Abstract
We report the molecular findings for the CTNS gene in 12 Turkish cystinosis patients aged 7–29 years. All presented initially with severe failure to thrive, polyuria, and polydipsia. Cystinosis was diagnosed at age 1 month to 9 years. Seven patients reached end-stage renal failure at ages ranging from 6.5 to 15 years. Whereas three of the remaining five have renal Fanconi syndrome with proteinuria, two have had kidney failure of varying degrees. Molecular analyses involved an initial multiplex polymerase chain reaction (PCR) to determine the presence or absence of the 57-kb northern European founder deletion in CTNS, followed by sequencing of the ten coding exons of CTNS. Comprehensive mutation analysis verified that none of the 12 patients carried the common 57-kb deletion. We identified four previously reported nucleotide variations associated with cystinosis and five new variants: a 10-kb deletion, three missense variants, and a nucleotide substitution in a potential branch point site of intron 4. This study is the first molecular analysis of Turkish cystinosis patients and provides guidance for the molecular diagnosis of cystinosis in this population.
Keywords: Cystinosis, CTNS gene, Turkish patients, Genetic basis, New mutations
Introduction
Cystinosis (CTNS; OMIM 219800) is an autosomal recessive disease caused by mutations of the CTNS gene (OMIM 606272; GenBank NM_004937.2) located on chromosome 17p13. CTNS codes for cystinosin, a protein that facilitates cystine egress from lysosomes. Defective lysosomal transport leads to widespread accumulation and crystallization of cystine in many organs, particularly the kidney, cornea, bone marrow, thyroid, lymph nodes, liver, and spleen [1–3]. The most common and severe phenotype is infantile nephropathic cystinosis in which renal signs and symptoms predominate; patients develop renal Fanconi syndrome in infancy and renal failure in the first decade. Other organs frequently affected are the cornea, causing painful photophobia, and the thyroid, associated with hypothyroidism. Myopathy, swallowing, pulmonary dysfunction, diabetes, male hypogonadism, and central nervous system involvement are seen in adolescents and young adults with nephropathic cystinosis [1, 4]. A milder cystinosis phenotype presents in adolescence with more slowly progressive renal failure and ocular disease, and an adult form (i.e., ocular) presents with only corneal disease [1, 5].
Management and treatment of infantile nephropathic cystinosis includes supportive treatment to maintain fluid and electrolyte balance in infancy and early childhood. Early diagnosis and specific treatment with orally administered cysteamine delays the onset of renal failure and prevents the occurrence of other serious organ involvement [6, 7]. Cysteamine eye drops may lessen visual symptoms by dissolving the corneal cystine crystals. Hemodialysis (HD), continuous ambulatory peritoneal dialysis (CAPD), and renal transplantation (Tx) are associated with increased survival rates in patients who progressed to end-stage renal failure (ESRF). Cystine accumulation has been reported in transplanted kidneys but with no clinical manifestations, as it reflects invasion of host cells containing cystine. However, after renal Tx, extrarenal manifestations of cystinosis continue to occur [1].
The CTNS gene consists of 12 exons; numbers 3–12 are coding exons. CTNS mutations result in either complete abolition of, or reduced cystine transport [1, 5]. Many different mutations have been identified throughout the entire gene in European and American cystinosis patients [8–18]. The most common mutation is a large 57-kb deletion [11, 13–16]. Here we report CTNS mutation analysis of 12 Turkish patients with cystinosis, representing the first such molecular diagnosis of cystinosis in this population.
Materials and methods
Patients
We studied 12 Turkish patients who were diagnosed with cystinosis and followed at Hacettepe University Faculty of Medicine Children’s Hospital Department of Pediatric Nephrology and Metabolic Diseases Unit. Informed consent was obtained from the patients and/or their parents. Blood samples were collected from all patients; DNA was extracted from leukocytes, as previously described [19].
