Abstract
Killer cell immunoglobulin-like receptors (KIR) are expressed on natural killer (NK) cells and subsets of T cells. The KIR genes are polymorphic and the KIR gene complex is polygenic with varying numbers of inhibitory and activating receptors. HLA class I molecules serve as ligands for the KIR. Interactions of the independently segregating KIR and HLA loci are important for recognition of targets by NK cells as well as NK cell ‘licensing’. Several disease association studies indicate a role for interactions between these loci in infectious diseases, autoimmune/inflammatory disorders, cancer and reproduction. Emerging functional data supports a mechanism based on a continuum of inhibition to activation through various compound KIR-HLA genotypes in diseases.
Keywords: KIR, HLA, Natural killer cells, disease associations
1. Introduction
Natural killer (NK) cells are part of the innate immune arsenal and have important roles in killing virally infected cells and neoplasms, and building vasculature for nourishing the fetus in eutherian mammals. To some extent, mammals have evolved species-specific NK cell receptors that belong to structurally disparate gene families, including C-type lectin receptors in mice (Ly49) and immunoglobulin-like receptors in primates (killer cell immunoglobulin-like receptors; KIR). Their products interact with MHC class I molecules and they appear to be restively adapting to and even surpassing MHC evolution [1–4].
Due to their spontaneous killing of targets that either lack self MHC or express allogeneic MHC, NK cells were initially thought to be non-MHC restricted. Karrë and colleagues observed, however, that NK cells were actively inhibited by targets expressing self MHC class I [5], whereas they were able to reject bone marrow cells from β2-microglobulin deficient mice [6], the so-called “missing self hypothesis”. This hypothesis suggested that NK cells recognized and killed targets lacking MHC class I, which is generally expressed on all healthy nucleated cells but can be downregulated as a result of viral infection or malignant transformation [7]. This led to a paradigm shift in the role of NK cells from being “null cells” to being the first line of defense. The missing self model does not entirely explain the mechanism of NK cell activation and the protection of some normal self cells with little/no expression of MHC against NK cell attack [8]. Therefore, the presence of activating receptors on NK cells and cognate ligands on infected or transformed cells was hypothesized in the ‘induced self recognition’ hypothesis [9]. Indeed, multiple activating receptors that recognize self protein (reviewed in [10]) have been implicated in NK cell killing of virally infected or transformed cells, supporting the hypothesized recognition of ‘induced self’ by NK cell receptors. Along these lines, the mouse activating NK cell receptor Ly49H was shown to recognize the virally encoded MHC-like protein produced by mouse cytomegalovirus, m157 [11].
Once NK cell receptors that recognize MHC class I molecules appeared in mammals, they evolved dramatically. Most of these receptors belong to two main families, the killer cell lectin-like receptor family (KLR) and the killer cell immunoglobulin-like receptor (KIR) family, which have been preferentially expanded across different species. The KLR, for example, have expanded in rodents (Ly49), while the KIRs have expanded in primates, a quintessential example of convergent evolution. Although these distinct sets of receptors do not share a common evolutionary ancestor, they nevertheless have remarkable similarities in that they are polygenic, highly diverse, variegated in expression on NK cell clones, and functionally equivalent. Ly49 and KIR haplotypes vary in both the number and types of genes present, some of which are inhibitory and some activating. The presence of remnants of KIR in rodents [12] and Ly49 in primates [13] [14] suggests that the two receptor gene families evolved independently since the existence of their last common ancestor, perhaps in response to species specific pathogens.
For simplicity in this review, we attribute the consequence of KIR variation on human disease to one cell type, NK cells. In some cases, functional data have employed purified NK cells showing that this cell type is at least mostly responsible for the observed genetic association. However, KIR are also expressed on subsets of T cells, so some of the disease associations mentioned below could actually be due to the effects of KIR in modulating T cell activity, and indeed we note the involvement of T cells when it is known (see sections 5–8 below).
2. KIR
The KIR gene cluster on chromosome 19q13.4 within the leukocyte receptor complex consists of a centromeric and telomeric region separated by KIR2DL4, which is present on virtually all haplotyes (Fig. 1). To date, 14 KIR genes and 2 psuedogenes have been described. The KIR gene cluster is flanked by KIR3DL3 at the centromeric end and KIR3DL2 at the telomeric end, both of which are present on virtually all haplotypes [15]. Recently, a novel and divergent KIR gene, termed KIR3DX1, of unknown function was identified approximately 180 kb centromeric to the KIR cluster and between the two LILR clusters (Fig. 1). This gene is conserved throughout primates, suggesting that it is the ancestral gene from which all other KIR were derived [16]. Interestingly, while it is maintained as a single copy in primates, it has been expanded in cattle [17]. Variability in gene content at the KIR locus appears largely due to gene duplication [18] and non-allelic homologous recombination [19]. Two basic haplotypes have been defined on the basis of gene content, and are termed haplotypes A and B. Haplotype A is uniform in terms of gene content and is composed of five inhibitory genes (KIR2DL1, 2DL3, 3DL1, 3DL2, 3DL3), one activating gene (KIR2DS4), and KIR2DL4, which may have both inhibitory and activating capacity. KIR2DL4 has a charged amino acid in the transmembrane domain that interacts with the activating motif (ITAM) of FCεRI-γ [20], as well as a long cytoplasmic tail containing a single inhibitory motif (ITIM). Interestingly, many A haplotypes possess null variants of both KIR2DS4 [21] and KIR2DL4 [22] that are not expressed on the cell surface. Thus, these haplotypes technically possess no functional activating KIR. The B haplotypes contain variable numbers of activating and inhibitory receptors and are the primary contributors to the extraordinary differences in gene profiles observed in distinct ethnic populations across the world [3].
Figure 1. Genomic organization of the KIR gene cluster.
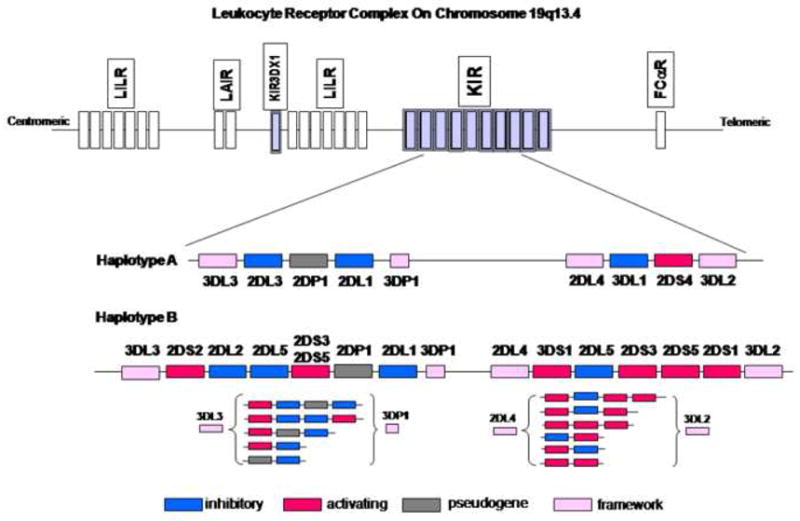
The KIR gene cluster is located on chromosome 19q13.4 within the Leukocyte Receptor Complex. KIR haplotypes vary extensively in gene content. The A haplotype is fixed in terms of gene content, but the B haplotypes are characterized by variable gene numbers (shown in brackets). Framework genes (pink boxes) are present on all haplotypes. The ancestral KIR gene 3DX1 is also shown.
3. KIR ligands
The inhibitory KIR2DL1, 2DL2, and 2DL3 recognize HLA-C ligands. Each HLA-C allotype belongs to one of two ligand groups based on a dimorphism at position 80 in the alpha helix: group 1 (HLA-C1), which has asparagine, and group 2 (HLA-C2), which has lysine at position 80 (Fig. 2). Position 44 in the D1 domain of KIRs appears to determine their ability to discriminate between the two groups of HLA-C allotypes [23], where KIR2DL1 binds HLA-C2 allotypes and KIR2DL2/2DL3 bind HLA-C1. Recent data, however, suggest that KIR2DL2/2DL3 can also bind weakly to some HLA-C2 allotypes in vitro [24]. The authors postulate an interesting evolutionary model whereby the interactions between HLA-C1 and KIR2DL2/2DL3 existed prior to the appearance of the HLA-C2 epitope. The eventual appearance of the HLA-C2 epitope then selected for novel KIR variants that interacted with HLA-C2. The current binding specificities are indicative of a continually evolving system where the newer KIR2DL1 receptors exclusively bind HLA-C2 and the older KIR2DL2 and KIR2DL3 have retained their functional, although weaker, interactions with HLA-C2. KIR3DL1 is known to bind HLA-B allotypes with the Bw4 motif [25] [26], although some low affinity binding with Bw6 has also been reported [27]. The dimorphic position 80 among the Bw4 allotypes affects its interaction with KIR3DL1 subtypes, where HLA-B Bw4- containing allotypes with isoluecine at position 80 (Bw4-80I) generally exhibit stronger inhibition through KIR3DL1 [25, 27, 28]. However, Bw4 allotypes with threonine at position 80 (Bw4-80T), such as HLA-B*2705, appear to be better ligands for certain 3DL1 subtypes [29].
Figure 2. HLA-ligand binding specificities for KIR.

Alleles belonging to the KIR ligand groups HLA-C1/C2 and Bw4 80I/80T are listed in the boxes. The activating receptors KIR2DS2, 2DS1 and 3DS1 are thought to exhibit ligand specificity similar to the corresponding inhibitory counterparts, although their interactions are much weaker (depicted as smaller red broken arrows). The interaction of KIR3DL1 with Bw4 80I (dark blue arrow) is thought to be stronger than that with Bw4 80T (light blue arrow). Ligands for KIR2DL5, 2DS3, 2DS4, 2DS5 and 3DL3 have not been identified.
