Abstract
In order to asses performance of the LDA in describing physisorption on graphene, adsorptions of TCNE, TCNQ, TNF, TTF, and DMPD as well as four benzene derivatives on C54H18 and C110H30 were explored with a variety of DFTs such as MPWB1K, M06-2X, PBE-D and LSDA. Although it is well known that the LDA considerably overestimate non-covalent interaction, the LSDA predicted adsorption energies except for TCNE on C110H30 are systematically lower than those from the M06-2X by 0.4–3.2 kcal/mol, and they are more significantly lower than those from the PBE-D for all the molecules by 3–6 kcal/mol. However, the LSDA adsorption energy sequence is consistent with that from the PBE-D, TNF~TCNQ>TCNE~DMPD>TTF. Moreover, the domain interaction between the electron donor and acceptor molecules with graphene through cooperative π···π, C-H···π and N-H···π were visualized with sign(λ2)×ρ, and the relationships between the binding energy with London force, molecular electronegativity, and frontier orbital level were extensively discussed.
Keywords: graphene, DFT, physisorption, electron donor and acceptor
Introduction
Adsorptions of electron donor and acceptor molecules on graphene have attracted wide attention since the electronic property of graphene can be considerably changed by the physical functionalization. Analogous to silicon semiconductors with electrons doped by germanium or gallium, it is realized that graphene can exhibit similar properties of the n- and p-type semiconductors through adsorption of electron donating and accepting organic molecules, respectively.1 Thus, it was proposed that graphene may become a new generation of semiconductor material in future.2 Some authors recently reported their experimental results for the functionalized graphene with several organic molecules. Chen et al. performed angle-resolved photoemission spectroscopy experiment and showed the p-type doping effect of tetrafluoro-tetracyanoquinodimethane (F4-TCNQ) molecule for the functionalized graphene.1a Varghese et al. investigated the interaction of electron donor and acceptor molecules with single-walled carbon nanotube (SWCNT) and graphene through isothermal titration calorimetry (ITC)3 and the trend in the relative binding energies was predicted for the molecules such as tetracyanoethylene (TCNE), tetracyanoquinodimethane (TCNQ), 2,4,7-trinitrofluorenone (TNF), tetrathiafulvalene (TTF), and N,N-dimethyl paraphenylenediamine (DMPD). Wang et al.1b employed Kelvin probe force microscopy (KPFM) to investigate both n-type and p-type surface doping of exfoliated graphene by organic molecules vanadyl phthalocyanine (VPOC) and tetrafluoro-tetracyanoquinodimethane (F4-TCNQ), respectively. Barja et al. studied the intermolecular interaction between two electron acceptor molecules on graphene surface with scanning tunneling microscope (STM) image of the moiré pattern.4 They showed that the molecular ordering of TCNQ molecules on graphene is dominated by intermolecular attraction while repulsion determines the ordering for the F4-TCNQ.
In addition to the above experimental investigations, in the most recent years this subject was also explored by density functional theories; however, the most applied local spin density approximation (LSDA) or LDA, is well known to considerably overestimate physisorption or non-covalent interaction systems. Lu et al. studied the adsorption of TCNE on graphene with the LSDA.5 They demonstrated that p-type graphene can be obtained through TCNE adsorption with the binding energy of approximately 0.9 eV for a variety of adsorption configurations. Saha et al. applied both the LDA and GGA type of DFT (PBE) to the adsorptions of aniline (PhNH2) and nitrobenzene (PhNO2) on graphene.6 They predicted strong adsorptions by the LDA, e.g. the respective adsorption energies of 0.341 and 0.412eV for PhNH2 and PhNO2 on 5×5 graphene supercell; however, similar to its performance on other non-covalent systems the PBE brings about rather small binding. The LDA predicted an adsorption energy for F4-TCNQ on graphene was around 1.26 eV by Pinto et al.,7 1.28eV by Sun et al.,8 yet 1.95eV by Tian et al.9 The binding energies obtained by the LDA for benzene (PhH) and TTF on graphene are 0.24 and 0.51 eV, respectively.10 For the adsorption of the TTF on graphene, its binding energy with the LDA obtained by Sun et al.8 is 0.72 eV, and those of TCNQ and F4-TCNQ on graphene are 1.15, and 1.28 eV, respectively. With the LDA the adsorption energies for PhH and naphthalene on graphene predicted by AlZahrani are 0.30 and 0.47 eV, respectively.11 Most recently, Chi et al still applied the PBE to the adsorption of TCNE and TCNQ on graphene, and it is not surprising that the adsorption energies are only one fifth of the LDA predicted ones, 0.233 and 0.255 eV.12 Besides the adsorption energies, the electron transfer between graphene and the organic molecules were also theoretically discussed. For example, the amounts of electron transferred from graphene are estimated to be 0.27–0.44e to TCNE,5,12 and 0.263-0.34e to TCNQ,8,12 and 0.07e to TTF.8 However, for the PhH and TTF on graphene Zhang et al.10 predicted that electrons are transferred from Ph-H and TTF to graphene with 0.02e and 0.26e, respectively, which is opposite to the previous result.8
Accordingly, there exists a controversy for both the binding energy and the electron transfer for the adsorptions of the electron donor and acceptor molecules on graphene. It is generally believed that the adsorptions are dominated by the non-covalent weak interactions, such as π···π and other weak C/N-H···π interactions, which are usually not well described by the LDA and GGA density functional theories. The novel hybrid meta-GGA functionals, M05-2X and M06-2x, developed by Truhlar et al 13 are proven to reasonably represent the non-covalent interactions. The Grimme’s empirical dispersion-corrected density functional theory (DFT-D), 14 has better performance on the noncovalent systems through directly accounting for long-range dispersion effects with a damped interatomic potential. Thus, one of the objectives of the present investigation is to assess the LDA method widely applied in aforementioned plane-wave calculations through comparing its results with those from the M06-2X and the DFT-D methods.
