Abstract
Insulin resistance, one of the major components of type 2 diabetes mellitus (T2DM), is a known risk factor for Alzheimer’s disease (AD), which is characterized by an abnormal accumulation of intra- and extracellular amyloid β peptide (Aβ). Insulin resistance is known to increase Aβ generation, but the underlying mechanism that links insulin resistance to increased Aβ generation is unknown. In this study, we examined the effect of high-fat diet–induced insulin resistance on amyloid precursor protein (APP) processing in mouse brains. We found that the induced insulin resistance promoted Aβ generation in the brain via altered insulin signal transduction, increased β- and γ-secretase activities, and accumulation of autophagosomes. These findings were confirmed in diabetic db/db mice brains. Furthermore, in vitro experiments in insulin-resistant SH-SY5Y cells and primary cortical neurons confirmed the alteration of APP processing by insulin resistance–induced autophagosome accumulation. Defects in insulin signal transduction affect autophagic flux by inhibiting the mammalian target of rapamycin pathway, resulting in altered APP processing in these cell culture systems. Thus, the insulin resistance that underlies the pathogenesis of T2DM might also trigger accumulation of autophagosomes, leading to increased Aβ generation, which might be involved in the pathogenesis of AD.
Alzheimer’s disease (AD) is sometimes referred to as type 3 diabetes because of the shared risk factors for the two disorders (1,2). Because insulin plays an important role in maintaining normal brain function and in peripheral glucose metabolism (3), insulin dysregulation has harmful effects on brain function as well as on peripheral glucose regulation. A number of epidemiological studies have suggested that insulin resistance, characterized by failed glucose utilization, confers an approximate two- to threefold relative risk for AD (4). Several factors could help to explain this link, including insulin degrading enzyme (IDE) activity, mitochondrial dysfunction, inflammation, and oxidative stress (5). Although type 2 diabetes mellitus (T2DM) may be linked to AD via these factors, our understanding of the underlying mechanisms is limited.
AD, the most common form of dementia, is characterized by senile plaques, neurofibrillary tangles, and neuronal loss (6). Senile plaques are extracellular deposits of amyloid-β peptide (Aβ); the deposits are associated with AD-related neurodegeneration (7). Aβ is a peptide of 40 or 42 amino acids derived predominantly from amyloid precursor protein (APP) upon sequential cleavage by β-secretase (β-site APP cleaving enzyme 1 [BACE1]) and the γ-secretase complex (8,9). The β- and γ-secretases reside predominantly in intracellular membrane compartments of the vacuolar apparatus, including autophagic vacuoles (AVs) (10). Several reports have shown that AVs exist in the brains of AD model mice and AD patients (10,11) and that they colocalize intimately with the γ-secretase complex, APP, and β-secretase–derived COOH-terminal fragment (β-CTF) (10,12). Several important signaling pathways, including the mammalian target of rapamycin (mTOR) pathway, AMP-activated protein kinase (AMPK) signaling, and the insulin/IGF-I signaling pathways, are reported to regulate AV formation (13,14). During activation of these signaling pathways, defects in the insulin signaling pathway (insulin resistance) induced abnormal autophagosome formation (15,16).
During macroautophagy (hereafter referred to as autophagy), the pivotal processes required for survival of long-lived cytoplasmic constituents are degraded; it is the principal means by which cellular organelles and protein aggregates are turned over (17). Autophagosome formation is induced by the inhibition of the mTOR signal pathway (17). Autophagosomes and their contents undergo clearance upon fusion with endosomes (amphisomes) or lysosomes (autolysosomes) that contain proteases (18) (autophagic maturation process).
Autophagosomes are one of the generation sites for Aβ, major toxic peptides in AD pathology (10). Because insulin resistance induces autophagosome accumulation and Aβ generation, and increased Aβ levels become the cause of AD, we wonder whether insulin resistance accelerates AD pathology via an autophagosome-induced increase in Aβ generation. To investigate accumulation of autophagosomes and altered APP processing under insulin-resistant conditions, we examined abnormalities in insulin signaling and in APP metabolism and expression levels in autophagy-related protein in mice fed a high-fat diet (HFD) and in diabetic db/db mice (19). To explore possible underlying links between insulin resistance, autophagosomes, and AD-like changes in vitro, human neuroblastoma SH-SY5Y cells and primary cortical neurons were subjected to prolonged exposure to high levels of insulin, leading to cellular insulin resistance (20). We present evidence that insulin resistance alters APP processing through autophagy activation and that insulin resistance–induced autophagosome accumulation is a potential link between AD and diabetes.
RESEARCH DESIGN AND METHODS
Animals and tissues.
C57BL/6 male mice were used for these experiments because this mouse strain is a good model of human obesity and T2DM (21). The mice were fed for 22 months with a Western diet (Harlan, TD.88137), comprising 21% (w/w) total lipid (42% calories from fat) and 0.2% (w/w) cholesterol, or a control diet (no. 5001, Test Diet), comprising 4% (w/w) total lipid (<12% calories from fat) and <0.04% (w/w) cholesterol. Body weight was recorded weekly. Food intake was measured for a 2- to 3-day period three times during the Western diet regimen. Mice were fasted overnight on the day before they were killed (24-month-old mice), and body fat contents were measured by dual-energy X-ray absorptiometry (PIXImus and Lunar Piximus 2 2.0 software, GE Lunar) and blood samples were collected.
Mice were maintained in the accredited pathogen-free facility at Seoul National University Bundang Hospital on a 12-h light/dark cycle and fed the indicated diet and water ad libitum. All procedures involving the use of laboratory animals were in accordance with the Guide for Standard Operation Procedures and approved by institutional animal care and use committee of the Clinical Research Institute, Seoul National University Bundang Hospital. Male C57BL/KsJ db/db mice (n = 10), a rodent model of T2DM (19), and their nondiabetic db/+ littermates (n = 9) were purchased from Jung Ang Laboratory Animal Inc. (Korea). The animals had free access to tap water and standard chow. Body weights and nonfasting blood glucose levels were determined at 14 weeks of age.
Serum analysis.
Serum cholesterol and triglyceride levels were determined by Beckman Coulter AU480 automatic biochemistry analysis system (Japan), with reagent kits provided by the manufacturer. Insulin concentration was measured using a mouse insulin radioimmunoassay kit (Millipore-Linco).
Cell culture and transfection.
Mouse primary cortical neurons were prepared as described previously (11). Experiments were conducted at days in vitro (DIV) 14. The human neuroblastoma cell line SH-SY5Y was cultured as described previously (11). The cells were transfected with several expression vectors using Lipofectamine Plus (Invitrogen) and small interfering (si)RNAs (Santa Cruz Biotechnology) using RNAimax (Invitrogen) according to the manufacturer’s instructions.
Antibodies.
Harvested cell pellets and mouse brain tissues were prepared as described (22). The antibodies for the Western blot analysis were anti-tumor necrosis factor-α converting enzyme (TACE), anti-mTOR, and anti-insulin receptor substrate 1 (IRS1) (Santa Cruz Biotechnology); anti-p62/sequestosome 1 (SQSTM1) and anti–β-actin (Sigma-Aldrich); anti-LC3B (microtubule-associated protein 1 light chain 3β), anti–phosphorylated (p)-AMPK (Thr172), anti-AMPK, anti–p-Akt (Thr308), anti-Akt, p-extracellular signal–related kinase (ERK)1/2, anti-ERK1/2, anti–BACE1, anti-matrix metalloproteinase (MMP)-9, anti–p-mTOR (S2448), anti–p-p70S6 kinase (p70S6K), and anti–p70S6K (Cell Signaling Technology); anti-phosphatidylinositol 3-kinases (PI3K) (p85α) and anti–p-IRS1 (Ser307) (Upstate Biotechnology); anti-APP (6E10) and anti-sAPPβ (Signet); anti-nicastrin and anti-PS1 loop (Millipore); and anti-beclin 1 and anti-IDE (Abcam). The immunoreactive bands were photographed and quantified on an LAS-3000 with MultiGauge (FujiFilm Inc.) and ImageJ software.
Live and dead cell assay.
For measuring cell viability, we performed calcein-acetoxymethyl ester assay and MTT assay as previously described (11).
