening at the posterior-most tip has the greatest number of specialized muscles and is the region that shows the strongest signal (Fig. 2F). Contrary to the fluorophore-tagged phalloidin, which has been successfully applied in S. roscoffensis to label muscles and other structures, our antibody does not recognize any kind of sensory structure and does not mark the muscle mantles of sagittocysts, neither in juveniles nor in adult specimens (Semmler et al., 2008; Fig. 2F, inset).
SrTrp Gene and Protein Expression During Muscle Regeneration in S. roscoffensis
Once cut transversally in a median plane, S. roscoffensis is able to regenerate both the anterior and posterior halves of its body. Generally, the head fragment regenerates the missing half slightly faster than the tail fragment does (data not shown). The process of muscle regeneration is similar in both cases. For simplicity, we describe only the regeneration of the posterior end of the animal, within the first 24 hr after amputation. During this time frame, the common pattern of the body-wall muscles is completely restored. However, the copulatory organs are not formed until much later (personal observations).
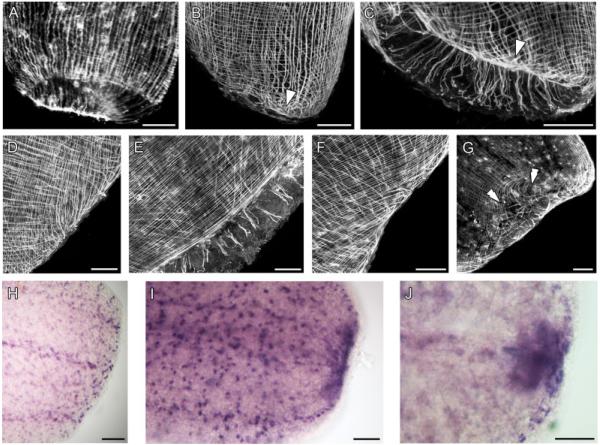
At the site of the wound, a translucent blastema becomes visible 12–16 hr after amputation (hpa) (Fig. 4I). Within 1 hpa, no major rearrangement of the muscles occurs, but a contraction of the circular muscles around the wound rim reduces its exposed surface (Fig. 4A). At 5 hpa, the preexisting body-wall muscles lose their regular arrangement, both at the dorsal and ventral sides of the animal but only in the region proximal to the wound (Fig. 4B and C). Along the rim of the wound, discrete regions of stronger signal are observed. At this time, the longitudinal muscles bend and converge to those regions (Fig. 4B and C, arrowhead). An outgrowth of dorsal longitudinal muscles occurs immediately after this local loss of the regular muscle pattern (about 9 hpa). The longitudinal fibers bend and grow toward the ventral side, thus provoking a ventral shift of the wound (Fig. 4D and E). By 16 hpa, the wound is closed and has completely shifted to the ventral side. In this area, the wound is covered by a faint web of young fibers, which are slender if compared with the old ones (Fig. 4G, double arrowheads). The dorsal side of the body wall has recovered, at this time, a more regular arrangement of the musclature (Fig. 4F). We have not observed any changes in the level of tropomyosin gene expression in connection with muscle rearrangement, growth, or differentiation within the first 16 hpa (Fig. 4H and I). At 16 hpa, a domain of increased tropomyosin expression appears next to the wound border (Fig. 4I). This domain of increased SrTrp expression persists until 24 hpa, even though it becomes progressively more restricted (Fig. 4J). At 24 hpa, the body-wall muscles have restored their original arrangement. The increased expression of the tropomyosin is a clear sign that new muscles are still differentiating in the posterior growing tip.
Figure 4.
Regeneration of the musculature in an adult S. roscoffensis revealed by anti-SrTrp antibody staining and in situ hybridization of the SrTrp gene. All specimens are anterior halves in the process of regenerating the posterior part of the animal. (A) 1 hpa. An initial contraction is visible. Anterior toward the top. (B) 5 hpa, dorsal view. (C) 5 hpa. Ventral view of the same specimen as in B. In both cases, disorganized muscles are evident at the wound border, both at the dorsal and ventral side (arrowhead). Anterior is up in both panels. (D) 9 hpa, dorsal view. Longitudinal muscles appear still disorganized, bending and growing toward the ventral side. Anterior is toward the top left. (E) 9 hpa, ventral view of the same specimen. In this case, no outgrowth of muscles is observed; thus, the wound is progressively shifted toward the ventral side. Anterior is toward the top left. (F) 16 hpa, dorsal view. The outgrowing muscles at the dorsal side of the animal seem to be more organized than in the previous stage. Anterior is toward the top left. (G) 16 hpa, ventral view of the same specimen as in F. The wound region (double arrowheads) has completely moved to the ventral side. The wound is now covered by a faint web of very thin muscles. Anterior is toward the top left. (H) SrTrp mRNA expression in a specimen 1 hpa. The levels of SrTrp transcript expression at the wound border are comparable to the expression level in the rest of the body. Anterior is toward the left. (I) 16 hpa. A translucent blastema has appeared in the region of the wound. Along the wound border, a domain of increased expression of the tropomyosin gene is now observed. Anterior is toward the left. (J) 24 hpa. The wound is already covered by the ciliated epidermis. A restricted domain of increased tropomyosin expression is still present in the former wound region, indicating that differentiation of new myocytes is still going on at this stage of regeneration. Anterior is toward the left. Scale bars: 50 μm in all aspects.
