Abstract
Cdx transcription factors regulate embryonic positional identities and have crucial roles in anteroposterior patterning (AP) processes of all three germ layers. Previously we have shown that the zebrafish homologues cdx1a and cdx4 redundantly regulate posterior mesodermal derivatives inducing embryonic blood cell fate specification and patterning of the embryonic kidney. Here we hypothesize that cdx factors restrict formation of anterior mesodermal derivatives such as cardiac cells by imposing posterior identity to developing mesodermal cells. We show that ectopic expression of Cdx1 or Cdx4 applied during the brief window of mesoderm patterning in differentiating murine embryonic stem cell (ESC) strongly suppresses cardiac development as assayed by expression of cardiac genes and formation of embryoid bodies (EB) containing “beating” cell clusters. Conversely, in loss-of-function studies performed in cdx-deficient zebrafish embryos, we observed a dose-dependent expansion of tbx5a+ anterior-lateral plate mesoderm giving rise to cardiac progenitors. However, further cardiac development of these mesodermal cells required additional suppression of the retinoic acid (RA) pathway, possibly due to differential activity of inhibitory RA signals in cdx mutants. Together, our data suggest that cdx proteins affect cardiogenesis by regulating the formation of cardiogenic mesoderm and together with the RA pathway control the early development of cardiac precursor cells.
Keywords: Cdx, retinoic acid, mesoderm, cardiac, embryonic stem cells, zebrafish
Introduction
The anteroposterior (AP) patterning of several embryonic tissues, including the developing mesoderm, is tightly regulated by Hox gene expression patterns, especially by precise positioning of their anterior expression boundaries (Kmita and Duboule, 2003). Although the mechanisms of Hox activation along the developing AP axis are not completely understood, one plausible model is the ‘instructional (morphogen) gradient hypothesis that proposes that retinoic acid (RA), FGF and Wnt establish Hox expression boundaries at threshold concentrations (Deschamps and van Nes, 2005; Gaunt, 2000). Members of the caudal-related family of homeobox (Cdx) proteins have been proposed to mediate positional information between morphogen pathways and downstream Hox genes (Allan et al., 2001).
The Cdx gene family derives from the ancestral ParaHox cluster and comprises Cdx1, Cdx2, and Cdx4 in mammals and cdx1a, cdx1b and cdx4 in zebrafish. In the developing embryo, Cdx expression is induced within the primitive streak/tailbud (Gaunt et al., 2003; Gaunt et al., 2005) and later, protein levels are distributed along a posterior-to-anterior concentration gradient, probably due to decay in protein concentration in cells moving out of this region (Beck et al., 1995; Gamer and Wright, 1993; Meyer and Gruss, 1993). Consistent with this expression pattern, Cdx genes play major roles during patterning of the AP axis and regulation of axial elongation during development (Chawengsaksophak et al., 2004; van den Akker et al., 2002). For example, loss- and gain-of-function studies performed in mice have identified roles for Cdx genes during the patterning of paraxial mesoderm and the development of the somites and vertebrae (reviewed by (Young and Deschamps, 2009). More recently, Cdx genes have been associated with the expansion and patterning of posterior tissues (Davidson et al., 2003; Davidson and Zon, 2006; Shimizu et al., 2005; Wingert et al., 2007), the embryonic kidney (Wingert et al., 2007), and the specification of hematopoietic cell fate, a function that can be rescued by specific hox genes (Davidson et al., 2003; Davidson and Zon, 2006; Lengerke et al., 2007; McKinney-Freeman et al., 2008). Molecularly, Cdx genes are well known as master regulators of Hox gene expression (Lohnes, 2003). Presumably due to similar effects of downstream Hox genes, redundancies between Cdx family members have been reported in different systems (Davidson and Zon, 2006; Lengerke et al., 2007; McKinney-Freeman et al., 2008). These redundant effects complicate in vivo loss of function studies in mice, where a Cdx-knockout mouse model has been challenging to create due to essential early roles of Cdx2 during placenta formation (Strumpf et al., 2005). During development, expression of Cdx genes is induced and maintained by morphogens such as Wnt, FGF and RA (Lengerke et al., 2008; Lohnes, 2003; Pilon et al., 2006). However, recent data suggest a more complex model, and shows that Cdx genes themselves can modulate morphogen expression levels (e.g. maintenance of posterior Wnt signaling and clearance of retinoic acid in the “posterior growth zone”) (Lengerke et al., 2008; Young et al., 2009a).
To date, there have been no reports implicating Cdx genes as regulators of heart development. At early gastrula stage, cardiac precursor cells are found at the anterior region of the primitive streak. During gastrulation, they leave the primitive streak and migrate anterolaterally to form the precardiac mesoderm within the left and right anterior lateral plate mesoderm. Here, commitment to the heart lineage occurs in response to endoderm-derived signals such as BMP, FGF and Wnt-antagonists (reviewed by (Nakajima et al., 2009)) and in accordance to retinoic acid exposure (Keegan et al., 2005). Given the prominent role of Cdx genes during early patterning processes, we hypothesized that they play roles in the development of anterior mesoderm derivatives such as cardiac cells. In this report we analyze the impact of Cdx genes on cardiac development from mouse ESC and during in vivo zebrafish embryo development by performing functional studies and analyzing expression of markers indicating commitment to the cardiac lineage such as Nkx2.5 and Mesp1 (Bondue et al., 2008; David et al., 2008).
Material and methods
Cell culture and differentiation
iCdx1, iCdx4 and parental Ainv15 murine ESC (Kyba et al., 2002; Lengerke et al., 2008; McKinney-Freeman et al., 2008; Wang et al., 2008) were cultured as reported on irradiated mouse embryonic fibroblasts in Dulbecco modified Eagle medium with 15% fetal calf serum (HyClone Laboratories, Logan, UT), 1000 U/ml leukemia inhibitory factor (Chemicon International, Temecula, CA), 2 mM penicillin/streptomycin/glutamine (Invitrogen, Carlsbad, CA), 0.1 mM nonessential amino acids (Invitrogen), and 0.1 mM β-mercaptoethanol (Sigma-Aldrich, St Louis, MO) at 37°C/5% CO2 (Kyba et al., 2002). Media was refreshed daily, and cultures were passaged with trypsine (Invitrogen) every 2 to 3 days.
Murine ESC were differentiated in embryoid bodies (EB) as described previously (Kyba et al., 2002; Lengerke et al., 2008). Briefly, confluent cultures were harvested and resuspended at a concentration of 100 cells/20 μL in EB differentiation media composed of Iscove modified Dulbecco medium (IMDM) plus 15% fetal calf serum (StemCell Technologies, Vancouver, BC), 2 mM penicillin/streptomycin/glutamine (Invitrogen), 4.5 mM monothioglycerol (Sigma-Aldrich), 200 μg/mL holo-transferrin (Sigma-Aldrich), and 50 μg/ml ascorbic acid (Sigma-Aldrich). EB were cultured in 20 μl hanging drops for 48 hours and then transferred and cultured in 10 cm2 Petri dishes for an additional 4 days at 37°C/5% CO2 while shaking at 50 rpm. For ectopic gene expression, doxycycline (1.0 μg/ml; Sigma-Aldrich) was added as indicated.
Flow cytometry and cell sorting
EB cells were stained with PE-conjugated anti-Flk1 (BD Pharmingen). Flow cytometry was performed on a FACSCalibur from Becton Dickinson. Flk1 positive and Flk1 negative cells were isolated with magnetic anti-PE microbeads (Miltenyi Biotech). Purity after sorting was >90%.
Functional cardiomyocyte assessment
At day 6 of EB differentiation 150–200 EB were transferred from the shaking Petri dishes onto gelatinized six-well dishes (Gelatine 1%, Biochrom AG) and further cultured in EB differentiation medium. At day 7, 9 and respectively 11, EB containing beating areas were scored by counting a total number of at least 50 to 100 EB for each condition.
Real-time RT-PCR
Cells were harvested in RLT Buffer (supplied with RNeasy Mini Kit, Qiagen), and total RNA was isolated according to manufacturer instructions, including on-column DNAse treatment. cDNAs were prepared according to the manufacturer’s protocol using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Real-time quantitative PCR was performed using SYBR Green reagent (Eurogentec) on a Light Cycler480 Real-time PCR instrument (Roche Applied Science). Primer sequences were used as previously reported (Lengerke et al., 2007) or as listed in Table 1. The annealing temperatures that were used are listed in table 1. All primers were used at 150 nM.