Aβ quantification.
Sandwich enzyme-linked immunosorbent assay (ELISA) was performed as described for Aβ quantification (23).
β- or γ-Secretases activity measurement.
In vitro peptide cleavage assay was performed as described for the measurement of β- or γ-secretases activity (22).
Examination of LC3-II translocation.
To analyze green fluorescent protein (GFP)-LC3, red fluorescent protein (RFP)-LC3, and tandem-fluorescence LC3 (TfLC3), plasmid encoding GFP-LC3, RFP-LC3, and TfLC3 (a gift of Dr. M.S. Lee, Sungkyunkwan University) was transfected into SH-SY5Y cells. The appearance of RFP-LC3 and GFP-LC3 puncta was visualized on a confocal laser scanning microscope (FV10i-w; Olympus).
Immunocytochemistry.
Immunocytochemical staining was performed as described (24). Images were taken on a confocal laser scanning microscope (FV10i-w; Olympus).
Trichloroacetic acid precipitation.
Cell medium was centrifuged at 4,000 rpm for 5 min to remove cell debris and subjected to trichloroacetic acid precipitation.
Electron microscopy.
For electron microscopy (EM) analysis, brain tissue and SH-SY5Y cells were prepared as described (11).
Immunogold electron microscopy.
Immunogold electron microscopy (ImmunoEM) was performed as described previously (25).
Data analysis.
All data are expressed as mean ± SEM. Differences between groups were analyzed by Tukey-Kramer multiple comparison test and t test. P values < 0.05 were considered to be statistically significant.
RESULTS
Confirmation of insulin resistance in HFD mice.
HFD-induced insulin resistance was confirmed in 24-month-old C57BL/6 mice on the basis of elevated circulating serum insulin, obesity (increased body weight and ependymal fat pad weight), and elevated levels of blood glucose and cholesterol relative to those of age- and sex-matched control mice (mice fed regular chow; Supplementary Fig. 1).
Alteration of APP processing in brains of HFD mice.
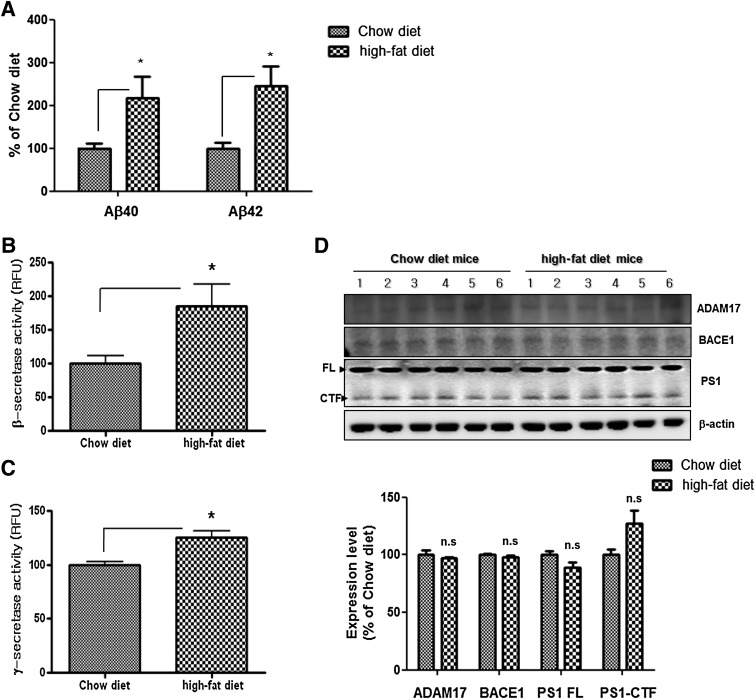
Abnormalities in insulin metabolism are among the central factors thought to mechanistically influence the onset of AD (5) via their influence on the synthesis and degradation of Aβ, held central to AD neuropathology. To identify the alteration in APP processing under insulin-resistant conditions, Aβ in the cortical areas of brains of HFD mice was quantified by ELISA. Levels of Aβ40 and Aβ42 were increased in HFD mice compared with levels in control mice (Fig. 1A), indicating that the insulin resistance directed APP processing toward the amyloidogenic pathway. To determine changes in the Aβ-generating pathway, we performed in vitro peptide cleavage assay to measure β- and γ-secretase activities. Their activities were increased in HFD mice compared with control mice (Fig. 1B and C). Western blot analysis revealed no difference in expression of a disintegrin and metallopeptidase domain 17 (ADAM17) (α-secretase), BACE1 (β-secretase), or presenilin 1 (γ-secretase component) (Fig. 1D), indicating that the brains of HFD mice are subject to increased β- and γ-secretase activities and Aβ expression without a change in protein expression.
FIG. 1.
Altered APP processing in the brains of HFD mice. A: The Aβ40 and Aβ42 levels were increased in the brains of HFD mice compared with levels in control mice (n = 6 each). The activities of β-secretase (B) and γ-secretase (C) were increased in the brains of HFD mice compared with levels in control mice. D: The expression levels of α-, β-, and γ-secretase in the brains of HFD mice compared with levels in control mice. *P < 0.05 vs. control mice.
Association between insulin resistance in HFD mice and decreased insulin signaling in the brain.
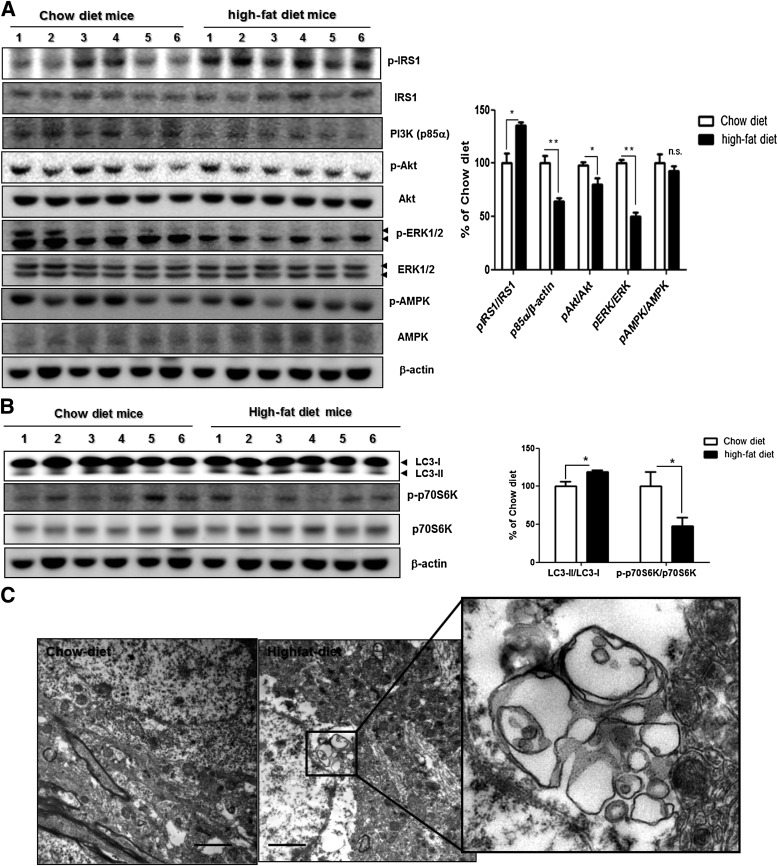
To explore the mechanisms by which insulin resistance promoted APP processing directed toward the amyloidogenic pathway, we looked for indices of altered insulin signaling in the brains of HFD mice relative to that of control mice. Previous studies have indicated that increased Ser/Thr phosphorylation of IRS proteins leads to an insulin-resistant state by various mechanisms (26,27). According to Western blot analysis, the phosphorylation level of IRS1 (Ser307) was increased in HFD mice compared with control mice (Fig. 2A). Also, the alteration in insulin signaling in the brain of HFD mice was examined. We found that the dietary condition leading to insulin resistance results in a significant reduction of PI3K p85 subunit expression (indicative of reduced PI3K signaling) (28) and phosphorylation of downstream Akt/protein kinase B in the cortex of HFD mice significantly (Fig. 2A). Considering that p-IRS1 and p-Akt were modified by diet, we evaluated other proteins involved in the insulin-signaling pathway (i.e., ERK1/2 and AMPK). The total level of ERK1/2 did not change, but the HFD significantly decreased p-ERK1/2 but not p-AMPK (Fig. 2A). These data show that the brains of HFD mice undergo altered insulin signaling.
