G-CSF stimulates the expression of the chemokine MIP-2 in mature neutrophils by a direct transcriptional mechanism dependent upon STAT3.
Keywords: chemotaxis, transcription, chromatin
Abstract
Neutrophil mobilization from the bone marrow is a critical aspect of the innate immune response, enabling a rapid deployment of phagocytes to infected or inflamed tissue. The cytokine G-CSF, which is induced rapidly during infection, elicits a swift and potent mobilizing response, yet its mechanisms of action remain poorly understood. Here, we studied the role of G-CSF and its principal signal transducer STAT3 in regulating expression of the neutrophil chemoattractant MIP-2. Our studies revealed Gr-1hi mature neutrophils as major sources of Cxcl2 (MIP-2) mRNA in bone marrow and G-CSF-responsive MIP-2 protein production. Induction of Cxcl2 was regulated directly by G-CSF-activated STAT3 via interaction at a STAT consensus element in the Cxcl2 promoter. G-CSF coordinately stimulated the association of STAT3, induction of the transcriptionally active H3K4me3 modification, and recruitment of RNA Pol II at the Cxcl2 proximal promoter, as well as the promoter region of Il8rb, encoding the MIP-2 receptor. These results suggest that the G-CSF–STAT3 pathway directly regulates transcriptional events that induce neutrophil mobilization.
Introduction
Neutrophils are an essential cell type in the innate immune system, as a result of their ability to migrate rapidly to inflamed tissues and mount antibacterial or antifungal responses through phagocytosis, degranulation, and production of ROS. Neutrophil development occurs in the bone marrow under the direction of G-CSF, a cytokine that promotes the survival, proliferation, and differentiation of granulocytic progenitor cells into mature neutrophils [1–4]. G-CSF circulating amounts become elevated during bacterial or fungal infections, stimulating an increase in neutrophil generation and a surge of neutrophils from the bone marrow to the blood in a process called mobilization [3, 5, 6]. rG-CSF is used to treat life-threatening congenital or acquired neutropenia (e.g., after chemotherapy) [7, 8]. However, G-CSF is also linked with driving destructive inflammation in arthritis, and clinical G-CSF use has been suggested to contribute to neoplastic transformation in a subset of patients with congenital neutropenia [9, 10]. Thus, it is beneficial to understand the molecular mechanisms of G-CSF action, as this knowledge may provide new approaches to regulate neutrophil production and function in humans.
G-CSF binds a high-affinity receptor, the G-CSFR, on myeloid progenitor cells and granulocytic precursors, resulting in activation of multiple signaling intermediates including Erk1/2, the Src-related kinase Lyn, and Jak–STAT proteins [11–17]. STAT3 is a predominant signaling molecule stimulated by the activated G-CSFR; hence, its function in granulopoiesis has been investigated in murine models. These include conditional Stat3 deletion in hematopoietic cells or replacement of Csf3r (G-CSFR) with a mutated gene encoding a receptor isoform unable to activate STAT3 [18–22]. These studies revealed that STAT3 controls inhibitory and stimulatory pathways that regulate granulopoiesis. For example, STAT3 is necessary for G-CSF-dependent expression of SOCS3, a critical feedback inhibitor of G-CSFR signal transduction [19, 23]. STAT3 also enhances granulocytic progenitor cell-cycle progression in response to G-CSF by controlling the expression of C/EBPβ and c-Myc [21]. Moreover, STAT3 is required for G-CSF-dependent neutrophil mobilization, as well as neutrophil chemotaxis [20, 22], indicating a function in regulating pathways involved in neutrophil migration. In agreement with its role in these critical neutrophil responses, STAT3 is necessary for effective clearance of Listeria monocytogenes from infected mice [21, 22], implying a central function in antibacterial immunity. This essential activity is also evident in humans, as loss-of-function STAT3 mutations underlie the primary human immunodeficiency, hyper-IgE syndrome [24, 25].
Whereas G-CSF administration results in neutrophil mobilization, this cytokine acts as a growth factor without apparent chemotactic activity [26]. Thus, the mechanism(s) by which G-CSF signals, via STAT3, elicit neutrophil mobilization remain poorly resolved. Recent work suggests that CXCR2, a key neutrophil chemoattractant receptor, participates in G-CSF-responsive neutrophil mobilization [22, 27]. CXCR2 belongs to the seven-transmembrane GPCR family and is best known for its ability to direct neutrophil migration in response to the chemokines KC (CXCL1) and MIP-2 (CXCL2) in mice or IL-8 in humans during inflammatory responses [28]. Deletion of murine Il8rb, encoding CXCR2, results in neutrophilia, which may be a result of an inability of neutrophils to migrate into peripheral tissues and/or undergo effective clearance [6, 28]. Il8rb deficiency also abrogates G-CSF-responsive neutrophil mobilization [27], indicating the importance of CXCR2 in regulating neutrophil efflux from the bone marrow. In previous work, we found that G-CSF stimulates CXCR2 cell-surface expression and Il8rb mRNA amounts in neutrophils via a STAT3-dependent pathway [22]; however, effects of the G-CSF–STAT3 pathway on other mobilization factors remain to be determined.
Despite the importance of STAT3 in immunity, there is surprisingly little information about the mechanisms of STAT3 action at target gene promoters, particularly within differentiating myeloid cells. Expression of protein-encoding genes is controlled by a complex orchestration of post-translational chromatin modifications, DNA methylation/demethylation, transcription factor and cofactor recruitment, and association, as well as activation of RNA Pol II [29–34]. STATs and their upstream activators, the Jak kinases, contribute to initiating and/or reinforcing chromatin modifications and association of regulatory molecules that coordinate gene expression [35–40]. For example, STAT3 interacts with the histone acetyltransferase p300/CBP, is involved in recruiting coactivators and histone-modifying enzymes to target gene promoters, and undergoes post-translational modifications that contribute to transcriptional regulation [38, 41–43]. These events have been studied in immortalized and cancer cell lines, providing important insight into potential mechanism(s) by which STAT3 may function at target promoters in primary myeloid cells.
To further extend our understanding of STAT3 mechanisms involved in maintaining an immunocompetent state and in light of the critical role of STAT3 in regulating G-CSF-responsive neutrophil mobilization and CXCR2-mediated neutrophil chemotaxis [20, 22], we investigated whether and how STAT3 controls the expression of Cxcl2, which encodes the CXCR2 ligand MIP-2. Our results show that STAT3 directly regulates G-CSF-responsive but not developmental induction of Cxcl2 expression in primary bone marrow neutrophils. Moreover, we found that G-CSF controls the abundance of activating H3K4me3 modifications in the vicinity of the STAT3-binding regions in the Cxcl2 and Il8rb promoters. STAT3 recruitment is accompanied by H3K4me3 induction and the presence of a post-translationally modified form of RNA Pol II associated with transcriptional initiation. These results suggest that STAT3 coordinates transcriptional and epigenetic events involved in G-CSF-responsive induction of neutrophil-mobilizing factors, revealing mechanisms by which the G-CSF–STAT3 pathway contributes to effective neutrophil mobilization, a critical aspect of the innate immune response.
MATERIALS AND METHODS
Bone marrow Stat3-deficient mice, neutrophil isolation
Hematopoietic Stat3-deficient [Tg(Tek-cre)12Flv, Stat3 f/Δ] mice were bred as described previously [20, 22]. Littermate or aged-matched, Stat3-sufficient mice were used as controls as indicated [20, 22]. Mice were used at ages 4–8 weeks. C57BL/6NCr mice were obtained from the National Cancer Institute (Frederick, MD, USA). Mice were maintained in a specific pathogen-free facility and used in accordance with the Institutional Animal Care and Use Committee guidelines at UT MD Anderson Cancer Center (Houston, TX, USA).
Peripheral blood samples were collected by retro-orbital puncture. Complete blood counts were determined by automated counting through the MD Anderson Department of Veterinary Medicine, as described previously [20]. Bone marrow cells were isolated from femurs and tibiae and labeled with FITC-conjugated Gr-1 (Becton Dickinson, Franklin Lakes, NJ, USA) and allophycocyanin-conjugated CD115 (eBioscience, San Diego, CA, USA). The Gr-1− CD115+, Gr-1lo CD115−, and Gr-1hi CD115− populations were sorted using a FACSAria (BD Biosciences, San Diego, CA, USA), as described [22]. Gr-1lo CD115− and Gr-1hi CD115− cells correspond to immature and mature neutrophil subsets, as described previously [20, 44]. For simplicity, these subsets are referred to as Gr-1lo immature and Gr-1hi mature neutrophils herein.
G-CSF and MIP-2 antibody administration in vivo
rhG-CSF (Amgen, Thousand Oaks, CA, USA) was diluted in sterile PBS containing 0.1% endotoxin-free BSA and administered s.c. (250 μg/kg). In some experiments, mouse MIP-2 or IgG2B control antibody (R&D Systems, Minneapolis, MN, USA) was administered i.v. (20 μg/mouse) 30 min prior to rhG-CSF treatment.
RNA isolation, real-time PCR
Total RNA was isolated using Tri-Reagent (Molecular Research Center, Cincinnati, OH, USA) and reverse-transcribed using the iScript cDNA synthesis kit (Bio-Rad Laboratories, Hercules, CA, USA). Real-time PCR was performed with iQ SYBR Green Supermix. PCR products were detected on the iQ5 or CFX96 real-time PCR detection systems and analyzed using iQ5 Optical System Software or CFX Manager Software, respectively (Bio-Rad Laboratories), following the manufacturer's recommendations. CT values for each gene were normalized to CT values for the Rpl13a housekeeping gene to determine relative mRNA expression (ΔCT). Relative expression was calculated with the following formula: 1.8(ΔCT), as described [21]. Real-time PCR primers were: Rpl13a forward 5′-GAGGTCGGGTGGAAGTACCA-3′, Rpl13a reverse 5′-TGCATCTTGGCCTTTTCCTT-3′; Cxcl2 forward 5′-AGACAGAAGTCATAGCCACTCTCAAG-3′, Cxcl2 reverse 5′-CCTCCTTTCCAGGTCAGTTAGC-3′.
ELISAs
Neutrophils were cultured in RPMI containing 1% BSA and stimulated with 25 ng/ml rhG-CSF, as indicated in the figure legends. Supernatants were isolated and analyzed for MIP-2 expression by ELISA using the mouse CXCL2/MIP-2 DuoSet (R&D Systems), following the manufacturer's instructions.