Table 1.
| Gene | Forward Primer | Reverse Primer | Annealing temperature |
|---|---|---|---|
| Cdx1 | GTA AGA CCCGAACCA AGGAC | GGA ACCAGATCTTTA CCTGC | 58°C |
| Cdx4 | GAGGAAGTCAGAGCTGGCAGTTA | GGCTCTGCGATTCTGAAACC | 60°C |
| Gata4 | AGGGTGAGCCTGTATGTA ATGCCT | AGGACCTGCTGGCGTCTTAGATTT | 63°C |
| Isl1 | GCA AGGACA AGA AACGCAGCATCA | ACTGGGTTAGCCTGTAAACCACCA | 63°C |
| Mesp1 | TACGCAGAA ACAGCATCCCAGGAA | CCAGGTTTCTAGAAGAGCCAGCAT | 63°C |
| Nkx2.5 | CAA GTGCTCTCCTGCTTTCC | CCAGCTCCACTGCCTTCTG | 60°C |
| MyoD1 | ATCCCTAAGCGACACAGA ACAGGGAA | TGCAGTCGATCTCTCAAAGCACCT | 63°C |
| Myogenin | ACAATCTGCACTCCCTTACGTCCA | TCTCAGTTGGGCATGGTTTCGTCT | 63°C |
| GAPDH | AATGCATCCTGCACCACCAACTGCTT | AGTGATGGCATGGACTGTGGTCAT | 64°C |
| Actin | TCTTGGGTATGGAATCCTGTGGCA | ACTCCTGCTTGCTGATCCACATCT | 59°C |
Zebrafish husbandry, genetic strains, chemical treatments and morpholinos
Zebrafish were maintained and staged as described (Kimmel, et al., 1995). The cdx4 (kggtv205) mutant allele was maintained on the Tübingen strain, and incrosses of heterozygous adults were used to obtain cdx4−/− embryos. Doubly deficient cdx1a/4 embryos were generated by injecting cdx4−/− embryos at the 1-cell stage with cdx1a morpholino (CAGCAGATAGCTCACGGACATTTTC) as described (Davidson, et al. 2003). Both retinoic acid signaling antagonists, diethylaminobenzaldehyde (DEAB) (Sigma) and the RARα antagonist R0-41-5253 (Biomol International), were dissolved in 100% dimethyl sulfoxide (DMSO) to make 0.1 M stock solutions, and aliquots were stored at −80°C. All-trans retinoic acid (RA) Sigma was dissolved in 100% DMSO to make a 1 M stock, and aliquots were stored at −80°C. For chemical treatments, embryos were incubated in 1.6 × 10−5 M DMSO (vehicle control), 1.6 × 10−5 M DEAB/DMSO in E3 embryo media, 1 × 10−7 M Ro-41-5253/DMSO in E3 embryo media, or 1.6 × 10−5 M DEAB with 1 × 10−8 M RA in E3 embryo media (DEAB rescue study), between 60% epiboly and the 15 somite stage. The cdx4 mutation, cdx1a morpholino, and RA antagonists produced fully penetrant effects as described (Wingert, et al. 2007). Following chemical incubation, embryos were rinsed in E3 media then fixed in 4% paraformaldehyde (PFA) in 1X Pbst for gene expression analysis. Whole-mount in situ hybridization of zebrafish embryos was performed as described (Davidson, et al. 2003), using established antisense probe construction as reported for krox20, mhc, myoD, pax2a (Wingert, et al. 2007), nkx2.5 (Serbedzija, et al. 1998), tbx5a (Begemann and Ingham, 2000) and cmlc2 (Yelon, Horne, Stainier, 1999). For each reported gene expression pattern, at least 15 embryos were examined. Embryos were deyolked and flat-mounted on glass slides, then photographed using a Nikon Eclipse 80i microscope and Nikon CoolPix 4500 digital camera. Nikon Elements Basic Research software was used with a Nikon DS-Fi1 color camera sytsem to measure the areas of nkx2.5 and cmlc2-expressing cells.
Statistics
For all experiments, error bars represent the standard error, and P values are derived via the application of a 2-tailed, unpaired Student t test. * p<0.01, ** p<0.005.
Results
Endogenous and ectopic Cdx1 and Cdx4 expression in differentiating murine ESC
ESC differentiation into EB recapitulates the commitment events of early embryonic development and can be documented as temporal waves of tightly controlled transcription factors such as Cdx genes (Figure 1A) and lineage-specific gene expression (Keller, 2005). Brachyury-positive, primitive-streak like mesodermal cells emerge sequentially between day 2 and 4 of EB development (Figure 2A). Gene expression indicative of mesodermal commitment to specific fates occurs slightly later, around day 3 to 4 of EB development for hematopoietic (not shown) and day 4 to 5 of EB development for cardiac cell fates with specified hematopoietic and cardiac cells peaking around day 6 of EB development (Keller, 2005) (Figure 2A). Focusing on day 0 to 11 of EB development, we assessed the expression pattern of murine Cdx1 and Cdx4 by performing gene expression analysis by real-time PCR (Figure 1A). As previously reported, undifferentiated mouse ESC express only very low levels of Cdx4 but higher levels of Cdx1 (McKinney-Freeman et al., 2008). When ESC are differentiated as EB, Cdx1 and Cdx4 expression increases showing enhanced expression around day 1 and 3 of differentiation for Cdx1 and slightly later, at day 3, for Cdx4. After day 3 and respectively day 4, Cdx1 and Cdx4 expression rapidly wanes (Figure 1A). Cdx1 sometimes shows a second peak of slightly enhanced expression at later time-points (e.g. day 6 to day 8, Figure 1A and data not shown), probably reflecting expression in developing endodermal tissues. Thus, as previously reported (McKinney-Freeman et al., 2008), Cdx genes are expressed in an overlapping temporal manner with Cdx1 slightly preceding Cdx4 and overall both genes active in the time-window when mesoderm is formed and patterned to specific fates. This in vitro expression pattern is in accordance with the in vivo expression data reporting most intense Cdx induction at the time of gastrulation in the primitive streak. The earliest mesodermal cells to adopt a cardiac fate are the cardioangioblasts which can be isolated as Flk1 positive cells at day 4 of EB development (Kattman et al., 2006). We detected Cdx1 and Cdx4 transcripts in this population, although at lower levels than in the corresponding Flk1 negative fraction containing progenitors of other tissues such as hematopoietic, renal and endodermal lineages (Figure 1B).
Fig. 1.
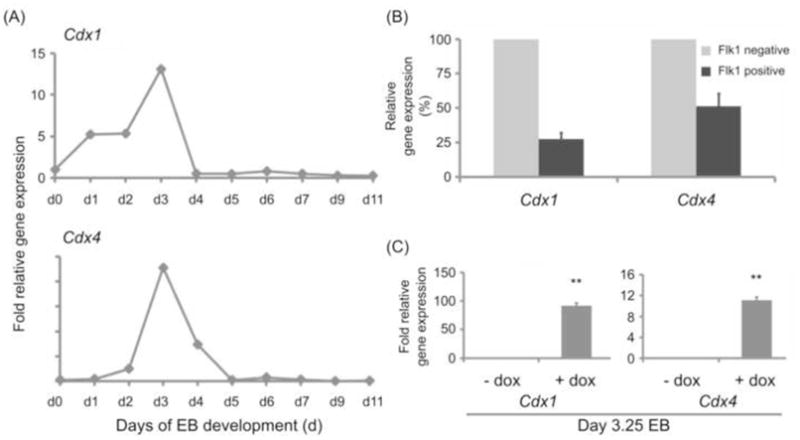
(A) Time course of Cdx1 and Cdx4 expression in differentiating EB show an expression peak around day 3 of EB development. Mouse ESC were differentiated in EB and samples collected between day 1 and day 11 of EB development were subjected to real-time PCR analysis. Results are shown as fold expression relative to expression in undifferentiated ES cells. Shown are data from one representative experiment. (B) Expression of Cdx1 and Cdx4 in Flk1 positive cardioangioblasts isolated at day 4 of EB development. Results are shown as % expression relative to Flk1 negative cells isolated at the same point in time. Shown are data from two biological experiments which showed a purity >90% in the sorted Flk1 positive population. (C) Ectopic Cdx1 and Cdx4 gene induction within the endogenous expression window. iCdx1 and respectively iCdx4 ES cells were differentiated until day 2.25 in basic differentiation medium to allow mesoderm induction and afterwards further cultured in basic differentiation medium +/− doxycycline. Induction of Cdx1 and respectively Cdx4 induction was analyzed by real-time PCR performed 24 hours later on day 3.25 EB. Results are shown as relative fold expression in comparison to non-induced controls collected at the same time-point. Gene expression levels summarize endogenous and if applicable ectopic levels. Shown are summarized results from three experiments.
