Abstract
The molecular diagnosis of mitochondrial disorders still remains elusive in a large proportion of patients, but advances in next generation sequencing are significantly improving our chances to detect mutations even in sporadic patients. Syndromes associated with mitochondrial DNA multiple deletions are caused by different molecular defects resulting in a wide spectrum of predominantly adult-onset clinical presentations, ranging from progressive external ophthalmoplegia to multi-systemic disorders of variable severity. The mutations underlying these conditions remain undisclosed in half of the affected subjects. We applied next-generation sequencing of known mitochondrial targets (MitoExome) to probands presenting with adult-onset mitochondrial myopathy and harbouring mitochondrial DNA multiple deletions in skeletal muscle. We identified autosomal recessive mutations in the DGUOK gene (encoding mitochondrial deoxyguanosine kinase), which has previously been associated with an infantile hepatocerebral form of mitochondrial DNA depletion. Mutations in DGUOK occurred in five independent subjects, representing 5.6% of our cohort of patients with mitochondrial DNA multiple deletions, and impaired both muscle DGUOK activity and protein stability. Clinical presentations were variable, including mitochondrial myopathy with or without progressive external ophthalmoplegia, recurrent rhabdomyolysis in a young female who had received a liver transplant at 9 months of age and adult-onset lower motor neuron syndrome with mild cognitive impairment. These findings reinforce the concept that mutations in genes involved in deoxyribonucleotide metabolism can cause diverse clinical phenotypes and suggest that DGUOK should be screened in patients harbouring mitochondrial DNA deletions in skeletal muscle.
Keywords: DGUOK, mitochondrial DNA instability, autosomal recessive progressive external ophthalmoplegia, multiple mitochondrial DNA deletions
Introduction
The replication of mitochondrial DNA relies on a set of proteins encoded by the nuclear genome (Wanrooij and Falkenberg, 2010). This is relevant for human health because mutations in genes encoding these factors have been described in familial and sporadic cases of autosomal mitochondrial diseases (Copeland, 2008). Further, mitochondrial disorders associated with mitochondrial DNA instability affect all stages of life.
A group of infantile or early-onset disorders, collectively termed mitochondrial DNA depletion syndromes, is characterized by a massive reduction of mitochondrial DNA content. Three main clinical presentations are known: myopathic, encephalomyopathic and hepatocerebral, depending on the tissues affected and their residual mitochondrial DNA levels (Rötig and Poulton, 2009).
The coexistence of mitochondrial DNA depletion and multiple deletions is a feature of mitochondrial neurogastrointestinal encephalopathy, which typically has juvenile onset; symptoms begin before age 20 years in ∼60% of patients (Hirano et al., 2004).
In adult life, the number of mitochondrial DNA multiple deletions in skeletal muscle increases with age because of defective or unreliable replication of mitochondrial genomes (Krishnan et al., 2008; Yu-Wai-Man and Chinnery, 2012). In patients with multiple mitochondrial DNA deletions, the first, as well as predominant symptom is progressive external ophthalmoplegia (Zeviani et al., 1989), but additional clinical features may include sensory axonal neuropathy, optic atrophy, ataxia, hypogonadism and parkinsonism (Suomalainen et al., 1992; Luoma et al., 2004).
The autosomal dominant form of progressive external ophthalmoplegia is genetically heterogeneous. Indeed, mutations have been disclosed in subunits of DNA polymerase gamma, POLG1 (Van Goethem et al., 2001) and POLG2 (Longley et al., 2006), the helicase Twinkle (PEO1, Spelbrink et al., 2001), the adenine nucleotide translocator ANT1 (SLC25A4, Kaukonen et al., 2000) and the protein p53R2, encoded by RRM2B gene and involved in the cytoplasmic pathway for de novo synthesis of deoxynucleotide triphosphates (dNTPs) (Tyynismaa et al., 2009). Multiple mitochondrial DNA deletions have also been observed in patients with autosomal dominant optic atrophy and multi-system clinical involvement (including progressive external ophthalmoplegia) because of defects in genes involved in the mitochondrial fusion pathway, such as OPA1 (Amati-Bonneau et al., 2008; Hudson et al., 2008) and MFN2 (Rouzier et al., 2012).
Conversely, the autosomal recessive form of progressive external ophthalmoplegia has only been associated with mutations in POLG1 and, recently, with mutations in TK2, which encodes the mitochondrial isoform of thymidine kinase that catalyses the rate-determining step of the pyrimidine salvage pathway (Tyynismaa et al., 2012).
Defects in the same gene might result in strikingly different manifestations. Indeed, mutations in RRM2B and TK2 have also been associated with encephalomyopathic or myopathic forms of mitochondrial DNA depletion syndrome (Saada et al., 2001; Galbiati et al., 2006; Bourdon et al., 2007). Similarly, recessive mutations in POLG1 (Isohanni et al., 2011) and PEO1 (Nikali et al., 2005; Hakonen et al., 2007) have been found in early-onset hepatocerebral forms of mitochondrial DNA depletion syndrome.
Molecular diagnosis is not achieved in a large proportion of patients with mitochondrial DNA instability (Virgilio et al., 2008; Spinazzola et al., 2009). Recent advances in next-generation sequencing hold the promise to overcome this limitation, and its application to mitochondrial disorders has already achieved genetic diagnoses in both familial and sporadic patients (Calvo et al., 2010, 2012).
Here, we have applied targeted next-generation sequencing of nuclear genes encoding either known or likely mitochondrial factors (MitoExome) in a cohort of adult-onset patients and found recessive mutations in DGUOK (Johansson and Karlsson, 1996) in three independent probands with cytochrome c oxidase (COX)-negative fibres and multiple mitochondrial DNA deletions in muscle. Mutational screening of DGUOK in a larger series of adult patients revealed pathogenic mutations in three more subjects. The effects of these mutations were investigated at a protein and biochemical level.
Mutated patients showed heterogeneous clinical presentations, including an uncommon adult-onset lower motor neuron disease with mild cognitive impairment.
Our study demonstrates the efficacy of MitoExome analysis in adult patients, expands the spectrum of disorders caused by mutations in DGUOK and suggests that DGUOK be screened in patients harbouring mitochondrial DNA multiple deletions.
Materials and methods
Patients
Clinical data are summarized in Table 1.
Table 1.
Clinical, histochemical and molecular data of affected subjects
| Patient | Sex | Family history | Age at exam | Age at onset | Primary neurological symptoms | Liver involvement | Additional clinical symptoms | Instrumental investigation | Muscle histology/histochemistry | mtDNA deletions (Southern blot) | mtDNA deletions (long-range PCR) | Allele 1 | Allele 2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | F | No | 69 | 58 | PEO | No | EMG: M | RRF, SDH (+), COX (−) | ++ | ++ | c.605_606delGA p.R202YfsX12 | c.462T>Abp.N154K | |
| 2 | F | No | 67 | 46 | PEO, ptosis, limb girdle weakness | No | Occasional dysphagia and cramps | EMG: M | COX (−), RRF | ++ | ++ | c.130 G>A p.E44K | c.462 T>A p.N154K |
| 3 | M | No | 80 | 69 | Lower limb girdle weakness, myalgia | HCC | Bilateral cataract, diabetes | EMG: M | COX (−), RRF | ++ | ++ | c.186 C>A p.Y62Xa | c.509 A>G p.Q170R |
| 4 | F | No | 23 | 20 | Lower limb weakness | Liver transplantation at 9 months | Rhabdomyolisis | EMG: M | COX (−), RRF | ++ | ++ | c.605_606delGA p.R202YfsX12 | c.137 A>G p.N46S |
| 5.1 | F | Yes | 48 | 40 | Limb weakness, dysphonia, dysphagia | No | Depression, dementia, ptosis, strabismus, sensorineural deafness | EMG: N BMRI: cortical-subcortical atrophy SPECT: decreased parietal-thalamus perfusion | RRF, SDH (+), COX (−), nuclear centralization | + | ++ | c.444-11 C>G | c.509 A>G p.Q170R |
| 5.2 | M | Yes | 46 | 44 | Asthenia, dysphonia, dysphagia | No | Sensorineural deafness, HIV positive, ptosis | EMG: N MRI: cortical atrophy Sural biopsy: A/D | SDH (+), COX (−) | + | ++ | c.444-11 C>G | c.509 A>G p.Q170R |
aNovel mutations.