Other receptor-ligand relationships among KIR and HLA include KIR2DL4 specificity for HLA-G, which is primarily expressed on fetal trophoblasts, thymic endothelial cells and cornea [30], and KIR3DL2 specificity for HLA-A3 and A11 [31]. The activating receptors KIR2DS1, 2DS2 and 3DS1 share sequence similarity in their extracellular domains with their corresponding inhibitory counterparts (KIR2DL1, 2DL2/2DL3 and 3DL1, respectively) and are thought to share HLA ligand binding specificities as well. KIR2DS1 has been shown to bind weakly to HLA-C2 allotypes [32], which appears to have functional significance [33–35], and KIR2DS2 may bind weakly to HLA-C1, though this has not been conclusively established [33]. Until recently, expression of KIR3DS1 was in doubt, but there is now convincing evidence that it is indeed expressed [36–39]. KIR3DS1 shares >95% similarity with KIR3DL1 in its extracellular domain, but there is no direct evidence of interactions between 3DS1 and Bw4 allotypes. Still, genetic epidemiological [40, 41], functional [42], and population genetic data [3] strongly support such an interaction. KIR2DS4 is thought to interact with HLA-Cw4 alleles [43] and possibly with a non-HLA ligand expressed on melanoma cells [44]. Ligands for KIR2DL5, 2DS5, 2DS3 have not yet been identified.
4. KIR/HLA interactions
HLA class I genes map to chromosome 6, unlinked to the KIR genes on chromosome 19. Thus, the inheritance and expression of the genes encoding the receptors and their ligands are physically independent of one another other. It is therefore possible that a certain KIR, its ligand, or both might be absent in a given individual, each of which results in a functionally null situation. KIR also exhibit variegated expression on NK cells (i.e. a given KIR gene is expressed on some, but not all NK cell clones within an individual), adding another dimension to the variability and complexity of the system. Once acquired, the pattern of expression of inherited KIR genes remains stable in NK cell clones under varying cell culture conditions and activation stimuli [45]. Expression of KIR is controlled at the transcriptional level by epigenetic changes, primarily by methylation of CpG islands surrounding the transcriptional start site [46]. Identification of bidirectional promoters for KIR establishes yet another parallel between mouse and human NK cell receptor genes. In humans, bidirectional promoters control expression of KIR by switching the direction of transcription during the development of NK cells. Reverse transcripts are found in immature NK cells not expressing KIR, but these transcripts are absent in mature KIR-expressing NK cells, suggesting that reverse transcription blocks gene activation in immature and precursor NK cells [47]. Thus, if the promoter for a given KIR gene is switched in a forward position during development of the NK cell clone, then that mature clone will express the gene, but if the promoter is switched in the reverse direction, the clone will not express that KIR gene. The switch is stochastic, resulting in variegated expression of KIR and a repertoire of subpopulations of NK cells expressing different combinations of cell surface activating and inhibitory receptors that together are capable of responding to a wide variety of stimuli.
It has become increasingly clear that the strength of HLA-KIR interactions has functional significance and can influence disease susceptibility. This is exemplified by the interactions between HLA-C and inhibitory KIR2D where KIR2DL1/HLA-C2 (and probably KIR2DL2/HLA-C1) appears to confer stronger inhibitory responses than does KIR2DL3/HLA-C1 [48, 49] (Fig. 3). Allelic variation also plays a role in determining the strength of the interaction. Some allotypes of KIR3DL1, for example, are expressed at high levels, while others are expressed at low levels, which in turn correlates with level of binding to HLA-B Bw4 ligands [50, 51]. Allelic variants can also differ in terms of the frequency of NK cells that express them [51]. Disease association data are now accumulating and indicate that these differences are very significant in determining the outcome to infection [2, 52].
Figure 3. Functional model for KIR-HLA mediated hierarchy of inhibition.

The cartoon shows possible KIR-HLA interactions in an individual homozygous for the A haplotype and homozygous for either HLA-C2 (left side) or HLA-C1 (right side), based on the findings of Ahlenstiel et al [49]. Interaction of KIR2DL1 with HLA-C2 results in strong inhibition that is difficult to overcome by simultaneous activating signals, and thus there is no killing of the target in this model. The weaker KIR2DL3-HLA-C1 interaction, on the other hand, can be overridden by signals through activating receptors upon appropriate ligand binding, resulting in lysis of the target.
Interactions between KIR and their cognate HLA ligands can also be affected by peptides present in the binding groove of the HLA molecule [53–55], particularly the residues at positions 7 and 8 [56]. KIR3DL2 was shown to bind to HLA-A3 and A-11 only when specific EBV peptides were used to fold HLA tetramers [57]. Binding of KIR3DL1 to some HLA-A and HLA-B allotypes containing the Bw4 motif has also been shown to be dependent on bound peptide [58].
While the contribution of HLA class I ligands in KIR gene expression is not perfectly clear, there is evidence that expressed HLA ligand does indeed modulate the frequency of NK cells expressing cognate inhibitory receptor and its level of expression [51]. Expressed HLA ligands are also known to be crucial to NK cell tolerance and education. The so-called “at least one model” assumed that each NK cell had at least one inhibitory receptor specific for self MHC class I that would account for self-tolerance [45, 59]. However, in view of the stochastic expression of KIR, this model seems unlikely. Indeed, NK cells from MHC class I deficient mice do not kill their MHC class I deficient targets [8]. Two models have recently been proposed to explain this self-tolerance. The first model purports that NK cells expressing at least one inhibitory receptor recognizing self MHC class I are allowed to mature and become functionally competent or “licensed”, while NK cells that do not possess at least one functional inhibitory interaction are hypo-responsive and therefore “unlicensed” [60]. The other model, referred to as the “disarming” model, proposes that NK cells that do not express inhibitory receptors for self-MHC are chronically stimulated and consequently become anergic [61]. Therefore, inhibition during NK cell maturation is a critical requirement for NK cells to acquire cytotoxic potential. Whether or not human NK cell activity is regulated in a similar manner is not entirely clear, but recent data suggest that this might be the case. Anfossi et al provided the first evidence of licensing in human NK cells by showing that peripheral NK cells lacking inhibitory KIR for self MHC class I molecules are present in human peripheral blood and are hyporesponsive to various stimuli [62]. The authors propose a role for KIR-MHC class I inhibitory interactions in the calibration of NK cell potency [62]. Kim et al have also shown that specific KIR and HLA alleles are associated with the level of NK cell responsiveness, suggesting a role for “licensing” of NK cells in humans [63].
A variety of HLA-KIR compound genotypes have been implicated in disease pathogenesis and resistance to viral infections, autoimmune diseases, inflammatory disorders and cancers (listed in Table 1). Generally, genotypes that theoretically lead to lower inhibition and higher activation appear to be beneficial in viral infections such as human immunodeficiency virus (HIV) and hepatitis C virus (HCV), whereas activating genotypes constitute a risk for susceptibility to autoimmunity and perhaps cancers that have an inflammatory component to the disease pathogenesis (Fig. 4).
Table 1.
KIR-HLAdisease associations
| Disease | KIR-HLA ligand pair | Effect | Ref. |
|---|---|---|---|
| INFECTIOUS DISEASES
| |||
| HIV | KIR3DS1/Bw4-80I | Slower progression | [40] |
| KIR3DL1*004/Bw4 | Slower progression | [69] | |
| KIR3DL1*h/ Bw4 80I | |||
| KIR3DS1 | Reduced risk of infection | [64, 65, 67] | |
| HCV | KIR2DL3/HLA-C1 homozygosity | Resolution of infection | [71] |
| Human cytomegalovirus (HCMV) | KIR2DL1 expression on all NK cells | Recurrent CMV infection | [75] |
| >1 activating KIR in donor in bone marrow transplantation | Protection from CMV reactivation in the recipient. | [76] | |
| Herpes simplex virus (HSV) | KIR3DS1 in absence of Bw4 | Reactivation of HSV during IRD in HIV | [77] |
| M. tuberculosis | KIR2DL1; KIR2DL3 | Susceptibility | [78] |
| P. falciparum | KIR3DL2*002 | High response to infected RBCs | [82] |
|
| |||
| AUTOIMMUNE AND INFLAMMATORY CONDITIONS
| |||
| Psoriatic Arthritis | KIR2DS1/2DS2; HLA-Cw group homozygosity | Susceptibility | [94, |
| Psoriasis | KIR2DS1/HLA-Cw06 | Susceptibility | [92] |
| KIR2DS1; KIR2DL5; KIR haplotype B | Susceptibility | [91] | |
| Rhuematoid Vasculitis | KIR2DS2/HLA-Cw03 | Susceptibility | [87] |
| Scleroderma | KIR2DS2+/2DL2− | Susceptibility | [90] |
| Acute Coronary Syndrome | De novo expression of KIR2DS2 on CD4+CD28nullcells | Susceptibility | [88] |
| IDDM | KIR2DS2/HLA-C1 | Susceptibility | [99] |
| Endometriosis | KIR3DS1/ Bw4 | Protection | [95] |
| Birdshort Chorioretinopathy | Weak inhibitory KIR/HLA combinations and activating KIR in HLA-A*29+ individuals | Susceptibility | [96] |
| Idiopathic Bronchiectasis | HLA-C1/C1 and 2DS1/2DS2 | Susceptibility | [97] |
| Primary Sclerosing Cholangitis | KIR3DL1/Bw4; KIR2DL1/HLA-C2 | Protection | [98] |
|
| |||
| CANCER
| |||
| Malignant Melanoma | KIR/2DL2/2DL3; HLA-C1 | Susceptibility | [101] |
| Leukemia | KIR2DL1; KIR2DL2; KIR2DL3 | Susceptibility | [103] |
| Hodgkin’s lymphoma | KIR2DS1; KIR3DS1 | Protection | [106] |
| Nasopharyngeal carcinoma | ≥5 activating KIR | Susceptibility | [108] |
| Cervical Cancer | KIR3DS1and absence of HLA-C2 &/or HLA-Bw4 | Susceptibility | [107] |
| T-LGL | Expression of inhibitory KIR in absence of ligands | More severe disease | [112] |
| NK-LGL | Expression of activating KIR | May contribute to disease pathogenesis | [111, |
| Sezary syndrome | Expression of KIR3DL2 | Useful diagnostic marker | [110] |
|
| |||
| REPRODUCTION
| |||
| Preeclampsia | Mothers with AA KIR genotype; fetus with HLA-C2 | Susceptibility | [117] |
| Recurrent miscarriages/spontaneous abortions | Lack of KIR2DS1 in mothers and increased frequency of HLA-C2 in both mother and male partner | Susceptibility | [120] |
| Increased KIR2DS2 and decreased HLA-C2 frequency, overall increased frequency of activating KIR. | Susceptibility | [121] | |
| Higher cell surface expression of KIR2DL4 | Susceptibility | [122] | |
Figure 4. Summary of KIR mediated NK cell activation/inhibition in diseases.