32D cell culture, retroviral transduction
32D.G-CSFR were generated by retroviral transduction as indicated [21]. Retrovirus was produced in 293T cells following calcium phosphate-mediated transfection, using the retroviral vector pMX-G-CSFR-IRES-GFP, as described previously [21]. After 48–72 h, GFP+ G-CSFR+ cells were enriched by FACS, and cells were maintained in RPMI containing 10% FCS and 10% WEHI-3B cell-conditioned media (source of IL-3).
Identification of the Cxcl2 promoter, reporter assays
Cxcl2 corresponds to genomic coordinates 5:90,903,899-90,905,909 within the Ensembl database. The proximal Cxcl2 promoter was identified as sequences 5′ to the predicted transcriptional start site of Cxcl2, corresponding to genomic coordinates 5:90902699:90903898 within the Ensembl database. STATx consensus elements were located using TFSEARCH (http://www.cbrc.jp/research/db/TFSEARCH.html). Promoter sequences were amplified from murine genomic DNA (C57Bl6) by PCR using AccuPrime Pfx DNA polymerase (Invitrogen, Carlsbad, CA, USA) and the following primers: Cxcl2 forward 5′-GGAGGTACCCTCAGACCCACAACTATC-3′, Cxcl2 reverse 5′-GGAAGATCTGGCTCTGAGGTCCCGAGA-3′. The amplified Cxcl2 promoter region was cloned into the pGL3-Basic plasmid (Promega, Madison, WI, USA) to generate pGL3-Cxcl2; the sequence was confirmed by the MD Anderson DNA Core Analysis Facility. Site-directed mutagenesis was performed using the QuikChange XL site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA, USA) to induce mutations in the −310-bp distal or −210-bp proximal STATx consensus elements within pGL3-Cxcl2 using mutant distal sense 5′-TTCTTCAGTCCTAATGACTAAAGCCACCCAAACTGTTAGGTCTCCACTG-3′ and antisense 5′-CAGTGGAGACCTAACAGTTTGGGTGGCTTTAGTCATTAGGACTGAAGAA-3′ or mutant proximal sense 5′-CTTCCTTTTGAGGATTTGGGTGGGGACATCCCAGGGTCCCATA-3′ and antisense 5′-TATGGGACCCTGGGATGTCCCCACCCAAATCCTCAAAAGGAAG-3′ primers, respectively (location of mutation is underlined).
Reporter assays were conducted with 32D.G-CSFR cells, electroporated using the Cell Line Nucleofector Kit V (Lonza, Switzerland) and the Nucleofector Device (Lonza) following the manufacturer's instructions. Electroporations were performed with 2 μg of the appropriate reporter construct, 4 ng pTK-Renilla, and 0.5–1 μg of additional plasmids, as indicated in the figure legends. Cells were plated in RPMI containing 10% FBS and 5–10% WEHI-conditioned media. After 18 h, cells were washed and treated with 25 ng/ml rhG-CSF (Amgen) for 6 or 12 h, as indicated, prior to measurement of luciferase activity using the Dual-Luciferase reporter assay system (Promega), according to the manufacturer's instructions. Data were analyzed as a ratio of firefly light units:renilla light units. Additional plasmids used were: pRc/CMV-STAT3, encoding WT STAT3 (from Dr. James Darnell, Rockefeller University), and pMX-STAT3-DN-IRES-GFP, encoding a dominant-negative form of STAT3, which contains mutations in critical DNA-binding residues (derived from pBABE-STAT3-DN from Dr. Curt Horvath, Northwestern University) [21].
EMSAs and ChIPs
For EMSAs, 32D.G-CSFR cells were starved for 5 h by incubation in RPMI/FCS and then stimulated with RPMI/FCS plus 25 ng/ml rhG-CSF or left untreated, as indicated in the figure legends. Nuclear extracts were purified and incubated with 32P-labeled double-stranded oligonucleotides corresponding to the −310-bp distal or −210-bp proximal STATx consensus elements within the Cxcl2 proximal promoter, as indicated [45, 46]. The oligonucleotide probe encompassing the −310-bp distal STATx site was generated by annealing 5′-TAAAGTTCCCCAAACTGTTAGG-3′ and 5′-CCTAACAGTTTGGGGAACTTTA-3′ oligonucleotides. The probe corresponding to the −210-bp proximal STATx site was generated by annealing 5′-TGAGGATTTGGGGAAGGACATC-3′ and 5′-GATGTCCTTCCCCAAATCCTCA-3′ oligonucleotides. Positions of the STATx elements are underlined. In some cases, EMSA binding reactions were incubated with an excess of nonradiolabeled competitor oligonucleotide or STAT3 antibody, as indicated in the figure legends. EMSA reactions were separated on nondenaturing polyacrylamide gels and exposed to X-ray film.
ChIPs were performed with the ChIP assay kit (Millipore, Bedford, MA, USA), according to the manufacturer's instructions. Following formaldehyde-induced cross-linking, DNA was sheared to 0.2- to 1.0-kb fragments prior to immunoprecipitation. Chromatin fragments were immunoprecipitated with the following antibodies: rabbit IgG (Santa Cruz Biotechnology, Santa Cruz, CA, USA), STAT3 (c-20; Santa Cruz Biotechnology), trimethyl H3 Lys4 (Millipore), trimethyl H3 Lys27 (Millipore), total H3 (Abcam, Cambridge, MA, USA), and Pol II-pSer5 (Active Motif, Carlsbad, CA, USA). Following reversal of cross-linking, STAT3 or H3 ChIP products were amplified by PCR using primers corresponding to the proximal promoter regions of Cxcl2 or Il8rb: Cxcl2 forward 5′-GGTCACTTCAGCGCAGAC-3′, Cxcl2 reverse 5′-TCTGAGGTCCCGAGAGCT-3′; Il8rb forward 5′-CTCCCAAGTTAGGTAGCATTTCCAC-3′, Il8rb reverse 5′-TACCTGTTTGCCTGTAGGCAGGTA-3′. As a result of the length of sheared DNA fragments used in the ChIP experiments, these primers will detect immunoprecipitated DNA fragments that contain the proximal promoter region, including −310-bp distal and/or −210-bp proximal STATx consensus elements. Pol II-pSer5 ChIP products were detected with primers that amplify the region in the vicinity of the transcriptional initiation site within the Cxcl2 or Il8rb promoters: Il8rb forward 5′-CCCAGAACAGCCTAGCCA-3′, Il8rb reverse 5′-GGCTCCCAACTCTCTGTG-3′; Cxcl2 forward 5′-GACCCTGAGCTCAGGGAA-3′, Cxcl2 reverse 5′-AGTGTGGCTGGAGTCTGG-3′. These primers will detect immunoprecipitated DNA fragments that contain the proximal promoter region, including the transcriptional initiation site. Input chromatin samples were also amplified by PCR; these samples were collected prior to formaldehyde-induced cross-linking. For each experiment, the PCR results from ChIPs were normalized to PCR data from input samples using the formula 1.8(CT input−CT sample) as described previously [21]; the factor 1.8 estimates PCR amplification efficiency. In some cases, the ChIP data were further analyzed to obtain STAT3/IgG, Pol II-pSer5/IgG, H3K4me3/H3, or H3K27me3/H3 ratios to control for nonspecific antibody binding (IgG) or nucleosome abundance (H3) or were analyzed to obtain G-CSF/nontreatment ratios to obtain fold induction with G-CSF, as indicated in the figure legends.
Immunoblotting
Whole cell lysates were separated by SDS-PAGE and detected with phospho-STAT3 (Y705; Cell Signaling Technology, Danvers, MA, USA) or total STAT3 (c-20; Santa Cruz Biotechnology) antibody, as described previously [47].
Statistical analyses
Shown are mean values ± sem. P values were determined by unpaired two-tailed Student's t test using GraphPad Prism version 5 for Mac OS X (GraphPad Software, San Diego, CA, USA; http://www.graphpad.com). P values <0.05 were considered statistically significant.
RESULTS
G-CSF stimulates MIP-2 production from Gr-1hi mature neutrophils
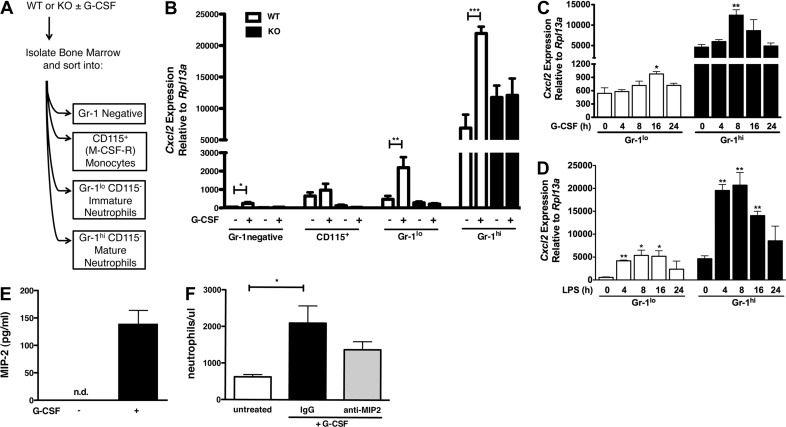
We reported previously that G-CSF up-regulates Cxcl2 (MIP-2) mRNA in total bone marrow [22], yet the predominant cell type producing MIP-2 was unknown. To address this and investigate the role for STAT3, we evaluated Cxcl2 mRNA expression in myeloid and nonmyeloid bone marrow populations isolated from WT or Stat3-deficient mice under steady-state conditions or 24 h after G-CSF administration. We purified bone marrow cells according to their expression of CD115 and Gr-1, markers that designate monocyte, immature, and mature neutrophil populations (Fig. 1A) [20, 44, 48]. We found that in vivo delivery of G-CSF induced Cxcl2 mRNA in WT Gr-1lo immature and Gr-1hi mature neutrophils. Cxcl2 mRNA amounts were approximately tenfold higher in Gr-1hi neutrophils compared with immature neutrophils (Fig. 1B). By contrast, G-CSF-responsive up-regulation of Cxcl2 mRNA was abrogated in Stat3-deficient immature and mature neutrophils (Fig. 1B). We also observed constitutive Cxcl2 mRNA expression in Gr-1hi neutrophils; however, this appeared to be controlled independently of STAT3 (Fig. 1B). Cxcl2 mRNA expression was detected in the Gr-1− bone marrow fraction and in CD115+ monocytes, yet mRNA amounts were reduced compared with those found in mature and immature neutrophils. These results indicate that Gr-1hi neutrophils are the major bone marrow cell type expressing Cxcl2 mRNA in basal conditions and following G-CSF administration in vivo, whereas STAT3 is required for G-CSF-responsive but not constitutive Cxcl2 mRNA expression.