Molecular analysis
The 57-kb deletion was tested using the method previously described [20]. Primers were designed to amplify the ten coding exons of CTNS, including their intronic boundaries (primer sequences available on request). Standard polymerase chain reaction (PCR) amplification procedures were employed. All PCR amplified products were directly sequenced using the Big Dye 3 Terminator chemistry (Applied Biosystems) employing an ABI 3130xl genetic analyzer (Applied Biosystems). Data were analyzed using Sequencher 4.8 (Gene Codes Corporation).
Mutation prediction analysis
The effect of missense variants and potential splice-site variants was evaluated, as described previously [21].
Results
Twelve patients (seven male; five female) from ten families were evaluated. Patients’ ages ranged from 7 to 29 years; age at diagnosis was 1 month to 9 years. Consanguinity was reported for six families, and in two families there were two affected siblings. All patients were put on cysteamine bitartrate treatment at diagnosis. Six patients reached ESRF; one is receiving PD, three are on HD, and two have undergone renal Tx. Of the remaining six patients, three had varying degree of renal failure and three had proteinuria at the time of the study (Table 1). Before sequencing, all patients were tested for the 57-kb deletion and it was not found in any of them. Analysis of the ten coding exons of CTNS revealed the presence of nucleotide variations in all patients. Thirteen different variants (Table 1) were identified in the 12 patients; four were already referenced in the dbSNP (http://www.ncbi.nlm.nih.gov/snp/) (rs161400, rs1800528, rs222753, and rs76153698). The others were one deletion (10 kb), one small deletion (4 bp), four missense variants, two potential splice-site modifications, and a nucleotide substitution in a potential branch-point site of intron 4.
Table 1.
Clinical data and CTNS variantsa of the patients
| Patient | Age (years) | Sex | Age at diagnosis (months) | Consanguinity | Sibling disease | Renal status | RRT | CTNS variant allele 1 | CTNS variant allele 2 | Other |
|---|---|---|---|---|---|---|---|---|---|---|
| TR1 | 12 | M | 15 | No | No | Proteinuria | c.451A>G;p.R151G | c.1015 G>A;p.G339R | rs222753 (h) | |
| TR2 | 11 | M | 9 | Yes | No | Proteinuria | c.18_21delGACT;p.T7X7 | c.18_21delGACT; p.T7FX7 | rs222753 (H) | |
| TR3 | 16 | M | 12 | No | No | ESRD at age 9 | CAPD, HDTx, HD | c.62-1083_c.551del10217 | c.62-1083_c.551del10217 | rs161400 (H) |
| TR4 | 21.5 | M | 12 | Yes | Yes | ESRD at age 9.5 | CAPD, HD | c.141-22a>g | c.681 G>A; p.E227E | rs1800528 (h), rs222753(h) |
| TR5 | 17.5 | M | 1 | Yes | Sib of TR4 | RI | c.141-22a>g | c.681 G>A; p.E227E | rs1800528 (h), rs222753(h) | |
| TR6 | 15.5 | F | 12 | Yes | No | Proteinuria | c.518A>G, p.Y173C | c.518A>G, p.Y173C | ||
| TR7 | 25.5 | M | 105 | Yes | No | RI | c.451A>G; p.R151G | c.451A>G; p.R151G | ||
| TR36 | 14 | F | 14 | No | No | ESRD at age 6 | CAPD, HD, TX | c.681 G>A; p.E227E | c.1015 G>A; p.G339R | rs222753 (h) |
| TR37 | 29 | F | 106 | Yes | No | ESRD at age 16 | CAPD | c.18_21delGACT;p.T7X7 | c.470 G>A; p.G157D | rs222753 (h) |
| TR121 | 20.5 | M | 13 | No | Only child | ESRD at age 11 | CAPD, HD | c.140+1 G>T | c.140+1 G>T | rs76153698 (H) |
| TR11 | 13 | F | 18 | Yes | Yes | ESRD at age 9 | Preemptive Tx | c.681 G>A; p.E227E | c.681 G>A; p.E227E | rs222753 (H) |
| TR12 | 7 | F | 24 | Yes | Sib of TR 12 | RI | c.681 G>A; p.E227E | c.681 G>A; p.E227E | rs222753 (H) |
CAPD continuous ambulatory peritoneal dialysis, ESRD end-stage renal disease, HD hemodialysis, RI renal insufficiency, Tx transplantation, H homozygote, h heterozygote, RRT renal replacement therapy
Position based on the GenBank NM_004937.2 sequence (HGVS nomenclature). The “rs” numbers correspond to dbSNP (http://www.ncbi.nlm.nih.gov/snp/) accession numbers
Newly identified missense variations c.451A>G, c.518A>G, and c.470 G>A were searched in 100 healthy Turkish controls by direct sequencing; none of these three variants was found in healthy Turkish controls (Figs. 1–3). We also demonstrated segregation in two families in which DNA was available (Figs. 2 and 3).