Distinct KIR and HLA ligand pairs generate a hierarchy of NK cell activation. Both increased activation and inhibition are associated with susceptibility to and protection against a variety of diseases.
5. VIRAL INFECTIONS
5.1. Human Immunodeficiency Virus (HIV)
KIR3DL1 and KIR3DS1 segregate as alleles of the same locus and share about 97% sequence similarity in their extracellular domain, suggesting that they may bind similar ligands. HLA -Bw4 molecules are ligands for KIR3DL1, particularly Bw4-80I [25, 27, 28]. We observed that in individuals infected with HIV, the combination of KIR3DS1 with its putative ligand HLA-B Bw4-80I was associated with slower progression to AIDS, lower mean viral load, and protection against opportunistic infections [40, 41]. Subsequently, Alter et al demonstrated that NK cells expressing KIR3DS1 strongly and significantly inhibited HIV-1 replication in cells expressing HLA-B Bw4-80I as compared to NK cells that do not express KIR3DS1, providing the first functional support for the influence of KIR3DL1/S1 and Bw4-80I on anti-HIV NK activity [42]. KIR3DS1 positive NK cell clones were preferentially activated by HIV-1 infected target cells expressing HLA-B Bw4-80I, resulting in lysis of these cells. These data indicate that NK cells identify HIV infected cells actively through KIR3DS1 and Bw4-80I interactions. The necessity to have KIR3DS1 on the NK cells and Bw4-80I on the infected target supports a model in which KIR3DS1 binds directly or indirectly to Bw4-80I, perhaps in complex with specific HIV peptides or stress-induced self peptides produced in response to HIV infection. In a more recent study involving 60 treatment-naïve individuals with early infection, Long and colleagues observed that presence of KIR3DS1 was associated with higher NK cell effector function as measured by IFN-γ production and CD107a expression in vitro [64]. This effect was partially, but not completely dependent on the presence of B*57 and B*58, both of which are Bw4-80I allotypes. The presence of KIR3DS1 was also associated with lower viral load levels and diminished CD8+ T cell activation, which is an important marker of disease progression.
A study of twenty five HIV exposed uninfected intravenous drug users from Vietnam found transcription of KIR3DS1 to be significantly higher than KIR3DL1 in KIR3DS1/3DL1 heterozygous individuals and there was expansion of NK cells expressing KIR2DL3 in HLA-C1/C1 individuals who were KIR2DS2−/2DL2− [65]. KIR3DS1 homozygosity was also found to be significantly increased in HIV exposed seronegative intravenous drug users and HIV negative partners of sero-discordant couples [66]. These results are somewhat in keeping with previous data showing that HIV exposed seronegative sex workers carried more inhibitory receptors in the absence of their cognate HLA ligands as compared to the seropositive sex workers [67]. These individuals also had an increase in KIR AB haplotypes which are characterized by increased numbers of activating KIR.
KIR3DL1 has remarkable allelic diversity and the alleles have been shown to vary in their expression patterns on NK cells to the extent that they can be grouped into high (referred to as KIR3DL1*h), low (referred to as KIR3DL1*l), or no expression groupings [50, 51] [68] (the latter of which involves a single allele, KIR3DL1*004). KIR3DL1*h allotypes show higher affinity for their Bw4 ligands, particularly with Bw4-80I, which probably leads to greater inhibition [51]. We recently analyzed our AIDS cohorts for effects of KIR3DL1 alleles in the presence of their Bw4 ligands and showed that individuals possessing the high inhibitory allotypes, KIR3DL1*h, along with their Bw4-80I ligands had the lowest mean viral loads and progressed at a slower rate to AIDS relative to other KIR3DL1/HLA-B genotypes (individuals with KIR3DS1/Bw4-80I were completely excluded from the study) [69]. While these results seem to contradict the model in which NK cell activation is protective (based on the protective effect of KIR3DS1/Bw4-80I in the same cohort), a model incorporating the importance of inhibition in NK cell education unifies consistently results from this study with previous studies that indicate a protective role for NK cell activation against HIV. Since interactions between inhibitory KIR and MHC class I are important in establishing tolerance to healthy cells as well as the activation potential of mature NK cells [60, 62, 63, 70], it follows that the stronger, more inhibitory interactions conferred by KIR3DL1 during NK cell development lead to a fiercer NK cell response when ligand is downregulated/lost during viral infection. Surprisingly, KIR3DL1*004, which is not expressed on the cell surface was the single most protective KIR3DL1 allele against HIV disease progression and viral load and this protection was completely dependent on the presence of Bw4. While there are no apparent explanations for this observation, it is possible that KIR3DL1*004 has an as yet undefined intracellular function or, alternatively, it may be in linkage disequilibrium with another locus that confers protection against the virus.
Overall, studies reported to date indicate that HLA-KIR combinations with strong activation potential are protective against HIV and point decidedly to the KIR3DL1/S1 locus as most important in this regard.
5.2. Hepatitis C Virus (HCV)
Along the lines observed for HIV, compound genotypes of KIR and HLA class I that are expected to be relatively activating have also been implicated in outcome to HCV infection. As mentioned above, previous data have suggested that KIR2DL1/HLA-C2 may confer stronger inhibitory responses than does KIR2DL3/HLA-C1 [48, 49]. In individuals infected with a low-dose viral inoculum, the genotypic combination of KIR2DL3/HLA-C1, which theoretically results in weaker inhibitory signals (and therefore a lower activation potential), was protective in terms of spontaneous clearance of the virus [71]. Another study has also implicated a role for KIR2DL3 in clearance of HCV [72].
Using an in vitro model of infection with influenza A virus, Ahlenstiel et al recently provided functional data that strongly supports a lower threshold for activation conferred by KIR2DL3/HLA-C1 interactions as compared to KIR2DL1/HLA-C2 interactions [49]. In order to compare the inhibitory effect of KIR2DL3/HLA-C1 with that of KIR2DL1/HLA-C2, individuals were selected who were homozygous for haplotype A and homozygous for either HLA-C1 (KIR2DL3 ligand) or HLA-C2 (KIR2DL1 ligand) (Figs. 1 and 3). NK cells from KIR2DL3/HLA-C1 individuals secreted more IFN-γ at earlier time points after infection and displayed greater degranulation than did NK cells from KIR2DL1/HLA-C2 individuals. These results provide direct evidence for the differential level of inhibition associated with the presence of the KIR2DL3/HLA-C1 genotype as compared to KIR2DL1/HLA-C2 and underscore the functional significance of such a difference in response to viral infection.
5.3. Herpes Viruses
NK cells are important in herpes virus infections since individuals deficient in NK cells are particularly susceptible to these infections [73]. In order to evade the immune system, cytomegaloviruses (CMV) encode several proteins that interfere with MHC class I expression [74], potentially rendering infected cells more susceptible to attack by NK cells. In a case study of a child with a novel immunodeficiency syndrome and recurrent CMV infection [75], the entire population of NK cells from this patient expressed KIR2DL1 and the child also possessed the KIR2DL1 ligand, HLA-C2, raising the possibility that the strongly inhibitory KIR2DL1/HLA-C2 combination crippled NK cell activity and prevented the cells from mounting a protective response against CMV. Further evidence for KIR-mediated protection in CMV infection stems from a report demonstrating that activating KIR are protective against CMV reactivation during T cell replete stem cell transplantation [76]. Thus, as in other viral infections, NK cell activation appears to be protective in CMV infection. On the other hand, NK cell activation might be detrimental in a situation where immune activation is undesirable. Indeed, Price et al found that activating KIR predisposed to immune restoration diseases (defined by reactivation of quiescent opportunistic infection after combination antiretroviral therapy) associated with herpes virus infections [77].
5.4. Other infections
Mycobacterium tuberculosis is highly endemic in some parts of the world. In a recent study, Mendes et al found a significantly higher frequency of KIR2DL1 and KIR2DL3 in patients, although after correction for multiple comparisons, only KIR2DL3 remained weakly significant [78].
The role for NK cell responses to protozoa remains poorly understood. In the case of malaria, P. falciparum erythrocyte membrane protein I (PfEMP) downregulates IFNγ production by NK cells, NKR+ γδ T cells and αβ T cells [79], as well as NK cell cytotoxicity [80]. So far, studies involving NK cell receptors and responses in malaria have been very contradictory and require validation (reviewed by Hansen, D et al) [81]. However, carriers of KIR3DL2*002 produced higher levels of IFNγ in response to activation by P. falciparum infected red blood cells (RBCs), which is intriguing since RBCs do not express HLA ligands [82]. NK cells are a major source of IFNγ in early malarial parasite infection and NK cell activation may be regulated by crosstalk with the myeloid component of the immune system [83].
