Abstract
Synovial joints are among the most important structures that give us complex motor abilities as humans. Degenerative joint diseases, such as arthritis, cause loss of normal joint functioning and affect over 40 million people in the USA and approximately 350 million people worldwide. Therapies based on regenerative medicine hold the promise of effectively repairing or replacing damaged joints permanently. Here, for the first time, we introduce a model for synovial joint regeneration utilizing the chick embryo. In this model, a block of tissue that contains the prospective elbow is excised, leaving a window with strips of anterior and posterior tissue intact (window excision, WE). In contrast, we also slice out the same area containing the elbow and the distal piece of the limb is pinned back onto the stump (slice excision, SE). Interestingly, when the elbow is removed via WE, regeneration of the joint takes place, whereas the elbow joint does not regenerate following SE. In order to investigate whether the regeneration response recapitulates the developmental program of forming joints, we used GDF-5 and Autotaxin (Atx) as joint tissue specific markers, and Sox-9 and Col-9 as cartilage markers for in situ hybridization on sections at different time points after WE and SE surgeries. Re-expression of GDF-5 and Atx is observed in the WE samples by 60 hours after surgery. In contrast, the majority of the samples that underwent SE surgery did not express GDF-5 and Atx. Also, in SE fusion of cartilage elements takes place and the joint interzone does not form. This is indicated by continuous Col-9 expression in SE limbs, whereas Col-9 is downregulated at the joint interzone in the regenerating WE samples. This order and pattern of gene expression observed in regenerates is similar to the development of a joint suggesting that regeneration recapitulates development at the molecular level. This model defines some of the conditions required for inducing joint regeneration in an otherwise nonregenerating environment. This knowledge can be useful for designing new therapeutic approaches for joint loss or for conditions affecting joint integrity in humans.
Keywords: limb regeneration, joint regeneration, joint development, elbow development, elbow regeneration, chick limb regeneration
Introduction
Our current knowledge of skeletal tissue regeneration comes from studies utilizing urodeles, such as axolotls and newts. Urodeles can regenerate a fully functional limb throughout their lives after the limb is amputated. Other vertebrates, such as birds and mammals, do not possess this type of robust limb regeneration ability. Instead, amputation injuries undergo a wound healing response with the formation of scar tissue covering the limb stump. During a regeneration response, the regenerating structure is re-patterned from a population of cells, including stem cells and/or dedifferentiated cells that migrate from the surrounding tissue. Several studies indicate that the process of remaking the lost structure from these cells follows the developmental program of that organ or tissue (re-development). For example, during limb regeneration, the blastema forms a bud that behaves like a developing limb (Muneoka and Bryant, 1982, 1984) and there are many studies that demonstrate similarities in spatial and temporal patterns of gene expression to development and regeneration (Imokawa and Yoshizato, 1997; Torok et al., 1998). However, there are also studies which suggest different mechanisms for regeneration wherein regeneration does not follow the re-development paradigm. Mouse digit tip regeneration of the terminal phalangeal bone occurs through intramembranous ossification as opposed to endochondral ossification, the developmental mechanism (Han et al., 2008). Also, in zebrafish, expression patterns of Msx genes and Fgf20a have been shown to be different during development and regeneration of the fins (Akimenko et al., 1995; Whitehead et al., 2005). Determining whether a particular regeneration event redeploys developmental mechanisms, or not, could be useful for designing therapeutic approaches for the regeneration of tissues or organs.
The chick limb has been thought to lack an endogenous regeneration response: limb amputation at any stage of its development fails to mount a regenerative response. Even though the chicken lacks regeneration ability, studies demonstrate that it has some regeneration potential. For example, chicken embryos regenerate amputated limb buds when the amputation wound is treated with FGFs (Kostakopoulou et al., 1996; Taylor et al., 1994). In addition, studies in the developing limb bud have shown that removal of a portion of mesenchyme, thereby creating a hole in the bud, results in normal development of these limbs (Hayamizu et al., 1994; Summerbell, 1981; Summerbell and Wolpert, 1973). This ability to regenerate following excisional injuries is limited by the limb stage and the size of the tissue removed (Stark and Searls, 1974). Also, studies demonstrate that the apical ectodermal ridge (AER, a structure required for limb development) in the embryonic chick limb can be induced to form ectopically (Satoh et al., 2010) or be reinduced following removal, and as a consequence, lead to a regeneration response (Kawakami et al., 2006). These studies indicate that the developing chick limb possesses regenerative potential that can be exploited to understand what is required for triggering an effective regenerative response in higher vertebrates.
Regeneration studies have been carried out extensively in many tissue/organ systems and in a variety of animals. However, the regeneration of a joint has not been studied specifically; a few studies have focused on the repair of articular cartilage, the type of cartilage that contributes to the joint (Duterloo and Jansen, 1969; Jolly, 1961; O'Driscoll, 1999). Joints play a critical role in limb pattern formation because they determine the shape, size and number of skeletal elements (Shubin and Alberch, 1986). The skeletal elements of the limb arise as an unsegmented mesenchymal condensation, which eventually begins differentiating into cartilage upon the expression of the transcription factor Sox-9 (Bi et al., 1999; Healy et al., 1999). Sox-9 drives the expression of chondrogenic genes Col-2α1 and Col-9 (Kimura et al., 1985; Nalin et al., 1995). At this stage, the tissue uniformly expresses these collagens. Next, a joint interzone begins to form, causing the uniform collagen expression to become segmented. The interzone is made of densely packed flattened cells and is indicative of the area where the joint will form (Mitrovic, 1977; 1978). The appearance of these noncartilagenous joint progenitors is also the time at which joint specific markers, such as Autotaxin (Atx) and GDF-5 become more restricted to the joint interzone (Bachner et al., 1999; Storm and Kingsley, 1999). Around the same time, cartilage-specific collagens at the joint interzone are downregulated. Sox-9-expressing mesenchymal cells are still present at this stage, and they will downregulate Sox-9 later than Col-9. The interzone finally differentiates into three layers of cells with different densities. Cells in higher density regions differentiate into articular cartilage, which covers the surfaces of the bones where they articulate. The inner region progressively disappears by a process called cavitation (Mitrovic, 1978). Thus, joint formation is marked by clear histological and molecular changes.
Here, we introduce a joint tissue regeneration model by utilizing the developing elbow joint of the chicken (Gallus gallus) embryo as a model system. We show that, excisional removal of the developing elbow joint at stage 26 results in the regeneration of a joint. This regeneration response does not require an external inducer; but it is affected by the method of excision. We provide histological and molecular evidence that, after wound healing, the mechanism of remaking the joint involves the reiteration of the joint development program.
Materials and Methods
Embryos and manipulations
Fertilized White Leghorn chicken eggs were obtained from Charles River Laboratories Avian Products and Services (Wilmington, MA) and incubated at 38°C in a humidified incubator. Embryos were staged according to Hamburger and Hamilton (1951).
Microsurgeries were performed by using sharpened tungsten needles (Ted Pella Inc., diameter: 0.005”). Measurements were done by using an eyepiece graticule. Window excision: Elbow tissue was removed by carving a 150 µm-wide region of tissue out, leaving a “window” in the limb with anterior and posterior tissues holding the distal limb attached to the proximal portion of the limb (Fig. 2A). Slice excision: Elbow tissue was removed by slicing out a 150 µm-wide region of tissue. The distal tissue was pinned to the proximal stump using a platinum pin (Fig. 2A’). After the surgery, embryos were incubated until desired time points and then fixed for further analysis.
Figure 2.
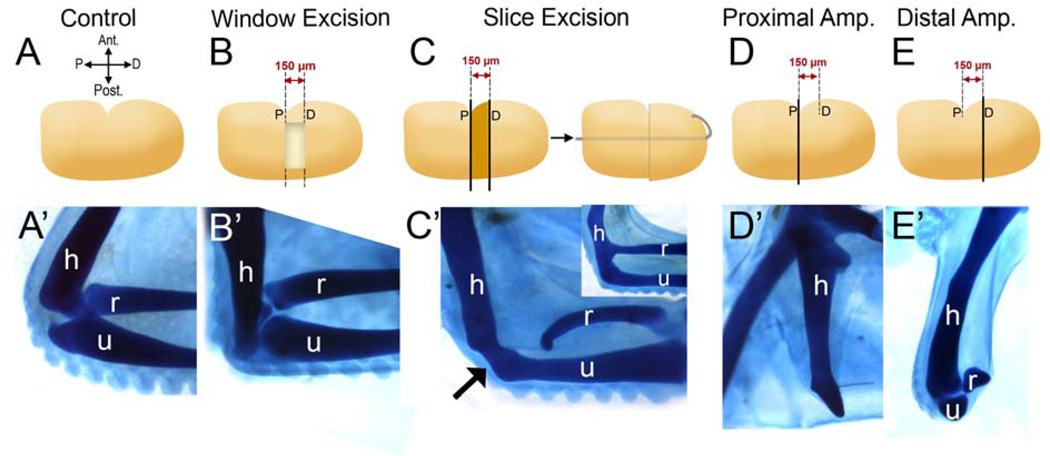
The elbow joint of the chicken embryo is able to regenerate depending on the method of joint removal. A) The contralateral limb served as a control. (A’–E’) All samples were stained with Victoria blue to visualize the cartilage morphology. B) Window Excision (WE). Using the anterior indentation as a landmark, a 150µm-wide block of tissue containing the prospective elbow is removed leaving a “window”. C) Slice Excision (SE). Using the anterior indentation as a landmark, a piece of the limb, 150µm-wide containing the prospective elbow is sliced out and discarded. The remaining distal piece of the forelimb is pinned onto the stump, leaving no space between the two wound surfaces. Both surgeries were performed at st. 26 and embryos were incubated for four days following surgery. B) After WE surgery, 10 out of 20 limbs displayed joint regeneration (B’). When the joint was removed by SE surgery (C), regeneration did not take place (n=6) and a fused cartilage was observed (C’, arrow). Fusion is observed mainly between the humerus (h) and the ulna (u); however, h-u and h-r (r: radius) fusion together is also observed (C’, inset). D and E) Proximal and distal amputations verify that the elbow was removed by both WE or SE methods. Limbs were amputated at the indentation, indicated by “P” for proximal (D) or 150 µm distal to the indentation (E) indicated by “D” for distal amputation (see lines in D, E). The proximal amputations did not include elbow tissue and only the humerus could be observed (D’). On the other hand, the distal amputations contained the elbow tissue (E’). h: humerus, r: radius, u: ulna.