In addition, although theoretical and experimental studies on the adsorptions of the electron donors and acceptors on graphene were carried out, a relationship between electron transfer characteristics of adsorbates with their electronic structure properties such as IP, EA, and frontier orbital level has not yet been well established. It should be an important issue how to estimate electron withdrawing or donating capacity for molecules adsorbed on graphene. A good semiconductor material should have significant electronic transfer between the organic molecule and graphene, while on the other hand the interaction between the organic molecule and graphene should not essentially change π orbital framework of graphene. In order to better understand the relationship and to find some regulations to identify electronic donor and acceptor molecules, we systematically investigated the adsorption of a series of some organic molecules, including PhH, PhNH2, PhOH, PhNO2, TCNE, TTF, DMPD, TCNQ, and TNF, on graphene in the current study.
Graphene Model and Computational Method
Full optimizations and property calculations for the adsorptions of the organic molecules on graphene were performed with the MPWB1K,15, LSDA and M06-2X16 methods implemented in Gaussian09.17 The basis set superposition error18 (BSSE) of the intermolecular interaction was applied to the energy correction of the binding complexes. In addition, an empirical dispersion-corrected density functional theory (DFT-D) of Grimme’s scheme14,19 was adopted for long-range dispersion effects in the noncovalent systems with the ORCA suite of programs.20 Because of employing the Perdew-Burke-Ernzerhof (PBE) exchange-correlation functional21 in the DFT part of the Grimme’s scheme, the DFT-D method was therefore referred to as PBE-D. The above four calculation methods were carried out with Dunning’s correlation-consistent polarized valence basis sets (cc-pVDZ and cc-pVTZ).22 The adsorption energy is calculated by the equation: Eads = EA + EB − EAB, where AB, A and B respectively represent the optimized adsorption complex and the separate monomers. Charges carried by the electron donor and acceptor molecules, such as PhH, PhNH2, PhOH, PhNO2, TCNE, TTF, DMPD, TCNQ, and TNF, were calculated with electrostatic potential fitting scheme(CHelpG).23 For the sake of comparison, London’s empirical formula24 may help understand the non-covalent interaction of the organic molecule with graphene. Here, α and I are the polarizability and ionization potential, respectively, for monomers A and B; R is the interplanar distance between A and B.
Two clusters C54H18 (G54) and C110H30 (G110) were used to model graphene layer. The cluster models together with the hybrid meta-GGA method (M06-2X) implemented in Gaussian 09 and the PBE-D in ORCA, which may not be available in the plane-wave based DFT packages, will allow assessing the accuracy of the LDA method for the non-covalent adsorptions on graphene.
Results and Discussions
Geometry and Binding Energy
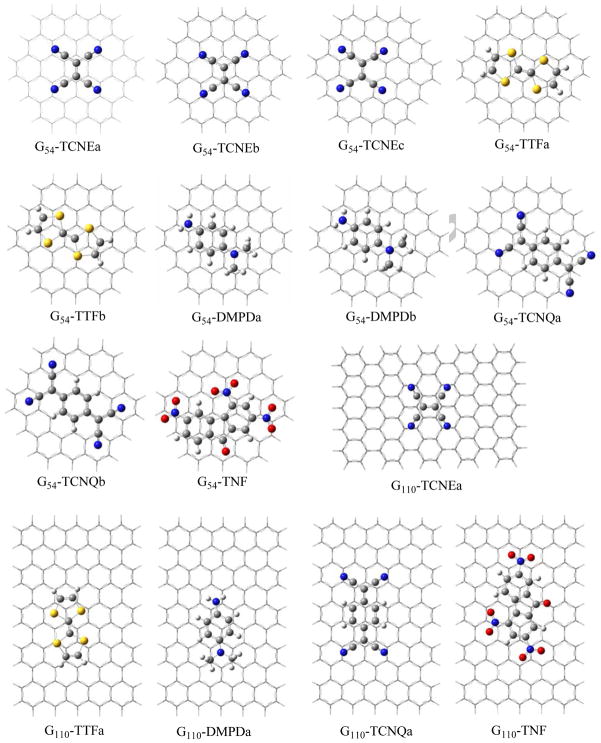
The fully optimized geometries for the adsorptions of TCNE, TTF, DMPD, TCNQ, and TNF on G54 and G110 with the MPWB1K, PBE-D, M06-2X, and LSDA methods are shown in Figure 1, and the adsorption energies are listed in Table 1. From the results of G54 at the MPWB1K/cc-pVDZ level, it can be seen that the bridge type complexes (a) are more stable than other structures. For example, the adsorption energy of G54-TCNE-a is greater than those of G54-TCNE-b and G54-TCNE-c by 0.8 and 2.2 kcal/mol, respectively; the adsorption energies of G54-TTF-a, G54-DMPD-a, and G54-TCNQ-a are also 4.1, 0.9 and 3.2 kcal/mol higher than those of G54-TTF-b, G54-DMPD-b, G54-TCNQ-b, respectively. Comparing with the result at the PBE-D/cc-pVDZ level, the MPWB1K only provides less than one half of the adsorption energy predicted by the PBE-D. Even after the BSSE correction is included at the PBE-D level, the adsorption energies of G54-TCNE, G54-TTF, and G54-DMPD are still one time higher than the corresponding ones from the MPWB1K method. The BSSE corrections at the PBE-D/cc-pVDZ level are 3.5, 3.8, 4.6, 5.0, and 10.5kcal/mol for G54-TCNE, G54-TTF, G54-DMPD, G54-TCNQ, and