Abstract
Histo-aspartic protease (HAP) from Plasmodium falciparum offers a promising target for the development of novel antimalarial drugs. HAP exhibits high sequence similarity to pepsin-like aspartic proteases, but one of the two catalytic aspartates, Asp32, is replaced by histidine. Crystal structures of the truncated zymogen of HAP and of the complex of the mature enzyme with inhibitor KNI-10395 have been determined at 2.1 and 2.5 Å resolution, respectively. As in other proplasmepsins, the propeptide of the zymogen interacts with the C-terminal domain of the enzyme, forcing the N- and C- terminal domains apart, thereby separating His32 and Asp215 and preventing formation of the mature active site. In the inhibitor complex the enzyme forms a tight domain-swapped dimer, not previously seen in any aspartic proteases. The inhibitor is found in an unprecedented conformation resembling the letter “U”, stabilized by two intramolecular hydrogen bonds. Surprisingly, the location and conformation of the inhibitor are similar to the fragment of helix 2 comprising residues 34p–38p in the prosegments of the zymogens of gastric aspartic proteases; a corresponding helix assumes a vastly different orientation in proplasmepsins. Each inhibitor molecule is in contact with two molecules of HAP, interacting with the carboxylate group of the catalytic Asp215 of one HAP protomer through a water molecule, while also making a direct hydrogen bond to Glu278A′ of the other protomer. A comparison of the shifts in the positions of the catalytic residues in the inhibitor complex presented here with those published previously gives further hints regarding the enzymatic mechanism of HAP.
The parasite Plasmodium falciparum is implicated in the of death of nearly two million people annually, most of them children (1, 2). With the rapid emergence of resistance to the current antimalarial therapy, development of novel therapeutic agents targeting this organism is necessary. During the growth phase of the parasite in the human erythrocytes it breaks down a significant amount of host hemoglobin in order to obtain amino acids for protein synthesis (3, 4), and also to reduce the colloid-osmotic pressure within the host cell, thus preventing its premature lysis (5). This degradation process takes place in the food vacuole of the parasite (6). Four pepsin-like aspartic proteases known as plasmepsins (PMs) are present in the food vacuole of P. falciparum. PMI, PMII, histo-aspartic protease (HAP), and PMIV have all been shown to be directly involved in the process of hemoglobin degradation (1, 7) and all have been characterized structurally (8). For such reasons, vacuolar plasmepsins are potential targets for developing novel drugs against malaria (9).
Although the amino acid sequence of HAP exhibits high level of identity compared to the other three vacuolar PMs (almost 60%), some of the differences are very significant, especially in the active site region (10). The most important change is that Asp32 (amino acids are numbered throughout according to the sequence of pepsin, with insertions marked by trailing characters A, B, etc. (8)), one of the two catalytic aspartates in pepsin-like aspartic proteases (11), is replaced in HAP by histidine. There are also other important substitutions, including strictly conserved Tyr75 and highly conserved Val/Gly76, which are replaced by Ser and Lys, respectively. The latter substitutions are found in the flexible loop named “flap” (residues 70–83), located in the N-terminal domain of the enzyme. The flap plays a major role in all pepsin-like aspartic proteases, changing its conformation upon ligand binding (11). Previously determined medium-resolution crystal structures of uncomplexed HAP (PDB ID: 3FNS; to abbreviate the nomenclature, here referred to as apoenzyme) and of complexes with pepstatin A (PDB ID: 3FNT) and KNI-10006 (PDB ID: 3FNU) (12) confirmed the pepsin-like fold of this enzyme. The observed binding mode of pepstatin A in the active site of HAP disproved the earlier hypothesis that HAP is a serine protease (13), but left open an alternative mechanistic proposal (14). Thus the catalytic mechanism of HAP is still not fully elucidated, in part because of the lack of higher resolution crystal structures of the complexes of this enzyme with peptidomimetic inhibitors.
Formation of an oligomeric structure is often critical for the ability of an enzyme to carry out its catalytic function and for its regulation. It has been noted that some pepsin-like aspartic proteases form homodimers and higher oligomers which may affect protein stability, but which are not required for catalytic activity (15). Several biochemical and crystallographic studies have attempted to determine the oligomeric state of plasmepsins (15, 16), but the significance of oligomerization of these enzymes has not been fully established. Crystals of PMII contain apparent dimers (17), although it is agreed that PMII exists in solution mainly as a monomer (15). Recent structural studies of PMI (18) and of the apoenzyme of HAP also indicate the presence of dimers in the crystals. A zinc ion found in the active site of apo-HAP was tetrahedrally coordinated by His32 and Asp215 from one monomer, Glu278A′ from the other monomer, and by a water molecule (12). In recent studies involving gel filtration chromatography, sedimentation velocity, and equilibrium ultracentrifugation (16) it was shown that HAP exists in a dynamic monomer-dimer equilibrium, with the dissociation constant increased by the addition of CHAPS. It has also been reported that HAP forms dimeric and higher oligomeric species at concentration higher than 0.05 mg/ml, with accompanying loss of activity.
Similarly to many other proteases (19), HAP is synthesized as an inactive zymogen and subsequently activated through cleavage and removal of the prosegment. It has been reported that the size of the prosegments of the zymogens of various proteases varies from two residues for some granzymes to more than 200 residues for some members of the thermolysin family (20). The average length of the prosegments of aspartic proteases is ~50 amino acids (19), but vacuolar plasmepsins provide an exception, since their prosegments are composed of ~120 amino acids (21). All prosegments of aspartic proteases are located at the N termini of the zymogens. Proteolytic cleavage and removal of the prosegment of the zymogen, followed by structural rearrangements, finally produce the mature, active enzyme. The processes involved in the conversion of the inactive zymogen to its active mature form are quite complicated (21). For aspartic proteases, activation utilizes three different mechanisms: (1) auto activation at acidic pH (for gastric zymogens); (2) self-processing, partially assisted by exogenous proteases (for lysosomal and vacuolar proteases); and (3) fully assisted processing (for prorenin) (19, 21, 22).
It has been shown that pro-PMII may be activated by two different mechanisms. Inside the acidic food vacuole of the parasite, pro-PMII is activated by a maturase, a probable cysteine protease, in a process that requires acidic pH (6). In vitro, activation of recombinant pro-PMII takes place at pH 4.7 by autolysis at the Phe112p-Leu113p bond (sequences within propeptides are identified by appended letter p), twelve residues upstream of the wild type N terminus (22, 23). The location of the cleavage site of the recombinant pro-PMII varies depending on the conditions used (24). It has been reported that in vitro autoactivation of recombinant HAP takes place at Lys119p-Ser120p, four residues upstream of the native cleavage site (Gly123p - Ser-1) (25). Crystal structures of the zymogens of several aspartic proteases have been determined in the past. High-resolution structure of porcine pepsinogen shows that part of the prosegment (Ser11p - Leu44p) occupies the substrate binding cleft of the enzyme and competitively blocks access to the two catalytic aspartates (26). It has been proposed that the salt bridge interactions that stabilize the position of prosegment across the active site of pepsin are disrupted at low pH, thus releasing the prosegment and opening the substrate binding cleft (26). The same mechanism of activation was also confirmed by the crystal structure of an activation intermediate of human gastricsin (27). However, two structures of plasmepsin zymogens indicated a significantly different mechanism of inactivation. Crystal structures of the zymogens of PMII from P. falciparum (PDB ID: 1PFZ) (22) and of pvPMIV (PDB ID: 1MIQ), its ortholog from P. vivax (28), have shown that the N- and C-terminal domains which contain the two catalytic aspartates were pushed apart, thus preventing the formation of a functional active site (22). In the zymogens of plasmepsins the prosegment interacts extensively with the C-terminal domain, resulting in its separation from the N-terminal domain. It has been suggested that lowering the pH disrupts the interdomain salt bridges, allowing the domains to come closer and form a catalytically competent active site (22, 28).
We have now determined the crystal structure of the truncated zymogen form of HAP (here referred to simply as zymogen, unless noted otherwise), as well as the structure of the mature HAP complexed with a potent inhibitor, KNI-10395 (29). These crystals diffracted to a higher resolution than the ones used in our previous studies (12), allowing us to elucidate the structural details with higher confidence. An analysis of the structure of HAP zymogen and the comparison with the mature HAP clarifies the inactivation mechanism by the propeptide. The structure of the HAP–KNI-10395 complex indicates a unique mode of binding of the inhibitor, as well as the presence in the crystals of domain-swapped dimers. The type of domain swapping reported here has not been seen in any other plasmepsins, or indeed in any other aspartic proteases.
EXPERIMENTAL PROCEDURES
Expression and purification
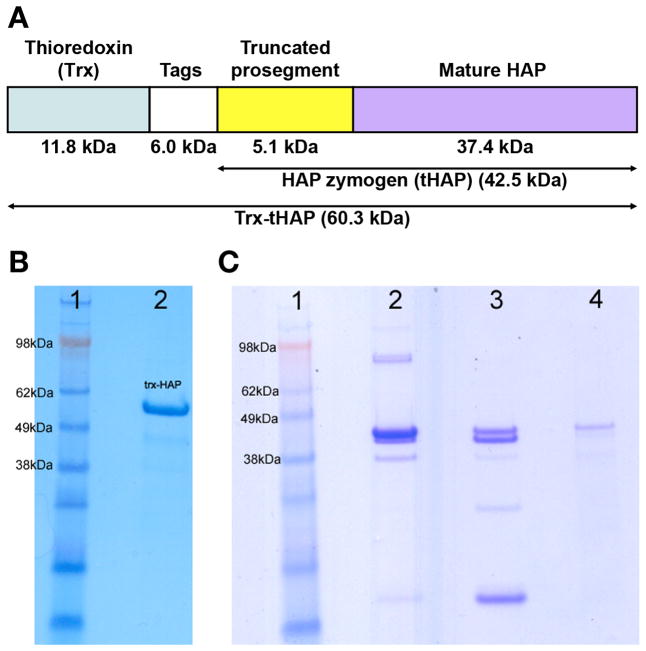
The fusion protein used in this study (here named Trx-tHAP) consists of thioredoxin (Trx), a thrombin cleavage site, internal 6His-tag, S-tag, and an enterokinase cleavage site, followed by the residues of the truncated prosegment (77p–123p) and mature HAP (-1 to 328) (Figure 1A). The logic behind preparation of this particular construct, cloned into pET32b (+) vector and expressed in E. coli Rosetta-gami B (DE3)pLysS cells, was presented before (30). A total of six liters of cell culture was grown. Purification of Trx-tHAP was performed at 4 °C. The cell pellets were resuspended in 1X BugBuster and incubated for 40 minutes. 1 ml of DNAse I (10 mg/ml) was added to the cell suspension and the sample was centrifuged at 34000 g for 30 minutes. The supernatant was loaded on to a HisTrap™ (GE Healthcare) nickel Sepharose column (10 ml) equilibrated with the buffer (50 mM Tris buffer (pH 8.0) containing 0.5 M NaCl,10 mM imidazole, 0.2% CHAPS). Next, the unbound protein was washed with 10% elution buffer (50 mM Tris buffer (pH 8.0) containing 0.5 M NaCl, 250 mM imidazole, 0.2% CHAPS), and finally eluted with 50% elution buffer. The eluted sample was concentrated and dialyzed overnight against 50 mM Tris buffer (pH 8.0) containing 0.2% CHAPS. The dialyzed sample was loaded on to a HiTrap™ (GE Healthcare) Q-Sepharose anion exchange column equilibrated with 50 mM Tris buffer (pH 8.0) containing 0.2% CHAPS. After washing the unbound protein, Trx-tHAP was eluted with 100 ml gradient from 0 to 0.5 M NaCl in 50 mM Tris buffer (pH 8.0) containing 0.2% CHAPS. The fractions containing Trx-tHAP were collected and concentrated to 2.0 ml. Trx-tHAP was obtained by purifying the protein using a Sephacyl S-400 gel filtration column equilibrated with 50 mM Tris buffer (pH 8.0) containing 0.3 M NaCl, 0.2% CHAPS. Purified protein migrates on the denaturing gels at the molecular mass of 60.3 kDa (Figure 1B), suggesting that all 540 amino acid residues of the construct are present.