DiI injections
Lipophilic tracer DiI (Molecular Probes) was injected by using thin-walled capillary glass needles (10 µl, Drummond Scientific Company), which were prepared using a hot glass needle puller. The needles were backfilled with DiI (3 mg/ml in dimethylformamide) (50 µg DiI + 5 µl 100% ethanol + 45 µl 0.3M sucrose) and injections were carried out using an automated microinjection apparatus (Micro4-IM-0800, World Precision Instrument Inc.) or mouth pipetting. For visualizing the fluorescent signal, the samples were paraffin embedded, sectioned and counterstained with DAPI (Andrade et al., 1996; Obara-Ishihara et al., 1999). Slides were mounted with Prolong Gold anti-fade agent.
Histological and skeletal staining
Histological analysis
Limbs were fixed in zinc-formalin (Z-FIX, Anatech LTD) at room temperature (RT) overnight, processed for paraffin embedding and then sectioned. For hematoxylin and eosin staining, standard protocols were followed. For Alcian blue on sections, Alcian blue 8GX solution (pH 2.5) was prepared in 3% glacial acetic acid. Deparaffinized sections were rehydrated and stained in Alcian blue for 30 mins. After rinsing in distilled water, sections were counterstained in nuclear fast red for 5 mins., dehydrated and mounted in Permount.
Skeletal staining
Embryos fixed in zinc-formalin were washed in PBS the next day. For Victoria blue staining, they were directly transferred to staining solution (1% Victoria blue in acidified ethanol [1% concentrated HCl in 70% ethanol]) and kept on a rotator for 2 hours. They were then destained in 70% ethanol, dehydrated in ethanol and cleared in methyl salicylate for visualizing skeletal elements (Bryant and Iten, 1974).
Section in situ hybridization
Embryos were fixed at the desired time point in 4% paraformaldehyde (PFA) at 4°C overnight. Each specimen was sectioned at 8 µm and individual sections were placed on four different slides to obtain serial sections on each slide. This allowed testing of four different markers for one sample embryo. For each time point and marker, a minimum number of five embryos was used. Section in situ hybridization was performed as previously described (Han et al., 2003). Briefly, dewaxed slides were rehydrated in an ethanol series, digested with 1 µg/ml Proteinase K for 20 mins., fixed in 4% PFA for 30 mins., and washed in 1x SSC before hybridization. Hybridization solution containing probe was added and slides were incubated at 70°C overnight. On the second day, AP-conjugated anti-digoxygenin antibody was used (overnight at 4°C). On the following day, embryos were washed in TBST (with 1 mM levamisole) and NTMT (with 2 mM levamisole) at RT and the color reaction was obtained by incubation in BM Purple (Roche) at 4°C. After the desired color intensity was obtained, slides were counterstained with safranin-O and coverslipped in Permount.
Antisense digoxigenin-labeled RNA probes for in situ hybridization were generated by in vitro transcription from linearized plasmids. The plasmids that were obtained from ARK Genomics EST database (Boardman et al., 2002) for making anti-sense RNA probes are: Autotaxin - ChEST265j18 (pBKS (+) - Sac2/T3), Sox-9 - ChEST378d23 (pBKS (+) - Sac1/T3), Col-9 - ChEST859o21 (pBKS (+) - Sac2/T3), Chordin - ChEST851k13 (pBSK (+) - Sac2/T3), Cux-1 - ChEST372n14 (pBKS (+) - Sac2/T3), Col-2α1 – ChEST212h23 (pBKS (+) - Sac1/T3), Noggin - ChEST228i9 (pBKS (+) - Sac1/T3), Wnt-9a - ChEST592n13 – (pBKS (+) - Sac1/T3), Hes-1 - ChEST900h16 (pBSK (+) - Sac1/T3). GDF-5 (pCRII-TOPO NotI/Sp6) was cloned by Jangwoo Lee (accession number: NM_204338). Prickle-1 and Prickle-2 plasmids were kindly provided by Dr. Andrea Münsterberg. BMP-2, BMP-4 and Gli-3 plasmids were kindly provided by the laboratory of Dr. Juan Carlos Izpisúa Belmonte. Wnt-4 plasmid was kindly provided by Dr. Cristophe Marcelle.
Results
Histological analysis and cell-fate mapping of the elbow joint
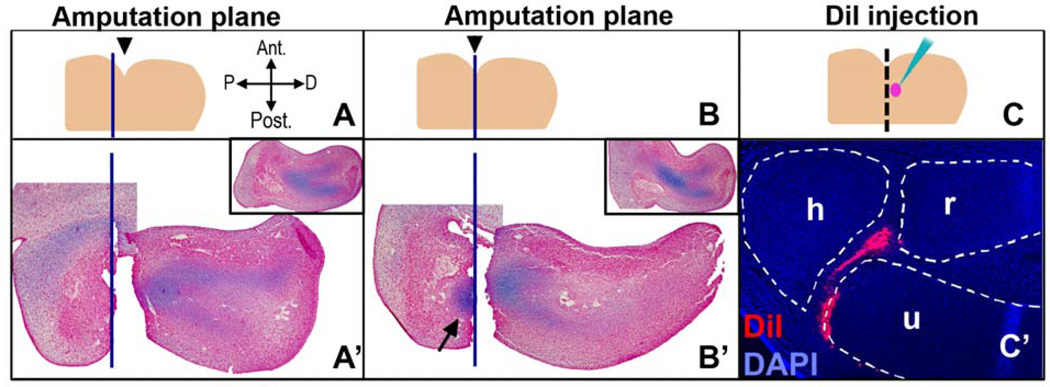
In order to surgically remove the prospective elbow tissue, it was first necessary to analyze the developing forelimb for its histology and anatomy, and to set up a technique whereby the elbow tissue could be precisely determined by using a morphological landmark. The elbow joint forms at the intersection of three skeletal elements: the humerus, the radius and the ulna. At stage 26 the skeletal system of the limb is a “Y-shaped” condensed mesenchyme that can be identified based on Alcian blue staining (Fig. 1A'). At this stage a morphological landmark is available: the anterior side of the limb forms an easily visible indentation that marks the initial curvature of the forming wing (Fig. 1A). We used a measurement graph provided by a previous study on elbow development in the chicken (Holder, 1977) and roughly predicted that the elbow tissue should be found near the level of this indentation. Next, in order to determine which part of the differentiating cartilage was located relative to the indentation, the limb was amputated either 75 µm proximal to the indentation (Fig. 1A, A’) or at the indentation (Fig. 1B, B’). The stumps and corresponding amputated pieces were fixed immediately after amputation, then paraffin sectioned and stained with Alcian blue (cartilage) and neutral red (cell nuclei). Note that because of the asymmetry of the limb along the dorsal-ventral axis and/or the plane of sectioning, as well as shrinkage during tissue preparation, indentations observed in sectioned limbs are not indicative of the indentation used to locate elbow tissue at the time of the surgery. Amputations proximal to the indentation removed most of the Y-shaped cartilage (Fig. 1A’), whereas amputations at the indentation left a piece of cartilage that would be the distal part of the humerus (Fig. 1B’, arrow). As a result, the indentation level marked the proximal border of the presumptive elbow tissue.
Figure 1.
Determination of the prospective elbow tissue by histological and fate mapping analyses. A landmark on a stage 26 limb was determined (the indentation indicated by black arrowhead in 1A and 1B) and used for locating the elbow tissue. Note that the indentation serves as a landmark at the time the limb is operated on. Indentations observed in sections of limbs are not necessarily reflective of the dorsal view used to initially pinpoint the elbow tissue since limbs are not symmetrical along the dorsal-ventral axis. Forelimbs of embryos at st. 26 were amputated either 75 µm proximal to the landmark (A-A’) or at the landmark (B-B’) level (blue lines indicate the amputation plane). After the limbs were amputated, the amputated piece and the stump were both fixed at t=0, sectioned and stained with Alcian blue and neutral red in order to visualize cartilage and the relative position of the amputation. Insets show the control contralateral limbs. When the amputation level was at the indentation, a part of the differentiating cartilage was left in the stump (B’, arrow). This indicated that the prospective elbow tissue was distal to the indentation. C-C’) In order to confirm this, limbs at stage 26 were injected with DiI for cell fate tracking and incubated until stage 35. The injected area was between 60–90 µm distal to the indentation (represented by pink dot in C). The sections of these limbs revealed that injections in this region result in cells positive for DiI in the elbow joint interzone (red). Blue signal is DAPI. h: humerus, r: radius, u: ulna.
