Abstract
To identify epigenetic patterns, which may predispose to type 2 diabetes (T2D) due to a family history (FH) of the disease, we analyzed DNA methylation genome-wide in skeletal muscle from individuals with (FH+) or without (FH−) an FH of T2D. We found differential DNA methylation of genes in biological pathways including mitogen-activated protein kinase (MAPK), insulin, and calcium signaling (P ≤ 0.007) and of individual genes with known function in muscle, including MAPK1, MYO18B, HOXC6, and the AMP-activated protein kinase subunit PRKAB1 in skeletal muscle of FH+ compared with FH− men. We further validated our findings from FH+ men in monozygotic twin pairs discordant for T2D, and 40% of 65 analyzed genes exhibited differential DNA methylation in muscle of both FH+ men and diabetic twins. We further examined if a 6-month exercise intervention modifies the genome-wide DNA methylation pattern in skeletal muscle of the FH+ and FH− individuals. DNA methylation of genes in retinol metabolism and calcium signaling pathways (P < 3 × 10−6) and with known functions in muscle and T2D including MEF2A, RUNX1, NDUFC2, and THADA decreased after exercise. Methylation of these human promoter regions suppressed reporter gene expression in vitro. In addition, both expression and methylation of several genes, i.e., ADIPOR1, BDKRB2, and TRIB1, changed after exercise. These findings provide new insights into how genetic background and environment can alter the human epigenome.
The prevalence of type 2 diabetes (T2D) is rapidly increasing worldwide. Although genome-wide association studies have identified polymorphisms contributing to the risk of T2D, a person’s lifestyle is a key factor in the development of the disease (1–3). Indeed, several studies have shown that the risk of T2D can be halved in high-risk groups through nonpharmacological lifestyle interventions involving exercise and diet (4,5). These studies show that the effect is rapid and does not require intensive interventions. Although little is known about the genes that convey the effects in these interventions, changes in DNA methylation have been suggested as a potential molecular mechanism through which exercise and diet mediate their effects on the transcriptome (6). Indeed, dietary factors can affect the degree of DNA methylation (7–11). However, whether an exercise intervention changes DNA methylation genome-wide in skeletal muscle is unknown. A family history (FH) of T2D increases the risk of developing the disease and may also affect the individual’s response to physical exercise (2,12–14). Yet, the impact of an FH of T2D on the genome-wide DNA methylation pattern in skeletal muscle is unknown. The objective of this study was therefore to study global DNA methylation patterns in skeletal muscle from individuals with or without an FH of T2D (FH+ and FH−, respectively) before and after an exercise intervention.
RESEARCH DESIGN AND METHODS
Cohorts.
Fifteen men with (FH+) and 13 men without (FH−) a first-degree FH of T2D were included in this study (Table 1). All FH+ men had at least one first-degree relative with T2D. At screening, 82 ± 29 days prior to the start of the study, the subjects underwent a physical examination and a 75-g oral glucose tolerance test, in which glucose levels were measured at 0 and 120 min (Table 1). At inclusion, the participants were healthy but sedentary. Based on self-report in which fitness level is rated on a scale of 1–5 (1 is the lowest level), the participants’ overall fitness level was 1.75 ± 0.58 prior to inclusion. A total of 25 of the participants were nonsmokers, and 3 were smokers (2 FH+ and 1 FH−). Anthropomorphic measurements and a max biking test using an ergometer bicycle (Marquette-Hellige Medical Systems 900ERG; Milwaukee, WI) were administered at the start of the exercise intervention (Table 1). The FH+ and FH− groups were groupwise matched for age, sex, BMI, and Vo2max at baseline, and there were no significant differences in weight, BMI, waist-to-hip ratio, blood pressure, pulse, and Vo2max between the FH+ and FH− men (Table 1). A muscle biopsy was taken from the vastus lateralis muscle in the fasting state under local anesthesia (1% lidocaine) using a 6-mm Bergström needle (Stille AB, Sweden). The participants were instructed to refrain from vigorous exercise for 48 h prior to the biopsy.
TABLE 1.
Clinical characteristics of men with or without an FH of T2D (FH+ and FH−, respectively) before and after a 6-month exercise intervention
All FH+ and FH− men participated in a 6-month supervised exercise intervention consisting of mainly endurance exercise. The participants were enrolled in a group training program including one session of 1-h spinning class and two sessions of 1-h aerobic class per week led by a certified instructor. On average, the participation level was 44.3 ± 3.5 sessions, which is slightly less than two sessions per week. After a 6-month exercise intervention and 48 h after the last bout of exercise, a second muscle biopsy and anthropomorphic measurements were taken, and Vo2max was analyzed with a max biking test (Table 1). Participants were invited 30 ± 11 days after the intervention for a second oral glucose tolerance test (follow-up).
Nine monozygotic twin pairs discordant for T2D were identified from the Swedish Twin Registry. They underwent clinical examinations, and muscle biopsies were taken in fasting state. Their characteristics are described in Supplementary Table 1.
The studies were approved by the local ethics committee, and written informed consent was obtained from all participants.
MeDIP-Chip analysis of muscle.
A total of 1.4 μg of genomic DNA was sonicated to an average of 500 bp by 13 cycles of 30 s on and 30 s off at high frequency with the BioRuptor (Diagenode, Liege, Belgium). For immunoprecipitation of methylated DNA, the mc-green-03 kit was used (Diagenode). A total of 1 μg of sonicated DNA was immunoprecipitated using the 5meC antibody with Sepharose beads overnight at 4°C. Immunoprecipitated DNA was purified with the QIAquick-PCR purification kit (Qiagen, Heidelberg, Germany) prior to whole-genome amplification of the DNA with the WGA-kit (Sigma-Aldrich, Stockholm, Sweden). A total of 15 ng of sonicated but not immunoprecipitated DNA (input) was also subjected to whole-genome amplification. A total of 6 μg of whole-genome amplified DNA was hybridized to the human 2.1 promoter DeLuxe tiling array (version 081229_HG18_Promoter_MeDIP_HX1) at the Roche-Nimblegen facility (Roche, Nimblegen, Iceland). Input and immunoprecipitated samples were labeled with Cy3 and Cy5, respectively, and hybridized to the same array. The human 2.1 promoter DeLuxe tiling array covers 10,000 bp of all known genes: 7,500 bp upstream of the transcription start sites (TSS) and 2,500 bp downstream of the TSS and all annotated cytosine guanine dinucleotide (CpG) islands. The total number of probes is 2.1 million per array. A GFF annotation file provided by Nimblegen was used for localization of the probes in relation to gene TSS, and the annotation file is based on build HG18 of the University of California Santa Cruz database. The output data from the MeDIP-Chip analysis consist of log2 ratios of immunoprecipitated (Cy5) versus input (Cy3) signals for each individual probe. The log2 ratio is computed and scaled to center the ratio data around zero. Scaling is performed by subtracting the biweight mean for the log2-ratio values for all features on the array from each log2-ratio value.