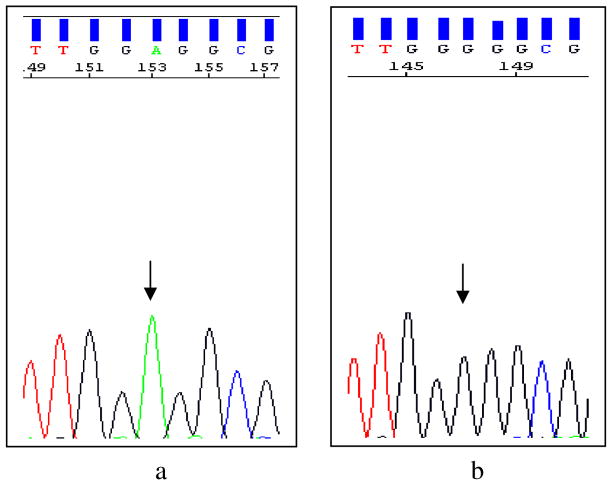
Fig. 1.
Sequence electropherograms of exon 5 of the CTNS gene for c.451 A>G variant. The wild-type sequence in a healthy control is shown in the left electropherogram (a, arrow); the affected individual has a homozygous c.451 A>G variation (b, arrow)
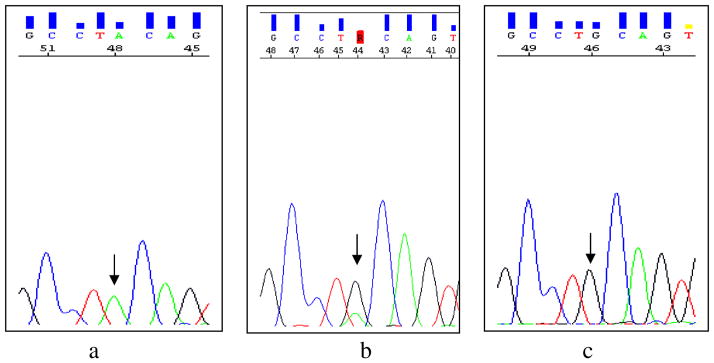
Fig. 3.
Sequence electropherograms of exon 6 of the CTNS gene for c.470 G>A variation. The wild-type sequence in a healthy control is shown in the left electropherogram (a, arrow); the father is heterozygous (b, arrow); the affected individual has a heterozygous c.470 G>A variation (c, arrow)
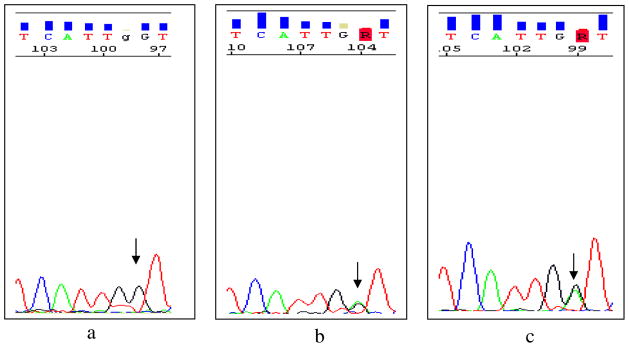
Fig. 2.