Fig. 2.
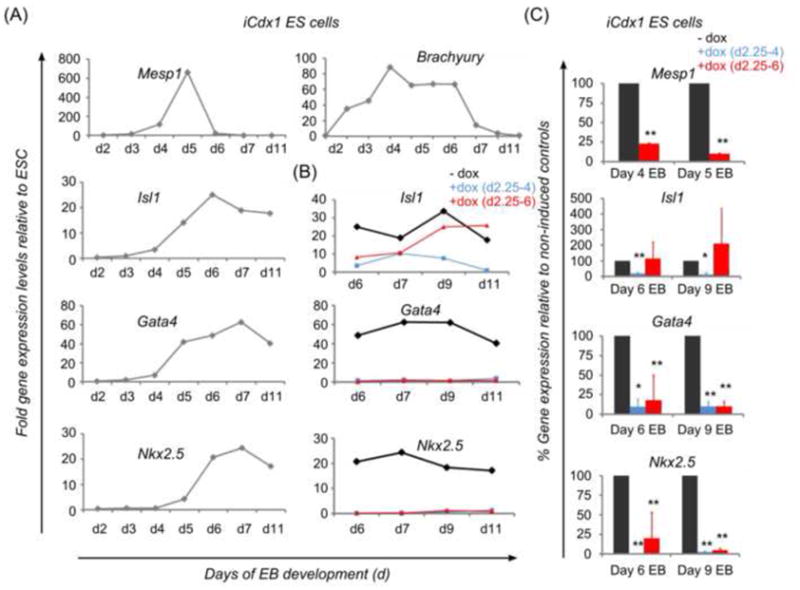
Expression pattern of genes involved in mesoderm and cardiac development during in vitro EB differentiation. Suppressive effects of Cdx induction on cardiac genes. (A) Markers of early cardiac development such as Mesp1, Isl1, Gata4 and Nkx2.5 are upregulated during EB differentiation shortly after establishment of the mesodermal marker Brachyury. Mesp1 shows a sharp peak of expression and extinguishes rapidly upon activation of other cardiac genes such as Nkx2.5. iCdx1 ES cells were differentiated in basic differentiation medium in the absence of doxycycline and samples collected at several time-points. Gene expression analysis was performed by real-time PCR analysis and results are shown as fold gene expression relative to expression levels in undifferentiated ES cells. Shown are data from one representative experiment for each marker. (B, C) Continuous suppression of Isl1, Gata4 and Nkx2.5 following early induction of Cdx1 by addition of doxycycline between day 2.25 to 6 (B, C) or as a brief pulse between day 2.25 to 4 only (C). A,B: Shown are data from one representative experiment; C: shown are summarized data from at least two independent experiments for each analyzed condition.
To test the effect of Cdx1 and Cdx4 expression on cardiogenesis, we used two previously generated mouse ESC lines with Cdx1 and Cdx4 under the control of a tetracycline inducible promoter, respectively (Davidson et al., 2003; Kyba et al., 2002; McKinney-Freeman et al., 2008). Doxycycline exposure of day 2.25 EB generated from inducible Cdx1 (iCdx1) and Cdx4 (iCdx4) ESC enhances Cdx gene transcripts (Figure 1C) and protein levels (McKinney-Freeman et al., 2008) within their endogenous expression window in a tightly regulated manner (Supplemental Figure 1A). Moreover, previously documented auto- and cross-regulation of Cdx genes (Beland et al., 2004; Lengerke et al., 2008; Young et al., 2009b; Savory et al., 2011) may contribute to the enhanced gene expression levels documented by PCR (Figure 1C). As expected, no induction of Cdx was reported by addition of doxycycline in parental cells (McKinney-Freeman et al., 2008; Supplemental Figure 1C).
Mesodermal and cardiac differentiation from iCdx1 and iCdx4 ES cells
Since individual ESC lines have been reported to behave differently with respect to cardiac development, we first examined cardiac development in our ESC lines by performing gene expression time-courses and analyzing the formation of functional cardiac cells at different time-points. The iCdx1 and iCdx4 ESC lines, which have been generated in the same Ainv15 ES cell background (Kyba et al., 2002), behaved similarly in these assays (data not shown) and experiments performed in these ESC are shown in summary (Figure 2A and 3A).
Fig. 3.
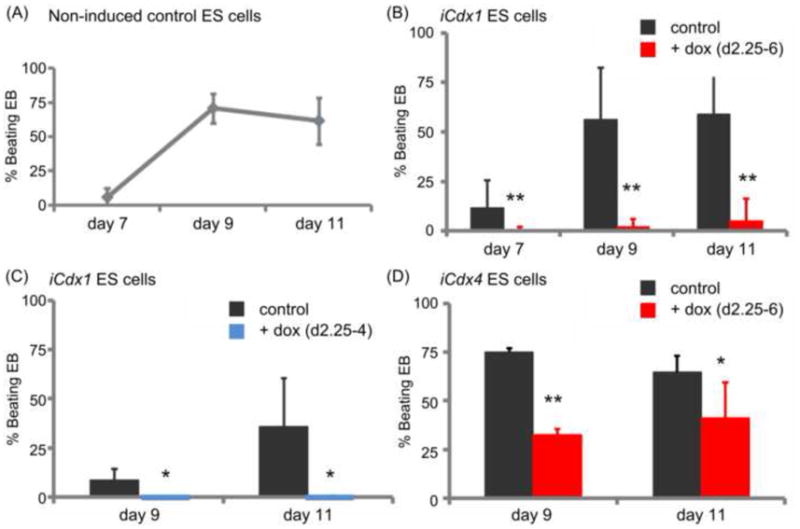
Cdx1 and Cdx4 suppress the formation of “beating” cardiomyocytes from differentiating EB. (A) Time-course of development of “beating” clusters in iCdx ES cells differentiated in basic differentiation medium without addition of doxycycline first detects functional cardiomyocytes around day 7 of EB development with reproducible development in high numbers at day 9 and 11 across experiments. Shown are summarized data on % beating EBs in non-induced iCdx1 and iCdx4 EB collected in nine independent experiments. (B, C, D) Exposure to doxycycline during EB development (from day 2.25 to 6 and respectively from day 2.25 to 4) strongly suppresses cardiomyocyte development from both iCdx1 and iCdx4 ES cells at all analyzed time-points. Results are shown as % beating EB in doxycycline induced cells in comparison to non-induced controls. Shown are data from at least three independent experiments for each analyzed condition.
As expected, cardiac gene expression was induced around day 4 of development, after mesoderm had been initiated as indicated by the expression of Brachyury. These developmental kinetics are congruent with previously reported results showing that cardiac progenitors first arise at day 4 of EB development in the form of a Flk1 positive population (Kattman et al., 2006; Kouskoff et al., 2005), slightly after the development of the blood and vascular lineages at day 3 (Fehling et al., 2003). Of note, the earliest marker reported to induce cardiac development, Mesp1, showed an induction peak around day 4–5, before extinguishing abruptly. Nkx2.5 transcripts, which initiate following cardiac progenitor specification, followed Mesp1 expression and showed strong up-regulation at day 6 of EB development. Isl-1 and Gata4 showed a rather early induction, consistent with their role in early cardiac specification, but expression did not wane at later time-points, indicating an on-going involvement in cardiac development and/or differentiation of other cell lineages.
Of note, doxycycline treatment alone did not impact differentiation of parental Ainv15 cells lacking transgenes (Supplemental Figure 1b and 1c).
Cdx1 or Cdx4 activation during mesoderm formation suppresses the formation of beating cardiomyocytes in differentiating murine ESC
To assess the role of Cdx during cardiac development, iCdx1 and iCdx4 ESC were cultured in the presence or absence of doxycycline. In the cultures supplemented with doxycycline, addition was performed between days 2.25–6 or days 2.25–4 of EB differentiation, in order to enhance Cdx expression within their endogenous expression window (Figure 1A). At day 6, EB were transferred onto gelatinized plates and cultured in basic differentiation medium without doxycycline for another 1 to 5 days. Development of cardiac cells was assessed by scoring the development of EB presenting “beating” areas (Wu et al., 2006). In some experiments, a few beating areas were observed as early as day 7 (Figure 3A). At day 9 consistent beating was observed in approximately 75% of EB throughout the experiments, with no progression occurring at later time-points such as day 11 (Figure 3A).