Normalization and statistical analysis of MeDIP-Chip data.
Within-array normalization was performed using model-based analysis of two-color arrays (MA2C), a normalization method for two-color tiling arrays incorporating sequence-specific probe effects (15). As MA2C standardizes probe intensities, dye bias and other nonbiological variations originating from array processing are removed. The normalized log2 ratios of immunoprecipitated versus input data were used for statistical comparisons using R software (16). The impact of an FH of T2D was analyzed using two-sample Mann-Whitney U tests for all probes, both at baseline and after exercise. The impact of exercise was analyzed for all probes using nonparametric paired tests: Wilcoxon signed-rank tests. The impact of exercise training was analyzed for the whole cohort (n = 28) and for each FH group separately.
To examine if an FH of T2D or exercise affects the degree of DNA methylation of individual genes, we calculated the mean level of DNA methylation for respective gene only including probes with P ≤ 0.01 due to either an FH of T2D or exercise. The impact of an FH of T2D or exercise on the mean level of methylation for respective gene was then analyzed using Mann-Whitney U tests or Wilcoxon signed-rank test, respectively. P values were then corrected for multiple testing using Bonferroni corrections and false discovery rate (FDR) analyses. Genes exhibiting differential DNA methylation with P ≤ 0.05 after Bonferroni corrections are presented in individual supplementary tables. Moreover, genes exhibiting differential DNA methylation with Q ≤ 0.005 after FDR were included in pathway analyses using Webgestalt (http://bioinfo.vanderbilt.edu/webgestalt/). Benjamini-Hochberg correction was used to determine the P values for the pathways within the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. For each comparison, the top significant KEGG pathways within Metabolism (1.1–1.8), Signal transduction (3.2), and Endocrine systems (5.2) are presented.
Microarray analysis.
RNA was isolated from muscle with the RNA fibrous tissue kit (Qiagen). Biotin-labeled cRNA was synthesized and hybridized to the Affymetrix Custom-Array NuGO-Hs1a520180-GeneChip (http://www.nugo.org; Affymetrix), which contains 23,941 probe sets. Images were analyzed using the GeneChip Operating-System (Affymetrix), and data were normalized using the robust multiarray average algorithm (17). The impact of an FH of T2D on expression was analyzed using a two-sample Mann-Whitney U test, and the impact of exercise training was tested using nonparametric paired tests. Genes showing nominally significant differences in expression with P ≤ 0.01 were included for further analysis. We next tried to identify genes displaying changes in DNA methylation (P ≤ 0.01) and expression (P ≤ 0.01) in the opposite direction (i.e., increased DNA methylation is associated with decreased expression or vice versa) in FH+ compared with FH− men or after compared with before exercise. Moreover, a mean centroid expression value was calculated for each of the biological pathways that are among the most significant. We first normalized the expression levels on the arrays to values between 0 and 1 across all analyzed samples, in which the highest expression value on the arrays is normalized to 1. The mean centroid expression value is then calculated as the mean expression of all genes included in respective pathway. Additionally, we examined if expression correlates negatively with DNA methylation of individual probes for respective gene by Spearman correlations.
Genetic analyses.
Single nucleotide polymorphisms (SNPs) were genotyped using HumanOmniExpress arrays according to the manufacturer’s instructions (Illumina, San Diego, CA). SNP data were extracted 7.5 kb upstream and 2.5 kb downstream of the TSS for each gene exhibiting differential DNA methylation in FH+ versus FH− men after Bonferroni corrections (Supplementary Table 2). SNPs were associated with DNA methylation of the respective gene based on an additive genetic model. A genetic risk score was generated for each individual by counting the number of risk alleles for SNPs previously associated with T2D (Supplementary Table 13).
Biological validation.
DNA methylation was analyzed in muscle of monozygotic twin pairs discordant for T2D using Infinium HumanMethylation450 BeadChip (Illumina) according to the manufacturer’s recommendations.
Technical validation.
Genes were selected for technical validation of the MeDIP-Chip data based on either their inclusion in biological pathways with differential DNA methylation, or that they show differential expression, and/or play a role in T2D and/or muscle physiology. For technical validation, DNA methylation levels were determined with bisulfite conversion and EpiTYPER (Sequenom, San Diego, CA) according to Sequenom’s protocol. The assays were designed with EpiDesigner (Supplementary Table 14). EpiTYPER data were generated for a subset of the included muscle samples because there was not enough DNA from some samples for technical validation. The EpiTYPER data were considered validated when both EpiTYPER and MeDIP show either increased or decreased DNA methylation of a gene with P ≤ 0.05. To technically validate the microarray expression data, quantitative RT-PCR was used to analyze expression of MSI2 with an ABI7900HT system and Assays-on-Demand (Hs00292670_m1; Applied Biosystems).
Luciferase assay.
Human promoter fragments containing 2,580 bp of THADA, 2,460 bp of MEF2A, 2,700 bp of RUNX1, or 2,500 bp of NDUFC2 were inserted into a CpG-free luciferase reporter vector (pCpGL-basic) provided by Klug and Rehli (18). These promoter fragments cover the gene regions analyzed in the MeDIP-Chip assay. The constructs were either mock-methylated or methylated using two different DNA methyltransferases: SssI and HhaI (New England Biolabs, Frankfurt, Germany). Human embryonic kidney (HEK) 293 cells were cotransfected with 100 ng pCpGL-vector either without (control) or with respective insert together with 2 ng of pRL renilla luciferase control reporter vector (pRL-CMV vector; Promega) as a control for transfection efficiency using the FuGENE-HD transfection reagent (Promega) according to the protocol. The luciferase signal was measured with the TD-20/20-Luminometer (Turner Designs).
Analyses of mitochondrial density and lipid content.
Muscle biopsies from 5 FH− and 10 FH+ men, taken before and after exercise, were sectioned and fixed in 4% formaldehyde. Immunostaining was performed as previously described (19) using a primary mouse monoclonal mitochondria marker antibody, dilution 1:200, code MTCO2 (Abcam), and a secondary mouse-IgG antibody coupled to Cy2 (Jackson ImmunoResearch Laboratories, West Grove, PA). Lipid content was analyzed by staining with Oil Red O (20). Images were captured with a digital camera (Nikon-DS-2Mv; Nikon, Tokyo, Japan). Areas of immunostaining and Oil Red O staining in digitized images were analyzed using Biopix iQ software (BioPix AB, Gothenburg, Sweden).
RESULTS
Impact of an FH of T2D on DNA methylation.
