Abstract
In this study we characterized phosphoribulokinase (PRK, EC 2.7.1.19) from the eukaryotic marine chromophyte Heterosigma carterae. Serial column chromatography resulted in approximately 300-fold purification of the enzyme. A polypeptide of 53 kD was identified as PRK by sequencing the amino terminus of the protein. This protein represents one of the largest composite monomers identified to date for any PRK. The native holoenzyme demonstrated by flow performance liquid chromatography a molecular mass of 214 ± 12.6 kD, suggesting a tetrameric structure for this catalyst. Because H. carterae PRK activity was insensitive to NADH but was stimulated by dithiothreitol, it appears that the enzyme may require a thioredoxin/ferredoxin rather than a metabolite mode of regulation. Kinetic analysis of this enzyme demonstrated Michaelis constant values of ribulose-5-phosphate (226 μm) and ATP (208 μm), respectively. In summary, H. carterae PRK is unique with respect to holoenzyme structure and function, and thus may represent an alternative evolutionary pathway in Calvin-cycle kinase development.
Three mechanisms have been described by which CO2 can be autotrophically processed. The reductive citric acid cycle and acetyl CoA condensation reaction are found exclusively in the green, methanogenic, and acetogenic bacteria (Hemming and Blotevogel, 1985; Fuchs, 1986; Schäfer et al., 1989). In contrast, the Calvin cycle is used by diverse organisms, including bacteria and eukaryotes, for CO2 processing (McFadden and Tabita, 1974).
Carbon isotope fractionation indicates that the Calvin cycle has been used to process CO2 for more than 3.5 billion years (Raven, 1997). The origin of this cycle is not well understood. Two enzymes, Rubisco and PRK, are unique to Calvin-cycle function and they probably provided the evolutionary “breakthrough” with respect to the biogenesis of this metabolic pathway, because the remaining Calvin-cycle enzymes have additional (i.e. non-Calvin cycle) metabolic responsibilities.
PRK catalyzes the reaction Ru5P + ATP → ribulose-1,5-bisphosphate + ADP. Because this reaction is essentially irreversible, the enzyme serves a critical role in regulating the flow of sugars through the CO2-fixation cycle. Some of the first catalysts to occur in primitive cells were similar to extant kinases (Loomis, 1988). These enzymes phosphorylated a wide range of molecules with ATP. Thus, unlike Rubisco, for which no other enzymatic analog exists (McFadden and Tabita, 1974), one may hypothesize that PRK was probably a “retrofitted” kinase. Any enzyme that was capable of moving a phosphate from ATP to an acceptor molecule can serve as a candidate for Calvin-cycle specialization.
Kinases tend to be monomers. However, PRK does not fit this profile, perhaps as a result of Calvin-cycle isolation (Loomis, 1988). The structure of PRK varies extensively among taxa. Terrestrial plants and algal representatives of the phylum Chlorophyta (chl a- and b-containing plants) have the simplest PRK design. In these taxa (Table I), 80- to 90-kD holoenzymes are homodimeric except in Selenastrum minutum, which is heterodimeric (Lin and Turpin, 1992). Spinach PRK exists in the chloroplast stroma in a multienzyme complex (Gontero et al., 1988). All enzymes of this complex are committed to Calvin-cycle function. A similar but smaller multienzyme complex has been described for pea (Sainis and Harris, 1986).
Table I.
Summary of PRK structure and regulatory control requirements among divergent taxa
| Organism | Holoenzyme | Monomer | Subunit | Activation
|
Ref. | |
|---|---|---|---|---|---|---|
| DTT | NADH | |||||
| D | n | |||||
| Chlorophyta | ||||||
| Arabidopsis | NDa | 38,400b | ND | ND | ND | Horsnell and Raines (1991) |
| Ice plant | ND | 44,000b | ND | ND | ND | Michaelowski et al. (1992) |
| Spinach | 90,000 | 44,000 | 2 | +c | ND | Porter et al. (1986) |
| Wheat | ND | 39,200b | ND | + | ND | Raines et al. (1989) |
| Bryopsis maxima | 90,000 | 41,000 | 2 | ND | ND | Satoh et al. (1985) |
| Chlamydomonas reinhardtii | 39,000 | ND | + | ND | Roesler and Oregen (1990) | |
| Chlorella sorokiniana | ND | ND | ND | −d | Tabita (1980) | |
| Scenedesmus obliquus | 83,000 | 42,000 | 2 | + | ND | Lazaro et al. (1986) |
| S. minutum | 83,000 | 41,000 | 2 | ND | ND | Lin and Turpin (1992) |
| 42,000 | ||||||
| Cyanobacteria | ||||||
| A. cylindrica | 72,000 | 43,000 | 2 | + | ND | Serra et al (1989) |
| 26,000 | ||||||
| C. fritschii | 230,000 | 40,000 | 6 | + | − | Marsden and Codd (1984) |
| Synechocystis sp. PCC 6803 | ND | 39,000 | ND | + | ND | Su and Bogorad (1991) |
| Synechococcus sp. PCC 7942 | 178,000 | 42,000 | 4 | + | ND | Wadano et al. (1995) |
| Chromophyta | ||||||
| Heterosigma carterae | 214,000 | 53,000 | 4 | + | − | This work |
| Odentella sp. | ND | 45,000b | ND | ND | ND | K. Weyrauch (1996) Genbank accession no. Y08610 |
| Proteobacteria | ||||||
| Alcaligenes eutrophus | 256,000 | 33,319e | 8 | ND | ND | Siebert et al. (1981); Kossmann et al. (1989) |
| 33,164f | ||||||
| Agmenellum quadruplicatum | ND | ND | ND | ND | + | Tabita (1980) |
| Chromatium sp. | 240,000 | ND | ND | ND | − | Hart and Gibson (1971) |
| Hydrogenomonas eutropha H16 | 237,000 | ND | ND | ND | ND | Abdelal and Schlegel (1974) |
| Nitrobacter winogradskyi | ND | ND | ND | ND | + | Kiesow et al. (1977) |
| Rhodobacter sphaeroides | 33,000 | ND | ND | + | Hallenbeck and Kaplan (1987) | |
| Rhodopseudomonas acidophila | 248,000 | 32,000 | 8 | ND | + | Rippel and Bowien (1984) |
| Rhodopseudomonas capsulata | 220,000 | 36,000 | 6 | ND | ND | Tabita (1980) |
| Rhodopseudomonas spaeroides | ND | 33,000 | ND | ND | + | Hallenbeck and Kaplan (1987) |
| Thiobpacillus neapolitanus | ND | ND | ND | ND | + | Tabita (1980) |
| Xanthobacter flavus H4-14 | ND | 33,409 | ND | ND | ND | Kossman et al. (1989); Meijer et al. (1990) |
a ND, Not determined. b Inferred from DNA sequence data. c +, Present. d −, Absent. e Chromosome encoded. f Plasmid encoded.