DISCUSSION
Muscular Genes in a Basal Bilaterian
Much of our knowledge on the physiology of muscular contraction comes from studies conducted in vertebrates (Gordon et al., 2000). Nevertheless, the three genes characterized in this study are known to interact in the striated muscles of both invertebrates and vertebrates (Bullard et al., ’73; Hooper and Thuma, 2005). The thin filaments of the muscles are double helices of F-actin (filaments of polymerized actin monomeres). In both smooth and striated muscles, the thin filaments interact with thick filaments made of myosin heavy chain, in order to produce the muscular contraction. Although in the striated muscles the thin and thick filaments are highly organized in a structure, sarcomere, giving these muscles their distinct appearance, they are arranged less strictly in the smooth muscles (Clark et al., 2002). Another difference between smooth and striated muscles is how they respond to variations in calcium concentration. When calcium levels increase in reaction to a stimulus, the tropomyosin is displaced from its resting position, allowing the myosin and the actin to interact and lead to the contraction (Lees-Miller and Helfman, ’91). Although in the smooth muscles the calcium response is mediated by a calmodulin–caldesmon complex, through the phosphorylation of a myosin light chain (Rasmussen et al., ’87; Kureishi et al., ’97; Morgan and Gangopadhyay, 2001), in the striated muscles the same mechanism is mediated by the proteins of the troponin complex (Galinska-Rakoczy et al., 2008; Lehman et al., 2009). This complex is formed by three different subunits: the calcium binding subunit (TnC), the tropomyosin-binding subunit (TnT), and the inhibitory subunit (TnI), whose main role is to inhibit the actomyosin ATPase by interacting at the same time with the actin, the tropomyosin, and the other two subunits of the complex (Farah and Reinach, ’95).
Actin belongs to a highly conserved multigene family present in all eukaryotes and is highly conserved. Each gene encodes for different isoforms which are classified into two main groups in the Metazoa: the muscular isoforms and the nonmuscular or cytoplasmic isoforms (Mounier and Sparrow, ’97). Although the differential usage of the isoforms is well understood in vertebrates and to a lower extent in the arthropods, this is far from being clear in other invertebrates (Mounier and Sparrow, ’97). The muscular isoforms arose independently from a cytoplasmic ancestor in the vertebrates (Vandekerckhove and Weber, ’84), the insects (most likely in the whole Ecdysozoa) (Mounier et al., ’92), and most probably in the lophotrochozoans as well (Carlini et al., 2000). Whether there are muscle- and nonmuscle-specific actins in cnidarians is not clear yet (Aerne et al., ’93).
In our phylogenetic analysis, SrAct resembles more the deuterostomes’ muscular actins. This result is surprising, because it has been shown that the chordates’ muscular actins arose independently from a cytoplasmic ancestor (Vandekerckhove and Weber, ’84). However, care should be taken evaluting these results owing to the extreme conservation and the frequent occurrence of adaptive substitutions of amino acids in this protein (Mounier and Sparrow, ’97).
In juveniles, SrAct is broadly expressed from the anterior to the posterior end of the animal. However, the presence of unstained cells and tissues, such as in the anterior-most region, suggests that SrAct is not ubiquitous (as expected for a cytoplasmic actin) and that it might be only specific of myoblasts and differentiated myocytes. Differentiating myoblasts and myocytes are expected to be widely distributed, especially in the body of a growing juvenile. Additionally, it is known for both invertebrates and vertebrates (Cox et al., ’86; Sassoon et al., ’88; Kelly et al., 2002) that actin transcripts accumulate in myoblasts before they differentiate into myocytes. Expression analysis of SrAct in adult worms further supports its muscular role. The highest signal is recovered around the male gonopore, which is the most muscular structure of adults and in the muscle mantles of sagittocysts (Semmler et al., 2008). The latter are extrusomes used for defense and copulation, and are produced by a specialized cell, the sagittocyte. The distal tip of the sagittocyte is wrapped by a muscular cell (the muscle mantle), the contraction of which causes the discharge of the sagittocyst (Gschwentner et al., 2002).
Although actin and tropomyosin proteins exist in different isoforms and are present both in muscular and nonmuscular cells (Lees-Miller and Helfman, ’91; Pittenger et al., ’94; Mounier and Sparrow, ’97), troponins are only known from muscles of bilateral animals, whereas no troponin orthologs have been found in the available nonbilaterian genomes (U. Technau, personal communication). Unquestionably, SrTnI is a muscular gene, as it is shown to be the homolog of the inhibitory subunit of the troponin complex in our phylogenetic analyses and by its expression in the male genital opening and in the adults’ muscle mantles of the sagittocysts. The expression of SrTnI is strikingly similar to the expression of SrAct in juveniles, thus the coexpression of the two genes is likely. Troponin proteins are key regulators of the muscular contraction in the sarcomeric muscles of both invertebrates and vertebrates, with the only exception known from the ascidian Ciona intestinalis, whose body-wall smooth muscles contain troponin and a striated muscle isoform of the tropomyosin (Meedel and Hastings, ’93; Endo et al., ’96). Interestingly, S. roscoffensis has exclusively smooth muscles (Semmler et al., 2008), a condition likely to be ancestral for all acoels, because no striated muscles have ever been described in any acoel species so far investigated (Rieger et al., ’91; Hooge, 2001).