FIG. 2.
Defects in insulin signaling pathway and autophagosome accumulation in the brains of HFD mice. A: Immunoblots for insulin signaling in the brain of control mice (lanes 1–6) and HFD mice (lanes 7–12). Western blot signals were quantified by densitometry and normalized to pan-form or β-actin. B: The levels of phosphorylated p70S6K and the LC3-II–to–LC3-I ratio in the cortical areas of mouse brains were determined by Western blot. The β-actin served as a loading control. Western blot signals were quantified by densitometry and normalized to pan-p70S6K or β-actin (n = 6 per group). Values are mean ± SE. *P < 0.05, **P < 0.01. C: EM analysis revealed autophagosome formation in cortical areas of mouse brains of 24-month-old mice fed chow diet (left) or HFD (right) for 22 months (from age 2 to 24 months). Scale bars represent 2 µm.
Autophagosome accumulation in the brains of HFD mice.
A greatly increased level of autophagosomes was reported in the pancreatic β-cells of diabetic db/db mouse and HFD mice (15). Another study demonstrated that autophagic vacuoles accumulate pathologically in the brains of AD patients, and autophagy acts as a major Aβ-generating factor (10,12). To explore the mechanisms of altered APP processing in the HFD condition, we examined whether HFD-induced insulin resistance increases autophagosome formation in mouse brains. Western blot analysis showed that the ratio of LC3-II to LC3-I, protein markers of autophagy (29), increased in cortical areas of the brains of HFD mice (Fig. 2B). EM of the cortex of HFD mice revealed the abnormal accumulation of large autophagosomes (Fig. 2C); the brains of control mice did not harbor autophagosomes. Previous studies have shown that autophagosome formation is induced by inhibition of mTOR (30). The brains of HFD mice showed decreased phosphorylation of p70S6K (Fig. 2B), a downstream component of the mTOR pathway (31), indicating that the HFD induced accumulation of autophagosomes via the mTOR pathway.
Defective insulin signaling, altered APP processing, and accumulation of autophagosomes in diabetic db/db mouse brains.
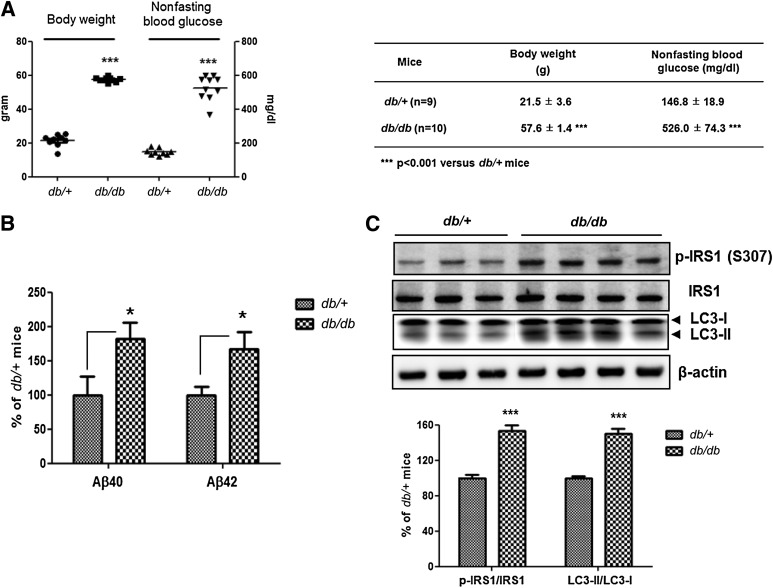
We further investigated whether insulin resistance induces autophagosome accumulation and altered APP processing in db/db mice, which showed typical characteristics of T2DM (19). At 14 weeks, body weights and blood glucose levels of db/db mice were increased significantly compared with those of their db/+ littermates (Fig. 3A). By ELISA, Aβ40 and Aβ42 levels were increased in db/db mice compared with levels in littermates (Fig. 3B). The phosphorylation level of IRS1 (Ser307) and the LC3-II–to–LC3-I ratio were increased in the brains of db/db mice (Fig. 3C), indicating defective insulin signaling and accumulation of autophagosomes in the db/db mice. Thus, as with HFD mice, db/db mice show insulin resistance, autophagosome accumulation, and increased Aβ levels in the brain.
FIG. 3.
Defective insulin signaling and accumulation of autophagosomes in diabetic db/db mouse brains. A: Body weight and nonfasting blood glucose levels of db/+ (n = 9) and db/db mice (n = 10). B: The Aβ40 and Aβ42 levels were increased in db/db mouse brains compared with db/+ mouse brains. Relative percentage values represent Aβ levels in db/db mouse brains per Aβ levels in db/+ mouse brains. C: Immunoblots for p-IRS1 (Ser307), IRS1, and LC3 from db/+ and db/db mouse brains. Values for the p-IRS1 (Ser307)-to-IRS1 ratio in db/db mouse brains represent altered insulin signaling, and values for the LC3-II–to–LC3-I ratio represent autophagic flux relative to db/+ mouse brains. Values are mean ± SE. *P < 0.05, ***P < 0.001.
Autophagosome accumulation in insulin-resistant SH-SY5Y cells.
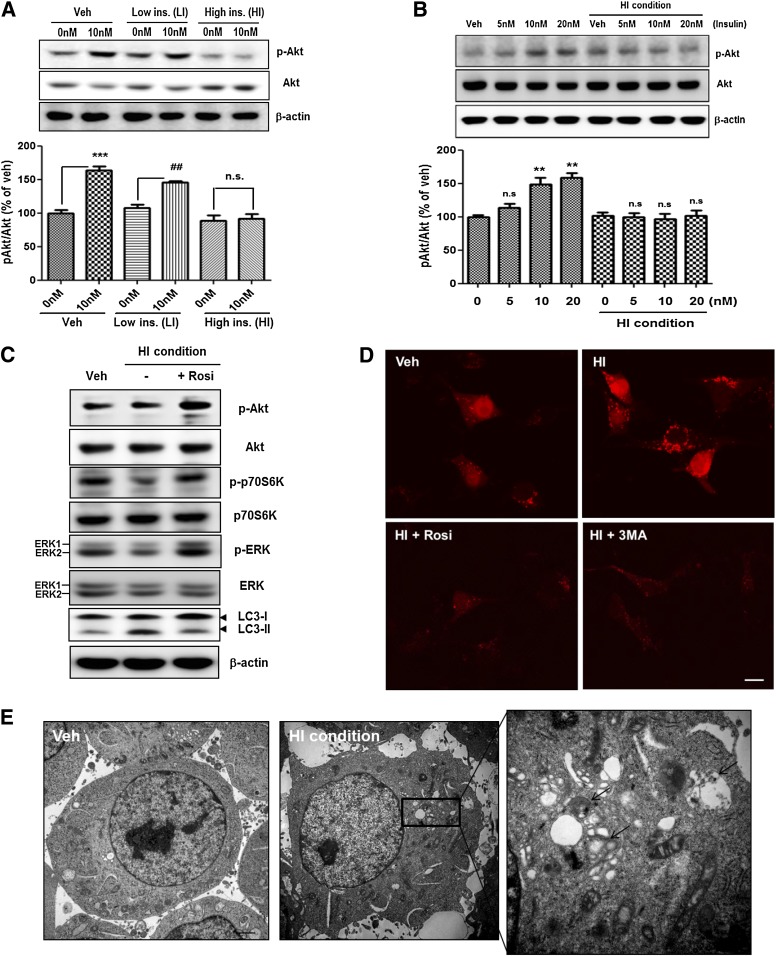
To determine the relationship between HFD-induced autophagosome accumulation and alteration in APP processing, we established a cell culture model of insulin resistance using SH-SY5Y cells. To make an in vitro model of insulin resistance based on previous study in skeletal muscle and neurons (20,32), we cultured SH-SY5Y cells in serum-free media in the absence or presence of chronic low insulin (LI; 10 nmol/L insulin) or chronic high insulin (HI; 1 μmol/L insulin) for 48 h, SH-SY5Y cells in the HI condition showed comparable attenuation of insulin-stimulated phosphorylation of Akt (Fig. 4A and B). Acute insulin treatment induced Akt phosphorylation in a concentration-dependent manner (5–20 nmol/L), whereas cells in the HI condition had no effect on Akt phosphorylation in response to acute insulin treatment (Fig. 4B).