6. Autoimmunity and Inflammatory disorders
KIR associations with susceptibility to autoimmune/inflammatory conditions invariably point to the short chain activating KIR. Activating KIR are evolutionarily younger, probably having arisen from inhibitory homologues [84]. Variation in frequencies of activating KIR across diverse ethnic populations is extensive [3], but allelic diversity is quite limited as compared to inhibitory receptors [85]. The phenotypic frequencies of activating KIR and their ligands (or putative ligands) show strong negative correlations across populations, in contrast to weak positive correlations between various inhibitory KIR genes and their ligands [3]. These data suggest that there are selection pressures to maintain a consistently low frequency of corresponding activating KIR receptors and their ligands, perhaps in part due to selection pressures from autoimmune diseases.
Several conditions involving vascular damage and inflammation have shown association with the activating KIR2DS2. CD4+CD28null T cells, which are expanded in rheumatoid arthritis (RA) and cause endothelial damage, were found to express KIR2DS2 in the absence of inhibitory KIR2DL2 in this condition [86]. Further, the frequency of KIR2DS2 was increased in RA patients with vasculitis in comparison to normal controls and RA patients without vasculitis [87]. HLA-Cw*03, an HLA-C1 allotype and therefore a putative ligand for KIR2DS2, was also increased in subjects with vasculitis, although this was not true for other C1 alleles [87]. Thus, it is possible that KIR2DS2 recognizes a specific HLA-Cw03-peptide complex generated during RA vasculitis. Similarly, CD4+CD28null T cells are also present in the inflammatory infiltrate of atherosclerotic plaques in acute coronary syndrome and they express KIR2DS2 [88]. Other KIR/HLA associations with distinct clinical manifestations of RA [89] include the KIR2DL3+/2DS3− genotype, present in patients that were diagnosed early, and KIR2DS1 and KIR3DS1, which are higher in patients with bone erosions. KIR2DL2/2DS2 were significantly increased in patients with extra-articular manifestations, including vasculitis as previously reported [89]. KIR2DS2 in the absence of KIR2DL2 (genes that are in strong positive LD) was also observed to be increased among scleroderma patients [90].
Activating B haplotypes of KIR [91] and KIR2DS1 alone [92] or in combination with HLA-Cw6 (a C2 ligand for KIR2DS1) have been reported to associate with psoriasis [93]. We were also able to delineate the effect of HLA and KIR compound genotypes in psoriatic arthritis [94]. Based on the data, a model was proposed in which a gradient of more activating to more inhibitory compound genotypes of KIR2D and HLA-C appear to influence susceptibility to psoriatic arthritis [94]. Genotypes conferring highest activation (KIR2DS1 and/or KIR2DS2 with either HLA-C1 or C2 homozygosity) associated with greatest susceptibility whereas the genotypes conferring maximum inhibition (absence of activating receptors KIR2DS1 and KIR2DS2 and presence of both the inhibitory ligands HLA-C1 and C2) were protective.
Activating KIR gene profiles have also been associated with other inflammatory conditions such as endometriosis [95], birdshot chorioretinopathy [96], idiopathic bronchiectasis [97] primary sclerosing cholangitis [98], and type I diabetes mellitus [99] [100]. No doubt the list will continue to grow, with the most reliable conclusions being based on precise clinical data and large sample sizes. As in all genetic epidemiological studies, functional evidence for interaction between the short chain KIRs and putative ligand is necessary to support the various genetic models of predisposition to autoimmune conditions.
7. Cancer
Loss of MHC class I molecules on tumors evokes a role for NK cells in elimination of the transformed cell. Higher levels of KIR-mediated inhibition may also facilitate tumor escape, as has been shown for melanoma where the compound genotype KIR2DL2/2DL3/HLA-C1 was more frequent in patients as compared to controls [101] and KIR3DL1/Bw4 80I was marginally higher in patients with metastatic melanoma [102]. A role for KIR2DS4 was proposed in melanoma through its binding to non-HLA ligands expressed on melanoma cell lines and primary melanoma [44], but this data contrasts with a study showing no difference in the frequency of KIR2DS4 in patients vs. controls [101]. Inhibitory KIR (KIR2DL1, 2DL2 and 2DL3) were also present at significantly higher frequencies among patients with leukemia [103], and it has been suggested that they may contribute to a lack of NK or CTL antitumor responses in renal cell carcinoma [104]. KIR2DL5A and 2DL5B genes were more frequent in patients with NK type lymphoproliferative disease of granular lymphocytes [105]. Along the same lines, the activating receptors KIR3DS1 and KIR2DS1 associate with protection in a familial study of Hodgkin’s lymphoma [106].
Activating KIR genotypes may have opposite effects on distinct malignancies depending on whether inflammation is or is not a major component of tumor pathogenesis. Unlike the susceptibility effects of inhibitory KIR-HLA genotypes on cancers in which an inflammatory component plays no apparent role in the pathogenesis, we have observed that strongly inhibitory KIR-HLA genotypes were actually protective against cervical neoplasia [107]. NK cell activation may contribute to a chronic inflammatory state in response to human papilloma virus, the causative agent of cervical cancer, setting the stage for carcinogenesis. An increased number of activating KIR was also found to be associated with nasopharyngeal carcinoma (NPC), a cancer that is strongly associated with EBV infection [108]. It will be important to determine whether consistent results are observed with other cancers that clearly involve inflammation in the pathogenesis, such as gastric cancer and colon cancer. On the other hand, KIR3DS1 with Bw4-80I allotypes protected against the development of hepatocellular carcinoma (HCC) in patients with chronic hepatitis C virus infection [109], in spite of a probably role for inflammation in the development of HCC in some cases.
Abnormal expression of KIR has been associated with various malignant conditions. Expression of KIR3DL2 (but not other KIR) on phenotypically abnormal T cells was observed in patients with Sézary syndrome. [110]. Inhibitory KIR are also expressed on subsets of T cell large granular lymphocytic leukemia (T-LGL) [111] and increased disease severity was associated with absence of HLA ligands for the expressed KIR [112]. KIR are also expressed on NK-LGLs [111] and activating KIR were prominent among those that were expressed [113].
8. Reproduction
Pre-elampsia is a condition caused by inadequate extravillous trophoblast invasion into the maternal spiral arteries, which results in poor placental perfusion [114]. Uterine NK (uNK) cells, which account for 50–90% of the leukocytes in the decidua, are CD56bright and produce cytokines, chemokines and angiogenic factors thought to be involved in the remodeling of the spiral arterioles during pregnancy [115]. They also express KIR2D that recognize HLA-C allotypes. The fetal trophoblast expresses HLA-G, HLA-E and HLA-C [116], only the latter of which is polymorphic. A predominance of homozygosity for haplotype A in pre-eclamptic mothers combined with the presence of HLA-C2 in the fetus [117] suggested that strong inhibition of decidual NK cells via KIR2DL1 and HLA-C2 leads to strong uNK cell inhibition, which in turn impairs the remodeling of maternal blood vessels. Correspondingly, activating KIR appeared to decrease the likelihood of pre-eclampsia in a cumulative manner. Given the extreme consequences of inappropriate maternal-fetal interactions, more selection pressure may be imposed on KIR loci through its effect on reproductive diseases than through that of any other type of disease, including infectious diseases.
A balance in the level of activation/inhibition may be necessary in reproductive success in that excessive activation, like excessive inhibition, of NK cells could be detrimental in the maintenance of pregnancy. NK-like large granular lymphocytes have been implicated in alloimmune reactions against the fetus and their numbers are increased in the uterus and periphery in mothers who tend to abort [118]. Decreased frequency of inhibitory KIR or increased numbers of activating KIR-HLA ligand combinations are suggested to associate with miscarriages [119] [120] and recurrent abortions [121]. These studies are small and need confirmation in larger cohorts.
KIR2DL4 has been of particular interest for its role in maternal-fetal interactions because it binds HLA-G, which is expressed on trophoblasts. Yan et al showed that cell surface expression of KIR2DL4 was significantly higher in normal controls as compared to those with recurrent spontaneous abortion [122]. In spite of these studies, the absolute necessity of KIR2DL4 in/on uNK cells has been ruled out, since women who are missing the KIR2DL4 gene altogether have had successful pregnancies with apparently healthy children [123, 124]
9. Summary
Models regarding the role of KIR in diseases have been as dynamic and fickle as the locus itself. Many of the models that have been generated based on genetic data are inconsistent with one another and require functional data to clarify the biological role of KIR in the pathogenesis of these diseases. Nevertheless, some consistent threads have been forthcoming, including the association of activating KIR genotypes with increased risk of autoimmune disease and decreased risk of some infectious disease outcomes. Further efforts in determining KIR ligand specificities and affinities will greatly enhance our ability to define the role of KIR in human disease, and in turn, to potentially apply this knowledge clinically. Even at this early point in our efforts to characterize KIR, the astonishing population genetic, evolutionary, and biological properties of this locus that have been uncovered so far have provided a daily dose of mental vitamins to those of us fortunate enough to have landed in this field.