A = axonal; BMRI = brain MRI; D = demyelinating; HCC = hepatocarcinoma; M = myogenic pattern; mtDNA = mitochondrial DNA; N = neurogenic pattern; PEO = progressive external ophthalmoplegia; SPECT = single-photon emission computed tomography; RRF = ragged-red fibres; EMG = electromyography; mtDNA = mitochondrial DNA; PCR = polymerase chain reaction.
Patient 1 is a 69-year-old woman who had suffered from progressive external ophthalmoplegia and ptosis for the past 11 years. Other skeletal muscles were unaffected. An electromyography (EMG) showed a myogenic pattern. Resting serum lactate was moderately increased, as was post-exercise creatine kinase. A sister was reported to suffer from exercise intolerance. Quadriceps muscle biopsy showed preserved structure but several COX-negative fibres, some of which were ragged-red.
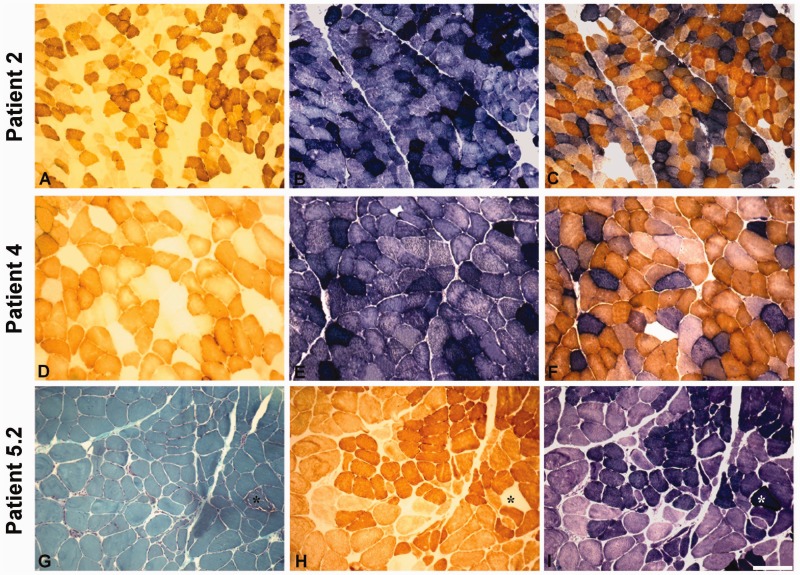
Patient 2 is a 72-year-old woman who had bilateral ptosis and progressive external ophthalmoplegia for >20 years and only in the later years started to complain of upper and lower limb girdle weakness with leg cramps. She also reported occasional dysphagia for liquids. Family history was negative for neuromuscular disorders. Neurological examination confirmed the presence of marked ptosis and ophthalmoplegia and disclosed limb girdle and proximal muscle weakness (Medical Research Council score between 3 and 4 in both upper and lower limbs). Serum creatine kinase levels were persistently increased (up to 2000 U/l), whereas both pyruvate and basal lactate were normal. EMG examination was myopathic in both proximal and distal muscles. A biopsy of the left biceps showed moderate fibre size variability, slight increase of endomysial connective tissue and several COX-negative ragged-red fibres (Fig. 1D–F).
Figure 1.
Histological and histochemical findings in muscle biopsies from Patients 2, 4 and 5.2. COX (A, D and H), SDH (B, E and I) and combined COX–SDH (C and F) histochemistry show severe and diffuse COX deficiency with evidence of mitochondrial proliferation in Patients 2 (A–C), 4 (D–F) and 5.2 (H and I). In Patient 5.2, Gomori thrichrome (G) histology and histochemistry (H and I) show one ragged-red fibre (asterisk) within an otherwise conserved muscle structure. Scale bar = 100 µm.
Patient 3 is an 80-year-old man who first sought medical attention at the age of 69 years because of exercise-induced muscle pain and lower limb girdle muscle weakness. He further presented non-insulin dependent diabetes mellitus and bilateral cataract. Blood examination showed mild increase of liver enzymes and creatine kinase levels (257 U/l). Electromyography disclosed a myogenic pattern with some lumbar neurogenic signs. A biceps brachii muscle biopsy showed focal COX deficiency and some ragged-red fibres. At age 78 years, he underwent surgery for the removal of a large hepatocarcinoma.
Patient 4 is a 23-year-old woman born after normal pregnancy and delivery, with negative family history. At 18 months of life, she presented with sub-icterus and hypoglycaemia, soon followed by ascites and hepatosplenomegaly. Liver insufficiency led to liver transplantation at 9 months of age. Histology of the affected liver did not disclose the cause of liver failure; the patient was then started on immunosuppressant therapy and developed normally. She was later diagnosed with mitochondrial disease at the age of 20 years, when a massive increase in serum creatine kinase (13 000 U/l) was observed during an episode of gastroenteritis associated with leg weakness. This presumably viral gastroenteritis resolved in a week with normalization of the creatine kinase level. Four months later, the patient had another episode of rhabdomyolisis. Neurological examination showed decreased muscle strength with scapulo-peroneal distribution. The EMG was myopathic. A muscle biopsy showed moderate myopathic signs (some hypotrophic fibres, rare central nuclei and splittings) associated with severe COX deficiency and the presence of many ragged-red fibres. (Fig. 1A–C).
Patient 5.1, a 48-year-old woman, had an 8-year history of slowly progressive, predominantly distal, upper and lower limb muscle weakness, mild dysphonia and dysphagia. Her mother had died at the age of 59 in a hospice after suffering for several years from an undefined neurological disorder (clinical records were not available). At the age of 46 years, our patient presented a major depressive episode, requiring hospitalization. When seen at age 48 years, neuropsychological evaluation demonstrated reduced performances with naming test and decreased short- and long-term spatial and verbal memory. Mini-Mental Status Examination was 23/30. She had mild bilateral ptosis with right convergent strabismus, dysphagia for solids, mild bilateral weakness of the trapezius and sterno-cleidomastoid muscles and tongue hypotrophy. Both proximal and distal limb muscles were wasted and showed diffuse fasciculations. Medical Research Council strength evaluation was as follows: biceps brachii: 3, triceps: 3, hand opponens: 3 and hand interossei: 4. The patient could not walk without support, climb stairs or stand on her heels or toes. Deep tendon reflexes were absent, and superficial and deep somatic sensations were normal.
At blood examination, liver function was normal, whereas serum creatine kinase levels were slightly increased (196 IU; normal values <180). Electrocardiography (EKG) was normal. An EMG study showed marked neurogenic signs in all four limb muscles, together with fibrillation and fasciculation potentials, with exception of cervical dorsal and lumbar paravertebral muscles. Sensory action potentials of the ulnar, median and sural nerves were normal, whereas nerve conduction velocities of the peroneus, ulnar, median and tibialis nerves were moderately decreased. Electroencephalography (EEG) and citrate synthase analysis were also normal. Brain Magnetic Resonance Imaging (MRI) showed cortico-subcortical atrophy. A Tc99m-HMPAO (Tc-hexamethylpropyleneamine oxime) cerebral single-photon emission computed tomography showed mild decrease of perfusion of the parietal cortex, anterior cingulus and left thalamus. A biopsy of the biceps brachii muscle showed variability of fibre size, fibre splitting, central nuclei, few succinate dehydrogenase (SDH)-intensely positive fibres (‘ragged-blue’ fibres) and scattered focal COX deficiency.
Patient 5.2 is the 46-year-old brother of Patient 5.1. He had noted weight loss since age 44 and came to medical attention after his sister was investigated. He complained of general asthenia, mild dysphonia and dysphagia. An EMG at age 45 years showed neurogenic signs and active denervation in distal upper and lower limb muscles. Serum creatine kinase was moderately increased (235 UI/l).