FIG. 4.
Autophagosome accumulation in insulin-resistant SH-SY5Y cells. A and B: Establishment of a cell culture model of insulin resistance on SH-SY5Y cells. LI indicates chronic low insulin condition (10 nmol/L for 48 h), and HI indicates chronic high insulin condition (1 μmol/L for 48 h). Veh indicates absence of insulin. After pretreatment, the cells were rechallenged with 5, 10, and 20 nmol/L insulin to determine responsiveness to insulin. Values for the p-Akt–to–pan-Akt ratio represent alteration in insulin signaling. Data were obtained from at least five replicates for each group. Values are mean ± SE. **P < 0.01 and ***P < 0.001; ##P < 0.01. C: Immunoblots are shown for insulin signaling and autophagosome formation from cells treated with 1 μmol/L insulin and cotreated with 1 μmol/L insulin and 10 μmol/L rosiglitazone (Rosi) for 48 h. D: Insulin-induced autophagosome accumulation in fluorescently tagged LC3-transfected cells. Cells were treated with 1 μmol/L insulin alone or both 1 μmol/L insulin and 10 μmol/L rosiglitazone (Rosi) for 48 h or preincubated with 3MA (1 mmol/L) for 2 h, then exposed to 1 μmol/L insulin for 48 h. The mRFP-positive puncta indicate autophagosome formation. Scale bars represent 20 µm. E: EM analysis revealed autophagosome formation in cells treated with 1 μmol/L insulin. The arrows indicate autophagosomes. Scale bars represent 2 µm. (A high-quality digital representation of this figure is available in the online issue.)
We next tested the effect of the HI condition on other insulin-signaling and autophagy-related proteins. We observed a marked reduction in the phosphorylation level of p70S6K and ERK and an increased LC3-II–to–LC3-I ratio (Fig. 4C), indicating that the HI condition induced altered insulin signaling and increased autophagosome formation. To determine whether the alteration in insulin signaling increased the LC3-II–to–LC3-I ratio, rosiglitazone, an insulin-sensitizing agent (33), was administered to SH-SY5Y cells in the HI condition. Rosiglitazone reversed changes in the phosphorylation states of p70S6K, ERK, and the LC3-II–to–LC3-I ratio (Fig. 4C). To visualize autophagosome formation, SH-SY5Y cells were transiently transfected with RFP-LC3. Treatment of cells with 1 μmol/L insulin increased the number of RFP-positive puncta (Fig. 4D). Conversely, rosiglitazone reversed the changes in RFP-LC3 puncta (Fig. 4D). The agents had no effect on cell survival (Supplementary Fig. 2). EM revealed an abnormal accumulation of autophagosomes in SH-SY5Y cells under the HI condition (Fig. 4E), indicating that altered insulin signaling induces upregulation of autophagosome formation.
The role of Akt-mTOR pathway on insulin-induced accumulation of autophagosomes in SH-SY5Y cells.
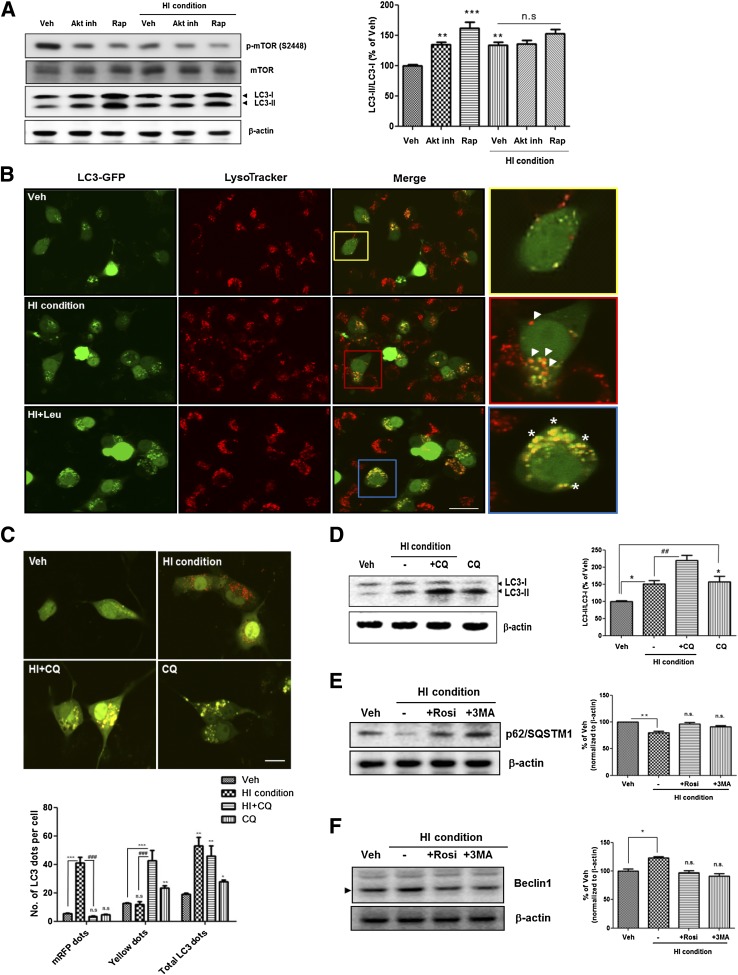
It is reported that the reduced insulin signaling can induce neuronal autophagy activation under nutrient-deprived conditions (16). To investigate how altered insulin signaling induces upregulation of autophagosome formation, Akt inhibitor and rapamycin, the mTOR inhibitor, were used to treat SH-SY5Y cells. Both inhibitors induced autophagosome accumulation under the normal condition but not in the HI condition (Fig. 5A). These data indicate that insulin-induced accumulation of autophagosome is mediated by the Akt-mTOR pathway in SH-SY5Y cells.
FIG. 5.
Insulin-induced accumulation of autophagosomes is associated with the Akt-mTOR pathway and is not caused by defective clearance of autophagosomes. A: SH-SY5Y cells were pretreated with 1 μmol/L Akt inhibitor IV (Akt inh) for 6 h before treatment with 1 μmol/L insulin for 48 h or treated with 1 μmol/L insulin alone or both 1 μmol/L insulin and 500 nmol/L rapamycin (Rap) for 48 h. B: Determination of autophagic flux using the GFP-LC3 construct and LysoTracker-red probe. Cells were pretreated with 25 μmol/L leupeptin (Leu) or X for 3 h before treatment with 1 μmol/L insulin for 48 h. Cells treated with 1 μmol/L insulin induced LC3 degradation in lysosome-like vacuoles. Arrowhead: only red signals (autolysosome). Asterisk: defects in autophagic maturation. Scale bars represent 20 µm. C: Determination of autophagic flux using the Tf-LC3 construct. SH-SY5Y cells were preincubated with 10 μmol/L CQ or X for 6 h, then exposed to 1 μmol/L insulin for 48 h. The mRFP-positive dots indicate only mRFP-LC3 signals (autolysosome), and the yellow signal indicates colocalization with GFP and mRFP signals (defects in autophagic maturation). Values represent the number of positive dots for red signals and yellow signals and are mean ± SE of 80 different cells in each experiment (n = 5). D: Immunoblot for LC3 from 1 μmol/L insulin-treated SH-SY5Y cells cultured in the presence or absence of 10 μmol/L CQ. Western blot signals were quantified by densitometry and normalized to β-actin. E: Immunoblot for p62/SQSTM1 from insulin-treated SH-SY5Y cells cultured in the presence or absence of 10 μmol/L rosiglitazone (Rosi) or 1 mmol/L 3MA for 48 h. Values represent net p62 flux (n = 5). F: Immunoblot for beclin1 from insulin-treated SH-SY5Y cells cultured in the presence or absence of 10 μmol/L rosiglitazone (Rosi) or 1 mmol/L 3MA for 48 h. Values represent autophagosome formation (n = 5). Values are mean ± SE. *P < 0.05, **P < 0.01, and ***P < 0.001; ##P < 0.01 and ###P < 0.001. (A high-quality digital representation of this figure is available in the online issue.)