Acknowledgments
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This Research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Khakoo SI, Rajalingam R, Shum BP, Weidenbach K, Flodin L, Muir DG, et al. Rapid evolution of NK cell receptor systems demonstrated by comparison of chimpanzees and humans. Immunity. 2000;12(6):687–98. doi: 10.1016/s1074-7613(00)80219-8. [DOI] [PubMed] [Google Scholar]
- 2.Khakoo SI, Carrington M. KIR and disease: a model system or system of models? Immunol Rev. 2006;214:186–201. doi: 10.1111/j.1600-065X.2006.00459.x. [DOI] [PubMed] [Google Scholar]
- 3.Single RM, Martin MP, Gao X, Meyer D, Yeager M, Kidd JR, et al. Global diversity and evidence for coevolution of KIR and HLA. Nat Genet. 2007;39(9):1114–9. doi: 10.1038/ng2077. [DOI] [PubMed] [Google Scholar]
- 4.Norman PJ, Abi-Rached L, Gendzekhadze K, Korbel D, Gleimer M, Rowley D, et al. Unusual selection on the KIR3DL1/S1 natural killer cell receptor in Africans. Nat Genet. 2007;39(9):1092–9. doi: 10.1038/ng2111. [DOI] [PubMed] [Google Scholar]
- 5.Karre K, Ljunggren HG, Piontek G, Kiessling R. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature. 1986;319(6055):675–8. doi: 10.1038/319675a0. [DOI] [PubMed] [Google Scholar]
- 6.Hoglund P, Ohlen C, Carbone E, Franksson L, Ljunggren HG, Latour A, et al. Recognition of beta 2-microglobulin-negative (beta 2m−) T-cell blasts by natural killer cells from normal but not from beta 2m− mice: nonresponsiveness controlled by beta 2m− bone marrow in chimeric mice. Proc Natl Acad Sci U S A. 1991;88(22):10332–6. doi: 10.1073/pnas.88.22.10332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ljunggren HG, Karre K. In search of the ‘missing self’: MHC molecules and NK cell recognition. Immunol Today. 1990;11(7):237–44. doi: 10.1016/0167-5699(90)90097-s. [DOI] [PubMed] [Google Scholar]
- 8.Liao NS, Bix M, Zijlstra M, Jaenisch R, Raulet D. MHC class I deficiency: susceptibility to natural killer (NK) cells and impaired NK activity. Science. 1991;253(5016):199–202. doi: 10.1126/science.1853205. [DOI] [PubMed] [Google Scholar]
- 9.Diefenbach A, Raulet DH. Strategies for target cell recognition by natural killer cells. Immunol Rev. 2001;181:170–84. doi: 10.1034/j.1600-065x.2001.1810114.x. [DOI] [PubMed] [Google Scholar]
- 10.Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–74. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 11.Arase H, Mocarski ES, Campbell AE, Hill AB, Lanier LL. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science. 2002;296(5571):1323–6. doi: 10.1126/science.1070884. [DOI] [PubMed] [Google Scholar]
- 12.Hoelsbrekken SE, Nylenna O, Saether PC, Slettedal IO, Ryan JC, Fossum S, et al. Cutting edge: molecular cloning of a killer cell Ig-like receptor in the mouse and rat. J Immunol. 2003;170(5):2259–63. doi: 10.4049/jimmunol.170.5.2259. [DOI] [PubMed] [Google Scholar]
- 13.Mager DL, McQueen KL, Wee V, Freeman JD. Evolution of natural killer cell receptors: coexistence of functional Ly49 and KIR genes in baboons. Curr Biol. 2001;11(8):626–30. doi: 10.1016/s0960-9822(01)00148-8. [DOI] [PubMed] [Google Scholar]
- 14.Westgaard IH, Berg SF, Orstavik S, Fossum S, Dissen E. Identification of a human member of the Ly-49 multigene family. Eur J Immunol. 1998;28(6):1839–46. doi: 10.1002/(SICI)1521-4141(199806)28:06<1839::AID-IMMU1839>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 15.Carrington M, Norman PJ. The KIR gene cluster. Bethesda, M.D: U.S. Natl. Library Med., National Centre for Biotechnology Information; 2003. [Google Scholar]
- 16.Sambrook JG, Bashirova A, Andersen H, Piatak M, Vernikos GS, Coggill P, et al. Identification of the ancestral killer immunoglobulin-like receptor gene in primates. BMC Genomics. 2006;7:209. doi: 10.1186/1471-2164-7-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guethlein LA, Abi-Rached L, Hammond JA, Parham P. The expanded cattle KIR genes are orthologous to the conserved single-copy KIR3DX1 gene of primates. Immunogenetics. 2007;59(6):517–22. doi: 10.1007/s00251-007-0214-x. [DOI] [PubMed] [Google Scholar]
- 18.Martin AM, Kulski JK, Gaudieri S, Witt CS, Freitas EM, Trowsdale J, et al. Comparative genomic analysis, diversity and evolution of two KIR haplotypes A and B. Gene. 2004;335:121–31. doi: 10.1016/j.gene.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 19.Martin MP, Bashirova A, Traherne J, Trowsdale J, Carrington M. Cutting edge: expansion of the KIR locus by unequal crossing over. J Immunol. 2003;171(5):2192–5. doi: 10.4049/jimmunol.171.5.2192. [DOI] [PubMed] [Google Scholar]
- 20.Kikuchi-Maki A, Catina TL, Campbell KS. Cutting edge: KIR2DL4 transduces signals into human NK cells through association with the Fc receptor gamma protein. J Immunol. 2005;174(7):3859–63. doi: 10.4049/jimmunol.174.7.3859. [DOI] [PubMed] [Google Scholar]
- 21.Hsu KC, Liu XR, Selvakumar A, Mickelson E, O’Reilly RJ, Dupont B. Killer Ig-like receptor haplotype analysis by gene content: evidence for genomic diversity with a minimum of six basic framework haplotypes, each with multiple subsets. J Immunol. 2002;169(9):5118–29. doi: 10.4049/jimmunol.169.9.5118. [DOI] [PubMed] [Google Scholar]
- 22.Witt CS, Martin A, Christiansen FT. Detection of KIR2DL4 alleles by sequencing and SSCP reveals a common allele with a shortened cytoplasmic tail. Tissue Antigens. 2000;56(3):248–57. doi: 10.1034/j.1399-0039.2000.560307.x. [DOI] [PubMed] [Google Scholar]
- 23.Winter CC, Long EO. A single amino acid in the p58 killer cell inhibitory receptor controls the ability of natural killer cells to discriminate between the two groups of HLA-C allotypes. J Immunol. 1997;158(9):4026–8. [PubMed] [Google Scholar]
- 24.Moesta AK, Norman PJ, Yawata M, Yawata N, Gleimer M, Parham P. Synergistic Polymorphism at Two Positions Distal to the Ligand-Binding Site Makes KIR2DL2 a Stronger Receptor for HLA-C Than KIR2DL3. J Immunol. 2008;180(6):3969–79. doi: 10.4049/jimmunol.180.6.3969. [DOI] [PubMed] [Google Scholar]
- 25.Cella M, Longo A, Ferrara GB, Strominger JL, Colonna M. NK3-specific natural killer cells are selectively inhibited by Bw4-positive HLA alleles with isoleucine 80. J Exp Med. 1994;180(4):1235–42. doi: 10.1084/jem.180.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gumperz JE, Litwin V, Phillips JH, Lanier LL, Parham P. The Bw4 public epitope of HLA-B molecules confers reactivity with natural killer cell clones that express NKB1, a putative HLA receptor. J Exp Med. 1995;181(3):1133–44. doi: 10.1084/jem.181.3.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carr WH, Pando MJ, Parham P. KIR3DL1 polymorphisms that affect NK cell inhibition by HLA-Bw4 ligand. J Immunol. 2005;175(8):5222–9. doi: 10.4049/jimmunol.175.8.5222. [DOI] [PubMed] [Google Scholar]
- 28.Gumperz JE, Barber LD, Valiante NM, Percival L, Phillips JH, Lanier LL, et al. Conserved and variable residues within the Bw4 motif of HLA-B make separable contributions to recognition by the NKB1 killer cell-inhibitory receptor. J Immunol. 1997;158(11):5237–41. [PubMed] [Google Scholar]
- 29.Luque I, Solana R, Galiani MD, Gonzalez R, Garcia F, Lopez de Castro JA, et al. Threonine 80 on HLA-B27 confers protection against lysis by a group of natural killer clones. Eur J Immunol. 1996;26(8):1974–7. doi: 10.1002/eji.1830260845. [DOI] [PubMed] [Google Scholar]
- 30.Rajagopalan S, Long EO. A human histocompatibility leukocyte antigen (HLA)-G-specific receptor expressed on all natural killer cells. J Exp Med. 1999;189(7):1093–100. doi: 10.1084/jem.189.7.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dohring C, Scheidegger D, Samaridis J, Cella M, Colonna M. A human killer inhibitory receptor specific for HLA-A1,2. J Immunol. 1996;156(9):3098–101. [PubMed] [Google Scholar]
- 32.Biassoni R, Pessino A, Malaspina A, Cantoni C, Bottino C, Sivori S, et al. Role of amino acid position 70 in the binding affinity of p50.1 and p58. 1 receptors for HLA-Cw4 molecules. Eur J Immunol. 1997;27(12):3095–9. doi: 10.1002/eji.1830271203. [DOI] [PubMed] [Google Scholar]
- 33.Stewart CA, Laugier-Anfossi F, Vely F, Saulquin X, Riedmuller J, Tisserant A, et al. Recognition of peptide-MHC class I complexes by activating killer immunoglobulin-like receptors. Proc Natl Acad Sci U S A. 2005;102(37):13224–9. doi: 10.1073/pnas.0503594102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Foley B, De Santis D, Lathbury L, Christiansen F, Witt C. KIR2DS1-mediated activation overrides NKG2A-mediated inhibition in HLA-C C2-negative individuals. Int Immunol. 2008;20(4):555–63. doi: 10.1093/intimm/dxn013. [DOI] [PubMed] [Google Scholar]