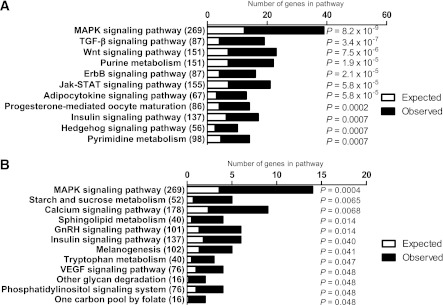
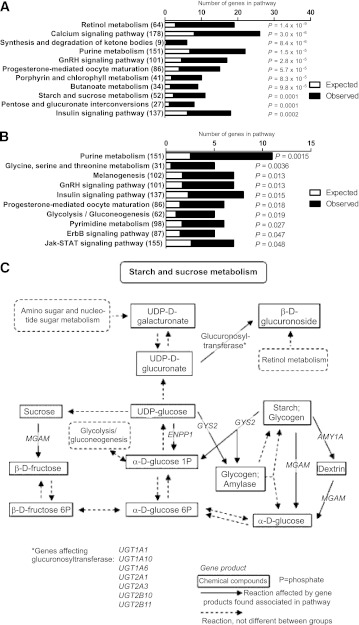
DNA methylation was analyzed in skeletal muscle of 15 FH+ and 13 FH− men using MeDIP-Chip (Table 1). The Chip comprised 2.1 million probes covering gene regions 7.5 kb upstream and 2.5 kb downstream of TSS. We identified 65 individual genes exhibiting differential DNA methylation in muscle of FH+ compared with FH− men at baseline after Bonferroni correction (Supplementary Table 2). Of these 65 genes, 60 genes had decreased and 5 genes had increased DNA methylation levels in the FH+ men. We next performed KEGG pathway analysis using Webgestalt (http://bioinfo.vanderbilt.edu/webgestalt/) to identify biological pathways with genes exhibiting different DNA methylation patterns in muscle of FH+ compared with FH− men. A total of 2,085 genes with decreased and 603 genes with increased DNA methylation in FH+ men at Q ≤ 0.005 were included in the pathway analysis. The top biological pathways of differentially methylated genes in FH+ men are shown in Fig. 1A and B and Supplementary Table 3. The mitogen-activated protein kinase (MAPK) and insulin-signaling pathways contain genes that exhibit both decreased and increased methylation (Fig. 1A and B). In contrast, the Wnt-signaling and adipocytokine-signaling pathways only include genes that exhibit decreased methylation, whereas the starch and sucrose metabolism, calcium signaling, as well as sphingolipid metabolism pathways contain genes that exhibit increased methylation in FH+ versus FH− men (Fig. 1A and B and Supplementary Table 3).
FIG. 1.
Comparison of DNA methylation in skeletal muscle of men with (FH+) vs. men without (FH−) a family history of T2D. The top KEGG pathways of genes, which exhibit decreased (A) and increased (B) methylation in skeletal muscle of FH+ (n = 15) vs. FH− (n = 13) men, respectively, with the expected number of genes (white), the observed number of genes (black), and the total number of genes in the pathway in parentheses. The P values were adjusted for multiple testing.
DNA methylation has been associated with transcriptional silencing (21). We hence examined if any of the genes with differential DNA methylation in FH+ compared with FH− men also showed different levels of expression. Using microarray data, we identified 46 genes in which differences in DNA methylation (P ≤ 0.01) were also associated with differential expression (P ≤ 0.01) (Supplementary Table 4). We further examined if there is any concordance between the top biological pathways of genes that are differentially methylated in FH+ compared with FH− men and differential expression of the mean centroid expression value of these pathways. The mean centroid expression value of adipocytokine signaling pathway showed differential expression in muscle of men with an FH of T2D (P = 0.007). Moreover, DNA methylation correlated negatively with the expression level for 534 genes at P ≤ 0.001 in the whole cohort at baseline.
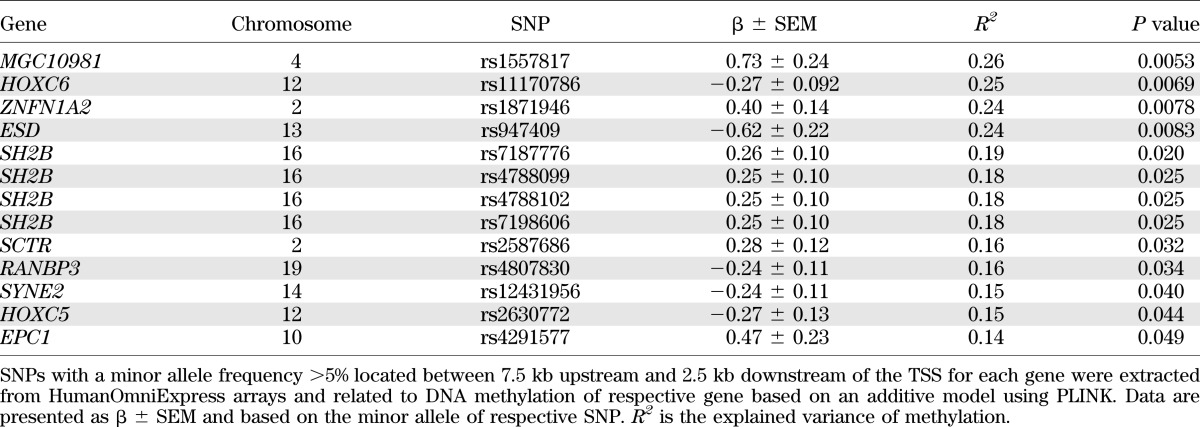
We next addressed whether genetic variation could influence DNA methylation in muscle of this cohort. SNPs located near the 65 genes that exhibit differential methylation in muscle of FH+ versus FH− men (Supplementary Table 2) were related to DNA methylation of respective gene (Table 2).
TABLE 2.