Tropomyosin is an elongated protein that assembles in dimers and forms a coiled-coil structure. In the vertebrates’ muscles, but very likely in the invertebrates’ muscles too (Bullard et al., ’73; Lehman et al., 2000), each tropomyosin dimer lies in the groove of the filamentous actin and its role is to inhibit the actin–myosin interaction, thus preventing contraction during resting conditions. Tropomyosin genes exist in multiple copies in the genomes of all metazoans as well as in other eukaryotes so far sequenced (Lees-Miller et al., ’90; Lees-Miller and Helfman, ’91; Irimia et al., 2010). In the Bilateria, one tropomyosin gene encodes for short (about 250 aa) and long (usually 284 aa) isoforms. Any tropomyosin gene can generate several different isoforms, either by alternative splicing or differential promoter usage (Lees-Miller and Helfman, ’91). The two forms (long and short) differ by a 38 amino acid-long peptide at the N-terminus of the protein (Greenfield et al., ’98), which is highly conserved among distantly related bilaterians, although it is never recovered in nonbilateral animals (green box in supplementary material, S7). This domain is necessary for head-to-tail interactions between two consecutive tropomyosins and for their stability in the F-actin furrow. Mutations in the N-terminal domain result in loss of affinity of the tropomyosin for the actin filaments (Greenfield et al., ’98). With a few exceptions (Weinberger et al., ’93; Pittenger et al., ’94; Perry, 2001), the long forms are expressed in muscles, whereas the short isoforms are expressed in other cell types (Lees-Miller and Helfman, ’91; Pittenger et al., ’94; Irimia et al., 2010). In line with these findings in other organisms, we show that the long tropomyosin SrTrp (supplementary material, S7) is muscular, using a specific anti-SrTrp antibody. However, it is possible that the ISH of this gene could be misleading, because the riboprobe generated against the full length of the tropomyosin clone might recognize transcripts of the short tropomyosin as well, thus labeling muscles and, perhaps, also other cell types (Pittenger et al., ’94). This would explain why SrTrp seems more widely expressed than SrAct and SrTnI.
Accordingly, one could interpret the longitudinal bands of cells strongly stained in the region frontal to the male genital opening as nerve cords (Fig. 2C”), a real possibility because some long isoforms of the tropomyosin are known to be present in the nervous system (Weinberger et al., ’93). However, we have to point out that S. roscoffensis has six longitudinal nerve cords (Bery et al., 2010; Semmler et al., 2010), meaning that four of the nerve cords would not express the gene. In our view, there are two ways to explain this domain of expression. First, these cells could be accessory muscles, because the region anterior to the male gonopore is rich with them (Semmler et al., 2008; this study), or alternatevely, they could be special gland cells, which we have found to be distributed in a paired manner that coincides with the observed pattern (unpublished data).
The genes SrAct, SrTnI, and SrTrp show mostly overlapping expression domains in both juvenile and adult S. roscoffensis, thus suggesting that in the acoel the three proteins might physically interact, as they do in the striated muscles of other bilaterians. However, this might be restricted only to a subset of muscles because tropomyosin is not transcribed (no ISH signal) or translated (no antibody signal), for instance, in the muscle mantles of sagittocysts (see below).
Tropomyosin Expression During Muscle Regeneration in the Acoel S. roscoffensis
In acoels, the dynamic pattern of muscular gene expression can be best studied in regenerating animals.
The first sign of a reorganization of the musculature is a constriction of the circular muscles along the rim of the wound. As seen in Macrostomum lignano, the initial constriction might help in reducing the wound surface (Salvenmoser et al., 2001). Subsequently, the muscles of the body wall lose their local regular organization, and some longitudinal fibers grow from the dorsal to the ventral side, causing a ventral shift of the wound. The wound closure by an initial layer of preexisting muscles takes approximately 12 hr. Right afterwards, an undifferentiated blastema becomes visible, the tropomyosin gene expression is upregulated in the region behind the blastema, and most likely new myocytes begin to differentiate. It is very likely (as is the case in M. lignano) that in addition to help in closing the wound, the old fibers also serve as scaffold for the growing blastema (Salvenmoser et al., 2001).
During regeneration of the platyhelminth planarian Girardia tigrina, muscles are formed anew in the blastema, without any previous wound closure. However, few of the old fibers are observed to invade the blastema having, probably, the function of guiding differentiating cells into it (Cebrià et al., ’97). Similarly, during the regeneration of the nervous system in Convolutriloba longifissura and S. roscoffensis, some of the existing nerve cords invade the blastema before the differentiation of new neurons starts (Gaerber et al., 2007; Bery and Martinez, 2011). It is, therefore, likely that old muscle fibers provide guidance cues for undifferentiated cells once they migrate into the blastema during the regeneration of S. roscoffensis.
The tropomyosin gene is up-regulated in the wound area, simultaneously or immediately after to the wound closure by preexisting muscles. The up-regulation of the gene indicates that differentiation of myocytes might occur in this area. In fact, accumulation of the tropomyosin transcripts has been observed in invertebrates and vertebrates during embryonic muscle development. In the gastropod Haliotis rufescens, the tropomyosin gene is expressed before myofibrillogenesis (Degnan et al., ’97), whereas in Xenopus laevis, tropomyosin transcripts accumulate in the somites and in the embryonic heart long before mature myocytes are formed (Gaillard et al., ’98).