The structural identity of PRK among prokaryotes is taxon dependent (for review, see Table I). Cyanobacterial PRK holoenzyme composition is quite variable. For example, the PRK of Anabaena cylindrica is slightly smaller (72 kD) than that found in terrestrial plants. Both 43- and 26-kD composite proteins were observed. It is not known whether the smaller protein represents a subunit of the holoenzyme or a tightly associated contaminating polypeptide. Synechococcus spp. and Chlorogleopsis fritschii PRK both display monomeric subunits of 40 kD. The Synechococcus spp. enzyme, however, is tetrameric in structure (178 kD), whereas the C. fritschii enzyme is hexameric (230 kD). Cyanobacterial PRK holoenzyme structural differences are not seen in proteobacterial PRK enzymes, which exhibit relatively uniform molecular masses. These bacteria contain a holoenzyme of 250 kD that is composed of six to eight subunits, each with a molecular mass of approximately 35 kD.
Although the data are not entirely representative of all cells that use the Calvin cycle for CO2 fixation, it appears that PRK regulation occurs by two different mechanisms. These differences in PRK regulation may reflect separate evolutionary origins or early evolutionary divergence of PRK. Many but not all bacterial PRK enzymes are strongly activated by NADH (Table I) (Gibson and Tabita, 1987). PRK activation via this type of allosteric modulation may be quite effective (Kiesow et al., 1977) because energy-linked NADH-generating reactions (e.g. via nitrate oxidation) are functional in both photosynthetic and chemoautotrophic bacteria. In contrast, chlorophytes (chl a- and b-containing plants) and cyanobacteria use thioredoxin in the modulation of Calvin-cycle enzymes, including PRK (Holmgren, 1985; Hartman et al., 1990). In the light, electrons from chl are transferred to Fd and then to thioredoxin. The reduced thioredoxin activates PRK by affecting the redox-active S-S bridge of the enzyme. Milanez et al. (1991) has shown that regulation of PRK activity involves Cys residues 16 and 55 in the protein. Sequence analysis has shown that equivalent Cys residues are not found in allosterically regulated proteobacterial PRK.
The Chromophyta (chl a- and c-containing plants) represent a large assemblage of organisms that rank as the primary producers in many aquatic ecosystems. Chromophytes have a significant effect on the global carbon budget. Approximately one-third of the total carbon fixed worldwide is processed by these organisms (Raven, 1997). To our knowledge, there has been no analysis of PRK structure and function in a marine eukaryote. Here we present information on the isolation and characterization of PRK from the toxic, unicellular marine alga H. carterae.
MATERIALS AND METHODS
All chemicals, enzymes, and column supports were obtained from Sigma except for MgCl2 and KCl (J.T. Baker), Tris base (GIBCO-BRL), Sephadex G-50 and DE-52 cellulose (Whatman), and a Superose-6 flow performance liquid chromatography column (Pharmacia). Antibodies to Alcaligenes eutrophus PRK were kindly provided by B. Bowien (Universität Göttingen, Germany).
Algal Culture
Heterosigma carterae (Taylor, 1992), isolate Carter, was grown in an artificial seawater medium (McIntosh and Cattolico, 1978) at 20°C on a 12-h light/12-h dark diel cycle. Cultures of 1 L were maintained in 2.8-L Fernbach flasks with continuous shaking at 60 rpm. Cultures were illuminated with cool-white fluorescent lamps at an intensity of 20 μE m−2 s−1. Cells were counted using a ZBI counter (Coulter, Hialeah, FL) with a 100-μm aperture.
Enzyme Purification
Unless otherwise indicated, all of the following procedures were done at 5°C. Cells were harvested when cultures had a density of approximately 105 cells mL−1 by centrifugation at 1,110g for 10 min. The cells were resuspended to a final concentration of 5 × 107 cells mL−1 in 50 mm Hepes buffer (pH 7.8) that contained 10 mm MgCl2, 0.1% β-mercaptoethanol, and 1 mm PMSF as a protease inhibitor. This mixture was stored at −70°C for later use. An atmosphere of N2 was maintained over the homogenate at all phases of cellular disruption and centrifugation. Approximately 4 × 109 stored cells (40 L of cells at 105 cells mL−1) were thawed on ice, diluted to a final concentration of 2 × 107 cells mL−1 using an equilibration buffer (50 mm Tris [pH 7.6], 0.5 mm EDTA, 100 mm KCl) that contained 0.1% β-mercaptoethanol, and further disrupted by a single passage through a precooled French pressure cell at 8,000 p.s.i. The broken cells were centrifuged at 10,900g for 15 min. Retrieved supernatant was then centrifuged at 100,000g for 40 min. A degassed solution that contained saturated (NH4)2SO4 (pH 7.6) and 0.1% β-mercaptoethanol was slowly added to the 100,000g supernatant to attain 35% final concentration. This mixture was stirred for 30 min. The resulting precipitate was removed by centrifugation at 8,700g for 30 min. Saturated (NH4)2SO4 was added to the supernatant to achieve a final concentration of 50%, after which solid (NH4)2SO4 was added to 80% final concentration. The solution was subject to continuous stirring for 30 min. The precipitate, retrieved by centrifuging at 8,700g for 30 min, was resuspended in 10 mL of equilibration buffer containing 0.1% β-mercaptoethanol. The suspension was flushed with N2. The 35 to 80% (NH4)2SO4 fraction was desalted by passage through a Sephadex G-50 column (approximately 60 mL) that had been swollen in water and washed with the same buffer. Fractions containing the enzyme were further desalted by dialysis against the same buffer for a minimum of 6 h. The dialyzed solution was then subject to chromatographic separation (at room temperature) using DE-52 microcellulose anion-exchange resin (Whatman) that had been swollen overnight in water, washed with equilibration buffer, and then activated for 60 min by adding 500 mm KCl to the equilibration buffer.
The column was packed to a bed volume of 25 mL in equilibration buffer containing 1 mm DTT using a pressure pump set at 100 mL h−1. The sample was loaded at 50 mL h−1 and the column was washed with six bed volumes of equilibration buffer. Protein was eluted at 50 mL h−1 using a linear salt gradient of 100 to 400 mm KCl in equilibration buffer containing 1 mm DTT. Fractions showing enzyme activity were pooled and concentrated to one-tenth the volume by high-pressure ultrafiltration using an Amicon (Beverly, MA) model 12 stirred cell equipped with a PM30 Diaflo membrane at a maximum pressure (60 p.s.i.) of N2. The sample was activated by adding 10 mm DTT, flushed with N2, and stored on ice for 2 h.