Figure 1.
Analysis of the purified Trx-HAP, crystallization drops, and a dissolved crystal. The two gels presented here are from different runs. (A) Block diagram of the expressed construct named Trx-HAP, with different major segments marked. The fusion protein (540 residues) consists of thioredoxin (Trx), a 6kDa sequence consisting of a thrombin cleavage site, internal 6His-tag, S-tag and an enterokinase cleavage site, followed by the truncated prosegment peptide with the N-terminal 76 amino residues removed, and the sequence corresponding to the mature HAP. (B) A gel showing the purified sample of Trx-HAP which was used for crystallization (lane 2). The apparent molecular mass is in agreement with the expectations for a protein containing 540 residues. Lane 1 contains the molecular mass markers. (C) A gel showing the contents of the crystal and crystallization drop. Lane 1: molecular mass marker, lane 2: crystals removed from the crystallization drop and dissolved in water, lane 3: crystallization drop after removing most of the crystals, lane 4: the single crystal used for data collection, dissolved in water.
The expression and purification of the mature form of HAP was performed starting from the same expression construct according to a previously described procedure (12).
Crystallization
A freshly purified sample of Trx-tHAP concentrated to 14.0 mg/ml was used for crystallization. The crystallization screens were set up using sitting-drop vapor-diffusion method at 293 K. The best crystals appeared in the drop containing 0.4 μl protein solution and 0.2 μl reservoir solution, equilibrated against 75 μl reservoir solution. The reservoir solution contained 20% (w/v) PEG 3350 and 0.2 M tri-potassium citrate (pH 8.1).
For crystallization of the HAP–KNI-10395 complex, HAP sample was first transferred to 0.1 M sodium acetate buffer (pH 5.0) and concentrated to 12.0 mg/ml. KNI-10395 was dissolved in DMSO and the resulting solution was mixed with the protein sample to yield the final inhibitor concentration of 0.3 mM in the mixture (1:1 protein:inhibitor molar ratio). The mixture was allowed to incubate for 24 h. Crystallization screens were set up using sitting-drop vapor-diffusion method at 293 K. The best crystals appeared in the drop containing 0.4 μl protein solution and 0.2 μl reservoir solution, equilibrated against 75 μl reservoir solution. The reservoir solution contained 0.1 M sodium chloride, 0.1 M sodium acetate (pH 4.5) and 30% (v/v) PEG 200.
Data collection
Diffraction data for the HAP zymogen crystal were collected to 2.1 Å resolution using Cu Kα radiation generated by a Rigaku MicroMax 007HF X-ray source operated at 40 kV and 30 mA, equipped with a MAR345dtb detector. A data set was collected at 100 K, using 30% (v/v) ethylene glycol added to the reservoir solution as cryoprotectant. Diffraction data for the HAP–KNI-10395 complex crystal were collected to 2.5 Å resolution using a MAR300CCD detector and the wavelength of 1.000 Å at the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory, Argonne, IL. A data set was collected at 100 K, using 30% (v/v) ethylene glycol added to reservoir solution as cryoprotectant. All data were indexed and integrated using the program XDS (31). Integrated intensities were converted to structure factors with modules F2MTZ and CAD of CCP4 (32). Crystal parameters and the statistics of data processing are presented in Table 1.
Table 1.
Data collection and refinement statistics
| A. Data collection statisticsa | HAP zymogen | HAP–KNI-10395 |
| Space group | C2 | P212121 |
| Unit cell parameters a, b, c (Å) | 121.5, 68.4, 72.8 | 88.4, 90.5, 192.4 |
| β (°) | 125.8 | 90 |
| Temperature (K) | 100 | 100 |
| Wavelength (Å) | 1.5418 | 1.0000 |
| Resolution (Å) | 40.0-2.10 (2.20-2.10) | 40.0-2.50 (2.60-2.50) |
| Rmerge (%)b | 7.9 (76.1) | 14.7 (93.4) |
| Completeness (%) | 98.8 (98.3) | 100.0 (100.0) |
| I/σ(I) | 15.8 (2.4) | 13.1 (2.2) |
| Unique reflections | 38189 (3625) | 54222 (5954) |
| Redundancy | 5.1 (5.0) | 7.4 (7.4) |
| No. of molecules/asymmetric unit | 1 | 4 |
| B. Refinement statistics | ||
| Resolution (Å) | 36.4-2.10 | 39.7-2.50 |
| Working set: number of reflections | 26780 | 51509 |
| Rfactor (%)b | 18.3 (26.3) | 17.5 (24.5) |
| Test set: number of reflections | 1410 | 2712 |
| Rfree (%)b | 23.9 (33.6) | 25.2 (33.8) |
| Protein atoms | 2971 | 10522 |
| No. of KNI-10395 molecules | - | 3 |
| No. of ethylene glycol molecules | 5 | 12 |
| No. of water molecules | 242 | 508 |
| C. Geometry statistics | ||
| rmsd (bond distance) (Å) | 0.023 | 0.018 |
| rmsd (bond angle) (deg) | 2.27 | 1.9 |
| Ramachandran plotc | ||
| Ramachandran favored (%) | 95.88 | 95.83 |
| Ramachandran outliers (%) | 0.82 | 0.31 |
| D. PDB code | 3QVC | 3QVI |
The values in parentheses are for the highest resolution shell
Rmerge = ΣhΣi |<I>h−I h,i|/ΣhΣi Ih,i.
As defined by MOLPROBITY (38).
Structure solution and refinement
The expressed fusion protein (Trx-tHAP) contains 540 amino acid residues (molecular mass, 60.3 kDa). Although the purified Trx-tHAP was used for crystallization, analysis of the crystals, as well as the crystallization drops, shows that the protein has been degraded during the crystallization process (Figure 1C). The thioredoxin and part of the tag region have been removed, leaving the remaining part of the HAP zymogen (tHAP) that contains 382 amino acid residues (molecular mass 43.2 kDa), with 7 N-terminal residues (AMAISDP) derived from the expression vector (determined by mass spectrometry and N-terminal sequencing). The Matthews coefficient (33) for these crystals is 2.84 Å 3 Da−1, assuming the presence of one molecule in the asymmetric unit. Automated search by PHASER (34) using the A chain of P. falciparum PMII zymogen (PDB ID: 1PFZ) revealed the correct placement of the molecule in the asymmetric unit. Initially, several cycles of refinement using REFMAC5 (35) and rebuilding using COOT (36) were performed. Solvent molecules were progressively introduced at peaks of electron density higher than 3σ in the Fo-Fc weighted maps while monitoring the decrease of Rfree. Proper hydrogen bonding was required for placement of solvent molecules. The overall anisotropy was modeled with the TLS parameters by dividing each molecule into three groups, comprising residues 77p-8, 9–193, and 194–328. The final model does not include the pro-mature junction (residues 120p to 1), which could not be built due to disorder.
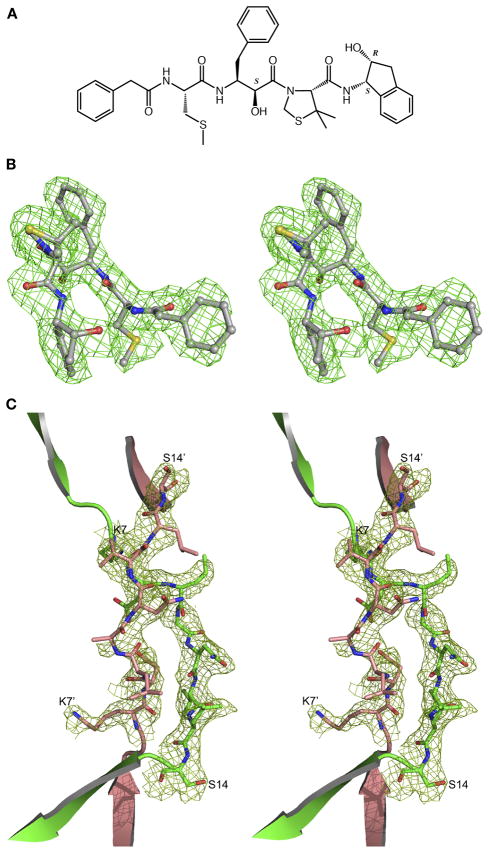
The structure of HAP–KNI-10395 (the chemical structure of the inhibitor is shown in Figure 2A) was solved using molecule A of the HAP–KNI-10006 complex (PDB ID: 3FNU) (12). Molecular replacement with PHASER allowed placement of the three out of four molecules in the asymmetric unit, with the fourth molecule placed manually into the resulting Fo-Fc electron density map. The first few cycles of refinement of the model without the inhibitor using REFMAC5 showed that the C-terminal loop region composed of residues 229–255 assumed a conformation different from the starting one. Several cycles of rebuilding with COOT and refinement with REFMAC5 were performed to complete an interim model. Subsequent analysis of the Fo-Fc electron density map (Figure 2B) indicated the presence of the inhibitors in only three active sites (molecules A, C, and D). After inhibitor modeling, iterative cycles of refinement in REFMAC5 and model building in the electron density maps using COOT were carried out. During the refinement and model analysis no positive 2Fo-Fc electron density was seen for residues Ala10 and Asn11, but positive Fo-Fc density connecting Leu9 of one monomer to Val12 of the other monomer was visible. To check the connectivity, the omit (Fo-Fc) electron density map (Figure 2C) was calculated after refining the model without residues 6–14. The resulting electron density map clearly shows that the N-terminal polypeptide chain (residues 0–10) has been exchanged between the two adjacent monomers. Very tight NCS restraints were used in the initial stages of refinement, but they were slowly released as the model was becoming more complete; medium NCS restraints were applied for the final refinement cycles. The overall anisotropy was modeled with TLS parameters by dividing each molecule into three TLS groups, comprising residues 0–149, 150–245, and 246–327. The final model also lacks residues −5 to −1 (corresponding to the cloning artifact and the first residue of the enzyme) in each monomer and residues 239–242 of the monomer A, which could not be built because of disorder. The final statistics for the refined structures are presented in Table 1. The structures were analyzed using PROCHECK (37), MOLPROBITY (38), and COOT (36). Structural superpositions were performed with SSM (39) and ALIGN (40) and the figures were generated with PYMOL (41).
Figure 2.
(A) Chemical structure of KNI-10395. Chiral carbon atoms are labeled S or R. (B) The initial Fo-Fc electron density omitmap contoured at 2.0σ level with the final model superimposed, showing KNI-10395 bound to molecule A of HAP. (C) The Fo-Fc electron density omitmap contoured at 2.0σ level after deleting the domain swap switch region residues 6–14 in molecules A and B (primed) of the HAP–KNI-10395 complex structure, with the final domain swapped polypeptide chain superimposed.