The great regenerative capacity of acoels is owing to their unique stem cell system. The stem cells, neoblasts, are located exclusively in the parenchyma and they are able to differentiate in any cell type. A subpopulation of neoblasts expresses the gene piwi, which in other bilaterians is a germ line specification factor (De Mulder et al., 2009a,b; Egger et al., 2009). During regeneration of I. pulchra, piwi-expressing cells appear in the blastema approximately 1 day after amputation. These cells have most likely migrated into the blastema from other regions of the body (De Mulder et al., 2009a). We propose here that in S. roscoffensis a similar migration of undifferentiated cells could occur after the wound has been closed by preexisting muscle fibers. These migrating cells would be guided into the newly formed blastema by the same fibers. After proliferating, the neoblasts would initiate their differentiation into various cell types, by expressing tissue-specific genes (e.g. SrTrp in differentiating myocytes).
Evolutionary Implications
Our data show that the smooth muscles of S. roscoffensis have a molecular architecture similar to the striated muscles of other bilaterians, although only smooth muscles have been reported in Acoela (Rieger et al., ’91; Hooge, 2001). In line with this and taking into account most recent animal phylogenies (Egger et al., 2009; Hejnol et al., 2009) that suggest the Acoela as the earliest offshoot of the Bilateria, it is tempting to suggest that the segregation of specialized muscle cells from “ancestral” epithelial muscle cells coincided with the diploblast–triploblast transition. Accordingly, the first bilateral animals possessed only smooth muscles with the molecular repertoire necessary to build a striated muscle.
Even though it has been proposed that the evolution of the long tropomyosin of the Bilateria might be linked to the evolution of the sarcomere (Irimia et al., 2010), and that the same reasoning could be applied to the evolution of the troponin which do not exist in nonbilaterians (U. Technau, personal communication), our data suggests that it is more parsimonious to regard striated muscle cells as a sister cell type to the smooth muscle cells. In this scenario, striated and smooth muscles would have arisen in the stem lineage that led to the Nephrozoa (i.e. all Bilateria exclusive the acoelomorphs) (Hejnol et al., 2009), from an “acoel-like” smooth muscle, by segregation and divergence of functions and through differential recruitment of additional genes (Arendt, 2008).
In myocytes, which express troponin genes, the myofibrils would have assembled eventually into a sarcomere, whereas the smooth muscles of the nephrozoans recruited new components, such as the calmodulin and the caldesmon among others, to regulate their contraction.
However, the case is far from being settled. Myocytes (true muscular cells ) are also present in Cnidaria and Ctenophora (Seipel and Schmid, 2005), and cnidarian muscles express genes that are also present in bilaterian muscles (Schuchert et al., ’93; Groger et al., ’99; Renfer et al., 2010). Striated muscular cells have been described for one species of the ctenophores and in all medusozoan cnidarians (cubozoans, hydrozoans, and scyphozoans). However, the latter are the most derived classes of cnidarians and their striated muscles have a different ultrastructure from the bilaterian ones, making homology very unlikely (Burton, 2008).
In most bilaterians, smooth and striated muscles coexist and in the invertebrates the distinction between striated and smooth muscles, on a molecular basis, is not as clearly defined as in the vertebrates (Hooper and Thuma, 2005). For example, the smooth body-wall muscles of the ascidian C. intestinalis are regulated by the troponin (Endo et al., ’96), or in the planarian Dugesia japonica, striated muscular isoforms of the myosin heavy chain are expressed in its smooth muscles (Kobayashi et al., ’98). The latter example would indicate, for instance, that most likely in turbellarian flatworms striated muscles have been reduced (Ruppert et al., 2004). All these cases suggest that a “striated muscle” molecular architecture in smooth muscles is not exceptional to acoels. The independent loss of the sarcomeric organization in the muscles of some lineages, such as the planarians or Ciona, would be easily explained as an adaptation to a lifestyle that does not require the presence of fast striated muscles. Admittedly, the same argument could be applied to acoels as well. In this case, the ancestor of all Bilateria could have had striated muscles and they have been lost in the Acoela, among other lineages.
Philippe et al. (2011) have recently proposed that the Acoelomorpha (i.e. acoels+nemertodermatids) and Xenoturbella group together within the deuterostomes (as already suggested in a previous article by Philippe et al., 2007) instead of being basal bilaterians (Hejnol et al., 2009). If this scenario would be true, referring to the Acoelomorpha condition (or Xenoacoelomorpha, sensu Philippe et al., 2011) as ancestral to all Bilateria would be unfounded. Now we would have to assume following the most parsimonious reasoning that the protostome–deuterostome ancestor would have had striated muscles that have been lost in some lineages of the protostome (e.g. in turbellarian flatworms) and most probably in the whole Xenoacoelomorpha.
Obviously, to better understand the evolution of muscles, a final settlement within the metazoan tree of pivotal groups, such as Ctenophora, Acoelomorpha, and Xenoturbella, would be critical. Additionally, the acquisition of molecular data from Xenoturbella, which also exhibits only smooth muscles (Ehlers and Sopott-Ehlers, ’97), and from the Nemertodermatida would be essential as well. However, it must be pointed out that we need further analyses of the connection between molecular composition of muscles and their morphology and function, before a final reconstruction of the stepwise evolution of musculature in the Metazoa is possible.
In summary, though we are far from a complete understanding of how the various types of muscles evolved over time, at present, three points may be considered:
Myocytes evolved from epithelial muscle cells (Rieger and Ladurner, 2003).