Agarose-Reactive Green 19 that had been swollen in water for several hours was equilibrated with 10 mm Bicine-KOH buffer (pH 8.0) and then reequilibrated with 10 mm Bicine-KOH (pH 6.8) buffer that contained 10 mm MgCl2. The pH of the PRK sample that had been activated was adjusted to 6.8 with Bicine-KOH (pH 5.0). The sample was then added to this affinity slurry and maintained at 4°C for 18 h to allow complete binding of the enzyme. The Reactive Green 19 slurry containing bound enzyme was brought to room temperature and packed into a column that was washed with 10 bed volumes of 20 mm Tris-HCl (pH 7.6), 10 mm DTT. Enzyme was eluted with 10 mm ATP in the same buffer. Retrieved enzyme was further concentrated by ultrafiltration as described above. The recovered protein was stored under N2 at −70°C after the addition of 10% glycerol, 1 mm leupeptin, and 10 mm DTT. This fraction was used for enzyme characterization and electrophoretic analyses.
Active fractions from the affinity column were chromatographed on a Superose-6 gel-filtration column that was equilibrated with buffer composed of 50 mm Tris (pH 7.6), 0.5 mm EDTA, 100 mm KCl, 10 mm DTT. For molecular mass determination of the native enzyme size, the Superose-6 column was calibrated with catalase (232 kD), aldolase (158 kD), BSA (68 kD), and Cyt c (12.5 kD). The following equation was used to calculate the elution constant (Kav) of both PRK and the protein standards: Kav = (Ve − Vo)/(Vt − Vo), where Ve is the elution volume of sample, Vo is the void volume, and (Vt − Vo) is the volume of gel-forming substance (Pharmacia Biotech, 1993). Protein profiles were spectrophotometrically monitored during chromatography at 280 nm. Protein was quantified using a Bio-Rad assay kit with BSA as a standard.
Protein Electrophoresis and Sequencing
Fractions showing PRK activity from the DE-52 column and the Reactive Green 19 column were pooled, concentrated, and resolved on 12% SDS-PAGE gels (Laemmli, 1970). Gels were either stained with Coomassie blue R-250 or silver stained. PRK subunit size was determined by comparison with known standards.
Proteins (10 μg) in the PRK-containing pool from a post-Reactive Green 19 column were separated on SDS-PAGE, transferred to a PVDF membrane, and stained with Coomassie blue R-250. The major band was sequenced by Edman degradation at the amino terminus (Protein Sequencing Facility, University of Washington, Seattle).
Enzyme Assays
Two coupled assays were used to assess PRK activity during the course of this study. In the radioactive procedure, the conversion of Ru5P to a three-carbon sugar was assessed by monitoring the incorporation of 14C into an acid-precipitable product (Paulsen and Lane, 1966). The reaction mixture consisted of 0.5 μg of PRK extract, 50 μg of spinach Rubisco, 50 mm Tris (pH 8.0), 20 mm DTT, 20% glycerol, 30 mm NaHCO3, and 10 mm MgCl2 in 50 μL, which was incubated on ice for 30 min. The volume of the reaction mixture was increased to 200 μL, and the mixture was adjusted to a final concentration of 100 mm Tris (pH 8.0), 5 mm ATP, 10 mm MgCl2, 20% glycerol, and 20 mm DTT. This new solution was incubated on ice for 15 min, after which 1 μL (0.06 Ci/mol) of NaH14CO3 was added. The reaction was initiated with 2 mm Ru5P and incubated for 10 min at 30°C. The assay mixture was then added to 200 μL of 2 n HCl that was contained in a scintillation vial, brought to dryness by heating in a 95°C water bath, dissolved in 200 μL of distilled water, and the product was counted in 3 mL of scintillation fluid (Ecolume, ICN) using a scintillation counter (Beckman). Parameters of pH and temperature were optimized for this assay as well as for the spectrophotometric assay.
In the spectrophotometric assay, the activity of PRK was analyzed via a coupled reaction (Kagawa, 1982). Use of ATP during the phosphorylation of Ru5P by PRK was coupled to the conversion of PEP to lactate. The oxidation of NADH in this scheme was monitored at 340 nm. The reaction mixture (1.0 mL) contained 100 mm Tris (pH 8.0), 3 mm MgCl2, 10 mm DTT, 2 mm PEP, 2 mm ATP, 4 mm Rib5P, phosphoriboisomerase (2 units), lactic dehydrogenase (8 units), pyruvate kinase (13 units), and 0.1 mm NADH. To eliminate effects of contaminating ADP, the assay mixture minus PRK and Rib5P was incubated at 25°C for 1 min. Background absorbance was observed by incubating the reaction mixture plus 5 μg of PRK enzyme. The reaction was initiated by the addition of Rib5P. Change in absorbance was monitored using a UV-160 spectrophotometer (Shimadzu Scientific, Columbia, MD) at 340 nm set on kinetic mode. One unit of activity was defined as the amount of enzyme that catalyzed the oxidation of 1 μmol of NADH per minute (or production of ADP). To determine the Km of substrate, commercially available Ru5P was added to the reaction mixture (instead of converting Rib5P to Ru5P with phosphoriboisomerase, as is routinely done for activity assays).
RESULTS
Enzyme Isolation and Physical Characteristics
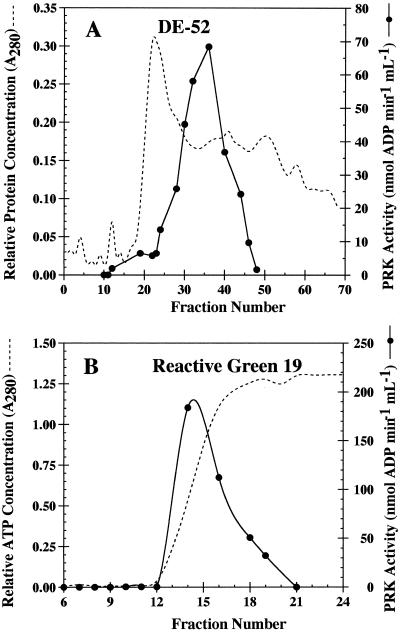
Although initial PRK isolations were done with isolated chloroplasts, subsequent work demonstrated that the use of whole cells provided an easier and more efficient approach for enzyme retrieval. H. carterae, which is naturally wall-less, is easily disrupted using a French pressure cell. A typical PRK purification profile is presented in Table II. Enzyme initially obtained from a 35 to 80% (NH4)2SO4 fractionation was desalted by exclusion chromatography on a Sephadex G-50 column. The enzyme was further purified by serial application of DE-52 chromatography (Fig. 1A) followed by affinity chromatography with agarose-Reactive Green 19 (Fig. 1B). This last step in PRK isolation resulted in a highly enriched preparation (218 units mg−1 protein) that was stored under N2 at −70°C in the presence of glycerol, leupeptin, and DTT for at least 1 month without appreciable loss of activity.