Sequence electropherograms of exon 6 of the CTNS gene for c.518A>G variation. The wild-type sequence in a healthy control is shown in the left electropherogram (a, arrow). Parents are heterozygous (b, arrow); the affected individual has a homozygous c.518A>G variation (c, arrow)
The potential effects of the nucleotide changes on cystinosin are listed in Table 2.
Table 2.
Predicted effects of CTNS variantsa
| Variantb | Prediction | Reference | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Deletions | |||||||||
| c.18_21delGACT; p.T7FX7 | Frameshift | [8] | |||||||
| c.62-1083_551del10217bp | Deletion from intron 3 to exon 8 | This study | |||||||
| Missense variants | Blossum 62 | PolyPhen
|
Panther
|
PMut
|
SIFT | ||||
| PSIC score | PHAT score | Prediction based | subPSEC | NN output | Reliability | ||||
| c.451A>G; p.R151G | −2 | 2.328 | NA | Align | −4.49143 | 0.7813 | 5 | Not Tolerated | This study |
| c.518A>G; p.Y173C | −2 | 3.048 | −1 | Annot | −6.882 | 0.6686 | 3 | Not Tolerated | This study |
| c.470 G>A; p.G157D | −2 | 2.267 | NA | Align | −5.63012 | 0.6911 | 3 | Not Tolerated | This study |
| c.779 C>T; pT260I | −1 | 1.072 | NA | Align | −2.13647 | 0.6573 | 3 | Tolerated | rs161400 |
| c.1015 G>A; p.G339R | −2 | 2.285 | −5 | Annot | −5.3956 | 0.8416 | 6 | Not Tolerated | [9] |
| Splicing variants | BDGP
|
NetGene2
|
|||||||
| Score | NN score | Confidence | |||||||
| c.140+1 G>T | 1>0 | 0.998>0 | 1>0 | [8] | |||||
| c.504 G>A | No effect | No effect | No effect | rs1800528 | |||||
| c.681 G>A | 0.99>0 | 0.992>0.845 | 1>0.904 | [24] | |||||
| c.970+15 G>A | No effect | No effect | No effect | rs76153698 | |||||
| c.970+70 C>T | No effect | No effect | No effect | rs222753 | |||||
Abbreviations: Align, Prediction based on the alignment; Annot, Prediction based on the gene annotation; BDGP, Berkeley Drosophila Gerome Project; NA, Not Available; NN, Neuronal Network; PHAT, Predicted Hydrophobic And Transmembrane; PSIC, Position-Specific Independent Court; subPSEC, substitution Position-Specific Evolutionary Conservation
Position based on the GenBank NM004937.2 sequence (HGVS nomenclature).The “rs” number corresponds to the dbSNP accession URLs:
dbSNP: http://www.ncbi.nlm.nih.gov/snp/)
NetGene2: http://www.cbs.dtu.dk/services/NetGene2/
PolyPhen: http://coot.embl.de/PolyPhen/
Discussion
The diagnosis of cystinosis is confirmed by demonstrating markedly elevated cystine levels in polymorphonuclear leucocytes [1, 22]. Detection of corneal crystals by slit-lamp examination is virtually diagnostic of cystinosis in patients with Fanconi syndrome. However, corneal crystals may not develop until 1–1.5 years of age, so this finding is not sensitive for early diagnosis that, along with diligent compliance with cystine-depleting treatment, determines prognosis. Another determinant is genetic predisposition. Infantile nephropathic cystinosis is associated with two severe CTNS mutations, including deletions, insertions, nonsense, missense, and splicing mutations, that cause complete abolition of cystine transport. The adolescent and ocular forms generally have one severe and one mild mutation, leading to reduced cystine transport [1, 5, 23]. Molecular studies have revealed that 50–60% of northern European and North American patients with cystinosis have a common 57-kb deletion that affects the first ten exons of the CTNS gene, consistent with a founder effect [11, 13–16]. On the other hand, the 57-kb deletion has been detected with a much lower frequency in Italians, French Canadians, and Mexicans [12, 18, 20]; the carrier rate was only 17% in Italians. HGMD (http://www.hgmd.cf.ac.uk/ac/index.php) describes approximately 100 mutations in the CTNS gene.