The first true myocytes were most likely of the smooth type, as it is hardly possible that such a complex and organized structure as the sarcomere evolved promptly from an epithelial muscle cell type.
The striated muscle cell type is not the sister of “the” smooth muscle type, but to one of numerous smooth cell types as there are cryptic subtypes of smooth muscles even in basal cnidarians (Renfer et al., 2010).
Supplementary Material
ACKNOWLEDGMENTS
P.M. and M.C. are supported by the Spanish Ministry of Science (BFU2006-00898/BMC), A.W. by the EU-funded Marie Curie Network MOLMORPH (contract grant number MEST-CT-2005-020542). M.C. was a recipient of a SYNTHESYS Fellowship (DK-TAF-4568), to cover the research stay in the laboratory of AW. The Austrian Science Fund (FWF) supported JGA by means of an Erwin Schrödinger Fellowship, grant number J 3029-B17. We thank Dr. Henrike Semmler and Alen Kristof for the help with confocal microscopy. We are also indebted to Arnau Sebé Pedrós and Dr. Ignacio Maeso Martín for helping with the phylogenetic analysis. Finally, we thank the two anonymous referees, whose insightful comments improved the final version of this manuscript.
Grant Sponsor: “Ministerio de Ciencia y Tecnologia” Spain; Grant number: BFU2006-00898; Grant Sponsor: Marie Curie Network MOLMORPH; Grant number: MEST-CT-2005-020542.
Footnotes
Additional Supporting Information may be found in the online version of this article.
LITERATURE CITED
- Abascal F, Zardoya R, Posada D. Prottest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Aerne BL, Schmid V, Schuchert P. Actin-encoding genes of the hydrozoan Podocoryne carnea. Gene. 1993;131:183–192. doi: 10.1016/0378-1119(93)90292-b. [DOI] [PubMed] [Google Scholar]
- Arendt D. The evolution of cell types in animals: emerging principles from molecular studies. Nat Rev. 2008;9:868–882. doi: 10.1038/nrg2416. [DOI] [PubMed] [Google Scholar]
- Bery A, Cardona A, Martinez P, Hartenstein V. Structure of the central nervous system of a juvenile acoel, Symsagittifera roscoffensis. Dev Genes Evol. 2010;220:61–76. doi: 10.1007/s00427-010-0328-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bery A, Martinez P. Acetylcholinesterase activity in the developing and regenerating nervous system of the acoel Symsagittifera roscoffensis. Acta Zoologica. 2011 In press. [Google Scholar]
- Bullard B, Dabrowska R, Winkelman L. The contractile and regulatory proteins of insect flight muscle. Biochem J. 1973;135:277–286. doi: 10.1042/bj1350277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton PM. Insights from diploblasts; the evolution of mesoderm and muscle. J Exp Zool B (Mol Dev Evol) 2008;310:5–14. doi: 10.1002/jez.b.21150. [DOI] [PubMed] [Google Scholar]
- Carlini DB, Reece KS, Graves JE. Actin gene family evolution and the phylogeny of coleoid cephalopods (mollusca: Cephalopoda) Mol Biol Evol. 2000;17:1353–1370. doi: 10.1093/oxfordjournals.molbev.a026419. [DOI] [PubMed] [Google Scholar]
- Cebrià F, Vispo M, Newmark P, Bueno D, Romero R. Myocyte differentiation and body wall muscle regeneration in the planarian Girardia tigrina. Dev Genes Evol. 1997;207:306–316. doi: 10.1007/s004270050118. [DOI] [PubMed] [Google Scholar]
- Clark KA, McElhinny AS, Beckerle MC, Gregorio CC. Striated muscle cytoarchitecture: an intricate web of form and function. Annu Rev Cell Dev Biol. 2002;18:637–706. doi: 10.1146/annurev.cellbio.18.012502.105840. [DOI] [PubMed] [Google Scholar]
- Cox KH, Angerer LM, Lee JJ, Davidson EH, Angerer RC. Cell lineage-specific programs of expression of multiple actin genes during sea urchin embryogenesis. J Mol Biol. 1986;188:159–172. doi: 10.1016/0022-2836(86)90301-3. [DOI] [PubMed] [Google Scholar]
- De Mulder K, Kuales G, Pfister D, Willems M, Egger B, Salvenmoser W, Thaler M, Gorny AK, Hrouda M, Borgonie G, Ladurner P. Characterization of the stem cell system of the acoel Isodiametra pulchra. BMC Dev Biol. 2009a;9:69. doi: 10.1186/1471-213X-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mulder K, Pfister D, Kuales G, Egger B, Salvenmoser W, Willems M. Stem cells are differentially regulated during development, regeneration and homeostasis in flatworms. Dev Biol. 2009b;334:198–212. doi: 10.1016/j.ydbio.2009.07.019. [DOI] [PubMed] [Google Scholar]
- Degnan BM, Degnan SM, Morse DE. Muscle-specific regulation of tropomyosin gene expression and myofibrillogenesis differs among muscle systems examined at metamorphosis of the gastropod Haliotis rufescens. Dev Genes Evol. 1997;206:464–471. doi: 10.1007/s004270050076. [DOI] [PubMed] [Google Scholar]
- Drummond A, Ashton B, Buxton S, Cheung M, Cooper A, Heled J, Kearse M, Moir R, Stones-Havas S, Sturrock S, Thierer T, Wilson A. Geneious v5.1. 2010 available from http://www.Geneious.com.