Table II.
H. carterae PRK purification procedure
| Step | Total Activity | Total Protein | Specific Activity | Purification | Units Recovered |
|---|---|---|---|---|---|
| units | mg | units mg−1 | -fold | % | |
| 35–80% (NH4)2SO4 fraction | 86.8 | 86.80 | 1.0 | 1.4 | 100 |
| (from crude) | |||||
| Post-G-50 (Sephadex) | 88.7 | 40.30 | 2.2 | 3.1 | 100 |
| Post-DE-52 (anion exchange) | 35.5 | 2.50 | 14.2 | 20.3 | 41 |
| Post-Reactive Green 19 (affinity) | 13.0 | 0.06 | 217.8 | 311.0 | 15 |
PRK activity was monitored using a spectrophotometric assay. Enzyme units are micromoles of ADP per minute. Protein was quantified using a protein assay kit (Bio-Rad) with BSA as a standard. Purification was determined using specific activity at each step compared with the specific activity of the crude enzyme (0.7 unit mg−1).
Figure 1.
Chromatographic profile of H. carterae PRK fractions collected after Whatman DE-52 anion-exchange (A) and Reactive Green 19 affinity column chromatography (B). PRK activity was determined using the spectrophotometric assay. Relative protein (post-DE-52) and relative ATP concentration (post-Reactive Green 19) were determined by monitoring A280. The A280 for ATP concentration contained less than 5% protein in each fraction.
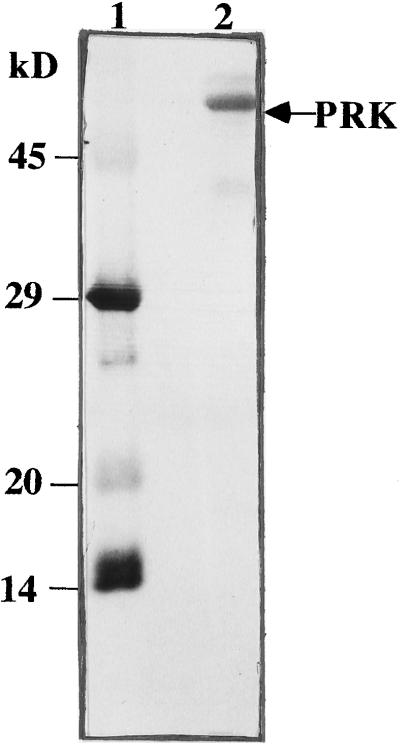
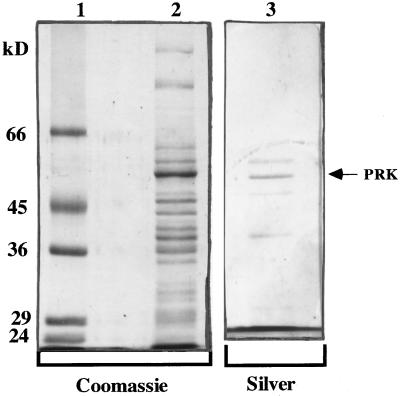
All PRK enzymes observed to date are composed of two to eight subunits. Subunit size for the H. carterae PRK enzyme was electrophoretically resolved by 12% SDS-PAGE. As seen in Figure 2, this chrysophytic enzyme is composed of subunits that have a molecular mass of 53 ± 1 kD. Additional minor protein bands were routinely observed in the enzyme preparation (Fig. 3). The sequence of the amino terminus of the 53-kD protein verified a PRK identity (Fig. 2; Table III). Because H. carterae Rubisco small subunit protein showed significant sequence homology to that of A. eutrophus (Boczar et al., 1989), and given the clustered arrangement of these two genes in proteobacteria (Gibson et al., 1990), we hypothesized that PRK and Rubisco may have been moved into the chloroplast as a laterally transferred cassette. However, antisera to A. eutrophus PRK failed to cross-react with this putative H. carterae PRK subunit (data not shown).
Figure 2.
H. carterae PRK protein eluted from the Reactive Green 19 affinity column was resolved on a 12% SDS-PAGE gel, transferred to a PVDF membrane, and stained with Coomassie blue. The band (10 μg, lane 2) marked with an arrow was cut from the membrane, sequenced at the amino terminus, and identified as PRK. Molecular mass standards (lane 1) are α-lactalbumin (14.2 kD), trypsin inhibitor (20.1 kD), carbonic anhydrase (29 kD), and ovalbumin (45 kD).
Figure 3.
Coomassie blue- and silver-stained 12% SDS-polyacrylamide gels at various protein purification steps. Lysed H. carterae cells containing PRK activity were fractionated with (NH4)2SO4, passed through a Sephadex G-50 column, and further purified on a DE-52 anion-exchange column followed by a Reactive-Green 19 affinity column. The fractions with PRK activity are shown after DE-52 anion-exchange (lane 2) and Reactive Green 19 affinity column chromatography (lane 3). The PRK band is marked to the right with an arrow. Molecular mass standards (lane 1) are trypsinogen (24 kD), carbonic anhydrase (29 kD), glyceraldehyde-3-phosphate dehydrogenase (36 kD), ovalbumin (45 kD), and BSA (66 kD).
Table III.
Alignment of amino N-terminal amino acid sequences of PRK from selected taxa
| Organism | Sequence | Ref. |
|---|---|---|
| Arabidopsis | A · · Q E T I V · I G L A A D S G C G K S T F | Horsnell and Raines (1991) |
| Ice plant | A G D S Q T I V · I G L A A D S G C G K S T F | Michaelowski et al. (1992) |
| Spinach | C S Q Q Q T I V · I G L A A D S G C G K S T F | Roesler and Ogren (1988) |
| Wheat | A · V E Q P I V · I G L A A D S G C G K S T F | Raines et al. (1989) |
| Chlamydomonasreinhardtii | A D K D K T V V · I G L A A D S G C G K S T F | Roesler and Ogren (1990) |
| Selenastrum minutum A | A D G L X · V V · I G L A A D | Lin and Turpin (1992) |
| Selenastrum minutum B | X G L X I V V · G L A A D | Lin and Turpin (1992) |
| H. carterae | K E G E E · V V L I G V A A D S G CG K | This work |
| Odontella spp. | K E G E K P I V · I G V A A D S G C G K S T F | K. Weygrauch (1996) (GenBank accession no. Y08610) |
| Synechococcus sp. PCC 7942 | S K P · D R V V L I G V A G D S G C G K S T F | Wadano et al. (1995) |
| Synechocystis sp. PCC 6803 | T T Q L D R V V L I G V A G D S G C G K S T F | Su and Bogorad (1991) |
| Xanthobacter flavus | M S I K H P I I V V T G S S G A G T T S V | Meijer et al. (199) |
| Rhodobacter sphaeroides | V S K K Y P I I S V V G S S G A G T S T V | Gibson et al. (1990) |
Residues in boldface letters represent the ATP-binding domain for oxygenic autotrophs. The shaded area indicates a conserved Cys. Dots represent amino acid deletion.