This report represents the first description of the genetic basis of cystinosis in Turkish patients. Our data reveal that the Turkish cystinosis population has novel genetic characteristics. Specifically, we did not detect the northern European 57-kb deletion in any Turkish patient. Of 13 different variants identified, five were novel, such as three missense variants [c.518A>G (p.Y173C), c.451A>G (p.R151G), c.470 G>A (p.G157D)], a new gross deletion (c.62-1083_551del10217bp), removing nucleotides between intron 3 and exon 8, and the sequence variation in a potential consensus branch point sequence (c.141-22a>g). Although no functional studies have been performed, the amino-acid changes p.Y173C, p.R151G, and p.G157D are very likely to be causative mutations based on formal criteria, evolutionary conservation, and absence in healthy controls, as well as prediction software (Figs. 1– 3 and Table 2). The sequence variation c.141-22a>g may disturb a consensus sequence for a branch point and interfere with correct or efficient splicing [24]. Regular software to predict efficiency of RNA splicing does not exist for branch sites to our knowledge. Confirmation that this variant has an effect on splicing analysis on the RNA level is required. [25].
Of the eight previously reported variants, four had been associated with cystinosis [8–10, 26] and four were referenced in the dbSNP. The variant c.140+1 G>T has been described as 479+1 G>T according to old nomenclature; it affects a canonical splice site and results in loss of cystinosin function [8]. The variant c.1015 G>A (p.G339R) abolishes cystine transport in an in vitro functional model [9, 10]. The variant 4-bp deletion c.18_21del GACT (p.T7FX7) was described by Town et al. [8] and reported in different populations with cystinosis. The variant c.681 G>A (p.E227E), described in an Arabic population, is located in an exon–intron boundary and disturbs normal splicing [26].
In our study, the most common variants identified were rs222753 at around 45%, as described in the dbSNP; and c.681 G>A; pE227E at around 29%, as a disease-causing variant [26]. Interestingly, the consanguinity of TR2, TR3, TR6, TR7, TR11, and TR12 is supported by the presence of homozygous variants. However, for patients TR4, TR5, and TR37, heterozygous variants are present despite parental consanguinity. Clinically, the patient homozygous for the 10-kb deletion (TR3) followed a severe course and progressed to ESRF at 10 years of age. Patients TR6 and TR7, homozygous for the new missense variants c.518A>G (p.Y173C) and c.451A>G (p.R151G), both had moderate disease courses, reaching adolescence with proteinuria but no renal failure. Patient TR121, homozygous for the known variant c.140+1 G>T that affects a canonical splice site, had a severe disease course [8]. The siblings (TR11 and TR12) homozygous for c.681 G>A (pE227E) exhibited a severe disease course but were the most noncompliant patients, which might be related to disease course. TR1, compound heterozygous for a new missense variant c.451A>G (p.R151G) and a known missense variant c.1015 G>A (p.G339R), had a mild disease course, with good compliance and largely intact renal function at age 12. On the other hand, TR36, compound heterozygous for the c.681 G>A (p.E227E) and the same known missense variant (c.1015 G>A; p.G339R), is noncompliant and has experienced severe disease, with ESRF at approximately 10 years of age; she was recently transplanted with her mother’s kidney.
The discrepancy in disease course in patients TR2 and TR 37 could be due to the different variants in the second allele as well as compliance to cysteamine treatment. TR2, homozygous for a known 4-bp deletion c.18_21delGACT; p.T7FX7, has severe disease with liver involvement. This deletion has been found in different populations and with widely variable disease severities [8, 9]. TR37 was compound heterozygous for a new missense mutation c. 470 G>A (p.G157D) inherited from her father (Fig. 3b) and a known deletion c.18_21delGACT (p.T7FX7) inherited from her mother. The patient had a fairly good course but progressed to end-stage renal disease (ESRD) at 16 years. Siblings TR4 and TR5 were compound heterozygous for a known splice-site variant c.681 G>A (p.E227E) and a new variant c.141-22a>g. These siblings had a completely different courses. Although TR4 was diagnosed at age 13 months, he was lost to follow-up for a long period and progressed to ESRD at age 9.5 years. On the other hand, TR5 had adequate management and follow-up and did not required renal replacement treatment as of age 17.5 years. These observations confirm once again the key role of early diagnosis and adequate cysteamine treatment in the course of cystinosis. From the genetic point of view, our findings support previous data on genotype–phenotype correlations of cystinosis patients [1, 5, 23].