Insulin-induced accumulation of autophagosomes was not caused by defective clearance of autophagosomes.
To determine whether the insulin-induced accumulation of autophagosomes was caused by activation of the induction for autophagosome formation or by defects in autolysosome maturation, we used LysoTracker probes to label lysosomes on GFP-LC3–transfected SH-SY5Y cells. Double-labeling with GFP-LC3 and LysoTracker Red revealed that treatment with 1 μmol/L insulin induced LC3 degradation in LysoTracker-positive vesicles (Fig. 5B; arrowhead). To assess the defects in autolysosome maturation, we incubated cells with leupeptin, a lysosomal inhibitor, to halt the digestion of autolysosomal contents before the insulin treatment. We found insulin-induced accumulation of LC3 in LysoTracker-labeled vesicles after pre-exposure to leupeptin (Fig. 5B; asterisk). To examine autophagosome clearance in greater detail, we used mRFP-GFP TfLC3 (18). The application of the TfLC3 construct in SH-SY5Y cells with 1 μmol/L insulin showed a significant accumulation of LC3 dots that were only RFP-positive signals but not GFP signals (Fig. 5C), suggesting that the cells were localized in the autolysosomes because the GFP signals were preferentially quenched in the acidic condition (18). When chloroquine (CQ), a lysosomal inhibiting agent, was used to treat SH-SY5Y cells with 1 μmol/L insulin, GFP and RFP signals were colocalized in most cells (Fig. 5C). Western blot analysis also showed that SH-SY5Y cells with 1 μmol/L insulin in the presence of CQ resulted in a significantly higher level of LC3-II–to–LC3-I ratio than with 1 μmol/L insulin alone (Fig. 5D). In an alternative approach, p62/SQSTM1 degradation was examined to evaluate impairment in autophagic protein degradation (34). Treatment with 1 μmol/L insulin induced p62 degradation (Fig. 5E), suggesting that the HI condition did not impair autolysosome maturation. We also checked levels of beclin1, a component of the autophagosome initiation complex. Under the HI condition, beclin1 expression was increased (Fig. 5F). Each of these findings supports the notion that the HI condition can induce autophagosome formation without defects in autolysosome maturation.
Alteration in APP processing under HI condition–induced insulin resistance: prevention of change in APP processing through autophagy inhibition.
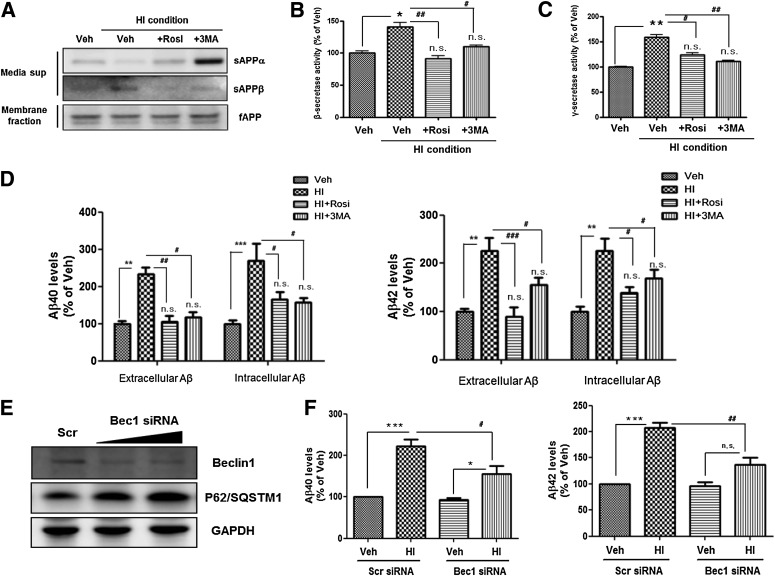
To determine whether HI condition–induced insulin resistance alters APP processing through autophagosome accumulation, we analyzed alteration of secretase activity by measuring levels of secreted forms of APP (sAPPα and sAPPβ) in conditioned medium. With 1 μmol/L insulin, the membrane-associated APP (full-length APP) level did not change, whereas the sAPPα level decreased but the sAPPβ level increased (Fig. 6A), indicating that the HI condition induced β-secretase activation. Cotreatment of 1 μmol/L insulin with rosiglitazone or 3-methyladenine (3MA) prevented the insulin-induced β-secretase activation (Fig. 6A). To assess the activation of β- and γ-secretase under the HI condition, we measured β- and γ-secretase activities by means of in vitro peptide cleavage assay under the HI condition (Fig. 6B and C). Both β- and γ-secretase activities were increased markedly, and cotreatment with 1 µmol/L of insulin and rosiglitazone or 3MA attenuated the β- and γ-secretase activation (Fig. 6B and C). Consistent with activation of β- and γ-secretases under the HI condition, the condition increased Aβ generation intra- and extracellularly (Fig. 6D). Conversely, rosiglitazone and 3MA prevented these changes in Aβ generation (Fig. 6D).
FIG. 6.
Alteration in APP processing under HI condition–induced insulin resistance. A: Immunoblots for sAPPα and sAPPβ in conditioned medium from insulin-treated SH-SY5Y cells cultured in the presence or absence of 10 μmol/L rosiglitazone (Rosi) or 1 mmol/L 3MA for 48 h. The full-length APP (fAPP) level was measured on membrane-fraction samples. The activities of β-secretase (B) and γ-secretase (C) were measured by in vitro cleavage assay. D: The levels of Aβ40 and Aβ42 in insulin-treated SH-SY5Y cells cultured in the presence or absence of 10 μmol/L rosiglitazone or 1 mmol/L 3MA for 48 h were determined by ELISA. The “extracellular” Aβ level was measured on conditioned medium, and “intracellular” Aβ level was measured on cell lysates. E: Immunoblots for beclin1 and p62/SQSTM1 on beclin1 siRNA-transfected SH-SY5Y cells. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. F: The levels of Aβ40 and Aβ42 on beclin1 siRNA-transfected SH-SY5Y cells were determined by ELISA. *P < 0.05, **P < 0.01, and ***P < 0.001 vs. vehicle-treated cells; #P < 0.05, ##P < 0.01, and ###P < 0.001 vs. cells treated with 1 μmol/L insulin.
To confirm that insulin-induced autophagosome accumulation induces increased Aβ generation, we transfected SH-SY5Y cells with beclin1-specific siRNA for 48 h. Beclin1 siRNA reduced total beclin1 levels by up to ∼60% without altering β-actin expression, and p62/SQSTM1 accumulated in the beclin1 knockdown condition (Fig. 6E). Consistent with the effects of 3MA, beclin1 siRNA blocked the significant insulin-induced increased Aβ generation (Fig. 6F). These data indicate that HI condition–induced insulin resistance promotes amyloidogenic pathway through accumulation of autophagosome.
The enrichment of secretase proteins in insulin resistance–induced autophagosomes.