To confirm the location of the prospective elbow tissue, we performed cell lineage tracking of the tissue that lies distal to the indentation. An area between 60 to 90 µm distal to the indentation was injected with DiI, a fluorescent and lipophilic dye that binds to cell membranes and labels the cells at the site of injection and their progeny (Fig. 1C, C’). Embryos were then incubated until stage 35, fixed in zinc-formalin, paraffin sectioned and processed for DAPI staining. The mounted slides were analyzed for the location of DiI positive cells. In all of the samples that were labeled this way, there were positive cells in the elbow joint interzone (Fig. 1C’). Some of these limbs also had positive cells in the humerus, the radius and the ulna (data not shown). This confirmed that the presumptive elbow joint lies distal to the indentation at stage 26, with the interzone located specifically between 60–90 µm distal to the indentation.
The elbow joint is able to regenerate after excisional removal but not slice removal
The chick limb bud lacks the ability to regenerate following amputation at all stages of its development. We verified this for stage 26; limbs were amputated at the proximal (Fig. 2D, D’) and distal (Fig. 2E, E’) borders of the prospective elbow region. After amputation, limbs were incubated for four more days and processed for skeletal staining. According to our histological and cell fate mapping analysis (Fig. 1), the proximal amputations should not contain any elbow tissue, and the distal amputations should include the elbow joint but should not display the complete radius and ulna. Consistent with this, in the proximal amputations only the humerus could be observed, but it lacked the distal tip that articulates with the radius and the ulna (Fig. 2D’). On the other hand, the distal amputations contained the elbow tissue (Fig. 2E’) with small pieces of radius and ulna present, but without any sign of significant regeneration of these two skeletal elements. These amputations verify that the developing chick limb is not able to regenerate upon amputation at stage 26. Also, here we verify that the area between the proximal and distal borders of the wound contains the cells that form the elbow, and elbow tissue can be removed from the developing limb by surgical excision of this area.
In addition to simple amputations, it has been previously shown that intercalary regeneration does not take place if the elbow tissue is removed by slicing it out at stages 24–26 and pinning the distal tip of the amputated bud to the stump (Holder, 1977; Summerbell, 1977). This type of tissue removal results in fusion of the humerus to the radius and to the ulna and failure of the elbow joint to form. We repeated Holder’s slicing method to remove developing elbow tissue at stage 26 (Slice Excision, SE; Fig. 2C). This and the following surgeries were designed based on the cell lineage tracking studies of the elbow; cutting a 150 µm-wide block of tissue starting from the indentation level at stage 26 successfully removes the elbow tissue (See Methods and Fig. 1). After the elbow was sliced out and the pieces of the limb were attached to each other by using platinum pins, embryos were incubated for four days. Then, the operated limb and the unoperated contralateral limb (Fig. 2A) as the control were fixed and processed for Victoria blue skeletal stain (Fig. 2A’–E’) in order to reveal overall morphology. In all limbs that underwent SE (n=6), regeneration did not take place; instead skeletal elements were fused (Fig. 2C’). This verifies Holder’s results. Intercalary regeneration does not take place if the elbow tissue is sliced out and the remaining limb pieces are pinned together without a space between the wound surfaces.
Next, we designed a surgical method for removing the prospective elbow at stage 26 by keeping the surrounding tissue intact (Window Excision, WE; Fig. 2B). In WE, the elbow forming region is specifically excised and this excision creates a hole (or “window”) in the dorso-ventral axis of the limb with anterior and posterior strips of tissue holding the distal piece attached to the proximal side of the limb (Fig. 2B). After the surgeries, embryos were incubated for four days and experimental and control limbs were processed the same way as in SE. Interestingly, this excisional surgery created a permissive environment for regeneration and limb buds operated in this way showed endogenous regenerative capability: in 50% of the limbs (n=20), elbow joint regeneration took place (Fig. 2B’). In the regenerates, the articulation between the humerus and the ulna was always present, while the articulation of humerus and radius sometimes failed to form (data not shown). However, with SE the regeneration response was inhibited, and this was indicated by fusion of skeletal elements (Fig. 2C’). In addition, the regeneration observed in WE was temporally restricted; the surgeries performed a day later (stage 28/29) did not regenerate (data not shown).
Regeneration is similar to development at the histological level
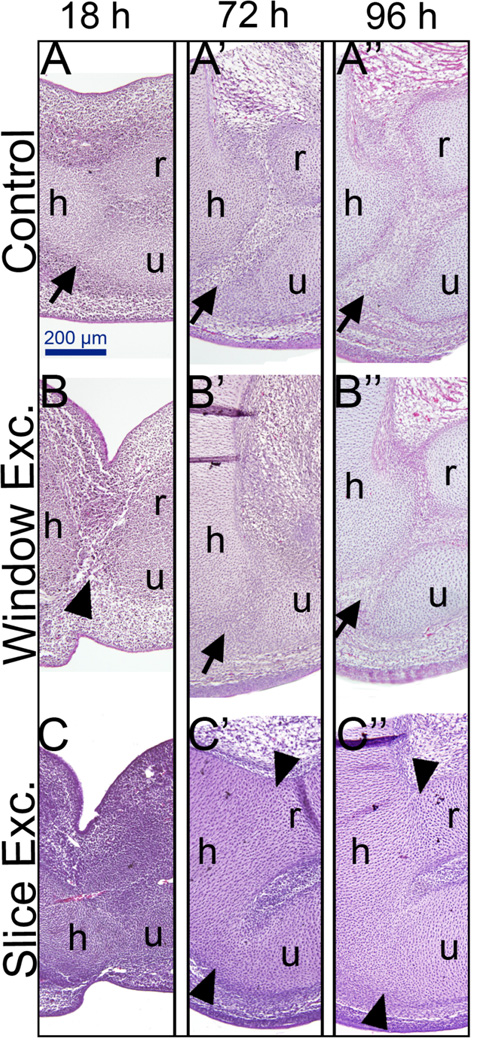
During joint development, the differentiating cartilage becomes segmented due to a group of cells that become flattened and mesenchymal. This region is called the joint interzone, and these cells eventually form the mature joint. We analyzed the regeneration response in WE as well as in nonregenerating SE limbs at different time points after surgery. The operated limbs and their contralateral unoperated controls were fixed at 18, 72 and 96 hours after the surgery, and sectioned and stained with hematoxylin and eosin. At 18 hours after WE surgery, the wound was completely closed, the anterior and posterior ectoderm surrounding the wound appeared constricted, and the hole that was created after tissue removal was filled by cells (Fig. 3B). By 72 hours, a structure that histologically looked similar to a joint interzone started to form (Fig. 3B’). We also observed that the regenerating joint looked less developed than the contralateral control. By 96 hours, the regenerating joint became even more evident with cells that appeared more differentiated and layered (Fig. 3B”).
Figure 3.
Histological analysis of joint regeneration. After either WE or SE, limbs were incubated until 18 hours, 72 hours or 96 hours, sectioned and stained with hematoxylin and eosin. A-A”) Controls. 18 hours after surgery corresponds to a 6 day old embryo in the control. By this time, cartilage is differentiating and the interzone can be observed (A, arrow). By 72 hours and 96 hours the control limbs have a developing joint with a distinct interzone (A’ and A”, arrows). B-B”) Window excision (WE). By 18 hours after surgery, the excised area is filled with cells (B, arrowhead). By 72 hours, the regeneration response can already be observed indicated by the presence of a joint interzone (B’, arrow). Note that this interzone does not look as developed as the control (compare A’ to B’). By 96 hours, the regeneration response can be observed by a distinct joint interzone (arrow in B”) comparable to the control (A”). C-C”) Slice Excision (SE). In contrast to WE, SE limbs show fusion of cartilage elements observable at all time points: fusion of the humerus and the ulna is evident at 18 hours. By 72 and 96 hours after surgery, a majority of SE limbs lack an interzone, and they all have completely fused elements (C’ and C”, arrowheads). h: humerus, r: radius, u: ulna, arrows: interzone
Limbs that had undergone SE surgery showed fusion of cartilage elements starting as early as 18 hours after the surgery (Fig. 3C), in contrast to WE where noncartilagenous cells are present between skeletal elements (Fig. 3B). By 72 and 96 hours after the SE surgery, two or three of the skeletal elements (either the humerus and ulna, or the humerus, radius and ulna) appear as a continuous cartilage without any evidence of joint interzone formation (Fig. 3C’, C”).
Regeneration follows a similar molecular program as joint development
Morphological and histological analysis of the developing elbow joint has been described thoroughly (Holder, 1977; Merida-Velasco et al., 2000). However, a gene expression analysis specifically for the elbow joint has not been performed; most of the molecular analyses of synovial joints have focused on phalangeal joints. The information available in the literature suggests that developmental mechanisms might be different for making a phalangeal joint versus an elbow joint (or other larger and more proximal joints of the limb such as the knee or shoulder). This is most dramatically reflected in gene knock-out (KO) and overexpression studies. For example, the GDF-5 KO displays a joint fusion phenotype in the digits; however, the elbow joint does not display such a phenotype (Storm and Kingsley, 1999). In order to describe the gene expression patterns for the developing elbow and select suitable markers among them for addressing whether regeneration of the elbow recapitulates development at the molecular level, we performed a gene expression analysis of previously described joint and cartilage markers as well as some additional markers of pathways that are implied to be involved in joint and cartilage development for the elbow joint (see Supplementary Fig. 1).