G54-TNF, respectively, which exhibits an increase with the size of the adsorbed organic molecule. Although the adsorption energies with the cc-pVTZ basis set without BSSE correction are improved about 1–5 kcal/mol over those with the cc-pVDZ basis set, there is only a slight variation of approximately 1 kcal/mol after the BSSE is included. Thus, with the BSSE correction the cc-pVDZ basis set is able to provide similar binding energies to those from the cc-pVTZ for the current systems. The most stable G110-molecule adsorption complexes, i.e., bridge adsorptions were therefore calculated at the PBE-D, M06-2X and LSDA levels with the cc-pVDZ basis set. Due to more van der Waals contact, Table 1 shows that the BSSE corrected adsorption energies of G110-TCNE-a, G110-TTF-a, G110-DMPD-a, G110-TCNQ-a, and G110-TNF are 3.1, 2.9, 1.1, 3.6, and 3.0 kcal/mol higher than those of G54-TCNE-a, G54-TTF-a, G54-DMPD-a, G54-TCNQa, and G54-TNF, respectively, at the PBE-D/cc-pVDZ level. Although the cluster model is rather extended from G54 to G110 the adsorption energy only changes less than 10%. This trend is similar to that of the adsorption of DNA bases on the clusters C24H12 and C54H18.25 The PBE-D predicted sequence of the adsorption energies on the both clusters is TNF (28.5 kcal/mol for G110) ~TCNQ (28.1) >DMPD (19.1)~TCNE (19.2)>TTF (17.4). However, the M06-2X method considerably underestimates the adsorption energy of G110-TCNE-a so that the order reverses to be TNF (25.3 kcal/mol for G110) > TCNQ (23.0) > DMPD (18.1) > TTF (16.6) >TCNE (13.2). This implies that the M06-2X may be insufficient to describe the dispersion for the non-ring conjugated systems like the TCNE.
Figure 1.
Structures of adsorption complexes of TCNE, TTF, DMPD, TCNQ and TNF with C54H18 and C110H30. G54 and G110 represent C54H18 and C110H30, respectively.
Table 1.
Adsorption energies (Eads, kcal/mol) with BSSE correction (Eads+BSSE) for adsorptions of TCNE, TTF, DMPD, TCNQ, and TNF on C54H18 (G54) and C110H30 (G110) from theoretical methods
| Complex | G54 | G110 | Literature LDA | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MPWB1K/cc-pVDZ | PBE-D/cc-pVDZ | PBE-D/cc-pVTZ | PBE-D/cc-pVDZ | M062x/cc-pVDZ | CPCM-M062x | LSDA/cc-pVDZ | |||||||
| Eads | Eads | Eads+BSSE | Eads | Eads+BSSE | Eads | Eads+BSSE | Eads | Eads+BSSE | Eads | Eads | Eads+BSSE | Eads | |
| Gx-TCNEa | 9.3 | 19.6 | 16.1 | 18.7 | 17.7 | 22.3 | 19.2 | 16.1 | 13.2 | 14.0 | 19.4 | 15.8 | 21.3,a 5.4d |
| Gx-TCNEb | 8.5 | ||||||||||||
| Gx-TCNEc | 7.1 | ||||||||||||
| Gx-TTFa | 10.6 | 18.3 | 14.5 | 15.9 | 14.8 | 21.3 | 17.4 | 20.2 | 16.6 | 17.7 | 17.9 | 13.5 | 16.6,b 11.8c |
| Gx-TTFb | 6.5 | ||||||||||||
| Gx-DMPDa | 9.4 | 22.6 | 18.0 | 19.5 | 17.7 | 24.7 | 19.1 | 22.8 | 18.1 | 24.1 | 20.8 | 15.3 | |
| Gx-DMPDb | 8.5 | ||||||||||||
| Gx-TCNQa | 14.2 | 29.5 | 24.5 | 25.9 | 24.5 | 33.6 | 28.1 | 27.6 | 23.0 | 25.6 | 28.5 | 22.6 | 26.5,b 5.9d |
| Gx-TCNQb | 11.0 | ||||||||||||
| Gx-TNF | 21.1 | 35.7 | 25.2 | 29.9 | 26.3 | 39.7 | 28.5 | 35.5 | 25.3 | 37.2 | 32.9 | 22.1 | |
Comparing the result of the PBE-D method with those of the M06-2X method for G110 in Table 1, the BSSE corrected adsorption energies of G110-TCNE-a, G110-TTF-a, G110-DMPD-a, G110-TCNQ-a, and G110-TNF at the M06-2X level are 6.0, 0.8, 1.0, 5.1, and 3.2 kcal/mol less than the corresponding ones at the PBE-D level. Obviously, the M06-2X estimation slightly deviates from the PBE-D result for G110-TTF-a, G110-DMPD-a; however, it provides considerably weaker binding for G110-TCNE-a, G110-TCNQ-a and G110-TNF so that the binding sequence is different from what is predicted by the PBE-D as aforementioned. The solvent effect in the aqueous solution on these five adsorption systems was estimated with the conductor-like polarizable continuum model (CPCM) at the M06-2X/cc-pVDZ level. Because of low polarity of the molecules, as expected the solvent effects only change the binding energies by 1–2 kcal/mol. For example, the dipole moments of the molecules TNF and DMPD at M06-2X/cc-pVDZ level are only 1.083 and 0.158 Debye, respectively; while the dipole moments of the high symmetric molecules, TCNE, TCNQ and TTF, are almost negligible.