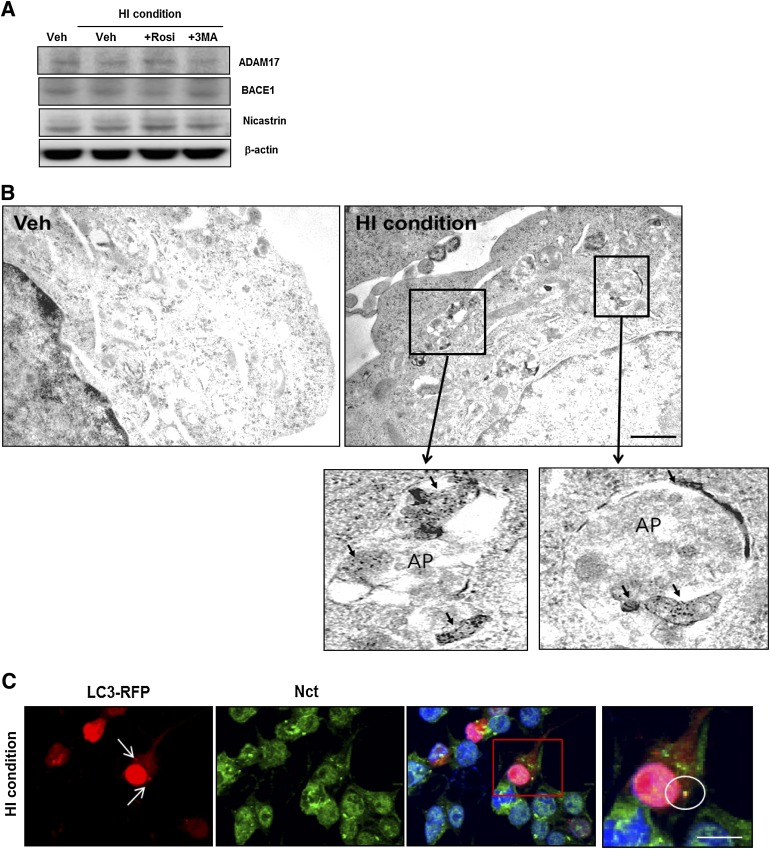
To explore the mechanisms through which insulin resistance–induced autophagosome accumulation alters APP processing, we analyzed alteration in secretase protein expression by treatment with 1 μmol/L insulin. Western blot analysis revealed no difference in the expression levels of ADAM17 (α-secretase), BACE1 (β-secretase), and nicastrin (γ-secretase component) between vehicle-treated and insulin-treated cells (Fig. 7A). Previous studies have indicated that autophagosome induction with rapamycin or Leu/His deprivation increases Aβ generation due to enrichment of APP processing-related proteins in autophagosomes (10). On the basis of this consideration, we explored whether secretase proteins enriched predominantly in insulin resistance–induced autophagosomes. Using ImmunoEM labeling of insulin-treated SH-SY5Y cells, we observed that BACE1 was preferentially localized to autophagosomes (Fig. 7B). Also, by immunostaining LC3-RFP–labeled cells with nicastrin antibody, the HI condition was shown to increase colocalization on autophagosomes and nicastrin (Fig. 7C). These data demonstrate that insulin-induced autophagosome induction activates β- and γ-secretases through enrichment of secretase proteins in autophagosomes, thus increasing Aβ generation.
FIG. 7.
The enrichment of BACE1 and nicastrin in insulin-induced autophagosomes. A: The expression levels of ADAM17, BACE1, and nicastrin in insulin-treated SH-SY5Y cells cultured in the presence or absence of 10 μmol/L rosiglitazone (Rosi) or 1 mmol/L 3MA for 48 h. B: Immunogold localization of BACE1 in vehicle-treated and HI-treated cells. The arrow indicates BACE1 signals in autophagosomes (AP). C: Enrichment of nicastrin protein in RFP-LC3–positive puncta. Immunostaining with nicastrin (Nct) antibody (pseudo green) on RFP-LC3–labeled cells showed that nicastrin and RFP-LC3–positive AVs were partially colocalized under the HI condition in SH-SY5Y. Circle: colocalization with nicastrin and RFP-LC3–positive puncta. Data for immunostaining are presented as a representative from more than three independent experiments. Scale bars represent 15 µm. (A high-quality digital representation of this figure is available in the online issue.)
Altered APP processing by insulin-induced autophagosome accumulation in primary cortical neurons.
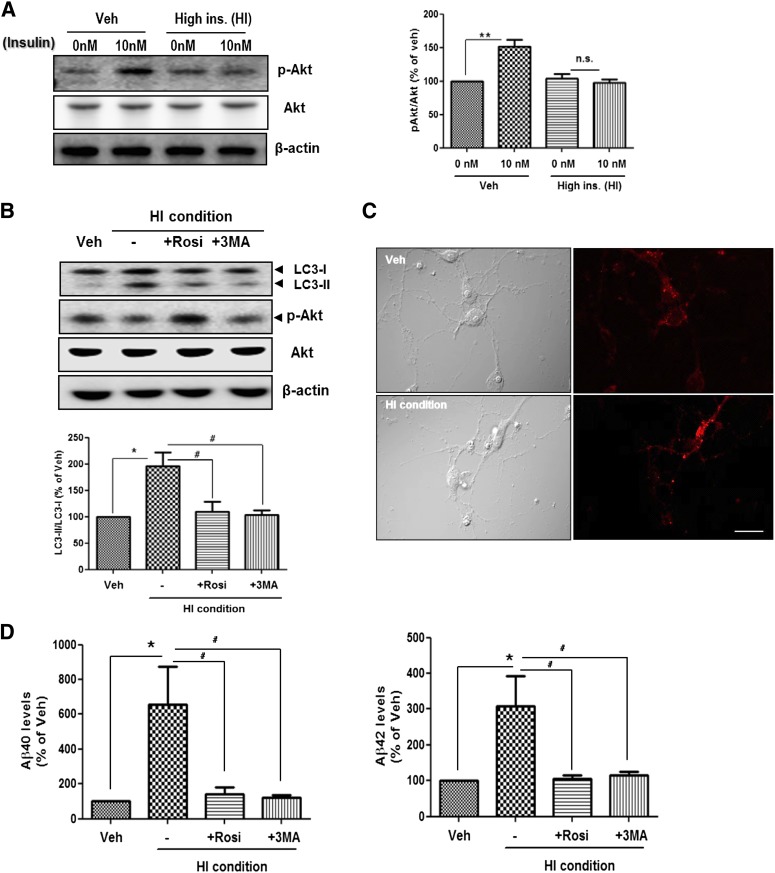
To confirm that the insulin-induced insulin resistance alters APP processing through autophagosome accumulation in primary cortical neurons, we established insulin resistance in primary cortical neurons (Fig. 8A). As in the SH-SY5Y cells, we cultured primary cortical neurons (DIV 14) in serum-free media in the absence or presence of insulin (1 μmol/L) for 24 h (HI condition) and found that the HI condition induced insulin resistance in primary neurons (Fig. 8A). We then observed a markedly increased autophagosome formation on primary neurons with 1 μmol/L insulin (Fig. 8B and C), indicating that the HI condition induced altered insulin signaling and increased autophagosome formation in primary neurons. Furthermore, the HI condition increased Aβ generation in primary neurons (Fig. 8D). Rosiglitazone and 3MA prevented these changes in autophagosome formation and Aβ generation (Fig. 8B and D). These data indicate that altered insulin signaling induces upregulation of autophagosome formation and that accumulated autophagosomes increase Aβ generation in primary cortical neurons.
FIG. 8.
Altered APP processing by insulin-induced autophagosome accumulation in primary cortical neurons. A: Establishment of a cell culture model of insulin resistance on primary cortical neurons (DIV 14). Veh indicates absence of insulin, and HI indicates high insulin condition (1 μmol/L for 24 h). After pretreatment, the cells were rechallenged with 10 nmol/L insulin to determine responsiveness to insulin. B: Immunoblots for LC3 from insulin-treated primary cortical neurons cultured in the presence or absence of 5 μmol/L rosiglitazone or 2 mmol/L 3MA for 24 h. C: Immunostaining with LC3 antibody (pseudo red) on vehicle-treated or 1 μmol/L insulin-treated primary cortical neurons. D: The levels of Aβ40 and Aβ42 in insulin-treated primary cortical neurons cultured in the presence or absence of 5 μmol/L rosiglitazone (Rosi) or 2 mmol/L 3MA for 24 h were determined by ELISA. *P < 0.05 and **P < 0.01; #P < 0.05. (A high-quality digital representation of this figure is available in the online issue.)