- Dunn C, Hejnol A, Matus D, Pang K, Browne W, Smith S. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452:745–749. doi: 10.1038/nature06614. [DOI] [PubMed] [Google Scholar]
- Egger B, Steinke D, Tarui H, De Mulder K, Arendt D, Borgonie G, Funayama N, Gschwentner R, Hartenstein V, Hobmayer B, Hooge M, Hrouda M, Ishida S, Kobayashi C, Kuales G, Nishimura O, Pfister D, Rieger R, Salvenmoser W, Smith J, Technau U, Tyler S, Agata K, Salzburger W, Ladurner P. To be or not to be a flatworm: the acoel controversy. PLoS ONE. 2009;4:e5502. doi: 10.1371/journal.pone.0005502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers U, Sopott-Ehlers B. Ultrastructure of the subepidermal musculature of Xenoturbella bocki, the adelphotaxon of the bilateria. Zoomorphology. 1997;117:71–79. [Google Scholar]
- Endo T, Matsumoto K, Hama T, Ohtsuka Y, Katsura G, Obinata T. Distinct troponin T genes are expressed in embryonic/larval tail striated muscle and adult body wall smooth muscle of ascidian. J Biol Chem. 1996;271:27855–27862. doi: 10.1074/jbc.271.44.27855. [DOI] [PubMed] [Google Scholar]
- Farah C, Reinach F. The troponin complex and regulation of muscle contraction. FASEB J. 1995;9:755–767. doi: 10.1096/fasebj.9.9.7601340. [DOI] [PubMed] [Google Scholar]
- Gaerber C, Salvenmoser W, Rieger R, Gschwentner R. The nervous system of Convolutriloba (Acoela) and its patterning during regeneration after asexual reproduction. Zoomorphology. 2007;126:73–87. [Google Scholar]
- Gaillard C, Theze N, Hardy S, Allo MR, Ferrasson E, Thiebaud P. Alpha-tropomyosin gene expression in Xenopus laevis: differential promoter usage during development and controlled expression by myogenic factors. Dev Genes Evol. 1998;207:435–445. doi: 10.1007/s004270050134. [DOI] [PubMed] [Google Scholar]
- Galinska-Rakoczy A, Engel P, Xu C, Jung H, Craig R, Tobacman LS, Lehman W. Structural basis for the regulation of muscle contraction by troponin and tropomyosin. J Mol Biol. 2008;379:929–935. doi: 10.1016/j.jmb.2008.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon AM, Homsher E, Regnier M. Regulation of contraction in striated muscle. Physiol Rev. 2000;80:853–924. doi: 10.1152/physrev.2000.80.2.853. [DOI] [PubMed] [Google Scholar]
- Greenfield NJ, Montelione GT, Farid RS, Hitchcock-DeGregori SE. The structure of the N-terminus of striated muscle alpha-tropomyosin in a chimeric peptide: nuclear magnetic resonance structure and circular dichroism studies. Biochemistry. 1998;37:7834–7843. doi: 10.1021/bi973167m. [DOI] [PubMed] [Google Scholar]
- Groger H, Callaerts P, Gehring WJ, Schmid V. Gene duplication and recruitment of a specific tropomyosin into striated muscle cells in the jellyfish podocoryne carnea. J Exp Zool B (Mol Dev Evol) 1999;285:378–386. [PubMed] [Google Scholar]
- Gschwentner R, Baric S, Rieger R. New model for the formation and function of sagittocysts: Symsagittifera corsicae n. Sp. (Acoela) Invertebr Biol. 2002;121:95–103. [Google Scholar]
- Haszprunar G. Plathelminthes and Plathelminthomorpha; paraphyletic taxa. J Zoo Syst Evol Res. 1996;34:41–48. [Google Scholar]
- Hejnol A, Martindale MQ. Acoel development indicates the independent evolution of the bilaterian mouth and anus. Nature. 2008;456:382–386. doi: 10.1038/nature07309. [DOI] [PubMed] [Google Scholar]
- Hejnol A, Martindale MQ. Coordinated spatial and temporal expression of Hox genes during embryogenesis in the acoel Convolutriloba Longifissura. BMC Biol. 2009;7:65. doi: 10.1186/1741-7007-7-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hejnol A, Obst M, Stamatakis A, Ott M, Rouse GW, Edgecombe GD, Martinez P, Baguna J, Bailly X, Jondelius U, Wiens M, Muller WE, Seaver E, Wheeler WC, Martindale MQ, Giribet G, Dunn CW. Assessing the root of bilaterian animals with scalable phylogenomic methods. Proc R Soc Lond [Biol] 2009;276:4261–4270. doi: 10.1098/rspb.2009.0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry J, Martindale M, Boyer B. The unique developmental program of the acoel flatworm, Neochildia fusca. Dev Biol. 2000;220:285–295. doi: 10.1006/dbio.2000.9628. [DOI] [PubMed] [Google Scholar]
- Hooge MD. Evolution of body-wall musculature in the Platyhelminthes (Acoelomorpha, Catenulida, Rhabditophora) J Morphol. 2001;249:171–194. doi: 10.1002/jmor.1048. [DOI] [PubMed] [Google Scholar]
- Hooge MD, Tyler S. New tools for resolving phylogenies: a systematic revision of the Convolutidae (Acoelomorpha, Acoela) J Zool Syst Evol Res. 2005;43:100–113. [Google Scholar]