The propositus had had several episodes of abdominal pain in the past 2 years. Neurological examination showed mild bilateral ptosis, decreased muscle bulk of hand interossei and bilateral lower limb muscle hypotrophy with predominant proximal weakness. Tendon reflexes were ubiquitously absent, and there was mild bilateral dysmetria. The patient tested positive for HIV1 infection. Citrate synthase analysis showed increased protein (72 mg/dl) and Link’s index; several oligoclonal bands were also observed. The EEG was unremarkable. Brain MRI showed mild cortical atrophy. Audiometry disclosed bilateral sensorineural hypoacusia. Abdominal echography showed normal liver size but moderately dyshomogeneous liver structure. Electroneurography revealed decreased nerve conduction velocities of right median and peroneal muscles and decreased sensory action potential amplitude of sural and median nerves. A triceps brachii muscle biopsy showed a severe neurogenic pattern with scattered SDH hyperintense COX-negative fibres (Fig. 1G–I). Sural nerve biopsy documented severe axonal neuropathy. Fibre density was decreased: 2538 fibres/mm2 (normal values: 5600 ± 900); teasing analysis showed total demyelination in 22% of fibres.
Histological studies of muscle
Muscle samples were frozen in cooled isopentane and stored in liquid nitrogen for histological and histochemical analyses, including modified Gomori Trichrome staining, COX activity, SDH activity and double COX/SDH staining according to standard protocols (Sciacco et al., 2001).
Mitochondrial DNA molecular analysis
Total DNA was extracted from muscle using standard phenol–chloroform procedure. Long-range PCR and Southern blot analysis were performed as previously described (Zeviani et al., 1990). Mitochondrial DNA quantification in muscle was performed by real-time quantitative PCR as previously described (Spinazzola et al., 2006), using primers and probes for the human cytb gene (mitochondrial DNA) and the APP gene (nuclear DNA). All determinations were carried out in triplicate using 25 ng of total DNA as a template. Primer sequences and PCR conditions are available on request. Levels of mitochondrial DNA were normalized to nuclear DNA and expressed in terms of relative quantification values, using as reference (relative quantification = 1) the amount of mitochondrial DNA detected in skeletal muscle from a pool of healthy age-matched controls. Results were analysed with the Student’s t-test.
MitoExome sequencing
We used an in-solution hybridization capture method (Gnirke et al., 2009) to isolate target DNA, which was sequenced on the Illumina GA-II platform (Bentley et al., 2008). The 4.1 Mb of targeted DNA included the 16 kb mitochondrial DNA and all coding and untranslated exons of 1381 nuclear genes, including 1013 mitochondrial genes from the MitoCarta database (Pagliarini et al., 2008), 21 genes with recent strong evidence of mitochondrial association and 347 additional genes. The sequencing was performed with a pair-end approach and an average coverage of 142×. All analyses were restricted to the mitochondrial DNA and coding exons of the 1034 genes with confident evidence of mitochondrial association (1.4 Mb), including those previously involved in paediatric and adult-onset presentations showing defective mitochondrial DNA maintenance (Calvo et al., 2012).
Sequence analysis, variant detection and variant prioritization were performed as previously described (Calvo et al., 2012).
Estimates of variant allele frequency were obtained from the Exome Variant Server (National Heart, Lung, and Blood Institute Exome Sequencing Project, http://evs.gs.washington.edu/EVS/) and by analysing available whole-exome data from 371 healthy individuals of European ancestry obtained with permission from the National Institute of Mental Health (NIMH) control sample collection.
Sequencing of nuclear genes
The coding regions of POLG1 (NM_002693.2), SLC25A4 (ANT1) (NM_001151.3), PEO1 (Twinkle) (NM_021830.3) and POLG2 (NM_007215.3) genes were sequenced as previously described (Virgilio et al., 2008). Standard Sanger sequencing ruled out OPA1 (NM_130833.1), RRM2B (NM_015713.3), TK2 (NM_004614.3) and MFN2 (NM_014874.3) genes in selected patients. For the analysis of DGUOK (NM_080916.2), all exons were amplified with intronic primers as described in Mandel et al. (2001).
PCR products were purified with ExoSAP-IT enzyme (USB), processed with a Big Dye® Terminator Sequencing protocol (Applied Biosystems) and electrophoresed on an ABI 3100 automated sequencer (Applied Biosystems).
Transcript analysis
Muscle from Patient 5.1 and unaffected controls was homogenized, and total RNA was extracted using the Eurozol protocol (Euroclone). First-strand complementary DNA was synthesized from total RNA with random hexamer primers using the First-Strand cDNA Synthesis kit (GE Healthcare). PCR was performed on muscle complementary DNA using primers that amplify the complete coding region of the DGUOK transcript and primers that amplify part of the transcript, from exon 3 to 6. Primer sequences are available on request. The resulting PCR products were sequenced to detect aberrant transcripts and to determine the number of transcribed alleles based on the presence of the c.509 A>G variant.
Protein analysis
Protein aliquots (40 μg) obtained from skeletal muscle lysates were separated on 12% polyacrylamide gel and blotted to nitrocellulose membrane (Whatman). Membranes were decorated with a monoclonal anti-DGUOK antibody (Sigma) and detected with secondary antibody and ECL plus chemiluminescent detection system (Amersham Biosciences). Quantitative data were normalized to the signal obtained for GAPDH (Santa Cruz). Densitometry analysis of normalized DGUOK protein levels was performed using ImageJ tool (http://rsbweb.nih.gov/ij/).
Enzymatic measurements
Frozen muscle samples (∼35 mg) were processed as described in Arner et al. (1992).
The DGUOK activity was measured radiochemically by a procedure described in Mousson de Camaret et al. (2007), using 2-[8-3H] dG (deoxyguanosine, Moravek Biochemicals) as substrate. Any interference of cytosolic deoxycytidine kinase was excluded by performing the reactions in the presence of 2 mM deoxycytidine. Assays were initiated by addition of 30 μg of proteins per assay and conducted at 37°C. Activities are expressed in picomoles of transformed substrate per minute per milligram of protein (pmol/min/mg). The determination of citrate synthase and phosphoglucose isomerase activities was performed as previously described (Bergmeyer and Bernt, 1974).
Results
Molecular analysis of mitochondrial DNA
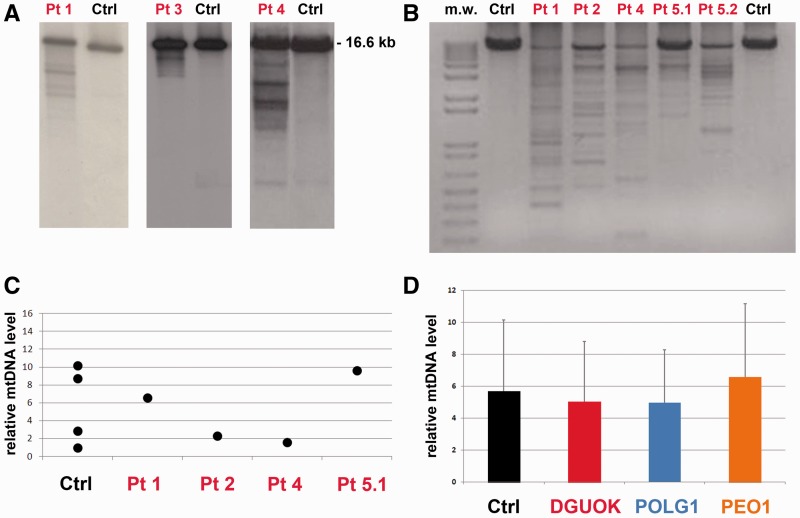
Mitochondrial DNA from muscle samples was analysed by Southern blot and long-range PCR analysis. A pattern of multiple mitochondrial DNA deletions was observed in Southern blots in all patients, and it was particularly evident in Patients 1, 3 and 4 (Fig. 2A). In all probands, multiple mitochondrial DNA deletions were also detected by long-range PCR analysis (Fig. 2B).
Figure 2.