Induction of either Cdx1 or Cdx4 under these conditions strongly suppressed beating at all points in time, suggesting that the inhibitory effects were not due to differences in ESC differentiation dynamics but rather a specific developmental effect on cardiogenesis (Figure 3B–D). A brief pulse of doxycycline between day 2.25 and 4 was sufficient to inhibit cardiogenesis (Figure 3C), consistent with an early effect of Cdx on cardiac development. The suppressive effect was stronger for Cdx1 than Cdx4, supporting data collected in other murine organs where Cdx4 showed less potent effects than Cdx1 (Deschamps and van Nes, 2005; Lengerke et al., 2008; McKinney-Freeman et al., 2008). However, the more pronounced effect of Cdx1 in our system could also be due to the fact that ectopic activation resulted in a greater increase in gene expression relative to endogenous levels documented at this developmental stage.
Cdx induction inhibits the expression of cardiac genes, including the early marker Mesp1
We next examined cardiac marker expression in iCdx1 and iCdx4 ESC and EB at several time-points. Up-regulation of Cdx expression at day 2.25 inhibited the induction of Gata4, Nkx2.5 and Mesp1 (Figure 2B and 2C, Figure 4A and 4B). Interestingly, modulation of Isl-1 expression was different when Cdx activation was performed in the time-window between days 2.5–4 or continued until day 6, suggesting that in addition to an early role at the level of mesoderm, Cdx may impact the differentiation of Isl-1 regulated tissues at later time-points (Figure 2B and 2C, Figure 4A and 4B). Overall, ectopic Cdx1 showed a stronger suppressive effect than ectopic Cdx4, confirming the results in functional assays (Figure 3B and 3D).
Fig. 4.
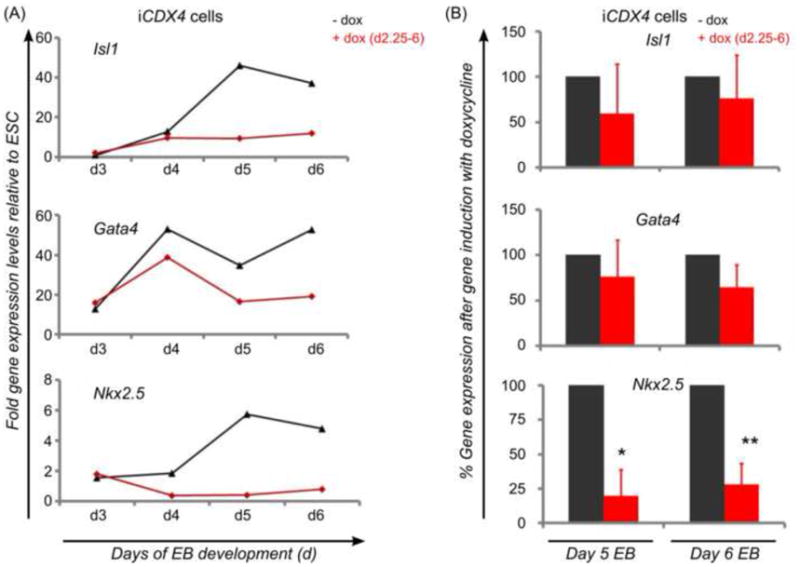
Effect of ectopic Cdx4 on the expression of cardiac genes in differentiating murine ES cells. (A) Time-course analysis in samples collected from iCdx4 ES cells differentiating as EB with or without exposure to doxycycline during days 2.25 to 6 of development shows continuous suppression of Gata4, Nkx2.5 and Isl-1. Results are shown as fold expression levels relative to gene expression in undifferentiated ES cells. Shown are data from one representative experiment showing more pronounced effects. (B) Gene expression analysis in day 5 and day 6 iCdx4 EB previously differentiated in basic medium with or without exposure to doxycycline as indicated shows strong suppression of Nkx2.5 while only mild, non-significant effects were seen on expression of Isl-1 and Gata4. Shown are data on % gene expression relative to non-induced controls collected at the same time-points in at least two independent experiments.
Cdx morphants display a dose-dependent enhancement in tbx5a+ cardiac mesoderm
Next, we examined whether Cdx genes function to restrict cardiac fates during heart development in vivo using the zebrafish embryo as a model. As cdx4 and cdx1a have established partially redundant roles in mesoderm patterning in the zebrafish (Davidson and Zon, 2006), we analyzed the expression of cardiac lineage markers in cdx4 deficient and cdx4/1a doubly-deficient embryos using whole mount in situ hybridization. The expression of tbx5a, a marker of the anterior lateral plate mesoderm that includes the precursor cardiomyocyte population, was expanded posteriorly in both cdx4 deficient embryos and cdx1a/4 doubly-deficient embryos (Figure 5). Conversely, the expression domain of pax2a, an intermediate mesoderm marker, was reduced and shifted posteriorly (Figure 5). These data suggest that broad mesodermal patterning was altered in the absence of cdx activity. However, the expression domains of nkx2.5 and cmlc2, which are specific to cardiac precursors, were not altered in cdx deficient embryos (Figure 6 and 7; Supplemental Figure 2). This finding was surprising given the changes in mesoderm gene expression and the expression changes subsequent to Cdx overexpression in ESC and EB cultures.
Fig. 5.
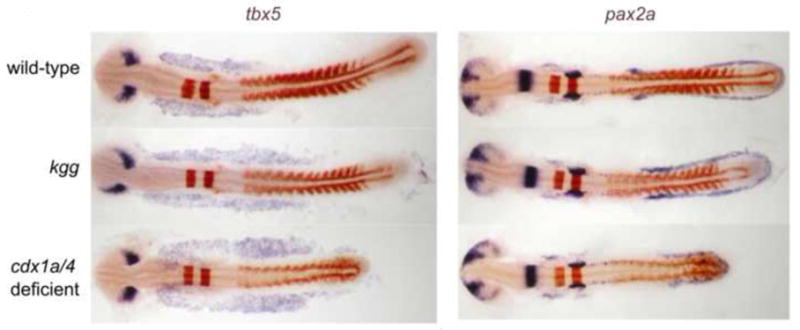
Whole mount in situ hybridization analysis of tbx5a expression in wildtype, cdx4 and cdx1a/4-deficient embryos shows expansion of cardiogenic anterior-lateral plate mesoderm. The intermediate mesoderm field, marked by pax2a expression, shows a posterior shift and reduction in cell number, as reported previously. The effect was more pronounced in cdx1/4 double deficient embryos than in cdx4−/− embryos. Shown are zebrafish embryos at 15-somite stage. Purple staining was used for tbx5 and pax2a and red staining for krox20 and myoD as landmarks of other tissues.
Fig. 6.
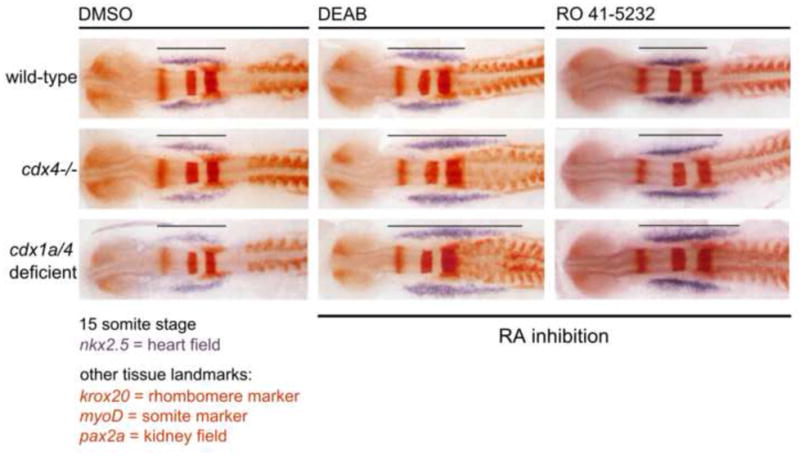
Excessive tbx5 positive anterior-lateral plate mesoderm formed in cdx-deficient embryos requires additional inhibition of the retinoic acid pathway in order to adopt cardiac fate and form nkx2.5 expressing cardiac cells. Shown are wildtype, cdx4−/− and cdx1/4 deficient embryos analyzed at 15-somite stage after treatment with the retinoic acid inhibitors DEAB, Ro-41-5253, or DMSO (vehicle) as a control. cdx-dose-dependent effects are observed with both inhibitors.
