Abstract
In this Commentary, we describe a cryptographic method for returning research results to individuals who participate in clinical studies. Controlled use of this method, which relaxes the typical anonymization guarantee, can ensure that clinically actionable results reach participants while also addressing most privacy concerns.
To protect privacy, the current regulatory framework for research on human subjects encourages the use of anonymization—a guarantee that a biological sample cannot be associated with the research participant who donated it. There are multiple approaches to anonymization used by biobanks—repositories for human biological samples—and many are based on the Health Insurance Portability and Accountability Act Privacy Rule (U.S. 65 FR 82462) as well as others (for example, the Vanderbilt Synthetic Derivative) (1). Although there are proposed alternatives to anonymization that provide more flexibility regarding return of information to participants (2), regulatory requirements associated with these frameworks drive most biobanks to choose anonymization. We propose a cryptographic mechanism that relaxes the anonymization guarantee under carefully controlled circumstances, addressing this harm at a modest cost to privacy.
A promise of anonymity is often a central aspect of the informed consent process (3–5). Although not its primary purpose, anonymity also prevents the return of research results to an individual participant; thus, a participant cannot be informed about any clinically actionable information obtained through the research. Only non-identifiable aggregate results of a research study are typically made available through publications in scientific journals or other public media (6).
On occasion, a specific research result can suggest clinical action, either preventive or therapeutic, that could benefit a particular participant—action that would unlikely be taken without specific knowledge of the result. This work was motivated in part by the experience of a colleague who ran a large proteomics study intended to validate a predictive proteomic approach for early diagnosis of lung cancer (7). The study involved ~1400 people, some of whom had been diagnosed with lung cancer and others acting as controls who were heavy smokers thought to be cancer-free. In work preliminary to that study, when fewer participants had been recruited, mathematicians at SomaLogic created a high-performance classifier that distinguished (retrospectively) the participants with non–small cell lung cancer (NSCLC) from the controls.
The annual rate of diagnosis of lung cancer in heavy smokers, such as the controls, is ~1% (8); 1 of the 63 controls in this preliminary analysis scored in a range that suggested NSCLC. That is, one of the putative control participants displayed results that were different from the others and similar to those of the cancer patients. Taken together with other information about the participant, these results strongly suggested an undiagnosed NSCLC. If that diagnosis were confirmed, the participant’s life could be saved because early-stage surgical intervention has a high likelihood of success, whereas later-stage diagnoses have no effective therapy (9). There are a variety of other clinical conditions, such as ovarian cancer or sudden cardiac death, in which early detection is likely to have positive therapeutic consequences; in fact, much diagnostic research addresses such conditions.
This proposal regards findings that have clear, immediate clinical utility and are unlikely to be screened or tested for in standard routine medical care. Such results could include both the intended product of the research and unanticipated incidental findings (10). The clinical actionability of a research finding for a participant depends on four factors: the validity, utility, risk, and benefit of the finding. First, the validity of the research findings should be confirmed—for example, by a clinical laboratory. Second, clinical utility means the results are relevant to treatment decisions (4, 6, 11) or entail risks that have effective preventive interventions (12, 13). Last, consideration of the risks and potential benefits of transmitting the result must demonstrate a reasonable chance of improving well-being, reducing harm, or both for the participant. The clinical utility of research results—in particular, those from new and as yet unapproved procedures—must be evaluated carefully in the context of reporting the results to study participants.
Currently, there is no universally recognized authoritative policy that addresses the ethical duty or lack thereof to return individual research results to participants. Concerns about shortcomings of existing international norms are driving additional ethics research (14). However, recommendations that researchers have an ethical obligation to communicate actionable individual results to participants are growing in the literature (4, 6, 15–19). In addition, international surveys are in progress (for example, http://www.genomethics.org/about_questionnaire.html) to assess public attitudes toward return of incidental findings.