Molecular studies in skeletal muscle. (A) Southern blot analysis of muscle mitochondrial DNA from Patients 1, 3 and 4 (independent electrophoresis runs and blots are shown) demonstrates the presence of multiple bands. Next to each patient’s lane is a lane showing the Southern blot of mitochondrial DNA obtained from age-matched healthy controls, which displays only a single band corresponding to wild-type mitochondrial DNA. (B) Long-range PCR. This panel shows an agarose electrophoresis separation of the wild-type mitochondrial DNA long-range PCR amplified fragment of about 8520 bp. Variably abundant additional bands because of partially deleted mitochondrial DNA molecules from muscle are observed in all the probands but not in age-matched controls (Ctrl). (C) Mitochondrial DNA copy number is normal in muscle samples of our probands (Patients 1, 2, 4 and 5.1) as determined by quantitative real-time PCR and expressed as the ratio of the mitochondrial MT-CYB gene to the nuclear APP gene. Values are shown relative to age-matched control samples. (D) The mitochondrial DNA copy number was determined by quantitative real-time PCR in skeletal muscle samples of our probands (n = 4), age-matched control individuals (n = 4) and two groups of patients with autosomal dominant or recessive progressive external ophthalmoplegia and harbouring mutations in POLG1 (n = 4) or PEO1 (n = 4): no significant difference was observed. All determinations were performed in triplicate. Values, after normalization to a control sample, are presented as mean values of each group. Error bars indicate standard deviations.
Assessment of relative mitochondrial DNA copy number by real-time quantitative PCR in muscle failed to show any evidence of depletion compared with healthy age-matched controls (Fig. 2C). We also evaluated the amount of mitochondrial DNA in muscle samples from patients with autosomal dominant or recessive progressive external ophthalmoplegia due to mutations in POLG1 or PEO1 genes (n = 4 for each group). There were no significant differences among these patients, our probands and age-matched control subjects (Fig. 2D).
Identification of causative mutations in the DGUOK gene
We performed next-generation sequencing of coding exons from 1034 nuclear-encoded mitochondrial-associated genes and the mitochondrial DNA (collectively termed ‘MitoExome’) in three adult patients with mitochondrial myopathy and multiple mitochondrial DNA deletions in skeletal muscle (Patients 1, 2 and 3). DNA was captured by an in-solution hybridization method (Gnirke et al., 2009) and sequenced on an Illumina GA-II platform (Bentley et al., 2008).
Analysis of genes involved in multiple mitochondrial DNA deletions disorders (POLG1, POLG2, SLC25A4, PEO1, RRM2B, OPA1, TK2 and MFN2) had already excluded the presence of pathogenic mutations in our patients.
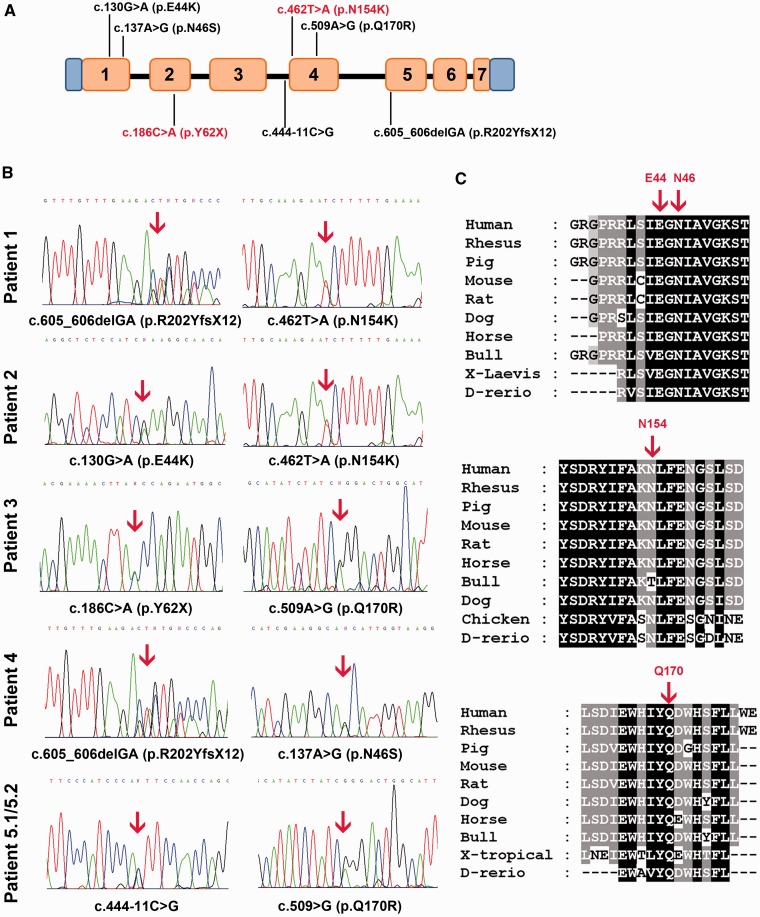
All of the probands harboured compound heterozygous mutations within the DGUOK coding sequence (Fig. 3A). We point out that heterozygous coding mutations in DGUOK are extremely rare, as none were detected in whole-exome data from 371 healthy individuals.
Figure 3.
DGUOK gene analysis. (A) Scheme of DGUOK gene showing the localization of all identified mutations along its coding sequence. Exons are numbered. Novel mutations are highlighted in red. (B) Identification of DGUOK mutations in our patients; sequence analysis of DGUOK disclosed heterozygous compound mutations in all the probands as indicated by arrows above the electropherograms. (C) Alignments of nine vertebrate amino acid sequences of DGUOK demonstrating that the residues predicted to be mutated in our probands (indicated by red arrows) are evolutionarily conserved across species.
Sequence analysis in Patient 1 disclosed the c.605_606delGA microdeletion, which produces a premature stop codon (p.R202YfsX12), and the missense variant c.462T>A resulting in the p.N154K change at the protein level. Two single-nucleotide variants, c.130 G>A (p.E44K) and c.462T>A, were identified in Patient 2. Patient 3 harboured the novel nonsense mutation c.186C>A, which results in a premature stop codon (p.Y62X), and the c.509A>G (p.Q170R) variant. These mutations were confirmed by standard sequencing.
We then performed mutational screening of DGUOK in a cohort of 51 undiagnosed patients in whom multiple mitochondrial DNA deletions in muscle had been detected by Southern blot and PCR analyses. We discovered the microdeletion c.605_606delGA in compound with the c.137A>G transition resulting in the p.N46S change in a fourth subject (Patient 4). In two siblings (Patients 5.1 and 5.2) with an adult-onset form of lower motor neuron disease, sequence analysis revealed the c.509A>G point mutation and a single base substitution in intron 3 (c.444-11C>G), which had been previously reported in a child with hepatocerebral mitochondrial DNA depletion syndrome (Dimmock et al., 2008a) (Fig. 3B).
All missense mutations described earlier, leading to amino acid changes in the DGUOK protein, affect residues that are evolutionarily conserved across species (Fig. 3C).
We checked the frequency of alleles carrying missense mutations by standard sequencing in a group of Italian controls: p.Q170R occurred four times in 198 alleles analysed (2%), while p.E44K, p.N46S and p.N154K were not found in 226 control alleles. Additionally, allele frequencies were assessed using the Exome Variant Server, which contains >10 000 analysed chromosomes: p.Q170R was detected in 171/10 587 chromosomes (1.6%); p.N154K was detected in 4/10 754 chromosomes (0.04%), whereas the remaining variants were not detected.
Analysis of single nucleotide variations
The amino acid substitutions p.N154K and p.Q170R were reported in the dbSNP database (http://www.ncbi.nlm.nih.gov/projects/SNP/) with a frequency of 0.002 and 0.029, respectively. The previously described p.E44K, p.N46S and p.Q170R changes were reported as causative mutations in several articles (Poulton et al., 2009).
Two software programs were used to predict the overall severity of the identified single-nucleotide mutations leading to amino acid substitutions: PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/, Sunyaev et al., 2001) and SIFT (Sorting Intolerant From Tolerant, http://sift.bii.a-star.edu.sg/, Ng and Henikoff, 2003). Default conditions were used for both programs. The prediction of the impact of these variants using PolyPhen-2 suggested that all of them would affect the structure and the function of DGUOK. According to SIFT analysis, all variants are predicted to be deleterious with the exception of p.Q170R, which appeared to be a tolerated change.
We also evaluated the effect of the c.444-11C>G mutation which had been found to result in abnormal splicing by bioinformatics analysis (Dimmock et al., 2008b). In silico prediction analysis using NetGene2 (http://www.cbs.dtu.dk/services/NetGene2/), NNSplice (http://www.fruitfly.org/seq_tools/splice.html) and Genio (http://biogenio.com/splice/) (Houdayer, 2011) tools confirmed the prediction of an abnormal splicing resulting from the loss of an acceptor splice site for exon 4.