Performance of LSDA/LDA
The five adsorption complexes were fully optimized at the LSDA level of theory implemented in Gaussian program with the cc-pVDZ basis set. Although it is well known that the LDA considerably overestimate non-covalent interaction, Table 1 shows that the BSSE corrected adsorption energies of G110-TCNE-a, G110-TTF-a, G110-DMPD-a, G110-TCNQ-a, and G110-TNF at the LSDA level are systematically less than the corresponding ones at the PBE-D level by 3.4, 3.9, 3.8, 5.5, and 6.4 kcal/mol, respectively. The deviation is in the range of 13–22%. It can be seen that the LSDA result has slightly bigger deviation than that of M06-2x except for the G110-TCNE-a adsorption complex, which is up to 31% at the M06-2X level. However, the sequence of the adsorption energies at the LSDA level is same as that at the PBE-D level, TNF (22.1 kcal/mol for G110)~TCNQ (22.6) >DMPD (15.3)~TCNE (15.8)>TTF (13.5).
Most of the previous adsorption energies for TCNE, TTF and TCNQ on graphene were achieved by the LSDA together with ultrasoft pseudopotential plane-wave basis and periodic boundary condition. In general, the binding energies from the periodic LSDA agree well in 1–2 kcal/mol with those from the present PBE-D with the cluster model, G110. For the TCNE-graphene the Lu’s binding energy 21.3 kcal/mol 5 is 2.1 kcal/mol bigger than the current PBE-D result (21.3 vs 19.2 kcal/mol), and 8.1 and 5.5 kcal/mol bigger than the M06-2X and LSDA values, respectively. For TTF-graphene, Zhang et al.10 and Sun et al.8 reported their theoretical values, 11.8 and 16.6 kcal/mol. The latter one is the same as the M06-2X result and only 0.8 kcal/mol less than the PBE-D result (17.4 kcal/mol). In the same publication, Sun et al.8 also reported the adsorption energy of TCNQ-graphene, 26.5 kcal/mol, which is 3.5 kcal/mol higher than the M062x value and 1.6 kcal/mol less than the PBE-D one. However, Chi et al. considerably underestimated the adsorption energies for TCNE and TCNQ on graphene (5.4 and 5.9kcal/mol) with the PBE method although a 8×8 graphene supercell (128 carbon atoms per graphene layer) was used.12
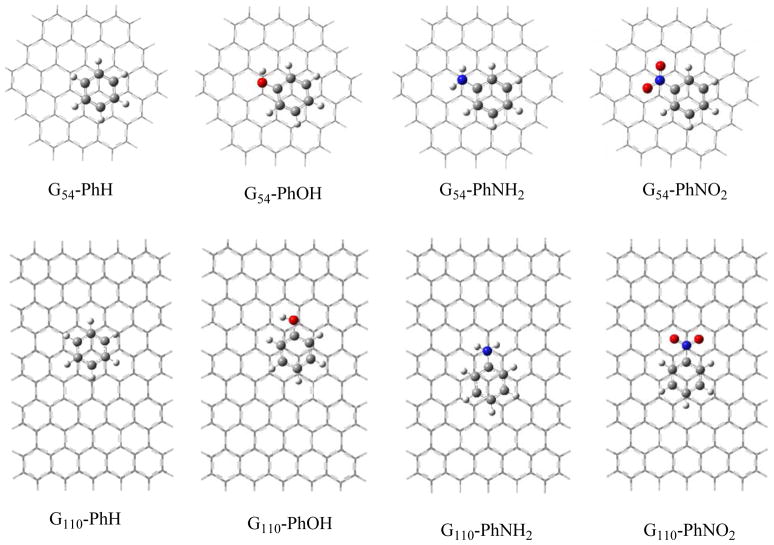
In addition to the above five organic molecules, the binding of benzene and three derivatives (PhNH2, PhOH, and PhNO2) with graphene clusters (G54 and G110) are also calculated at MPWB1K and M06-2X levels, respectively. The geometries of these adsorption complexes are shown in Figure 2 and their adsorption energies are listed in Table 2. It can be seen that the adsorption energies at the MPWB1K level are only a half of those at the M06-2X level, similar to the case of the above five adsorption complexes. At the M062X level the BSSE corrected adsorption energies of G110-PhH, G110-PhOH, G110-PhNH2, and G110-PhNO2 are 8.0, 8.9, 8.6, and11.0 kcal/mol, respectively. According to Table 2 the binding energies are slightly higher than the previous values predicted by the periodic LDA density functional theory with pseudopotential. For example, for PhH-graphene Zhang’s 10 and AlZahrani’s11 binding energies are lower than the current one from the M06-2X by 2.5 and 1.1 kcal/mol, respectively (5.5 and 6.9 vs 8.0 kcal/mol); Saha’s 6 respective values for the PhNH2-graphene and PhNO2-graphene are 7.9 and 9.5 kcal/mol, which deviate from the current M06-2X results in the range of 0.7–2.5 kcal/mol.