Figure 1. G-CSF stimulates Cxcl2 and MIP-2 expression in Gr-1hi mature neutrophils.
(A) Schematic diagram of the cell-sorting strategy. (B) WT (white bars) and Stat3-deficient [knockout (KO); black bars] mice were treated with G-CSF (250 μg/kg) or left untreated. Bone marrow cells were harvested after 24 h, purified by cell sorting as indicated, and analyzed by qPCR for expression of Cxcl2. Shown are mean expression levels relative to the housekeeping gene Rpl13a ± sem (n≥3). (C and D) WT Gr-1lo immature and Gr-1hi mature neutrophils were purified by cell sorting and cultured with 25 ng/ml G-CSF (C) or 0.1 μg/ml LPS (D) for 0, 4, 8, 16, or 24 h. Cxcl2 mRNA amounts were determined by qPCR as indicated in B (n=3). (E) WT Gr-1hi mature neutrophils were purified by cell sorting and cultured in the absence (−) or presence (+) of 25 ng/ml G-CSF for 24 h, as indicated. MIP-2 expression was analyzed in culture supernatants by ELISA. Results show mean values ± sem (n≥3). (F) WT C57BI6 mice were treated with 20 μg anti-MIP-2 antibody (gray bar), IgG isotype control (black bar), or left untreated (white bar). After 30 min, mice were treated with G-CSF as indicated in B. Results show mean neutrophil levels/ml blood ± sem, determined 6 h after G-CSF treatment (n≥3). *P < 0.05; **P < 0.01; ***P < 0.001.
We next assessed whether G-CSF stimulated Cxcl2 expression in neutrophils in vitro as one approach toward determining if the effects of G-CSF upon Cxcl2 were direct or indirect. For these experiments, we measured Cxcl2 mRNA amounts in purified Gr-1lo and Gr-1hi neutrophils immediately after isolation or following culture in G-CSF-containing medium for 4–24 h. These assays showed that Cxcl2 mRNA was induced by G-CSF in immature and mature neutrophils by 16 or 8 h of G-CSF treatment, respectively (Fig. 1C). In addition, these experiments confirmed that Gr-1hi mature neutrophils show increased expression of Cxcl2 mRNA relative to Gr-1lo immature cells under nonstimulation conditions (Figs. 1B and C). By contrast, the TLR4 agonist LPS enhanced Cxcl2 mRNA amounts in Gr-1lo immature and Gr-1hi mature neutrophils within 4 h (Fig. 1D). These results collectively suggest that G-CSF stimulates a rapid but not immediately early gene expression response, relative to LPS-mediated Cxcl2 induction.
To determine whether MIP-2 protein production was regulated by G-CSF, we evaluated MIP-2 secretion from immature and mature neutrophil subsets. We purified Gr-1lo and Gr-1hi neutrophils by FACS, cultured cells −/+ G-CSF for 24 h in vitro, and examined MIP-2 amounts in culture supernatants by ELISA. These experiments showed that G-CSF stimulated MIP-2 secretion from Gr-1hi neutrophils (Fig. 1E). By contrast, MIP-2 amounts were below the detection limit in Gr-1hi neutrophil cultures lacking G-CSF or Gr-1lo neutrophil cultures with or without G-CSF (Fig. 1E; data not shown). Therefore, although Gr-1hi neutrophils contain abundant Cxcl2 mRNA amounts in homeostatic conditions, and Gr-1lo and Gr-1hi neutrophil populations up-regulate Cxcl2 mRNA in response to G-CSF, only Gr-1hi mature neutrophils are effective producers of MIP-2 protein upon G-CSF stimulation. These data suggest that transcriptional and post-transcriptional mechanisms regulate the production of MIP-2.
We determined whether MIP-2 contributes to G-CSF-responsive neutrophil mobilization using an antibody-blocking strategy in vivo. We delivered a MIP-2-neutralizing antibody or an isotype-matched control to mice 30 min prior to administration of G-CSF and then analyzed circulating neutrophil levels 6 h after G-CSF treatment. We found a three- to four-fold increase in peripheral neutrophil amounts in animals that received G-CSF and the IgG control antibody compared with untreated animals (Fig. 1F), consistent with G-CSF-dependent neutrophil-mobilizing responses observed in previous studies [20]. By contrast, treatment with the MIP-2-neutralizing antibody blunted the response to G-CSF; mice that received MIP-2 antibody showed a 1.5- to twofold increase in circulating neutrophils relative to untreated controls (Fig. 1F). Whereas the difference between MIP-2 and IgG antibody treatments was not statistically significant, these results are consistent with a role for MIP-2, as well as other factors in the G-CSF-induced neutrophil mobilization response [26, 49, 50].
STAT3 mediates G-CSF-dependent induction of Cxcl2 by interaction with the Cxcl2 promoter
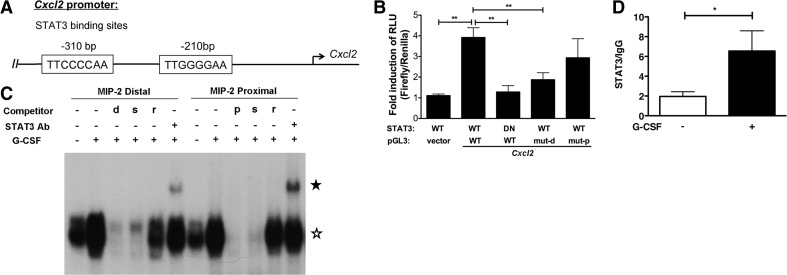
As STAT3 is best-characterized as a transcriptional regulator, we focused our attention on whether STAT3 contributed to transcriptional activation of Cxcl2 in response to G-CSF stimulation. To examine this, we identified the proximal Cxcl2 promoter and used software analysis to scan for putative STAT-binding sites. We located two STATx consensus elements ∼310 bp and ∼210 bp upstream (5′) of the Cxcl2 transcriptional initiation site (Fig. 2A). For clarity, we refer to these elements as the −310-bp distal and −210-bp proximal STAT sites, respectively, within the Cxcl2 proximal promoter region. To assess the function of these elements, we used 32D.G-CSFR cells, a murine myeloid progenitor line that responds to G-CSF as a result of engineered expression of G-CSFR [47, 51]. We used gene reporter assays with a construct containing the proximal Cxcl2 promoter encompassing the STATx sites, fused to the firefly luciferase gene, to examine whether G-CSF induced Cxcl2 transcriptional activity. These experiments demonstrated that G-CSF stimulated a three- to fourfold increase in Cxcl2 reporter activity, in agreement with G-CSF-responsive induction of endogenous Cxcl2 mRNA expression (Figs. 1B and 2B). By contrast, cotransfection with a STAT3 mutant that contains substitutions in critical DNA-binding residues inhibited G-CSF-induced Cxcl2 gene reporter activity (Fig. 2B). Moreover, mutation of the −310-bp distal STATx consensus element significantly inhibited G-CSF-responsive reporter activity, whereas mutation of the −210-bp proximal element showed modest but nonsignificant repression (Fig. 2B). These assays suggest that G-CSF stimulates Cxcl2 transcription directly via STAT3 interaction with sequences in the proximal Cxcl2 promoter.
Figure 2. G-CSF-activated STAT3 induces Cxcl2 expression by direct interaction at the proximal promoter.
(A) Schematic diagram of the Cxcl2 proximal promoter, indicating the locations of STATx consensus elements at −310-bp distal and −210-bp proximal sites, as well as the transcriptional initiation site (arrow). (B) Luciferase assays in 32D.G-CSFR cells electroporated with WT or mutant (mut) forms of pGL3-Cxcl2 (pGL3), pGL3 empty vector (vector), pTK-Renilla, and plasmids encoding WT or DN STAT3, following incubation in the presence (+) or absence (−) of 25 ng/ml G-CSF for 6 h, as indicated. Mutant pGL3-Cxcl2 constructs contain substitutions in the −310-bp distal (mut-d) or −210-bp proximal (mut-p) STATx consensus elements. The ratio of firefly:renilla relative light units (RLU) was calculated for each experiment. Results represent mean values ± sem of ratios of RLU from G-CSF-treated cells normalized to RLU from nontreated cells (n=3–6). (C) EMSAs were performed using nuclear extracts purified from 32D.G-CSFR cells treated with (+) or without (−) 25 ng/ml G-CSF for 30 min, as indicated, following initial growth factor starvation for 5 h. 32P-labeled, double-stranded oligonucleotide probes corresponding to the −310-bp distal or −210-bp proximal STATx consensus elements in the Cxcl2 promoter were incubated with nuclear extracts in the presence or absence of excess unlabeled competitor oligonucleotides (d, −310-bp Cxcl2 STATx oligonucleotide; p, −210-bp proximal Cxcl2 STATx oligonucleotide; s, oligonucleotide encoding STAT3 site from murine Socs3 promoter; r, random probe; probe sequences are provided in Materials and Methods). The locations of STAT3:DNA and STAT3:DNA:STAT3 antibody complexes in nondenaturing gels are indicated by open and closed stars, respectively. Results represent four independent experiments. (D) qPCR analysis of STAT3 ChIPs from 32D.G-CSFR cells. 32D.G-CSFR cells were treated −/+ 25 ng/ml G-CSF for 30 min. STAT3 or IgG control antibodies were used to immunoprecipitate cross-linked, sheared chromatin fragments from cells. PCR analysis was performed with primers corresponding to the Cxcl2 proximal promoter region, as indicated in Materials and Methods. PCR assays were conducted on ChIP samples and input chromatin; ChIP results were normalized to input values as described in Materials and Methods. STAT3 ChIP data were then normalized to IgG ChIP data to obtain a STAT3/IgG ratio, as indicated. Error bars represent ± sem (n=3). *P < 0.05; **P < 0.01.