Fig. 7.
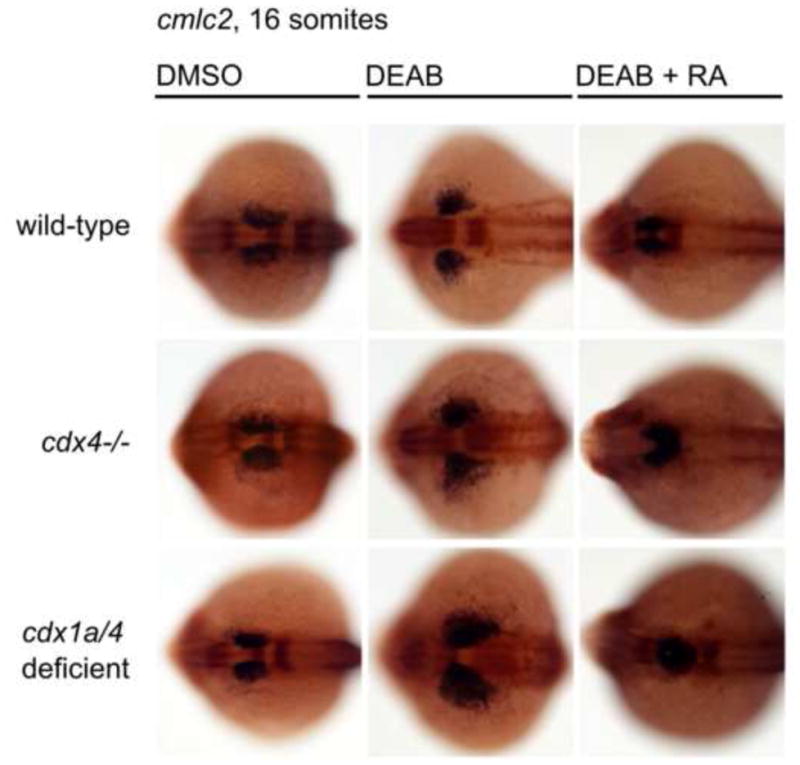
Suppression of RA levels expands cardiac myosin expressing-cells (cmlc2) in cdx-deficient embryos, and can be rescued by exogenous RA treatment. Expression of cmlc2 expression in wildtype, cdx4 and cdx1a/4-deficient embryos incubated with DMSO (control), DEAB, or DEAB and RA, was assayed by whole mount in situ hybridization at the 16 somite stage. cdx mutants evinced dramatic expansion of cardiac mesoderm when RA production was abrogated, and this phenotype was rescued by concomitant DEAB and RA treatment.
Expansion of nkx2.5 and cmlc2 expressing cardiac cells in cdx mutants requires simultaneous inhibition of the RA pathway
During cardiac specification in the zebrafish embryo, retinoic acid (RA) signaling plays an essential role in restricting the number of cardiac progenitors (Keegan et al., 2005). RA is synthesized during development through the sequential action of enzymes that include retinaldehyde dehydrogenases (raldhs or aldhs), and RA signals are conferred by binding to complexes of retinoic acid receptors (RARs, RXRs) (Duester, 2008). Chemical modulation of RA signaling by exposure to RA antagonists causes expansion of the cardiomyocyte pool (Keegan, et al., 2005). We hypothesized that the presence of endogenous RA signaling was sufficient to limit cardiac expansion in the setting of cdx deficiency. To test this idea, we treated cdx4-deficient and cdx1a/4-deficient embryos with diethylaminobenzaldehyde (DEAB), a pan-raldh inhibitor, or RO-41-5253, a RARα-antagonist. Interestingly, exposure to DEAB or RO-41-5253 induced a significant expansion in the nkx2.5-expressing in cdx4-deficient embryos, and cdx1a/4-deficient embryos showed an even more dramatic expansion (Figure 6; Supplemental Figure 2A and 2C). Next, we examined the expression of the cardiac myosin gene cmlc2 in cdx-deficient animals when RA levels were reduced. Both cdx4-deficient and cdx1a/4-deficient embryos exposed to DEAB displayed significant expansions of cmlc2-expressing cells, as compared to wild-type embryos similarly treated with DEAB (Figure 7; Supplemental Figure 2B and 2D). In addition, the expansion of the cmlc2 field in cdx-deficient embryos was rescued when they were treated with a combination of DEAB and RA (Figure 7; Supplemental Figure 2B and 2D). This finding provides evidence that the restoration of RA levels is sufficient to reduce the expansion of cardiac progenitors. Taken together, these data confirm our hypothesis that RA is the critical signaling molecule that restricts the expansion of cardiac fate in a cdx1a/4-deficient background.
Discussion
Cdx transcription factors regulate positional identities and have crucial roles in anteroposterior patterning processes during embryonic development, where they have been especially studied as regulators of Hox gene expression in the paraxial mesoderm (reviewed by (Deschamps et al., 1999)). More recent data collected in zebrafish indicate an additional involvement of Cdx genes in the patterning and formation of posterior mesodermal tissues such as embryonic blood and kidney (Davidson et al., 2003; Wingert et al., 2007). Studies in mouse and human pluripotent stem cells suggest conservation of pathways and redundant roles for Cdx1 and Cdx4 during mesoderm specification to blood in mammalian cells (Lengerke et al., 2009; Lengerke et al., 2007; Lengerke et al., 2008b; McKinney-Freeman et al., 2008). Generation of blood formation has been intensively studied on the molecular level in different models and Cdx genes shown to regulate blood formation by the induction of posterior Hox genes (Davidson et al., 2003; Davidson and Zon, 2006; Lengerke et al., 2008; Wang et al., 2008), in a partially Wnt-dependent fashion (Lengerke et al., 2008), as well as via regulation of RA production (de Jong et al., 2010).
The current report analyzes whether the regulatory effects of Cdx genes on early mesoderm development extend to cardiac cells, which are derived from more anterior mesoderm. We show in mouse EB in vitro and zebrafish embryos in vivo that Cdx genes in cooperation with RA may restrict the development of mesoderm prone to undergo cardiac differentiation and the early development of cardiac cells. In EB, a pulse of Cdx expression, during the window of mesoderm patterning, is sufficient to suppress cardiac development and marker expression, whereas loss-of-function studies in zebrafish show that cdx genes restrict the size of the tbx5a+ ‘pre-cardiac’ anterior lateral plate mesoderm domain. Both results argue for an early effect of Cdx genes on heart development, at the level of mesoderm.
The effects of Cdx overexpression on heart development is unlikely to be due to non-specific toxicity as we have previously shown that EB morphology and cell number are unaffected by Cdx overexpression and treatment with doxycycline alone had no impact on cardiac differentiation in our system (Supplemental Figure 1b and 1c). In addition, we found that induction of Cdx1 and Cdx4 during a similar time-window to that reported here promotes hematopoietic development (Lengerke et al., 2008). We explored the effects of Cdx induction on other mesodermal derivatives, such as muscle but could find no consistent alteration (Supplemental Figure 3). However, possible shifts in formation of posterior rather than anterior muscle would not be detectable by our method of whole EB assessment by PCR. Together, these findings argue that the inhibitory effect on the cardiac lineage is specific and not the result of general toxic effects of ectopic Cdx expression.
The in vitro development of ESC is highly dynamic and gene expression as well as the functional capacity of the cells depends on their developmental stage. The early cardiac-suppressing effect of Cdx genes was not due to impaired developmental dynamics, as our time-course of marker gene expression and assessment of beating areas showed that suppression could be detected at all analyzed time-points. In contrast, activation of Cdx genes at later developmental time-points (e.g. after gastrulation and the formation of Nkx2.5 expressing cardiac progenitor cells) cannot reverse the developmental program and moreover does not show any inhibitory effect on cardiac cell development (Ehrman and Yutzey, 2001).