RESULTS AND DISCUSSION
The structures of a truncated form of the HAP zymogen and of the complex of the mature enzyme with a potent peptidomimetic inhibitor have been solved with data extending to the resolution of 2.1 Å and 2.5 Å, respectively. The resolution of data for the inhibitor complex is higher than in the previously reported structures of HAP (12), whereas the resolution of data for the HAP zymogen is comparable with data for other proplasmepsins. The structure of the zymogen will be presented and discussed first, followed by the structure of the inhibitor complex.
The structure of the zymogen of HAP
Although the full length fusion protein containing 540 residues (Trx-tHAP - Figure 1A,B) was used for crystallization, neither the thioredoxin nor the tags were present in the crystals. An analysis of a dissolved crystal (Figure 1C) and of the electron density maps indicated that the fragment of HAP zymogen that was crystallized contained only residues 77p – 123p of the propeptide, followed by the sequence corresponding to the mature enzyme. (Following the accepted usage (28) and for simplicity, the term “mature enzyme” is also used here to denote the part of the zymogen that corresponds to the active protease). The reasons for such truncation are not obvious. The expression construct was designed for production of the mature HAP through cleavage of the thioredoxin tag with enterokinase, followed by autoactivation of the enzyme at low pH. No enterokinase was used to prepare the zymogen and the protein was kept at elevated pH 8.0 in order to prevent autolysis. It is thus not clear how the observed cleavage was accomplished – whether through the presence of small amounts of a contaminating protease, or by autoactivation of a minor fraction of HAP. However, no traces of the mature HAP were seen in the crystal (Figure 1C, lane 4).
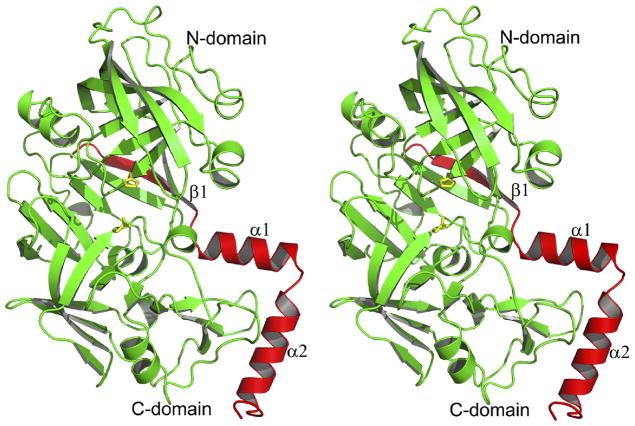
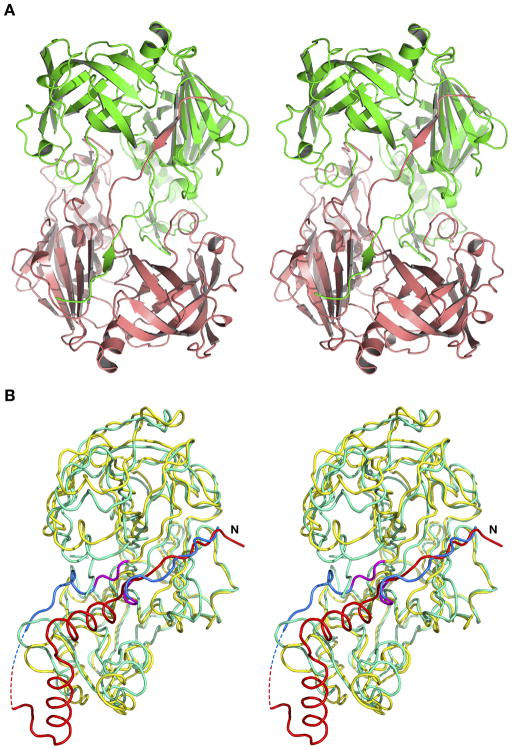
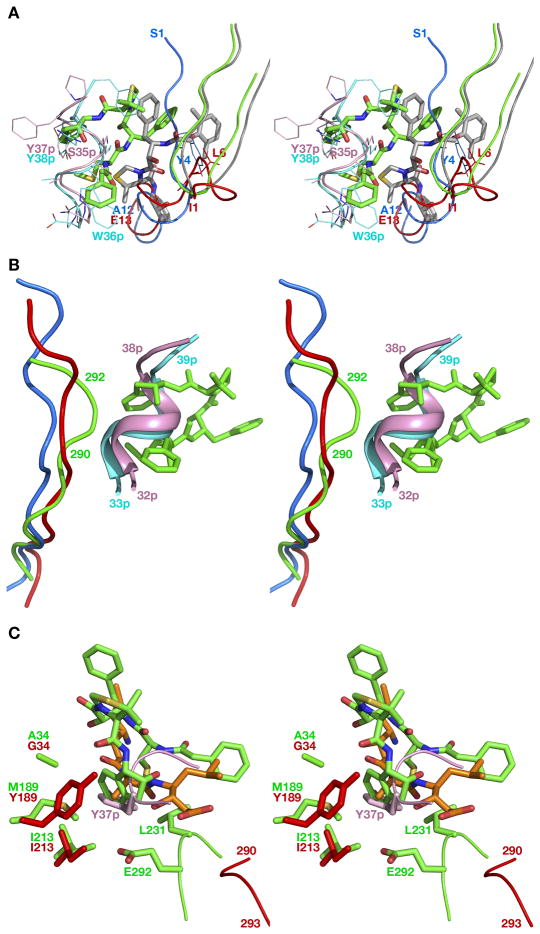
The overall fold of the truncated HAP zymogen (pro-HAP) is quite similar to the previously reported zymogens of PMII from P. falciparum (pro-PMII) (22) and PMIV ortholog from P. vivax (pro-pvPMIV) (28). The bean-shaped HAP zymogen is composed of two topologically similar N- and C-terminal domains (Figure 3), parts of which are also referred to as a central motif. The N-terminal domain consists of residues 13–148, the central motif consists of residues 77p–85p, 149–184, and 309–328, whereas residues 86p-12 and 185–308 comprise the C-terminal domain.
Figure 3.
The fold of the truncated zymogen of HAP. The prosegment fragment is colored red and the mature enzyme is colored green. The catalytic residues (His32 and Asp215) are shown in stick representation and are colored yellow.
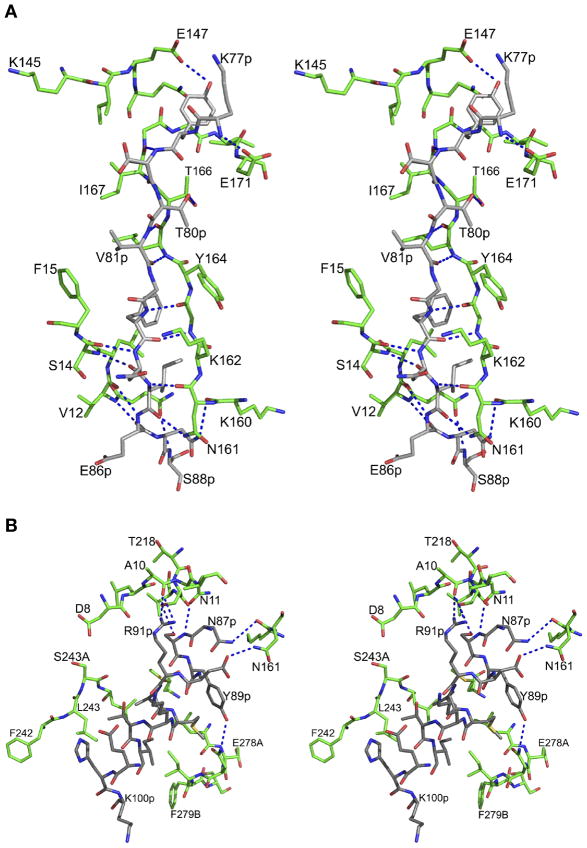
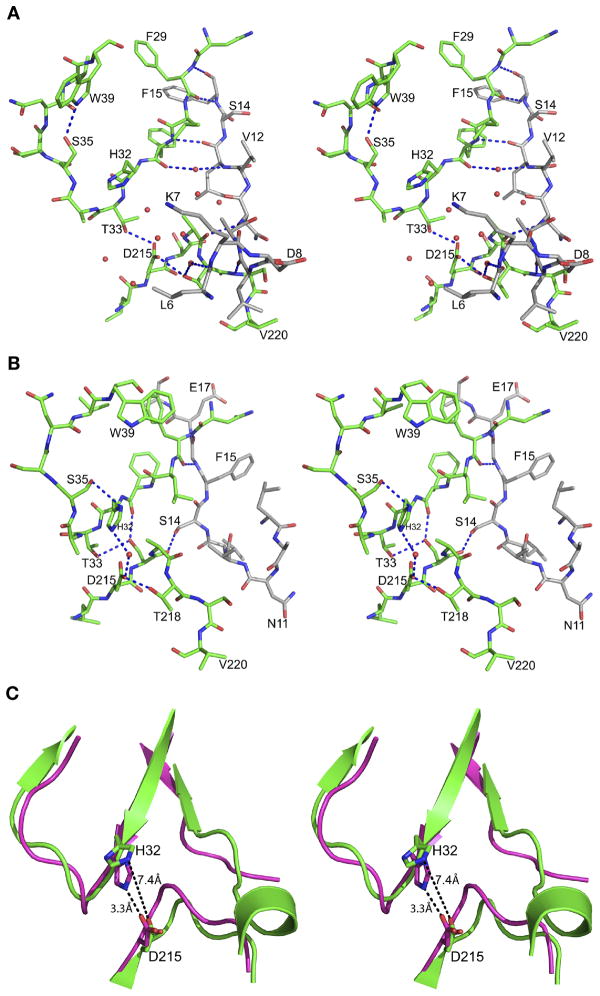
The prosegment has a clearly defined secondary structure (Figure 3), folding into a β-strand (79p–86p) followed by the first α-helix (88p–98p), a helical turn, a second α-helix (101p–113p), and a coil connection to the mature segment. The junction containing residues 120p to 1 could not be modeled due to disorder. The prosegment makes 25 hydrogen bonds and 9 hydrophobic interactions with the parts of the molecule that correspond to mature HAP (Figure 4). The main chain amide group of Lys77 makes a hydrogen bond with OE2 of Glu171, whereas the hydroxyl group of Tyr78p forms a hydrogen bond with OE1 of Glu147. The residues which form the first β-strand of the prosegment (Ser79p to Glu86p) are involved in formation of an antiparallel β-sheet, with a string of hydrogen bonds with the residues Ile167 through Asn161. After activation, the latter strand is replaced by residues Ser-1 to Leu9. The C-terminal residues (Asn84p to Glu86p) of the first β-strand also form an antiparallel sheet connected through hydrogen bonds with Ser14 to Val12. These hydrogen bonded interactions participate in forming the central motif of the HAP zymogen. The side chains of Val81p, Phe83p, and Ile85p are placed in a hydrophobic pocket formed by the mature part of HAP. Atoms OG1 of Thr80p and O of Ile85p make hydrogen bonds with OG1 of Thr166 and ND2 of Asn161, respectively. The side chain of Asn87p, the residue connecting the β-strand and the α-helix 1 of the prosegment, forms hydrogen bonds with the main chain carbonyl of Lys160.
Figure 4.
Interactions between the prosegment and the mature HAP. (A) Interactions of the first β-strand of the HAP prosegment with the mature polypeptide of HAP. The carbon atoms of the prosegment and the mature enzyme are colored gray and green, respectively. The hydrogen bonds are represented as blue dotted lines. (B) Interactions of the first α-helix of the HAP prosegment with the mature polypeptide of HAP. The color scheme is the same as in panel B.