Reinitiation of molecular developmental programs during regeneration is indicated as a common mechanism in the literature (Bely and Wray, 2001; Callaerts et al., 1999; Devarajan et al., 2003; Duggal et al., 1997; Imokawa and Yoshizato, 1997; Muneoka and Bryant, 1982, 1984; Torok et al., 1998). During limb development, Sox-9 is one of the initial transcription factors that defines the bauplan of the future skeletal tissue; it is expressed in the cells of the condensed mesenchyme (Supplementary Fig. 1). Sox-9 expression marks the beginning of mesenchymal differention into cartilage because Sox-9 directly regulates the expression of cartilage differentiation genes, among which is Col-9 (Bi et al., 1999). Initially, both Sox-9 and Col-9 have uniform expression throughout the differentiating cartilage. However, once the joint tissue starts to form and is evident as a joint interzone, the uniform Col-9 expression becomes segmented by becoming downregulated at the joint interzone. Sox-9 expression persists longer than Col-9 in the joint interzone, but eventually becomes downregulated. The downregulation of Col-9 is accompanied by Autotaxin (Atx) and GDF-5 expression becoming more specific to the joint interzone (Supplementary Fig. 1).
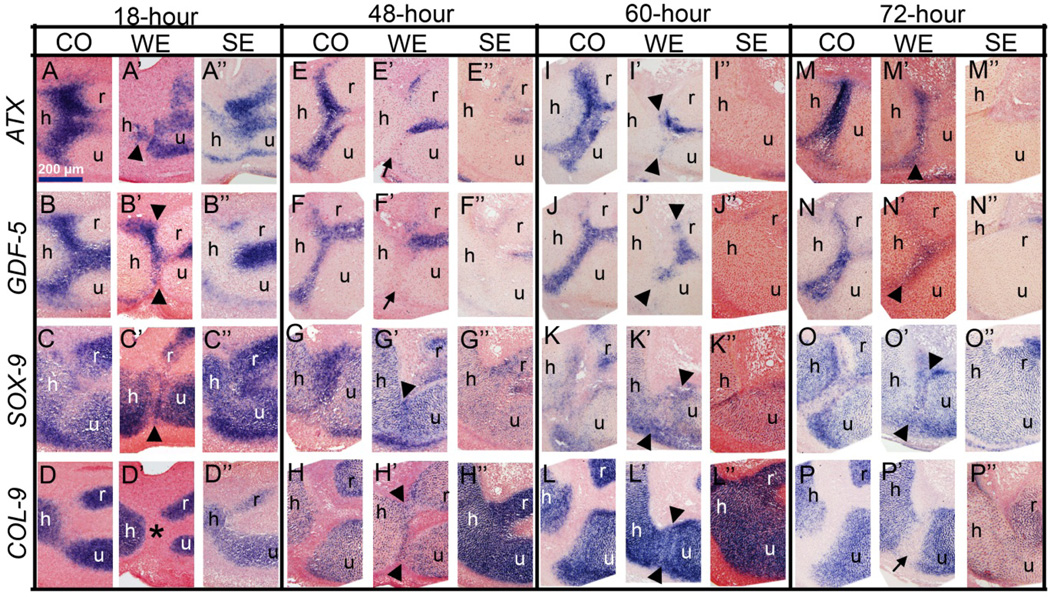
In order to determine whether the regeneration response observed after WE surgery reinitiates this developmental program, we analyzed Sox-9, Col-9, GDF-5 and Atx as the markers of developing cartilage (Sox-9, Col-9) and the joint (GDF-5, Atx) by in situ hybridization on sections. For each time point and marker, a minimum of five samples were used (see Materials and Methods) and a representative picture was selected for the panel in Figure 4. In the samples that underwent WE, the regenerating joint followed similar temporal and spatial patterns of gene expression to development, whereas nonregenerating samples of SE did not (Fig. 4). In WE embryos, 18 hours after the surgery, the cells that had filled the wound site between the humerus, radius and ulna expressed Atx, GDF-5 and Sox-9 (Fig. 4A’, B’ and C’, respectively) but not Col-9 (Fig. 4D’). Interestingly, the expression of joint genes (Atx and GDF-5) disappeared by 48 hours after surgery in WE embryos (Fig. 4E’, F’). However, these samples expressed both Sox-9 and Col-9 (Fig. 4G’, H’). Sometime between 18 and 48 hours, the cells that filled the wound site had downregulated joint specific genes and reinitiated Col-9 expression. By 60 hours after WE surgery, the joint interzone started to become evident with cells that appeared slightly flattened, compared to the surrounding cartilage (Fig. 4I’, J’, K’, L’, arrowheads). In addition, cells of the joint interzone expressed Atx and GDF-5 (Fig. 4I’, J’). Finally, 72 hours after surgery, the joint interzone could be clearly distinguished with cells that appeared mesenchymal and that expressed Atx, GDF-5 and Sox-9 (Fig. 4M’, N’, O’, respectively), but downregulated Col-9 (Fig. 4P’). In contrast to WE, after the SE surgery, Atx and GDF-5 gene expression patterns were absent at both 60 hours (Fig. 4I”, J”) and 72 hours (Fig. 4M”, N”). In addition, the skeletal elements appeared fused in a uniformly cartilaginous pattern. Consistent with this observation is the finding that Sox-9 and Col-9 expression was continuous between all three limb elements (Fig. 4K”, L”, O”, P”). In summary, molecular events in regenerating WE samples is comparable to the events that take place during joint development, and this strongly suggests that joint regeneration recapitulates development.
Figure 4.
Gene expression analysis of window excision (WE) and slice excision (SE) limbs at different time points after surgery. 18 hours: A–D”) By this time, in WE the wound is closed and cells that fill in between the skeletal elements appear mesenchymal and express Atx (A’), GDF-5 (B’), Sox-9 (C’) but not Col-9 (asterix in D’). In contrast, SE samples do not display such a mesenchymal cell population and the skeletal elements appear fused (A”, B”, C”, D”). 48 hours: E–H”) At this time point, in WE, cells at the interzone downregulate Atx (E’, arrow) and GDF-5 (F’, arrow) and start expressing Col-9 (H’, arrowheads). SE skeletal elements still look fused (G”, H”) without Atx (E”) and GDF-5 (F”) expression. 60 hours: I–L”) In WE, the appearance of a histologically prominent interzone is accompanied by re-expression of Atx (I’, arrowheads) and GDF-5 (J’, arrowheads). At this stage, Sox-9 (K’, arrowheads) and Col-9 (L’, arrowheads) are still expressed at the regenerating interzone. SE elements still appear fused without joint gene expression (I”, J”, K”, L”) 72 hours: M-P”) In WE, the regenerating joint is even more evident, interzone cells express Atx ( M’, arrowhead), GDF-5 (N’, arrowhead) and Sox-9 (O’, arrowhead) but Col-9 is downregulated (P’, arrow). Compare these expression patterns of the regenerating joint to the control joint in Fig. 4E, F, G, H for similarity. In contrast to WE, SE limbs still lack the expression of joint genes Atx (M”) and GDF-5 (N”). However, consistent with lack of regeneration, fused cartilage cells express Sox-9 (N”) and Col-9 (P”). In all sections, safranin-O was used for the counterstain. For each time point and each marker, a minimum number of five samples were analyzed and a representative sample was used in this figure. Arrowheads indicate gene expression at the interzone and arrows indicate the lack of gene expression. h: humerus, r: radius, u: ulna.
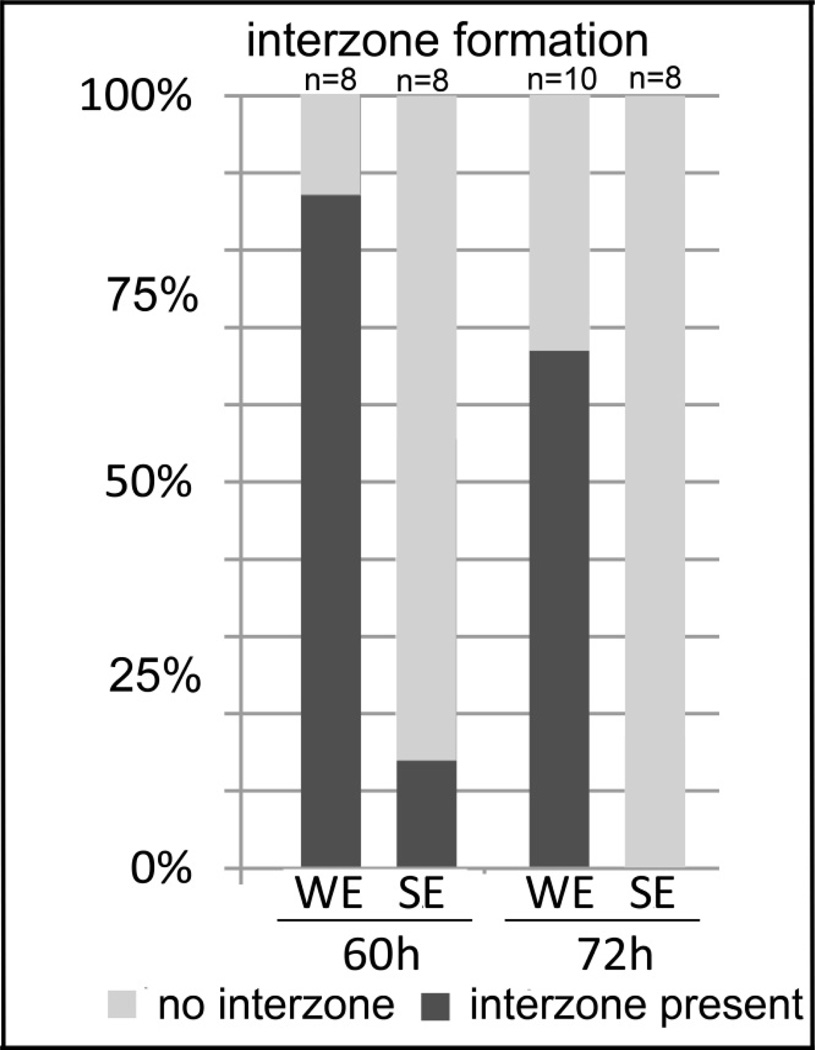
In order to determine whether gene expression correlated with joint regeneration or its failure at the histological level, we scored all the 60-hour and 72-hour in situ hybridization samples for interzone formation and cartilage fusion. Here, every sample was analyzed for the presence or absence of joint interzone formation and cartilage fusion between the humerus and the ulna. The percentages for interzone formation was summarized in a bar graph (Fig. 5). Among the 60-hour samples, 88% of WE surgeries resulted in interzone formation while only 12.5% of SE limbs displayed interzone formation. By 72 hours after surgery, none of the SE samples had any evidence of an interzone while interzone formation was significantly higher in WE samples (67%). The WE samples are able to regenerate, have a higher percentage of interzone formation and a lower percentage of cartilage fusion (data not shown) correlating with the presence of joint gene expression. In addition, the absence of joint-specific gene expression in SE correlates with a lower percentage of interzone formation and a higher percentage of cartilage fusion (data not shown).