Associations between SNPs and DNA methylation with P < 0.05 in gene and promoter regions exhibiting differential DNA methylation in muscle of FH+ vs. FH− men
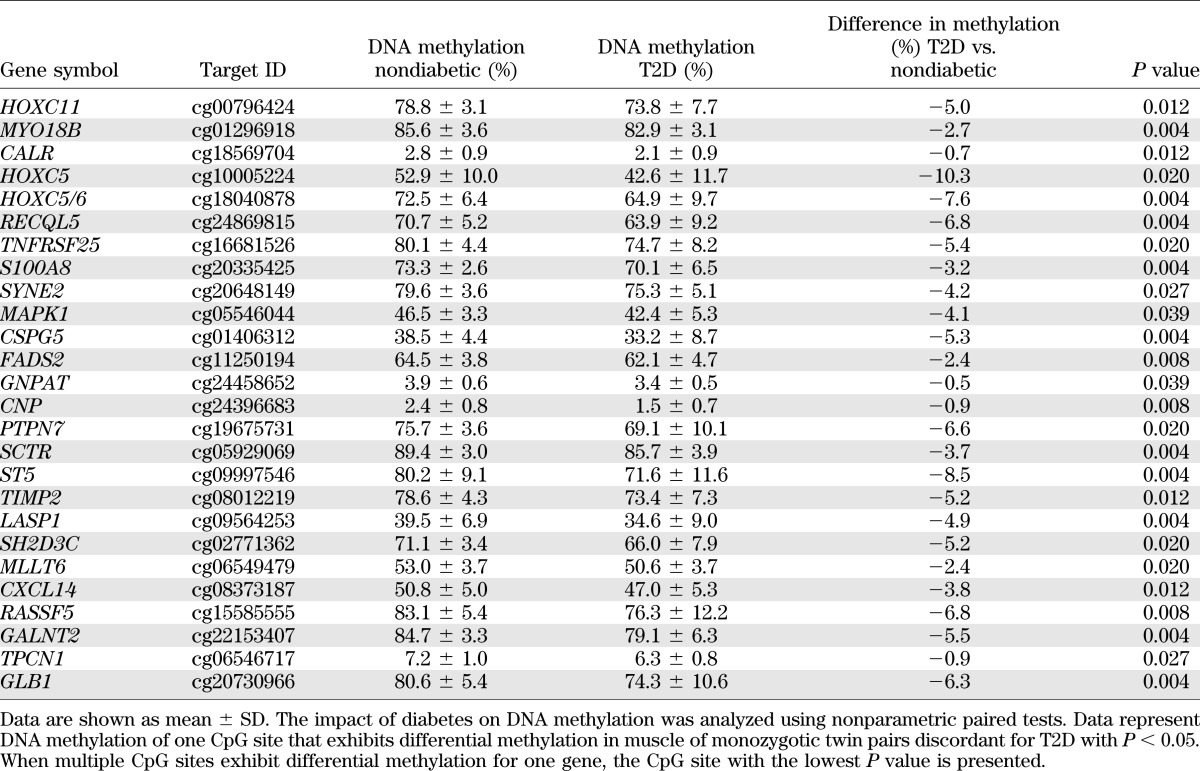
To test if DNA methylation found in muscle of FH+ men is associated with overt T2D, we analyzed DNA methylation of 65 genes included in Supplementary Table 2 in muscle of monozygotic twin pairs discordant for T2D. Forty percent of the 65 analyzed genes exhibit differential methylation in both diabetic twins and FH+ men (Table 3 and Supplementary Table 5).
TABLE 3.
Genes exhibiting differential DNA methylation in muscle of 15 FH+ vs. 13 FH− men as well as of 9 monozygotic twin pairs discordant for T2D
Impact of exercise on DNA methylation.
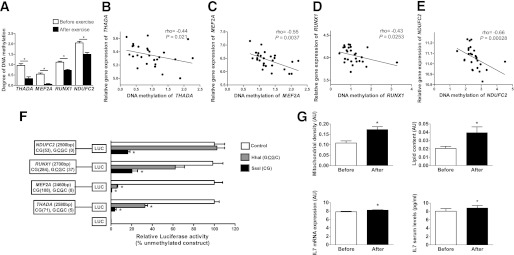
We proceeded to test if a 6-month exercise intervention was associated with genome-wide changes in DNA methylation in muscle of all 28 FH+ and FH− men included in the study. After Bonferroni corrections, we identified 134 individual genes that changed in the degree of DNA methylation after exercise in all men independent of FH status (Supplementary Table 6). Of these 134 genes, 115 showed decreased and 19 genes showed increased methylation after exercise. Exercise-induced changes in methylation of four selected genes are illustrated in Fig. 2A. The expression of these four genes correlates negatively with DNA methylation (Fig. 2B–E). To functionally test if promoter DNA methylation of these genes is associated with reduced expression, we produced reporter gene constructs in which the human promoter sequences of THADA, MEF2A, RUNX1, and NDUFC2 were inserted into a luciferase expression plasmid that completely lacks CpG dinucleotides (Fig. 2F). The constructs could hence be used to study the effect of promoter DNA methylation on luciferase activation in transfection assays. Each construct was mock-methylated or methylated with two methyltransferases. While SssI methylates all CpG sites, HhaI only methylates the internal cytosine residue in a GCGC sequence. Hence, SssI results in totally methylated constructs, and Hha1 gives point methylated constructs in which only a fraction of the CpG sites are methylated. The number of CpG sites that may be methylated in a respective construct is shown in Fig. 2F. HEK293 cells were transfected with the mock-methylated or methylated constructs. The highest reporter gene expression was generated by the mock-methylated constructs including the promoter regions (Fig. 2F). Furthermore, methylation of the human THADA, MEF2A, RUNX1, and NDUFC2 promoter regions suppressed reporter expression. While total methylation of the promoter regions by SssI suppressed reporter gene expression to 3.6 ± 2.7%, P = 0.004 for THADA; 0.8 ± 0.01%, P = 0.006 for MEF2A; 20.4 ± 9.4%, P = 0.005 for RUNX1; and 16.3 ± 3.7%, P = 0.009 for NDUFC2, point methylation by Hha1 suppressed the reporter expression to 32.8 ± 4.5%, P = 0.01 for THADA; 5.9 ± 1.5%, P = 0.007 for MEF2A; and 62.2 ± 15.4%, P = 0.08 for RUNX1 (Fig. 2F). The NDUFC2 promoter contains no GCGC sequence and could thus not be methylated by Hha1.
FIG. 2.
Impact of a 6-month exercise intervention on DNA methylation in human skeletal muscle. A: Exercise-induced changes in DNA methylation of THADA, MEF2A, RUNX1, and NDUFC2. Data are presented as mean ± SEM, and P values were corrected for multiple testing using Bonferroni corrections. Gene expression of THADA (B), MEF2A (C), RUNX1 (D), and NDUFC2 (E) correlates negatively with DNA methylation of respective gene. F: A diagram of the four luciferase reporter plasmids used to test the effect of DNA methylation on THADA, MEF2A, RUNX1, and NDUFC2 promoter activity and the empty vector are visualized. The four plasmids contain either 2,580 bp of the human THADA promoter, 2,460 bp of the human MEF2A promoter, 2,700 bp of the human RUNX1 promoter, or 2,500 bp of the human NDUFC2 promoter region inserted into a pCpGL-basic vector. Methylated (gray and black bars) or mock-methylated (white bars) promoter constructs were transfected into HEK293 cells for 48 h prior to luciferase assay. The data were normalized with cotransfected renilla luciferase control vector and are the average from three separate experiments of five replicates each. In each experiment, cells were transfected with an empty pCpGL-vector as a background control. A Student t test was used for statistical comparisons, and data are presented as relative expression compared with the nonmethylated construct including the promoter regions. G: Mitochondrial density, lipid content, and IL-7 mRNA expression in skeletal muscle as well as serum levels of IL-7 before and after exercise. Results are expressed as mean ± SEM. The analyses of mitochondrial density and lipid content were performed in 10 images covering at least 50 muscle fiber profiles. *P < 0.05.