The residues Ser88p through Glu98p form the α-helix 1 of the HAP prosegment. This helix is involved in several interactions with the C-terminal domain of the mature HAP (Figure 4B). The side chain hydroxyl group of the Ser88p forms a hydrogen bond with OG of Ser219, whereas the hydroxyl of Tyr89p makes a hydrogen bond with ND2 of Asn279. A number of other hydrogen bonds tie α-helix 1 to the mature part of HAP, including interactions between OD2 of Asp90p and ND2 of Asn161, as well as NH2 of Arg91p and the carbonyl oxygen of Ala10. The hydrophobic residues of helix 1 (Leu92p and Ile96p) are involved in the interactions with the residues from the C-terminal domain of mature HAP polypeptide. Leu92p makes hydrophobic interactions with the side chains of Met283 and Leu244, whereas Ile96p is involved in a hydrophobic contact with Ile279A. The two connecting residues (His99p and Lys100p) of α-helix 1 and 2 are solvent exposed.
The α-helix 2 of the HAP prosegment consists of 13 residues (Leu101p to Asn113p) and is positioned in a hydrophobic groove formed by two loops (238–245 and 276–283). Helix 2 interacts with the mature HAP through a single hydrogen bond formed between the hydroxyl of Tyr104p and the carbonyl of Pro241. Six residues (Lys114p to Lys119p) of the prosegment connecting helix 2 form a coil, whereas Ser120p-Phe1 were disordered.
The first 13 residues of the mature HAP (Ser-1 to Asn11) fold mainly as a coil, with the exception of a single 310 helical turn involving Leu6 to Leu9. This one turn helix is present at one side of the active site cleft and occupies the S2 and S4 substrate binding pockets of the enzyme. This first segment of the mature enzyme makes two hydrogen bonded interactions, the first one between the carbonyl oxygen of Leu9 and the amide nitrogen of Ser219 and the second one between the amide nitrogen of Asn11 and the carbonyl of Ala217. The flap (residues 70–83) which covers the active site of HAP is well defined in the zymogen and is present in an open conformation.
A comparison of the zymogens of plasmepsins and gastric aspartic proteases
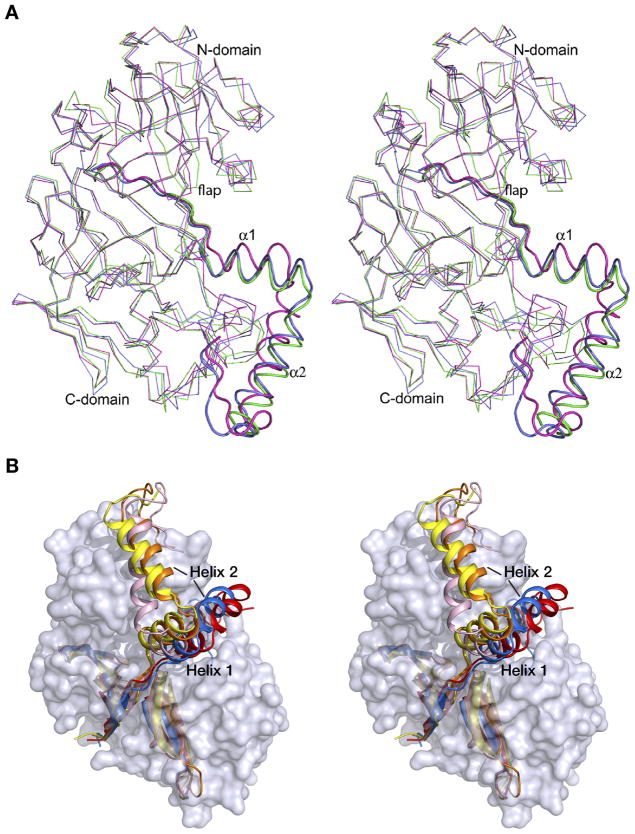
Crystal structures of only two proplasmepsins, pro-PMII (22) and pro-pvPMIV (28), have been reported previously, thus the high resolution crystal structure of pro-HAP is the third solved structure of plasmepsin zymogens. The coordinates of pro-HAP have been superimposed onto pro-PMII and pro-pvPMIV (Figure 5A), with the resulting r.m.s.d. of 1.39 and 1.38 Å, respectively. The overall fold of the progsegments in these three structures is similar, including the N-terminal β-strand followed by two α-helices and a coil. Helix 1 is considerably shorter than helix 2 in pro-pvPMIV, whereas both helices are of almost similar length in the zymogens of HAP and PMII. The relative positions of helices 1 and 2 are much closer in the structures of pro-HAP and pro-PMII compared to their positions in pro-pvPMIV (Figure 5A). The connecting residues of pro-pvPMIV assume different conformation compared to their counterparts in the other two structures. Although there are slight conformational differences among helix 2 in the three structures, the relative positions of this helix are comparable. In all structures helix 2 is positioned in a hydrophobic groove formed by two loops (238–245 and 276–283), maintaining a hydrogen bond to the mature portion through Tyr104p, conserved in all three proplasmepsins.
Figure 5.
(A) Superposition of the zymogen structures of HAP (green), PMII (blue; PDB ID: 1PFZ), and pvPMIV (magenta; PDB ID: 1MIQ). The enzyme is shown as Cα trace and the prosegment fragments are shown in a cartoon representation. (B) Prosegments of gastric aspartic proteases and plasmepsins in the context of the surface model of the mature enzyme portion of pro-HAP. The prosegment fragments of the zymogens of HAP (yellow), PMII (orange), pvPMIV (pink), pepsin (red), and gastricsin (blue) are shown in a cartoon representation.
The pro-mature junction, composed of a “Tyr-Asp” loop (defined in Ref. (28) based on the presence of a hydrogen bond between Tyr121p and Asp2), is visible in pro-PMII and pro-pvPMIV, whereas this junction is disordered in the structure of pro-HAP. A sequence comparison (18) shows that both residues forming the “Tyr-Asp” loop are conserved in all plasmepsin zymogens except PMI, where tyrosine is replaced by histidine. It is also important to note that the C-terminal loop (residues 237–247) which interacts with the “Tyr-Asp” loop has a similar conformation in the other two PM zymogens but a different conformation in pro-HAP. The single 310 helical turn of the N-terminal mature segment occupies an almost identical position in the three zymogen structures.
An important difference in the conformation of the flap is observed in the three zymogens. In pro-PMII the flap is disordered, whereas the flaps in proHAP and pro-pvPMIV are visible, assuming an open and closed conformation, respectively (Figure 5A). Because of the closed conformation of the flap in the pro-pvPMIV, Tyr75 comes close to the active site and forms a hydrogen bond with Trp39, which is flipped into the flap pocket (28). In this structure the active site serine residue is pointing towards the catalytic aspartic acid (Asp32) and forms a hydrogen bond. In the zymogens of HAP and PMII Trp39 is facing away from the flap pocket and forms a hydrogen bond with the active site Ser35.
The helix in the N-terminal domain consisting of residues 109–114 assumes a different conformation in the HAP zymogen compared to the other two structures (Figure 5A). The C-terminal domain loop consisting of residues 278A to 279C is found in an open conformation in the structure of pro-pvPMIV.
A comparison of the zymogens of plasmepsins with those of gastric pepsins shows a striking difference in the mode of inhibition of their catalytic activity. In all these zymogens the first strand of the prosegment forms part of the central β-sheet and has to be replaced upon activation by the N terminus of the mature enzyme. Similarly, the position of the first helix in gastric zymogens, although slightly shifted, is close to its location in the zymogens of plasmepsins. Major differences in the structures of the prosegments start after the first helix due to changes in the directionality of the second helix, which runs in gastric zymogens at an angle of ~70° to that in proplasmepsins (Figure 5B) and directly blocks the active site cleft. Therefore, while in the gastric zymogens the prosegment inactivates the enzyme by preventing the entry of substrates to the active site, in proplasmepsins it leaves the active site open, but not functional, due to distortions caused by the interactions between the prosegment and the rest of the enzyme.
The structure of the HAP–KNI-10395 complex
The asymmetric unit of the HAP–KNI-10395 complex contains four molecules of the enzyme, engaged in extensive intermolecular contacts. The four monomers are quite similar and a superposition of monomer A using the corresponding Cα atoms onto monomers B, C, and D results in r.m.s.d. of 0.7, 0.5, and 0.7 Å, respectively. However, only the active sites of the A, C, and D monomers contain bound molecules of KNI-10395 (Figure 2A), clearly defined by the Fo-Fc electron density map (Figure 2B). Four watermolecules are found in the active site area of molecule B. The overall fold of each monomer in the HAP–KNI-10395 complex is quite similar to what was observed in the previously described inhibitor complexes of HAP (12). Formation of tight dimers leads to displacement of the helix containing residues 225–235, and the following loop composed of residues 238–245, from their positions assumed in other pepsin-like aspartic proteases (11), but their conformation is similar to the one found in the HAP apoenzyme. The C-terminal loop consisting of residues 276–283 of one monomer is packed in the active site of the other molecule of the dimer. Despite the presence of an inhibitor, the flap is found in an open conformation.
The most intriguing feature of the HAP–KNI-10395 dimer is swapping of the first β-strand of the central motif (residues 0–10) between molecules A and B, as well as between C and D. This phenomenon is clearly seen in the Fo-Fc omit electron density map (Figure 2C). Such domain swapping (Figure 6A) is unique in this family, not reported for any other plasmepsins (or, for that matter, any other aspartic proteases). The first β-strand of one monomer forms a part of an antiparallel β-sheet in the second monomer, making a number of hydrogen bonds with residues 164–167. At the crossover point the amide group of Ala10 is hydrogen bonded to the side chain of Asp116.
Figure 6.
Structure of the inhibited form of HAP and its comparison with the structure of the zymogen. (A) A dimer consisting of molecules A (green) and B (salmon) of the HAP–KNI-10395 complex, showing domain swapping involving the N termini of both molecules. (B) Superposition of the zymogen of HAP (yellow) and the complex of the mature enzyme with pepstatin A (cyan, PDB ID: 3FNT). The prosegment is colored red, and the N-terminal fragments (residues 2–9) of the mature enzyme that undergo major rearrangement during activation are colored blue in both models. The hinge region (10–12) is colored magenta and the dashed lines represent the disordered residues at the junction of the propeptide and the N terminus of the mature enzyme in the zymogen.
Although the phenomenon of domain swapping described here has not been seen in any other aspartic proteases, other plasmepsins were reported to form tight dimers (17). HAP, in particular, was previously reported to form the tightest dimer among all plasmepsins, with a buried surface area of 2284 Å 2 for the apoenzyme (12). Because of the domain swapping, the surface area buried upon formation of the domain swapped dimer in the HAP–KNI-10395 complex is even much larger, 3789 Å 2 for each monomer.
Comparisons of the structures of the zymogen and mature HAP
The structure of pro-HAP has been compared with the structure of the apoenzyme and with HAP complexed with pepstatin A, KNI-10006, and KNI-10395. The overall superposition of the coordinates of the HAP zymogen onto apo-HAP (308 Cα pairs), HAP-pepstatin A (304 Cα pairs), HAP–KNI-10006 (311 Cα pairs), and HAP–KNI-10395 (291 Cα pairs) complexes resulted in r.m.s.d. of 1.5, 1.85, 1.5, and 1.7 Å, respectively. When the N-terminal and C-terminal domains of the respective pairs were superimposed individually, the resulting r.m.s.d. values were 1.8 and 3.6 Å for the apoenzyme, 2.1 and 1.5 Å for the pepstatin A complex, 1.7 and 1.1 Å for the KNI-10006 complex, and 2.0 and 2.3 Å for the complex with KNI-10395. Unusually high deviations between the C-terminal domains of the zymogen and the apoenzyme are due to extensive shifts caused by dimer formation in the latter (12). In the inhibitor complexes, these values correlate well with the binding mode of the inhibitors. The N- and C-terminal domains of the zymogen are shifted away from each other compared to their positions in the inhibitor complexes, with the rigid-body shift of the N-terminal domain being more prominent (Figure 6B). In this section, we utilize the structure of the pepstatin A complex for detailed structural comparison with the zymogen (Figure 6B), since this structure of HAP is most similar to other active plasmepsins.