Anonymization prevents any information derived from biological samples provided by the participant from ever being associated with the participant for any reason. So despite the apparently compelling reason to contact the participant in the NSCLC case, there is no way to reconnect the sample to a specific research participant. Alternative approaches for reaching this participant are problematic; for example, contacting all participants involved in the study and informing them of the finding would cause unjustified concern among the vast majority of participants, who are not the ones who had the actionable result (20). In addition, conveying that one person out of potentially many thousands of possible participants has an actionable problem might not be motivating enough to lead to action by the individual that could benefit from the knowledge. Furthermore, the NSCLC study described above was conducted under an exemption (anonymization) from investigational device regulation (21 CFR 812), making any attempt to provide results to a patient—no matter how indirect—a violation of U.S. law.
The very possibility of a guarantee of anonymity for genomic data has been called into question by recent technical advances. Genomic data itself is an indicator of identity and can be decoded, even when samples are provided anonymously. Individual participants have been identified from pooled genotypic data (21) and through the use of bioinformatics approaches (22). Current regulation of anonymized research samples and the communication about anonymization provided during the informed consent process probably overstate the guarantee of privacy protection and can lead to clinical or psychosocial harm. The failure to return clinically actionable information clearly harms research participants; we’ve been making this case above. There are other harms (which we find less compelling) that are covered in the citations. (23, 24).
CRYPTEX SECURITY
As a much-needed replacement for anonymization of biomedical research samples, we recommend a cryptographic approach that is based on the idea of “secret sharing,” which involves several parties, each of whom holds a share of a secret (25). The secret can be reconstructed only when a sufficient number of shares are combined. The individual shares are of no use in reconstructing the secret until the sufficient number is reached. By sharing the secret of a research participant’s identity among the researcher and other responsible parties, most advantages of anonymization can be retained while making possible the identification of participants who have clinically actionable results. Although there are previous proposals to use secret sharing in genomics (26), we are not aware of any biobanks implementing a secret-sharing approach for delivering research results to participants.
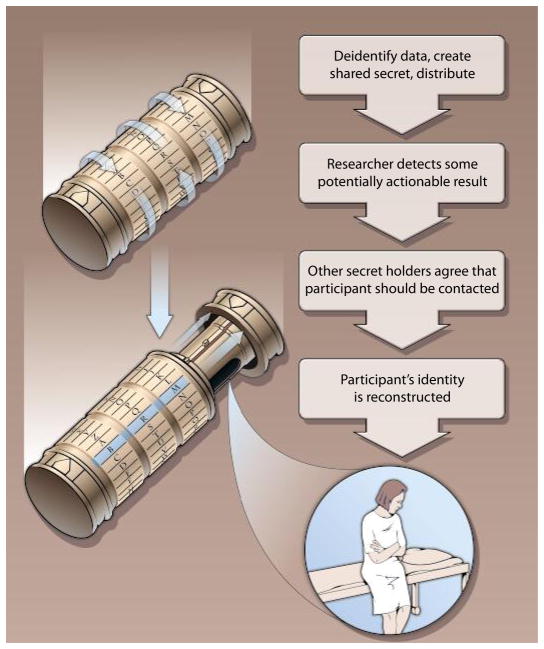
An illustrative scenario for the proposed approach is a case in which a research participant consents to donate a tissue sample to a biobank (Fig. 1). The informed consent process would ascertain whether a participant desires to be contacted in the unlikely event that a researcher discovers a result with actionable implications for the participant’s health. For participants who consent, the biobank would assign a random identifier to each donated sample and use secret sharing to encrypt the link between that random key and the donor’s identity. The sample or data from it would be distributed by the biobank using only the random identifier, but each researcher using it would also be given part of the shared secret for each participant’s identity. The data and safety monitoring board overseeing the experiment would also be given a share of the secret. The biobank would keep only its part of the shared secret and destroy any copies it has of the researcher’s and the board’s parts.
Fig. 1. Code read.