We next tried to identify some functional changes related to the genes that exhibit differential DNA methylation in muscle after exercise (Supplementary Table 6 and Fig. 2A). Because NDUFC2 is part of the respiratory chain, we studied mitochondria in muscle. With morphological analyses, we found that mitochondrial density and lipid content increased in muscle after exercise (Fig. 2G). Moreover, because IL7 belongs to the genes that exhibit decreased methylation after exercise (Supplementary Table 6), we analyzed mRNA expression in muscle and serum levels of interleukin-7 (IL-7). Both muscle expression and serum levels of IL-7 increased after exercise (Fig. 2G).
To identify biological pathways among genes that exhibit differences in DNA methylation after exercise, we performed a KEGG pathway analysis. A total of 2,051 genes with decreased and 766 genes with increased methylation at Q ≤ 0.005 after exercise were included in the pathway analysis. The most significant pathways of genes with altered DNA methylation due to exercise are shown in Fig. 3A and B and Supplementary Table 7. Genes involved in retinol metabolism, calcium-signaling pathway, starch and sucrose metabolism, and the insulin-signaling pathway exhibit decreased methylation after exercise. The genes in the starch and sucrose metabolism pathway exhibiting decreased DNA methylation in muscle after exercise are further shown in Fig. 3C. Moreover, genes involved in purine metabolism, glycine, serine, threonine metabolism, insulin signaling, and glycolysis/gluconeogenesis exhibit increased methylation after exercise.
FIG. 3.
The top KEGG pathways of genes, which are differentially methylated in skeletal muscle after exercise. KEGG pathways of genes, which exhibit decreased (A) and increased (B) methylation in skeletal muscle of all men (n = 28) after a 6-month exercise intervention with the expected number of genes (white), the observed number of genes (black), and the total number of genes in the pathway in parentheses. The P values were adjusted for multiple testing. C: Diagram showing the genes in the starch and sucrose metabolism pathway with decreased DNA methylation in skeletal muscle of all men (n = 28) after a 6-month exercise intervention. P, phosphate.
We further identified 111 genes that showed changes in both the degree of DNA methylation and the level of expression in muscle after exercise with P ≤ 0.01 (Supplementary Table 8). Additionally, four of the biological pathways of genes that alter methylation due to exercise exhibited increased mean centroid expression values after exercise (purine, metabolism, P = 0.016; insulin, signaling, P = 0.012; ErbB, signaling, P = 0.032; and progesterone-mediated oocyte maturation, P = 0.029).
Because both FH− and FH+ men were included in the exercise intervention, we further tested if their response to exercise differed with regard to changes in DNA methylation. However, because no individual genes changed DNA methylation significantly after exercise in the FH− or FH+ men after Bonferroni corrections, we did not perform any further analyses of these data.
Differences between FH+ and FH− men in DNA methylation after exercise.
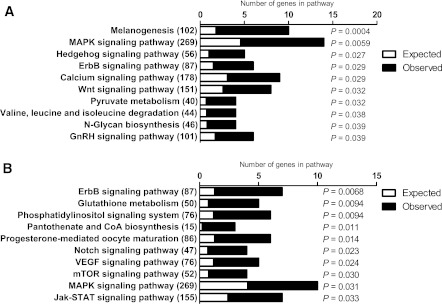
We then examined if differences in DNA methylation between FH− and FH+ men persist after 6 months of exercise. We identified 38 genes with significant differences after Bonferroni corrections, with 18 genes showing decreased and 20 genes showing increased DNA methylation in FH+ compared with FH− men (Supplementary Table 9) compared with 65 genes prior to the exercise intervention. A KEGG pathway analysis was further performed including 779 genes with decreased and 689 genes with increased DNA methylation at Q < 0.005 in FH+ compared with FH− men after exercise. The most significant pathways are shown in Fig. 4A and B and Supplementary Table 10. In agreement with the result prior to exercise, genes in the MAPK signaling pathway exhibited both decreased and increased DNA methylation in muscle of FH+ and FH− men after exercise. Moreover, we identified 10 genes in which differences in DNA methylation were also associated with differential expression in FH+ compared with FH− men (Supplementary Table 11). However, none of these 10 genes showed both differential DNA methylation and expression in FH+ compared with FH− men before exercise (Supplementary Table 4). After exercise, 239 genes showed inverse correlations between the degree of DNA methylation and the level of expression at P ≤ 0.001 in the whole cohort.
FIG. 4.
Comparison of DNA methylation in skeletal muscle of men with (FH+) vs. men without (FH−) a family history of T2D after a 6-month exercise intervention. The top KEGG pathways of genes, which exhibit decreased (A) and increased (B) methylation in skeletal muscle of FH+ (n = 15) vs. FH− (n = 13) men after exercise, with the expected number of genes (white), the observed number of genes (black), and the total number of genes in the pathway in parentheses. The P values were adjusted for multiple testing.
Candidate genes for T2D and DNA methylation.
We proceeded to test if any of 39 candidate genes for T2D, identified using genome-wide association studies (1), show differential DNA methylation in muscle of FH+ compared with FH− men or due to exercise. A total of 21 T2D candidate genes show nominally differential DNA methylation in FH+ compared with FH− men (Supplementary Table 12). Exercise changed DNA methylation significantly of 2 (THADA and RBMS1) and nominally of 18 T2D candidate genes (Supplementary Table 12). The number of observed candidate genes for T2D with differences in methylation at P < 0.01 in FH+ compared with FH− men or after exercise were more than expected (for an FH of T2D, expected number of genes = 12.64, χ2 = 5.5, and P = 0.02; for exercise, expected number of genes = 12.44, χ2 = 4.6, and P = 0.03).
A genetic risk score, generated by counting the number of risk alleles for SNPs previously associated with T2D (Supplementary Table 13), was further composed. However, the genetic risk score was similar for FH+ and FH− men (P = 0.56).
Technical validation.