Figure 5.
Percentage summary of interzone formation and cartilage fusion. All the samples analyzed for in situ hybridization were individually scored for the presence and absence of the interzone and cartilage fusion. A) The percentage of samples that display interzone formation after window excision (WE) and slice excision (SE) at 60h and 72h after surgery. At both time points, a significantly higher number of samples after WE have interzone formation, 88% at 60h and 67% at 72h. In contrast, only 12.5% of samples have interzone formation at 60h and none of them display a joint interzone at 72h. Interzone formation indicates joint regeneration. B) A significantly higher percentage of samples after SE surgery display fusion of the humerus and the ulna at 60h (75%) and 72h (100%), in contrast to WE samples which do not show fusion at 60h or a much lower percentage of samples at 72h (22%). Skeletal fusion indicates failure of regeneration.
Cell tracking after window excision
In order to identify the source of cells that participate in the regeneration response we performed a cell tracking analysis on window excised (WE) limbs. Limbs were injected with lipophilic DiI in several different ways. In experiments A, B and C, first the WE was performed and then the limbs were injected with DiI (Fig. 6F). Once the tissue is excised, the remaining anterior and posterior tissue strips are very thin and, as a result, hard to label by DiI injection without the dye leaking into the “window” or without the tissue being torn. In order to overcome this, for marking the anterior or posterior sides of the wound, DiI was injected first and then the window excision surgery was performed immediately after this injection in experiments D and E (Fig. 6F’).
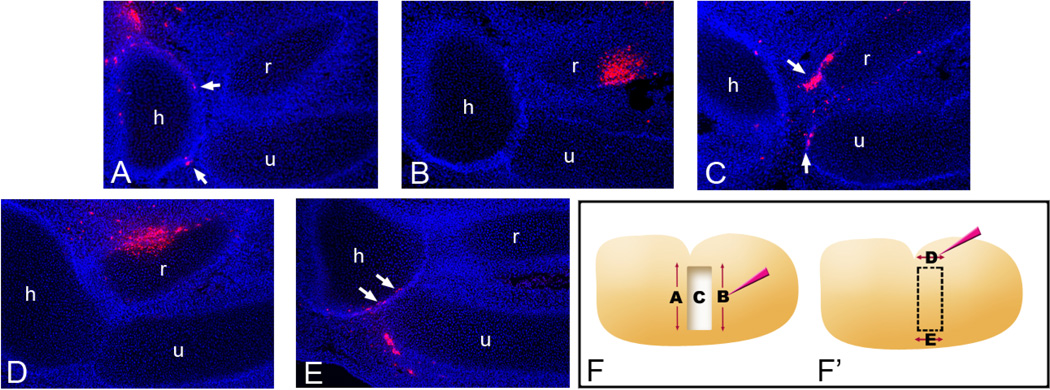
Figure 6.
DiI injections to determine contribution of cell populations to the regenerated joint. (A) DiI injections to the proximal side of the window (A), the distal side of the window (B) and application of DiI to the wound surface (C) were made following window excision. DiI injections to the anterior side of the window (D) and the posterior side of the window (E) were made immediately before window excision. All the experiments are shown schematically in F and F’. (A) DiI injections to the proximal side of the window excision result in a few labeled cells located in the joint interzone with most located near the mid to proximal humerus. (B) Injections to the distal side of the excision show labeled cells in the radius. (C) Application of DiI to the wound surface results in labeled cells in the regenerated joint interzone (arrows). (D) Anterior DiI injections mark a large population of cells near the radius but not in the interzone. (E) Injections to the posterior margin of the window excision results in a number of labeled cells in the regenerated joint interzone (arrows). Each picture is a representative of several injections carried out along the corresponding axis. The blue signal is DAPI. h: humerus, r: radius, u: ulna.
After proximal injections were carried out along the anterior-posterior axis, only a few DiI-labeled cells were observed at the regenerating joint interzone (Fig. 6A). In most of the samples that received distal injections along the anterior-posterior axis, DiI-positive cells were not observed in the regenerating joint interzone (Fig. 6B). On the other hand, when the wound surface cells were marked by slowly injecting DiI towards the wound surface, large clusters of DiI-labeled cells were observed at the regenerating joint interzone in contrast to proximal and distal injections (Fig. 6C). Anterior cell markings with DiI made along the proximal-distal axis also did not reveal a significant amount of cells in the regenerating interzone, although there were very occasionally a few positive cells (Fig. 6D). Lastly, the samples that received posterior injections along the proximal-distal axis displayed a significant number of DiI-positive cells in the regenerating interzone (Fig. 6E), indicating that this tissue, along with the cells of the wound surface (Fig. 6C), is one of the major sources of cells that contribute to the regeneration response.
To verify the significance of the posterior cell contribution to the regenerated joint, DiI injections were made along the posterior margin of the unoperated developing limb at stage 26. None of these injections (n = 14) resulted in labeled cells in the joint (data not shown). DiI-labeled cells were found along the proximal-distal axis in a linear trajectory toward the distal limb. In addition, labeling cells at the posterior margin of slice excisions (n = 8), whether proximally in the stump or distally in the pinned tissue, resulted in labeled cells confined to the posterior margin of the limb (data not shown). Our results suggest that the responsiveness of posterior cells to migrate to the injury site dictates the ability to regenerate.
Discussion
Using the urodele limb regeneration model, researchers have determined that regeneration can be conceptually divided into two phases: a regeneration-specific phase and a redevelopment phase (reviewed in Bryant et al., 2002 and Gardiner et al., 2002; Han et al., 2005). The regeneration-specific phase includes wound healing, dedifferentiation and cell migration to form the regeneration blastema; the redevelopment phase involves growth and patterning events of the blastema to form the regenerate (Bryant et al., 2002; Gardiner et al., 2002). Although urodele regeneration has enabled researchers to identify the key requirements for successful regeneration, it is becoming apparent that higher vertebrate limb regeneration follows a similar pattern of events. Indeed, many of these events have been demonstrated to occur in the regenerating digit tips of adult mice (Fernando et al., 2011). In the work we present here, our initial characterization of the window excision model in the chick embryo forelimb confirms that regeneration following excision exhibits many of the hallmarks of urodele limb regeneration, including cell migration during wound healing and re-initiation of the developmental program.
Regeneration permissive wounding
One of the key factors required to mount a successful regeneration response is the creation of a regeneration permissive wound (Bryant et al., 2002; Fernando et al., 2011; Han et al., 2005; Yokoyama, 2008). Our experimental paradigm, window excision (WE), is essential to achieve elbow joint regeneration. We have confirmed that the chick limb is not regenerative after simple amputation and that the chick elbow cannot regenerate if the presumptive elbow “slice” is removed (SE) and the distal tip of the bud is pinned back onto the stump, sandwiching the tissues together so as to minimize a wound healing response (Holder, 1977; Summerbell, 1977). We find that the elbow joint is able to regenerate when the removal method is altered: if the prospective elbow joint tissue is removed using an excision method creating a “window” with anterior and posterior strips of tissues holding the limb together, elbow regeneration takes place (Fig. 2). This observation suggests that varying the way that the wound is healed (tissues in close association versus cells filling in a space) results in dramatic differences in the regenerative response of the tissues. In addition, our results indicate that the wound environment itself is developmentally regulated. The lack of 100% regenerative success in our st. 26 WE limbs (50% regenerate) is likely a temporal effect. This idea is supported by the complete loss of regenerative repair by st. 29. Indeed, previous studies have demonstrated that in the developing chick limb, regeneration of excisional wounds is limited by the limb stage (Stark and Searls, 1974). Thus, these findings, combined with our own, suggest that we have defined a temporal window that marks the decline of regenerative ability.