To determine PRK holoenzyme size, affinity column-purified protein was subject to Superose-6 flow performance liquid chromatography (Fig. 4). The molecular mass of 214 ± 12.6 kD observed in these experiments suggested that H. carterae PRK is composed of four subunits.
Figure 4.
Determination of H. carterae PRK native size. Kav was calculated as described in Methods.
Catalytic and Regulatory Properties
The kinetic characteristics of H. carterae PRK were analyzed using post-Reactive Green 19 affinity column-purified enzyme. Classic Michaelis-Menten kinetics were seen for both ATP and Ru5P, suggesting that no positive cooperativity occurs between the enzyme and those substrates. Lineweaver-Burk plots resulted in an observed Km(ATP) of 208 μm (Fig. 5A) and an observed Km(Ru5P) of 226 μm (Fig. 5B) for the enzyme.
Figure 5.
Rate of ADP production by partially purified H. carterae PRK at various concentrations of ATP (0–1.5 mm, radioactive assay) (A) and Ru5P (0–1 mm, spectrophotometric assay) (B). Insets show Lineweaver-Burk plots of each data set. Km(ATP) = 208 μm and Km(Ru5P) = 226 μm.
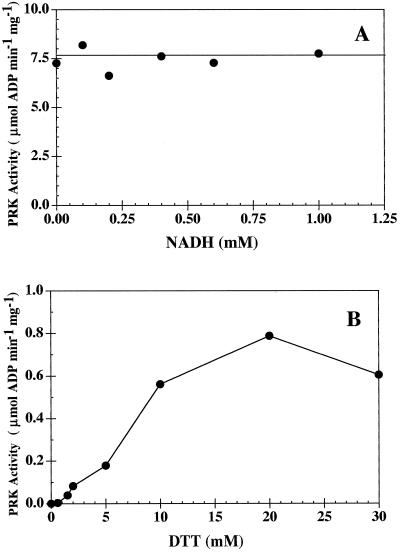
To further assess whether H. carterae PRK was allosterically regulated by NADH (as found in many proteobacteria) or affected by a thioredoxin/Fd system (as seen in cyanobacteria and terrestrial plants), the PRK was incubated at 4°C for at least 30 min in the presence of varying concentrations of either NADH (0–1.0 mm) or DTT (0–30 mm). The enzyme was dialyzed against equilibration buffer at 4°C for 2 h before incubation.
Partially purified PRK (post-Reactive Green 19) was incubated with increasing concentrations of NADH before the radioactive assay was initiated with Ru5P. Enzyme activity was not affected by NADH (Fig. 6A). In contrast, DTT caused a significant increase in enzymatic activity (Fig. 6B). Partially purified PRK (post-DE-52) was incubated with increasing concentrations of DTT before the spectrophotometric assay was initiated with Ru5P. The concentration of DTT in the spectrophotometric reaction mixture was kept constant to ensure that the other enzymes in the assay were fully active.
Figure 6.
Rate of ADP production by partially purified H. carterae PRK at increasing concentrations of NADH (0–1 mm, radioactive assay, se less than 15%) (A) and DTT (0–30 mm, spectrophotometric assay, se less than 5%) (B).
Amino-Terminal Amino Acid Sequencing
Proteobacterial PRK polypeptides have amino acid sequences that are completely divergent from those reported for cyanobacteria and terrestrial plants (Porter et al., 1988; Kossman et al., 1989). The amino termini of these two kinase variants have signature sequences (Table III) that identify each PRK type as either allosterically or thioredoxin/Fd regulated. Using Edman degradation, the amino-terminal 21 amino acids of H. carterae PRK were determined using protein obtained from the Reactive Green 19 affinity column. Its sequence demonstrates that the H. carterae polypeptide has a consensus ATP-binding domain that shows almost perfect homology to that observed for PRK in both cyanobacteria and terrestrial plants (thioredoxin/Fd PRK). Also evident in this short sequence is a cysteinyl residue (Cys-19) located within the nucleotide-binding domain. This amino acid serves as a regulatory disulfide in thioredoxin/Fd PRK enzymes (Milanez et al., 1991). The Edman method released a single amino acid at each cycle, demonstrating homogeneity in the H. carterae PRK polypeptide.
DISCUSSION
Enzyme Purification
Obtaining sufficient biomass provided an early challenge in PRK isolation even though H. carterae was harvested at h 6 a point in the 12-h light/12-h dark cell cycle when the events of chloroplast biogenesis (transcription initiation, photosynthetic capacity, protein accumulation) are maximal (Reith and Cattolico, 1985; Reynolds et al., 1993; Doran and Cattolico, 1997). In the laboratory Heterosigma spp. grow poorly in volumes of more than 1.5 L, and, like many chromophytes, their cells fill with polysaccharide material and other secondary metabolites as they enter later phases of exponential cell growth (R.A. Cattolico, unpublished data). This factor precluded cell harvest at high cell densities. Enzyme could be successfully retrieved from accumulated cells (40 L of culture was needed per enzyme purification procedure) when stored at −70° in the presence of PMSF. This observation may be valuable to studies of PRK in other unicellular chromophytes.
Treatment with 35 to 80% (NH4)2SO4 followed by Sephadex G-50 and ion-exchange chromatography provided a small (20-fold) but effective initial step in H. carterae PRK isolation. Most productive in the recovery of this enzyme was the use of Reactive Green affinity chromatography (300-fold purification). Triazine-based dyes that mimic coenzyme binding have been widely used as adsorbents for the purification of dehydrogenases and kinases (Clonis and Lowe, 1980). Although Cibacron Blue and Reactive Red columns have been successfully used to isolate PRK from both bacterial and eukaryotic sources (Siebert et al., 1981; Ashton, 1984; Satoh et al., 1985; Porter et al., 1986; Serra et al., 1989), these columns did not bind the H. carterae enzyme efficiently.