- Hooper SL, Thuma JB. Invertebrate muscles: muscle-specific genes and proteins. Physiol Rev. 2005;85:1001–1060. doi: 10.1152/physrev.00019.2004. [DOI] [PubMed] [Google Scholar]
- Hopp TP, Woods KR. Prediction of protein antigenic determinants from amino acid sequences. PNAS. 1981;78:3824–3828. doi: 10.1073/pnas.78.6.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia M, Maeso I, Gunning PW, Garcia-Fernandez J, Roy SW. Internal and external paralogy in the evolution of tropomyosin genes in metazoans. Mol Biol Evol. 2010;27:1504–1517. doi: 10.1093/molbev/msq018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly KK, Meadows SM, Cripps RM. Drosophila mef2 is a direct regulator of actin57b transcription in cardiac, skeletal, and visceral muscle lineages. Mech Dev. 2002;110:39–50. doi: 10.1016/s0925-4773(01)00586-x. [DOI] [PubMed] [Google Scholar]
- Kobayashi C, Kobayashi S, Orii H, Watanabe K, Agata K. Identification of two distinct muscles in the planarian Dugesia japonica by their expression of myosin heavy chain genes. Zool Sci. 1998;15:861–869. [Google Scholar]
- Kureishi Y, Kobayashi S, Amano M, Kimura K, Kanaide H, Nakano T, Kaibuchi K, Ito M. Rho-associated kinase directly induces smooth muscle contraction through myosin light chain phosphorylation. J Biol Chem. 1997;272:12257–12260. doi: 10.1074/jbc.272.19.12257. [DOI] [PubMed] [Google Scholar]
- Ladurner P, Rieger R. Embryonic muscle development of Convoluta pulchra (Turbellaria-Acoelomorpha, Platyhelminthes) Dev Biol. 2000;222:359–375. doi: 10.1006/dbio.2000.9715. [DOI] [PubMed] [Google Scholar]
- Lees-Miller JP, Goodwin LO, Helfman DM. Three novel brain tropomyosin isoforms are expressed from the rat alpha-tropomyosin gene through the use of alternative promoters and alternative RNA processing. MCB. 1990;10:1729–1742. doi: 10.1128/mcb.10.4.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees-Miller JP, Helfman DM. The molecular basis for tropomyosin isoform diversity. Bioessays. 1991;13:429–437. doi: 10.1002/bies.950130902. [DOI] [PubMed] [Google Scholar]
- Lehman W, Hatch V, Korman V, Rosol M, Thomas L, Maytum R, Geeves MA, Van Eyk JE, Tobacman LS, Craig R. Tropomyosin and actin isoforms modulate the localization of tropomyosin strands on actin filaments. J Mol Biol. 2000;302:593–606. doi: 10.1006/jmbi.2000.4080. [DOI] [PubMed] [Google Scholar]
- Lehman W, Galinska-Rakoczy A, Hatch V, Tobacman LS, Craig R. Structural basis for the activation of muscle contraction by troponin and tropomyosin. J Mol Biol. 2009;388:673–681. doi: 10.1016/j.jmb.2009.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martindale MQ, Henry JQ. Intracellular fate mapping in a basal metazoan, the ctenophore Mnemiopsis leidyi, reveals the origins of mesoderm and the existence of indeterminate cell lineages. Dev Biol. 1999;214:243–257. doi: 10.1006/dbio.1999.9427. [DOI] [PubMed] [Google Scholar]
- Martindale MQ, Pang K, Finnerty JR. Investigating the origins of triploblasty: “mesodermal” gene expression in a diploblastic animal, the sea anemone Nematostella vectensis (phylum, Cnidaria; class, Anthozoa) Development. 2004;131:2463–2474. doi: 10.1242/dev.01119. [DOI] [PubMed] [Google Scholar]
- Meedel TH, Hastings KE. Striated muscle-type tropomyosin in a chordate smooth muscle, ascidian body-wall muscle. J Biol Chem. 1993;268:6755–6764. [PubMed] [Google Scholar]
- Moreno E, Nadal M, Baguna J, Martinez P. Tracking the origins of the bilaterian Hox patterning system: insights from the acoel flatworm Symsagittifera roscoffensis. Evol Dev. 2009;11:574–581. doi: 10.1111/j.1525-142X.2009.00363.x. [DOI] [PubMed] [Google Scholar]
- Morgan KG, Gangopadhyay SS. Signal transduction in smooth muscle: invited review: cross-bridge regulation by thin filament-associated proteins. J Appl Physiol. 2001;91:953–962. doi: 10.1152/jappl.2001.91.2.953. [DOI] [PubMed] [Google Scholar]
- Mounier N, Gouy M, Mouchiroud D, Prudhomme JC. Insect muscle actins differ distinctly from invertebrate and vertebrate cytoplasmic actins. J Mol Evol. 1992;34:406–415. doi: 10.1007/BF00162997. [DOI] [PubMed] [Google Scholar]
- Mounier N, Sparrow JC. Structural comparisons of muscle and nonmuscle actins give insights into the evolution of their functional differences. J Mol Evol. 1997;44:89–97. doi: 10.1007/pl00006125. [DOI] [PubMed] [Google Scholar]