A regeneration permissive wound environment is dependent on the formation of a wound epidermis, which forms from the migration of epidermal cells across the wound surface that eventually thickens (the apical epithelial cap, AEC, in amphibian limb regeneration). This specialized epidermis forms in a matter of hours following amputation (reviewed in Gardiner et al., 2002; Yokoyama, 2008). Following wound closure, fibroblasts from the loose connective tissue of the dermis migrate across the amputation plane to accumulate and form the blastema (Endo et al., 2004; Gardiner et al., 1986; Muneoka et al., 1986). In WE, we have observed that the wound site is filled in by cells in less than 18 hours, suggesting a rapid cellular response, i.e., proliferation and/or migration, which could promote wound closure. Both of these mechanisms might be prevented in SE because of the immediate contact and fusion of the wound surfaces.
Contrasting the two methodologies, WE and SE, suggests that the the anterior and posterior strips of tissue found at the window excision margins are important for the regeneration response to occur. One explanation for the regenerative success of the WE method is that these tissues could contain joint progenitor cells that are not removed during WE. While our DiI fatemapping of the normal (unoperated control) stage 26 limb does not support this (see last paragraph of the Results section), we cannot be certain that all joint forming cells are removed in WE. However, despite this possibility, the regeneration response observed in WE is in striking contrast to SE and the model we present here provides a useful assay for joint repair. Another plausible explanation that could account for regeneration of the joint in WE is that the cells remaining near the window excision comprise part of a joint field that are then recruited via wounding to participate in the regeneration response (see below).
Posterior cells migrate in response to WE
The ability of cells to migrate in response to injury has been characterized in amphibian limb regeneration. Cell migration across the amputation surface is an essential component of blastema formation in the regenerating urodele limb (Gardiner et al., 1986; Muneoka et al., 1986). Our cell tracking analysis indicates that in WE limbs, cells from the wound surface are found in the joint interzone, placed there presumably through migration and/or proliferation. In addition, analysis indicates that cells at the posterior margin of the excision migrate and participate in joint regeneration. This data indicates that like urodele limb regeneration, a migratory response to wounding is initiated. Unlike urodele limb regeneration, a posterior bias to cellular contribution to the regenerate is observed. While the role of posterior cell migration to joint regeneration is unclear, it is an additional distinction between SE and WE. SE limbs do not manifest a posterior cell migration to the wound excision site.
The movement of posterior cells in an anterior direction is highly irregular in the chick limb during normal development (Gros et al., 2010; S. Li and Muneoka, 1999; Vargesson et al., 1997; Wyngaarden et al., 2010). In our own hands, Di-labeled posterior cells in the developing stage 26 limb show a clear trajectory from proximal to distal toward the apical ectodermal ridge (AER). However, posterior cells are responsive to FGF-4 or WNT5A signaling and their migratory trajectory during limb outgrowth can be modified (Gros et al., 2010, Li and Muneoka, 1999; Wyngaarden et al., 2010). Thus, these two signaling pathways represent potential candidates for regulating the migration response during WE joint regeneration.
The contribution of posterior cells to the regenerated joint is consistent with the demonstrated versatility of the posterior limb bud during development. Fate maps indicate that posterior cells are largely responsible for the bulk of limb formation (Saunders, 1948; Stark and Searls, 1973; Bowen et al., 1989; Muneoka et al., 1989; Vargesson et al., 1997; Harfe et al., 2004), demonstrating a developmental bias of posterior cell contribution to the limb. Shubin and Alberch (1986) have suggested a strong posterior bias in the evolution of the limb with posterior branching of the metapterygial limb axis forming the digital arch (reviewed in Hinchliffe, 2002). These observations along with the responsiveness of posterior cells to chemoattractive signals (Gros et al., 2010; Li and Muneoka, 1999; Vargesson et al., 1997; Wyngaarden et al., 2010) are consistent with the hypothesis that posterior cells display a level of plasticity that allow them to participate in a regenerative response.
A recapitulation of the joint developmental program during joint regeneration
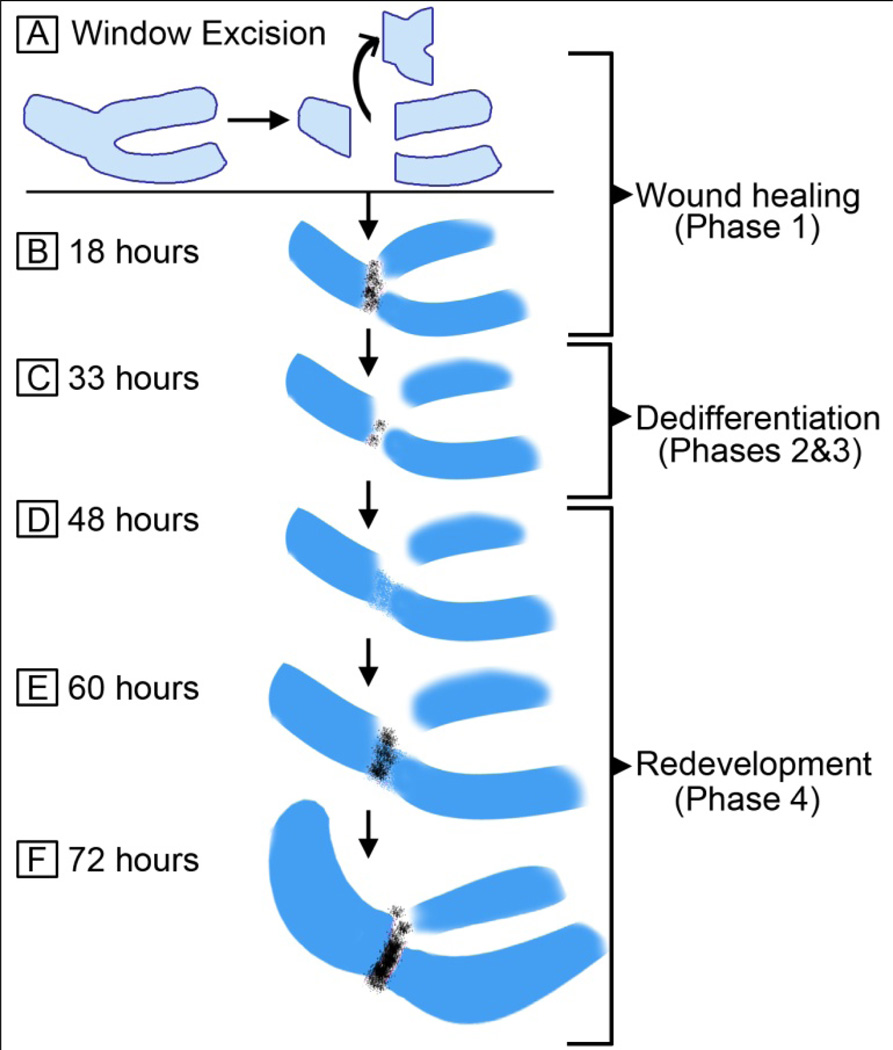
Recapitulation of developmental pathways has been documented in a number of regenerating systems and contexts (Akimenko et al., 1995; Bely and Wray, 2001; Burton and Finnerty, 2009; Callaerts et al., 1999; Devarajan et al., 2003; Duggal et al., 1997; Goetsch et al., 2003; Han et al., 2008; Imokawa and Yoshizato, 1997; Muneoka and Bryant, 1982, 1984; Torok et al., 1998; Whitehead et al., 2005; Yu et al., 2010). In our joint tissue regeneration model, a pattern of gene expression that reflects both a dedifferentiation or reversal of a prescribed molecular program and a recapitulation of the developmental program is observed. We have modeled our findings on the described phases of regeneration (reviewed in Bryant et al., 2002; Gardiner et al., 2002; Han et al., 2005). Eighteen hours after WE in the regenerating samples, cells fill the wound site (Fig. 7B). At this time they are positive for the joint markers GDF-5 and Atx, but not for Col-9. This time frame can be regarded as the wound healing phase (phase 1, Fig. 7B). By 33 hours, the cells have downregulated joint-specific genes and appear mesenchymal and dedifferentiated (Fig. 7C). These are phases 2 and 3 of regeneration, and correspond to dedifferentiation and blastema formation, respectively. Next, 48 hours after the surgery, cells express Sox-9 and Col-9, laying out the “bauplan” of the missing part of the skeleton, marking the initiation of redevelopment (phase 4). There is no significant joint gene expression at this time (Fig. 7D). By 60 hours after the surgery, cells start to express joint genes (Fig. 7E), and the interzone starts to form, which is evident by flattened cells. Finally, by 72 hours, similar to development, Col-9 becomes downregulated at the joint interzone during regeneration in WE and interzone cells continue to express joint specific genes (Fig. 7F). In contrast to the WE samples, limbs that have undergone SE fail to regenerate joints. This lack of regeneration is correlated with the absence of expression of joint specific genes Atx and GDF-5 and the absence of interzone formation (see Fig. 5), resulting in cartilage fusion. In accordance with this data, GDF-5 has been shown to be necessary and sufficient for rescuing joint development (Storm and Kingsley, 1999). The failure to re-express joint genes and induce interzone formation after SE strongly suggests that initiation of the joint development program is critical to the success of regenerating a joint.
Figure 7.
The model for elbow joint regeneration after window excision. A) Scheme of window excision: Elbow tissue is cut out and discarded. B) 18 hours after surgery: Cells fill the wound site in 18 hours. They are positive for joint markers GDF-5 and Atx (black). C) 33 hours after surgery: GDF-5 and Atx positive cells at the joint area are dramatically reduced. The tissue between the three cartilage elements look largely dedifferentiated and mesenchymal. D) 48 hours after surgery: Sox-9 and Col-9 positive cells layout the “bauplan” of the missing part, marking the initiation of redevelopment. There is no significant joint gene expression at this time. E) 60 hours after surgery: Cells start to express joint genes. The interzone starts to form, which is evident by flattened cells. F) 72 hours after surgery: Col-9 becomes downregulated at the joint interzone (similar to development). The regenerating joint is more prominent with GDF-5 and Atx positive cells.
