Abstract
Carrot (Daucus carota) extracellular protein 3 (EP3) class IV endochitinases were previously identified based on their ability to rescue somatic embryos of the temperature-sensitive cell line ts11. Whole-mount in situ hybridization revealed that a subset of the morphologically distinguishable cell types in embryogenic and nonembryogenic suspension cultures, including ts11, express EP3 genes. No expression was found in somatic embryos. In carrot plants EP3 genes are expressed in the inner integumentary cells of young fruits and in a specific subset of cells located in the middle of the endosperm of mature seeds. No expression was found in zygotic embryos. These results support the hypothesis that the EP3 endochitinase has a “nursing” function during zygotic embryogenesis and that this function can be mimicked by suspension cells during somatic embryogenesis.
Carrot (Daucus carota) cell cultures secrete many different proteins into the medium, a process that contributes to the conditioning of the medium. Conditioned media are reported to have a promoting effect on the initiation of somatic embryogenesis (Hari, 1980; Smith and Sung, 1985). The initiation of somatic embryogenesis results in major changes of the extracellular protein pattern compared with that of intracellular proteins (De Vries et al., 1988). Some of the secreted proteins are thought to be related to the formation of embryogenic cells and somatic embryos. To investigate whether a causal relationship between certain secreted proteins and embryogenic potential exists, secreted proteins that were differentially expressed under different culture conditions were analyzed and their localization was studied.
The extracellular protein EP1 is secreted only by nonembryogenic cells (Van Engelen et al., 1991), whereas the extracellular protein EP2, identified as a lipid-transfer protein, is secreted only by embryogenic cells and somatic embryos (Sterk et al., 1991). These results showed that different cell types contribute to the total pattern of proteins secreted into the culture medium (Van Engelen and De Vries, 1993). Another way of studying a causal relationship between secreted proteins and embryogenic potential led to the identification of extracellular protein 3 (EP3), which was identified as a chitinase.
EP3 was originally purified as a protein capable of rescuing somatic embryos in the mutant carrot cell line ts11 at the nonpermissive temperature (De Jong et al., 1992). The acidic endochitinase EP3 was found to be a member of a small family of class IV chitinase genes (Kragh et al., 1996). These highly homologous isoenzymes are encoded by at least four EP3 genes. Two of these proteins, EP3–1 and EP3–3, were purified and shown to have subtly different effects on the formation of somatic embryos in newly initiated ts11 embryo cultures (Kragh et al., 1996).
Since the effect of the chitinases was mimicked by Rhizobium sp.-produced nodulation (Nod) factors (De Jong et al., 1993), it was proposed that the chitinases are involved in the generation of signal molecules essential for embryogenesis in ts11 (De Jong et al., 1993). The EP3 proteins produced by ts11 at the permissive and nonpermissive temperatures did not show any difference in biochemical characteristics compared with the ones produced in wild-type cultures and were capable of rescuing ts11 somatic embryos. It was also shown that the sensitivity of ts11 to chitinases coincided with a transient decrease in the amount present of this otherwise functional set of proteins (De Jong et al., 1995).
The roots of leguminous plants are known to produce chitinases, and during the interaction with Rhizobium sp., these plant-produced chitinases have been suggested to control the biological activity of Nod factors by cleaving and inactivating them. In this way, chitinases are proposed to have the potential to control plant morphogenesis and cell division (Staehelin et al., 1994).
Many chitinase genes are induced upon infection, wounding, and treatment with elicitors. Infection can lead to a 600-fold induction of chitinase activity (Métraux and Boller, 1986). The molecular mechanisms and signaling pathways responsible for this induction remain largely unclear (Graham and Sticklen, 1994). The response of a plant to infection involves nonspecific responses, because challenge by different pathogens can induce production of the same set of pathogen-response proteins (Meins and Ahl, 1989). The induction is not restricted to the infection zone, since uninfected areas of infected leaves and even uninfected second leaves show an increase in chitinase activity (Métraux and Boller, 1986).
We aimed to establish in carrot, first, which cells in wild-type and ts11 suspension cultures express the EP3 genes, and second, whether (and if so, where) the same chitinases are expressed during zygotic embryogenesis. The results show that the carrot EP3 class IV endochitinases are expressed in a subset of (most likely) nonembryogenic suspension cells and in carrot plants in the integument cells and endosperm during zygotic embryogenesis.
MATERIALS AND METHODS
Plant Material and Culture Conditions
Cell-suspension cultures of carrot (Daucus carota cv Autumn King/Trophy) and ts11 were initiated and maintained as described previously (De Vries et al., 1988). One-week-old suspension cultures were used for gene-expression analysis and protein-localization experiments. Hypocotyl sections were prepared as described by Guzzo et al. (1994). The hypocotyls of 1-week-old plantlets were divided into segments of 3 to 5 mm and incubated for 10 d in B5 medium with 2 μm 2,4-D and then returned to B5 medium without 2,4-D.
One-week-old cell-suspension cultures of an embryogenic cell line were grown in the presence of Phytophthora infestans or Botrytis cinerea or supplemented with 0.002 or 0.02 mg mL−1 chitosan. After 2 d of incubation the cells and fungi were removed by filtration and the resulting cell-free conditioned media were assayed for the presence of EP3 by western analysis, as described by Sterk et al. (1991).
Cell-Suspension Fractionation Analysis
Suspension cultures were sieved to obtain cells and cell clusters between 50 and 125 μm. The cells were loaded onto a discontinuous gradient of 10, 20, 30, and 40% Percoll (Van Engelen et al., 1995). After centrifugation the cells at the interfaces between the different densities of Percoll were recovered with a syringe, washed in B5 medium containing 2 μm 2,4-D, and allowed to grow in B5 medium with 2 μm 2,4-D. After 5 d the proteins in the medium were analyzed.
Partial Purification of Seed Proteins
Dry, mature seeds were ground in liquid nitrogen with a mortar and pestle. Ten milliliters of 25 mm Tris-HCl, pH 8.5, was added to 2 g of ground seeds. After incubation for 15 min on a rotary shaker at 4°C, the resulting slurry was filtrated through Whatman filter paper and supplemented with 0.5 mL of DEAE-Sepharose FF resin, equilibrated in 25 mm Tris-HCl, pH 8.5. After incubation on a rotary shaker for 1 h at 4°C a column of the DEAE-Sepharose FF resin was poured and washed twice with 25 mm Tris-HCl, pH 8.5, and the bound proteins were eluted with 0.2 m KCl in 25 mm Tris-HCl, pH 8.5. The protein sample was desalted with dialysis.
Chitinase Activity Determination
Chitinase activity was determined essentially as described by Trudel and Asselin (1989).
In Situ Hybridization
For whole-mount in situ hybridizations, 1-week-old suspension cultures grown in B5 medium in the presence of 2 μm 2,4-D were washed in B5 medium, run through nylon sieves with pore sizes of 125 or 30 μm, and allowed to grow for another week in the absence of 2,4-D. The cells were concentrated by centrifugation and immobilized onto poly-l-Lys-coated glass slides by mixing 0.5 mL of cell suspension with an equal volume of fixation buffer (130 mm NaCl, 10 mm NaPO4 buffer, pH 6.4 [PBS] with 0.1% Tween 20 [PBT], 70 mm EGTA, 4% paraformaldehyde, 0.25% glutaraldehyde, and 10% DMSO). For fixation the slides were placed on a heated plate (30°C) for 30 min and then washed with methanol and then ethanol. Postfixations were performed after xylene and proteinase K treatments. Fixed cells and embryos were prehybridized in a solution containing PBT, 50% deionized formamide, 0.33 m NaCl, and 50 μg/mL heparin.