DISCUSSION
Although several epidemiological studies have identified a relationship between AD and T2DM in the central nervous system (1,2,4), the molecular mechanisms underlying this comorbidity are not fully understood (35). In this study, we found altered insulin signaling, autophagosome accumulation, and an enhanced amyloidogenic pathway in HFD mice and in db/db mice, two widely used animal models of diabetes. The db/db mice carry a homozygous mutation in the leptin receptor and suffer severe glucose intolerance, obesity, hyperglycemia, and hyperinsulinemia (19). In contrast, HFD mice have milder metabolic symptoms, which is reversible with a low-fat diet (36). To investigate the relationship among diet-induced or genetically induced insulin resistance, autophagosome accumulation, and alteration in APP processing, we established cell culture models of insulin resistance using SH-SY5Y cells and primary cortical neurons. In insulin-resistant SH-SY5Y cells and primary cortical neurons, insulin resistance induced autophagosome accumulation via inhibition of the Akt-mTOR pathway, and this accumulation provided for greater Aβ peptide generation, resulting in accelerated Aβ generation by modulation of β- and γ-secretase activities. Finally, we found that secretase proteins enriched predominantly in insulin resistance–induced autophagosomes, thus increasing Aβ generation.
These findings point to autophagosome accumulation as at least one link between AD and T2DM. We also found that the level of IDE, an Aβ-degrading protease (37), was decreased in the brains of HFD mice, resulting in accumulation of Aβ in the brains, whereas MMP-9, another Aβ-degrading enzyme (38), was not changed (Supplementary Fig. 3). Increased Aβ levels in the brains of HFD mice may be affected by enhanced APP processing due to autophagosome accumulation and by partially decreased IDE levels. We chose SH-SY5Y cells for an in vitro study based on previous reports that showed effects of insulin on AD pathology and alteration in brain metabolism (AD-like phenotype) under diabetic conditions (39,40). When we repeated several experiments on autophagosome formation and altered APP processing with primary cortical neurons, similar results were obtained (Figs. 4 and 6 vs. 8).
An HFD-induced increase in cholesterol is reported to enhance the levels of membrane lipid rafts, which are implicated in Aβ production (41). With insulin-resistant SH-SY5Y cells, cotreatment of simvastatin or methyl-β-cyclodextrin (MβCD), cholesterol-lowering drugs, with 1 μmol/L insulin had no effect on Aβ generation (Supplementary Fig. 4). These results indicate autophagosome accumulation, not cholesterol or lipid raft, is a major factor in Aβ generation, at least in our cell culture system.
There are many reports about the role of autophagosomes in the pathology of AD and T2DM. Yu et al. (10) reported that AVs accumulated in the brains of AD model mice and that AVs were highly enriched in the components and activities of γ-secrease complexes, APP and βCTF, and were one of major sources of intracellular Aβ in the AD brain. A recent study reported that the brains of mice with streptozotocin-induced diabetes showed upregulated autophagosome and increased Aβ levels (42). Although a relation between increased Aβ levels in the brains of diabetic mice and autophagosome accumulation has not been established, there is indirect evidence that increased Aβ levels in diabetic mice are associated with autophagosome accumulation. Previous studies showed that insulin resistance–induced Aβ accumulation is caused by alteration in Aβ generation via certain insulin-signaling pathways, such as via glycogen synthase kinase-3 (GSK-3) (43), and inhibition of GSK-3 signaling decreased Aβ generation and reduced autophagosome formation (44).
There is also evidence that insulin resistance mediates pathological conditions such as oxidative stress and inflammation (5,45). These pathological conditions have been shown to induce both abnormal autophagosome accumulation and Aβ over-production (46–48). Moreover, rosiglitazone reduces the Aβ level and thus rescues memory impairment in AD model mice (49). We found that rosiglitazone reduced autophagosome accumulation by sensitizing the insulin-signaling pathway (Fig. 4C and D); thus, it is possible that decreased Aβ level by rosiglitazone is associated with reduced autophagosome accumulation. However, these reports do not provide sufficient direct evidence that increased Aβ levels in the insulin-resistant conditions are associated with autophagy activation through altered insulin-signaling pathways. We found a direct relation between increased Aβ levels and autophagosome accumulation in diabetic mouse brains and insulin-resistant cells. In particular, we established in vitro models of insulin resistance and found that insulin-resistant neurons show autophagosome accumulation and increased Aβ generation. These effects were blocked by treatment with autophagy inhibitor and by beclin1 knockdown, indicating that exacerbation of Aβ generation under insulin-resistant conditions might be associated with autophagosome accumulation.
In conclusion, this study provides in vitro and in vivo evidence that hyperinsulinemia and insulin resistance induced by genetic or dietary factors enhanced Aβ generation through autophagosome accumulation. Our findings suggest that insulin resistance–induced autophagosome accumulation might be a potential link between AD and diabetes.
ACKNOWLEDGMENTS
This work was supported by grants from the National Research Foundation (2012R1A2A1A01002881, 2008-05943), the Medical Research Council (2011-0030738), the World Class University (R32-10084), and the Korea National Institute of Health ROAD R&D Program Project (A092058) to I.M.-J., and by the Hi Seoul Science/Humanities Fellowship from Seoul Scholarship Foundation to S.M.S.
No potential conflicts of interest relevant to this article were reported.
S.M.S. wrote the manuscript and researched data. H.S. researched data. J.B. researched data and contributed to discussion. K.S.P, H.C.J., and Y.J.P. researched animal data. I.M.-J. supervised the study and reviewed and edited the manuscript. I.M.-J. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db11-1735/-/DC1.
REFERENCES
- 1.Ott A, Stolk RP, van Harskamp F, Pols HA, Hofman A, Breteler MM. Diabetes mellitus and the risk of dementia: The Rotterdam Study. Neurology 1999;53:1937–1942 [DOI] [PubMed] [Google Scholar]
- 2.Steen E, Terry BM, Rivera EJ, et al. Impaired insulin and insulin-like growth factor expression and signaling mechanisms in Alzheimer’s disease—is this type 3 diabetes? J Alzheimers Dis 2005;7:63–80 [DOI] [PubMed] [Google Scholar]
- 3.Benedict C, Hallschmid M, Hatke A, et al. Intranasal insulin improves memory in humans. Psychoneuroendocrinology 2004;29:1326–1334 [DOI] [PubMed] [Google Scholar]
- 4.Schrijvers EM, Witteman JC, Sijbrands EJ, Hofman A, Koudstaal PJ, Breteler MM. Insulin metabolism and the risk of Alzheimer disease: the Rotterdam Study. Neurology 2010;75:1982–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de la Monte SM. Insulin resistance and Alzheimer’s disease. BMB Rep 2009;42:475–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walsh DM, Selkoe DJ. Deciphering the molecular basis of memory failure in Alzheimer’s disease. Neuron 2004;44:181–193 [DOI] [PubMed] [Google Scholar]
- 7.Selkoe DJ. Cell biology of protein misfolding: the examples of Alzheimer’s and Parkinson’s diseases. Nat Cell Biol 2004;6:1054–1061 [DOI] [PubMed] [Google Scholar]
- 8.Edbauer D, Winkler E, Regula JT, Pesold B, Steiner H, Haass C. Reconstitution of gamma-secretase activity. Nat Cell Biol 2003;5:486–488 [DOI] [PubMed] [Google Scholar]
- 9.Vassar R, Bennett BD, Babu-Khan S, et al. Beta-secretase cleavage of Alzheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science 1999;286:735–741 [DOI] [PubMed] [Google Scholar]