From a comparison of the zymogen and mature forms of HAP it is evident that the first 13 residues of the latter undergo a significant structural rearrangement upon maturation. In the zymogen, these residues are packed against the surface of the C-terminal domain forming a coil, except for a single 310 helical turn between Leu6 and Leu9. After activation, these residues change their conformation and become part of a β-sheet. The residues that form the 310 helix in the zymogen move away from the active site after maturation. The main chain carbonyl oxygen of Leu6 forms a water-mediated hydrogen bond with the side chain hydroxyl of Thr218. Three hydrogen bonded interactions of the main chain are present in this region of the zymogen, involving Leu9 and Ser219, Asn11 and Ala217, and Leu13 and Phe31 (Figure 7A). These hydrogen bonds are absent in the mature enzyme since Leu9-Leu13 are folded differently.
Figure 7.
Interactions in the active site regions of the HAP zymogen and HAP-pepstatin A complex. (A) The region corresponding to the immature active site of HAP in the zymogen. Residues Leu6-Phe15 that belong to the N-terminal segment of mature HAP are shown with carbon atoms gray. (B) Active site of HAP in its complex with pepstatin A (PDB ID: 3FNT). The carbon atoms of Leu9-Ala18 are colored gray. The central hydroxyl oxygen of the statine residue in pepstatin A is shown as a sphere. (C) Schematic diagram showing the main chains of the zymogen (green) and of the mature enzyme (magenta) in its complex with pepstatin and the active site residues, emphasizing the rearrangement that resulted in the increase of the distance between the latter (marked in black dashed lines).
The active site cleft is wider in the zymogen compared to the mature enzyme. The distances between NE2 of His32 and OD1 of Asp215 in the zymogen and in the mature active enzyme are 7.4 and 3.3 Å, respectively (Figure 7C). In the zymogen, several water molecules are hydrogen bonded to the carboxylate of Asp215 (Figure 7A). Wat44, located close to OD2 of Asp215 in the zymogen, is replaced by the central statine hydroxyl group of pepstatin A in the HAP-pepstatin A complex (Figure 7B). This water molecule is likely involved in the catalytic activity of the enzyme and its equivalent is also present in the active sites of apo-HAP, HAP–KNI-10006, and HAP–KNI-10395 complexes. A hydrogen bond between OG1 of Thr33 and OD1 of Asp215 that is present in the zymogen is absent in the mature enzyme. The N-terminal domain active site loop (His32-Ser35) assumes a different conformation in the zymogen compared to the mature enzyme. In the zymogen, Trp39 is removed from the flap pocket and makes a hydrogen bond with Ser35. Trp39 maintains the same conformation in apo-HAP and in the complexes with KNI-10006 and KNI-10395, but its conformation is different in the HAP-pepstatin A complex.
Binding of KNI-10395 to HAP
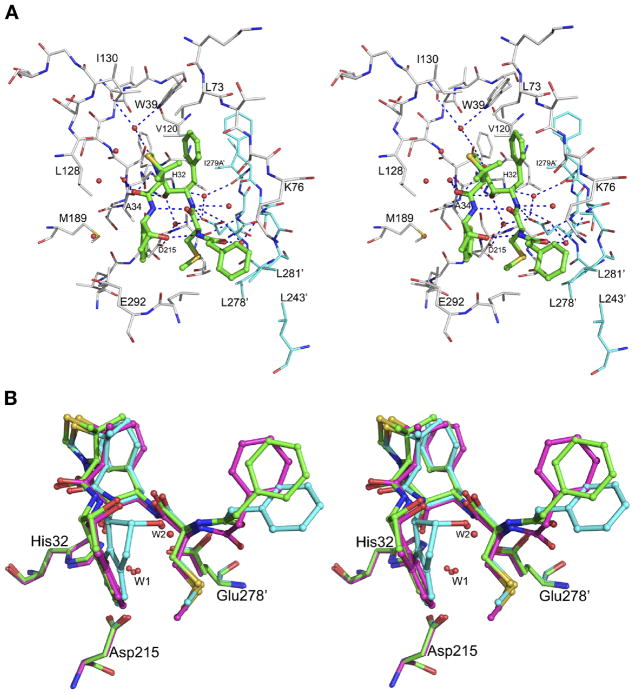
KNI compounds are peptidomimetic inhibitors that have been extensively developed during the last 20 years. The original aim was to create chemotherapeutic anti-HIV agents targeting retroviral proteases (42, 43)), although they have been later shown to be also active against plasmepsins (44). Many KNI compounds utilize a common molecular scaffold containing an allophenylnorstatine moiety followed by a thioproline ring (44). Different chemical functional groups are added to this main scaffold to generate a variety of KNI compounds. Our recent crystal structure of the complex of HAP with KNI-10006 (12) showed a unique mode of binding of the inhibitor in the active site, quite different from the previously observed mode of binding of other peptidomimetic inhibitors to plasmepsins, thus raising the interest in further exploring these compounds as potential candidates for the development of anti-malaria drugs. Biochemical studies using several KNI compounds show that KNI-10395 (Figure 2A) is a potent inhibitor of HAP (99.2% inhibition at 5.0 μM), and thus it was chosen for crystallographic studies.
Structure of the HAP–KNI-10395 complex was determined at 2.5 Å resolution (Table 1). The crystals contain four molecules of the complex in an asymmetric unit, with inhibitors clearly seen in the active sites of three molecules (A, C, and D). The mode of binding of KNI-10395 in these sites is similar, but differs substantially from the way in which other peptidomimetic inhibitors bind to aspartic proteases. The conformation of the inhibitor is considerably deformed, with the main chain turning back on itself, creating a “U”-shaped structure. This structure is internally stabilized by two hydrogen bonds, one between the peptide amino group of the 2-aminoindanol moiety and the peptide carbonyl of the methylthioalanine, and the other between the peptide amino group of the methylthioalanine and the hydroxyl group of the 2-aminoindanol moiety. Although the central hydroxyl group of the inhibitor is bound close to the active site residues His32 and Asp215, it is not positioned directly in between them, but is hydrogen bonded to the OD2 of Asp215 via water molecule Wat243, as well as to the main chain carbonyl of Ala34. The inhibitor is bound to the enzyme by 25 hydrogen bonds, either direct or through water molecules (Figure 8A). The carbonyl group of the allophenylnorstatine moiety is within a hydrogen bond distance to the side chain hydroxyl of Ser35 and to Wat607. The phenyl group of the allophenylnorstatine moiety is packed in a hydrophobic pocket formed by the side chains of Trp39, Leu73, and Val120 from one molecule, and Phe279B′ (prime denotes the other molecule of the dimer). The amide group of the allophenylnorstatine moiety forms hydrogen bonds with the side chain carboxylate oxygen atoms of Glu278A′ and with the main chain carbonyl oxygen atom of Phe279B′ via Wat233. The carbonyl oxygen atom of the methylthioalanine is hydrogen bonded to the side chain hydroxyl of Ser75 via Wat865. The 2-methylsulfanylethyl group is bound in a hydrophobic pocket formed by Leu291, Val300, and Leu278′. The carbonyl oxygen atom of the terminal phenylacetyl group is hydrogen bonded to Glu278A′, side chain hydroxyl of Ser279′, and main chain amide of Leu281′ through Wat41. The terminal phenyl group of the inhibitor is involved in hydrophobic interactions with the side chains of Leu291, Leu281′, and Leu243′ and one of the methyl groups of the thioproline ring is involved in hydrophobic interactions with the side chain of Leu73. The carbonyl group of the thioproline is hydrogen bonded to Wat607. The 2-aminoindanol group is placed in a hydrophobic pocket formed by the side chains of Ala34, Ile213, Met189, and Glu292. Nine water molecules are also present in the active site of the complex. Because of formation of a tight domain-swapped dimer, the flap pocket is filled by the loop consisting of residues 276′–283′ from the other protomer. The flap assumes an open conformation and the side chain of Trp39 is flipped away from the flap pocket, forming a hydrogen bond with Ser35 through Wat377. The NE2 atom of His32 is hydrogen bonded to the main chain carbonyl oxygen of Ile279A′ and to the side chain carboxyl oxygen (OE2) of Glu278A′ through Wat38. This mode of binding is completely different from the mode of binding observed for other KNI compounds in the complexes with HIV-1 protease (45, 46), PMI (18), and pmPMIV (47).
Figure 8.
The complex between HAP and KNI-10395. (A) The active site of molecule A in the HAP–KNI-10395 complex. The inhibitor (green) is shown in ball-and-stick representation. Protein residues are shown as thinner sticks with carbons colored gray for monomer A and cyan for monomer B (in which residue numbers are primed). Hydrogen bonds between the inhibitor and the molecule are shown as dotted lines. (B) Superposition of the KNI-10395 molecules bound to three molecules of HAP. The inhibitors bound to monomers A (cyan), C (green), and D (magenta) are shown in ball-and-stick representation, whereas the active site residues are shown as sticks. The important water molecules are shown as spheres.
Superposition of molecules A and B of the AB dimer, with the inhibitor bound only in the active site of the former, shows that the side chains of Thr218 and Glu278A′ assume different conformation. In the active site of molecule B the OD2 atom of Asp215 is hydrogen bonded to OE2 of Glu278A′, as well as to OG1 of Thr218. Equivalent hydrogen bonds are absent in the active site of molecule A, in which KNI-10395 is present. The water molecule which is close to the carboxyl oxygen OD2 of Asp215 in the active site of molecule A is absent in molecule B. A comparison of the active sites of the three molecules that contain bound KNI-10395 shows that there are some relatively minor, nevertheless important differences in the conformation of the inhibitors (Figure 8B). It is clear that the inhibitors in the C and D active site assume a very similar conformation, with a water molecule mediating hydrogen bonds between the hydroxyl group of the 2-aminoindanol moiety and the NH group of the terminal methylthioalanine. This water molecule is replaced by the hydroxyl group of the 2-aminoindanol moiety of the inhibitor in molecule A. The 2-aminoindanol moiety of the inhibitor in the A active site is in a more constrained conformation and the terminal phenyl group is further away from the flap. The tips of the flap are similar in molecules A and C, and slightly different in molecule D. The lack of significant conformational differences among the four monomers, as well as the absence of crystal contacts in the vicinity of the active site of molecule B, do not allow us to rationalize the absence of the inhibitor in this molecule. However, similar phenomena have been reported earlier for other enzyme-inhibitor complexes (for example, see Refs. (48, 49)).