To assess whether STAT3 bound Cxcl2 promoter STATx elements, we performed EMSAs with double-stranded oligonucleotides corresponding to the −310-bp distal or −210-bp proximal STATx elements and nuclear extracts from G-CSF-treated or nontreated 32D.G-CSFR cells. These experiments showed G-CSF-responsive STAT3 binding to the −310-bp distal and −210-bp proximal STATx elements, as judged by comparison of protein:oligonucleotide complexes −/+ G-CSF treatment and supershift assays with STAT3 antibody (Fig. 2C). Furthermore, the EMSAs revealed that G-CSF-responsive STAT3 binding was abrogated by a surplus of self-oligonucleotide or by an excess of an oligonucleotide that encodes the STAT3 consensus element in the murine Socs3 gene promoter, an established STAT3 target gene [46, 52], but not by a random oligonucleotide probe (Fig. 2C). These results indicate specificity in the interaction of STAT3 with Cxcl2 promoter STATx consensus elements. With the use of ChIPs, we found that STAT3 was rapidly recruited to the endogenous Cxcl2 promoter in the vicinity of the STATx elements upon G-CSF stimulation, with approximately threefold enrichment of STAT3 detected 30 min following G-CSF treatment (Fig. 2D). These data collectively suggest that STAT3 interacts directly with the Cxcl2 proximal promoter to enhance Cxcl2 transcription in response to G-CSF stimulation.
Chromatin modifications at the Cxcl2 and Il8rb proximal promoters in immature and mature neutrophils in steady-state conditions
Previously, we found that G-CSF-responsive STAT3 directly activates Il8rb expression, similar to Cxcl2 [22] (Fig. 2). Moreover, like Cxcl2, Il8rb expression is regulated developmentally, with higher constitutive expression in Gr-1hi mature neutrophils relative to Gr-1lo immature cells [22]. However, G-CSF–STAT3-responsive Il8rb expression is detectable in Gr-1lo immature neutrophils but not Gr-1hi cells, whereas Cxcl2 is inducible in both subsets, with significantly higher expression in mature neutrophils [22] (Fig. 1). Thus, Cxcl2 and Il8rb share common and distinct features of transcriptional control, raising the question of whether their promoters exhibit similar or unique mechanisms of regulation during neutrophil development and/or in response to G-CSF stimulation.
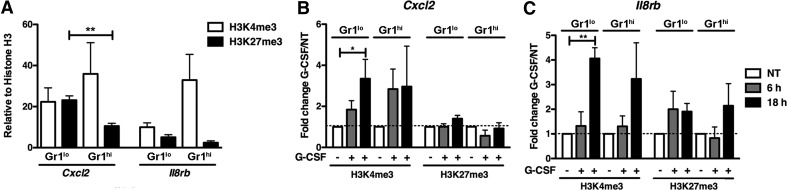
Chromatin configuration has an essential function in transcriptional control by controlling the accessibility of DNA regulatory elements to the transcriptional machinery [33, 53–55]. Thus, we evaluated the status of H3 modifications associated with transcriptional activity (H3K4me3) or repression (H3K27me3) [56] in the vicinity of the STAT3-binding elements at the Cxcl2 and Il8rb proximal promoters using ChIP analysis of FACS-purified Gr-1lo immature and Gr-1hi mature neutrophils from bone marrow. These experiments were performed on freshly isolated cells in the absence of exogenous G-CSF stimulation to capture the endogenous H3 marks in steady-state. Our assays revealed that the Cxcl2 promoter in Gr-1lo immature granulocytes contained a comparable abundance of H3K4me3 and H3K27me3 modifications, relative to total H3 amounts (Fig. 3A). We found a similar pattern at the Il8rb promoter in Gr-1lo immature granulocytes, with roughly equivalent amounts of H3K4me3 and H3K27me3 relative to total H3 (Fig. 3A). These data are consistent with transcriptionally silent or low-activity promoters that can be readily induced, as observed at poised genes of embryonic stem cells [57], and in agreement with our results showing modest basal yet G-CSF-inducible expression of Cxcl2 (Fig. 1B) or Il8rb [22] in Gr-1lo immature granulocytes. By contrast, we found that Gr-1hi mature neutrophils contained increased amounts of H3K4me3 marks compared with H3K27me3 at Cxcl2 and Il8rb promoters (Fig. 3A). In addition, the abundance of H3K27me3 was decreased significantly at the Cxcl2 promoter in Gr-1hi mature neutrophils relative to Gr-1lo immature granulocytes (Fig. 3A). These results suggest a greater degree of chromatin accessibility and/or elevated basal transcription rates of Cxcl2 and Il8rb in mature neutrophils relative to immature granulocytes, consistent with the high constitutive expression of Cxcl2 (Fig. 1B) and Il8rb [22] in Gr-1hi mature neutrophils. Moreover, the decrease in Cxcl2 H3K27me3 modifications at the proximal promoter in mature versus immature neutrophils suggests that this mark might be regulated developmentally, reflecting changes in constitutive gene expression patterns observed during neutrophil differentiation (Fig. 1B).
Figure 3. H3 modifications at the Cxcl2 and Il8rb proximal promoters in immature and mature neutrophils.
(A) Gr-1lo immature and Gr-1hi mature neutrophil subsets were purified from the bone marrow of WT mice and analyzed by ChIPs using antibodies against H3K4me3 (white bars), H3K27me3 (black bars), or total H3 (not shown). qPCR analysis was performed with primers corresponding to the Cxcl2 (left) or Il8rb (right) proximal promoter regions, as indicated in Materials and Methods. Following normalization to input, results for H3K4me3 and H3K27me3 ChIPs were normalized to total H3; mean amounts ± sem are shown (n=3). (B and C) Gr-1lo immature and Gr-1hi mature neutrophil subsets were purified from the bone marrow of WT mice and stimulated with G-CSF in vitro for 6 h (gray bars) or 18 h (black bars) or left untreated [not treated (NT); white bars]. ChIPs were performed using antibodies against H3K4me3, H3K27me3, or total H3. qPCR analysis was performed with primers corresponding to the Cxcl2 (B) or Il8rb (C) proximal promoter regions. H3K4me3 and H3K27me3 ChIP data were analyzed as described in A; data from G-CSF-treated samples were then normalized to untreated samples to determine G-CSF/NT fold change (shown). Data shown are mean values ± sem (n=3). *P < 0.05; **P < 0.01.
G-CSF enhances the abundance of H3K4me3 at the proximal Cxcl2 and Il8rb promoters in Gr-1lo immature neutrophils
To assess mechanisms by which G-CSF regulates transcription from the Cxcl2 and Il8rb promoters, we evaluated whether H3K4me3 and H3K27me3 modifications were affected by G-CSF treatment. For these experiments, we purified Gr-1lo and Gr-1hi neutrophils by FACS, treated cells with G-CSF in vitro, and performed ChIPs with antibodies against H3K4me3, H3K27me3, or total H3. We found that G-CSF had relatively little effect on H3K27me3 abundance relative to total H3 amounts at either promoter (Fig. 3B and C). By contrast, G-CSF stimulated a two- to fourfold enhancement of H3K4me3 abundance relative to total H3 at the Cxcl2 and Il8rb promoters in Gr-1lo immature granulocytes, compared with untreated controls (Fig. 3B and C). These results suggest that H3 marks associated with active transcription are enhanced at the Cxcl2 and Il8rb promoters in immature neutrophils by G-CSF signaling, consistent with the ability of G-CSF to stimulate Cxcl2 and Il8rb mRNA expression in Gr-1lo immature granulocytes.