- Perry SV. Vertebrate tropomyosin: distribution, properties and function. J Muscle Res Cell Motil. 2001;22:5–49. doi: 10.1023/a:1010303732441. [DOI] [PubMed] [Google Scholar]
- Philippe H, Brinkman H, Martinez P, Riutort M, Baguña J. Acoel flatworms are not platyhelminthes: evidences from phylogenomics. PLoS ONE. 2007;2:e717. doi: 10.1371/journal.pone.0000717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H, Brinkman H, Copley RR, Moroz LL, Nakano H, Poustka AJ, Wallberg A, Peterson KJ, Teldford MJ. Acoelomorph flatworms are deuterostomes related to Xenoturbella. Nature. 2011;470:255–258. doi: 10.1038/nature09676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittenger MF, Kazzaz JA, Helfman DM. Functional properties of non-muscle tropomyosin isoforms. Curr Opin Cell Biol. 1994;6:96–104. doi: 10.1016/0955-0674(94)90122-8. [DOI] [PubMed] [Google Scholar]
- Rasmussen H, Takuwa Y, Park S. Protein kinase C in the regulation of smooth muscle contraction. FASEB J. 1987;1:177–185. [PubMed] [Google Scholar]
- Renfer E, Amon-Hassenzahl A, Steinmetz PRH, Technau U. A muscle-specific transgenic reporter line of the sea anemone, Nematostella vectensis. PNAS. 2010;107:104–108. doi: 10.1073/pnas.0909148107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieger RM, Ladurner P. The significance of muscle cells for the origin of mesoderm in bilateria. Integr Comp Biol. 2003;43:47–54. doi: 10.1093/icb/43.1.47. [DOI] [PubMed] [Google Scholar]
- Rieger RM, Tyler S, Smith JPS, III, Rieger GE. Platyhelminthes: Turbellaria. In: Harrison B, editor. Microscopic anatomy of invertebrates. Wiley-Liss, Inc.; New York: 1991. pp. 7–140. [Google Scholar]
- Ruppert EE, Fox RS, Barnes RD. Invertebrate zoology. Brooks/Cole-Thomson Learning; Belmont, CA: 2004. [Google Scholar]
- Salvenmoser W, Riedl D, Ladurner P, Rieger R. Early steps in the regeneration of the musculature in Macrostomum sp. (Macrostomorpha) Belgian J Zool. 2001;131:105–109. [Google Scholar]
- Sassoon DA, Garner I, Buckingham M. Transcripts of alpha-cardiac and alpha-skeletal actins are early markers for myogenesis in the mouse embryo. Development. 1988;104:155–164. doi: 10.1242/dev.104.1.155. [DOI] [PubMed] [Google Scholar]
- Scholz C, Technau U. The ancestral role of brachyury expression nembra1 in the basal cnidarian Nematostella vectensis. Dev Genes Evol. 2003;212:563–570. doi: 10.1007/s00427-002-0272-x. [DOI] [PubMed] [Google Scholar]
- Schuchert P, Reber-Muller S, Schmid V. Life stage specific expression of a myosin heavy chain in the hydrozoan Podocoryne carnea. Differentiation. 1993;54:11–18. doi: 10.1111/j.1432-0436.1993.tb00654.x. [DOI] [PubMed] [Google Scholar]
- Seipel K, Schmid V. Evolution of striated muscle: jellyfish and the origin of triploblasty. Dev Biol. 2005;282:14–26. doi: 10.1016/j.ydbio.2005.03.032. [DOI] [PubMed] [Google Scholar]
- Semmler H, Bailly X, Wanninger A. Myogenesis in the basal bilaterian Symsagittifera roscoffensis (Acoela) Front Zool. 2008;5:14. doi: 10.1186/1742-9994-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semmler H, Chiodin M, Bailly X, Martinez P, Wanninger A. Steps towards a centralized nervous system in basal bilaterians: insights from neurogenesis of the acoel Symsagittifera roscoffensis. Dev Growth Differ. 2010;52:701–713. doi: 10.1111/j.1440-169X.2010.01207.x. [DOI] [PubMed] [Google Scholar]
- Sikes J, Bely A. Radical modification of the a-p axis and the evolution of asexual reproduction in Convolutriloba acoels. Evol Dev. 2008;10:619–631. doi: 10.1111/j.1525-142X.2008.00276.x. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Technau U, Scholz CB. Origin and evolution of endoderm and mesoderm. Int J Dev Biol. 2003;47:531–539. [PubMed] [Google Scholar]
- Vandekerckhove J, Weber K. Chordate muscle actins differ distinctly from invertebrate muscle actins: the evolution of the different vertebrate muscle actins. J Mol Biol. 1984;179:391–413. doi: 10.1016/0022-2836(84)90072-x. [DOI] [PubMed] [Google Scholar]
- Wanninger A. Shaping the things to come: ontogeny of Lophotrochozoan Neuromuscular systems and the Tetraneuralia concept. Biol Bull. 2009;216:293–306. doi: 10.1086/BBLv216n3p293. [DOI] [PubMed] [Google Scholar]
- Weinberger RP, Henke RC, Tolhurst O, Jeffrey PL, Gunning P. Induction of neuron-specific tropomyosin mRNAs by nerve growth factor is dependent on morphological differentiation. J Cell Biol. 1993;120:205–215. doi: 10.1083/jcb.120.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.