Binding of the KNI compounds to plasmepsins
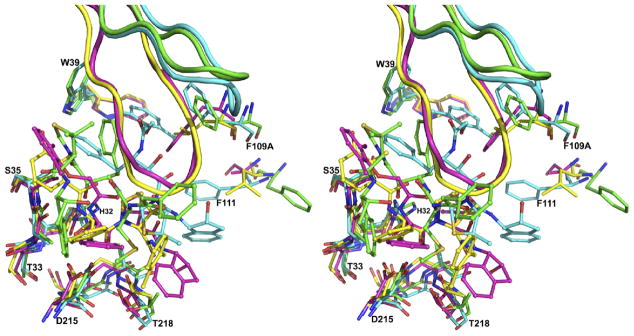
Four structures of three plasmepsins (pmPMIV, PMI, and HAP) complexed with three compounds from the KNI series (KNI-764, KNI-10006, and KNI-10395) are now available for comparisons (PDB IDs: 2ANL, 3QS1, 3FNU, and 3QVI). Superposition of the structures of HAP–KNI-10006, PMI–KNI-10006 and pmPMIV–KNI-764 complexes on the HAP–KNI-10395 complex, based on Cα atoms, produced r.m.s.d. of 1.05, 1.3, and 1.4, respectively. Whereas a vast majority of peptidomimetic inhibitors binds to various aspartic proteases in extended conformation with the side chains on both sides of the central core (…P2, P1–P1′, P2′…) properly docked into the corresponding binding pockets (…S2, S1–S1′, S2′…) (50), all KNI inhibitors are bound to plasmepsins in non-traditional ways (Figure 9). Although KNI-10006 and KNI-764 bind to PMI and pmPMIV, respectively, in an extended conformation with the central hydroxyl group located in between the two catalytic aspartate residues (18), the directionality of the main chain in both inhibitors is opposite to that found in the structures of aspartic proteases with peptidomimetic inhibitors (18). The mode of binding of KNI-10006 in the active site of HAP (12) is substantially different from the mode of binding of the same compound in PMI (18). In both KNI complexes of HAP the flap is in the open conformation. In the HAP–KNI-10006 complex, the 2,6-dimethylphenyloxyacetyl moiety of the inhibitor is positioned in the “flap pocket” and interacts with the residues from the flap (12) (Figure 10A). Although the “flap pocket” is recognized as being specific to the plasmepsins, it is notable that in the gastric zymogens the equivalent pocket is occupied by the residues from the N terminus of proenzyme, Tyr4 (PDB ID: 1HTR) or Leu6 (PDB ID: 3PSG) (Figure 10A).
Figure 9.
Active sites of the superimposed complexes of plasmepsins. The HAP–KNI-10395 complex is colored green, HAP–KNI-10006 (cyan; PDB ID: 3FNU), PMI–KNI-10006 (magenta; PDB ID: 3QS1) and pmPMIV–KNI-764 (yellow; PDB ID: 2ANL). Individual amino acid residues are shown in stick representation, the inhibitors in ball-and-stick, and the flaps as cartoons. Selected residues belonging to the HAP–KNI-10395 complex are labeled.
Figure 10.
(A) Structures of HAP–KNI-10006 (gray; PDB ID: 3FNU) and HAP–KNI-10395 (green) superimposed on the structures of gastric zymogens. Prosegments of pepsinogen (PDB ID: 3PSG) and progastricsin (PDB ID: 1HTR) are pink and cyan, whereas the N-terminal fragments of pro-mature enzymes are colored red and blue, respectively. The inhibitors are shown as sticks and selected residues as lines. (B) A superposition of the HAP–KNI-10395 complex onto pepsinogen and progastricsin showing the concomitant shift of the loop 290–292 and the inhibitor in HAP. The colors are the same as in panel A. (C) A comparison of the S1′ binding pocket in HAP with bound KNI-10395 (green) and pepstatin A (orange; PDB ID: 3FNT) with its counterpart in pepsinogen (red). The side chain of Tyr37p in the prosegment of pepsinogen is shown in pink.
The orientation of KNI-10395 in the active site of HAP is substantially different from the binding mode of the KNI compounds to other plasmepsins (Figure 9). Because of formation of the tight domain swapped dimer in the HAP–KNI-10395 complex, the “flap pocket” is occupied by the loop consisting of residues 276–283 from the other molecule. The position of the 2,6-dimethylthioproline moiety of KNI-10006 in its complex with HAP is occupied by Glu278A′ from the other molecule in the HAP–KNI-10395 complex. In the HAP–KNI-10006 complex the aminoindanol moiety is located in a hydrophobic pocket formed by Ala10, Val12, Phe15, and Phe111 (12). In the HAP–KNI-10395 complex, Phe15 and Phe111 adopt different conformation and the loop containing Ala10 and Val12 is present in the crossover region of the domain swapped dimer. The side chains of His32 assume different conformation in the complexes with KNI-10006 and KNI-100395, whereas the putative nucleophilic water molecules and Asp215 are located in similar positions.
Due to the inaccessibility of the “flap pocket” in the crystals of HAP complexed to KNI-10395, the inhibitor adopts a very unusual conformation of an U-shaped δ turn, occupying the space which is taken by the residues 35p–38p in the helical turn of the prosegment in the complexes of the gastric zymogens. However, a shift in plasmepsins of the loop containing residues 290–292 towards the active site leads to a concomitant shift of the inhibitor (Figure 10B). The C-terminal 2-aminoindanol moiety of the inhibitor occupies the S1′ binding pocket of mature HAP, where the side chain Tyr37p in the pepsinogen structure (PDB ID: 3PSG - (51)) is found, as well as the side chain of P2′ Ala of pepstatin in the complexes with HAP (Figure 10C). The differences between the binding mode of the C-terminal half of pepstatin in HAP and the other plasmepsins, as well as pepsin-like enzymes, have been discussed previously (12). In all known complexes of pepstatin bound to aspartic proteases the residue at the P2′ position of the inhibitor occupies the S2′ pocket, but HAP is an exception, since in its complex the orientation of the C-terminal part of the inhibitor has changed, bringing the side chain of P2′ Ala into the S1′ pocket. HAP contains two substitutions of the residues comprising the S1′ pocket that are unique to plasmepsins family. Gly34, which is conserved in a vast majority of other aspartic proteases, is substituted by Ala, and an aromatic group of Tyr/Phe189 present in other plasmepsins is replaced by a flexible side chain of Met. The S2′ pocket is occupied by the dimethylthioproline moiety, thus switching the order for binding S1 – S2 pockets by two C-terminal groups in the inhibitor molecule. The allophenylnorstatine moiety takes the space occupied in other plasmepsins by the side chain of Tyr75 (that residue is substituted by Ser in HAP) and is close to that in the KNI-10006 bound to HAP. The phenylacetyl group on the N-terminus of KNI-10395 assumes a position that is structurally equivalent to Ser35p and Trp36p in the prosegments of pepsinogen (PDB ID: 3PSG) and progastricsin (PDB ID: 1HTR), respectively (Figure 10A).
Inactivation and activation mechanism of HAP
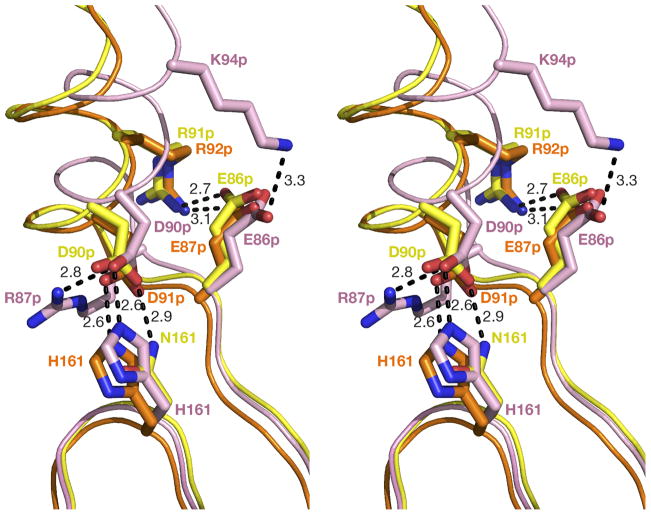
The zymogen structures of PMII (22) and pvPMIV (28) have been determined previously, making the structure of the HAP zymogen the third one. The previously described structures of plasmepsin zymogens led to the elucidation of the autoinhibition of PMII (22) and pvPMIV (28). Bernstein et al. (22) noted major differences in the mode of autoinhibition of plasmepsins compared to autoinhibition of such aspartic proteases as porcine pepsinogen A and human progastricsin. In the latter enzymes, the prosegment blocks the substrate binding sites (51), whereas inactivation of the zymogens of PMII (22) and pvPMIV (28) is caused by enforcing separation of the two catalytic aspartic acids. A comparison of the zymogen structure of HAP with those of pro-PMII and pro-pvPMIV shows that in HAP the two catalytic residues are also far apart (His32CG-Asp215CG: 7.9 Å). Two important hydrogen bonded interactions between the C-terminal active site ψ loop and the N-terminal segment of the mature HAP in the HAP zymogen are involved in separating Asp215 from His32. The first hydrogen bond is found between the carbonyl oxygen of Leu9 and the amide nitrogen of Ser219, whereas the second one is formed between the amide nitrogen of Asn11 and the carbonyl of Ala217 (Figure 7A). The N-terminal segment of the mature HAP in the zymogen structure is locked by hydrogen bonding interactions with the prosegment. Similar hydrogen bonding interactions have also been observed in the pro-PMII (22) and pro-pvPMIV (28) and were described as a “harness” which helps to separate the two domains of the molecules. It is clear that separation of the active site residues also provides the basis of the inactivation of HAP.
Similar to other proplasmepsins, autoactivation of HAP zymogen takes place as a result of lowering the pH (25). It has been reported that several hydrogen bonds between the prosegment and the mature segment of pro-PMII (22) and pro-pvPMIV (28) involve Asp or Glu residues. These hydrogen bonds keep the prosegment attached to the C-terminal domain of proplasmepsin and thus keep the active site residues separated. The most important hydrogen bonding interactions are visible in the pro-mature junction “Tyr-Asp” loop region. Lower pH causes protonation of the aspartate residue of that loop (22), resulting in disruption of the hydrogen bonds made by its side chain and opening of the loop (22). Other hydrogen bonding interactions between the prosegment and the C-terminal domain of the mature polypeptide chain are also disrupted at low pH (22, 28), allowing the two domains of the mature polypeptide chain to approach each other. Although the pro-mature junction containing the “Tyr-Asp” loop is disordered in the HAP zymogen structure, a sequence comparison (18) shows conservation of both residues critical to forming this loop, as well as the other residues in the vicinity that are involved in this extensive hydrogen bonded network. The other hydrogen bonding interactions involving the prosegment and the C-terminal domain of the mature part of HAP zymogen are also similar to those observed in pro-PMII (22) and pro-pvPMIV (28). In particular, an important network of ion pairs and hydrogen bonds, stabilizing the N terminus of the first helix in the propart, is much more similar between the pro-PMII (22) and pro-HAP, and slightly modified in pro-pvPMIV (28) due to variation in sequence. As a result, the position of the first helix is identical in the pro-PMII (22) and pro-HAP, but shifted in pro-pvPMIV (Figure 11). It is thus likely that autoactivation of HAP is also triggered by lowering the pH, as reported for the zymogens of other plasmepsins (22, 28).
Figure 11.
A comparison of helix 1 in the prosegments of the zymogens of HAP (yellow), PMII (orange; PDB ID: 1PFZ) and pvPMIV (pink; 1MIQ). Selected residues are shown as sticks and hydrogen bonds as black dashed lines.