A number of studies have shown that the Cdx genes display both gene-specific as well as redundant activities. Knock-out experiments in the mouse have demonstrated that individual Cdx genes have slightly different impacts on vertebral patterning. Cdx1-null mice mainly show abnormalities in the cervical and thoracic region, partially following the rules defined for anterior transformations (van den Akker et al., 2002, and references therein), while Cdx2 and Cdx4 give rise to transformations of posterior cervical vertebrae and elements of the thoracic region, with much higher severity and penetrance for Cdx2 compared to Cdx4 (van Nes et al., 2006). Defects in sternal ribs suggest additional disruption of contributions to the paraxial mesoderm in Cdx mutants (van den Akker et al., 2002; Kato and Aoyama, 1998; Pinot, 1969). In zebrafish, cdx4-deficient fish display diminished blood development, and further suppression of cdx1a results in complete ablation of posterior embryonic blood, consistent with redundancy (Davidson et al., 2003; Davidson and Zon, 2006). In the mouse, the most severe Cdx loss-of-function allelic combination possible, the Cdx1/2/4 null embryo, has not been examined as Cdx2 is necessary for early placenta development (Strumpf et al., 2005). More recently, conditional Cdx2 knockout mice have been generated to study functions in specific lineages such as the enterocytes and the intestinal epithelium (Gao and Kaestner, 2010; Grainger et al. 2010). Redundant roles of Cdx1 and Cdx4 are found in hematopoietic development of murine ESC (Lengerke et al., 2008; McKinney-Freeman et al., 2008). Our data is consistent with Cdx1 and Cdx4 having similar, and redundant, suppressive effects on cardiac development.
Despite cdx-deficiency having a widespread effect on mesoderm patterning in zebrafish embryos, including a posterior expansion in tbx5a+ anterior lateral mesoderm, there was no significant change in the number of nkx2.5 and respectively cmlc2 expressing cardiac cells. During gastrulation, Keegan and colleagues have demonstrated that blocking RA signaling leads to an excess of cardiac precursors being specified, most likely at the expense of other tissues such as the pectoral fin mesenchyme (Keegan et al., 2005). Since RA was shown to restrict cardiac development and interactions between cdx and RA have been previously reported in the development of other organs, we tested whether the development of cardiac cells in cdx-deficient embryos was inhibited by RA signaling. A significant increase in nkx2.5 and respectively cmlc2 expressing cells was observed in cdx-deficient fish concomitantly treated with RA inhibitors, in comparison to untreated fish or treated wildtype controls. This dual requirement for cdx-deficiency and RA inhibition in order to reveal a cardiac phenotype is most likely due to the raldh2 expression domain being posteriorly expanded in cdx-deficient embryos (de Jong et al., 2010; Shimizu et al., 2006; Wingert et al., 2007). Thus, the increase in tbx5a+ ‘pre-cardiac’ anterior lateral plate mesoderm tissue in cdx-deficient embryos is offset by a concomitant increase in the cardiac-suppressive effects of RA signaling.
The work presented here suggests that the Cdx genes act to localize specific tissues within the developing mesoderm (e.g. posterior derivatives such as the blood and kidney at the expense of cardiac cells in the anterior lateral plate). This hypothesis is also supported by studies of other germ layers. In the zebrafish ectoderm, cdx4 is required to establish the boundary between the hindbrain and spinal cord territories. Loss of cdx4 results in posterior expansion of the segmented hindbrain at the expense of the spinal cord and, conversely, overexpression of cdx4 has a posteriorizing effect (Shimizu et al., 2006; Skromne et al., 2007). As seen in our studies, cdx1a/4-deficient embryos show enhanced phenotypes consistent with functional redundancy between the genes (Shimizu et al., 2006; Skromne et al., 2007). Within the endoderm, cdx4 confers posterior identity and regulates the localization of the developing pancreas (Kinkel et al., 2008).
Interestingly, important interactions between the cdx genes and retinoic acid (RA) were also found. While several reports demonstrated that RA can specify the pancreas in vertebrates (Chen et al., 2004; Martín et al., 2005; Molotkov et al., 2005; Stafford et al., 2004; Stafford and Prince, 2002), treatment of zebrafish embryos with RA only induces insulin-producing cells in the anterior endoderm, whereas simultaneous inhibition of Cdx function is required to induce this effect posteriorly. These data in the endoderm support our results in the developing mesoderm, and suggest that the normal (posterior) expression of Cdx genes may in general regulate cell fate by modulating inhibitory RA signals activity.
Taken together, our study has identified a strong Cdx-mediated suppression of cardiac development, reinforcing a general regulatory role of Cdx genes within the developing mesoderm. Moreover, our data reveal novel interactions between Cdx genes and the RA pathway, and suggest that Cdx genes regulate activity of inhibitory RA signals, a function which may be conserved across germ layers.
Conclusions
This study performed in zebrafish embryo and mouse embryonic stem cells suggests that Cdx genes modulate early cardiogenesis presumably by acting on the level of mesoderm and regulating together with RA the development of cardiac precursors. In zebrafish embryos, loss of cdx4 and/or cdx1a induced a profound, dose-dependent expansion of the anterior lateral plate mesoderm. Interestingly, further development of nkx2.5+ cardiac progenitors and cmlc2+ cells in cdx deficient fish requires suppression of the RA pathway, indicating close interactions between cdx genes and the retinoic acid pathway during early cardiogenesis.
Supplementary Material
Doxycycline exposure induces ectopic Cdx1 expression in iCdx1 cells in a tightly regulated manner (a) and does not affect the induction of beating EB (b) or the expression of lineage markers (c) in day 6 EB derived from Ainv15 parental ESC.
(a) ESC derived cells were cultured for 7.5 hours in medium containing doxycycline and then analysed for Cdx1 expression. Afterwards, doxycycline was removed for 16 hours from the medium and Cdx1 expression was compared to control non-exposed cells at the same time-point.
(b) Ainv15 ES cells were differentiated in EB with or without exposure to doxycycline between day 2.25 and day 6 of EB development. EB were analysed at day 7 and 9 for beating areas and (c) at day 6 by qPCR for expression of lineage specific markers. Shown are summarized data from three biological replicates.
Measurement of areas of cells expressing nkx2.5 (a) and cmlc2 (b) for wildtype, cdx4−/− and cdx1a/4 deficient zebrafish exposed to DMSO, DEAB or DEAB+RA (as presented in Figure 6 and 7) reveal significant cdx dose dependent enhancement of cardiogenesis in cdx4−/− and respectively cdx1a/4 deficient fish. The numbers of fish presenting the phenotype out of total numbers of fish scored for each condition are listed (c,d) as presented in Figures 6 and 7. Nikon Elements Basic Research software with a Nikon DS-Fi1 color camera system were used for the measurements. Measurements and statistical analysis (a,b) were done on three representative animals for each condition.
Expression of skeletal muscle markers in differentiating iCdx1 EB cultured with or without doxycycline. Induction of Cdx1 during day 2.25 to 6 of differentiation shows no significant alteration of skeletal muscle development by this method. Shown are fold relative gene expression levels in day 6 and day 9 EB after doxycycline induction in comparison to non-induced controls analyzed at the same time-point.
Research highlights.
Cdx1/4 suppresses early cardiac development from murine embryonic stem cells
In zebrafish embryos, loss of cdx1a/4 expands the tbx5 expressing anterior lateral plate mesoderm but does not enhance nkx2.5 and cmlc2 expression
Concomitant suppression of retinoic acid and cdx enhances cardiogenesis in zebrafish embryos
Acknowledgments
This study was supported by the Max-Eder-Program of the Deutsche Krebshilfe and grants from the Deutsche Forschungsgemeinschaft SFB773 and the fortüne Program of the University of Tuebingen for C.L.; A.J.D is supported by the NIH/NIDDK (DK077186); G.Q.D. is supported by grants from the United States National Institutes of Health, the NIH Director’s Pioneer Award of the NIH Roadmap for Medical Research, Clinical Scientist Awards in Translational Research from the Burroughs Wellcome Fund and the Leukemia and Lymphoma Society, and the Howard Hughes Medical Institute.
Footnotes
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Claudia Lengerke, Email: claudia.lengerke@med.uni-tuebingen.de.
Rebecca Wingert, Email: rwingert@nd.edu.
Michael Beeretz, Email: beeretz@gmx.de.
Matthias Grauer, Email: Mat.Grauer@gmx.de.
Martina Konantz, Email: martina.konantz@tuebingen.mpg.de.
George Q. Daley, Email: George.Daley@childrens.harvard.edu.