Single-stranded RNA probes 230 nucleotides in length were transcribed from pAJ41, a plasmid containing the NruI-StyI fragment from EP3B cDNA (Kragh et al., 1996). Digoxygenin sense and antisense probes were synthesized (Boehringer Mannheim). For each slide 100 ng of RNA probe was denatured in a solution containing 15 μg of yeast tRNA and 50 μg of poly(A+) RNA, and this was mixed with prehybridization solution and applied to the slides. The hybridization was for 16 h at 42°C. The slides were washed and incubated with anti-digoxigenin fragment, antigen-binding alkaline phosphatase diluted 1:2500 in PBT containing 50 μg of BSA and 25 μg of plant-protein extract to reduce nonspecific staining. The nonbound antibodies were removed by several washes with PBT, and the cells were stained in 100 mm NaCl, 5 mm MgCl2, 100 mm Tris-HCl, pH 9.5, and 0.1% Tween 20 supplemented with 9 μL/mL nitroblue tetrazolium and 7 μL/mL 5-bromo-4-chloro-3-indolyl phosphate. Slides were mounted in PBS and 0.8% glutaraldehyde and analyzed using Nomarski optics.
In situ hybridization on sections was carried out essentially according to the protocol of Cox and Goldberg (1988) except for the use of digoxigenin-labeled RNA probes.
Immunolocalization
Immunofluorescence labeling on suspension-cultured cells was carried out as described previously (Van Engelen et al., 1991). The anti-EP3 serum was obtained by immunizing a rabbit with the purified protein (De Jong et al., 1995). Both fluorescein isothiocyanate- and alkaline phosphatase-conjugated antibodies were used. For the latter, the staining was performed as described above. In control experiments preimmune serum was used.
Tissue Printing
Fresh plant material was cut and pressed onto PVDF membranes (Millipore). These membranes were washed in methanol and incubated in PBS. They were either directly stained in an amido black solution (Gershoni and Palade, 1982) or treated as immunoblots and incubated with rabbit EP3 polyclonal antibodies.
RT-PCR
Total RNA was isolated from different plant tissues as previously described (De Vries et al., 1982). From each sample 2 μg was used for reverse transcription. A mixture of RNA, 20 units of RNA Guard (Pharmacia), 1 mm deoxyribonucleotide triphosphates, 5 mm MgCl2, 1× RT buffer (10 mm Tris, pH 8.8, 50 mm KCl, and 0.1% Triton X-100), and 50 ng of oligo(dT)12–18 in a volume of 20 μL was incubated at 83°C for 3 min. The mixture was subsequently incubated at 42°C for 10 min, after which time 4 units of avian myeloblastosis virus RT was added and the incubation was continued for 1 h. The samples were denaturated at 95°C for 5 min and diluted to a final volume of 100 μL. A PCR reaction for the amplification of the ubiquitin cDNA was carried out, using 5 μL from the RT mixture, 1× Taq polymerase buffer (Boehringer Mannheim), 100 μm deoxyribonucleotide triphosphates, 1 unit of Taq polymerase, 100 ng of downstream ubiquitin primer (5′-TATGGATCCACCACCACGG/AAGACGGAG-3′), and 100 ng of upstream ubiquitin primer (5′-TAGAAGCTTATGCAGATC/TTTTGTGAAGAC-3′) (Horvath et al., 1993) in a total volume of 50 μL.
After denaturation for 30 s at 94°C the samples were subjected to 15, 20, or 30 cycles of 30 s at 94°C, 30 s at 48°C, and 2 min at 72°C. The PCR products were run on a 1% agarose gel and blotted on Nytran+ membranes (Schleicher & Schuell). For Southern analysis, a full-length ubiquitin probe was used. The concentration of cDNA was standardized by comparing the signals on the Southern blot. Equal amounts of cDNA were used for a control PCR reaction with ubiquitin primers, and one was used with EP3 primers.
Both PCR reactions were done simultaneously, using the same PCR conditions. In database searches the downstream primer (5′-ATGGCACGGATGGTTGCCCCGAAACCTTG-3′) for EP3 showed homology only to class IV chitinases. Therefore, amplification of class I chitinase cDNAs cannot occur, despite their high homology to EP3s. A PCR product with a length of 201 nucleotides was amplified using this primer and an upstream primer (5′-GTATTTTGGCCGCGGCCCTCTTCAGC-3′). The probe used for the detection of the EP3 PCR product was the 230-nucleotide insert from pAJ-41. All Southern blots were washed three times in 0.1% SSC and 1% SDS at 65°C before being exposed to radiographic film.
RESULTS
Cell-Specific Expression of the EP3 Genes and Localization of the Encoded Proteins in Suspension Cultures
To identify the suspension cells that express the EP3 genes, whole-mount in situ mRNA localization was used on entire, immobilized suspension cultures. Several cell lines differing in embryogenic potential were used to obtain a reliable indication of cell specificity in EP3 expression. It was not possible to distinguish between the different members of the family of EP3 genes because of their very high homology (Kragh et al., 1996). For in situ hybridization EP3 mRNA probes spanning the main class IV chitinase-specific deletions were used. This makes it unlikely that related class I chitinases were also detected. Several other probes were used to show that cell-specific expression patterns in plant-tissue cultures can be obtained by whole-mount in situ hybridization (Schmidt et al., 1996, 1997).
Figure 1A shows a single EP3-expressing suspension cell from an embryogenic suspension culture in the midst of cells that show no staining above background. This representative sample suggests that EP3 mRNAs could be detected only in a subpopulation of the total embryogenic culture. Nonembryogenic cultures showed essentially the same EP3 expression pattern (data not shown). Counting the number of stained cells in several cultures revealed that between 4 and 6% of the total number of cells in an embryogenic culture express the EP3 genes. The number of embryo-forming cells in a comparable culture does not exceed 1% (Toonen et al., 1994), suggesting that there is no quantitative relation to EP3-expressing cells.
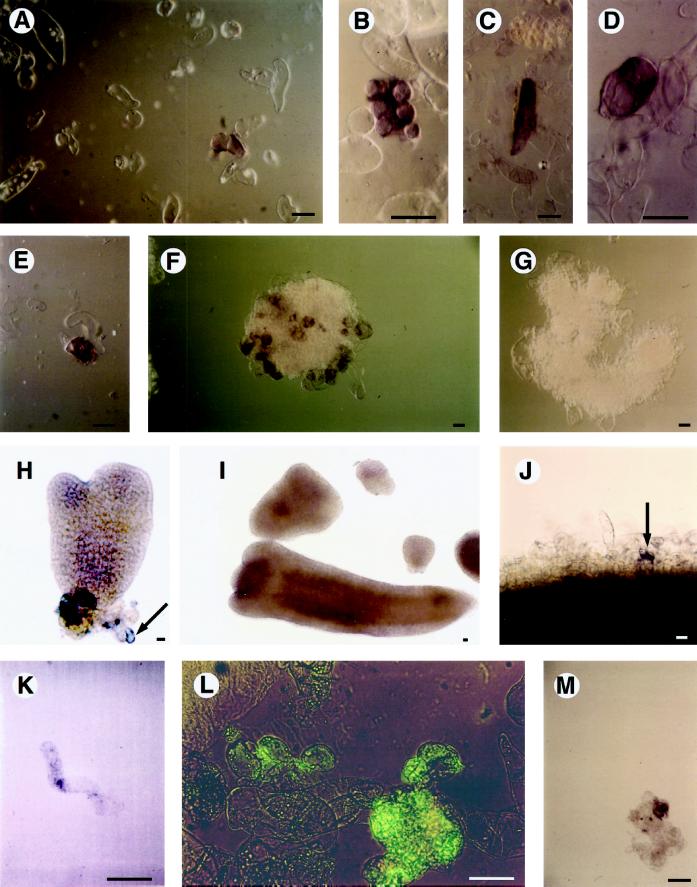
Figure 1.