Active site of HAP
HAP is unique among vacuolar plasmepsins from P. falciparum because of several amino acid substitutions in the active site region (12). The most important substitution is replacement by histidine of Asp32, one of the two catalytic aspartates in pepsin-like aspartic proteases (11). Other substitutions include strictly conserved Tyr75 and highly conserved Val/Gly76, which are replaced by Ser and Lys, respectively. The unique nature of the active site of HAP led to difficulties in elucidating its catalytic mechanism. Two different catalytic mechanisms of HAP have been previously proposed, each based only on modeling and/or computational studies. Andreeva et al. (13) suggested a serine protease-like catalytic mechanism, but recent structural studies (12) disproved this hypothesis. In an alternative mechanism proposed by Bjelic and Åqvist (14), Asp215 acts as the catalytic base as well as acid and the role of His32 is only to stabilize the reaction pathway through a strong interaction with the developing positive charge, since its position is not optimal to function as either acid or base catalyst. In addition, any possible role of the neighboring Ser35 in modifying the protonation state of His32 during catalysis has not been considered.
Ser35 is conserved in all plasmepsins (18) as well as in other pepsin-like enzymes (52), (11). In the catalytic mechanism of pepsin-like aspartic proteases, Ser35 and Thr/Ser218 play a crucial role in maintaining proper protonation states of Asp32 and Asp215, respectively (53). In the food vacuole of the P. falciparum, the reaction catalyzed by HAP takes place in an acidic environment. The pH optimum for the HAP-assisted catalysis is around 5.5 (1, 30), thus the role of the surrounding residues must be considered in order to understand the protonation state of His32. Previous biochemical investigations indicating no involvement of Ser35 and His32 in the catalytic mechanism of HAP were based on the kinetic data (25) at pH 7.5, much higher than the optimum, thus they cannot be considered to be final.
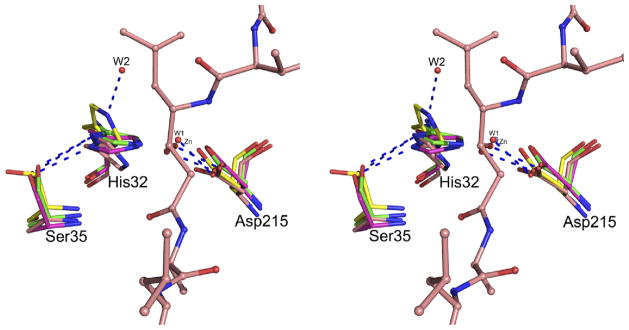
Crystal structures of HAP (12) proved that its fold is the same as in typical pepsin-like aspartic proteases. Based on the conservation of the structural fold and of the arrangement of the putative catalytic residues, His32 and Asp215, it might be expected that HAP might utilize a modified mechanism of typical pepsin-like enzymes. However, the exact role of the active site residues needs to be further elucidated. A comparison of the crystal structures of apo-HAP and three inhibitor complexes (with pepstatin A, KNI-10006, and KNI-10395) indicates that the conformation of the side chain of Asp215 is very similar in all of them (Figure 12). However, positions of the side chains of His32 and Ser35 in the HAP–KNI-10395 complex are quite different from the positions of these residues in the other three structures. The putative nucleophilic water molecules (W1) assume very similar positions in the HAP–KNI-10006 and HAP–KNI-10395 complexes, corresponding to the location of the hydroxyl group of the P1 statine residue in the pepstatin A complex (Figure 12). The hydroxyl group of Ser35 is hydrogen bonded to the ND1 atom of His32 in apo-HAP and in the complexes with pepstatin A and KNI-10006. However, in the HAP–KNI-10395 structure the hydroxyl of Ser35 is pointing away from His32, with the side chain of the latter residue flipped in such a way that its ND1 atom is close to the main chain amide of Ala34, thus Ser35 and His32 are no longer hydrogen bonded. Instead, the NE2 atom of His32 side chain forms a hydrogen bond with a neighboring water molecule (W2) (Figure 12). A similar water molecule is found in the close proximity of the carboxylate group of Asp32 in the apoenzyme of PMII (1LF4 - (17)).
Figure 12.
The active site of HAP in the superimposed structures of apo-HAP (green; PDB ID: 3FNS), HAP-pepstatin A (salmon; PDB ID: 3FNT), HAP–KNI-10006 (magenta; PDB ID: 3FNU), and HAP–KNI-10395 (yellow). The active site residues are shown in stick representation. The nucleophilic water molecules (W1) from the HAP–KNI-10006 and HAP–KNI-10395 structures are shown as red spheres. Another water molecule (W2) from the HAP–KNI-10395 structure is also shown. The Zn2+ ion bound in the active site of apo-HAP, very close behind W1, is gray. Pepstatin A from its HAP complex is shown in ball-and-stick representation. Hydrogen bonds are shown as blue dashed lines.
It is clear that, in HAP, Asp215 must act as a catalytic base activating the nucleophilic water molecule (W1) by formation of a hydroxyl anion capable of attacking the carbonyl carbon of the peptide linkage, leading to formation of a tetrahedral intermediate. In pepsin-like aspartic proteases, the tetrahedral intermediate accepts a proton from Asp32 (54). Andreeva and Rumsh (53) proposed that, at acidic pH, the acidity of Asp32 is increased by the proton relay mechanism involving the side chains of Trp39, Tyr75, and Ser35, and a water molecule situated between the hydroxyl groups of the latter two residues. However, the actual role of Tyr75 and Trp39 in the catalytic mechanism of pepsin-like aspartic protease is still unclear, as the replacement of Tyr75 by Asn in R. pusillus pepsin (55) increases the catalytic efficiency of the enzyme. Tyr75 is also not conserved in all plasmepsins (18). Based on the analysis of the structures of several pepsin-like aspartic proteases, Andreeva and Rumsh (53) noted different conformations of hydroxyl of Ser35 and postulated that a hydrogen bond between Ser35 and Asp32 is important to maintaining the proper protonation state of Asp32. An analogous change in the conformations of the side chain of Ser35 has also been observed in the HAP structures. Based on the arrangements of the His32 and Ser35 side chain in the HAP active site (Figure 12), we postulate that His32 might be directly involved in providing the electrophilic component for the catalytic mechanism. The role of Ser35 could be to maintain proper protonation state of His32. However, more structural, computational, as well as biochemical studies are necessary to support this hypothesis.
In summary, crystal structure of the HAP zymogen revealed a conformation of the prosegment similar to its counterparts in pro-PMII and pro-pvPMIV, suggesting a common mechanism for inctivation/activation. While the conformation and the interactions of the prosegments are different in proplasmepsins and in gastric zymogens, some of the interactions of KNI-10006 and KNI-10395 with HAP resemble those seen in the structures of gastric zymogens. Although both inhibitors are bound to HAP in a non-traditional way, their binding modes are very different. In the HAP–KNI-10006 complex the plasmepsin-specific “flap pocket” is filled by the N-terminal group of the inhibitor, whereas the C-terminal half follows the pathway of residues 7–10 of the N terminus of pro-mature enzyme in the structures of gastric zymogens. The internally folded, unusual δ turn conformation of KNI-10395 stabilized by two intramolecular hydrogen bonds, as well as the location of the inhibitor, are similar to the helical turn comprising the residues 34p–38p in the prosegments of pepsins. We postulate that the combination of factors such as the lack of access to the “flap pocket”, the unique nature of the S1′ pocket in HAP, combined with the presence of the long side chain of Lys76 at the tip of the flap, contribute to the unusual binding mode adopted by KNI-10395. The structure of the HAP–KNI-10395 complex also revealed a novel mode of dimerization involving domain swapping, previously not seen in any aspartic proteases.
Acknowledgments
Diffraction data were collected at the Southeast Regional Collaborative Access Team (SER- CAT) beamline 22-ID, located at the Advanced Photon Source, Argonne National Laboratory.
Abbreviations
- HAP
histo-aspartic protease
- PM
plasmepsin
- PEG
polyethylene glycol
- Trx
thioredoxin
- CHAPS
3-[(3-Cholamidopropyl)dimethylammonio]propanesulfonic acid
- PDB
Protein Data Bank
- NCS
non-crystallographic symmetry
- TLS
translation-libration-screw
Footnotes
Accession Codes
The atomic coordinates and structure factors for the HAP zymogen and HAP–KNI-10395 complex have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/), with the accession codes 3QVC and 3QVI, respectively.