Figure 2.
Top views of structures for adsorption complexes of PhH, PhOH, PhNH2 and PhNO2 with C54H18 and C110H30. G54 and G110 represent C54H18 and C110H30, respectively.
Table 2.
Adsorption energies (Eads, kcal/mol) with BBSE correction (Eads+BSSE) for adsorptions of PhH, PhOH, PhNH2, and PhNO2 on C54H18 (G54) with MPWB1K nd C110H30 (G110) with M06-2X.
Correlation of binding energy with London dispersion, and polarizability
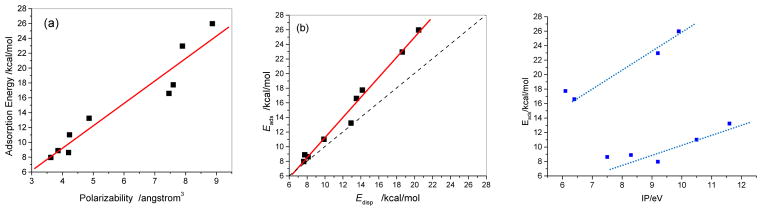
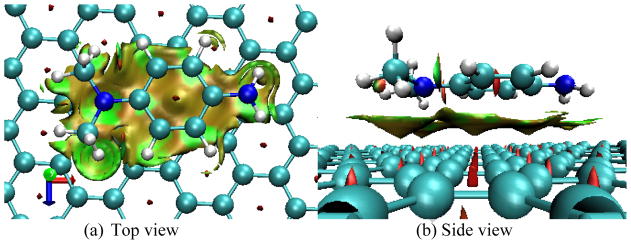
Table 3 lists the interplanar distances between adsorbed molecules and graphene, adsorption energies, London dispersion energies, and the charges carried by the adsorbed molecules in the adsorption complexes at the M062x/cc-pVDZ level. According to Table 3, all of the interplanar distances are approximately 3.2 Å. For G110-PhNO2, G110-TCNE, G110-TCNQ, and G110-TNF the distances are slightly less than 3.2 Å while the others are slightly larger than 3.2 Å. This small variation may be caused by the electron accepting and donating groups in the adsorbed organic molecules. Comparing the charges carried by the adsorbed molecules, PhNO2, TCNE, TCNQ, and TNF carry considerable negative charge with −0.0680e, −0.1337e, −0.2144e, and −0.1477e, respectively, implying that partial electrons are transferred from graphene to these molecules and the basic planar structure of graphene keeps almost invariance. The similar Eads and Edisp in Table 3 imply that the adsorption interaction dominantly arises from the dispersion. One can further see that the plot of Eads versus Edisp in Figure 4b exhibits a good linear relationship with a slope of 1.2 and a correlation coefficient of 0.9918. The deviation from the diagonal line (dashed line in Figure 4b) becomes gradually significant with Edisp increasing. Especially for G110-TTF-a, G110-DMPD-a, G110-TCNQ-a, and G110-TNF Eads are 3.1, 3.6, 4.3, and 5.5 kcal/mol higher than Edisp, respectively, which indicates that the strong adsorbed molecule enhances its electrostatic interaction with graphene. Figure 3, the isosurface of sign(λ2)×ρ defined by Yang et al.,26 of the G110-DMPD adsorption complex provides a clear visualization of the van der Waals interaction between graphene and DMPD. According to the color code that green shows vdW interaction and red strong nonbonded overlap, vdW interaction between DMPD and graphene is very obvious via cooperative π···π, N-H···π as well as C-H···π. The sign(λ2)×ρ isosurfaces for G110-TCNE and G110-TTF were provided as the supporting information.
Table 3.
Vertical separation (R), adsorption energies (Eads), and dispersion energies ( ) of adsorption complexes and CHelpG net charges (q) of adsorbed monomer parts on graphene (C110H30) at M062x/cc-pVDZ level.
| Complex | R/Ǻ | Eads/kcal/mol | Edisp/kcal/mol | q/e |
|---|---|---|---|---|
| G110-PhH | 3.230 | 7.96 | 7.63 | −0.0547 |
| G110-PhOH | 3.237 | 8.89 | 7.74 | −0.0516 |
| G110-PhNH2 | 3.238 | 8.62 | 8.11 | −0.0092 |
| G110-PhNO2 | 3.196 | 11.01 | 9.88 | −0.0680 |
| G110-TCNE | 3.144 | 13.23 | 12.90 | −0.1337 |
| G110-TTF | 3.237 | 16.59 | 13.51 | −0.0450 |
| G110-DMPD | 3.210 | 17.74 | 14.17 | 0.0712 |
| G110-TCNQ | 3.170 | 22.95 | 18.62 | −0.2144 |
| G110-TNF | 3.193 | 25.97 | 20.45 | −0.1477 |
Figure 4.
Correlation of adsorption energy with polarizability (a), dispersion energy (b), and adiabatic IP (c). Solid squares are the calculated points and the red lines are linear fitting.
Figure 3.