EP3 gene expression in suspension cultures. A to D, Examples of EP3-expressing cells in an embryogenic cell culture. E and F, Cell clusters present in an embryogenic cell culture. G, Cell cluster present in an embryogenic cell culture hybridized with an EP3 sense probe. H, Heart-stage somatic embryo; the arrow points to an attached EP3-expressing single cell. I, Torpedo-stage somatic embryo devoid of EP3 expression. J, Cells present at the periphery of the proliferated cell mass on a section of a hypocotyl explant that was cultured in the presence of 2,4-D for 10 d; the arrow points to a single Ep3-expressing cell. K, An elongated single cell released from a hypocotyl explant that was cultured in the presence of 2,4-D for 20 d. L, Cell clusters present in an embryogenic cell suspension subjected to immersion immunofluorescence; the presence of the EP3 protein is indicated by the green fluorescence. M, Cell cluster in a suspension of the mutant cell line ts11; the presence of the EP3 protein is indicated by a purple precipitate. Plant material was analyzed by whole-mount in situ hybridization with antisense EP3 RNA probes (A–F and H–K) or with a sense EP3 RNA probe (G). Light microscopy (coupled with Nomarski optics for A–E and M) was used for visualization of the purple precipitate (the use of Nomarski optics resulted in a change from purple to brown in A–F and J). In H, EP3 gene expression is visible as a purple precipitate in an individual cell. Immunolocalization of EP3 proteins in suspension cultures (L and M) was done by immersion immunofluorescence using either fluorescein isothiocyanate (L) or alkaline phosphatase as a second antibody. Bars = 50 μm. {/ANNT;152064n;3872n;center;704n}
The highest concentration of EP3 mRNA in both embryogenic and nonembryogenic cultures was found in single cells that were elongated and often strongly curved or coiled (Fig. 1B) or in single, vacuolated cells that were elongated (Fig. 1C) or rounded (Fig. 1D). Staining was also often seen in small clusters of cells (Fig. 1E), consisting of no more than about 30 cells. Cells that were loosely attached to large clusters present in embryogenic cultures occasionally showed intense staining (Fig. 1F), whereas the sense control hybridizations were always negative (Fig. 1G).
Figure 1H shows a single EP3-expressing cell attached to a heart-stage somatic embryo completely devoid of purple staining. Whole-mount in situ localization on globular-, heart-, or torpedo-shaped somatic embryos did not reveal EP3 mRNAs (Fig. 1I). To determine when the first EP3-expressing cells appeared during embryogenic cell formation, hypocotyl explants were treated with 2,4-D for 10 d, during which time embryogenic cell formation occurs (Guzzo et al., 1994). In this system cell division was reinitiated in cells of the vascular tissue and generated a mass of rapidly proliferating cells. Only after 10 d were a very small number of cells at the periphery of the proliferating mass found to contain EP3 mRNA (Fig. 1J). This is 3 d later than the appearance of the first cells competent to form somatic embryos in this system (Schmidt et al., 1997).
Subsequently, hypocotyls were transferred to hormone-free medium and after another 5 d, some of the peripheral cells started to enlarge. The EP3-positive cells were in some cases elongated. In a population of single cells released from these explants after 20 d, elongated, curved cells were the only cells that stained positively (Fig. 1K). Single cells shown to be competent to produce embryogenic cells never have this morphology, as was determined by cell tracking (Schmidt et al., 1997). We conclude, therefore, that EP3 gene expression is not directly correlated with embryogenic cell formation or somatic embryo development.
It was found previously that EP3-encoded chitinases occur in the medium (De Jong et al., 1992). To investigate whether all of these chitinases are secreted in the medium, whether some remain in the cell walls of the suspension cells that produced them, or whether they are present in somatic embryos, immersion immunofluorescence was used (Van Engelen et al., 1991).
In embryogenic suspension cultures a relatively high amount of the protein could be detected on the surface of a subset of cells. The chitinase was identified on single cells as well as on small clusters (Fig. 1L). Because EP3 chitinases are secreted proteins and contain a signal sequence (Kragh et al., 1996), we assume that the EP3 chitinases (Fig. 1L) are present in a component of the extracellular matrix. The protein could not be detected on the surface of somatic embryos, but cells that were loosely connected with somatic embryos often contained EP3 (data not shown). EP3 proteins were also observed on cells of the mutant cell line ts11 (Fig. 1M), which is also capable of producing EP3 proteins (De Jong et al., 1995). The localization of the EP3 proteins therefore appears to correspond to the pattern of expression of their encoding genes, suggesting that EP3 proteins are retained in the extracellular matrix of the cells that produced them, even though a substantial amount is secreted into the medium.
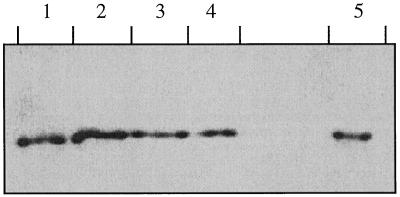
The production of chitinases can be induced by pathogens and elicitors, and in carrot suspension cultures many chitinases are induced after treatment with fungal wall components (Kurosaki et al., 1990). Suspension-cultured cells were exposed to chitosan as an elicitor or to P. infestans or B. cinerea. The medium of these cultures was isolated and the amount of EP3 produced was determined with western analysis. Scanning of the absorbance of the bands revealed that the constitutive production of the protein was not affected by the fungi or the elicitor (Fig. 2).
Figure 2.
Presence of EP3 chitinases in media of cultures treated with fungi and elicitors. Media of suspension cultures grown in the presence of: lane 1, 0.002 mg/mL chitosan; lane 2, 0.02 mg/mL chitosan; lane 3, P. infestans; lane 4, B. cinerea; and lane 5, control suspension culture. Proteins were separated by denaturing SDS-PAGE and detected by immunoblotting.