- 35.Chewning JH, Gudme CN, Hsu KC, Selvakumar A, Dupont B. KIR2DS1-positive NK cells mediate alloresponse against the C2 HLA-KIR ligand group in vitro. J Immunol. 2007;179(2):854–68. doi: 10.4049/jimmunol.179.2.854. [DOI] [PubMed] [Google Scholar]
- 36.Trundley A, Frebel H, Jones D, Chang C, Trowsdale J. Allelic expression patterns of KIR3DS1 and 3DL1 using the Z27 and DX9 antibodies. Eur J Immunol. 2007;37(3):780–7. doi: 10.1002/eji.200636773. [DOI] [PubMed] [Google Scholar]
- 37.Carr WH, Rosen DB, Arase H, Nixon DF, Michaelsson J, Lanier LL. Cutting Edge: KIR3DS1, a gene implicated in resistance to progression to AIDS, encodes a DAP12-associated receptor expressed on NK cells that triggers NK cell activation. J Immunol. 2007;178(2):647–51. doi: 10.4049/jimmunol.178.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pascal V, Yamada E, Martin MP, Alter G, Altfeld M, Metcalf JA, et al. Detection of KIR3DS1 on the cell surface of peripheral blood NK cells facilitates identification of a novel null allele and assessment of KIR3DS1 expression during HIV-1 infection. J Immunol. 2007;179(3):1625–33. doi: 10.4049/jimmunol.179.3.1625. [DOI] [PubMed] [Google Scholar]
- 39.O’Connor GM, Guinan KJ, Cunningham RT, Middleton D, Parham P, Gardiner CM. Functional polymorphism of the KIR3DL1/S1 receptor on human NK cells. J Immunol. 2007;178(1):235–41. doi: 10.4049/jimmunol.178.1.235. [DOI] [PubMed] [Google Scholar]
- 40.Martin MP, Gao X, Lee JH, Nelson GW, Detels R, Goedert JJ, et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002;31(4):429–34. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 41.Qi Y, Martin MP, Gao X, Jacobson L, Goedert JJ, Buchbinder S, et al. KIR/HLA pleiotropism: protection against both HIV and opportunistic infections. PLoS Pathog. 2006;2(8):e79. doi: 10.1371/journal.ppat.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alter G, Martin MP, Teigen N, Carr WH, Suscovich TJ, Schneidewind A, et al. Differential natural killer cell-mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J Exp Med. 2007;204(12):3027–36. doi: 10.1084/jem.20070695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katz G, Markel G, Mizrahi S, Arnon TI, Mandelboim O. Recognition of HLA-Cw4 but not HLA-Cw6 by the NK cell receptor killer cell Ig-like receptor two-domain short tail number 4. J Immunol. 2001;166(12):7260–7. doi: 10.4049/jimmunol.166.12.7260. [DOI] [PubMed] [Google Scholar]
- 44.Katz G, Gazit R, Arnon TI, Gonen-Gross T, Tarcic G, Markel G, et al. MHC class I-independent recognition of NK-activating receptor KIR2DS4. J Immunol. 2004;173(3):1819–25. doi: 10.4049/jimmunol.173.3.1819. [DOI] [PubMed] [Google Scholar]
- 45.Valiante NM, Uhrberg M, Shilling HG, Lienert-Weidenbach K, Arnett KL, D’Andrea A, et al. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity. 1997;7(6):739–51. doi: 10.1016/s1074-7613(00)80393-3. [DOI] [PubMed] [Google Scholar]
- 46.Chan HW, Miller JS, Moore MB, Lutz CT. Epigenetic control of highly homologous killer Ig-like receptor gene alleles. J Immunol. 2005;175(9):5966–74. doi: 10.4049/jimmunol.175.9.5966. [DOI] [PubMed] [Google Scholar]
- 47.Davies GE, Locke SM, Wright PW, Li H, Hanson RJ, Miller JS, et al. Identification of bidirectional promoters in the human KIR genes. Genes Immun. 2007;8(3):245–53. doi: 10.1038/sj.gene.6364381. [DOI] [PubMed] [Google Scholar]
- 48.Winter CC, Gumperz JE, Parham P, Long EO, Wagtmann N. Direct binding and functional transfer of NK cell inhibitory receptors reveal novel patterns of HLA-C allotype recognition. J Immunol. 1998;161(2):571–7. [PubMed] [Google Scholar]
- 49.Ahlenstiel G, Martin MP, Gao X, Carrington M, Rehermann B. Distinct KIR/HLA compound genotypes affect the kinetics of human antiviral natural killer cell responses. J Clin Invest. 2008;118(3):1017–26. doi: 10.1172/JCI32400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gardiner CM, Guethlein LA, Shilling HG, Pando M, Carr WH, Rajalingam R, et al. Different NK cell surface phenotypes defined by the DX9 antibody are due to KIR3DL1 gene polymorphism. J Immunol. 2001;166(5):2992–3001. doi: 10.4049/jimmunol.166.5.2992. [DOI] [PubMed] [Google Scholar]
- 51.Yawata M, Yawata N, Draghi M, Little AM, Partheniou F, Parham P. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J Exp Med. 2006;203(3):633–45. doi: 10.1084/jem.20051884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol. 2005;5(3):201–14. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- 53.Malnati MS, Peruzzi M, Parker KC, Biddison WE, Ciccone E, Moretta A, et al. Peptide specificity in the recognition of MHC class I by natural killer cell clones. Science. 1995;267(5200):1016–8. doi: 10.1126/science.7863326. [DOI] [PubMed] [Google Scholar]
- 54.Rajagopalan S, Long EO. The direct binding of a p58 killer cell inhibitory receptor to human histocompatibility leukocyte antigen (HLA)-Cw4 exhibits peptide selectivity. J Exp Med. 1997;185(8):1523–8. doi: 10.1084/jem.185.8.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zappacosta F, Borrego F, Brooks AG, Parker KC, Coligan JE. Peptides isolated from HLA-Cw*0304 confer different degrees of protection from natural killer cell-mediated lysis. Proc Natl Acad Sci U S A. 1997;94(12):6313–8. doi: 10.1073/pnas.94.12.6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boyington JC, Motyka SA, Schuck P, Brooks AG, Sun PD. Crystal structure of an NK cell immunoglobulin-like receptor in complex with its class I MHC ligand. Nature. 2000;405(6786):537–43. doi: 10.1038/35014520. [DOI] [PubMed] [Google Scholar]
- 57.Hansasuta P, Dong T, Thananchai H, Weekes M, Willberg C, Aldemir H, et al. Recognition of HLA-A3 and HLA-A11 by KIR3DL2 is peptide-specific. Eur J Immunol. 2004;34(6):1673–9. doi: 10.1002/eji.200425089. [DOI] [PubMed] [Google Scholar]
- 58.Thananchai H, Gillespie G, Martin MP, Bashirova A, Yawata N, Yawata M, et al. Cutting Edge: Allele-specific and peptide-dependent interactions between KIR3DL1 and HLA-A and HLA-B. J Immunol. 2007;178(1):33–7. doi: 10.4049/jimmunol.178.1.33. [DOI] [PubMed] [Google Scholar]
- 59.Raulet DH, Held W, Correa I, Dorfman JR, Wu MF, Corral L. Specificity, tolerance and developmental regulation of natural killer cells defined by expression of class I-specific Ly49 receptors. Immunol Rev. 1997;155:41–52. doi: 10.1111/j.1600-065x.1997.tb00938.x. [DOI] [PubMed] [Google Scholar]
- 60.Kim S, Poursine-Laurent J, Truscott SM, Lybarger L, Song YJ, Yang L, et al. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 2005;436(7051):709–13. doi: 10.1038/nature03847. [DOI] [PubMed] [Google Scholar]
- 61.Fernandez NC, Treiner E, Vance RE, Jamieson AM, Lemieux S, Raulet DH. A subset of natural killer cells achieves self-tolerance without expressing inhibitory receptors specific for self-MHC molecules. Blood. 2005;105(11):4416–23. doi: 10.1182/blood-2004-08-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anfossi N, Andre P, Guia S, Falk CS, Roetynck S, Stewart CA, et al. Human NK cell education by inhibitory receptors for MHC class I. Immunity. 2006;25(2):331–42. doi: 10.1016/j.immuni.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 63.Kim S, Sunwoo JB, Yang L, Choi T, Song YJ, French AR, et al. HLA alleles determine differences in human natural killer cell responsiveness and potency. Proc Natl Acad Sci U S A. 2008;105(8):3053–8. doi: 10.1073/pnas.0712229105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Long BR, Ndhlovu LC, Oksenberg JR, Lanier LL, Hecht FM, Nixon DF, et al. KIR3DS1 Conferral of Enhanced Natural Killer Cell Function in Early HIV-1 Infection. J Virol. 2008 doi: 10.1128/JVI.02449-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ravet S, Scott-Algara D, Bonnet E, Tran HK, Tran T, Nguyen N, et al. Distinctive NK-cell receptor repertoires sustain high-level constitutive NK-cell activation in HIV-exposed uninfected individuals. Blood. 2007;109(10):4296–305. doi: 10.1182/blood-2006-08-040238. [DOI] [PubMed] [Google Scholar]
- 66.Boulet S, Sharafi S, Simic N, Bruneau J, Routy JP, Tsoukas CM, et al. Increased proportion of KIR3DS1 homozygotes in HIV-exposed uninfected individuals. Aids. 2008;22(5):595–9. doi: 10.1097/QAD.0b013e3282f56b23. [DOI] [PubMed] [Google Scholar]