Alan J. Davidson, Email: a.davidson@auckland.ac.nz.
References
- Allan D, Houle M, Bouchard N, Meyer BI, Gruss P, Lohnes D. RARgamma and Cdx1 interactions in vertebral patterning. Dev Biol. 2001;240:46–60. doi: 10.1006/dbio.2001.0455. [DOI] [PubMed] [Google Scholar]
- Beck F, Erler T, Russell A, James R. Expression of Cdx-2 in the mouse embryo and placenta: possible role in patterning of the extra-embryonic membranes. Dev Dyn. 1995;204:219–27. doi: 10.1002/aja.1002040302. [DOI] [PubMed] [Google Scholar]
- Beland M, Pilon N, Houle M, Oh K, Sylvestre JR, Prinos P, Lohnes D. Cdx1 autoregulation is governed by a novel Cdx1-LEF1 transcription complex. Mol Cell Biol. 2004;24:5028–38. doi: 10.1128/MCB.24.11.5028-5038.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondue A, Lapouge G, Paulissen C, Semeraro C, Iacovino M, Kyba M, Blanpain C. Mesp1 acts as a master regulator of multipotent cardiovascular progenitor specification. Cell Stem Cell. 2008;3:69–84. doi: 10.1016/j.stem.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Chawengsaksophak K, de Graaff W, Rossant J, Deschamps J, Beck F. Cdx2 is essential for axial elongation in mouse development. Proc Natl Acad Sci USA. 2004;101:7641–5. doi: 10.1073/pnas.0401654101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Pan FC, Brandes N, Afelik S, Sölter M, Pieler T. Retinoic acid signaling is essential for pancreas development and promotes endocrine at the expense of exocrine cell differentiation in Xenopus. Dev Biol. 2004;271:144–60. doi: 10.1016/j.ydbio.2004.03.030. [DOI] [PubMed] [Google Scholar]
- David R, Brenner C, Stieber J, Schwarz F, Brunner S, Vollmer M, Mentele E, Müller-Höcker J, Kitajima S, Lickert H, Rupp R, Franz WM. MesP1 drives vertebrate cardiovascular differentiation through Dkk-1-mediated blockade of Wnt-signalling. Nat Cell Biol. 2008;10:338–45. doi: 10.1038/ncb1696. [DOI] [PubMed] [Google Scholar]
- Davidson AJ, Ernst P, Wang Y, Dekens MPS, Kingsley PD, Palis J, Korsmeyer SJ, Daley GQ, Zon LI. cdx4 mutants fail to specify blood progenitors and can be rescued by multiple hox genes. Nature. 2003;425:300–6. doi: 10.1038/nature01973. [DOI] [PubMed] [Google Scholar]
- Davidson AJ, Zon LI. The caudal-related homeobox genes cdx1a and cdx4 act redundantly to regulate hox gene expression and the formation of putative hematopoietic stem cells during zebrafish embryogenesis. Dev Biol. 2006;292:506–18. doi: 10.1016/j.ydbio.2006.01.003. [DOI] [PubMed] [Google Scholar]
- de Jong JLO, Davidson AJ, Wang Y, Palis J, Opara P, Pugach E, Daley GQ, Zon LI. Interaction of retinoic acid and scl controls primitive blood development. Blood. 2010;116:201–9. doi: 10.1182/blood-2009-10-249557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschamps J, van den Akker E, Forlani S, De Graaff W, Oosterveen T, Roelen B, Roelfsema J. Initiation, establishment and maintenance of Hox gene expression patterns in the mouse. Int J Dev Biol. 1999;43:635–50. [PubMed] [Google Scholar]
- Deschamps J, van Nes J. Developmental regulation of the Hox genes during axial morphogenesis in the mouse. Development. 2005;132:2931–42. doi: 10.1242/dev.01897. [DOI] [PubMed] [Google Scholar]
- Ehrman LA, Yutzey KE. Anterior expression of the caudal homologue cCdx-B activates a posterior genetic program in avian embryos. Dev Dyn. 2001;221:412–21. doi: 10.1002/dvdy.1151. [DOI] [PubMed] [Google Scholar]
- Gamer LW, Wright CV. Murine Cdx-4 bears striking similarities to the Drosophila caudal gene in its homeodomain sequence and early expression pattern. Mech Dev. 1993;43:71–81. doi: 10.1016/0925-4773(93)90024-r. [DOI] [PubMed] [Google Scholar]
- Gao N, Kaestner KH. Cdx2 regulates endo-lysosomales function and epithelial cell polarity. Genes & Dev. 2010;24:1295–305. doi: 10.1101/gad.1921510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaunt SJ. Evolutionary shifts of vertebrate structures and Hox expression up and down the axial series of segments: a consideration of possible mechanisms. Int J Dev Biol. 2000;44:109–17. [PubMed] [Google Scholar]
- Gaunt SJ, Drage D, Cockley A. Vertebrate caudal gene expression gradients investigated by use of chick cdx-A/lacZ and mouse cdx-1/lacZ reporters in transgenic mouse embryos: evidence for an intron enhancer. Mech Dev. 2003;120:573–86. doi: 10.1016/s0925-4773(03)00023-6. [DOI] [PubMed] [Google Scholar]
- Gaunt SJ, Drage D, Trubshaw RC. cdx4/lacZ and cdx2/lacZ protein gradients formed by decay during gastrulation in the mouse. Int J Dev Biol. 2005;49:901–8. doi: 10.1387/ijdb.052021sg. [DOI] [PubMed] [Google Scholar]
- Grainger S, Savory JGA, Lohnes D. Cdx2 regulates patterning of the intestinal epithelium. Dev Biol. 2010;339:155–165. doi: 10.1016/j.ydbio.2009.12.025. [DOI] [PubMed] [Google Scholar]
- Kato N, Aoyama H. Dermomyotomal origin of the ribs as revealed by extirpation and transplantation experiments in chick and quail embryos. Development. 1998;125:3437–43. doi: 10.1242/dev.125.17.3437. [DOI] [PubMed] [Google Scholar]
- Kattman SJ, Huber TL, Keller GM. Multipotent flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev Cell. 2006;11:723–32. doi: 10.1016/j.devcel.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Keegan BR, Feldman JL, Begemann G, Ingham PW, Yelon D. Retinoic acid signaling restricts the cardiac progenitor pool. Science. 2005;307:247–9. doi: 10.1126/science.1101573. [DOI] [PubMed] [Google Scholar]
- Keller G. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev. 2005;19:1129–55. doi: 10.1101/gad.1303605. [DOI] [PubMed] [Google Scholar]
- Kinkel MD, Eames SC, Alonzo MR, Prince VE. Cdx4 is required in the endoderm to localize the pancreas and limit beta-cell number. Development. 2008;135:919–29. doi: 10.1242/dev.010660. [DOI] [PubMed] [Google Scholar]
- Kmita M, Duboule D. Organizing axes in time and space; 25 years of colinear tinkering. Science. 2003;301:331–3. doi: 10.1126/science.1085753. [DOI] [PubMed] [Google Scholar]
- Kyba M, Perlingeiro RC, Daley GQ. HoxB4 confers definitive lymphoid-myeloid engraftment potential on embryonic stem cell and yolk sac hematopoietic progenitors. Cell. 2002;109:29–37. doi: 10.1016/s0092-8674(02)00680-3. [DOI] [PubMed] [Google Scholar]
- Lengerke C, Grauer M, Niebuhr NI, Riedt T, Kanz L, Park IH, Daley GQ. Hematopoietic development from human induced pluripotent stem cells. Ann N Y Acad Sci. 2009;1176:219–27. doi: 10.1111/j.1749-6632.2009.04606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengerke C, McKinney-Freeman S, Naveiras O, Yates F, Wang Y, Bansal D, Daley GQ. The cdx-hox pathway in hematopoietic stem cell formation from embryonic stem cells. Ann N Y Acad Sci. 2007;1106:197–208. doi: 10.1196/annals.1392.006. [DOI] [PubMed] [Google Scholar]
- Lengerke C, Schmitt S, Bowman TV, Jang IH, Maouche-Chretien L, McKinney-Freeman S, Davidson AJ, Hammerschmidt M, Rentzsch F, Green JB, Zon LI, Daley GQ. BMP and Wnt specify hematopoietic fate by activation of the Cdx-Hox pathway. Cell Stem Cell. 2008;2:72–82. doi: 10.1016/j.stem.2007.10.022. [DOI] [PubMed] [Google Scholar]
- Lohnes D. The Cdx1 homeodomain protein: an integrator of posterior signaling in the mouse. Bioessays. 2003;25:971–80. doi: 10.1002/bies.10340. [DOI] [PubMed] [Google Scholar]