An illustrative scenario for the Cryptex security approach. Step 1. Codes created. Participant gives sample to biobank and consent to be contacted to report actionable results. Biobank deidentifies the participant’s record, creates shared secret codes, keeps one shared secret code (B), and destroys links to participant’s identity. Researcher gets a set of deidentified records and one part of the shared secret code (R); data and safety monitoring board gets one part of the shared secret code. Step 2. Actionable research result detected. Researcher detects an actionable research result for a participant and reports this to the biobank and monitoring board. Biobank confirms result and reports the confirming results to the monitoring board. Step 3. Agreement to share. All three groups agree that the result should be shared with participant. Secret codes are shared to reveal participant’s identity (B, biobank; R, researcher; M, monitoring board). Biobank contacts participant and his or her physician and delivers the result.
In the event that a researcher believes a clinically actionable result has been found, a request is sent to the biobank to identify the participant, along with justifying documentation. The biobank then independently evaluates the result by, for example, sending the sample to a clinical laboratory for a confirmatory test. If the biobank concurs that the result is actionable, then the biobank sends the findings to the board for assessment. If all parties agree that the result is actionable and the rationale for contacting the participant is justified, then the combination of the three entities’ shares of the secret can reconstruct the identity of the participant and facilitate contact. The biobank would have the responsibility of contacting the participant, who would then be asked to designate a physician to receive the actionable information or decline. We propose to involve the patient’s physician because effective use of the actionable information requires that it be integrated into the patient’s care, which conforms to the traditional practices of delivering other sorts of clinically relevant information. As with any other sort of personally identifiable health information, all parties involved in the secret sharing have a clear obligation to protect that information by not sharing it with anyone except as authorized by law. Costs and other managerial challenges to implementing this approach are likely to be modest.
There are variations on the above scenario that might be appropriate in different circumstances. For example, when the researcher collects samples without a biobank as an intermediary, an independent data- and safety-monitoring panel and the researcher would hold shares of the secret and must agree to cooperate in order to establish a participant’s identity. In some cases, the research participants may be given a share of the secret; therefore, this approach may also be a suitable technology to support efforts to confer on research participants rights of ownership of their research results (27).
Our proposal is based on four balancing ethical principles: beneficence, justice, trust, and privacy. Beneficence entails the duty of researchers to act on behalf of the study participants’ welfare by maximizing the benefits and minimizing the harms of research. In the case of this proposal, the benefit is a meaningful improvement in health. Although the benefit is limited to the small number of people who participate in research and have an actionable result materialize, the number of people affected may grow with advances in proteomics and genomics research. There can conceivably be negative consequences to providing actionable results to research participants (28), including anxiety and the costs of further evaluation; these would be disclosed in the consent process. As with any other aspect of consent, a participant could opt out of the potential to be informed of actionable results at any time.
Moreover, this benefit provides a way to reward those who volunteer for biomedical research and increase the public trust in researchers and the scientific process. Because of the long-term nature of clinical and translational research, the benefits typically are reaped by society rather than by the study participants. Therefore, when the opportunity arises for the participants to obtain benefit from the research, the principle of justice is served (29). Likewise, research policies that place the health of volunteers as a paramount concern promote the trust that sustains the research enterprise (30).
The benefits in beneficence and justice are balanced by some cost to privacy. Individuals may have good reasons for not wanting to share personal health information with researchers. Genetic research may carry social risks to individuals or members of specific racial or ethnic groups (31). Furthermore, participants have not only a right to know but also a right not to know information about themselves (32).
However, there are degrees of invasion of privacy. For example, disclosure of private information to a small number of trusted people is different in degree from broad disclosure to the general public, and the potential disclosure of information about a single research participant is different in its effect on society from disclosure of information about a large number of research participants. Thus, our proposal carries a low degree of privacy invasion. Not even the monitoring board of a clinical study needs to know the identity of the research participant; the only disclosure of identifiable information is to a representative of the biobank for the purpose of contacting the participant and the designated physician. Studies indicate that most participants endorse the return of individual research results despite these possible negative implications (18, 33, 34).