Enzyme Size and Structure
As seen in Table II, PRK subunit size appears to vary significantly among taxa. Terrestrial plant, chlorophytic algae, and cyanobacterial PRK monomers (40 kD) are approximately 20% larger than those found in proteobacteria (32 kD). The H. carterae monomer (53 kD) is more than 50% greater in size than those in the proteobacterial cluster. It is premature to speculate on the significance of this size difference until the sequence of the H. carterae protein has been completed and the PRK monomer size in a larger number of non-chl b-containing plants has been assessed.
If the H. carterae enzyme is consistently found to be tetrameric in future studies, then the question concerning variability in quaternary structure among Calvin-cycle kinases must be addressed. It has been suggested by Meijer et al. (1990) that changes in holoenzyme subunit number may be related to differential conservation of domains critical to subunit interaction. One could argue that these changes would be rare events, and thus be taxonomically isolated. Alternatively, such changes could occur frequently and, if so, numerous variations in PRK holoenzyme structure would be observed within close phylogenetic lineages.
Extensive analysis seems to verify a consistent octameric PRK within all proteobacteria (see Tabita, 1980). Crystallographic analysis of R. sphaeroides shows the monomers to be stacked in two planar tetramers (Roberts et al., 1995). In contrast, sequence analysis of rRNA places the two genera Anabaena (dimeric PRK) and Chlorogleopsis (hexameric PRK) into sister groups within one of the eight phylogenetic clusters described for cyanobacteria (Wilmotte, 1994). Both of these prokaryotes are filamentous and fix N2. Given the phylogenetic proximity of these algae, one might propose that the difference in PRK size results from a simple rather than a complex evolutionary change.
Obviously, studies of other eukaryotic PRK enzymes are needed. Although it is well established that chlorophytic and chromophytic/rhodophytic taxa represent a major divergence in the evolution of autotrophic organisms, the dimeric PRK structure routinely accepted for chlorophytes (spinach, Scenedesmus, Selenastrum), and the tetrameric enzyme structure for chromophytes (Heterosigma), represent an insufficient data set for good comparative analysis. Alternatively, one may argue that the shift in enzyme structure (i.e. dimeric ⇆ tetrameric state) may simply reflect a means of regulating catalytic efficiency. Additional data will also allow comparison of the evolutionary channeling that has occurred for PRK (sequences and holoenzyme structure) with that of Rubisco. At least for Rubisco, organelle coding site and ancestral proteobacterial identity appear to be tightly correlated with either the chlorophyte or chromophyte/rhodophyte lineages (for review, see Delaney et al., 1995).
Microcompartmentation of Calvin-cycle enzymes occurs in terrestrial plant and green algal chloroplasts. As many as five enzymes, including PRK, can be found in large (500–900 kD) arrays (Gontero et al., 1988; Süss et al., 1993; Wedel et al., 1997) that theoretically enhance progression of catalytic intermediates from the active site of one enzyme to the active site of another within the complex, enhancing enzymatic efficiency. No similar association of PRK in carboxysomes has been observed for either Thiobacillus neapolitanus (Holthuijzen et al., 1986) or Chlorogleopsis fritschii (Marsden et al., 1984). Suc-gradient analysis of disrupted H. carterae cells suggests that the PRK of this alga may not exist in a large complex. These data are consistent with observations made by Mangeney et al. (1987), who failed to find PRK within carboxysome-like structures in the cyanelles of Cyanophora paradoxa and Glaucocystis nostochinearum, which had been immunocytochemically labeled with C. fritschii PRK antiserum.
Enzyme Regulation
Protein sequencing of the H. carterae PRK amino terminus has revealed a distinct similarity between the kinase of this chromophytic alga and those of cyanobacteria and chlorophytes, but low sequence identity to proteobacterial enzymes. All amino acids of the H. carterae ATP-binding domain are homologous to those seen in cyanobacteria and chlorophytic plants.
If H. carterae PRK is similar to enzymes found in oxygenic photoautotrophs, then one would expect that the enzyme would be regulated by a thioredoxin/Fd cascade rather than the allosteric control used by proteobacteria. Sequence data (the presence of Cys-16) and the stimulatory effect of DTT support this hypothesis. Like many thioredoxin/Fd PRK enzymes, H. carterae PRK quickly loses activity in an oxidized state, and compounds that have been implicated in the alteration of proteobacterial PRK configuration (e.g. NADH [Fig. 6A]) appear to have no effect on the DTT-activated catalytic efficiency of this enzyme.
Data show that H. carterae PRK displays hyperbolic kinetics similar to those seen for terrestrial plants and cyanobacteria for both ATP (Fig. 5A) and Ru5P (Fig. 5B). This response is unlike the sigmoidal kinetics observed for proteobacteria, which indicate positive cooperativity for both substrates (Abdelal and Schlegel, 1974). The Km values calculated for H. carterae PRK were 208 μm (ATP) and 226 μm (Ru5P). A literature review of PRK function from photooxygenic species provides no clear answer to whether Km(ATP) (53–1420 μm) or Km(Ru5P) (36–330 μm) values are associated with holoenzyme structure or taxonomic affiliation (Gardemann et al., 1983; Satoh et al., 1985; Roesler and Ogren, 1990, Milanez et al., 1991; Su and Bogorad, 1991).
ACKNOWLEDGMENTS
We thank Carrine Blank for initiating these studies, Laurie Connell for technical advice, William Hatheway for extensive support in this effort, and M. Kay Suiter for help in manuscript preparation. R.A.C. dedicates this work to the memory of her father, A.J. Cattolico.
Abbreviations:
- chl
chlorophyll
- DE-52
diethylaminoethyl cellulose
- PRK
phosphoribulokinase
- Rib5P
ribose-5-phosphate
- Ru5P
ribulose-5-phosphate
Footnotes
Supported by National Science Foundation grant no. MCB-9305923.