- Martín M, Gallego-Llamas J, Ribes V, Kedinger M, Niederreither K, Chambon P, Dollé P, Gradwohl G. Dorsal pancreas agenesis in retinoic acid-deficient Raldh2 mutant mice. Dev Biol. 2005;284:399–411. doi: 10.1016/j.ydbio.2005.05.035. [DOI] [PubMed] [Google Scholar]
- McKinney-Freeman SL, Lengerke C, Jang IH, Schmitt S, Wang Y, Philitas M, Shea J, Daley GQ. Modulation of murine embryonic stem cell-derived CD41+c-kit+ hematopoietic progenitors by ectopic expression of Cdx genes. Blood. 2008;111:4944–53. doi: 10.1182/blood-2007-11-124644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer BI, Gruss P. Mouse Cdx-1 expression during gastrulation. Development. 1993;117:191–203. doi: 10.1242/dev.117.1.191. [DOI] [PubMed] [Google Scholar]
- Molotkov A, Molotkova N, Duester G. Retinoic acid generated by Raldh2 in mesoderm is required for mouse dorsal endodermal pancreas development. Dev Dyn. 2005;232:950–7. doi: 10.1002/dvdy.20256. [DOI] [PubMed] [Google Scholar]
- Nakajima Y, Sakabe M, Matsui H, Sakata H, Yanagawa N, Yamagishi T. Heart development before beating. Anat Sci Int. 2009;84:67–76. doi: 10.1007/s12565-009-0025-2. [DOI] [PubMed] [Google Scholar]
- Pilon N, Oh K, Sylvestre JR, Bouchard N, Savory J, Lohnes D. Cdx4 is a direct target of the canonical Wnt pathway. Dev Biol. 2006;289:55–63. doi: 10.1016/j.ydbio.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Pinot M. Experimental study of the morphogenesis of the thoracic cage of the chick embryo: mechanisms and origins of the material. J Embryol Exp Morphol. 1969;21:146–94. [PubMed] [Google Scholar]
- Savory JG, Mansfield M, St Louis C, Lohnes D. Cdx2 is a Cdx4 target gene. Mech Dev. 2011;128:41–8. doi: 10.1016/j.mod.2010.09.004. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Bae YK, Hibi M. Cdx-Hox code controls competence for responding to Fgfs and retinoic acid in zebrafish neural tissue. Development. 2006;133:4709–19. doi: 10.1242/dev.02660. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Bae YK, Muraoka O, Hibi M. Interaction of Wnt and caudal-related genes in zebrafish posterior body formation. Dev Biol. 2005;279:125–41. doi: 10.1016/j.ydbio.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Skromne I, Thorsen D, Hale M, Prince VE, Ho RK. Repression of the hindbrain developmental program by Cdx factors is required for the specification of the vertebrate spinal cord. Development. 2007;134:2147–58. doi: 10.1242/dev.002980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafford D, Hornbruch A, Mueller PR, Prince VE. A conserved role for retinoid signaling in vertebrate pancreas development. Dev Genes Evol. 2004;214:432–41. doi: 10.1007/s00427-004-0420-6. [DOI] [PubMed] [Google Scholar]
- Stafford D, Prince VE. Retinoic acid signaling is required for a critical early step in zebrafish pancreatic development. Curr Biol. 2002;12:1215–20. doi: 10.1016/s0960-9822(02)00929-6. [DOI] [PubMed] [Google Scholar]
- Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, Rossant J. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development. 2005;132:2093–102. doi: 10.1242/dev.01801. [DOI] [PubMed] [Google Scholar]
- van den Akker E, Forlani S, Chawengsaksophak K, de Graaff W, Beck F, Meyer BI, Deschamps J. Cdx1 and Cdx2 have overlapping functions in anteroposterior patterning and posterior axis elongation. Development. 2002;129:2181–93. doi: 10.1242/dev.129.9.2181. [DOI] [PubMed] [Google Scholar]
- van Nes J, de Graaff W, Lebrin F, Gerhard M, Beck F, Deschamps J. The Cdx4 mutation affects axial development and reveals an essential role of Cdx genes in the ontogenesis of the placental labyrinth in mice. Development. 2006;133:419–28. doi: 10.1242/dev.02216. [DOI] [PubMed] [Google Scholar]
- Wang Y, Yabuuchi A, McKinney-Freeman S, Ducharme DMK, Ray MK, Chawengsaksophak K, Archer TK, Daley GQ. Cdx gene deficiency compromises embryonic hematopoiesis in the mouse. Proc Natl Acad Sci USA. 2008;105:7756–61. doi: 10.1073/pnas.0708951105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingert RA, Selleck R, Yu J, Song HD, Chen Z, Song A, Zhou Y, Thisse B, Thisse C, McMahon AP, Davidson AJ. The cdx genes and retinoic acid control the positioning and segmentation of the zebrafish pronephros. PLoS Genet. 2007;3:1922–38. doi: 10.1371/journal.pgen.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SM, Fujiwara Y, Cibulsky SM, Clapham DE, Lien CL, Schultheiss TM, Orkin SH. Developmental origin of a bipotential myocardial and smooth muscle cell precursor in the mammalian heart. Cell. 2006;127:1137–50. doi: 10.1016/j.cell.2006.10.028. [DOI] [PubMed] [Google Scholar]
- Young T, Deschamps J. Hox, Cdx, and anteroposterior patterning in the mouse embryo. Curr Top Dev Biol. 2009;88:235–55. doi: 10.1016/S0070-2153(09)88008-3. [DOI] [PubMed] [Google Scholar]
- Young T, Rowland JE, van de Ven C, Bialecka M, Novoa A, Carapuco M, van Nes J, de Graaff W, Duluc I, Freund JN, Beck F, Mallo M, Deschamps J. Cdx and Hox genes differentially regulate posterior axial growth in mammalian embryos. Dev Cell. 2009a;17:516–26. doi: 10.1016/j.devcel.2009.08.010. [DOI] [PubMed] [Google Scholar]
- Young T, Rowland JE, van de Ven C, Bialecka M, Novoa A, Carapuco M, van Nes J, de Graaff W, Duluc I, Freund JN, Beck F, Mallo M, Deschamps J. Cdx and Hox genes differentially regulate posterior axial growth in mammalian embryos. Dev Cell. 2009b;17:516–26. doi: 10.1016/j.devcel.2009.08.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Doxycycline exposure induces ectopic Cdx1 expression in iCdx1 cells in a tightly regulated manner (a) and does not affect the induction of beating EB (b) or the expression of lineage markers (c) in day 6 EB derived from Ainv15 parental ESC.
(a) ESC derived cells were cultured for 7.5 hours in medium containing doxycycline and then analysed for Cdx1 expression. Afterwards, doxycycline was removed for 16 hours from the medium and Cdx1 expression was compared to control non-exposed cells at the same time-point.
(b) Ainv15 ES cells were differentiated in EB with or without exposure to doxycycline between day 2.25 and day 6 of EB development. EB were analysed at day 7 and 9 for beating areas and (c) at day 6 by qPCR for expression of lineage specific markers. Shown are summarized data from three biological replicates.
Measurement of areas of cells expressing nkx2.5 (a) and cmlc2 (b) for wildtype, cdx4−/− and cdx1a/4 deficient zebrafish exposed to DMSO, DEAB or DEAB+RA (as presented in Figure 6 and 7) reveal significant cdx dose dependent enhancement of cardiogenesis in cdx4−/− and respectively cdx1a/4 deficient fish. The numbers of fish presenting the phenotype out of total numbers of fish scored for each condition are listed (c,d) as presented in Figures 6 and 7. Nikon Elements Basic Research software with a Nikon DS-Fi1 color camera system were used for the measurements. Measurements and statistical analysis (a,b) were done on three representative animals for each condition.
Expression of skeletal muscle markers in differentiating iCdx1 EB cultured with or without doxycycline. Induction of Cdx1 during day 2.25 to 6 of differentiation shows no significant alteration of skeletal muscle development by this method. Shown are fold relative gene expression levels in day 6 and day 9 EB after doxycycline induction in comparison to non-induced controls analyzed at the same time-point.