Expression of the EP3 Isoenzymes in Suspension Cultures
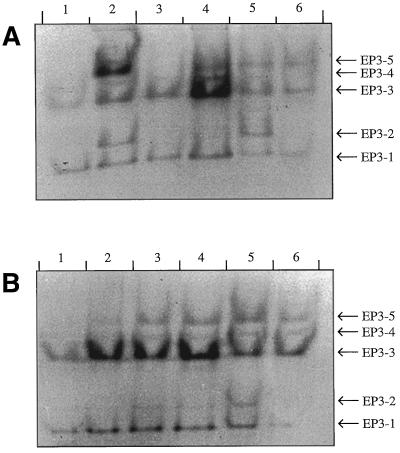
With the methods used so far it was not possible to discriminate between different EP3 isoenzymes or the mRNAs belonging to them. Five different EP3 chitinase isoenzymes have been detected in the medium of suspension-cultured cells of different cell lines by using native PAGE (Kragh et al., 1996). Given the subtle differences of the isoenzymes EP3–1 and EP3–3 in their biological effect on ts11 embryo formation, it was of interest to determine whether the different isoenzymes were produced by the same or different cell types. No correlation was found between the embryogenic capacity of a given cell line and the EP3 isoenzymes produced (Fig. 3A). However, fractionation of suspension cultures showed that different cell types produced different relative amounts of isoenzymes. A population of cells <50 μm consisted almost solely of single cells. A western blot of the proteins in the medium showed that only the isoenzymes EP3–1 and EP3–3 were produced by this cell population (Fig. 3B). Cells that were aggregated in large clusters did not pass through the 125-μm sieve. This fraction produced the isoenzymes EP3–1, EP3–3, EP3–4, and EP3–5.
Figure 3.
Presence of EP3 chitinases in media of nonembryogenic, embryogenic, and density-fractionated embryogenic cultures. A, Detection of EP3 isoenzymes in the medium of several different suspension cultures. Lanes 1 and 2, Nonembryogenic suspension cultures; and lanes 3 to 6, embryogenic suspension cultures. B, Detection of EP3 isoenzymes in the medium of a Percoll-fractionated suspension culture: lane 1, cells < 50 μm; lane 2, 0 to 10% Percoll; lane 3, 10 to 20% Percoll; lane 4, 20 to 30% Percoll; lane 5, 30 to 40% Percoll; and lane 6, cells > 125 μm. The proteins were separated by nondenaturing PAGE and detected by immunoblotting.
Density fractionation of a 50- to 125-μm cell fraction on a Percoll block gradient resulted in four subfractions of cells with different densities. The subfractions with relatively high densities contained more clusters compared with the low-density subfractions, which contained mainly single cells. After the cells were cultured for another 5 d the original difference in cell types was retained. Comparable amounts of EP3 proteins were produced in all cell populations; however, a shift in the relative amounts of the different isoenzymes was observed. The pattern of isoenzymes produced by the cells with the lowest density resembled that of the cell population that passed through the 50-μm sieve. A relative increase of the isoenzymes EP3–2 and EP3–5 was observed in cell populations with higher densities. This is in agreement with the pattern of isoenzymes produced by other cell lines (Fig. 3A). Cell lines that produce isoenzymes EP3–2 and EP3–5 always contain clusters of small, usually cytoplasm-rich cells. Presumably, EP3 chitinase isoenzymes 2 and 5 are produced by these cell clusters, whereas single cells are preferentially responsible for the production of EP3 chitinase isoenzymes 1 and 3.
Localization of EP3 Expression in Plants
The more sensitive method of RT-PCR was used instead of northern analysis to study EP3 gene expression. The primers allowed us to discriminate between EP3 and chitinases of class I, since the downstream primer has homology with only class IV chitinases, and the primers span a region that contains deletions characteristic of class IV chitinases. However, discrimination of the different EP3 isoenzymes was not possible because of their very high homology (Kragh et al., 1996).
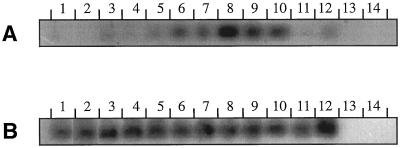
In leaves, stems, storage roots, and normal roots (Fig. 4A, lanes 1–4), very little expression of EP3 could be detected. EP3 mRNA was found in developing seeds at 3, 7, 10, and 20 DAP (Fig. 4A, lanes 6–9). Only very low levels of EP3 gene expression occurred prior to pollination (Fig. 4A, lane 5). The highest expression was found in seeds 10 DAP, which roughly corresponds to the early globular-stage embryo. In mature, dry seeds the amount of EP3 mRNA had decreased (Fig. 4A, lane 10), whereas during germination EP3 mRNA was hardly detectable (Fig. 4A, lanes 11 and 12). The presence of EP3 mRNA in developing seeds at 3, 7, 10, and 20 DAP could be confirmed by hybridizing a northern blot with the end-labeled downstream primer that had been designed for the RT-PCR. The length of the EP3 transcripts was approximately 950 nucleotides (data not shown).
Figure 4.
Expression of EP3 genes in carrot plants in the leaf (lane 1), stem (lane 2), storage root (lane 3), root (lane 4), flower (lane 5), fruit harvested 3 DAP (lane 6), fruit harvested 7 DAP (lane 7), fruit harvested 10 DAP (lane 8), fruit harvested 20 DAP (lane 9), mature seed (lane 10), seed 12 h after imbibition (lane 11), seed 60 h after imbibition (lane 12), genomic DNA (lane 13), and water control (lane 14). B, Ubiquitin control. Gene expression was determined by RT-PCR, followed by transfer to membranes and Southern hybridizaton.
In situ mRNA localization using sections of different plant tissues such as flowers, roots, root tips, hypocotyls, cotyledons, shoot apices, and shoot meristems confirmed that EP3 mRNA was not detectable in these tissues (data not shown). Only in developing and mature seeds could EP3 mRNAs be found. In the early stages of seed development (approximately 5–6 DAP), EP3 mRNA was detected in the inner integument cells lining the surface of the embryo sac in which the zygote or the early embryo is located (Fig. 5, A and B).
Figure 5.
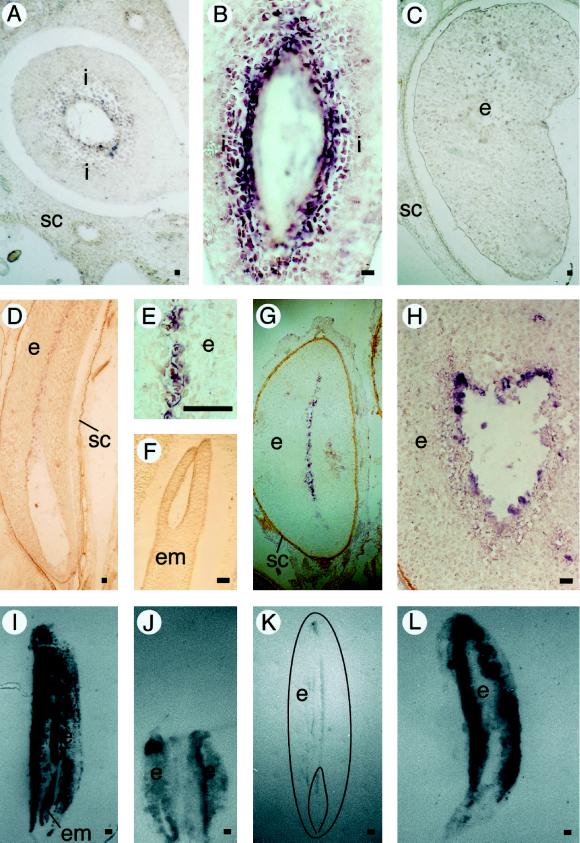
EP3 gene expression in seeds. A, Cross-section of a fruit 7 DAP. B, Longitudinal section of a fruit 7 DAP. C, Cross-section of a fruit 20 DAP. D, Longitudinal section of a mature seed. E, Longitudinal section of a mature seed. F, Zygotic embryo in a longitudinal section of a mature seed. G, Transverse section of a seed that soaked in water for 60 h. H, Transverse section of a seed that soaked in water for 60 h. In the immunolocalizations, the presence of EP3 proteins is visible as a dark precipitate. Bar = 100 μm. I, Control tissue print of a mature seed stained with amido black. J, Immunolocalization of EP3 on a tissue print of a fruit containing two developing seeds 20 DAP. K, Immunolocalization of EP3 on a tissue print of a mature seed (a drawing of the printed seed is superimposed on the picture). L, Immunolocalization of EP3 on a tissue print of a seed that soaked in water for 60 h. Plant material was analyzed by in situ hybridization on sectioned carrot seeds with antisense EP3 RNA probes (A–H). Light microscopy (coupled to Nomarski optics for A–E) was used for visualization of the purple precipitate (the use of Nomarski optics resulted in a change from purple to brown in D). Immunolocalization of the EP3 protein was done by tissue printing followed by immunostaining (I–L). i, Integuments surrounding the developing embryo and endosperm; SC, seed coat; e, endosperm; em, embryo. Bar = 50 μm.