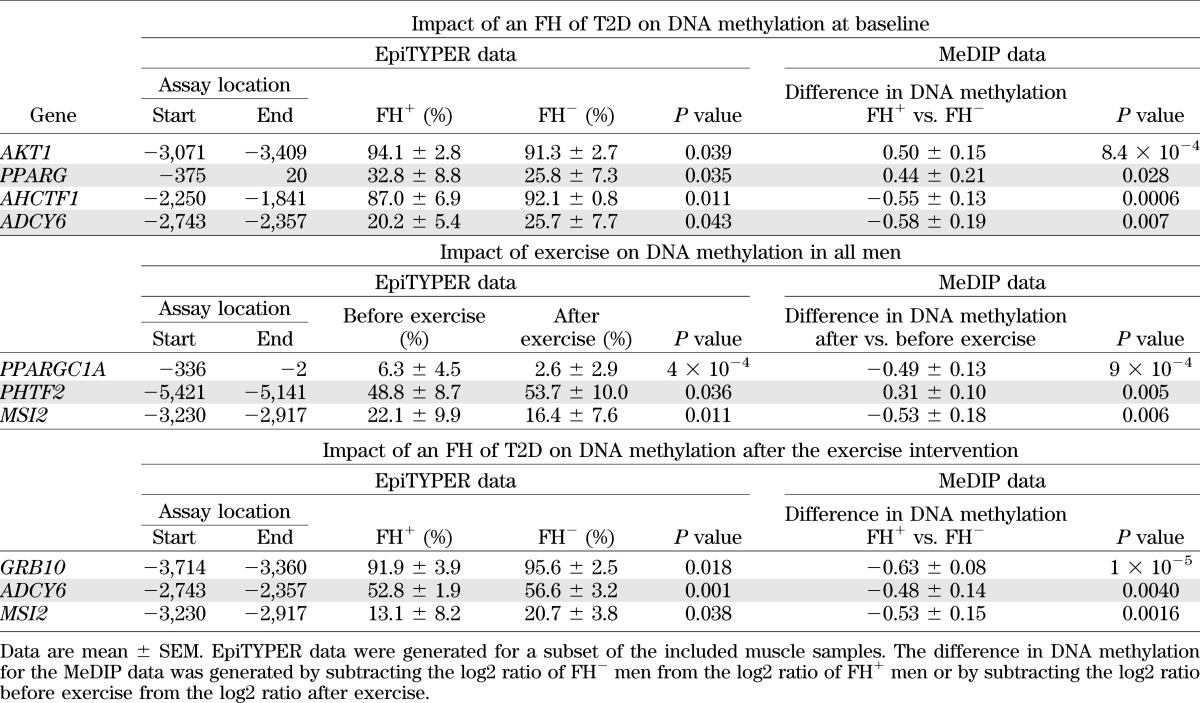
Finally, we technically validated three to four genes for each statistical comparison of the MeDIP-Chip data using EpiTYPER (Table 4). We also validated the microarray expression data for MSI2 using quantitative RT-PCR and the expression of MSI2 increased after exercise (before 1.83 ± 0.088 vs. after 2.09 ± 0.11; P = 0.038).
TABLE 4.
Technical validation of MeDIP-Chip data using EpiTYPER
DISCUSSION
An FH of T2D is an independent predictor of future risk for the disease (2,12,13,22). Moreover, epigenetic modifications of single genes have been shown to affect the pathogenesis of T2D (6,23–29). To our knowledge, this study presents the first global analysis of DNA methylation in muscle of humans with or without an FH of T2D. Our study has identified epigenetic differences in muscle of FH+ compared with FH− individuals. These include differential DNA methylation of genes in biological pathways with key functions in muscle such as MAPK, insulin, and calcium signaling and of individual genes including PRKAB1 and MAPK1. The protein encoded by PRKAB1 is a regulatory subunit of AMP-activated protein kinase, which is an enzyme that monitors cellular energy status and regulates metabolism in muscle (30). MAPK1 is also known to have important physiological and metabolic roles in human muscle (31). Pathway analyses provide overviews, not detailed descriptions of individual members. The KEGG analyses summarize our genome-wide methylation data in biological pathways. These results were followed up, and mean centroid expression values were calculated showing that with decreased methylation, there was increased overall expression of some pathways.
To test if DNA methylation in muscle of FH+ men is associated with T2D, we related our epigenetic findings in FH+ men with methylation in muscle of monozygotic twin pairs discordant for T2D. Forty percent of 65 studied genes exhibit differential methylation in both FH+ men and diabetic twins, suggesting that the epigenetic differences found in FH+ men may play a role in the development of T2D. Nevertheless, future prospective studies are needed to test if DNA methylation predicts T2D. In plants, it is well-established that epigenetic modifications can be inherited between generations (32). There are also reports describing transgenerational inheritance of epigenetic traits in mammals (33–35). Moreover, recent studies propose that both genetic and environmental factors may affect the epigenome in human muscle (7,23,24,26). In this study, we find associations between polymorphisms and DNA methylation of individual genes. Although the epigenetic differences we find between FH+ and FH− individuals may be due to genetic factors, future studies are needed to determine whether the observed differences are inherited or if they are simply due to a shared environment within families. Although the epigenome may be dynamic and change due to environmental exposures, once epigenetic modifications are introduced they may be both stable and inherited (29,35–37). Although epigenetics is strongly linked to certain disease states, including Retts syndrome, Prader-Willi syndrome, and transient neonatal diabetes (38–40), there are few studies that describe associations between epigenetic modifications and metabolic disease in humans (6). Epigenetic modifications have, however, been linked to metabolic disorders in animal models (29,37,41).
This is also the first study examining the impact of an exercise intervention on DNA methylation genome-wide in human muscle. We demonstrate that exercise for 6 months is associated with epigenetic changes, e.g., decreased DNA methylation of RUNX1 and MEF2A, two key transcription factors involved in exercise training (42–44), of THADA, previously associated with T2D (1), and of NDUFC2, which is part of the respiratory chain (45) was observed after exercise. MEF2A is a transcription factor involved in the exercise-induced regulation of GLUT4 expression, and hence it may influence glucose uptake in muscle (46). Moreover, exercise changed both DNA methylation and expression of a number of genes, including ADIPOR1, ADIPOR2, and BDKRB2, encoding receptors for adiponectin and bradykinin, respectively, which both regulate metabolism in muscle (47,48). Interestingly, IL-7 was recently found to be expressed and secreted from human skeletal muscle cells, and expression of IL-7 increased during differentiation of human myotubes (49). In this study, we found decreased DNA methylation in parallel with increased mRNA and serum levels of IL-7 in muscle after exercise, further supporting a role for IL-7 in human muscle. Although we find associations between increased DNA methylation and decreased expression for some genes in vivo and increased methylation was associated with reduced transcriptional activity in vitro, we cannot draw a conclusion as to whether differential expression is a consequence rather than a cause of changes in methylation (50).
Our group has previously shown that ageing is associated with increased DNA methylation and decreased expression of genes involved in oxidative phosphorylation in human muscle (23,24). In this study, we found that a gene from the respiratory chain NDUFC2 exhibited decreased methylation after exercise. We further showed that increased methylation of the NDUFC2 promoter reduced its transcriptional activity in vitro, indicating a role for DNA methylation in the regulation of NDUFC2 expression. Moreover, exercise increased Vo2max and the mitochondrial density in muscle.