Role for STAT3 in G-CSF-responsive recruitment of transcriptionally active RNA Pol II to the proximal Cxcl2 and Il8rb promoters
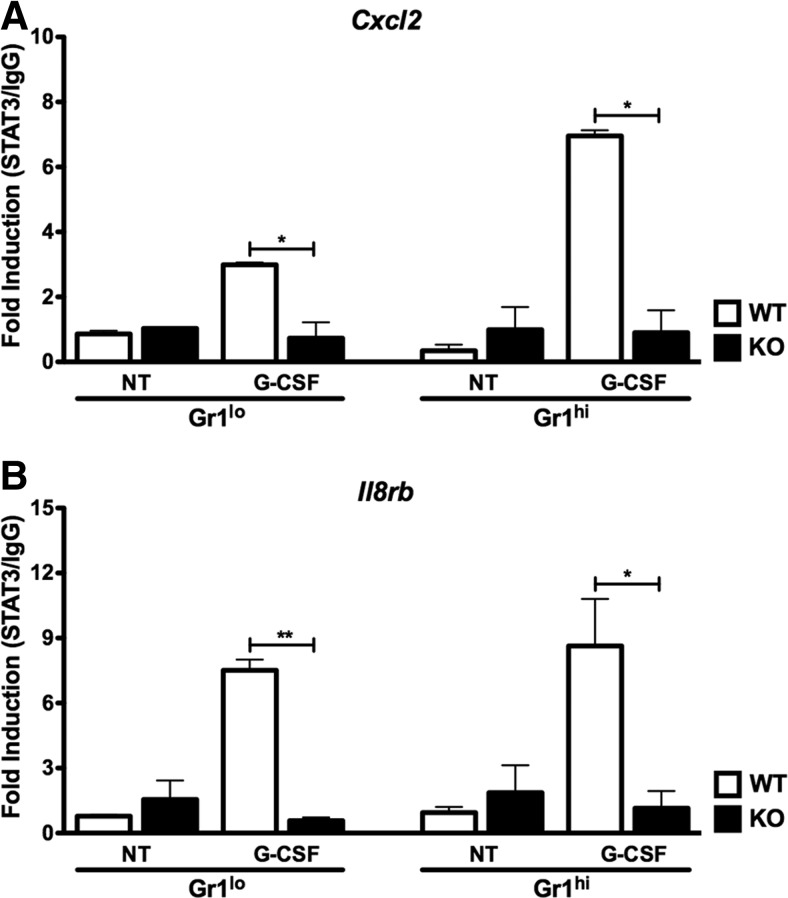
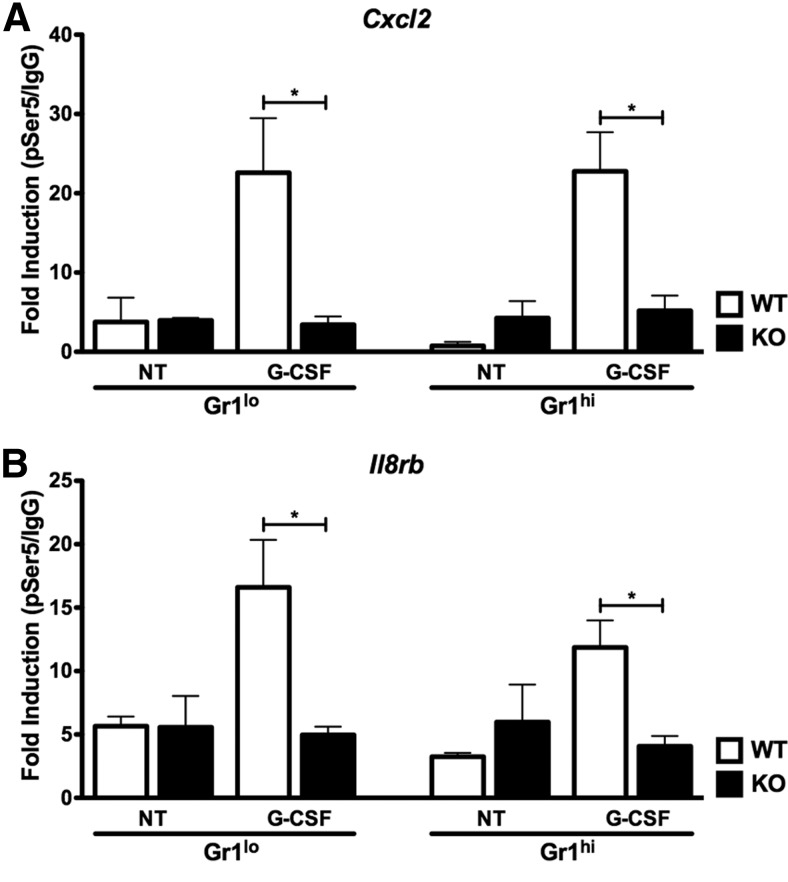
We next examined whether G-CSF-responsive STAT3 binding at the Cxcl2 and Il8rb promoters (Fig. 2) [22] correlated with recruitment of Pol II-pSer5, a form of Pol II associated with assembly of the transcriptional preinitiation complex [31]. For these studies, we performed STAT3 and Pol II-pSer5 ChIPs with Gr-1lo immature and Gr-1hi mature neutrophils purified from WT or Stat3-deficient mice following G-CSF treatment in vitro. Our analysis of STAT3 ChIPs showed that STAT3 accumulated at the Cxcl2 and Il8rb promoters in Gr-1lo immature and Gr-1hi mature neutrophils from WT mice in response to G-CSF stimulation, whereas STAT3 association was not detectable in neutrophils from Stat3-deficient animals or in untreated WT cells (Fig. 4A and B). G-CSF also enriched Pol II-pSer5 amounts at both promoters in Gr-1lo immature and Gr-1hi mature neutrophils from WT animals (Fig. 5A and B). By contrast, Pol II-pSer5 did not accumulate further at the Cxcl2 and Il8rb promoters in Gr-1lo immature or Gr-1hi mature neutrophils from Stat3-deficient mice (Fig. 5A and B). Thus, G-CSF appears to stimulate a coordinated induction of STAT3, H3K4me3, and Pol II-pSer5 at the Cxcl2 and Il8rb promoters in Gr-1lo immature or Gr-1hi mature neutrophils. These results suggest potential STAT3 involvement in regulating chromatin accessibility and additional recruitment of activated RNA Pol II to the Cxcl2 and Il8rb promoters.
Figure 4. G-CSF-responsive changes in STAT3 occupancy at the Cxcl2 and Il8rb proximal promoters.
Gr-1lo immature and Gr-1hi mature neutrophil subsets were purified from the bone marrow of WT or Stat3-deficient mice and treated with G-CSF in vitro or left untreated for 2 h, as indicated. ChIPs were performed with STAT3 or IgG control antibody. qPCR analysis was conducted with primers corresponding to the Cxcl2 (A) or Il8rb (B) proximal promoter regions, as indicated. ChIP results were analyzed as described in Fig. 2D, and STAT3/IgG ratios are shown. Mean values ± sem are shown (n=2–3). *P < 0.05; **P < 0.01.
Figure 5. Role for STAT3 in recruitment of RNA Pol II to the Cxcl2 and Il8rb proximal promoters.
Gr-1lo immature and Gr-1hi mature granulocytes were purified from the bone marrow of WT or Stat3-deficient mice and treated with G-CSF in vitro or left untreated for 2 h, as indicated. ChIPs were performed with an antibody against Pol II-pSer5 or IgG control antibody. qPCR analysis was performed with primers corresponding to the Cxcl2 (A) or Il8rb (B) proximal promoter regions, as indicated. ChIP results were analyzed as described in Fig. 2D, and Pol II-pSer5/IgG ratios are shown. Mean values ± sem are shown (n=2–3). *P < 0.05.
DISCUSSION
Neutrophil mobilization from the bone marrow occurs rapidly upon G-CSF treatment or bacterial infection, which elicits G-CSF production; however, the molecular events that mediate this mobilizing response are poorly defined. Here, we demonstrate that G-CSF stimulates production of the neutrophil chemotactic protein MIP-2 in mature bone marrow neutrophils. G-CSF also up-regulates Cxcl2 mRNA expression by a direct pathway involving STAT3, a principal signal transducer of the activated G-CSFR [18–20]. Our results suggest that STAT3 induces Cxcl2 transcription in response to G-CSF by binding STAT consensus element(s) in the Cxcl2 proximal promoter region. Furthermore, we find that STAT3 accumulation at the Cxcl2 promoter, as well as the Il8rb promoter, coincides with G-CSF-dependent increases in Pol II-pSer5 and H3K4me3 modifications. Collectively, these results imply a role for STAT3 in stimulating the assembly and/or activity of the transcriptional machinery at the Cxcl2 and Il8rb promoters in neutrophils upon G-CSF stimulation. These data suggest a direct mechanism of gene regulation by which the G-CSF–STAT3 pathway promotes the expression of factors involved in neutrophil mobilization.
The induction of Cxcl2 expression by G-CSF and STAT3 is consistent with previous results demonstrating that G-CSF does not stimulate neutrophil chemotaxis directly, as well as data that indicate that G-CSF-induced neutrophil mobilization requires the action of trans-acting factors [26, 58]. Additionally, patients receiving rG-CSF therapy show an increase in serum levels of IL-8, the human counterpart to MIP-2, whereas IL-8 gene induction is regulated by STAT3 in human endothelial cells [59, 60]. Recently, MIP-2 production was reported from bone marrow cells following G-CSF administration; however, distinct mechanisms were implicated [50, 61]. In one study, G-CSF-responsive MIP-2 accumulation in the bone marrow was attributed to expression from endothelial cells [50]. Separately, G-CSF was reported to stimulate thrombopoietin expression, which in turn, was found to activate production of MIP-2 and KC from megakaryocytes [61]. In agreement, we found that Cxcl2 mRNA was induced by G-CSF in the bone marrow Gr-1− population, which may include endothelial and megakaryocyte lineage cells. Furthermore, despite the abundant amounts of Cxcl2 mRNA in untreated and G-CSF-stimulated Gr-1hi mature neutrophils, MIP-2 protein was only produced in detectable amounts by Gr-1hi neutrophils upon G-CSF treatment in vitro. These results collectively suggest that MIP-2 production may be regulated by several mechanisms. Here, we uncover one pathway that mediates Cxcl2 transcriptional induction, namely, G-CSF–STAT3-dependent gene activation; however, new protein synthesis and/or regulated transport of the nascent, as well as previously synthesized MIP-2, may also be activated upon G-CSF stimulation. Additional work will be needed to discriminate among these mechanisms. Nonetheless, the collective body of evidence suggests that G-CSF stimulates MIP-2 expression in multiple bone marrow cell types via distinct mechanisms. The ability of G-CSF to induce MIP-2, coupled with high constitutive and G-CSF-responsive CXCR2 expression on neutrophils, may explain in part how this cytokine elicits neutrophil migration from the bone marrow without having direct chemotactic properties [20, 22, 26].
Whereas STAT transcription factors have been studied extensively for the last 20 years [62–65], we still have few details about the mechanisms by which they regulate gene expression. In particular, with the exception of elegant genome-wide associations in lymphoid subsets [66], studies in primary cells are lacking. To address these issues, we investigated molecular events that occur as STAT3 binds the Cxcl2 and Il8rb promoters in immature and mature neutrophils upon G-CSF treatment. Our experiments revealed a close coordination among G-CSF-responsive STAT3 binding, an accumulation of Pol II-pSer5, and increases in G-CSF-responsive H3K4me3 abundance at both proximal promoter regions. Specifically, STAT3 occupancy correlated with G-CSF-responsive increases in H3K4me3 abundance at the Cxcl2 and Il8rb promoters in Gr-1lo immature neutrophils. Furthermore, STAT3 and Pol II-pSer5 coordinately accumulated at the Cxcl2 and Il8rb promoters in Gr-1lo and Gr-1hi neutrophils, and studies with Stat3-deficient cells showed a requirement for STAT3 in the induction of Pol II-pSer5 association by G-CSF. These results suggest a role for STAT3 in assembly of an activated transcriptional initiation complex at the Cxcl2 and Il8rb promoters upon G-CSF treatment. Whereas our data from primary neutrophils do not distinguish between whether there is active transcription from both promoters at the time of cell purification, which is enhanced by G-CSF treatment, or whether G-CSF stimulation causes renewed transcription, our results from gene reporter assays suggest that G-CSF-dependent stimulation of STAT3 leads to transcriptional activation of Cxcl2 and Il8rb. Furthermore, G-CSF-responsive accumulation of the activating H3K4me3 modification and Pol II-pSer5 at both promoters is consistent with the idea of renewed transcription in primary neutrophils upon G-CSF treatment; however, additional studies are required to examine this hypothesis. With regard to Cxcl2, gene reporter assays in 32D.G-CSFR cells suggested a major contribution of the −310-bp distal STATx element, whereas EMSAs showed that −310-bp distal and −210-bp proximal sites interacted with STAT3. ChIPs do not provide sufficient resolution to distinguish between STAT3 occupancy in vivo, thus the question of whether one or both STAT3 elements are required for G-CSF-responsive Cxcl2 expression in primary neutrophils will require targeted mutations in vivo.