Since the highest amount of EP3 mRNAs was found 10 DAP, and the in situ hybridizations showed their presence in the integument cells, we conclude that the EP3 chitinases are produced by the integument cells shortly before they are degraded. At 20 DAP the degradation of the integuments is almost completed (data not shown). During the time that the number of integument cells declines and the volume of endosperm cells increases (until 35 d after anthesis), a decline in the level of seed EP3 mRNAs was observed. In the developing endosperm at 20 DAP, EP3 mRNA could not yet be detected (Fig. 5C). In the endosperm of mature seeds, EP3 mRNA was restricted to a narrow zone of endosperm cells starting at the cavity in which the embryo is located up to almost the opposite end of the endosperm (Fig. 5, D and E). Transverse sections showed that the EP3-expressing zone has a width of two to three cells and is located in the middle of the endosperm. In zygotic embryos no EP3 mRNA could be detected (Fig. 5F).
During seed development, cellularization of the endosperm starts with the outer layers of endosperm and is completed in the middle of the endosperm, which implies that EP3 genes are expressed in a place where the cellularization of the endosperm has been completed. The cells that contained the EP3 mRNAs also have protein bodies, which could be made visible using Nomarski optics (data not shown).
When carrot seeds are soaked, germination is initiated, the embryo starts to develop, and the central cavity is enlarged because of degradation of the endosperm. The cells in which EP3 mRNA is seen are thought to be the first ones to be degraded. The expression of the EP3 genes then shifts outward to the next cell layer that now lines the central cavity, as shown by in situ localization of EP3 mRNA in germinating seeds (Fig. 5, G and H). Because EP3 chitinases are secreted proteins, it was of interest to localize these enzymes during the development of seeds and in germinating seeds. For this purpose tissue printing of cut seeds onto nitrocellulose was applied. Protein transfer was ensured by staining with amido black, revealing a protein-distribution pattern in which the major seed tissues could be clearly observed (Fig. 5I). In the tissue prints the EP3 chitinase protein was localized using EP3 antibodies. The antibodies strongly react with all EP3 class IV chitinases but have a weak affinity for the 34-kD class I chitinase from carrot (Kragh et al., 1996).
Control experiments were performed using preimmune serum or antibodies raised against the related barley class I chitinase K that recognizes only the 34-kD class I chitinase and not the EP3 chitinases. No seed proteins present on the tissue prints were recognized by the barley class I chitinase K-reacting serum or by the preimmune serum (data not shown). We therefore conclude that the tissue prints almost exclusively show EP3 chitinases.
The EP3 protein was uniformly spread in the integuments at 10 DAP and in the developing endosperm at 20 DAP (Fig. 5J). The presence of the chitinase proteins in the developing seeds is therefore only partially in agreement with the EP3 gene-expression data. This points to transport of the EP3 proteins from the maternal integument tissues toward the endosperm. However, we cannot completely rule out that a very low level of EP3 gene expression that was below detection limits occurred in the peripheral endosperm cells. In mature seeds the protein was restricted to the inner cell layer of the cavity that surrounds the embryo and to a zone of cells that starts at this cavity and ends almost at the opposite side of the endosperm (Fig. 5K), which corresponds precisely with the expression pattern of the EP3 genes as determined by in situ hybridization (Fig. 5D).
In seeds in which imbibition had occurred the protein was again found to be spread uniformly in the endosperm. This pattern was observed in seeds soaked for 16 h as well as in seeds soaked for 60 h (Fig. 5L). In the 60-h-soaked seeds, enlargement of the embryo and the beginning of the degradation of the endosperm were already visible. Seeds soaked in water containing 1 mm cycloheximide did not show any enlargement of the embryo or endosperm degradation. Tissue prints of the cycloheximide-treated seeds showed that the localization of the EP3 chitinase protein was the same as in the seeds soaked in water only (data not shown). This suggests that there is no de novo synthesis of EP3 in the outer layers of the endosperm upon imbibition but that the water uptake is responsible for the observed outward spread and results in a uniform presence of EP3 protein in the endosperm.
The EP3 genes are expressed in the maternal integument cells. The EP3 chitinases, however, were detected in the developing endosperm at a time that EP3 chitinase gene products cannot be detected in the endosperm and as a consequence they must be largely of maternal origin. In mature seeds the integuments are completely degraded and then the EP3 proteins are produced in a subset of the endosperm cells. During germination the secreted EP3 chitinases diffuse in the water that is present in the cell walls to the outer region of the endosperm and around the embryo, in contrast to the inward-directed transport that occurs in developing seeds.
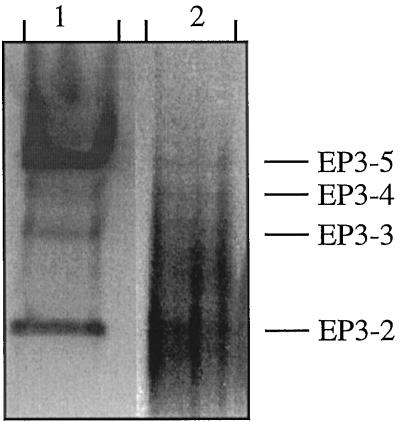
A western blot of partially purified seed proteins showed that isoenzymes EP3–2, EP3–3, EP3–4, and EP3–5 were present in dry, mature seeds (Fig. 6). EP3–1 could not be detected in seeds because of its low affinity for the DEAE-Sepharose FF resin that was used in the purification method. Isoenzymes EP3–2 to EP3–5 were all capable of degrading chitin (data not shown), as determined using glycol-chitin overlay gels. Except for isoenzyme EP3–1, all EP3 isoenzymes found in suspension cultures were present in developing seeds but at different relative amounts (Fig. 3). In Figure 6, both in the conditioned-medium sample and in the seed extract, additional bands are visible between EP3–2 and EP3–3. These bands are in a position corresponding to that of the carrot class I chitinases (Kragh et al., 1996). In both seed and conditioned-medium samples they represent a minor component of the total amount of chitinase present.