In summary, we show that Turkish cystinosis patients have different disease-causing variants of CTNS compared with European and North American patients. In light of our results, it seems that toward to Mediterranean region, the frequency of the 57-kb deletion decreases. In northern Europe and North American studies, the frequency approximates 50–70%, whereas it decreases to 17% in Italy [18] and is absent in this Turkish cohort. Our data once more confirm the wide mutational spectrum of the CTNS gene. Data provide a basis for further molecular diagnosis in the Turkish population.
Acknowledgments
We gratefully acknowledge Prof. Dr. Elena Levtchenko for her advice and help in completing the genetic analysis and the critical reading of the manuscript. Hacettepe University Nephrogenetics Laboratory was established by Hacettepe University Infrastructure Project (Project No: 06A101008)
Contributor Information
Rezan Topaloglu, Email: rezantopaloglu@hacettepe.edu.tr, Dept of Pediatric Nephrology, Hacettepe University Faculty of Medicine, Ankara 06100, Turkey.
Thierry Vilboux, NIH, NHGRI, MGB, Bldg. 10, Rm. 10 C107, 10 Center Drive, Bethesda, MD 20892, USA.
Turgay Coskun, Department of Pediatric Metabolic Diseases, Hacettepe University Faculty of Medicine, Ankara 06100, Turkey.
Fatih Ozaltin, Dept of Pediatric Nephrology, Hacettepe University Faculty of Medicine, Ankara 06100, Turkey.
Brad Tinloy, NIH, NHGRI, MGB, Bldg. 10, Rm. 10 C107, 10 Center Drive, Bethesda, MD 20892, USA.
Meral Gunay-Aygun, NIH, NHGRI, MGB, Bldg. 10, Rm. 10 C107, 10 Center Drive, Bethesda, MD 20892, USA.
Aysin Bakkaloglu, Dept of Pediatric Nephrology, Hacettepe University Faculty of Medicine, Ankara 06100, Turkey.
Nesrin Besbas, Dept of Pediatric Nephrology, Hacettepe University Faculty of Medicine, Ankara 06100, Turkey.
Lambert van den Heuvel, Department of Pediatrics Academic Hospital Leuven, Herestraat 49, 3000 Leuven, Belgium.
Robert Kleta, NIH, NHGRI, MGB, Bldg. 10, Rm. 10 C107, 10 Center Drive, Bethesda, MD 20892, USA.