Reduced density gradient isosurface (0.5 au) for G110-DMPD. Green and yellow indicates vdW interaction and red strong nonbonded overlap.
In addition, the correlation between Eads and adiabatic ionization potential IP and polarizability (α) of the electron donors and acceptors were searched for. As shown in Figure 4c, it seems that the Eads respectively correlates with the IP of four benzene derivates and another five organic molecules. However, it is found that the Eads directly relates to α of the organic molecules perpendicular to the molecular plane. Figure 4(a) well demonstrates that Eads linearly correlates (correlation coefficient 0.9584) with the polarizability of the organic molecule, positively depending on the polarizability of the organic molecule.
Electron transfer with molecular electronegativity and frontier orbital level
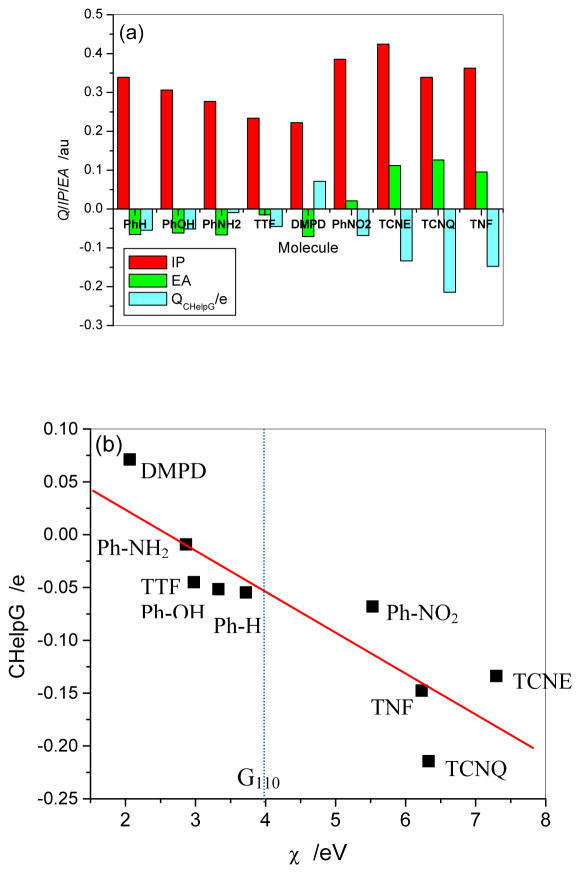
Table 4 summarizes the IP, electron affinity (EA), and HOMO and LUMO calculated at the M06-2X/cc-pVDZ level. For the IP of a molecule, the larger the ionization potential, the more difficult the molecule loses its electron to form a cation. However, the positive electron affinity makes a molecule obtain an electron easily to form an anion. The four molecules PhNO2, TCNE, TCNQ, and TNF clearly exhibit electron acceptor property because of their larger IP values and positive EA values. From the IP and EA of C110H30, one can see that the graphene would have dual properties as electron donor as well as acceptor because of its low IP value (4.5270 eV) and positive EA (3.4630 eV). Figure 5(a) intuitively shows the CHelpG charges carried by the adsorbed molecules in adsorption complexes, the IP and EA of the organic molecules. It is obvious to see that partial electrons are transferred from graphene to the four molecules with positive EA, PhNO2, TCNE, TCNQ, and TNF. DMPD exhibits electron donor property because of high negative EA and relative low IP, only transferring a small amount of electron to graphene. Because of high positive IP and negative EA, other molecules (PhH, PhOH, PhNH2, and TTF) on graphene appear a negligible electron withdrawing property, which shows they hardly modify graphene to become n-type or p-type semiconductor. This prediction is consistent with the previous theoretical results,5,8 but it is different from Zhang’s theoretical result for TTF,10 which showed that TTF is a good electron donator.
Table 4.
Ionization potential (IP), electron affinity (EA), polarizability (α), electronegativity (χ=(IP+EA)/2), and HOMO and LUMO of interacting monomers calculated at M062x/cc-pVDZ level.
| species | IP/eV | EA/eV | χ/eV | α/Å3 | εHOMO/au | εLUMO/au |
|---|---|---|---|---|---|---|
| PhH | 9.2288 | −1.7910 | 3.7189 | 3.6236 | −0.30382 | 0.03115 |
| PhOH | 8.3321 | −1.6782 | 3.3269 | 3.8562 | −0.27477 | 0.02901 |
| PhNH2 | 7.5354 | −1.8091 | 2.8631 | 4.1986 | −0.25340 | 0.03623 |
| PhNO2 | 10.4878 | 0.5676 | 5.5277 | 4.2298 | −0.33466 | −0.04646 |
| TCNE | 11.5503 | 3.0390 | 7.2946 | 4.8651 | −0.39017 | −0.14781 |
| TTF | 6.3633 | −0.4133 | 2.9750 | 7.4591 | −0.21126 | 0.00752 |
| DMPD | 6.0516 | −1.9320 | 2.0598 | 7.6000 | −0.19928 | 0.04506 |
| TCNQ | 9.2256 | 3.4317 | 6.3286 | 7.8983 | −0.31521 | −0.15073 |
| TNF | 9.8615 | 2.5976 | 6.2295 | 8.8690 | −0.34291 | −0.11547 |
| C110H30 | 4.5270 | 3.4630 | 3.9950 | 45.5361 | −0.15497 | −0.13978 |
Figure 5.