Figure 6.
Comparison of EP3 chitinases in suspension culture and in seeds. Lane 1, Proteins obtained from the conditioned medium of a suspension culture; and lane 2, proteins obtained from mature, dry carrot seeds. The proteins were separated by nondenaturing PAGE, and the EP3 chitinases were detected by immunoblotting.
DISCUSSION
In this study carrot suspension cells that produce EP3 chitinases were identified. Using whole-mount in situ hybridization, we found that a subset of the cells present in an embryogenic culture contains EP3 mRNA. On the basis of the number, cell type, and presence of EP3 mRNA-producing cells in embryogenic and nonembryogenic cultures, we found that there was no correlation with the ability to produce somatic embryos and the presence of EP3 mRNA. No EP3 gene expression in somatic embryos was found. The occurrence of the EP3 chitinase protein in the walls of suspension-cultured cells corresponds to the presence of the EP3 mRNA in the cells.
In plants EP3 mRNA was found in the inner integument, whereas EP3 proteins were found in the endosperm. Later, cells in the center of the endosperm do express the EP3 genes and this is likely to be responsible for the presence of EP3 chitinases during imbibition and germination. No EP3 gene expression was found in zygotic embryos. Together with the absence of a pathogen- or elicitor-induced response in EP3 gene expression, the results support the earlier conclusion that the carrot EP3 class IV chitinases are primarily involved in embryogenesis (De Jong et al., 1992).
The EP3 chitinases that are produced by single cells were identified as isoenzymes EP3–1 and EP3–3, whereas isoenzymes EP3–2 and EP3–5 were predominantly produced by clusters. Purified EP3–1 and EP3–3 chitinases are both able to rescue the carrot somatic embryo variant ts11 (Kragh et al., 1996); other EP3 isoenzymes were not tested for this capability. The arrest in somatic embryo development in this carrot variant ts11 has been shown to be due to a transient reduction in secreted EP3 (De Jong et al., 1995). On the basis of the expression pattern of the EP3 genes it is likely that cells that produce EP3 do not develop into embryos themselves. Thus, it appears that EP3 chitinases, or products of their enzymatic activity, diffuse via the conditioned medium to cells that are able to help somatic embryos develop and in this way play a “nursing” role in the process of somatic embryogenesis.
Since EP3 expression was also found in developing carrot seeds, it could be expected that the chitinases play a similar role in zygotic embryogenesis. In carrot, flowers are pollinated directly after anthesis and endosperm development starts soon after fertilization. Division of the primary endosperm nucleus leads to a large number of endosperm nuclei before the first division of the zygote has taken place. These nuclei are located in the upper part of the embryo sac, around the zygote, and at the periphery of the embryo sac (Borthwick, 1931). Cell wall formation in the carrot endosperm starts 7 d after anthesis (Gray et al., 1984), at about the time that the embryo is in the two-cell stage (Borthwick, 1931), and is complete 21 d after anthesis. The integuments that surround the embryo sac start to break down 6 to 7 DAP, at the moment that the endosperm becomes cellular (Lackie and Yeung, 1996; E.C. Yeung, personal communication). Maximum endosperm volume is reached about 35 d after anthesis. Mature seeds can be harvested about 80 d after anthesis.
The endosperm of mature carrot seeds consists of two different cell types, peripheral cell layers containing calcium oxalate crystals and more centrally located cells containing protein bodies with globular inclusions (Menon and Dave, 1988). The outermost cell layer of the endosperm has a high content of ER, whereas the central cells contain large protein bodies and lipid droplets or only lipid droplets. The endosperm cells that directly surround the embryo have very thick cell walls, contain lipid droplets, and have a large vacuole (Timmers, 1993).
The EP3 chitinase proteins found in the endosperm of 20 DAP seeds are most likely produced by integument cells. This indicates a maternal contribution to the proteins that are present in the endosperm.
Recent evidence for a role of maternal tissues in endosperm formation comes from the analysis of gametophytic mutations in Arabidopsis. In this species, fie (fertilization-independent endosperm) mutations (Ohad et al., 1996) that are female gametophytic and specifically affect endosperm formation are found. These data demonstrate a maternal component in endosperm formation. FIE/fie integuments that surround a mutant fie female gametophyte are degraded during development and give rise to the seed coat in the absence of fertilization, suggesting that Arabidopsis integument and seed coat development are initiated in response to a signal produced by the female gametophyte (Ohad et al., 1996).
In mature, dry seeds the EP3 mRNAs are located in the middle of the endosperm. In several other plant species, chitinases have been found in seeds. For example, in cucumber an endochitinase was found that is present only in mature seeds and in seeds during the early stages of germination (up to 2 d; Majeau et al., 1990). In barley a class I (Chi26) and a probable class II chitinase were found primarily in endosperm and aleurone tissues (Swegle et al., 1992). Chitinase mRNAs accumulated in barley endosperm and aleurone during seed development and were present from 15 d after anthesis until the end of germination (Leah et al., 1991; Swegle et al., 1992). For Chi26 an enhancer sequence has been identified that directs the aleurone-specific expression of this gene (Leah et al., 1994).
Cells of the starchy endosperm of cereals are dead at maturity and incapable of synthetic processes (Bewley and Black, 1994); therefore, the synthetic processes in the aleurone layer may resemble those that take place in the endosperm in species that do not contain aleurone tissue but rather live endosperm cells. In maize two 28-kD chitinases have been found that show high levels of expression in soaked seeds. These chitinases were reported as class I chitinases, but based on current definitions they should be reclassified as class IV chitinases (Huynh et al., 1992). Our results show that the localization of EP3-producing cells is restricted to the inner tissues of both the young fruits and the mature seeds. This pattern of localization suggests that the EP3 chitinases do not function primarily as enzymes that protect seeds against pathogens, as is commonly assumed for seed chitinases (Graham and Sticklen, 1994).
EP3 mRNAs are found in integuments and in endosperm cells that surround the central space in which the embryo is located. Both tissues are destined for degradation. In barley, apoptosis in the aleurone layer has been demonstrated during seed germination (M. Wang et al., 1996b), and fungal infection has been shown to induce apoptosis (H. Wang et al., 1996a). Since pathogen-induced chitinases are also induced prior to cell death (Kurosaki et al., 1987), this may suggest a more general role for chitinases in apoptosis.
The EP3 gene-expression pattern in seeds supports the hypothesis that chitinases are not only essential for the ts11 mutant but play a more general role in plant embryogenesis. Another, indirect indication for the importance of chitinases during seed development comes from a recent study of tobacco plants with low chitinase levels because of silencing caused by transgene expression of a tobacco class I chitinase. The silenced chitinases were transiently reset to a high-expressing state 8 to 11 DAP (Kunz et al., 1996). An explanation for the resetting of the silenced chitinases was not given, but it cannot be excluded that the requirement for chitinases during this period in seed development is a driving force for the resetting.
The finding that, in addition to the EP3 chitinases, Rhizobium sp.-produced Nod factors are capable of rescuing embryo formation in ts11 (De Jong et al., 1993) has led to the hypothesis that the presence of EP3s may result in the generation of GlcNAc-containing molecules that have a positive effect on the development of somatic embryos, although the correlation between EP3s and the Rhizobium sp. Nod factors remains to be elucidated (De Jong et al., 1995). Chitinases are capable of hydrolyzing Nod factors (Staehelin et al., 1994), whereas labeling studies indicate that plants may produce molecules that are analogous to Nod factors (Spaink et al., 1993).