Conclusion
From the amphibian limb regeneration studies, we know the key requirements for a successful regeneration response: first, a regeneration permissive environment is created through a wound healing response, second cells at the wound site dedifferentiate, then innervation takes place, and finally the patterning programs for making a limb are reinitiated. Among these key requirements, the wound healing response seems to be one of the differentiating aspects between the window versus slice excision models presented here. Clearly, the absence of a regenerative response in the slice excision does not mean that the tissue lacks regenerative potential, because in window excision regeneration is observed. There seem to be barriers that prevent the regeneration response from taking place in slice excision limbs at this stage, whereas window excision provides factors that overcome the barrier (or barriers). Through understanding the conditions that permit a joint regeneration response (such as the conditions that the window excision system provides), therapeutic approaches can be developed for joint injuries and degenerative joint diseases.
Supplementary Material
Supplementary Figure 1: In situ hybridization analysis of genes that are indicated in joint and/or cartilage development. Previous studies did not address the expression of most of these markers specifically at the elbow joint for the time points we used in this study. Limbs were fixed at Day 5, Day 7 and Day 8, sectioned and processed for in situ hybridization for the following markers: GDF-5, Atx, Sox-9, Col-9, BMP-2, BMP-4, Chordin, Noggin, Wnt-9a, Wnt-4, Hes-1, Col-2α1, Prickle-1, Prickle-2, Cux-1, Gli-3. Based on the expression patterns, we selected Atx and GDF-5 as joint specific markers and Sox-9 and Col-9 as cartilage markers for our study.
Highlights.
For the first time a model for joint regeneration using the chick embryo has been developed.
Joint regeneration in the developing chick recapitulates developmental pathways.
The injury response following window excision is conducive to regeneration of the joint.
Acknowledgements
We thank members of the Muneoka and Anderson Laboratories for critical reading and comments on the manuscript. This work was supported by funding from NIH-P01HD022610 (KM), ARO-W911NF0910305 (KM) and the John L. and Mary Wright Ebaugh Endowment Fund at Tulane University and NIHR15 HD060012-01 (RA) and 3R15HD060012-01S1 (RA), and the Loyola University Mullahy Biology Endowed Fund for Undergraduate Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Akimenko MA, Johnson SL, Westerfield M, Ekker M. Differential induction of four msx homeobox genes during fin development and regeneration in zebrafish. Development. 1995;121:347–357. doi: 10.1242/dev.121.2.347. [DOI] [PubMed] [Google Scholar]
- Andrade W, Seabrook TJ, Johnston MG, Hay JB. The use of the lipophilic fluorochrome CM-DiI for tracking the migration of lymphocytes. J. Immunol. Methods. 1996;194:181–189. doi: 10.1016/0022-1759(96)00083-x. [DOI] [PubMed] [Google Scholar]
- Bachner D, Ahrens M, Betat N, Schroder D, Gross G. Developmental expression analysis of murine autotaxin (ATX) Mech. Dev. 1999;84:121–125. doi: 10.1016/s0925-4773(99)00048-9. [DOI] [PubMed] [Google Scholar]
- Bely AE, Wray GA. Evolution of regeneration and fission in annelids: insights from engrailed- and orthodenticle-class gene expression. Development. 2001;128:2781–2791. doi: 10.1242/dev.128.14.2781. [DOI] [PubMed] [Google Scholar]
- Bi W, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Sox9 is required for cartilage formation. Nat. Genet. 1999;22:85–89. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- Boardman PE, Sanz-Ezquerro J, Overton IM, Burt DW, Bosch E, Fong WT, Tickle C, Brown WR, Wilson SA, Hubbard SJ. A comprehensive collection of chicken cDNAs. Curr. Biol. 2002;12:1965–1969. doi: 10.1016/s0960-9822(02)01296-4. [DOI] [PubMed] [Google Scholar]
- Bowen J, Hinchliffe JR, Horder TJ, Reeve AM. The fate map of the chick forelimb-bud and its bearing on hypothesized developmental control mechanisms. Anat. Embryol. (Berl.) 1989;179:269–283. doi: 10.1007/BF00326592. [DOI] [PubMed] [Google Scholar]
- Bryant SV, Endo T, Gardiner DM. Vertebrate limb regeneration and the origin of limb stem cells. Int. J. Dev. Biol. 2002;46:887–896. [PubMed] [Google Scholar]
- Bryant SV, Iten L. The regulative ability of the limb regeneration blastema of Notophthalmus viridescens: Experiments in situ. Wilhelm Roux’s Archives of Developmental Biology. 1974;174:90–101. doi: 10.1007/BF00577059. [DOI] [PubMed] [Google Scholar]
- Burton PM, Finnerty JR. Conserved and novel gene expression between regeneration and asexual fission in Nematostella vectensis. Dev. Genes Evol. 2009;219:79–87. doi: 10.1007/s00427-009-0271-2. [DOI] [PubMed] [Google Scholar]
- Callaerts P, Munoz-Marmol AM, Glardon S, Castillo E, Sun H, Li WH, Gehring WJ, Salo E. Isolation and expression of a Pax-6 gene in the regenerating and intact Planarian Dugesia(G)tigrina. Proc. Natl. Acad. Sci. USA. 1999;96:558–563. doi: 10.1073/pnas.96.2.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devarajan P, Mishra J, Supavekin S, Patterson LT, Potter SS. Gene expression in early ischemic renal injury: clues towards pathogenesis, biomarker discovery, and novel therapeutics. Mol. Genet. Metab. 2003;80:365–376. doi: 10.1016/j.ymgme.2003.09.012. [DOI] [PubMed] [Google Scholar]
- Duggal N, Schmidt-Kastner R, Hakim AM. Nestin expression in reactive astrocytes following focal cerebral ischemia in rats. Brain Res. 1997;768:1–9. doi: 10.1016/s0006-8993(97)00588-x. [DOI] [PubMed] [Google Scholar]
- Duterloo HS, Jansen HW. Chondrogenesis and osteogenesis in the mandibular condylar blastema. Rep. Congr. Eur. Orthod. Soc. 1969:109–118. [PubMed] [Google Scholar]
- Endo T, Bryant SV, Gardiner DM. A stepwise model system for limb regeneration. Dev. Biol. 2004;270:135–145. doi: 10.1016/j.ydbio.2004.02.016. [DOI] [PubMed] [Google Scholar]
- Fernando WA, Leininger E, Simkin J, Li N, Malcom CA, Sathyamoorthi S, Han M, Muneoka K. Wound healing and blastema formation in regenerating digit tips of adult mice. Dev. Biol. 2011;350:301–310. doi: 10.1016/j.ydbio.2010.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner DM, Muneoka K, Bryant SV. The migration of dermal cells during blastema formation in axolotls. Dev. Biol. 1986;118:488–493. doi: 10.1016/0012-1606(86)90020-5. [DOI] [PubMed] [Google Scholar]
- Gardiner DM, Endo T, Bryant SV. The molecular basis of amphibian limb regeneration: integrating the old with the new. Semin. Cell Dev. Biol. 2002;13:345–352. doi: 10.1016/s1084952102000903. [DOI] [PubMed] [Google Scholar]
- Goetsch SC, Hawke TJ, Gallardo TD, Richardson JA, Garry DJ. Transcriptional profiling and regulation of the extracellular matrix during muscle regeneration. Physiol. Genomics. 2003;14:261–271. doi: 10.1152/physiolgenomics.00056.2003. [DOI] [PubMed] [Google Scholar]
- Gros J, Hu JK-H, Vinegoni C, Feruglio PF, Weissleder R, Tabin CJ. WNT5A/Jnk and FGF/Mapk pathways regulate the cellular events shaping the vertebrate limb bud. Curr. Biol. 2010;20:1993–2002. doi: 10.1016/j.cub.2010.09.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger V, Hamilton HC. A series of normal stages in the development of the chick embryo. J. Morph. 1951;88:49–92. [PubMed] [Google Scholar]
- Han M, Yang X, Farrington JE, Muneoka K. Digit regeneration is regulated by Msx1 and BMP4 in fetal mice. Development. 2003;130:5123–5132. doi: 10.1242/dev.00710. [DOI] [PubMed] [Google Scholar]
- Han M, Yang X, Lee J, Allan CH, Muneoka K. Development and regeneration of the neonatal digit tip in mice. Dev. Biol. 2008;315:125–135. doi: 10.1016/j.ydbio.2007.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M, Yang X, Taylor G, Burdsal CA, Anderson RA, Muneoka K. Limb regeneration in higher vertebrates: developing a roadmap. Anat. Rec. B New Anat. 2005;287:14–24. doi: 10.1002/ar.b.20082. [DOI] [PubMed] [Google Scholar]
- Harfe BD, Scherz PJ, Nissim S, Tian H, McMahon AP, Tabin CJ. Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell. 2004;118:517–528. doi: 10.1016/j.cell.2004.07.024. [DOI] [PubMed] [Google Scholar]