The requirements of a shared secret decrease the likelihood that accidents or mistakes, such as lost disk drives or hacker break-ins, will cause the release of research participants’ identities. Furthermore, the risk of privacy loss accrues only to the person who benefits: the participant who receives actionable health information. If adopted by regulators, this proposal for cryptographic secret sharing would allow research participants to decide for themselves whether or not their actionable research results are returnable.
Acknowledgments
A workshop on Privacy Risks on Data Sharing in Genomics held at the University of Colorado Anschutz Medical Campus in August 2010 started the discussions that led to this Commentary. We gratefully acknowledge M. Yarborough and K. Edwards, who helped organize the workshop, as well as all of the participants. During the workshop, L. Gold of SomaLogic expressed his alarm at not being able to contact a participant of the research, which sparked our approach. We thank J. Sakai, M. Yarborough, B. Blankenship, and S. Williams for close readings and helpful suggestions.
Funding: M.E.C. and C.H. are supported in part by NIH grant 5R01DA029258002. M.E.C. is also supported in part by NIH grants UL1-RR025780 and 5P60DA011015-13. L.E.H. is supported in part by NIH grants 5R01LM000811-07 and 2R01LM009254-04.
Footnotes
Competing interests: The authors declare that they have no competing interests.
REFERENCES AND NOTES
- 1.Roden DM, Pulley JM, Basford MA, Bernard GR, Clayton EW, Balser JR, Masys DR. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther. 2008;84:362–369. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kohane IS, Mandl KD, Taylor PL, Holm IA, Nigrin DJ, Kunkel LM. Reestablishing the researcher-patient compact. Science. 2007;316:836–837. doi: 10.1126/science.1135489. [DOI] [PubMed] [Google Scholar]
- 3.Ries NM, LeGrandeur J, Caulfield T. Handling ethical, legal and social issues in birth cohort studies involving genetic research: Responses from studies in six countries. BMC Med Ethics. 2010;11:4. doi: 10.1186/1472-6939-11-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho MK. Understanding incidental findings in the context of genetics and genomics. J Law Med Ethics. 2008;36:280–285. 212. doi: 10.1111/j.1748-720X.2008.00270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shalowitz DI, Miller FG. Disclosing individual results of clinical research: Implications of respect for participants. JAMA. 2005;294:737–740. doi: 10.1001/jama.294.6.737. [DOI] [PubMed] [Google Scholar]
- 6.Fabsitz RR, McGuire A, Sharp RR, Puggal M, Beskow LM, Biesecker LG, Bookman E, Burke W, Burchard EG, Church G, Clayton EW, Eckfeldt JH, Fernandez CV, Fisher R, Fullerton SM, Gabriel S, Gachupin F, James C, Jarvik GP, Kittles R, Leib JR, O’Donnell C, O’Rourke PP, Rodriguez LL, Schully SD, Shuldiner AR, Sze RK, Thakuria JV, Wolf SM, Burke GL. National Heart, Lung, and Blood Institute working group, Ethical and practical guidelines for reporting genetic research results to study participants: Updated guidelines from a National Heart, Lung, and Blood Institute working group. Circ Cardiovasc Genet. 2010;3:574–580. doi: 10.1161/CIRCGENETICS.110.958827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ostroff RM, Bigbee WL, Franklin W, Gold L, Mehan M, Miller YE, Pass HI, Rom WN, Siegfried JM, Stewart A, Walker JJ, Weissfeld JL, Williams S, Zichi D, Brody EN. Unlocking biomarker discovery: Large scale application of aptamer proteomic technology for early detection of lung cancer. PLoS ONE. 2010;5:e15003. doi: 10.1371/journal.pone.0015003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bach PB, Kattan MW, Thornquist MD, Kris MG, Tate RC, Barnett MJ, Hsieh LJ, Begg CB. Variations in lung cancer risk among smokers. J Natl Cancer Inst. 2003;95:470–478. doi: 10.1093/jnci/95.6.470. [DOI] [PubMed] [Google Scholar]