- 67.Jennes W, Verheyden S, Demanet C, Adje-Toure CA, Vuylsteke B, Nkengasong JN, et al. Cutting edge: resistance to HIV-1 infection among African female sex workers is associated with inhibitory KIR in the absence of their HLA ligands. J Immunol. 2006;177(10):6588–92. doi: 10.4049/jimmunol.177.10.6588. [DOI] [PubMed] [Google Scholar]
- 68.Pando MJ, Gardiner CM, Gleimer M, McQueen KL, Parham P. The protein made from a common allele of KIR3DL1 (3DL1*004) is poorly expressed at cell surfaces due to substitution at positions 86 in Ig domain 0 and 182 in Ig domain 1. J Immunol. 2003;171(12):6640–9. doi: 10.4049/jimmunol.171.12.6640. [DOI] [PubMed] [Google Scholar]
- 69.Martin MP, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F, et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet. 2007;39(6):733–40. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cooley S, Xiao F, Pitt M, Gleason M, McCullar V, Bergemann TL, et al. A subpopulation of human peripheral blood NK cells that lacks inhibitory receptors for self-MHC is developmentally immature. Blood. 2007;110(2):578–86. doi: 10.1182/blood-2006-07-036228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Khakoo SI, Thio CL, Martin MP, Brooks CR, Gao X, Astemborski J, et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305(5685):872–4. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- 72.Romero V, Azocar J, Zuniga J, Clavijo OP, Terreros D, Gu X, et al. Interaction of NK inhibitory receptor genes with HLA-C and MHC class II alleles in Hepatitis C virus infection outcome. Mol Immunol. 2008;45(9):2429–36. doi: 10.1016/j.molimm.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Biron CA, Byron KS, Sullivan JL. Severe herpesvirus infections in an adolescent without natural killer cells. N Engl J Med. 1989;320(26):1731–5. doi: 10.1056/NEJM198906293202605. [DOI] [PubMed] [Google Scholar]
- 74.Lin A, Xu H, Yan W. Modulation of HLA expression in human cytomegalovirus immune evasion. Cell Mol Immunol. 2007;4(2):91–8. [PubMed] [Google Scholar]
- 75.Gazit R, Garty BZ, Monselise Y, Hoffer V, Finkelstein Y, Markel G, et al. Expression of KIR2DL1 on the entire NK cell population: a possible novel immunodeficiency syndrome. Blood. 2004;103(5):1965–6. doi: 10.1182/blood-2003-11-3796. [DOI] [PubMed] [Google Scholar]
- 76.Cook M, Briggs D, Craddock C, Mahendra P, Milligan D, Fegan C, et al. Donor KIR genotype has a major influence on the rate of cytomegalovirus reactivation following T-cell replete stem cell transplantation. Blood. 2006;107(3):1230–2. doi: 10.1182/blood-2005-03-1039. [DOI] [PubMed] [Google Scholar]
- 77.Price P, Witt C, de Santis D, French MA. Killer immunoglobulin-like receptor genotype may distinguish immunodeficient HIV-infected patients resistant to immune restoration diseases associated with herpes virus infections. J Acquir Immune Defic Syndr. 2007;45(3):359–61. doi: 10.1097/QAI.0b013e31805b82a1. [DOI] [PubMed] [Google Scholar]
- 78.Mendez A, Granda H, Meenagh A, Contreras S, Zavaleta R, Mendoza MF, et al. Study of KIR genes in tuberculosis patients. Tissue Antigens. 2006;68(5):386–9. doi: 10.1111/j.1399-0039.2006.00685.x. [DOI] [PubMed] [Google Scholar]
- 79.D’Ombrain MC, Voss TS, Maier AG, Pearce JA, Hansen DS, Cowman AF, et al. Plasmodium falciparum erythrocyte membrane protein-1 specifically suppresses early production of host interferon-gamma. Cell Host Microbe. 2007;2(2):130–8. doi: 10.1016/j.chom.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 80.Mavoungou E, Held J, Mewono L, Kremsner PG. A Duffy binding-like domain is involved in the NKp30-mediated recognition of Plasmodium falciparum-parasitized erythrocytes by natural killer cells. J Infect Dis. 2007;195(10):1521–31. doi: 10.1086/515579. [DOI] [PubMed] [Google Scholar]
- 81.Hansen DS, D’Ombrain MC, Schofield L. The role of leukocytes bearing Natural Killer Complex receptors and Killer Immunoglobulin-like Receptors in the immunology of malaria. Curr Opin Immunol. 2007;19(4):416–23. doi: 10.1016/j.coi.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 82.Artavanis-Tsakonas K, Eleme K, McQueen KL, Cheng NW, Parham P, Davis DM, et al. Activation of a subset of human NK cells upon contact with Plasmodium falciparum-infected erythrocytes. J Immunol. 2003;171(10):5396–405. doi: 10.4049/jimmunol.171.10.5396. [DOI] [PubMed] [Google Scholar]
- 83.Roetynck S, Baratin M, Johansson S, Lemmers C, Vivier E, Ugolini S. Natural killer cells and malaria. Immunol Rev. 2006;214:251–63. doi: 10.1111/j.1600-065X.2006.00446.x. [DOI] [PubMed] [Google Scholar]
- 84.Abi-Rached L, Parham P. Natural selection drives recurrent formation of activating killer cell immunoglobulin-like receptor and Ly49 from inhibitory homologues. J Exp Med. 2005;201(8):1319–32. doi: 10.1084/jem.20042558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hou L, Steiner NK, Chen M, Belle I, Kubit AL, Ng J, et al. Limited allelic diversity of stimulatory two-domain killer cell immunoglobulin-like receptors. Hum Immunol. 2008;69(3):174–8. doi: 10.1016/j.humimm.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 86.Namekawa T, Snyder MR, Yen JH, Goehring BE, Leibson PJ, Weyand CM, et al. Killer cell activating receptors function as costimulatory molecules on CD4+CD28null T cells clonally expanded in rheumatoid arthritis. J Immunol. 2000;165(2):1138–45. doi: 10.4049/jimmunol.165.2.1138. [DOI] [PubMed] [Google Scholar]
- 87.Yen JH, Moore BE, Nakajima T, Scholl D, Schaid DJ, Weyand CM, et al. Major histocompatibility complex class I-recognizing receptors are disease risk genes in rheumatoid arthritis. J Exp Med. 2001;193(10):1159–67. doi: 10.1084/jem.193.10.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nakajima T, Goek O, Zhang X, Kopecky SL, Frye RL, Goronzy JJ, et al. De novo expression of killer immunoglobulin-like receptors and signaling proteins regulates the cytotoxic function of CD4 T cells in acute coronary syndromes. Circ Res. 2003;93(2):106–13. doi: 10.1161/01.RES.0000082333.58263.58. [DOI] [PubMed] [Google Scholar]
- 89.Majorczyk E, Pawlik A, Luszczek W, Nowak I, Wisniewski A, Jasek M, et al. Associations of killer cell immunoglobulin-like receptor genes with complications of rheumatoid arthritis. Genes Immun. 2007;8(8):678–83. doi: 10.1038/sj.gene.6364433. [DOI] [PubMed] [Google Scholar]
- 90.Momot T, Koch S, Hunzelmann N, Krieg T, Ulbricht K, Schmidt RE, et al. Association of killer cell immunoglobulin-like receptors with scleroderma. Arthritis Rheum. 2004;50(5):1561–5. doi: 10.1002/art.20216. [DOI] [PubMed] [Google Scholar]
- 91.Suzuki Y, Hamamoto Y, Ogasawara Y, Ishikawa K, Yoshikawa Y, Sasazuki T, et al. Genetic polymorphisms of killer cell immunoglobulin-like receptors are associated with susceptibility to psoriasis vulgaris. J Invest Dermatol. 2004;122(5):1133–6. doi: 10.1111/j.0022-202X.2004.22517.x. [DOI] [PubMed] [Google Scholar]
- 92.Luszczek W, Manczak M, Cislo M, Nockowski P, Wisniewski A, Jasek M, et al. Gene for the activating natural killer cell receptor, KIR2DS1, is associated with susceptibility to psoriasis vulgaris. Hum Immunol. 2004;65(7):758–66. doi: 10.1016/j.humimm.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 93.Holm SJ, Sakuraba K, Mallbris L, Wolk K, Stahle M, Sanchez FO. Distinct HLA-C/KIR genotype profile associates with guttate psoriasis. J Invest Dermatol. 2005;125(4):721–30. doi: 10.1111/j.0022-202X.2005.23879.x. [DOI] [PubMed] [Google Scholar]
- 94.Nelson GW, Martin MP, Gladman D, Wade J, Trowsdale J, Carrington M. Cutting edge: heterozygote advantage in autoimmune disease: hierarchy of protection/susceptibility conferred by HLA and killer Ig-like receptor combinations in psoriatic arthritis. J Immunol. 2004;173(7):4273–6. doi: 10.4049/jimmunol.173.7.4273. [DOI] [PubMed] [Google Scholar]
- 95.Kitawaki J, Xu B, Ishihara H, Fukui M, Hasegawa G, Nakamura N, et al. Association of killer cell immunoglobulin-like receptor genotypes with susceptibility to endometriosis. Am J Reprod Immunol. 2007;58(6):481–6. doi: 10.1111/j.1600-0897.2007.00533.x. [DOI] [PubMed] [Google Scholar]
- 96.Levinson RD, Du Z, Luo L, Monnet D, Tabary T, Brezin AP, et al. Combination of KIR and HLA gene variants augments the risk of developing birdshot chorioretinopathy in HLA-A*29-positive individuals. Genes Immun. 2008 doi: 10.1038/gene.2008.13. [DOI] [PubMed] [Google Scholar]