- Hayamizu TF, Wanek N, Taylor G, Trevino C, Shi C, Anderson R, Gardiner DM, Muneoka K, Bryant SV. Regeneration of HoxD expression domains during pattern regulation in chick wing buds. Dev. Biol. 1994;161:504–512. doi: 10.1006/dbio.1994.1048. [DOI] [PubMed] [Google Scholar]
- Healy C, Uwanogho D, Sharpe PT. Regulation and role of Sox9 in cartilage formation. Dev. Dyn. 1999;215:69–78. doi: 10.1002/(SICI)1097-0177(199905)215:1<69::AID-DVDY8>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Hinchliffe JR. Developmental basis of limb evolution. Int. J. Dev. Biol. 2002;46:835–845. [PubMed] [Google Scholar]
- Holder N. An experimental investigation into the early development of the chick elbow joint. J. Embryol. Exp. Morphol. 1977;39:115–127. [PubMed] [Google Scholar]
- Imokawa Y, Yoshizato K. Expression of Sonic hedgehog gene in regenerating newt limb blastemas recapitulates that in developing limb buds. Proc. Natl. Acad. Sci. USA. 1997;94:9159–9164. doi: 10.1073/pnas.94.17.9159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly MT. Condylectomy in the rat. An investigation of the ensuing repair processes in the region of the temporomandibular articulation. Aust. Dent. J. 1961:243–256. [Google Scholar]
- Kawakami Y, Rodriguez Esteban C, Raya M, Kawakami H, Marti M, Dubova I, Izpisua Belmonte JC. Wnt/β-catenin signaling regulates vertebrate limb regeneration. Genes Dev. 2006;20(23):3232–3237. doi: 10.1101/gad.1475106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T, Yasui N, Ohsawa S, Ono K. Biosynthesis of type IX collagen during chick limb development. Biochem. Biophys. Res. Commun. 1985;130:746–752. doi: 10.1016/0006-291x(85)90479-6. [DOI] [PubMed] [Google Scholar]
- Kostakopoulou K, Vogel A, Brickell P, Tickle C. 'Regeneration' of wing bud stumps of chick embryos and reactivation of Msx-1 and Shh expression in response to FGF-4 and ridge signals. Mech. Dev. 1996;55:119–131. doi: 10.1016/0925-4773(95)00492-0. [DOI] [PubMed] [Google Scholar]
- Li S, Muneoka K. Cell migration and chick limb development: chemotactic action of FGF-4 and the AER. Dev. Biol. 1999;211:335–347. doi: 10.1006/dbio.1999.9317. [DOI] [PubMed] [Google Scholar]
- Merida-Velasco JA, Sanchez-Montesinos I, Espin-Ferra J, Merida-Velasco JR, Rodriguez-Vazquez JF, Jimenez-Collado J. Development of the human elbow joint. Anat. Rec. 2000;258:166–175. doi: 10.1002/(SICI)1097-0185(20000201)258:2<166::AID-AR6>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Mitrovic D. Development of the diarthrodial joints in the rat embryo. Am. J. Anat. 1978;151:475–485. doi: 10.1002/aja.1001510403. [DOI] [PubMed] [Google Scholar]
- Mitrovic DR. Development of the metatarsophalangeal joint of the chick embryo: morphological, ultrastructural and histochemical studies. Am. J. Anat. 1977;150:333–347. doi: 10.1002/aja.1001500207. [DOI] [PubMed] [Google Scholar]
- Muneoka K, Bryant SV. Evidence that patterning mechanisms in developing and regenerating limbs are the same. Nature. 1982;298:369–371. doi: 10.1038/298369a0. [DOI] [PubMed] [Google Scholar]
- Muneoka K, Bryant SV. Cellular contribution to supernumerary limbs resulting from the interaction between developing and regenerating tissues in the axolotl. Dev. Biol. 1984;105:179–187. doi: 10.1016/0012-1606(84)90273-2. [DOI] [PubMed] [Google Scholar]
- Muneoka K, Fox WF, Bryant SV. Cellular contribution from dermis and cartilage to the regenerating limb blastema in axolotls. Dev. Biol. 1986;116:256–260. doi: 10.1016/0012-1606(86)90062-x. [DOI] [PubMed] [Google Scholar]
- Muneoka K, Wanek N, Bryant SV. Mammalian limb bud development: in situ fate maps of early hindlimb buds. J. Exp. Zool. 1989;249:50–54. doi: 10.1002/jez.1402490110. [DOI] [PubMed] [Google Scholar]
- Nalin AM, Greenlee TK, Jr, Sandell LJ. Collagen gene expression during development of avian synovial joints: transient expression of types II and XI collagen genes in the joint capsule. Dev. Dyn. 1995;203:352–362. doi: 10.1002/aja.1002030307. [DOI] [PubMed] [Google Scholar]
- O'Driscoll SW. Articular cartilage regeneration using periosteum. Clin. Orthop. Relat. Res. 1999:S186–S203. doi: 10.1097/00003086-199910001-00020. [DOI] [PubMed] [Google Scholar]
- Obara-Ishihara T, Kuhlman J, Niswander L, Herzlinger D. The surface ectoderm is essential for nephric duct formation in intermediate mesoderm. Development. 1999;126:1103–1108. doi: 10.1242/dev.126.6.1103. [DOI] [PubMed] [Google Scholar]
- Satoh A, Makanae A, Wada N. The apical ectodermal ridge (AER) can be re-induced by wounding, wnt-2b, and fgf-10 in the chicken limb bud. Dev. Biol. 2010;342:157–168. doi: 10.1016/j.ydbio.2010.03.018. [DOI] [PubMed] [Google Scholar]
- Saunders JW. The proximo-distal sequence of origin of the parts of the chick wing and the role of the ectoderm. J. Exp. Zool. 1948;108:363–403. doi: 10.1002/jez.1401080304. [DOI] [PubMed] [Google Scholar]
- Shubin NH, Alberch P. A morphogenetic approach to the origin and basic organization of the tetrapod limb. Evol. Biol. 1986;20:319–387. [Google Scholar]
- Stark RJ, Searls RL. A description of chick wing bud development and a model of limb morphogenesis. Dev. Biol. 1973;33:138–153. doi: 10.1016/0012-1606(73)90170-x. [DOI] [PubMed] [Google Scholar]
- Stark RJ, Searls RL. The establishment of the cartilage pattern in the embryonic chick wing, and evidence for a role of the dorsal and ventral ectoderm in normal wing development. Dev. Biol. 1974;38:51–63. doi: 10.1016/0012-1606(74)90258-9. [DOI] [PubMed] [Google Scholar]
- Storm EE, Kingsley DM. GDF5 coordinates bone and joint formation during digit development. Dev. Biol. 1999;209:11–27. doi: 10.1006/dbio.1999.9241. [DOI] [PubMed] [Google Scholar]
- Summerbell D. Regulation of the deficiencies along the proximal distal axis of the chick wing-bud: a quantitative analysis. J. Embryol. Exp. Morphol. 1977;41:137–159. [PubMed] [Google Scholar]
- Summerbell D. Evidence for regulation of growth, size and pattern in the developing chick limb bud. J. Embryol. Exp. Morphol. 1981;65(Suppl):129–150. [PubMed] [Google Scholar]
- Summerbell D, Wolpert L. Precision of development in chick limb morphogenesis. Nature. 1973;244:228–230. doi: 10.1038/244228a0. [DOI] [PubMed] [Google Scholar]
- Taylor GP, Anderson R, Reginelli AD, Muneoka K. FGF-2 induces regeneration of the chick limb bud. Dev. Biol. 1994;163:282–284. doi: 10.1006/dbio.1994.1144. [DOI] [PubMed] [Google Scholar]
- Torok MA, Gardiner DM, Shubin NH, Bryant SV. Expression of HoxD genes in developing and regenerating axolotl limbs. Dev. Biol. 1998;200:225–233. doi: 10.1006/dbio.1998.8956. [DOI] [PubMed] [Google Scholar]
- Vargesson N, Clarke JDW, Vincent K, Coles C, Wolpert L, Tickle C. Cell fate in the chick limb bud and relationship to gene expression. Development. 1997;124:1909–1918. doi: 10.1242/dev.124.10.1909. [DOI] [PubMed] [Google Scholar]
- Whitehead GG, Makino S, Lien CL, Keating MT. fgf20 is essential for initiating zebrafish fin regeneration. Science. 2005;310:1957–1960. doi: 10.1126/science.1117637. [DOI] [PubMed] [Google Scholar]
- Wyngaarden LA, Vogeli KM, Ciruna BG, Wells M, Hadjantonakis A-K, Hopyan S. Oriented cell motility and division underlie early limb bud morphogenesis. Development. 2010;137:2551–2558. doi: 10.1242/dev.046987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama H. Initiation of limb regeneration: the critical steps for regenerative capacity. Dev. Growth Differ. 2008;50:13–22. doi: 10.1111/j.1440-169X.2007.00973.x. [DOI] [PubMed] [Google Scholar]
- Yu L, Han M, Yan M, Lee EC, Lee J, Muneoka K. BMP signaling induces digit regeneration in neonatal mice. Development. 2010;137:551–559. doi: 10.1242/dev.042424. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: In situ hybridization analysis of genes that are indicated in joint and/or cartilage development. Previous studies did not address the expression of most of these markers specifically at the elbow joint for the time points we used in this study. Limbs were fixed at Day 5, Day 7 and Day 8, sectioned and processed for in situ hybridization for the following markers: GDF-5, Atx, Sox-9, Col-9, BMP-2, BMP-4, Chordin, Noggin, Wnt-9a, Wnt-4, Hes-1, Col-2α1, Prickle-1, Prickle-2, Cux-1, Gli-3. Based on the expression patterns, we selected Atx and GDF-5 as joint specific markers and Sox-9 and Col-9 as cartilage markers for our study.