The CTD of RNA Pol II has tandem repeats of a 7-aa motif that includes serine 5 (YSPTSPS); mammalian RNA Pol II contains 52 repeats of this motif [67]. CTD hyperphosphorylation is associated with transcriptional initiation and elongation. Pol II-pSer5 is generally enriched at initiation regions, whereas the serine-5 modification is lost during elongation [29, 31, 68]. Kinase activity within the preinitiation complex is known to phosphorylate serine 5 in the CTD [69]. This would suggest that association of STAT3 with the Cxcl2 and Il8rb promoters in neutrophils is accompanied by recruitment and/or stabilization of preinitiation complex components as well as RNA Pol II, leading to Pol II-pSer5. One scenario we envisage is STAT3 DNA binding as an initial event upon G-CSF stimulation, coincident with or followed by increased recruitment or stabilization of RNA Pol II and preinitiation factors, directly or indirectly, via associated cofactors. For example, STAT3 has been shown to interact with p300/CBP, a histone acetyltransferase that can associate with Pol II and components of the preinitiation complex, whereas the STAT1–STAT2-containing IFN-stimulated gene factor 3 complex recruits Pol II and initiates serine 5 phosphorylation [34, 36, 42, 70]. To fully understand the mechanism by which STAT3 coordinates Pol II-pSer5 and transcriptional activation, it will be necessary to define STAT3-associated cofactors and the role for specific domains of STAT3 in recruitment of coregulators to the Cxcl2 and Il8rb promoters.
Interestingly, we also observed apparent discrepancies among G-CSF-mediated gene expression, the presence of Pol II-pSer5, or kinetic responses to other agonists. For example, G-CSF induced STAT3-dependent enrichment of Pol II-pSer5 at the Il8rb promoter in mature neutrophils, a subset that does not demonstrate significant up-regulation of Il8rb mRNA [22]. This may imply that the Il8rb promoter remains accessible, with G-CSF–STAT3 stimulating a preinitiation complex (e.g., reflected by Pol II-pSer5 accumulation), but the complex is unable to initiate transcription and/or elongation. An inability to proceed with active transcription from the Il8rb promoter in Gr-1hi mature neutrophils could result from changes in coactivator or repressor expression upon maturation. Furthermore, the kinetics of G-CSF-responsive Cxcl2 expression in purified neutrophils in vitro, relative to the time course of LPS-responsive Cxcl2 induction, suggested that additional factors or events beyond STAT3 association with promoter sequences might be required for full activation of G-CSF-dependent Cxcl2 expression. Thus, more detailed mechanistic studies are needed to establish the role for STAT3 in mediating formation of a preinitiation complex, RNA Pol II CTD phosphorylation, and transcriptional initiation/elongation at the Cxcl2 and Il8rb promoters in neutrophils in response to G-CSF stimulation.
In addition to Pol II-pSer5, we observed striking accumulation of H3K4me3 modifications at the Cxcl2 and Il8rb promoters in Gr-1lo immature neutrophils upon G-CSF stimulation. This suggests recruitment of a member of the Set domain-containing methyltransferase family, a group of enzymes that mediates H3 Lys 4 methylation [71, 72]. Set methyltransferases can be recruited upon phosphorylation of serine 5 in the Pol II CTD [73]. Moreover, regions containing H3K4me3 can be acetylated rapidly by p300 [74]. Thus, STAT3 association at the Cxcl2 and Il8rb promoters may directly or indirectly (e.g., via Pol II-pSer5 activation) recruit Set methyltransferase activity, resulting in chromatin modifications that reflect and possibly reinforce transcriptional activity. STAT3 is likely to be one of many components required for G-CSF-responsive H3K4me3 induction and activation of transcription from the Cxcl2 and Il8rb promoters.
The H3K27me3 modifications at the Cxcl2 and Il8rb promoters were less responsive or nonresponsive to G-CSF treatment. Furthermore, H3K27me3 modifications relative to total H3 decreased at the Cxcl2 promoter as granulocytes developed from Gr-1lo immature to Gr-1hi mature neutrophils. The H3K27me3 mark is often broadly distributed and is indicative of heterochromatin and silenced genes [56, 75]. Maintenance of H3K27me3 in vivo is attributed to the actions of PRC2, which contains the active methyltransferase subunit EZH2 that mediates the H3K27me3 modification [76–78]. PRC2 has important roles in stem and progenitor function in the hematopoietic system [79, 80], yet little is known about its regulation or activity throughout neutrophil development. PRC2 can be recruited by H3K27me3 motifs at promoter regions to maintain gene silencing [78]. Moreover, H3K27me3 modifications are removed by histone demethylases, including Lys-specific demethylase 1 and members of the Jumonji family [32, 81]. Therefore, the loss of H3K27me3 modifications at the Cxcl2 promoter during granulopoiesis may reflect cell-division events and a decline in PRC2 activity that results in gradual dilution of this repressive mark as neutrophils mature, global or promoter-specific histone demethylase activity, or a combination of these events. Interestingly, an opposite pattern was found at polycomb group target promoters during macrophage development, which showed induction of H3K27me3 modifications with differentiation as a result of up-regulation of the Jumonji C domain protein Jmjd3 [82]. Thus, developmental control of the repressive H3K27me3 mark may be regulated by distinct mechanisms in a promoter- and cell type-specific manner.
In conclusion, our experiments reveal mature neutrophils as major producers of Cxcl2 mRNA and MIP-2 protein upon G-CSF stimulation, suggesting that their ability to secrete and respond to MIP-2 contributes to G-CSF-responsive neutrophil mobilization from the bone marrow. Furthermore, our data suggest that STAT3 orchestrates RNA Pol II recruitment and/or activation as well as chromatin modifications at the Cxcl2 and Il8rb promoters that are needed for effective transcription upon G-CSF stimulation. These results show a direct pathway by which the G-CSF–STAT3 signaling cascade regulates transcription of important neutrophil-mobilizing factors.
ACKNOWLEDGMENTS
The DNA Analysis and Flow Core Facilities at UT MD Anderson are supported by National Cancer Institute Core grant P30CA16672. This study was supported by a pilot grant from the Center for Cancer Epigenetics at MD Anderson and grants from the U.S. National Institutes of Health (T32-CA-09598-16, H.T.N-J.; AI073587, S.S.W.). We thank Drs. Judy Layton, Jim Darnell, and Curt Horvath for providing plasmid reagents; Drs. Michelle Barton, Sharon Dent, and Valeria Facchinetti for helpful discussions; Dr. Michelle Barton for critical review of the manuscript; and Karen Ramirez, Z. David He, and Kimberlyn Acklin for assistance with cell sorting.
Footnotes
- 32D.G-CSFR
- 32D cells expressing human G-CSFR
- ChIPs
- chromatin immunoprecipitation
- CT values
- comparative threshold cycle values
- CTD
- carboxy terminal domain
- DN
- DNA-binding defective
- h
- human
- H3
- histone 3 protein
- H3K4me3
- histone 3 lysine 4 trimethylation
- H3K27me3
- histone 3 lysine 27 trimethylation
- KC
- keratinocyte chemoattractant
- Lys
- lysine
- Pol II-pSer5
- RNA polymerase II phosphorylated on serine 5 in the carboxy terminal domain
- PRC2
- polycomb repressor complex 2
- qPCR
- quantitative PCR
- RPMI/FCS
- RMPI containing 10% FCS
- SOCS3
- suppressor of cytokine signaling 3
- UT
- The University of Texas
- WEHI
- Walter and Eliza Hall Institute
AUTHORSHIP
H.T.N-J. and H.S.L. designed and performed experiments, analyzed data, and wrote the manuscript; H.Z. performed experiments and contributed reagents; E.O. performed experiments and edited the manuscript; S.S.W. supervised the project, designed experiments, analyzed data, and wrote and edited the manuscript.