Several examples in different systems appear to support the hypothesis that GlcNAc-containing oligosaccharides are important in development (Ioffe and Stanley, 1994). During Xenopus sp. embryonic development the DG42 gene is found to be expressed only between the late midblastula and neurulation stages (Rosa et al., 1988). This gene has some similarity with fungal chitin synthases and an even stronger homology with the Rhizobium sp. NodC gene (Bulawa and Wasco, 1991). The DG42 protein has been shown to catalyze the synthesis of short chitin oligosaccharides in vitro. The array of chitin oligosaccharides formed bears a striking resemblance to the oligosaccharides produced by NodC (Semino and Robbins, 1995). Homologs of this Xenopus sp. DG42 are present in zebrafish and mouse (Semino et al., 1996).
Regarding the biological function of the EP3 chitinases in plant embryogenesis, we propose that they are involved in reinitiating cell division in embryogenic cells and embryos as part of a nursing cell system that is required for but not restricted to embryogenesis. This hypothesis is based on the following observations: (a) the EP3 chitinases promote embryogenic cell formation and the number of somatic embryos as well as their progression in development when added to ts11 cultures (De Jong et al., 1992); (b) the expression of EP3 genes in cells that do not develop into embryos in culture and an absence of expression in somatic embryos; (c) the expression of EP3 genes in maternal tissue and the subsequent secretion of the encoded chitinase proteins, resulting in the presence of EP3 in the extracellular matrix of the endosperm surrounding globular-stage zygotic embryos; and (d) the absence of expression in zygotic embryos and the expression in endosperm cells prior to and during germination. The role of EP3 could be direct and involve a structural property such as the chitin-binding domain of the protein. It appears more likely that its function involves the catalytic properties of the enzyme through the release or modification of a GlcNAc-containing signal molecule, but this awaits the identification of such a molecule in plants. Whether such molecules are themselves soluble or attached to the extracellular matrix will then help to further unravel the role of chitin-based signaling molecules in plant embryogenesis.
Abbreviations:
- DAP
days after pollination
- RT
reverse transcriptase
Footnotes
This work was supported by The Netherlands Organization for Scientific Research (A.J.v.H.) and by the European Commission Biotechnology Program PTP-Biotech (F.G.).
LITERATURE CITED
- Bewley JD, Black M (1994) Seeds: Physiology of Development and Germination, Ed 2. Plenum Press, New York
- Borthwick HA. Development of the macrogametophyte and embryo of Daucus carota. Bot Gaz. 1931;92:23–44. [Google Scholar]
- Bulawa CE, Wasco W. Chitin and nodulation. Nature. 1991;353:710. doi: 10.1038/353710b0. [DOI] [PubMed] [Google Scholar]
- Cox KH, Goldberg RB. Analysis of plant gene expression. In: Shaw CH, editor. Plant Molecular Biology: A Practical Approach. Oxford, UK: IRL Press; 1988. pp. 1–34. [Google Scholar]
- De Jong AJ, Cordewener J, Lo Schiavo F, Terzi M, Vandekerckhove J, Van Kammen A, De Vries SC. A carrot somatic embryo mutant is rescued by chitinase. Plant Cell. 1992;4:425–433. doi: 10.1105/tpc.4.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jong AJ, Heidstra R, Spaink HP, Hartog MV, Meijer EA, Hendriks T, Lo Schiavo F, Terzi M, Bisseling T, Van Kammen A and others. Rhizobium lipooligosaccharides rescue a carrot somatic embryo mutant. Plant Cell. 1993;5:615–620. doi: 10.1105/tpc.5.6.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jong AJ, Hendriks T, Meijer EA, Penning M, Lo Schiavo F, Terzi M, Van Kammen A, De Vries SC. Transient reduction in secreted 32 kD chitinase prevents somatic embryogenesis in the carrot (Daucus carota L.) variant ts11. Dev Genet. 1995;16:332–343. [Google Scholar]
- De Vries SC, Booij H, Meyerink P, Huisman G, Wilde HD, Thomas TL, Van Kammen A. Acquisition of embryogenic potential in carrot cell-suspension cultures. Planta. 1988;176:196–204. doi: 10.1007/BF00392445. [DOI] [PubMed] [Google Scholar]
- De Vries SC, Springer J, Wessels JHG. Diversity of abundant mRNA sequences and patterns of protein synthesis in etiolated and greened pea seedlings. Planta. 1982;156:129–135. doi: 10.1007/BF00395427. [DOI] [PubMed] [Google Scholar]
- Gershoni JM, Palade GE. Electrophoretic transfer of proteins from sodium dodecyl sulfate polyacrylamide gels to a positively charged membrane filter. Anal Biochem. 1982;124:396–405. doi: 10.1016/0003-2697(82)90056-2. [DOI] [PubMed] [Google Scholar]
- Graham LS, Sticklen MB. Plant chitinases. Can J Bot. 1994;72:1057–1083. [Google Scholar]
- Gray D, Ward JA, Steckel JRA. Endosperm and embryo development in Daucus carota L. J Exp Bot. 1984;35:459–465. [Google Scholar]
- Guzzo F, Baldan B, Mariani P, LoSchiavo F, Terzi M. Studies on the origin of totipotent cells in explants of Daucus carota L. Exp Bot. 1994;45:1427–1432. [Google Scholar]
- Hari V. Effect of cell density changes and conditioned media on carrot cell embryogenesis. Z Pflanzenphysiol. 1980;96:227–231. [Google Scholar]
- Horvath B, Heidstra R, Lados M, Moerman M, Spaink HP, Promé JC, Van Kammen A, Bisseling T. Lipo-oligosaccharides of Rhizobium induce infection-related early nodulin gene expression in pea root hairs. Plant J. 1993;4:727–733. doi: 10.1046/j.1365-313x.1993.04040727.x. [DOI] [PubMed] [Google Scholar]
- Huynh QK, Hironaka CM, Levine EB, Smith CE, Borgmeyer JR, Shah DM. Antifungal proteins from plants. J Biol Chem. 1992;267:6635–6640. [PubMed] [Google Scholar]
- Ioffe E, Stanley P. Mice lacking N-acetylglucoseaminyltransferase I activity die at mid-gestation, revealing an essential role for complex or hybrid N-linked carbohydrates. Proc Natl Acad Sci USA. 1994;91:728–732. doi: 10.1073/pnas.91.2.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kragh KM, De Jong AJ, Hendriks T, Bucherna N, Højrup P, Mikkelsen JD, De Vries SC. Characterization of chitinases able to rescue somatic embryos of the temperature-sensitive carrot variant ts11. Plant Mol Biol. 1996;31:631–645. doi: 10.1007/BF00042235. [DOI] [PubMed] [Google Scholar]
- Kunz C, Schöb H, Stam M, Kooter JM, Meins F., Jr Developmentally regulated silencing and reactivation of tobacco chitinase transgene expression. Plant J. 1996;10:437–450. [Google Scholar]