- 10.Yu WH, Cuervo AM, Kumar A, et al. Macroautophagy—a novel Beta-amyloid peptide-generating pathway activated in Alzheimer’s disease. J Cell Biol 2005;171:87–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Son SM, Jung ES, Shin HJ, Byun J, Mook-Jung I. Aβ-induced formation of autophagosomes is mediated by RAGE-CaMKKβ-AMPK signaling. Neurobiol Aging 2012;33:1006.e11–e23 [DOI] [PubMed] [Google Scholar]
- 12.Nixon RA, Wegiel J, Kumar A, et al. Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J Neuropathol Exp Neurol 2005;64:113–122 [DOI] [PubMed] [Google Scholar]
- 13.Kanazawa T, Taneike I, Akaishi R, et al. Amino acids and insulin control autophagic proteolysis through different signaling pathways in relation to mTOR in isolated rat hepatocytes. J Biol Chem 2004;279:8452–8459 [DOI] [PubMed] [Google Scholar]
- 14.Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem 2000;275:992–998 [DOI] [PubMed] [Google Scholar]
- 15.Ebato C, Uchida T, Arakawa M, et al. Autophagy is important in islet homeostasis and compensatory increase of beta cell mass in response to high-fat diet. Cell Metab 2008;8:325–332 [DOI] [PubMed] [Google Scholar]
- 16.Young JE, Martinez RA, La Spada AR. Nutrient deprivation induces neuronal autophagy and implicates reduced insulin signaling in neuroprotective autophagy activation. J Biol Chem 2009;284:2363–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol 2007;8:931–937 [DOI] [PubMed] [Google Scholar]
- 18.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 2007;3:452–460 [DOI] [PubMed] [Google Scholar]
- 19.Hummel KP, Dickie MM, Coleman DL. Diabetes, a new mutation in the mouse. Science 1966;153:1127–1128 [DOI] [PubMed] [Google Scholar]
- 20.Kim B, McLean LL, Philip SS, Feldman EL. Hyperinsulinemia induces insulin resistance in dorsal root ganglion neurons. Endocrinology 2011;152:3638–3647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Surwit RS, Kuhn CM, Cochrane C, McCubbin JA, Feinglos MN. Diet-induced type II diabetes in C57BL/6J mice. Diabetes 1988;37:1163–1167 [DOI] [PubMed] [Google Scholar]
- 22.Hong HS, Hwang JY, Son SM, Kim YH, Moon M, Inhee MJ. FK506 reduces amyloid plaque burden and induces MMP-9 in AβPP/PS1 double transgenic mice. J Alzheimers Dis 2010;22:97–105 [DOI] [PubMed] [Google Scholar]
- 23.Cho HJ, Son SM, Jin SM, et al. RAGE regulates BACE1 and Abeta generation via NFAT1 activation in Alzheimer’s disease animal model. FASEB J 2009;23:2639–2649 [DOI] [PubMed] [Google Scholar]
- 24.Cho HJ, Jin SM, Youn HD, Huh K, Mook-Jung I. Disrupted intracellular calcium regulates BACE1 gene expression via nuclear factor of activated T cells 1 (NFAT 1) signaling. Aging Cell 2008;7:137–147 [DOI] [PubMed] [Google Scholar]
- 25.Takuma K, Fang F, Zhang W, et al. RAGE-mediated signaling contributes to intraneuronal transport of amyloid-beta and neuronal dysfunction. Proc Natl Acad Sci USA 2009;106:20021–20026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paz K, Hemi R, LeRoith D, et al. A molecular basis for insulin resistance. Elevated serine/threonine phosphorylation of IRS-1 and IRS-2 inhibits their binding to the juxtamembrane region of the insulin receptor and impairs their ability to undergo insulin-induced tyrosine phosphorylation. J Biol Chem 1997;272:29911–29918 [DOI] [PubMed] [Google Scholar]
- 27.Mothe I, Van Obberghen E. Phosphorylation of insulin receptor substrate-1 on multiple serine residues, 612, 632, 662, and 731, modulates insulin action. J Biol Chem 1996;271:11222–11227 [DOI] [PubMed] [Google Scholar]
- 28.Ho L, Qin W, Pompl PN, et al. Diet-induced insulin resistance promotes amyloidosis in a transgenic mouse model of Alzheimer’s disease. FASEB J 2004;18:902–904 [DOI] [PubMed] [Google Scholar]
- 29.Kabeya Y, Mizushima N, Ueno T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 2000;19:5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmelzle T, Hall MN. TOR, a central controller of cell growth. Cell 2000;103:253–262 [DOI] [PubMed] [Google Scholar]
- 31.Krause U, Bertrand L, Hue L. Control of p70 ribosomal protein S6 kinase and acetyl-CoA carboxylase by AMP-activated protein kinase and protein phosphatases in isolated hepatocytes. Eur J Biochem 2002;269:3751–3759 [DOI] [PubMed] [Google Scholar]
- 32.Gupta A, Bisht B, Dey CS. Peripheral insulin-sensitizer drug metformin ameliorates neuronal insulin resistance and Alzheimer’s-like changes. Neuropharmacology 2011;60:910–920 [DOI] [PubMed] [Google Scholar]
- 33.Mayerson AB, Hundal RS, Dufour S, et al. The effects of rosiglitazone on insulin sensitivity, lipolysis, and hepatic and skeletal muscle triglyceride content in patients with type 2 diabetes. Diabetes 2002;51:797–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bjørkøy G, Lamark T, Brech A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 2005;171:603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Profenno LA, Porsteinsson AP, Faraone SV. Meta-analysis of Alzheimer’s disease risk with obesity, diabetes, and related disorders. Biol Psychiatry 2010;67:505–512 [DOI] [PubMed] [Google Scholar]
- 36.Parekh PI, Petro AE, Tiller JM, Feinglos MN, Surwit RS. Reversal of diet-induced obesity and diabetes in C57BL/6J mice. Metabolism 1998;47:1089–1096 [DOI] [PubMed] [Google Scholar]
- 37.Vekrellis K, Ye Z, Qiu WQ, et al. Neurons regulate extracellular levels of amyloid beta-protein via proteolysis by insulin-degrading enzyme. J Neurosci 2000;20:1657–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan P, Hu X, Song H, et al. Matrix metalloproteinase-9 degrades amyloid-beta fibrils in vitro and compact plaques in situ. J Biol Chem 2006;281:24566–24574 [DOI] [PubMed] [Google Scholar]
- 39.Magrané J, Rosen KM, Smith RC, Walsh K, Gouras GK, Querfurth HW. Intraneuronal beta-amyloid expression downregulates the Akt survival pathway and blunts the stress response. J Neurosci 2005;25:10960–10969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Solano DC, Sironi M, Bonfini C, Solerte SB, Govoni S, Racchi M. Insulin regulates soluble amyloid precursor protein release via phosphatidyl inositol 3 kinase-dependent pathway. FASEB J 2000;14:1015–1022 [DOI] [PubMed] [Google Scholar]
- 41.Grimm MO, Kuchenbecker J, Grösgen S, et al. Docosahexaenoic acid reduces amyloid beta production via multiple pleiotropic mechanisms. J Biol Chem 2011;286:14028–14039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang T, Yan W, Li Q, et al. 3-n-Butylphthalide (NBP) attenuated neuronal autophagy and amyloid-β expression in diabetic mice subjected to brain ischemia. Neurol Res 2011;33:396–404 [DOI] [PubMed] [Google Scholar]
- 43.Phiel CJ, Wilson CA, Lee VM, Klein PS. GSK-3alpha regulates production of Alzheimer’s disease amyloid-beta peptides. Nature 2003;423:435–439 [DOI] [PubMed] [Google Scholar]
- 44.Li Q, Li H, Roughton K, et al. Lithium reduces apoptosis and autophagy after neonatal hypoxia-ischemia. Cell Death Dis 2010;1:e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krogh-Madsen R, Plomgaard P, Keller P, Keller C, Pedersen BK. Insulin stimulates interleukin-6 and tumor necrosis factor-alpha gene expression in human subcutaneous adipose tissue. Am J Physiol Endocrinol Metab 2004;286:E234–E238 [DOI] [PubMed] [Google Scholar]
- 46.Papassotiropoulos A, Hock C, Nitsch RM. Genetics of interleukin 6: implications for Alzheimer’s disease. Neurobiol Aging 2001;22:863–871 [DOI] [PubMed] [Google Scholar]
- 47.Shen C, Chen Y, Liu H, et al. Hydrogen peroxide promotes Abeta production through JNK-dependent activation of gamma-secretase. J Biol Chem 2008;283:17721–17730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinez-Outschoorn UE, Whitaker-Menezes D, Lin Z, et al. Cytokine production and inflammation drive autophagy in the tumor microenvironment: role of stromal caveolin-1 as a key regulator. Cell Cycle 2011;10:1784–1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Escribano L, Simón AM, Gimeno E, et al. Rosiglitazone rescues memory impairment in Alzheimer’s transgenic mice: mechanisms involving a reduced amyloid and tau pathology. Neuropsychopharmacology 2010;35:1593–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]