REFERENCES
- 1. Lord B. I., Bronchud M. H., Owens S., Chang J., Howell A., Souza L., Dexter T. M. (1989) The kinetics of human granulopoiesis following treatment with granulocyte colony-stimulating factor in vivo. Proc. Natl. Acad. Sci. USA 86, 9499–9503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lord B. I., Molineux G., Pojda Z., Souza L. M., Mermod J. J., Dexter T. M. (1991) Myeloid cell kinetics in mice treated with recombinant interleukin-3, granulocyte colony-stimulating factor (CSF), or granulocyte-macrophage CSF in vivo. Blood 77, 2154–2159 [PubMed] [Google Scholar]
- 3. Lieschke G. J., Grail D., Hodgson G., Metcalf D., Stanley E., Cheers C., Fowler K. J., Basu S., Zhan Y. F., Dunn A. R. (1994) Mice lacking granulocyte colony-stimulating factor have chronic neutropenia, granulocyte and macrophage progenitor cell deficiency, and impaired neutrophil mobilization. Blood 84, 1737–1746 [PubMed] [Google Scholar]
- 4. Nicola N. A., Metcalf D., Matsumoto M., Johnson G. R. (1983) Purification of a factor inducing differentiation in murine myelomonocytic leukemia cells. Identification as granulocyte colony-stimulating factor. J. Biol. Chem. 258, 9017–9023 [PubMed] [Google Scholar]
- 5. Cheers C., Haigh A. M., Kelso A., Metcalf D., Stanley E. R., Young A. M. (1988) Production of colony-stimulating factors (CSFs) during infection: separate determinations of macrophage-, granulocyte-, granulocyte-macrophage-, and multi-CSFs. Infect. Immun. 56, 247–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Furze R. C., Rankin S. M. (2008) Neutrophil mobilization and clearance in the bone marrow. Immunology 125, 281–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bhana N. (2007) Granulocyte colony-stimulating factors in the management of chemotherapy-induced neutropenia: evidence based review. Curr. Opin. Oncol. 19, 328–335 [DOI] [PubMed] [Google Scholar]
- 8. Welte K., Gabrilove J., Bronchud M. H., Platzer E., Morstyn G. (1996) Filgrastim (r-metHuG-CSF): the first 10 years. Blood 88, 1907–1929 [PubMed] [Google Scholar]
- 9. Eyles J. L., Hickey M. J., Norman M. U., Croker B. A., Roberts A. W., Drake S. F., James W. G., Metcalf D., Campbell I. K., Wicks I. P. (2008) A key role for G-CSF-induced neutrophil production and trafficking during inflammatory arthritis. Blood 112, 5193–5201 [DOI] [PubMed] [Google Scholar]
- 10. Beekman R., Touw I. P. (2010) G-CSF and its receptor in myeloid malignancy. Blood 115, 5131–5136 [DOI] [PubMed] [Google Scholar]
- 11. Nicola N. A., Metcalf D. (1985) Binding of 125I-labeled granulocyte colony-stimulating factor to normal murine hemopoietic cells. J. Cell. Physiol. 124, 313–321 [DOI] [PubMed] [Google Scholar]
- 12. Nicholson S. E., Oates A. C., Harpur A. G., Ziemiecki A., Wilks A. F., Layton J. E. (1994) Tyrosine kinase JAK1 is associated with the granulocyte-colony-stimulating factor receptor and both become tyrosine-phosphorylated after receptor activation. Proc. Natl. Acad. Sci. USA 91, 2985–2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tian S. S., Lamb P., Seidel H. M., Stein R. B., Rosen J. (1994) Rapid activation of the STAT3 transcription factor by granulocyte colony-stimulating factor. Blood 84, 1760–1764 [PubMed] [Google Scholar]
- 14. Shimoda K., Feng J., Murakami H., Nagata S., Watling D., Rogers N. C., Stark G. R., Kerr I. M., Ihle J. N. (1997) Jak1 plays an essential role for receptor phosphorylation and Stat activation in response to granulocyte colony-stimulating factor. Blood 90, 597–604 [PubMed] [Google Scholar]
- 15. Tian S. S., Tapley P., Sincich C., Stein R. B., Rosen J., Lamb P. (1996) Multiple signaling pathways induced by granulocyte colony-stimulating factor involving activation of JAKs, STAT5, and/or STAT3 are required for regulation of three distinct classes of immediate early genes. Blood 88, 4435–4444 [PubMed] [Google Scholar]
- 16. Chakraborty A., Dyer K. F., Cascio M., Mietzner T. A., Tweardy D. J. (1999) Identification of a novel Stat3 recruitment and activation motif within the granulocyte colony-stimulating factor receptor. Blood 93, 15–24 [PubMed] [Google Scholar]
- 17. De Koning J. P., Soede-Bobok A. A., Schelen A. M., Smith L., van Leeuwen D., Santini V., Burgering B. M., Bos J. L., Lowenberg B., Touw I. P. (1998) Proliferation signaling and activation of Shc, p21Ras, and Myc via tyrosine 764 of human granulocyte colony-stimulating factor receptor. Blood 91, 1924–1933 [PubMed] [Google Scholar]
- 18. McLemore M. L., Grewal S., Liu F., Archambault A., Poursine-Laurent J., Haug J., Link D. C. (2001) STAT-3 activation is required for normal G-CSF-dependent proliferation and granulocytic differentiation. Immunity 14, 193–204 [DOI] [PubMed] [Google Scholar]
- 19. Lee C. K., Raz R., Gimeno R., Gertner R., Wistinghausen B., Takeshita K., DePinho R. A., Levy D. E. (2002) STAT3 is a negative regulator of granulopoiesis but is not required for G-CSF-dependent differentiation. Immunity 17, 63–72 [DOI] [PubMed] [Google Scholar]
- 20. Panopoulos A. D., Zhang L., Snow J. W., Jones D. M., Smith A. M., El Kasmi K. C., Liu F., Goldsmith M. A., Link D. C., Murray P. J., Watowich S. S. (2006) STAT3 governs distinct pathways in emergency granulopoiesis and mature neutrophils. Blood 108, 3682–3690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang H., Nguyen-Jackson H., Panopoulos A. D., Li H. S., Murray P. J., Watowich S. S. (2010) STAT3 controls myeloid progenitor growth during emergency granulopoiesis. Blood 116, 2462–2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nguyen-Jackson H., Panopoulos A. D., Zhang H., Li H. S., Watowich S. S. (2010) STAT3 controls the neutrophil migratory response to CXCR2 ligands by direct activation of G-CSF-induced CXCR2 expression and via modulation of CXCR2 signal transduction. Blood 115, 3354–3363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Croker B. A., Metcalf D., Robb L., Wei W., Mifsud S., DiRago L., Cluse L. A., Sutherland K. D., Hartley L., Williams E., Zhang J. G., Hilton D. J., Nicola N. A., Alexander W. S., Roberts A. W. (2004) SOCS3 is a critical physiological negative regulator of G-CSF signaling and emergency granulopoiesis. Immunity 20, 153–165 [DOI] [PubMed] [Google Scholar]
- 24. Minegishi Y., Saito M., Tsuchiya S., Tsuge I., Takada H., Hara T., Kawamura N., Ariga T., Pasic S., Stojkovic O., Metin A., Karasuyama H. (2007) Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature 448, 1058–1062 [DOI] [PubMed] [Google Scholar]
- 25. Holland S. M., DeLeo F. R., Elloumi H. Z., Hsu A. P., Uzel G., Brodsky N., Freeman A. F., Demidowich A., Davis J., Turner M. L., Anderson V. L., Darnell D. N., Welch P. A., Kuhns D. B., Frucht D. M., Malech H. L., Gallin J. I., Kobayashi S. D., Whitney A. R., Voyich J. M., Musser J. M., Woellner C., Schaffer A. A., Puck J. M., Grimbacher B. (2007) STAT3 mutations in the hyper-IgE syndrome. N. Engl. J. Med. 357, 1608–1619 [DOI] [PubMed] [Google Scholar]
- 26. Wengner A. M., Pitchford S. C., Furze R. C., Rankin S. M. (2008) The coordinated action of G-CSF and ELR + CXC chemokines in neutrophil mobilization during acute inflammation. Blood 111, 42–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pelus L. M., Horowitz D., Cooper S. C., King A. G. (2002) Peripheral blood stem cell mobilization. A role for CXC chemokines. Crit. Rev. Oncol. Hematol. 43, 257–275 [DOI] [PubMed] [Google Scholar]
- 28. Cacalano G., Lee J., Kikly K., Ryan A. M., Pitts-Meek S., Hultgren B., Wood W. I., Moore M. W. (1994) Neutrophil and B cell expansion in mice that lack the murine IL-8 receptor homolog. Science 265, 682–684 [DOI] [PubMed] [Google Scholar]
- 29. O'Brien T., Hardin S., Greenleaf A., Lis J. T. (1994) Phosphorylation of RNA polymerase II C-terminal domain and transcriptional elongation. Nature 370, 75–77 [DOI] [PubMed] [Google Scholar]
- 30. Svejstrup J. Q. (2004) The RNA polymerase II transcription cycle: cycling through chromatin. Biochim. Biophys. Acta 1677, 64–73 [DOI] [PubMed] [Google Scholar]
- 31. Komarnitsky P., Cho E. J., Buratowski S. (2000) Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14, 2452–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Agger K., Christensen J., Cloos P. A., Helin K. (2008) The emerging functions of histone demethylases. Curr. Opin. Genet. Dev. 18, 159–168 [DOI] [PubMed] [Google Scholar]
- 33. Berger S. L. (2007) The complex language of chromatin regulation during transcription. Nature 447, 407–412 [DOI] [PubMed] [Google Scholar]
- 34. Chan H. M., La Thangue N. B. (2001) p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J. Cell Sci. 114, 2363–2373 [DOI] [PubMed] [Google Scholar]
- 35. Gray M. J., Zhang J., Ellis L. M., Semenza G. L., Evans D. B., Watowich S. S., Gallick G. E. (2005) HIF-1α, STAT3, CBP/p300 and. Ref-1/APE are components of a transcriptional complex that regulates Src-dependent hypoxia-induced expression of VEGF in pancreatic and prostate carcinomas. Oncogene. 24, 3110–3120 [DOI] [PubMed] [Google Scholar]
- 36. Farlik M., Reutterer B., Schindler C., Greten F., Vogl C., Muller M., Decker T. (2010) Nonconventional initiation complex assembly by STAT and NF-κB transcription factors regulates nitric oxide synthase expression. Immunity 33, 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dawson M. A., Bannister A. J., Gottgens B., Foster S. D., Bartke T., Green A. R., Kouzarides T. (2009) JAK2 phosphorylates histone H3Y41 and excludes HP1α from chromatin. Nature 461, 819–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang J., Huang J., Dasgupta M., Sears N., Miyagi M., Wang B., Chance M. R., Chen X., Du Y., Wang Y., An L., Wang Q., Lu T., Zhang X., Wang Z., Stark G. R. (2010) Reversible methylation of promoter-bound STAT3 by histone-modifying enzymes. Proc. Natl. Acad. Sci. USA 107, 21499–21504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wei L., Vahedi G., Sun H. W., Watford W. T., Takatori H., Ramos H. L., Takahashi H., Liang J., Gutierrez-Cruz G., Zang C., Peng W., O'Shea J. J., Kanno Y. (2010) Discrete roles of STAT4 and STAT6 transcription factors in tuning epigenetic modifications and transcription during T helper cell differentiation. Immunity 32, 840–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Griffiths D. S., Li J., Dawson M. A., Trotter M. W., Cheng Y. H., Smith A. M., Mansfield W., Liu P., Kouzarides T., Nichols J., Bannister A. J., Green A. R., Gottgens B. (2011) LIF-independent JAK signalling to chromatin in embryonic stem cells uncovered from an adult stem cell disease. Nat. Cell Biol. 13, 13–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Saxena N. K., Vertino P. M., Anania F. A., Sharma D. (2007) Leptin-induced growth stimulation of breast cancer cells involves recruitment of histone acetyltransferases and mediator complex to CYCLIN D1 promoter via activation of Stat3. J. Biol. Chem. 282, 13316–13325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Paulson M., Pisharody S., Pan L., Guadagno S., Mui A. L., Levy D. E. (1999) Stat protein transactivation domains recruit p300/CBP through widely divergent sequences. J. Biol. Chem. 274, 25343–25349 [DOI] [PubMed] [Google Scholar]