- 9.Henschke CI, Yankelevitz DF, Libby DM, Pasmantier MW, Smith JP, Miettinen OS. International Early Lung Cancer Action Program Investigators, Survival of patients with stage I lung cancer detected on CT screening. N Engl J Med. 2006;355:1763–1771. doi: 10.1056/NEJMoa060476. [DOI] [PubMed] [Google Scholar]
- 10.Wolf SM, Crock BN, Van Ness B, Lawrenz F, Kahn JP, Beskow LM, Cho MK, Christman MF, Green RC, Hall R, Illes J, Keane M, Knoppers BM, Koenig BA, Kohane IS, Leroy B, Maschke KJ, McGeveran W, Ossorio P, Parker LS, Petersen GM, Richardson HS, Scott JA, Terry SF, Wilfond BS, Wolf WA. Managing incidental findings and research results in genomic research involving biobanks and archived data sets. Genet Med. 2012;14:361–384. doi: 10.1038/gim.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murphy TE, Agostini JV, Van Ness PH, Peduzzi P, Tinetti ME, Allore HG. Assessing multiple medication use with probabilities of benefits and harms. J Aging Health. 2008;20:694–709. doi: 10.1177/0898264308321006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hodgkinson K, Pullman D. Duty to warn and genetic disease. Can J Cardiovasc Nurs. 2010;20:12–15. [PubMed] [Google Scholar]
- 13.Offit K, Groeger E, Turner S, Wadsworth EA, Weiser MA. The “duty to warn” a patient’s family members about hereditary disease risks. JAMA. 2004;292:1469–1473. doi: 10.1001/jama.292.12.1469. [DOI] [PubMed] [Google Scholar]
- 14.Zawati MH, Knoppers BM. International normative perspectives on the return of individual research results and incidental findings in genomic biobanks. Genet Med. 2012;14:484–489. doi: 10.1038/gim.2012.13. [DOI] [PubMed] [Google Scholar]
- 15.Hens K, Nys H, Cassiman JJ, Dierickx K. The return of individual research findings in paediatric genetic research. J Med Ethics. 2011;37:179–183. doi: 10.1136/jme.2010.037473. [DOI] [PubMed] [Google Scholar]
- 16.Ravitsky V, Wilfond BS. Disclosing individual genetic results to research participants. Am J Bioeth. 2006;6:8–17. doi: 10.1080/15265160600934772. [DOI] [PubMed] [Google Scholar]
- 17.Knoppers BM, Joly Y, Simard J, Durocher F. The emergence of an ethical duty to disclose genetic research results: International perspectives. Eur J Hum Genet. 2006;14:1170–1178. doi: 10.1038/sj.ejhg.5201690. [DOI] [PubMed] [Google Scholar]
- 18.Dressler LG. Disclosure of research results from cancer genomic studies: State of the science. Clin Cancer Res. 2009;15:4270–4276. doi: 10.1158/1078-0432.CCR-08-3067. [DOI] [PubMed] [Google Scholar]
- 19.Miller FA, Hayeems RZ, Bytautas JP. What is a meaningful result? Disclosing the results of genomic research in autism to research participants. Eur J Hum Genet. 2010;18:867–871. doi: 10.1038/ejhg.2010.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forsberg JS, Hansson MG, Eriksson S. Changing perspectives in biobank research: From individual rights to concerns about public health regarding the return of results. Eur J Hum Genet. 2009;17:1544–1549. doi: 10.1038/ejhg.2009.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Homer N, Szelinger S, Redman M, Duggan D, Tembe W, Muehling J, Pearson JV, Stephan DA, Nelson SF, Craig DW. Resolving individuals contributing trace amounts of DNA to highly complex mixtures using high-density SNP genotyping microarrays. PLoS Genet. 