Our study may point to some of the molecular mechanisms explaining the results seen in previous exercise intervention studies (4,5). It is further possible that the epigenetic modifications induced by exercise reduce the future risk of T2D among FH+ men. The two FH groups were matched for age, sex, BMI, and Vo2max at baseline in order to reduce the impact of lifestyle factors on our study. However, while exercise significantly improved a number of phenotypes, including waist circumference, diastolic blood pressure, and Vo2max in both FH groups, weight and BMI were only significantly reduced in FH+ men. A possible explanation for this phenomenon could be that the participation in the intervention study reminded the FH+ men that they are at greater disease risk and although they were requested not to change their overall lifestyle during the exercise intervention, one cannot exclude that they have changed their diet or other parts of their lifestyle.
Overall, this study provides novel insights into how exercise can induce genome-wide epigenetic changes in human muscle and that the response may differ in people with different genetic predispositions to metabolic disease.
ACKNOWLEDGMENTS
This work was supported by grants from the Swedish Research Council, including a Linnaeus grant (Lund University Diabetes Center, 349-2008-6589) and a strategic research area grant (Excellence of Diabetes Research in Sweden, 2009-1039), as well as equipment grants from the Knut and Alice Wallenberg Foundation (2009-0243), the Lundberg Foundation (Grant 359), Avtal om Läkarutbildning och Forskning, the Novo Nordisk Foundation, Universitetssjukhuset Malmö Allmäna Sjukhus Fonder, Tore Nilsson, Syskonen Svenssons Fond, Diabetes Foundation, Kungliga Fysiografiska Sällskapet in Lund, European Foundation for the Study of Diabetes-Lilly Foundation, Svenska stiftelsen för medicinsk forskning, and Påhlsson. The group of Swedish twins was recruited from the Swedish Twin Registry, which is supported by grants from the Swedish Department of Higher Education and the Swedish Research Council. No other potential conflicts of interest relevant to this article were reported.
M.D.N. and C.L. were responsible for study design, researched data, performed data analyses, and wrote the manuscript. T.D., E.H., E.N., and T.R. researched data, performed data analyses, and reviewed and edited the manuscript. P.V., B.T.Y., S.L., H.P., H.W., J.A., and P.A. performed data analyses and reviewed and edited the manuscript. T.E., K.-F.E., and L.G. were responsible for study design, collected clinical material, and reviewed and edited the manuscript. Y.W. collected clinical material, performed data analyses, and reviewed and edited the manuscript. M.A., N.W., P.-A.J., and O.H. researched data and reviewed and edited the manuscript. C.L. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
The authors thank the Swegene Centre for Integrative Biology at Lund University for analyzing DNA methylation using the Infinium HumanMethylation450 BeadChip (Illumina).
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db11-1653/-/DC1.
REFERENCES
- 1.McCarthy MI. Genomics, type 2 diabetes, and obesity. N Engl J Med 2010;363:2339–2350 [DOI] [PubMed] [Google Scholar]
- 2.Lyssenko V, Almgren P, Anevski D, et al. Botnia study group Predictors of and longitudinal changes in insulin sensitivity and secretion preceding onset of type 2 diabetes. Diabetes 2005;54:166–174 [DOI] [PubMed] [Google Scholar]
- 3.Hu FB, Manson JE, Stampfer MJ, et al. Diet, lifestyle, and the risk of type 2 diabetes mellitus in women. N Engl J Med 2001;345:790–797 [DOI] [PubMed] [Google Scholar]
- 4.Tuomilehto J, Lindström J, Eriksson JG, et al. Finnish Diabetes Prevention Study Group Prevention of type 2 diabetes mellitus by changes in lifestyle among subjects with impaired glucose tolerance. N Engl J Med 2001;344:1343–1350 [DOI] [PubMed] [Google Scholar]
- 5.Knowler WC, Barrett-Connor E, Fowler SE, et al. Diabetes Prevention Program Research Group Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Engl J Med 2002;346:393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ling C, Groop L. Epigenetics: a molecular link between environmental factors and type 2 diabetes. Diabetes 2009;58:2718–2725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brøns C, Jacobsen S, Nilsson E, et al. Deoxyribonucleic acid methylation and gene expression of PPARGC1A in human muscle is influenced by high-fat overfeeding in a birth-weight-dependent manner. J Clin Endocrinol Metab 2010;95:3048–3056 [DOI] [PubMed] [Google Scholar]
- 8.Bouchard L, Rabasa-Lhoret R, Faraj M, et al. Differential epigenomic and transcriptomic responses in subcutaneous adipose tissue between low and high responders to caloric restriction. Am J Clin Nutr 2010;91:309–320 [DOI] [PubMed] [Google Scholar]
- 9.Lillycrop KA, Phillips ES, Jackson AA, Hanson MA, Burdge GC. Dietary protein restriction of pregnant rats induces and folic acid supplementation prevents epigenetic modification of hepatic gene expression in the offspring. J Nutr 2005;135:1382–1386 [DOI] [PubMed] [Google Scholar]
- 10.Plagemann A, Harder T, Brunn M, et al. Hypothalamic proopiomelanocortin promoter methylation becomes altered by early overfeeding: an epigenetic model of obesity and the metabolic syndrome. J Physiol 2009;587:4963–4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Milagro FI, Campión J, Cordero P, et al. A dual epigenomic approach for the search of obesity biomarkers: DNA methylation in relation to diet-induced weight loss. FASEB J 2011;25:1378–1389 [DOI] [PubMed] [Google Scholar]
- 12.Köbberling J, Tillil H. Genetic and nutritional factors in the etiology and pathogenesis of diabetes mellitus. World Rev Nutr Diet 1990;63:102–115 [DOI] [PubMed] [Google Scholar]
- 13.Isomaa B, Forsén B, Lahti K, et al. A family history of diabetes is associated with reduced physical fitness in the Prevalence, Prediction and Prevention of Diabetes (PPP)-Botnia study. Diabetologia 2010;53:1709–1713 [DOI] [PubMed] [Google Scholar]
- 14.Kacerovsky-Bielesz G, Chmelik M, Ling C, et al. Short-term exercise training does not stimulate skeletal muscle ATP synthesis in relatives of humans with type 2 diabetes. Diabetes 2009;58:1333–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song JS, Johnson WE, Zhu X, et al. Model-based analysis of two-color arrays (MA2C). Genome Biol 2007;8:R178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.R Development Core Team R: A Language and Environment for Statistical Comp. Vienna, Austria, R Foundation for Statistical Computing, 2011 [Google Scholar]