- 43. Zhang Q., Wang H. Y., Marzec M., Raghunath P. N., Nagasawa T., Wasik M. A. (2005) STAT3- and DNA methyltransferase 1-mediated epigenetic silencing of SHP-1 tyrosine phosphatase tumor suppressor gene in malignant T lymphocytes. Proc. Natl. Acad. Sci. USA 102, 6948–6953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hestdal K., Ruscetti F. W., Ihle J. N., Jacobsen S. E., Dubois C. M., Kopp W. C., Longo D. L., Keller J. R. (1991) Characterization and regulation of RB6-8C5 antigen expression on murine bone marrow cells. J. Immunol. 147, 22–28 [PubMed] [Google Scholar]
- 45. Esashi E., Wang Y. H., Perng O., Qin X. F., Liu Y. J., Watowich S. S. (2008) The signal transducer STAT5 inhibits plasmacytoid dendritic cell development by suppressing transcription factor IRF8. Immunity 28, 509–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang L., Badgwell D. B., Bevers J. J., III, Schlessinger K., Murray P. J., Levy D. E., Watowich S. S. (2006) IL-6 signaling via the STAT3/SOCS3 pathway: functional analysis of the conserved STAT3 N-domain. Mol. Cell. Biochem. 288, 179–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wooten D. K., Xie X., Bartos D., Busche R. A., Longmore G. D., Watowich S. S. (2000) Cytokine signaling through Stat3 activates integrins, promotes adhesion, and induces growth arrest in the myeloid cell line 32D. J. Biol. Chem. 275, 26566–26575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sudo T., Nishikawa S., Ogawa M., Kataoka H., Ohno N., Izawa A., Hayashi S. (1995) Functional hierarchy of c-kit and c-fms in intramarrow production of CFU-M. Oncogene 11, 2469–2476 [PubMed] [Google Scholar]
- 49. Martin C., Burdon P. C., Bridger G., Gutierrez-Ramos J. C., Williams T. J., Rankin S. M. (2003) Chemokines acting via CXCR2 and CXCR4 control the release of neutrophils from the bone marrow and their return following senescence. Immunity 19, 583–593 [DOI] [PubMed] [Google Scholar]
- 50. Eash K. J., Greenbaum A. M., Gopalan P. K., Link D. C. (2010) CXCR2 and CXCR4 antagonistically regulate neutrophil trafficking from murine bone marrow. J. Clin. Invest. 120, 2423–2431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Panopoulos A. D., Bartos D., Zhang L., Watowich S. S. (2002) Control of myeloid-specific integrin α Mβ 2 (CD11b/CD18) expression by cytokines is regulated by Stat3-dependent activation of PU. 1. J. Biol. Chem. 277, 19001–19007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Auernhammer C. J., Bousquet C., Melmed S. (1999) Autoregulation of pituitary corticotroph SOCS-3 expression: characterization of the murine SOCS-3 promoter. Proc. Natl. Acad. Sci. USA 96, 6964–6969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Strahl B. D., Allis C. D. (2000) The language of covalent histone modifications. Nature 403, 41–45 [DOI] [PubMed] [Google Scholar]
- 54. Jenuwein T., Allis C. D. (2001) Translating the histone code. Science 293, 1074–1080 [DOI] [PubMed] [Google Scholar]
- 55. Rice J. C., Allis C. D. (2001) Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr. Opin. Cell Biol. 13, 263–273 [DOI] [PubMed] [Google Scholar]
- 56. Mikkelsen T. S., Ku M., Jaffe D. B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T. K., Koche R. P., Lee W., Mendenhall E., O'Donovan A., Presser A., Russ C., Xie X., Meissner A., Wernig M., Jaenisch R., Nusbaum C., Lander E. S., Bernstein B. E. (2007) Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bernstein B. E., Mikkelsen T. S., Xie X., Kamal M., Huebert D. J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Jaenisch R., Wagschal A., Feil R., Schreiber S. L., Lander E. S. (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 [DOI] [PubMed] [Google Scholar]
- 58. Semerad C. L., Liu F., Gregory A. D., Stumpf K., Link D. C. (2002) G-CSF is an essential regulator of neutrophil trafficking from the bone marrow to the blood. Immunity 17, 413–423 [DOI] [PubMed] [Google Scholar]
- 59. Watanabe T., Kawano Y., Kanamaru S., Onishi T., Kaneko S., Wakata Y., Nakagawa R., Makimoto A., Kuroda Y., Takaue Y., Talmadge J. E. (1999) Endogenous interleukin-8 (IL-8) surge in granulocyte colony-stimulating factor-induced peripheral blood stem cell mobilization. Blood 93, 1157–1163 [PubMed] [Google Scholar]
- 60. Gharavi N. M., Alva J. A., Mouillesseaux K. P., Lai C., Yeh M., Yeung W., Johnson J., Szeto W. L., Hong L., Fishbein M., Wei L., Pfeffer L. M., Berliner J. A. (2007) Role of the Jak/STAT pathway in the regulation of interleukin-8 transcription by oxidized phospholipids in vitro and in atherosclerosis in vivo. J. Biol. Chem. 282, 31460–31468 [DOI] [PubMed] [Google Scholar]
- 61. Kohler A., De Filippo K., Hasenberg M., van den Brandt C., Nye E., Hosking M. P., Lane T. E., Mann L., Ransohoff R. M., Hauser A. E., Winter O., Schraven B., Geiger H., Hogg N., Gunzer M. (2011) G-CSF-mediated thrombopoietin release triggers neutrophil motility and mobilization from bone marrow via induction of Cxcr2 ligands. Blood 117, 4349–4357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Darnell J. E., Jr., (1997) STATs and gene regulation. Science 277, 1630–1635 [DOI] [PubMed] [Google Scholar]
- 63. Darnell J. E., Jr., Kerr I. M., Stark G. R. (1994) Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264, 1415–1421 [DOI] [PubMed] [Google Scholar]
- 64. Cheon H., Yang J., Stark G. R. (2011) The functions of signal transducers and activators of transcriptions 1 and 3 as cytokine-inducible proteins. J. Interferon Cytokine Res. 31, 33–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Stark G. R. (2007) How cells respond to interferons revisited: from early history to current complexity. Cytokine Growth Factor Rev. 18, 419–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. O'Shea J. J., Lahesmaa R., Vahedi G., Laurence A., Kanno Y. (2011) Genomic views of STAT function in CD4+ T helper cell differentiation. Nat. Rev. Immunol. 11, 239–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Dahmus M. E. (1996) Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J. Biol. Chem. 271, 19009–19012 [DOI] [PubMed] [Google Scholar]
- 68. Weeks J. R., Hardin S. E., Shen J., Lee J. M., Greenleaf A. L. (1993) Locus-specific variation in phosphorylation state of RNA polymerase II in vivo: correlations with gene activity and transcript processing. Genes Dev. 7, 2329–2344 [DOI] [PubMed] [Google Scholar]
- 69. Feaver W. J., Svejstrup J. Q., Henry N. L., Kornberg R. D. (1994) Relationship of CDK-activating kinase and RNA polymerase II CTD kinase TFIIH/TFIIK. Cell 79, 1103–1109 [DOI] [PubMed] [Google Scholar]
- 70. Vo N., Goodman R. H. (2001) CREB-binding protein and p300 in transcriptional regulation. J. Biol. Chem. 276, 13505–13508 [DOI] [PubMed] [Google Scholar]
- 71. Santos-Rosa H., Schneider R., Bannister A. J., Sherriff J., Bernstein B. E., Emre N. C., Schreiber S. L., Mellor J., Kouzarides T. (2002) Active genes are tri-methylated at K4 of histone H3. Nature 419, 407–411 [DOI] [PubMed] [Google Scholar]
- 72. Ruthenburg A. J., Allis C. D., Wysocka J. (2007) Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol. Cell. 25, 15–30 [DOI] [PubMed] [Google Scholar]
- 73. Ng H. H., Robert F., Young R. A., Struhl K. (2003) Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell. 11, 709–719 [DOI] [PubMed] [Google Scholar]
- 74. Crump N. T., Hazzalin C. A., Bowers E. M., Alani R. M., Cole P. A., Mahadevan L. C. (2011) Dynamic acetylation of all lysine-4 trimethylated histone H3 is evolutionarily conserved and mediated by p300/CBP. Proc. Natl. Acad. Sci. USA 108, 7814–7819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sims R. J., 3rd, Nishioka K., Reinberg D. (2003) Histone lysine methylation: a signature for chromatin function. Trends Genet. 19, 629–639 [DOI] [PubMed] [Google Scholar]
- 76. Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R. S., Zhang Y. (2002) Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298, 1039–1043 [DOI] [PubMed] [Google Scholar]
- 77. Kuzmichev A., Nishioka K., Erdjument-Bromage H., Tempst P., Reinberg D. (2002) Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 16, 2893–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hansen K. H., Bracken A. P., Pasini D., Dietrich N., Gehani S. S., Monrad A., Rappsilber J., Lerdrup M., Helin K. (2008) A model for transmission of the H3K27me3 epigenetic mark. Nat. Cell Biol. 10, 1291–1300 [DOI] [PubMed] [Google Scholar]
- 79. Majewski I. J., Blewitt M. E., de Graaf C. A., McManus E. J., Bahlo M., Hilton A. A., Hyland C. D., Smyth G. K., Corbin J. E., Metcalf D., Alexander W. S., Hilton D. J. (2008) Polycomb repressive complex 2 (PRC2) restricts hematopoietic stem cell activity. PLoS Biol. 6, e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Majewski I. J., Ritchie M. E., Phipson B., Corbin J., Pakusch M., Ebert A., Busslinger M., Koseki H., Hu Y., Smyth G. K., Alexander W. S., Hilton D. J., Blewitt M. E. (2010) Opposing roles of polycomb repressive complexes in hematopoietic stem and progenitor cells. Blood 116, 731–739 [DOI] [PubMed] [Google Scholar]
- 81. Cloos P. A., Christensen J., Agger K., Helin K. (2008) Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev. 22, 1115–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. De Santa F., Totaro M. G., Prosperini E., Notarbartolo S., Testa G., Natoli G. (2007) The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 130, 1083–1094 [DOI] [PubMed] [Google Scholar]