(a) CHelpG charges of adsorbed molecule in adsorption complex and adiabatic inonization potential (IP) and electron affinity (EA) for monomers of organic molecules (b) CHelpG charges of adsorbed molecule in adsorption complex with electronegativities (χ) of organic molecules.
Moreover, to unify the effects of IP and EA on electron transfer the molecular electronegativity was defined as χ IP+EA 2. It is interesting to see that the four electron acceptors (TCNE, TCNQ, TNF and Ph-NO2) have higher χ than the graphene, and oppositely other electron donators (Ph-H, Ph-OH, TTF, Ph-NH2, and DMPD) have lower χ than the graphene. As shown in Figure 5b, the charge carried by the molecules also shows a good linear relationship with the electronegativity (correlation coefficient of 0.8709). Thus, molecular electronegativity χ can be used as an index to predict electron transfer property, and further n- and p-type semiconductors for the functionalized graphenes.
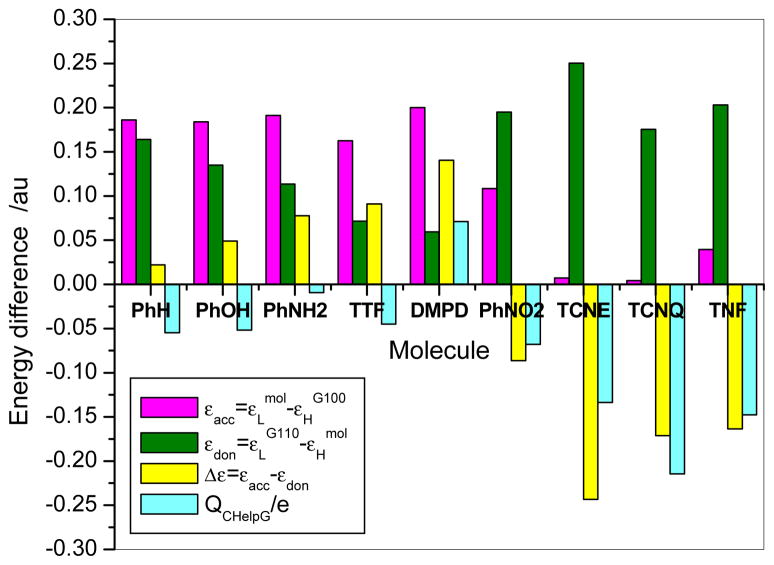
Figure 6 presents the molecular accepting and donating capacities (q/e), and the defined factors: and . Here, εLmol and εHmol respectively are the HOMO and LUMO orbital energies of an organic molecule, while εLG110 and εHG110 are those of graphene C110. Apparently, the electron transfer is determined by the competence of εacc with εdon. If εacc < εdon, the organic molecule will be an electron acceptor, otherwise it will be a donator. Thus, to simply correlate electron transfer and frontier orbital energy we define Δε = εacc − εdon. The more negative Δε, the easier a molecule accepts electrons; while the more positive Δε, the easier a molecule donates electrons. Figure 6 clearly shows that the considerable negative Δε of Ph-NO2, TCNE, TCNQ, and TNF agrees excellently with the charge (q) accepting from the graphene. In contrary, the high positive Δε value of DMPD is also good consistent with the charge (q) donating to the graphene. It is apparently to see that the former four molecules might make graphene become a p-type semiconductor and the latter one could make graphene an n-type one.
Figure 6.
CHelpG charges of adsorbed molecule in adsorption complex and difference of frontier orbital energy levels for monomers of organic molecules and graphene at M06-2x/cc-pVDZ level. εLmol and εHmol, LUMO and HOMO orbital energy of organic molecule; εLG110 and εHG110, LUMO and HOMO orbital energy of graphene.
Conclusion
Using a variety of DFT methods such as MPWB1K, M06-2X, PBE-D and LSDA, the adsorptions of TCNE, TCNQ, TNF, TTF, DMPD as well as four benzene derivatives on C54H18 and C110H30 were extensively investigated. Although it is well known that the LSDA considerably overestimate non-covalent interaction, the LSDA predicted adsorption energies for the above organic molecules except for TCNE on C110H30 are systematically lower than those from the M06-2X by 0.4–3.2 kcal/mol, and they are more significantly lower than those from the PBE-D for TCNE, TTF, DMPD, TCNQ, and TNF by 3.4, 3.9, 3.8, 5.5, and 6.4 kcal/mol, respectively. However, the LSDA adsorption energy sequence is consistent with that from the PBE-D, TNF~TCNQ>TCNE~DMPD>TTF. Thus, regarding the DFT methods for describing physisorption or noncovalent interaction on pristine graphene layer it can be concluded that the GGA methods are indeed improper; the LDA or LSDA method underestimates the weak interactions to some extent; M06-2X performs slightly better than the LDA; however, the dispersion correction is essential to further quantitatively improve the theoretical results. Remarkably, it was found that the adsorption energies well linearly correlate with London dispersion and polarizability rather than ionization potential, indicating that the dominant interaction between the electron donor and acceptor molecules with graphene is van der Waals interaction through cooperative π···π, C-H···π and N-H···π, which are well visualized with sign(λ2)×ρ. In addition, molecular electronegativity χ can be used as an index to predict electron transfer property, and further n- and p-type semiconductors for the functionalized graphenes. The bigger the electronegativity χ, the more electron the organic molecules will accept from graphene. As for Δε, the higher the negative Δε, the easier the organic molecules will gain electron from graphene. The PhNO2, TCNE, TCNQ, and TNF molecules therefore exhibit strong electron acceptors, while the DMPD molecule acts as an electron donator. However, the PhH, PhOH, PhNH2, and TTF molecules only show rather low electron withdrawing property.