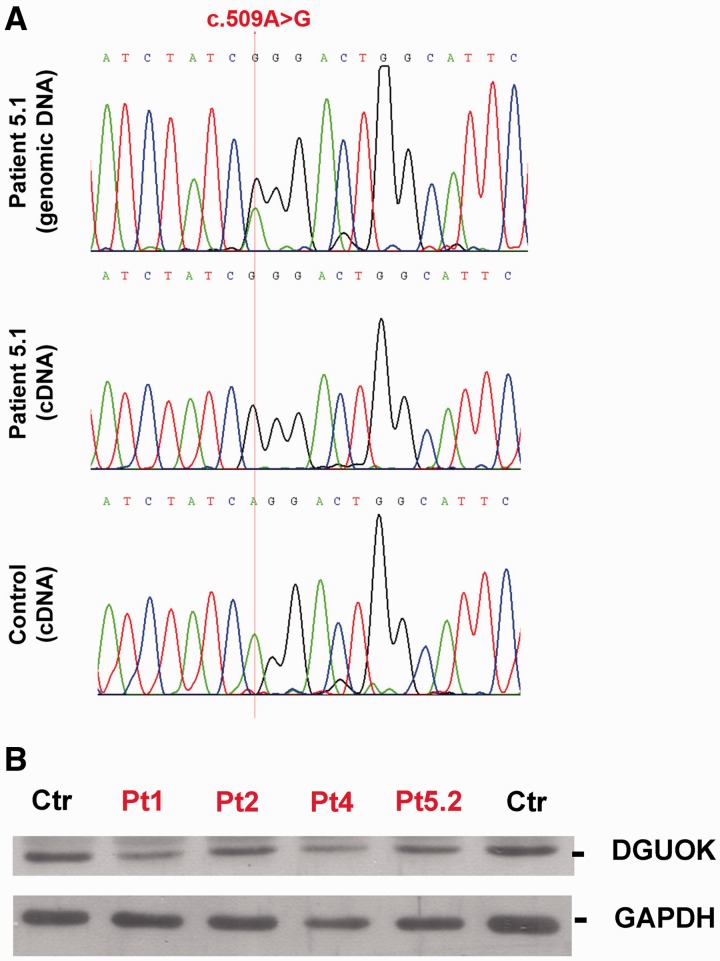
To document this deleterious effect on DGUOK transcript, we performed reverse transcriptase PCR analysis on muscle-derived messenger RNA from Patient 5.1 and from healthy controls: no aberrant transcript was identified when the full-length DGUOK complementary DNA was considered (data not shown). However, Patient 5.1, who was heterozygous (A>G) for the single-nucleotide mutation c.509A>G on genomic DNA, showed only the G allele when we sequenced her muscle DGUOK complementary DNA (Fig. 4A). These data support the absence of DGUOK messenger RNA transcript derived from the allele harbouring the c.444-11C>G mutation, most likely because of nonsense-mediated messenger RNA decay.
Figure 4.
Transcript and protein analysis. (A) Transcript analysis in Patient 5.1 sequence of c.509A>G position located in exon 4 of DGUOK in genomic and complementary DNA prepared from muscle of Patient 5.1 and an unaffected control. Patient 5.1 was heterozygous for the c.509A>G variant at genomic level. Conversely, transcript analysis showed only the G allele, suggesting loss of the allele harbouring the c.444-11C>G mutation. Amplicons were prepared from first-strand complementary DNA using PCR primers designed on exon 3 and 6. (B) Analysis of DGUOK protein normalized to Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) from muscle lysates of Patients 1, 2, 4, 5.2 and unaffected controls. Densitometric analysis showed that the normalized DGUOK staining in affected subjects was reduced by 20–45% compared with healthy controls.
Western blot analysis of skeletal muscle protein extracts from Patients 1, 2, 4 and 5.2 demonstrated a reduction of DGUOK protein levels (Fig. 4B). Normalized DGUOK staining in Patients 1 and 4 was compatible with the loss of a transcribed allele because of the heterozygous nonsense mutation disclosed at genomic level. Conversely, a significant amount of DGUOK protein, albeit less than in control muscles, was retained in Patient 2, who harboured two missense changes, and in Patient 5.2, who harboured a splicing mutation.
Biochemical studies
To confirm the pathogenicity of the mutations identified in our patients, we measured DGUOK activity in muscle extracts (Table 2). No significant difference was seen in measurements conducted in the presence or absence of non-radioactive deoxycytidine. As an internal control, we also measured the activities of citrate synthase, a matrix mitochondrial enzyme, and the cytosolic phosphoglucose isomerase.
Table 2.
Biochemical analysis in muscle tissues of affected and control subjects
| Sample | DGUOK activity pmol/min/mg | Citrate synthase activity nmol/min/mg | Phosphoglucose isomerase activity μmol/min/mg | DGUOK/citrate synthase |
|---|---|---|---|---|
| Controls (n = 3) | 19.7 ± 0.1 | 98.8 ± 22.8 | 1.50 ± 0.30 | 0.21 ± 0.04 |
| Patient 1 | 6.5 ± 1.1 | 164.1 | 1.80 | 0.04 (19%) |
| Patient 2 | 7.9 ± 1.0 | 152.1 | 1.77 | 0.05 (25%) |
| Patient 4 | 13.2 ± 3.0 | 139.1 | 1.46 | 0.09 (45%) |
| Patient 5.2 | 10.2 ± 1.9 | 162.7 | 1.74 | 0.06 (30%) |
Activity of DGUOK was measured in muscle extracts from affected and control individuals using 2-[8-3H] dG as substrate and are given in pmol/min/mg of protein. Values are the means of five independent determinations and are expressed as the mean ± SD. Mitochondrial citrate synthase and cytosolic phosphoglucose isomerase activities were assayed, and citrate synthetase was used for normalization purposes. Percentages indicate the residual DGUOK activity after citrate synthetase normalization compared with controls.
Normalized DGUOK/citrate synthase activities demonstrated a significant reduction in muscle from Patients 1, 2, 4 and 5.1. No muscle samples were available for biochemical studies from Patient 3. Residual activity ranged from 19 to 45% when compared with three unaffected controls. Phosphoglucose isomerase and citrate synthase activities were similar in all samples analysed, arguing against a spuriously low DGUOK activity in patients’ muscle because of poor sample preservation.
Discussion
DGUOK encodes the mitochondrial deoxyguanosine kinase that catalyses the first reaction of the purine salvage pathway in the mitochondrial matrix, which is essential for mitochondrial DNA maintenance (Jullig and Eriksson, 2000).
To date, autosomal recessive DGUOK mutations had been described in infants or young children presenting with severe encephalopathy associated with liver failure and liver mitochondrial DNA depletion (hepatocerebal mitochondrial DNA depletion syndrome phenotype), often leading to premature death (Mandel et al., 2001). Exceptionally, moderate muscle mitochondrial DNA depletion was documented in a 14-year-old patient with muscle weakness, who harboured compound heterozygous DGUOK mutations (Buchaklian et al., 2012).
We report an unexpected and previously unreported causative role of DGUOK in the pathogenesis of heterogeneous adult-onset disorders associated with mitochondrial DNA instability reflected by multiple mitochondrial DNA deletions in skeletal muscle. Patients 1 and 2 developed late-onset progressive external ophthalmoplegia sparing other muscle groups. Both of them presented the same DGUOK gene missense mutation c.462T>A (p.N154K) in association with two different variants whose deleterious effects were previously documented (Dimmock et al., 2008a; Spinazzola et al., 2009). Conversely, the functional consequence of N154K substitution had not been previously assessed. In our study, biochemical analysis performed in muscle from Patients 1 and 2 revealed a severe defect in DGUOK activity (the lowest residual activity in our cohort) supporting the pathogenicity of the N154K change.
Patient 3, who had a mitochondrial myopathy without progressive external ophthalmoplegia or ptosis, harboured the unpublished nonsense mutation p.Y62X and the missense change p.Q170R. While the deleterious effect of the first variant is self-evident, the significance of the p.Q170R mutation is still uncertain because it has been found in several control chromosomes in previous studies (Dimmock et al., 2008a). In a group of healthy Italian subjects, we identified this variant with an allelic frequency of 1.98%.