William A. Gahl, NIH, NHGRI, MGB, Bldg. 10, Rm. 10 C107, 10 Center Drive, Bethesda, MD 20892, USA
References
- 1.Gahl WA, Thoene JG, Schneider JA. Cystinosis. N Engl J Med. 2002;347:111–121. doi: 10.1056/NEJMra020552. [DOI] [PubMed] [Google Scholar]
- 2.Schneider JA, Rosenbloom FM, Bradley KH, Seegmiller JE. Increased free-cystine content of fibroblasts cultured from patients with cystinosis. Biochem Biophys Res Commun. 1967;29:527–531. doi: 10.1016/0006-291x(67)90516-5. [DOI] [PubMed] [Google Scholar]
- 3.Gahl WA, Bashan N, Tietze F, Bernardini I, Schulman JD. Cystine transport is defective in isolated leukocyte lysosomes from patients with cystinosis. Science. 1982;217:1263–1265. doi: 10.1126/science.7112129. [DOI] [PubMed] [Google Scholar]
- 4.Sonies BC, Almajid P, Kleta R, Bernardini I, Gahl WA. Swallowing dysfunction in 101 patients with nephropathic cystinosis: benefit of long-term cysteamine therapy. Medicine (Baltimore) 2005;84:137–146. doi: 10.1097/01.md.0000164204.00159.d4. [DOI] [PubMed] [Google Scholar]
- 5.Kalatzis V, Antignac C. New aspects of the pathogenesis of cystinosis. Pediatr Nephrol. 2003;18:207–215. doi: 10.1007/s00467-003-1077-5. [DOI] [PubMed] [Google Scholar]
- 6.Gahl WA, Balog JZ, Kleta R. Nephropathic cystinosis in adults: natural history and effects of oral cysteamine therapy. Ann Intern Med. 2007;147:242–250. doi: 10.7326/0003-4819-147-4-200708210-00006. [DOI] [PubMed] [Google Scholar]
- 7.Gahl WA, Reed GF, Thoene JG, Schulman JD, Rizzo WB, Jonas AJ, Denman DW, Schlesselman JJ, Corden BJ, Schneider JA. Cysteamine therapy for children with nephropathic cystinosis. N Engl J Med. 1987;316:971–977. doi: 10.1056/NEJM198704163161602. [DOI] [PubMed] [Google Scholar]
- 8.Town M, Jean G, Cherqui S, Attard M, Forestier L, Whitmore SA, Callen DF, Gribouval O, Broyer M, Bates GP, van’t Hoff W, Antignac C. A novel gene encoding an integral membrane protein is mutated in nephropathic cystinosis. Nat Genet. 1998;18:319–324. doi: 10.1038/ng0498-319. [DOI] [PubMed] [Google Scholar]
- 9.Shotelersuk V, Larson D, Anikster Y, McDowell G, Lemons R, Bernardini I, Guo J, Thoene J, Gahl WA. CTNS mutations in an American-based population of cystinosis patients. Am J Hum Genet. 1998;63:1352–1362. doi: 10.1086/302118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaletzis V, Nevo N, Cherqui S, Gasnier B, Antignac C. Molecular pathogenesis of cystinosis: effect of CTNS mutations on the transport activity and subcellular localization of cystinosin. Hum Mol Genet. 2004;13:1361–1371. doi: 10.1093/hmg/ddh152. [DOI] [PubMed] [Google Scholar]
- 11.Anikster Y, Shotelersuk V, Gahl WA. CTNS mutations in patients with cystinosis. Hum Mutat. 1999;14:454–458. doi: 10.1002/(SICI)1098-1004(199912)14:6<454::AID-HUMU2>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 12.McGowan-Jordan J, Stoddard K, Podolsky L, Orrbine E, McLaine P, Town M, Goodyer P, MacKenzie A, Heick H. Molecular analysis of cystinosis: probable Irish origin of the most common French Canadian mutation. Eur J Hum Genet. 1999;7:671–678. doi: 10.1038/sj.ejhg.5200349. [DOI] [PubMed] [Google Scholar]
- 13.Heil SG, Levtchenko E, Monnens LA, Trijbels FJ, Van der Put NM, Blom HJ. The molecular basis of Dutch infantile nephropathic cystinosis. Nephron. 2001;89:50–55. doi: 10.1159/000046043. [DOI] [PubMed] [Google Scholar]
- 14.Kalatzis V, Cherqui S, Jean G, Cordier B, Cochat P, Broyer M, Antignac C. Characterization of a putative founder mutation that accounts for the high incidence of cystinosis in Brittany. J Am Soc Nephrol. 2001;12:2170–2174. doi: 10.1681/ASN.V12102170. [DOI] [PubMed] [Google Scholar]