2008;4:e1000167. doi: 10.1371/journal.pgen.1000167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacobs KB, Yeager M, Wacholder S, Craig D, Kraft P, Hunter DJ, Paschal J, Manolio TA, Tucker M, Hoover RN, Thomas GD, Chanock SJ, Chatterjee N. A new statistic and its power to infer membership in a genome-wide association study using genotype frequencies. Nat Genet. 2009;41:1253–1257. doi: 10.1038/ng.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trinidad SB, Fullerton SM, Ludman EJ, Jarvik GP, Larson EB, Burke W. Research ethics. Research practice and participant preferences: The growing gulf. Science. 2011;331:287–288. doi: 10.1126/science.1199000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fullerton SM, Anderson NR, Guzauskas G, Freeman D, Fryer-Edwards K. Meeting the governance challenges of next-generation biorepository research. Sci Transl Med. 2010;2:15cm3. doi: 10.1126/scitranslmed.3000361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shamir A. How to share a secret. Commun ACM. 1979;22:612–613. [Google Scholar]
- 26.Kantarcioglu M, Jiang W, Liu Y, Malin B. A cryptographic approach to securely share and query genomic sequences. IEEE Trans Inf Technol Biomed. 2008;12:606–617. doi: 10.1109/TITB.2007.908465. [DOI] [PubMed] [Google Scholar]
- 27.Terry SF, Terry PF. Power to the people: Participant ownership of clinical trial data. Sci Transl Med. 2011;3:69cm3. doi: 10.1126/scitranslmed.3001857. [DOI] [PubMed] [Google Scholar]
- 28.Wolf SM, Lawrenz FP, Nelson CA, Kahn JP, Cho MK, Clayton EW, Fletcher JG, Georgieff MK, Hammerschmidt D, Hudson K, Illes J, Kapur V, Keane MA, Koenig BA, Leroy BS, McFarland EG, Paradise J, Parker LS, Terry SF, Van Ness B, Wilfond BS. Managing incidental findings in human subjects research: Analysis and recommendations. J Law Med Ethics. 2008;36:219–248. 211. doi: 10.1111/j.1748-720X.2008.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nestler G, Steinert R, Lippert H, Reymond MA. Using human samples in proteomics-based drug development: bioethical aspects. Expert Rev Proteomics. 2004;1:77–86. doi: 10.1586/14789450.1.1.77. [DOI] [PubMed] [Google Scholar]
- 30.Yarborough M, Fryer-Edwards K, Geller G, Sharp RR. Transforming the culture of biomedical research from compliance to trustworthiness: Insights from nonmedical sectors. Acad Med. 2009;84:472–477. doi: 10.1097/ACM.0b013e31819a8aa6. [DOI] [PubMed] [Google Scholar]
- 31.Goldenberg AJ, Hull SC, Wilfond BS, Sharp RR. Patient perspectives on group benefits and harms in genetic research. Public Health Genomics. 2011;14:135–142. doi: 10.1159/000317497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson J. To know or not to know? Genetic ignorance, autonomy and paternalism. Bioethics. 2005;19:492–504. doi: 10.1111/j.1467-8519.2005.00460.x. [DOI] [PubMed] [Google Scholar]
- 33.Roberts JS, Shalowitz DI, Christensen KD, Everett JN, Kim SY, Raskin L, Gruber SB. Returning individual research results: Development of a cancer genetics education and risk communication protocol. J Empir Res Hum Res Ethics. 2010;5:17–30. doi: 10.1525/jer.2010.5.3.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaufman DJ, Murphy-Bollinger J, Scott J, Hudson KL. Public opinion about the importance of privacy in biobank research. Am J Hum Genet. 2009;85:643–654. doi: 10.1016/j.ajhg.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]