- 17.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003;4:249–264 [DOI] [PubMed] [Google Scholar]
- 18.Klug M, Rehli M. Functional analysis of promoter CpG methylation using a CpG-free luciferase reporter vector. Epigenetics 2006;1:127–130 [DOI] [PubMed] [Google Scholar]
- 19.Wierup N, Kuhar M, Nilsson BO, Mulder H, Ekblad E, Sundler F. Cocaine- and amphetamine-regulated transcript (CART) is expressed in several islet cell types during rat development. J Histochem Cytochem 2004;52:169–177 [DOI] [PubMed] [Google Scholar]
- 20.Koopman R, Schaart G, Hesselink MK. Optimisation of oil red O staining permits combination with immunofluorescence and automated quantification of lipids. Histochem Cell Biol 2001;116:63–68 [DOI] [PubMed] [Google Scholar]
- 21.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 2007;447:425–432 [DOI] [PubMed] [Google Scholar]
- 22.Lyssenko V, Jonsson A, Almgren P, et al. Clinical risk factors, DNA variants, and the development of type 2 diabetes. N Engl J Med 2008;359:2220–2232 [DOI] [PubMed] [Google Scholar]
- 23.Ling C, Poulsen P, Simonsson S, et al. Genetic and epigenetic factors are associated with expression of respiratory chain component NDUFB6 in human skeletal muscle. J Clin Invest 2007;117:3427–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rönn T, Poulsen P, Hansson O, et al. Age influences DNA methylation and gene expression of COX7A1 in human skeletal muscle. Diabetologia 2008;51:1159–1168 [DOI] [PubMed] [Google Scholar]
- 25.Ling C, Del Guerra S, Lupi R, et al. Epigenetic regulation of PPARGC1A in human type 2 diabetic islets and effect on insulin secretion. Diabetologia 2008;51:615–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barrès R, Osler ME, Yan J, et al. Non-CpG methylation of the PGC-1alpha promoter through DNMT3B controls mitochondrial density. Cell Metab 2009;10:189–198 [DOI] [PubMed] [Google Scholar]
- 27.Yang BT, Dayeh TA, Kirkpatrick CL, et al. Insulin promoter DNA methylation correlates negatively with insulin gene expression and positively with HbA(1c) levels in human pancreatic islets. Diabetologia 2011;54:360–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bell CG, Finer S, Lindgren CM, et al. International Type 2 Diabetes 1q Consortium Integrated genetic and epigenetic analysis identifies haplotype-specific methylation in the FTO type 2 diabetes and obesity susceptibility locus. PLoS ONE 2010;5:e14040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sandovici IS, Dekker-Nitert NH, Ackers-Johnson M, et al. M. and Ozanne, SE.: Dynamic epigenetic regulation by early-diet and aging of the type 2 diabetes susceptibility gene Hnf4a in pancreatic islets. Proc Natl Acad Sci USA 2011;108:5449–5454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hardie DG. AMPK: a key regulator of energy balance in the single cell and the whole organism. Int J Obes (Lond) 2008;32(Suppl. 4):S7–S12 [DOI] [PubMed] [Google Scholar]
- 31.Wojtaszewski JF, Lynge J, Jakobsen AB, Goodyear LJ, Richter EA. Differential regulation of MAP kinase by contraction and insulin in skeletal muscle: metabolic implications. Am J Physiol 1999;277:E724–E732 [DOI] [PubMed] [Google Scholar]
- 32.Cubas P, Vincent C, Coen E. An epigenetic mutation responsible for natural variation in floral symmetry. Nature 1999;401:157–161 [DOI] [PubMed] [Google Scholar]
- 33.Chong S, Whitelaw E. Epigenetic germline inheritance. Curr Opin Genet Dev 2004;14:692–696 [DOI] [PubMed] [Google Scholar]
- 34.Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science 2005;308:1466–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ng SF, Lin RC, Laybutt DR, Barres R, Owens JA, Morris MJ. Chronic high-fat diet in fathers programs β-cell dysfunction in female rat offspring. Nature 2010;467:963–966 [DOI] [PubMed] [Google Scholar]
- 36.El-Osta A, Brasacchio D, Yao D, et al. Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia. J Exp Med 2008;205:2409–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park JH, Stoffers DA, Nicholls RD, Simmons RA. Development of type 2 diabetes following intrauterine growth retardation in rats is associated with progressive epigenetic silencing of Pdx1. J Clin Invest 2008;118:2316–2324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kriaucionis S, Bird A. DNA methylation and Rett syndrome. Hum Mol Genet 2003;12(Spec No 2):R221–R227 [DOI] [PubMed] [Google Scholar]
- 39.Cassidy SB, Schwartz S, Miller JL, Driscoll DJ. Prader-Willi syndrome. Genet Med 2012;14:10–26 [DOI] [PubMed] [Google Scholar]
- 40.Mackay DJ, Temple IK. Transient neonatal diabetes mellitus type 1. Am J Med Genet C Semin Med Genet 2010;154C:335–342 [DOI] [PubMed] [Google Scholar]
- 41.Morgan HD, Sutherland HG, Martin DI, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nat Genet 1999;23:314–318 [DOI] [PubMed] [Google Scholar]
- 42.Keller P, Vollaard NB, Gustafsson T, et al. A transcriptional map of the impact of endurance exercise training on skeletal muscle phenotype. J Appl Physiol 2011;110:46–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith JA, Collins M, Grobler LA, Magee CJ, Ojuka EO. Exercise and CaMK activation both increase the binding of MEF2A to the Glut4 promoter in skeletal muscle in vivo. Am J Physiol Endocrinol Metab 2007;292:E413–E420 [DOI] [PubMed] [Google Scholar]
- 44.Smith JA, Kohn TA, Chetty AK, Ojuka EO. CaMK activation during exercise is required for histone hyperacetylation and MEF2A binding at the MEF2 site on the Glut4 gene. Am J Physiol Endocrinol Metab 2008;295:E698–E704 [DOI] [PubMed] [Google Scholar]
- 45.Olsson AH, Rönn T, Ladenvall C, et al. Two common genetic variants near nuclear-encoded OXPHOS genes are associated with insulin secretion in vivo. Eur J Endocrinol 2011;164:765–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McGee SL, Sparling D, Olson AL, Hargreaves M. Exercise increases MEF2- and GEF DNA-binding activity in human skeletal muscle. FASEB J 2006;20:348–349 [DOI] [PubMed] [Google Scholar]
- 47.Yamauchi T, Kamon J, Ito Y, et al. Cloning of adiponectin receptors that mediate antidiabetic metabolic effects. Nature 2003;423:762–769 [DOI] [PubMed] [Google Scholar]
- 48.Taguchi T, Kishikawa H, Motoshima H, et al. Involvement of bradykinin in acute exercise-induced increase of glucose uptake and GLUT-4 translocation in skeletal muscle: studies in normal and diabetic humans and rats. Metabolism 2000;49:920–930 [DOI] [PubMed] [Google Scholar]
- 49.Haugen F, Norheim F, Lian H, et al. IL-7 is expressed and secreted by human skeletal muscle cells. Am J Physiol Cell Physiol 2010;298:C807–C816 [DOI] [PubMed] [Google Scholar]
- 50.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev 2011;25:1010–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]