- 15.Kiehntopf M, Schickel J, Gonne B, Koch HG, Superti-Furga A, Steinmann B, Deufel T, Harms E. Analysis of the CTNS gene in patients of German and Swiss origin with nephropathic cystinosis. Hum Mutat. 2002;20:237. doi: 10.1002/humu.9063. [DOI] [PubMed] [Google Scholar]
- 16.Kleta R, Anikster Y, Lucero C, Shotelersuk V, Huizing M, Bernardini I, Park M, Thoene J, Schneider J, Gahl WA. CTNS mutations in African American patients with cystinosis. Mol Genet Metab. 2001;74:332–337. doi: 10.1006/mgme.2001.3218. [DOI] [PubMed] [Google Scholar]
- 17.Kalatzis V, Cohen-Solal L, Cordier B, Frishberg Y, Kemper M, Nuutinen EM, Legrand E, Cochat P, Antignac C. Identification of 14 novel CTNS mutations and characterization of seven splice site mutations associated with cystinosis. Hum Mutat. 2002;20:439–446. doi: 10.1002/humu.10141. [DOI] [PubMed] [Google Scholar]
- 18.Mason S, Pepe G, Dall’Amico R, Tartaglia S, Casciani S, Greco M, Bencivenga P, Murer L, Rizzoni G, Tenconi R, Clementi M. Mutational spectrum of the CTNS gene in Italy. Eur J Hum Genet. 2003;11:503–508. doi: 10.1038/sj.ejhg.5200993. [DOI] [PubMed] [Google Scholar]
- 19.Huizing M, Anikster Y, Fitzpatrick DL, Jeong AB, D’Souza M, Rausche M, Toro JR, Kaiser-Kupfer MI, White JG, Gahl WA. Hermansky-Pudlak syndrome type 3 in Ashkenazi Jews and other non-Puerto Rican patients with hypopigmentation and platelet storage-pool deficiency. Am J Hum Genet. 2001;69:1022–1032. doi: 10.1086/324168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anikster Y, Lucero C, Touchman JW, Huizing M, McDowell G, Shotelersuk V, Green ED, Gahl WA. Identification and detection of the common 65-kb deletion breakpoint in the nephropathic cystinosis gene (CTNS) Mol Genet Metab. 1999;66:111–116. doi: 10.1006/mgme.1998.2790. [DOI] [PubMed] [Google Scholar]
- 21.Vilboux T, Kayser M, Introne W, Suwannarat P, Bernardini I, Fischer R, O’Brien K, Kleta R, Huizing M, Gahl WA. Mutation spectrum of homogentisic acid oxidase (HGD) in alkaptonuria. Hum Mutat. 2009;30:1611–1619. doi: 10.1002/humu.21120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Attard M, Jean G, Forestier L, Cherqui S, van’t Hoff W, Broyer M, Antignac C, Town M. Severity of phenotype in cystinosis varies with mutations in the CTNS gene: predicted effect on the model of cystinosin. Hum Mol Genet. 1999;8:2507–2514. doi: 10.1093/hmg/8.13.2507. [DOI] [PubMed] [Google Scholar]
- 23.Alcantara-Ortigoza MA, Belmont-Martinez L, Vela-Amieva M, Gonzalez-Del Angel A. Analysis of the CTNS gene in nephropathic cystinosis Mexican patients: report of four novel mutations and identification of a false positive 57-kb deletion genotype with LDM-2/exon 4 multiplex PCR assay. Genet Test. 2008;12:409–414. doi: 10.1089/gte.2008.0014. [DOI] [PubMed] [Google Scholar]
- 24.Baralle D, Baralle M. Splicing in action: assessing disease causing sequence changes. J Med Genet. 2005;42:737–748. doi: 10.1136/jmg.2004.029538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taranta A, Wilmer MJ, van den Heuvel LP, Bencivengo F, Bellomo F, Levtchenko EN, Emma F. Analysis of CTNS gene transcripts in nephropathic cystinosis. Pediatr Nephrol. 2010;25:1263–1267. doi: 10.1007/s00467-010-1502-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aldahmesh MA, Humeidan A, Almojalli HA, Khan AO, Rajab M, AL-A AA, Meyer BF, Alkuraya FS. Characterization of CTNS mutations in Arab patients with cystinosis. Ophthalmic Genet. 2009;30:185–189. doi: 10.3109/13816810903200953. [DOI] [PubMed] [Google Scholar]