References
- 1.Banerjee R, Liu J, Beatty W, Pelosof L, Klemba M, Goldberg DE. Four plasmepsins are active in the Plasmodium falciparum food vacuole, including a protease with an active-site histidine. Proc Natl Acad Sci USA. 2002;99:990–995. doi: 10.1073/pnas.022630099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greenwood BM, Bojang K, Whitty CJ, Targett GA. Malaria. Lancet. 2005;365:1487–1498. doi: 10.1016/S0140-6736(05)66420-3. [DOI] [PubMed] [Google Scholar]
- 3.Kolakovich KA, Gluzman IY, Duffin KL, Goldberg DE. Generation of hemoglobin peptides in the acidic digestive vacuole of Plasmodium falciparum implicates peptide transport in amino acid production. Mol Biochem Parasitol. 1997;87:123–135. doi: 10.1016/s0166-6851(97)00062-5. [DOI] [PubMed] [Google Scholar]
- 4.Xiao H, Tanaka T, Ogawa M, Yada RY. Expression and enzymatic characterization of the soluble recombinant plasmepsin I from Plasmodium falciparum. Protein Eng Des Sel. 2007;20:625–633. doi: 10.1093/protein/gzm066. [DOI] [PubMed] [Google Scholar]
- 5.Esposito A, Tiffert T, Mauritz JM, Schlachter S, Bannister LH, Kaminski CF, Lew VL. FRET imaging of hemoglobin concentration in Plasmodium falciparum-infected red cells. PLoS ONE. 2008;3:e3780. doi: 10.1371/journal.pone.0003780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Francis SE, Banerjee R, Goldberg DE. Biosynthesis and maturation of the malaria aspartic hemoglobinases plasmepsins I and II. J Biol Chem. 1997;272:14961–14968. doi: 10.1074/jbc.272.23.14961. [DOI] [PubMed] [Google Scholar]
- 7.Coombs GH, Goldberg DE, Klemba M, Berry C, Kay J, Mottram JC. Aspartic proteases of Plasmodium falciparum and other parasitic protozoa as drug targets. Trends Parasitol. 2001;17:532–537. doi: 10.1016/s1471-4922(01)02037-2. [DOI] [PubMed] [Google Scholar]
- 8.Bhaumik P, Gustchina A, Wlodawer A. Structural studies of vacuolar plasmepsins. Biochim Biophys Acta. 2011 doi: 10.1016/j.bbapap.2011.04.008. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gardiner DL, Skinner-Adams TS, Brown CL, Andrews KT, Stack CM, McCarthy JS, Dalton JP, Trenholme KR. Plasmodium falciparum: new molecular targets with potential for antimalarial drug development. Expert Rev Anti Infect Ther. 2009;7:1087–1098. doi: 10.1586/eri.09.93. [DOI] [PubMed] [Google Scholar]
- 10.Berry C, Humphreys MJ, Matharu P, Granger R, Horrocks P, Moon RP, Certa U, Ridley RG, Bur D, Kay J. A distinct member of the aspartic proteinase gene family from the human malaria parasite Plasmodium falciparum. FEBS Lett. 1999;447:149–154. doi: 10.1016/s0014-5793(99)00276-8. [DOI] [PubMed] [Google Scholar]
- 11.Dunn BM. Structure and mechanism of the pepsin-like family of aspartic peptidases. Chem Rev. 2002;102:4431–4458. doi: 10.1021/cr010167q. [DOI] [PubMed] [Google Scholar]
- 12.Bhaumik P, Xiao H, Parr CL, Kiso Y, Gustchina A, Yada RY, Wlodawer A. Crystal structures of the histo-aspartic protease (HAP) from Plasmodium falciparum. J Mol Biol. 2009;388:520–540. doi: 10.1016/j.jmb.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andreeva N, Bogdanovich P, Kashparov I, Popov M, Stengach M. Is histoaspartic protease a serine protease with a pepsin-like fold? Proteins. 2004;55:705–710. doi: 10.1002/prot.20078. [DOI] [PubMed] [Google Scholar]
- 14.Bjelic S, Åqvist J. Computational prediction of structure, substrate binding mode, mechanism, nd rate for a malaria protease with a novel type of active site. Biochemistry. 2004;43:14521–14528. doi: 10.1021/bi048252q. [DOI] [PubMed] [Google Scholar]
- 15.Liu J, Istvan ES, Goldberg DE. Hemoglobin-degrading plasmepsin II is active as a monomer. J Biol Chem. 2006;281:38682–38688. doi: 10.1074/jbc.M608535200. [DOI] [PubMed] [Google Scholar]
- 16.Xiao H, Briere LA, Dunn SD, Yada RY. Characterization of the monomer-dimer equilibrium of recombinant histo-aspartic protease from Plasmodium falciparum. Mol Biochem Parasitol. 2010;173:17–24. doi: 10.1016/j.molbiopara.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 17.Asojo OA, Gulnik SV, Afonina E, Yu B, Ellman JA, Haque TS, Silva AM. Novel uncomplexed and complexed structures of plasmepsin II, an aspartic protease from Plasmodium falciparum. J Mol Biol. 2003;327:173–181. doi: 10.1016/s0022-2836(03)00036-6. [DOI] [PubMed] [Google Scholar]
- 18.Bhaumik P, Horimoto Y, Xiao H, Miura T, Hidaka K, Kiso Y, Wlodawer A, Yada RY, Gustchina A. Crystal structures of the free and inhibited forms of plasmepsin I (PMI) from Plasmodium falciparum. J Struct Biol. 2011;175:73–84. doi: 10.1016/j.jsb.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koelsch G, Mares M, Metcalf P, Fusek M. Multiple functions of pro-parts of aspartic proteinase zymogens. FEBS Lett. 1994;343:6–10. doi: 10.1016/0014-5793(94)80596-2. [DOI] [PubMed] [Google Scholar]
- 20.Gao X, Wang J, Yu DQ, Bian F, Xie BB, Chen XL, Zhou BC, Lai LH, Wang ZX, Wu JW, Zhang YZ. Structural basis for the autoprocessing of zinc metalloproteases in the thermolysin family. Proc Natl Acad Sci U S A. 2010;107:17569–17574. doi: 10.1073/pnas.1005681107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horimoto Y, Dee DR, Yada RY. Multifunctional aspartic peptidase prosegments. New Biotechnol. 2009;25:318–324. doi: 10.1016/j.nbt.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 22.Bernstein NK, Cherney MM, Loetscher H, Ridley RG, James MN. Crystal structure of the novel aspartic proteinase zymogen proplasmepsin II from Plasmodium falciparum. Nature Struct Biol. 1999;6:32–37. doi: 10.1038/4905. [DOI] [PubMed] [Google Scholar]
- 23.Dame JB, Reddy GR, Yowell CA, Dunn BM, Kay J, Berry C. Sequence, expression and modeled structure of an aspartic proteinase from the human malaria parasite Plasmodium falciparum. Mol Biochem Parasitol. 1994;64:177–190. doi: 10.1016/0166-6851(94)90024-8. [DOI] [PubMed] [Google Scholar]
- 24.Moon RP, Tyas L, Certa U, Rupp K, Bur D, Jacquet C, Matile H, Loetscher H, Grueninger-Leitch F, Kay J, Dunn BM, Berry C, Ridley RG. Expression and characterisation of plasmepsin I from Plasmodium falciparum. Eur J Biochem. 1997;244:552–560. doi: 10.1111/j.1432-1033.1997.00552.x. [DOI] [PubMed] [Google Scholar]
- 25.Parr CL, Tanaka T, Xiao H, Yada RY. The catalytic significance of the proposed active site residues in Plasmodium falciparum histoaspartic protease. FEBS J. 2008;275:1698–1707. doi: 10.1111/j.1742-4658.2008.06325.x. [DOI] [PubMed] [Google Scholar]
- 26.James MNG, Sielecki AR. Molecular structure of an aspartic proteinase zymogen, porcine pepsinogen, at 1.8 Å resolution. Nature. 1986;319:33–38. doi: 10.1038/319033a0. [DOI] [PubMed] [Google Scholar]
- 27.Khan AR, Cherney MM, Tarasova NI, James MNG. Structural characterization of activation ‘intermediate 2’ on the pathway to human gastricsin. Nature Struct Biol. 1997;4:1010–1015. doi: 10.1038/nsb1297-1010. [DOI] [PubMed] [Google Scholar]
- 28.Bernstein NK, Cherney MM, Yowell CA, Dame JB, James MN. Structural insights into the activation of P. vivax plasmepsin. J Mol Biol. 2003;329:505–524. doi: 10.1016/s0022-2836(03)00444-3. [DOI] [PubMed] [Google Scholar]
- 29.Hidaka K, Kimura T, Tsuchiya Y, Kamiya M, Ruben AJ, Freire E, Hayashi Y, Kiso Y. Additional interaction of allophenylnorstatine-containing tripeptidomimetics with malarial aspartic protease plasmepsin II. Bioorg Med Chem Lett. 2007;17:3048–3052. doi: 10.1016/j.bmcl.2007.03.052. [DOI] [PubMed] [Google Scholar]
- 30.Xiao H, Sinkovits AF, Bryksa BC, Ogawa M, Yada RY. Recombinant expression and partial characterization of an active soluble histo-aspartic protease from Plasmodium falciparum. Protein Expr Purif. 2006;49:88–94. doi: 10.1016/j.pep.2006.02.022. [DOI] [PubMed] [Google Scholar]
- 31.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Cryst. 1993;26:795–800. [Google Scholar]
- 32.CCP4. ollaborative Computational Project, Number 4, 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 33.Matthews BW. Solvent content of protein crystals. J Mol Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 34.Read RJ. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr. 2001;D57:1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- 35.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. 2011;D67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 37.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 38.Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–W619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. 2004;D60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 40.Cohen GE. ALIGN: a program to superimpose protein coordinates, accounting for insertions and deletions. J Appl Crystallogr. 1997;30:1160–1161. [Google Scholar]
- 41.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- 42.Mimoto T, Imai J, Tanaka S, Hattori N, Takahashi O, Kisanuki S, Nagano Y, Shintani M, Hayashi H, Sakikawa H, Akaji K, Kiso Y. Rational design and synthesis of a novel class of active site-targeted HIV protease inhibitors containing a hydroxymethylcarbonyl isostere. Use of phenylnorstatine or allophenylnorstatine as a transition-state mimic. Chem Pharm Bull (Tokyo) 1991;39:2465–2467. doi: 10.1248/cpb.39.2465. [DOI] [PubMed] [Google Scholar]
- 43.Mimoto T, Imai J, Tanaka S, Hattori N, Kisanuki S, Akaji K, Kiso Y. KNI-102, a novel tripeptide HIV protease inhibitor containing allophenylnorstatine as a transition-state mimic. Chem Pharm Bull (Tokyo) 1991;39:3088–3090. doi: 10.1248/cpb.39.3088. [DOI] [PubMed] [Google Scholar]
- 44.Nezami A, Kimura T, Hidaka K, Kiso A, Liu J, Kiso Y, Goldberg DE, Freire E. High-affinity inhibition of a family of Plasmodium falciparum proteases by a designed adaptive inhibitor. Biochemistry. 2003;42:8459–8464. doi: 10.1021/bi034131z. [DOI] [PubMed] [Google Scholar]
- 45.Hidaka K, Kimura T, Abdel-Rahman HM, Nguyen JT, McDaniel KF, Kohlbrenner WE, Molla A, Adachi M, Tamada T, Kuroki R, Katsuki N, Tanaka Y, Matsumoto H, Wang J, Hayashi Y, Kempf DJ, Kiso Y. Small-sized human immunodeficiency virus type-1 protease inhibitors containing allophenylnorstatine to explore the S2′ pocket. J Med Chem. 2009;52:7604–7617. doi: 10.1021/jm9005115. [DOI] [PubMed] [Google Scholar]
- 46.Vega S, Kang LW, Velazquez-Campoy A, Kiso Y, Amzel LM, Freire E. A structural and thermodynamic escape mechanism from a drug resistant mutation of the HIV-1 protease. Proteins. 2004;55:594–602. doi: 10.1002/prot.20069. [DOI] [PubMed] [Google Scholar]
- 47.Clemente JC, Govindasamy L, Madabushi A, Fisher SZ, Moose RE, Yowell CA, Hidaka K, Kimura T, Hayashi Y, Kiso Y, Agbandje-McKenna M, Dame JB, Dunn BM, McKenna R. Structure of the aspartic protease plasmepsin 4 from the malarial parasite Plasmodium malariae bound to an allophenylnorstatine-based inhibitor. Acta Crystallogr. 2006;D62:246–252. doi: 10.1107/S0907444905041260. [DOI] [PubMed] [Google Scholar]
- 48.Nagpal A, Valley MP, Fitzpatrick PF, Orville AM. Crystal structures of nitroalkane oxidase: insights into the reaction mechanism from a covalent complex of the flavoenzyme trapped during turnover. Biochemistry. 2006;45:1138–1150. doi: 10.1021/bi051966w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fu Z, Runquist JA, Montgomery C, Miziorko HM, Kim JJ. Functional insights into human HMG-CoA lyase from structures of Acyl-CoA-containing ternary complexes. J Biol Chem. 2010;285:26341–26349. doi: 10.1074/jbc.M110.139931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schechter I, Berger A. On the size of the active site in proteases. I Papain. Biochem Biophys Res Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 51.Sielecki AR, Fujinaga M, Read RJ, James MN. Refined structure of porcine pepsinogen at 1.8 Å resolution. J Mol Biol. 1991;219:671–692. doi: 10.1016/0022-2836(91)90664-r. [DOI] [PubMed] [Google Scholar]
- 52.Davies DR. The structure and function of the aspartic proteinases. Annu Rev Biophys Biophys Chem. 1990;19:189–215. doi: 10.1146/annurev.bb.19.060190.001201. [DOI] [PubMed] [Google Scholar]
- 53.Andreeva NS, Rumsh LD. Analysis of crystal structures of aspartic proteinases: On the role of amino acid residues adjacent to the catalytic site of pepsin-like enzymes. Protein Sci. 2001;10:2439–2450. doi: 10.1110/ps.25801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Erskine PT, Coates L, Mall S, Gill RS, Wood SP, Myles DA, Cooper JB. Atomic resolution analysis of the catalytic site of an aspartic proteinase and an unexpected mode of binding by short peptides. Protein Sci. 2003;12:1741–1749. doi: 10.1110/ps.0305203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park YN, Aikawa J, Nishiyama M, Horinouchi S, Beppu T. Involvement of a residue at position 75 in the catalytic mechanism of a fungal aspartic proteinase, Rhizomucor pusillus pepsin. Replacement of tyrosine 75 on the flap by asparagine enhances catalytic efficiency. Protein Eng. 1996;9:869–875. doi: 10.1093/protein/9.10.869. [DOI] [PubMed] [Google Scholar]