- Kurosaki F, Tashiro N, Nishi A. Induction, purification and possible function of a chitinase in cultured carrot cells. Physiol Mol Plant Pathol. 1987;31:201–210. [Google Scholar]
- Kurosaki F, Tashiro N, Nishi A. Chitinase induction in carrot cell cultures treated with various fungal components. Biochem Int. 1990;20:99–106. [Google Scholar]
- Lackie S, Yeung EC. Zygotic embryo development in Daucus carota. Can J Bot. 1996;74:990–998. [Google Scholar]
- Leah R, Skriver K, Knudsen S, Ruud-Hansen J, Raikel NV, Mundy J. Identification of an enhancer/silencer sequence directing the aleurone-specific expression of a barley chitinase gene. Plant J. 1994;6:579–589. doi: 10.1046/j.1365-313x.1994.6040579.x. [DOI] [PubMed] [Google Scholar]
- Leah R, Tommerup H, Svendsen I, Mundy J. Biochemical and molecular characterization of three barley seed proteins with antifungal properties. J Biol Chem. 1991;266:1564–1573. [PubMed] [Google Scholar]
- Majeau N, Trudel J, Asselin A. Diversity of cucumber chitinase isoforms and characterization of one seed basic chitinase with lysozyme activity. Plant Sci. 1990;68:9–16. [Google Scholar]
- Meins F, Ahl P. Induction of chitinase and β-1,3-glucanase in tobacco plants infected with Pseudomonas tabaci and Phytophthora parasitica var. nicotianae. Plant Sci. 1989;61:155–161. [Google Scholar]
- Menon ARS, Dave Y. Structural design of the endosperm of carrot (Daucus carota L. var. sativa) Curr Sci. 1988;57:445–446. [Google Scholar]
- Métraux JP, Boller T. Local and systemic induction of chitinase in cucumber plants in response to viral, bacterial and fungal infections. Physiol Mol Plant Pathol. 1986;28:161–169. [Google Scholar]
- Ohad N, Margossian L, Hsu Y, Williams C, Repetti P, Fischer RL. A mutation that allows endosperm development without fertilization. Proc Natl Acad Sci USA. 1996;93:5319–5324. doi: 10.1073/pnas.93.11.5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa F, Sargent TD, Rebbert ML, Michaels GS, Jamrich M, Grunz H, Jonas E, Winkles JA, Dawid IB. Accumulation and decay of DG42 gene products follow a gradient pattern during Xenopus embryogenesis. Dev Biol. 1988;129:114–123. doi: 10.1016/0012-1606(88)90166-2. [DOI] [PubMed] [Google Scholar]
- Schmidt EDL, Guzzo F, Toonen MAJ, De Vries SC. A leucine-rich repeat containing receptor-like kinase marks somatic plant cells competent to form embryos. Development. 1997;124:2049–2062. doi: 10.1242/dev.124.10.2049. [DOI] [PubMed] [Google Scholar]
- Schmidt EDL, Van Hengel AJ, De Vries SC. A rapid method for localizing cell-specific transcripts in plant cell cultures. Biochemica. 1996;4:25–28. [Google Scholar]
- Semino CE, Robbins PW. Synthesis of “Nod”-like chitin oligosaccharides by the Xenopus developmental protein DG42. Proc Natl Acad Sci USA. 1995;92:3498–3501. doi: 10.1073/pnas.92.8.3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semino CE, Specht CA, Raimondi A, Robbins PW. Homologs of the Xenopus developmental gene DG42 are present in zebrafish and mouse and are involved in the synthesis of Nod-like chitin oligosaccharides during early embryogenesis. Proc Natl Acad Sci USA. 1996;93:4548–4553. doi: 10.1073/pnas.93.10.4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JA, Sung ZR (1985) Increase in regeneration of plant cells by cross feeding with regenerating Daucus carota cells. In M Terzi, L Pitto, ZR Sung, eds, Somatic Embryogenesis. Iucremento Produttivita Risorse Agricole, Rome, pp 133–137
- Spaink HP, Wijfjes AHM, Van Vliet TB, Kijne JW, Lugtenberg BJJ. Rhizobial lipo-oligosaccharide signals and their role in plant morphogenesis: are analogous lipophilic chitin derivatives produced by the plant? Aust J Plant Physiol. 1993;20:381–392. [Google Scholar]
- Staehelin C, Schultze M, Kondorosi E, Mellor RB, Boller T, Kondorosi A. Structural modifications in Rhizobium meliloti Nod factors influence their stability against hydrolysis by root chitinases. Plant J. 1994;5:319–330. [Google Scholar]
- Sterk P, Booij H, Schellekens GA, Van Kammen A, De Vries SC. Cell-specific expression of the carrot EP2 lipid transfer protein gene. Plant Cell. 1991;3:907–921. doi: 10.1105/tpc.3.9.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swegle M, Kramer KJ, Muthukrishnan S. Properties of barley seed chitinases and release of embryo-associated isoforms during early stages of imbibition. Plant Physiol. 1992;99:1009–1014. doi: 10.1104/pp.99.3.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmers ACJ (1993) A comparative structural study of somatic and zygotic embryogenesis of Daucus carota L. in imaging of polarity during zygotic and somatic embryogenesis of carrot (Daucus carota L.). PhD thesis. Agricultural University Wageningen, Wageningen, The Netherlands
- Toonen MAJ, Hendriks T, Schmidt EDL, Verhoeven HA, Van Kammen A, De Vies SC. Description of somatic-embryo-forming single cells in carrot suspension cultures employing video cell tracking. Planta. 1994;194:565–572. [Google Scholar]
- Trudel J, Asselin A. Detection of chitinase activity after polyacrylamide gel electrophoresis. Anal Biochem. 1989;178:362–366. doi: 10.1016/0003-2697(89)90653-2. [DOI] [PubMed] [Google Scholar]
- Van Engelen FA, De Jong AJ, Meijer EA, Kuil CW, Meyboom K, Dirkse WG, Booij H, Hartog MV, Vandekerckhove J, De Vries SC and others. Purification, immunological characterization and cDNA cloning of a 47kDa glycoprotein secreted by carrot suspension cells. Plant Mol Biol. 1995;27:901–910. doi: 10.1007/BF00037018. [DOI] [PubMed] [Google Scholar]
- Van Engelen FA, De Vries SC (1993) Secreted proteins in plant cell cultures. In KA Roubelakis-Angelakis, K Tran Thanh Van, eds, Markers of Plant Morphogenesis. Plenum Press, New York, pp 181–200
- Van Engelen FA, Sterk P, Booij H, Cordewener JHG, Rook W, Van Kammen A, De Vries SC. Heterogeneity and cell-type specific localization of a cell wall glycoprotein from carrot suspension cells. Plant Physiol. 1991;96:705–712. doi: 10.1104/pp.96.3.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Li J, Bostock RM, Gilchrist DG. Apoptosis: a functional paradigm for programmed plant cell death induced by a host-selective phytotoxin and invoked during development. Plant Cell. 1996a;8:375–391. doi: 10.1105/tpc.8.3.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Oppedijk BJ, Lu X, Van Duijn B, Schilperoort RA. Apoptosis in barley aleurone during germination and its inhibition by abscisic acid. Plant Mol Biol. 1996b;32:1125–1134. doi: 10.1007/BF00041396. [DOI] [PubMed] [Google Scholar]