- 97.Boyton RJ, Smith J, Ward R, Jones M, Ozerovitch L, Wilson R, et al. HLA-C and killer cell immunoglobulin-like receptor genes in idiopathic bronchiectasis. Am J Respir Crit Care Med. 2006;173(3):327–33. doi: 10.1164/rccm.200501-124OC. [DOI] [PubMed] [Google Scholar]
- 98.Karlsen TH, Boberg KM, Olsson M, Sun JY, Senitzer D, Bergquist A, et al. Particular genetic variants of ligands for natural killer cell receptors may contribute to the HLA associated risk of primary sclerosing cholangitis. J Hepatol. 2007;46(5):899–906. doi: 10.1016/j.jhep.2007.01.032. [DOI] [PubMed] [Google Scholar]
- 99.van der Slik AR, Koeleman BP, Verduijn W, Bruining GJ, Roep BO, Giphart MJ. KIR in type 1 diabetes: disparate distribution of activating and inhibitory natural killer cell receptors in patients versus HLA-matched control subjects. Diabetes. 2003;52(10):2639–42. doi: 10.2337/diabetes.52.10.2639. [DOI] [PubMed] [Google Scholar]
- 100.Middleton D, Halfpenny I, Meenagh A, Williams F, Sivula J, Tuomilehto-Wolf E. Investigation of KIR gene frequencies in type 1 diabetes mellitus. Hum Immunol. 2006;67(12):986–90. doi: 10.1016/j.humimm.2006.08.295. [DOI] [PubMed] [Google Scholar]
- 101.Naumova E, Mihaylova A, Stoitchkov K, Ivanova M, Quin L, Toneva M. Genetic polymorphism of NK receptors and their ligands in melanoma patients: prevalence of inhibitory over activating signals. Cancer Immunol Immunother. 2005;54(2):172–8. doi: 10.1007/s00262-004-0575-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Naumova E, Mihaylova A, Ivanova M, Mihailova S. Impact of KIR/HLA ligand combinations on immune responses in malignant melanoma. Cancer Immunol Immunother. 2007;56(1):95–100. doi: 10.1007/s00262-006-0151-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Verheyden S, Bernier M, Demanet C. Identification of natural killer cell receptor phenotypes associated with leukemia. Leukemia. 2004;18(12):2002–7. doi: 10.1038/sj.leu.2403525. [DOI] [PubMed] [Google Scholar]
- 104.Gati A, Da Rocha S, Guerra N, Escudier B, Moretta A, Chouaib S, et al. Analysis of the natural killer mediated immune response in metastatic renal cell carcinoma patients. Int J Cancer. 2004;109(3):393–401. doi: 10.1002/ijc.11730. [DOI] [PubMed] [Google Scholar]
- 105.Scquizzato E, Teramo A, Miorin M, Facco M, Piazza F, Noventa F, et al. Genotypic evaluation of killer immunoglobulin-like receptors in NK-type lymphoproliferative disease of granular lymphocytes. Leukemia. 2007;21(5):1060–9. doi: 10.1038/sj.leu.2404634. [DOI] [PubMed] [Google Scholar]
- 106.Besson C, Roetynck S, Williams F, Orsi L, Amiel C, Lependeven C, et al. Association of killer cell immunoglobulin-like receptor genes with Hodgkin’s lymphoma in a familial study. PLoS ONE. 2007;2(5):e406. doi: 10.1371/journal.pone.0000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Carrington M, Wang S, Martin MP, Gao X, Schiffman M, Cheng J, et al. Hierarchy of resistance to cervical neoplasia mediated by combinations of killer immunoglobulin-like receptor and human leukocyte antigen loci. J Exp Med. 2005;201(7):1069–75. doi: 10.1084/jem.20042158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Butsch Kovacic M, Martin M, Gao X, Fuksenko T, Chen CJ, Cheng YJ, et al. Variation of the killer cell immunoglobulin-like receptors and HLA-C genes in nasopharyngeal carcinoma. Cancer Epidemiol Biomarkers Prev. 2005;14(11 Pt 1):2673–7. doi: 10.1158/1055-9965.EPI-05-0229. [DOI] [PubMed] [Google Scholar]
- 109.Lopez-Vazquez A, Rodrigo L, Martinez-Borra J, Perez R, Rodriguez M, Fdez-Morera JL, et al. Protective effect of the HLA-Bw4I80 epitope and the killer cell immunoglobulin-like receptor 3DS1 gene against the development of hepatocellular carcinoma in patients with hepatitis C virus infection. J Infect Dis. 2005;192(1):162–5. doi: 10.1086/430351. [DOI] [PubMed] [Google Scholar]
- 110.Bahler DW, Hartung L, Hill S, Bowen GM, Vonderheid EC. CD158k/KIR3DL2 is a useful marker for identifying neoplastic T-cells in Sezary syndrome by flow cytometry. Cytometry B Clin Cytom. 2007;74B(3):156–62. doi: 10.1002/cyto.b.20395. [DOI] [PubMed] [Google Scholar]
- 111.Morice WG, Kurtin PJ, Leibson PJ, Tefferi A, Hanson CA. Demonstration of aberrant T-cell and natural killer-cell antigen expression in all cases of granular lymphocytic leukaemia. Br J Haematol. 2003;120(6):1026–36. doi: 10.1046/j.1365-2141.2003.04201.x. [DOI] [PubMed] [Google Scholar]
- 112.Nowakowski GS, Morice WG, Phyliky RL, Li CY, Tefferi A. Human leucocyte antigen class I and killer immunoglobulin-like receptor expression patterns in T-cell large granular lymphocyte leukaemia. Br J Haematol. 2005;128(4):490–2. doi: 10.1111/j.1365-2141.2004.05341.x. [DOI] [PubMed] [Google Scholar]
- 113.Zambello R, Falco M, Della Chiesa M, Trentin L, Carollo D, Castriconi R, et al. Expression and function of KIR and natural cytotoxicity receptors in NK-type lymphoproliferative diseases of granular lymphocytes. Blood. 2003;102(5):1797–805. doi: 10.1182/blood-2002-12-3898. [DOI] [PubMed] [Google Scholar]
- 114.Moffett A, Hiby SE. How Does the maternal immune system contribute to the development of pre-eclampsia? Placenta. 2007;28 (Suppl A):S51–6. doi: 10.1016/j.placenta.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 115.Hanna J, Goldman-Wohl D, Hamani Y, Avraham I, Greenfield C, Natanson-Yaron S, et al. Decidual NK cells regulate key developmental processes at the human fetal-maternal interface. Nat Med. 2006;12(9):1065–74. doi: 10.1038/nm1452. [DOI] [PubMed] [Google Scholar]
- 116.Trundley A, Moffett A. Human uterine leukocytes and pregnancy. Tissue Antigens. 2004;63(1):1–12. doi: 10.1111/j.1399-0039.2004.00170.x. [DOI] [PubMed] [Google Scholar]
- 117.Hiby SE, Walker JJ, O’Shaughnessy KM, Redman CW, Carrington M, Trowsdale J, et al. Combinations of maternal KIR and fetal HLA-C genes influence the risk of preeclampsia and reproductive success. J Exp Med. 2004;200(8):957–65. doi: 10.1084/jem.20041214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Paparistidis N, Papadopoulou C, Chioti A, Papaioannou D, Tsekoura C, Keramitsoglou T, et al. How Valuable is Measurement of Peripheral Blood Natural Killer Cells at the Time of Abortion? Am J Reprod Immunol. 2008;59(4):306–15. doi: 10.1111/j.1600-0897.2007.00547.x. [DOI] [PubMed] [Google Scholar]
- 119.Varla-Leftherioti M, Spyropoulou-Vlachou M, Keramitsoglou T, Papadimitropoulos M, Tsekoura C, Graphou O, et al. Lack of the appropriate natural killer cell inhibitory receptors in women with spontaneous abortion. Hum Immunol. 2005;66(1):65–71. doi: 10.1016/j.humimm.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 120.Hiby SE, Regan L, Lo W, Farrell L, Carrington M, Moffett A. Association of maternal killer-cell immunoglobulin-like receptors and parental HLA-C genotypes with recurrent miscarriage. Hum Reprod. 2008;23(4):972–6. doi: 10.1093/humrep/den011. [DOI] [PubMed] [Google Scholar]
- 121.Wang S, Zhao YR, Jiao YL, Wang LC, Li JF, Cui B, et al. Increased activating killer immunoglobulin-like receptor genes and decreased specific HLA-C alleles in couples with recurrent spontaneous abortion. Biochem Biophys Res Commun. 2007;360(3):696–701. doi: 10.1016/j.bbrc.2007.06.125. [DOI] [PubMed] [Google Scholar]
- 122.Yan WH, Lin A, Chen BG, Zhou MY, Dai MZ, Chen XJ, et al. Possible roles of KIR2DL4 expression on uNK cells in human pregnancy. Am J Reprod Immunol. 2007;57(4):233–42. doi: 10.1111/j.1600-0897.2007.00469.x. [DOI] [PubMed] [Google Scholar]
- 123.Witt CS, Whiteway JM, Warren HS, Barden A, Rogers M, Martin A, et al. Alleles of the KIR2DL4 receptor and their lack of association with pre-eclampsia. Eur J Immunol. 2002;32(1):18–29. doi: 10.1002/1521-4141(200201)32:1<18::AID-IMMU18>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 124.Witt CS, Goodridge J, Gerbase-Delima MG, Daher S, Christiansen FT. Maternal KIR repertoire is not associated with recurrent spontaneous abortion. Hum Reprod. 2004;19(11):2653–7. doi: 10.1093/humrep/deh483. [DOI] [PubMed] [Google Scholar]
- 125.Martin MP, Nelson G, Lee JH, Pellett F, Gao X, Wade J, et al. Cutting edge: susceptibility to psoriatic arthritis: influence of activating killer Ig-like receptor genes in the absence of specific HLA-C alleles. J Immunol. 2002;169(6):2818–22. doi: 10.4049/jimmunol.169.6.2818. [DOI] [PubMed] [Google Scholar]