LITERATURE CITED
- Abdelal ATH, Schlegel HG. Purification and regulatory properties of phosphoribulokinase from Hydrogenomonas eutropha H 16. Biochem J. 1974;139:481–489. doi: 10.1042/bj1390481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashton AR. An affinity label for the regulatory dithiol of ribulose-5-phosphate kinase from maize (Zea mays) Biochem J. 1984;217:79–84. doi: 10.1042/bj2170079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boczar BA, Delaney TP, Cattolico RA. Gene for the ribulose-1,5-bisphosphate carboxylase small subunit protein of the marine chromophyte Olisthodiscus luteus is similar to that of a chemoautotrophic bacterium. Proc Natl Acad Sci USA. 1989;86:4996–4999. doi: 10.1073/pnas.86.13.4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clonis YD, Lowe CR. Triazine dyes, a new class of affinity labels for nucleotide-dependent enzymes. Biochem J. 1980;191:247–251. doi: 10.1042/bj1910247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney T, Hardison L, Cattolico RA. Evolution of plastid genomes: inferences from discordant molecular phylogenies. In: Sandgren C, Smol J, Kristiansen J, editors. Chrysophytic Algae Ecology, Physiology and Development. Cambridge, UK: Cambridge University Press; 1995. pp. 25–45. [Google Scholar]
- Doran E, Cattolico RA. Photoregulation of chloroplast gene transcription in the chromophytic alga Heterosigma carterae. Plant Physiol. 1997;115:773–781. doi: 10.1104/pp.115.2.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs G. CO2 fixation in acetogenic bacteria: variations on a theme. FEMS Microbiol Rev. 1986;39:181–213. [Google Scholar]
- Gardemann A, Stitt M, Heldt HW. Regulation of spinach ribulose-5-phosphate kinase by stromal metabolite levels. Biochim Biophys Acta. 1983;722:51–60. [Google Scholar]
- Gibson JL, Chen J-H, Tower PA, Tabita FR. The form II fructose 1,6-bisphosphatase and phosphoribulokinase genes form part of a large operon in Rhodobacter sphaeroides: primary structure and insertional mutagenesis analysis. Biochemistry. 1990;29:8085–8093. doi: 10.1021/bi00487a014. [DOI] [PubMed] [Google Scholar]
- Gibson JL, Tabita R. Organization of phosphoribulokinase and ribulose bisphosphate carboxylase/oxygenase genes in Rhodopseudomonas (Rhodobacter) sphaeroides. J Bacteriol. 1987;169:3685–3690. doi: 10.1128/jb.169.8.3685-3690.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gontero B, Cárdenas ML, Ricard J. A functional five-enzyme complex of chloroplasts involved in the Calvin cycle. Eur J Biochem. 1988;173:437–443. doi: 10.1111/j.1432-1033.1988.tb14018.x. [DOI] [PubMed] [Google Scholar]
- Hallenbeck PL, Kaplan S. Cloning of the gene for phosphoribulokinase activity from Rhodobacter sphaeroides and its expression in Escherichia coli. J Bacteriol. 1987;169:3669–3678. doi: 10.1128/jb.169.8.3669-3678.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart BA, Gibson J. Ribulose 5-phosphate kinase from Chromatium sp. strain D. Arch Biochem Biophys. 1971;144:308–321. doi: 10.1016/0003-9861(71)90483-8. [DOI] [PubMed] [Google Scholar]
- Hartman H, Syyanen M, Buchanan BB. Contrasting evolutionary histories of chloroplast thioredoxins f and m. Mol Biol Evol. 1990;7:247–254. doi: 10.1093/oxfordjournals.molbev.a040602. [DOI] [PubMed] [Google Scholar]
- Hemming A, Blotevogel KH. A new pathway for CO2 fixation in methanogenic bacteria. Trends Biochem Sci. 1985;10:198–200. [Google Scholar]
- Holmgren A. Thioredoxin. Annu Rev Biochem. 1985;54:237–271. doi: 10.1146/annurev.bi.54.070185.001321. [DOI] [PubMed] [Google Scholar]
- Holthuijzen YA, van Breeman JFL, Kuenen JG, Konings WN. Protein composition of the carboxysomes of Thiobacillus neapolitanus. Arch Microbiol. 1986;144:398–404. [Google Scholar]
- Horsnell PR, Raines CA. Nucleotide sequence of a cDNA clone encoding chloroplast phosphoribulokinase from Arabidopsis thaliana. Plant Mol Biol. 1991;17:183–184. doi: 10.1007/BF00036828. [DOI] [PubMed] [Google Scholar]
- Kagawa T (1982) Isolation and purification of ribulose-5-phosphate kinase from Nicotiana glutinosa. In M Edelman, RB Hallick, N-H Chua, eds, Methods in Chloroplast Molecular Biology. Elsevier Press, Amsterdam, The Netherlands, pp 695–705
- Kiesow LA, Lindsley BF, Bless JW. Phosphoribulokinase from Nitrobacter winogradskyi: activation by reduced nicotinamide adenine dinucleotide and inhibition by pyridoxal phosphate. J Bacteriol. 1977;130:20–25. doi: 10.1128/jb.130.1.20-25.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kossmann J, Klintworth R, Bowien B. Sequence analysis of the chromosomal and plasmid genes encoding phosphoribulokinase from Alcaligenes eutrophus. Gene. 1989;85:247–252. doi: 10.1016/0378-1119(89)90490-3. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lazaro JJ, Sutton CW, Nicholson S, Powls R. Characterization of two forms of phosphoribulokinase isolated from the green alga, Scenedesmus obliquus. Eur J Biochem. 1986;156:423–429. doi: 10.1111/j.1432-1033.1986.tb09599.x. [DOI] [PubMed] [Google Scholar]
- Lin M, Turpin DH. Purification and molecular and immunological characterization of a unique phosphoribulokinase from the green alga Selenastrum minutum. Plant Physiol. 1992;98:82–88. doi: 10.1104/pp.98.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis WF. Four Billion Years: An Essay on the Evolution of Genes and Organisms. Sunderland, MA: Sinauer Associates; 1988. [Google Scholar]
- Mangeney E, Hawthornthwaite AM, Codd GA, Gibbs SP. Immunocytochemical localization of phosphoribulose kinase in the cyanelles of Cyanophora paradoxa and Glaucocystis nostochinearum. Plant Physiol. 1987;84:1028–1032. doi: 10.1104/pp.84.4.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden WJN, Codd GA. Purification and molecular and catalytic properties of phosphoribulokinase from the cyanobacterium Chlorogleopsis fritschii. J Gen Microbiol. 1984;130:999–1006. [Google Scholar]
- Marsden WJN, Lanaras T, Codd GA. Subcellular segregation of phosphoribulokinase and ribulose-1,5-bisphosphate carboxylase/oxygenase in the cyanobacterium Chlorogleopsis fritschii. J Gen Microbiol. 1984;130:2089–2093. [Google Scholar]