Supplementary Material
Acknowledgments
This work was supported by the National Institute of General Medical of the National Institute of Health (SC3GM082324) and the American Recovery and Reinvestment Act (3SC3GM082324-02S1).
Footnotes
Supporting Information Available: Figure S1 shows the sign(λ2)×ρ isosurfaces for G110-TCNE and G110-TTF. This material is available free of charge via the Internet at http://pubs.acs.org.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.(a) Chen W, Chen S, Qi DC, Gao XY, Wee ATS. J Am Chem Soc. 2007;129:10418. doi: 10.1021/ja071658g. [DOI] [PubMed] [Google Scholar]; (b) Wang X, Xu JB, Xie W, Du J. J Phys Chem C. 2011;115:7596. [Google Scholar]; (c) Matis BR, Burgess JS, Bulat FA, Friedman AL, Houston BH, Baldwin JW. ACS NANO. 2012 doi: 10.1021/nn2034555. [DOI] [PubMed] [Google Scholar]
- 2.Geim AK. Science. 2009;324:1530. doi: 10.1126/science.1158877. [DOI] [PubMed] [Google Scholar]
- 3.Varghese N, Ghosh A, Voggu R, Ghosh S, Rao CNR. J Phys Chem C. 2009;113:16855. [Google Scholar]
- 4.Barja S, Garnica M, Hinarejos JJ, Parga ALVd. Chem Commun. 2010;46:8198. doi: 10.1039/c0cc02675a. [DOI] [PubMed] [Google Scholar]
- 5.Lu YH, Chen W, Feng YP, He PM. J Phys Chem B. 2009;113:2. doi: 10.1021/jp806905e. [DOI] [PubMed] [Google Scholar]
- 6.Saha SK, Chandrakanth RC, Krishnamurthy HR, Waghmare UV. Phys Rev B. 2009;80:155414/1. [Google Scholar]
- 7.Pinto H, Jones R, Goss JP, Briddon PR. J Phys: Condens Matter. 2009;21:402001/1. doi: 10.1088/0953-8984/21/40/402001. [DOI] [PubMed] [Google Scholar]
- 8.Sun JT, Lu YH, Chen W, Feng YP, Wee ATS. Phys Rev B. 2010;81:155403/1. [Google Scholar]
- 9.Tian X, Xu J, Wang X. J Phys Chem B. 2010;114:11377. doi: 10.1021/jp102800v. [DOI] [PubMed] [Google Scholar]
- 10.Zhang YH, Zhou KG, Xie KF, Zeng J, Zhang HL, Yong P. Nanotech. 2010;21:065201/1. doi: 10.1088/0957-4484/21/6/065201. [DOI] [PubMed] [Google Scholar]
- 11.AlZahran AZ. Appl Sur Sci. 2010;257:807. [Google Scholar]
- 12.Chi M, Zhao YP. Comput Mater Sci. 2012;56:79. [Google Scholar]
- 13.Zhao Y, Schultz NE, Truhlar DG. J Chem Theory Comput. 2006;2:364. doi: 10.1021/ct0502763. [DOI] [PubMed] [Google Scholar]
- 14.Grimme S. J Comput Chem. 2006;27:1787. doi: 10.1002/jcc.20495. [DOI] [PubMed] [Google Scholar]
- 15.Zhao Y, TDG J Phys Chem A. 2004;108:6908. [Google Scholar]
- 16.Zhao Y, Schultz NE, Truhlar DG. J Chem Theory Comput. 2006;2:364. doi: 10.1021/ct0502763. [DOI] [PubMed] [Google Scholar]
- 17.Frisch MJ, Trucks GW, Schlegel HB, et al. Gaussian 09 B1. Gaussian, Inc; Pittsburgh PA: 2009. [Google Scholar]
- 18.Boys SF, Bernard F. Mol Phys. 1970;19:553. [Google Scholar]
- 19.Grimme S. J Comput Chem. 2004;25:1463. doi: 10.1002/jcc.20078. [DOI] [PubMed] [Google Scholar]
- 20.Neese F. Max-Planck-Institut fur Bioanorganische Chemie: Mulheim an der Ruhr. 2010 [Google Scholar]
- 21.Perdew JP, Burke K, Ernzerhof M. Phys Rev Lett. 1996;77:3865. doi: 10.1103/PhysRevLett.77.3865. [DOI] [PubMed] [Google Scholar]
- 22.Dunning TH. J Chem Phys. 1989;90:1007. [Google Scholar]
- 23.Breneman CM, Wiberg KB. J Comp Chem. 1990;11:361. [Google Scholar]
- 24.London F. Z Phys A Hadrons Nuclei. 1930;63:245. [Google Scholar]
- 25.Grimme S. Phys Chem Chem Phys. 2008;10:2722. doi: 10.1039/b718788b. [DOI] [PubMed] [Google Scholar]
- 26.Johnson ER, Keinan S, Mori-Sanchez P, Contreras-Garcia J, Cohen AJ, Yang W. J A Chem Soc. 2010;132:6498. doi: 10.1021/ja100936w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.