Patient 4 is unique in our cohort because she had flagrant liver involvement as a neonate, leading to organ transplantation at 9 months of age. DGUOK screening revealed the c.605_606delGA microdeletion and the p.N46S mutation. The latter mutation was found in four children with hepatopathy and liver mitochondrial DNA depletion syndrome associated to a second missense substitution (Mousson de Camaret et al., 2007; Sarzi et al., 2007; Dimmock et al., 2008a). Data collected by other groups had suggested that the p.N46S variant in association with another missense mutation may manifest as isolated hepatopathy with a more benign course (Dimmock et al., 2008a). The residual DGUOK activity detected in mitochondria isolated from patients harbouring two missense mutations (Salviati et al., 2002; Mousson de Camaret et al., 2007) may contribute to sustain replication of mitochondrial DNA in the critical foetal and perinatal periods. As mitochondrial DNA synthesis decreases progressively after the neonatal period, even a low residual DGUOK activity may suffice to prevent further disease development and allow regeneration of damaged liver. Our patient indirectly confirms this hypothesis: the allele bearing the N46S mutation is the only one able to result in a full, albeit impaired, DGUOK protein. The resulting severe liver failure required organ transplantation, which was successful in correcting liver dysfunction, but could not prevent the development of muscle pathology in the second decade.
The diagnosis of mitochondrial disorder was considered after two episodes of rhabdomyolsis triggered by intercurrent febrile illnesses. Although this is apparently a case of adult-onset mitochondrial metabolic myopathy, we think the myopathy was the long-term clinical outcome of a DGUOK-related liver failure successfully treated by liver transplantation early in life. A study addressing the long-term outcome of liver transplantation in DGUOK-mutated patients showed that neurological dysfunction in the first months of life seems to be the only negative predictor of long-term survival (Dimmock et al., 2008a).
Patient 4 is the first known DGUOK case who initially developed isolated liver disease and, after transplantation, reached adult life when she developed a mitochondrial myopathy with accumulation of multiple mitochondrial DNA deletions. DGUOK activity in skeletal muscle was markedly decreased, which may explain the accumulation of damaged mitochondrial DNA molecules leading to muscle disease. However, the residual activity was unexpectedly high, considering the early muscle involvement experienced by this patient.
Only longitudinal studies of more DGUOK patients transplanted in childhood will clarify if the late-onset development of neurological symptoms and the involvement of skeletal muscle are consistent features or uncommon complications.
Finally, we identified DGUOK mutations in two siblings with an unusual adult-onset form of lower motor neuron disease and mild cognitive impairment. Both harboured the p.Q170R mutation and the intronic nucleotide substitution c.444-11C>G. Both siblings showed COX-negative fibres and multiple mitochondrial DNA deletions in skeletal muscle. We performed protein and biochemical analysis in muscle from Patient 5.2 confirming the detrimental effect of the mutant genotype on protein stability and residual activity. The reduction of DGUOK activity by 70% compared with controls supports the pathogenetic role of Q170R mutation, although further confirmation will be provided by biochemical assays performed in Q170R carriers or using the purified recombinant protein.
Although we cannot exclude the coexistence of an unknown molecular defect causing the lower motor neuron disease in these patients, it is unlikely that the effects of DGUOK mutations are limited to the oxidative defect revealed in skeletal muscle by histochemical analysis. Lower motor neuron disorders compatible with spinal muscular atrophy or amyotrophic lateral sclerosis have been described in several paediatric and adult patients with primary mitochondrial disorders due to mutations either in mitochondrial DNA or in nuclear genes such as SCO2 and TK2 (see Hirano et al., 2008 for a review). In particular, recessive mutations in TK2 have been associated with heterogeneous paediatric presentations including a Spinal Muscular Atrophy (SMA)-like phenotype with mitochondrial DNA instability in skeletal muscle (Mancuso et al., 2002).
Recessively inherited syndromes characterized by multiple mitochondrial DNA deletions in post-mitotic tissues have been associated with mutations in POLG1 (Stewart et al., 2009) and TK2 (Tyynismaa et al., 2012). They are clinically heterogeneous but usually feature progressive external ophthalmoplegia and ptosis, or, in the case of TYMP mutations, result in the more complex mitochondrial neurogastrointestinal encephalopathy phenotype (Garone et al., 2011). Finally, in addition to causing juvenile-onset disorders with mitochondrial DNA depletion, recessive MPV17 mutations have been described in two patients presenting adult-onset multi-systemic disorders with neuropathy and leukoencephalopathy, both featuring multiple mitochondrial DNA deletions in muscle (Blakely et al., 2012; Garone et al., 2012).
Our study has identified recessive DGUOK mutations as an unexpected cause of multiple mitochondrial DNA deletions associated with late-onset mitochondrial disorders. After the discovery of RRM2B (Tyynismaa et al., 2009) and TK2 (Tyynismaa et al., 2012) mutations in patients with adult-onset mitochondrial DNA instability disorders, the involvement of DGUOK highlights the importance of both the cytoplasmic de novo synthesis and the mitochondrial nucleoside salvage pathway in the maintenance of balanced nucleotide pools and adequate mitochondrial DNA replication in adult skeletal muscle. Together, these studies bolster the notion that the crosstalk between nuclear and mitochondrial DNA is more dynamic than expected: the impairment of nuclear-encoded factors controlling replication and repair of mitochondrial DNA gives rise to a spectrum of clinical phenotypes affecting both infants and adults.
The biochemical evaluation of DGUOK activity in our patients clearly shows that loss-of-function mutations may impair enzyme activity in skeletal muscle. The degree of residual activity does not correlate with age at onset of muscle involvement or with the abundance of multiple mitochondrial DNA deletions in this tissue. However, residual DGUOK activities are higher than those observed in liver of children, which are generally <15% of controls. The long exposure of post-mitotic skeletal muscle cells to reduced (albeit not dramatically low) levels of DGUOK activity may increase the proportion of deleted mitochondrial DNA molecules over time and result in a clinical phenotype only in adult life.
Mutations in DGUOK gene occurred in five independent patients of our cohort. These account for 9.8% of genetically undiagnosed subjects (n = 51) carrying multiple mitochondrial DNA deletions and for 5.6% of all probands (n = 90) carrying multiple mitochondrial DNA deletions confirmed by Southern blot and long-range PCR analyses. Mutations in POLG1 remain the major cause of disorders with multiple mitochondrial DNA deletion, accounting for ∼20–25% of all patients with mitochondrial disease, followed by mutations in PEO1, which account for another 15–18% (Virgilio et al., 2008; Fratter et al., 2011).
Our observation was made possible by the application of next-generation sequencing restricted to mitochondrial DNA and nuclear genes involved in mitochondrial structure and function. It is likely that other defects leading to the accumulation of multiple mitochondrial DNA deletions in skeletal muscle will be identified in nuclear genes not comprised in our platform because their mitochondrial localization/function is currently unknown. The pathogenicity of the many novel candidate variants disclosed with exomic approach (400–500 12per patient) has to be carefully evaluated. Strong genetic (co-segregation studies and multiple reports of the same variants in independent subjects) and functional data are needed to support their causative role, especially in autosomal dominant and sporadic forms of mitochondrial disorders.
In conclusion, we have identified DGUOK mutations as a novel cause of mitochondrial disease with multiple mitochondrial DNA deletions.
DGUOK mutations were previously reported in 17% of patients with combined respiratory chain deficiency and hepatocerebral presentation (Slama et al., 2005). Our findings suggest that DGUOK mutational screening in adult patients with ascertained instability of muscle mitochondrial DNA may disclose the molecular defect in many of them.
Funding
This work was supported by a grant from the National Institutes of Health (GM077465 and GM097136 to V.K.M.; HD32062 to S.D.M.), by the Marriott Mitochondrial Disorder Clinical Research Fund to S.D.M. and by a contribution of the Italian Telethon (grant GUP09004 to G.P.C.) ‘Constructing a database for a nation-wide Italian collaborative network of mitochondrial diseases’. The financial support of Associazione Amici del Centro Dino Ferrari, University of Milan; the Telethon project GTB07001ER; the Eurobiobank project QLTR-2001-02769 and R.F. 02.187 Criobanca Automatizzata di Materiale Biologico is gratefully acknowledged.