- McFadden BA, Tabita FR. D-Ribulose-1,5-diphosphate carboxylase and the evolution of autotrophy. Biosystems. 1974;6:93–112. doi: 10.1016/0303-2647(74)90002-1. [DOI] [PubMed] [Google Scholar]
- McIntosh L, Cattolico RA. Preservation of algal and higher plant ribosomal RNA integrity during extraction and electrophoretic quantitation. Anal Biochem. 1978;91:600–612. doi: 10.1016/0003-2697(78)90546-8. [DOI] [PubMed] [Google Scholar]
- Meijer WG, Enequist HG, Terpstra P, Dijkhuizen L. Nucleotide sequences of the genes encoding fructosebisphosphatase and phosphoribulokinase from Xanthobacter flavus H4–14. J Gen Microbiol. 1990;136:2225–2230. doi: 10.1099/00221287-136-11-2225. [DOI] [PubMed] [Google Scholar]
- Michaelowski CB, DeRocher EJ, Bohnert H, Salvucci ME. Phosphoribulokinase from ice plant: transcription, transcripts and protein expressions during environmental stress. Photosynth Res. 1992;31:127–138. doi: 10.1007/BF00028789. [DOI] [PubMed] [Google Scholar]
- Milanez S, Mural RJ, Hartman FC. Roles of cysteinyl residues of phosphoribulokinase as examined by site-directed mutagenesis. J Biol Chem. 1991;266:10694–10699. [PubMed] [Google Scholar]
- Paulsen JM, Lane MD. Spinach ribulose disphosphate carboxylase I. Purification and properties of the enzyme. Biochemistry. 1966;5:2350–2357. doi: 10.1021/bi00871a025. [DOI] [PubMed] [Google Scholar]
- Pharmacia Biotech (1993) Gel Filtration Principles and Methods, Ed 6. Rahms i, Lund, Sweden, pp 6–13
- Porter MA, Milanez S, Stringer CD, Hartman FC. Purification and characterization of ribulose-5-phosphate kinase from spinach. Arch Biochem Biophys. 1986;245:14–23. doi: 10.1016/0003-9861(86)90185-2. [DOI] [PubMed] [Google Scholar]
- Porter MA, Stringer CD, Hartman FC. Characterization of the regulatory thioredoxin site of phosphoribulokinase. J Biol Chem. 1988;263:123–129. [PubMed] [Google Scholar]
- Raines CA, Longstaff M, Lloyd JC, Dyer TA. Complete coding sequence of wheat phosphoribulokinase: developmental and light-dependent expression of the mRNA. Mol Gen Genet. 1989;220:43–48. [PubMed] [Google Scholar]
- Raven JA. Putting the C in phycology. Eur J Phycol. 1997;32:319–333. [Google Scholar]
- Reith M, Cattolico RA. Chloroplast protein synthesis in the chromophytic alga Olisthodiscus luteus. Plant Physiol. 1985;79:231–236. doi: 10.1104/pp.79.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds AE, McConaughy B, Cattolico RA. Chloroplast genes of the marine alga Heterosigma carterae are transcriptionally regulated during a light/dark cycle. Mol Mar Biol Biotech. 1993;2:121–128. [Google Scholar]
- Rippel S, Bowien B. Phosphoribulokinase from Rhodopseudomonas acidophila. Arch Microbiol. 1984;139:207–212. [Google Scholar]
- Roberts DL, Runquist JA, Miziorko HM, Kim J-JP. Crystallization and preliminary X-ray crystallographic analysis of phosphoribulokinase from Rhodobacter sphaeroides. Protein Sci. 1995;4:2442–2443. doi: 10.1002/pro.5560041126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesler KR, Ogren WL. Nucleotide sequence of spinach cDNA encoding phosphoribulokinase. Nucleic Acids Res. 1988;14:7192. doi: 10.1093/nar/16.14.7192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesler KR, Ogren WL. Chlamydomonas reinhardtii phosphoribulokinase sequence, purification, and kinetics. Plant Physiol. 1990;93:188–193. doi: 10.1104/pp.93.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainis JK, Harris GC. The association of ribulose-1,5-bisphosphate carboxylase with phosphoriboisomerase and phosphoribulokinase. Biochem Biophys Res Commun. 1986;139:947–954. doi: 10.1016/s0006-291x(86)80269-8. [DOI] [PubMed] [Google Scholar]
- Satoh H, Okada M, Nakayama K, Murata T. Purification of ribulose 5-phosphate kinase and minor polypeptides of pyrenoid from the green alga Bryopsis maxima. Plant Cell Physiol. 1985;26:931–940. [Google Scholar]
- Schäfer S, Götz M, Eisenreich W, Bacher A, Fuchs G. 13C-NMR study of autotrophic CO2 fixation in Thermoproteus neutrophilus. Eur J Biochem. 1989;184:151–156. doi: 10.1111/j.1432-1033.1989.tb15001.x. [DOI] [PubMed] [Google Scholar]
- Serra JL, Llama MJ, Rowell P, Stewart WDP. Purification and characterization of phosphoribulokinase from the N1-fixing cyanobacterium Anabaena cylindrica. Plant Sci. 1989;59:1–9. [Google Scholar]
- Siebert K, Schobert P, Bowien B. Purification, some catalytic and molecular properties of phosphoribulokinase from Alcaligenes eutrophus. Biochim Biophys Acta. 1981;658:35–44. doi: 10.1016/0005-2744(81)90247-3. [DOI] [PubMed] [Google Scholar]
- Su X, Bogorad L. A residue substitution in phosphoribulokinase of Synechocystis PCC 6803 renders the mutant light-sensitive. J Biol Chem. 1991;266:23698–23705. [PubMed] [Google Scholar]
- Süss K-H, Arkona C, Manteuffel R, Adler K. Calvin cycle multienzyme complexes are bound to chloroplast thylakoid membranes of higher plants in situ. Proc Natl Acad Sci USA. 1993;90:5514–5518. doi: 10.1073/pnas.90.12.5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabita FR. Pyridine nucleotide control and subunit structure of phosphoribulokinase from photosynthetic bacteria. J Bacteriol. 1980;143:1275–1280. doi: 10.1128/jb.143.3.1275-1280.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor FGR. The taxonomy of harmful marine phytoplankton. Giorn Bot Ital. 1992;126:209–219. [Google Scholar]
- Wadano A, Kamata Y, Iwaki T, Nishikawa K, Hirahashi T. Purification and characterization of phosphoribulokinase from the cyanobacterium Synechococcus PCC7942. Plant Cell Physiol. 1995;36:1381–1385. [PubMed] [Google Scholar]
- Wedel N, Soll J, Paap BK. CP12 provides a new mode of light regulation of Calvin cycle activity in higher plants. Proc Natl Acad Sci USA. 1997;94:10479–10484. doi: 10.1073/pnas.94.19.10479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmotte A. Molecular evolution and taxonomy of the cyanobacteria. In: Bryant DA, editor. The Molecular Biology of the Cyanobacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 1–25. [Google Scholar]