Glossary
Abbreviations
- COX
cytochrome c oxidase
- SDH
succinate dehydrogenase
- mtDNA
mitochondrial DNA
- PCR
polymerase chain reaction
- dNTP
deoxyribonucleotide triphosphate
References
- Amati-Bonneau P, Valentino ML, Reynier P, Gallardo ME, Bornstein B, Boissière A, et al. OPA1 mutations induce mitochondrial DNA instability and optic atrophy ‘plus’ phenotypes. Brain. 2008;131:338–51. doi: 10.1093/brain/awm298. [DOI] [PubMed] [Google Scholar]
- Arnér ES, Spasokoukotskaja T, Eriksson S. Selective assays for thymidine kinase 1 and 2 and deoxycytidine kinase and their activities in extracts from human cells and tissues. Biochem Biophys Res Commun. 1992;188:712–18. doi: 10.1016/0006-291x(92)91114-6. [DOI] [PubMed] [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–9. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmeyer HU, Bernt E. Methods of enzymatic analysis. I–II. New York: Academic Press; 1974. [Google Scholar]
- Blakely EL, Butterworth A, Hadden RD, Bodi I, He L, McFarland R, et al. MPV17 mutation causes neuropathy and leukoencephalopathy with multiple mtDNA deletions in muscle. Neuromuscul Disord. 2012;22:587–91. doi: 10.1016/j.nmd.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdon A, Minai L, Serre V, Jais JP, Sarzi E, Aubert S, et al. Mutation of RRM2B, encoding p53-controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nat Genet. 2007;39:776–80. doi: 10.1038/ng2040. [DOI] [PubMed] [Google Scholar]
- Buchaklian AH, Helbling D, Ware SM, Dimmock DP. Recessive deoxyguanosine kinase deficiency causes juvenile onset mitochondrial myopathy. Mol Genet Metab. 2012 doi: 10.1016/j.ymgme.2012.04.019. Apr 26. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Calvo SE, Compton AG, Hershman SG, Lim SC, Lieber DS, Tucker EJ, et al. Molecular diagnosis of infantile mitochondrial disease with targeted next-generation sequencing. Sci Transl Med. 2012;4:118ra10. doi: 10.1126/scitranslmed.3003310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo SE, Tucker EJ, Compton AG, Kirby DM, Crawford G, Burtt NP, et al. High-throughput, pooled sequencing identifies mutations in NUBPL and FOXRED1 in human complex I deficiency. Nat Genet. 2010;42:851–8. doi: 10.1038/ng.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland WC. Inherited mitochondrial diseases of DNA replication (Review) Annu Rev Med. 2008;59:131–46. doi: 10.1146/annurev.med.59.053006.104646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimmock DP, Dunn JK, Feigenbaum A, Rupar A, Horvath R, Freisinger P, et al. Abnormal neurological features predict poor survival and should preclude liver transplantation in patients with deoxyguanosine kinase deficiency. Liver Transpl. 2008a;14:1480–5. doi: 10.1002/lt.21556. [DOI] [PubMed] [Google Scholar]
- Dimmock DP, Zhang Q, Dionisi-Vici C, Carrozzo R, Shieh J, Tang LY, et al. Clinical and molecular features of mitochondrial DNA depletion due to mutations in deoxyguanosine kinase. Hum Mutat. 2008b;29:330–1. doi: 10.1002/humu.9519. [DOI] [PubMed] [Google Scholar]
- Fratter C, Raman P, Alston CL, Blakely EL, Craig K, Smith C, et al. RRM2B mutations are frequent in familial PEO with multiple mtDNA deletions. Neurology. 2011;76:2032–4. doi: 10.1212/WNL.0b013e31821e558b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbiati S, Bordoni A, Papadimitriou D, Toscano A, Rodolico C, Katsarou E, et al. New mutations in TK2 gene associated with mitochondrial DNA depletion. Pediatr Neurol. 2006;34:177–85. doi: 10.1016/j.pediatrneurol.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Garone C, Tadesse S, Hirano M. Clinical and genetic spectrum of mitochondrial neurogastrointestinal encephalomyopathy. Brain. 2011;134:3326–32. doi: 10.1093/brain/awr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garone C, Rubio JC, Calvo SE, Naini A, Tanji K, DiMauro S, et al. MPV17 mutations causing adult-onset multisystemic disorder with multiple mitochondrial DNA deletions. Arch Neurol. doi: 10.1001/archneurol.2012.405. Published online September 10, 2012. doi:10.1001/archneurol.2012.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–9. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakonen AH, Isohanni P, Paetau A, Herva R, Suomalainen A, Lönnqvist T. Recessive Twinkle mutations in early onset encephalopathy with mtDNA depletion. Brain. 2007;130:3032–40. doi: 10.1093/brain/awm242. [DOI] [PubMed] [Google Scholar]
- Hirano M, Angelini C, Montagna P, Hays AP, Tanji K, Mitsumoto H, et al. Amyotrophic lateral sclerosis with ragged-red fibers (Review) Arch Neurol. 2008;65:403–6. doi: 10.1001/archneurol.2007.65. [DOI] [PubMed] [Google Scholar]
- Hirano M, Nishigaki Y, Marti R. Mitochondrial neurogastrointestinal encephalomyopathy (MNGIE): a disease of two genomes. Neurologist. 2004;10:8–17. doi: 10.1097/01.nrl.0000106919.06469.04. [DOI] [PubMed] [Google Scholar]
- Houdayer C. In silico prediction of splice-affecting nucleotide variants. Methods Mol Biol. 2011;760:269–81. doi: 10.1007/978-1-61779-176-5_17. [DOI] [PubMed] [Google Scholar]
- Hudson G, Amati-Bonneau P, Blakely E, Stewart J, He L, Schaefer A, et al. Mutation of OPA1 causes dominant optic atrophy with external ophtalmoplegia, ataxia, deafness and multiple mitochondrial DNA deletions: a novel disorder of mtDNA maintenance. Brain. 2008;131:329–37. doi: 10.1093/brain/awm272. [DOI] [PubMed] [Google Scholar]
- Isohanni P, Hakonen AH, Euro L, Paetau I, Linnankivi T, Liukkonen E, et al. POLG1 manifestations in childhood. Neurology. 2011;76:811–15. doi: 10.1212/WNL.0b013e31820e7b25. [DOI] [PubMed] [Google Scholar]
- Johansson M, Karlsson A. Cloning and expression of human deoxyguanosine kinase cDNA. Proc Natl Acad Sci USA. 1996;93:7258–62. doi: 10.1073/pnas.93.14.7258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullig M, Eriksson S. Mitochondrial and submitochondrial localization of human deoxyguanosine kinase. Eur J Biochem. 2000;267:5466–72. doi: 10.1046/j.1432-1327.2000.01607.x. [DOI] [PubMed] [Google Scholar]
- Kaukonen J, Juselius JK, Tiranti V, Kyttälä A, Zeviani M, Comi GP, et al. Role of adenine nucleotide translocator 1 in mtDNA maintenance. Science. 2000;289:782–5. doi: 10.1126/science.289.5480.782. [DOI] [PubMed] [Google Scholar]
- Krishnan KJ, Reeve AK, Samuels DC, Chinnery PF, Blackwood JK, Taylor RW, et al. What causes mitochondrial DNA deletions in human cells? Nat Genet. 2008;40:275–9. doi: 10.1038/ng.f.94. [DOI] [PubMed] [Google Scholar]
- Longley MJ, Clark S, Yu Wai Man C, Hudson G, Durham SE, Taylor RW, et al. Mutant POLG2 disrupts DNA polymerase gamma subunits and causes progressive external ophthalmoplegia. Am J Hum Genet. 2006;78:1026–34. doi: 10.1086/504303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luoma P, Melberg A, Rinne JO, Kaukonen JA, Nupponen NN, Chalmers RM, et al. Parkinsonism, premature menopause, and mitochondrial DNA polymerase gamma mutations: clinical and molecular genetic study. Lancet. 2004;364:875–82. doi: 10.1016/S0140-6736(04)16983-3. [DOI] [PubMed] [Google Scholar]
- Mancuso M, Salviati L, Sacconi S, Otaegui D, Camaño P, Marina A, et al. Mitochondrial DNA depletion: mutations in thymidine kinase gene with myopathy and SMA. Neurology. 2002;59:1197–202. doi: 10.1212/01.wnl.0000028689.93049.9a. [DOI] [PubMed] [Google Scholar]
- Mandel H, Szargel R, Labay V, Elpeleg O, Saada A, Shalata A, et al. The deoxyguanosine kinase gene is mutated in individuals with depleted hepatocerebral mitochondrial DNA. Nat Genet. 2001;29:337–41. doi: 10.1038/ng746. [DOI] [PubMed] [Google Scholar]
- Mousson de Camaret B, Taanman JW, Padet S, Chassagne M, Mayençon M, Clerc-Renaud P, et al. Kinetic properties of mutant deoxyguanosine kinase in a case of reversible hepatic mtDNA depletion. Biochem J. 2007;402:377–85. doi: 10.1042/BJ20060705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–14. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikali K, Suomalainen A, Saharinen J, Kuokkanen M, Spelbrink JN, Lönnqvist T, et al. Infantile onset spinocerebellar ataxia is caused by recessive mutations in mitochondrial proteins Twinkle and Twinky. Hum Mol Genet. 2005;14:2981–90. doi: 10.1093/hmg/ddi328. [DOI] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–23. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulton J, Hirano M, Spinazzola A, Arenas Hernandez M, Jardel C, Lombès A, et al. Collated mutations in mitochondrial DNA (mtDNA) depletion syndrome (excluding the mitochondrial gamma polymerase, POLG1) Biochim Biophys Acta. 2009;1792:1109–12. doi: 10.1016/j.bbadis.2009.08.016. [DOI] [PubMed] [Google Scholar]
- Rötig A, Poulton J. Genetic causes of mitochondrial DNA depletion in humans. Biochim Biophys Acta. 2009;1792:1103–8. doi: 10.1016/j.bbadis.2009.06.009. [DOI] [PubMed] [Google Scholar]
- Rouzier C, Bannwarth S, Chaussenot A, Chevrollier A, Verschueren A, Bonello-Palot N, et al. The MFN2 gene is responsible for mitochondrial DNA instability and optic atrophy ‘plus’ phenotype. Brain. 2012;135:23–34. doi: 10.1093/brain/awr323. [DOI] [PubMed] [Google Scholar]
- Saada A, Shaag A, Mandel H, Nevo Y, Eriksson S, Elpeleg O. Mutant mitochondrial thymidine kinase in mitochondrial DNA depletion myopathy. Nat Genet. 2001;29:342–4. doi: 10.1038/ng751. [DOI] [PubMed] [Google Scholar]
- Salviati L, Sacconi S, Mancuso M, Otaegui D, Camaño P, Marina A, et al. Mitochondrial DNA depletion and dGK gene mutations. Ann Neurol. 2002;52:311–17. doi: 10.1002/ana.10284. [DOI] [PubMed] [Google Scholar]
- Sarzi E, Goffart S, Serre V, Chrétien D, Slama A, Munnich A, et al. Twinkle helicase (PEO1) gene mutation causes mitochondrial DNA depletion. Ann Neurol. 2007;62:579–87. doi: 10.1002/ana.21207. [DOI] [PubMed] [Google Scholar]
- Sciacco M, Prelle A, Comi GP, Napoli L, Battistel A, Bresolin N. Retrospective study of a large population of patients affected with mitochondrial disorders: clinical, morphological and molecular genetic evaluation. J Neurol. 2001;248:778–88. doi: 10.1007/s004150170094. [DOI] [PubMed] [Google Scholar]
- Slama A, Giurgea I, Debrey D, Bridoux D, de Lonlay P, Levy P, et al. Deoxyguanosine kinase mutations and combined deficiencies of the mitochondrial respiratory chain in patients with hepatic involvement. Mol Genet Metab. 2005;86:462–5. doi: 10.1016/j.ymgme.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Spelbrink JN, Li FY, Tiranti V, Nikali K, Yuan QP, Tariq M, et al. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat Genet. 2001;28:223–31. doi: 10.1038/90058. [DOI] [PubMed] [Google Scholar]
- Spinazzola A, Invernizzi F, Carrara F, Lamantea E, Donati A, Dirocco M, et al. Clinical and molecular features of mitochondrial DNA depletion syndromes. J Inherit Metab Dis. 2009;32:143–58. doi: 10.1007/s10545-008-1038-z. [DOI] [PubMed] [Google Scholar]
- Spinazzola A, Viscomi C, Fernandez-Vizarra E, Carrara F, D’Adamo P, Calvo S, et al. MPV17 encodes an inner mitochondrial membrane protein and is mutated in infantile hepatic mitochondrial DNA depletion. Nat Genet. 2006;38:570–5. doi: 10.1038/ng1765. [DOI] [PubMed] [Google Scholar]
- Stewart JD, Tennant S, Powell H, Pyle A, Blakely EL, He L, et al. Novel POLG1 mutations associated with neuromuscular and liver phenotypes in adults and children. J Med Genet. 2009;46:209–14. doi: 10.1136/jmg.2008.058180. [DOI] [PubMed] [Google Scholar]
- Sunyaev S, Ramensky V, Koch I, Lathe W, Kondrashov AS, Bork P. Prediction of deleterious human alleles. Hum Mol Genet. 2001;10:591–7. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- Suomalainen A, Majander A, Haltia M, Somer H, Lonnqvist J, Savontaus ML, et al. Multiple deletions of mitochondrial DNA in several tissues of a patient with severe retarded depression and familial progressive external ophthalmoplegia. J Clin Invest. 1992;90:61–6. doi: 10.1172/JCI115856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyynismaa H, Sun R, Ahola-Erkkilä S, Almusa H, Pöyhönen R, Korpela M, et al. Thymidine kinase 2 mutations in autosomal recessive progressive external ophthalmoplegia with multiple mitochondrial DNA deletions. Hum Mol Genet. 2012;21:66–75. doi: 10.1093/hmg/ddr438. [DOI] [PubMed] [Google Scholar]
- Tyynismaa H, Ylikallio E, Patel M, Molnar MJ, Haller RG, Suomalainen A. A heterozygous truncating mutation in RRM2B causes autosomal-dominant progressive external ophthalmoplegia with multiple mtDNA deletions. Am J Hum Genet. 2009;85:290–5. doi: 10.1016/j.ajhg.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Goethem G, Dermaut B, Löfgren A, Martin JJ, Van Broeckhoven C. Mutation of POLG is associated with progressive external ophthalmoplegia characterized by mtDNA deletions. Nat Genet. 2001;28:211–12. doi: 10.1038/90034. [DOI] [PubMed] [Google Scholar]
- Virgilio R, Ronchi D, Hadjigeorgiou GM, Bordoni A, Saladino F, Moggio M, et al. Novel Twinkle (PEO1) gene mutations in mendelian progressive external ophthalmoplegia. J Neurol. 2008;255:1384–91. doi: 10.1007/s00415-008-0926-3. [DOI] [PubMed] [Google Scholar]
- Wanrooij S, Falkenberg M. The human mitochondrial replication fork in health and disease. Biochim Biophys Acta. 2010;1797:1378–88. doi: 10.1016/j.bbabio.2010.04.015. [DOI] [PubMed] [Google Scholar]
- Yu-Wai-Man P, Chinnery PF. Dysfunctional mitochondrial maintenance: what breaks the circle of life? Brain. 2012;135:9–11. doi: 10.1093/brain/awr352. [DOI] [PubMed] [Google Scholar]
- Zeviani M, Gellera C, Pannacci M, Uziel G, Prelle A, Servidei S, et al. Tissue distribution and transmission of mitochondrial DNA deletions in mitochondrial myopathies. Ann Neurol. 1990;28:94–7. doi: 10.1002/ana.410280118. [DOI] [PubMed] [Google Scholar]
- Zeviani M, Servidei S, Gellera C, Bertini E, DiMauro S, DiDonato S. An autosomal dominant disorder with multiple deletions of mitochondrial DNA starting at the D-loop region. Nature. 1989;339:309–11. doi: 10.1038/339309a0. [DOI] [PubMed] [Google Scholar]