Abstract
Three MADS-box genes isolated from Monterey pine (Pinus radiata), PrMADS1, PrMADS2, and PrMADS3, are orthologs to members of the AGL2 and AGL6 gene subfamilies in Arabidopsis. These genes were expressed during early stages of pine shoot development in differentiating seed- and pollen-cone buds. Their transcripts were found within a group of cells that formed ovuliferous scale and microsporophyll primordia. Expression of PrMADS3 was also detected in a group of cells giving rise to needle primordia within differentiated vegetative buds, and in needle primordia.
Formation of flowers involves at least three initial steps: transition to flowering, initiation of individual flowers, and floral patterning (Ma, 1994). The first step is the switch from the vegetative phase, during which shoots and leaves are produced, to the reproductive stage, during which flowers are initiated. Environmental stimuli such as day length (photoperiod), temperature, light quality, and availability of water and nutrients influence the transition to the reproductive phase.
Upon initiation of the reproductive phase, the development of floral meristems is promoted by flower meristem-identity genes LEAFY (LFY) (Weigel et al., 1992), APETALA1 (AP1) (Mandel et al., 1992), CAULIFLOWER (CAL) (Kempin et al., 1995), and AGAMOUS (AG) (Yanofsky et al., 1990). In Arabidopsis, shoot meristem-identity genes such as TERMINAL FLOWER 1 (TFL1) repress the activity of genes such as LFY, AP1, and AG, which have roles in specifying floral meristem identity (Shannon and Meeks-Wagner, 1991; Bradley et al., 1997; Mizukami and Ma, 1997).
Subsequent flower organ initiation is dependent on a set of homeotic, or organ-identity, genes, which fall into three classes: A, B, and C. Most of these genes belong to the MADS-domain protein gene family. The MADS-box genes encode a family of highly conserved transcription factors that play important roles in signal transduction and developmental control in plants, animals, and fungi. It has been shown that MADS-box genes exist in gymnosperms and angiosperms, as well as in ferns, which are the common ancestors of contemporary seed plants (Münster et al., 1997). Thus, these gene families were already established before conifer and angiosperm lineages separated more than 300 million years ago.
As in angiosperms, the “flowering” of Monterey pine (Pinus radiata) starts with the transformation of an indeterminate axillary apex into a determinate reproductive apex, which forms the strobili (cones). A new LSTB is formed during springtime at the tip of the rapidly elongating shoot. The axillary apices that emerge on the sides of the apical meristem can later differentiate as the following: (a) vegetative DSBs, which become anatomically differentiated, resulting in the initiation of three to five needle primordia; (b) reproductive PCBs, in which procambium tissue gradually develops into the microsporophylls. The appearance of microsporagial initials in the peripheral zone of the axis was designated stage 1 (mid-March). The size of the apex diminishes and finally disappears with the development of each sporophyll. At stage 2, microsporophyll initiation is complete and developed pollen mother cells are visible; or (c) SCBs, which become anatomically differentiated with the initiation of bracts formed directly from conspicuous cell pockets in the peripheral zone of the SCB apex (stage 1, late March–early April). Ovuliferous scale primordia are then initiated from hypodermal cells on the adaxial base of bracts (stage 2). A fused bract-ovuliferous scale complex is formed, and the scale becomes displaced from the cone-bud axis (stage 3). Thus, LSTBs can be classified as vegetative, male, or female (Harrison and Slee, 1992).
Although MADS-box genes have been extensively investigated in different angiosperms, few homologous genes have been isolated and characterized from gymnosperms. MADS-box genes have been previously reported in the gymnosperms spruce (Tandre et al., 1995) and red pine (Liu and Podila, 1997), and in their evolutionary ancestor, ferns (Münster et al., 1997). However, a detailed analysis of the possible role of MADS-box genes in the development of the shoot apical meristem and differentiated cone buds has not been reported. Here we describe isolation and characterization of three MADS-box genes from Monterey pine.
MATERIALS AND METHODS
Monterey Pine (Pinus radiata) Samples
Female, male, and vegetative LSTBs were collected from an adult tree (about 20 years of age and 30 m in height) in Victoria, Australia, from early March through June. Immature SCBs, PCBs, and DSBs were collected, placed on ice, dissected, and frozen in liquid N2 or fixed for the in situ-hybridization experiments. Elongated needles were also collected during this period of time, and tissues were dissected and frozen in liquid N2. Roots and stems were collected from seedlings that were grown for 3 to 4 weeks in growth cabinets.
Isolation of RNA
The various tissues were each homogenized in buffer (5.5 m guanidinium thiocyanate, 25 mm sodium citrate, 0.5% sarcosyl, 0.2 m β-mercaptoethanol, and 4% [w/v] PVP [40,000 Mr]). The DNA released was sheared by passing the homogenate through a 16-gauge needle, and the debris were pelleted. The RNA was pelleted through Cs-trifluoro-acetate (Pharmacia) in a swinging bucket rotor in a bench-top ultracentrifuge (55,000 rpm for 3 h; model TLX, Beckman). The RNA pellets were resuspended in 10 mm Tris-HCI (pH 8.0) and 1 mm EDTA, extracted with chloroform:butanol (4:1, v/v), and precipitated with potassium acetate and ethanol. Isolation of mRNA was performed using the Poly ATtract kit (Promega). For northern-hybridization experiments, 30 μg of total RNA was run on a 1% formaldehyde/agarose gel, blotted onto nylon membranes, and hybridized to the 32P-labeled 3′ end of the cDNA clones.
cDNA Library Construction and Screening
A cDNA library was constructed using a HybriZAP Two-Hybrid cDNA cloning kit (Stratagene), from mRNA extracted from a mixture of immature (stages 1 and 2) female and male cones. The cDNA library was screened under low-stringency conditions with a mixture of MADS-box domain sequences amplified from Arabidopsis thaliana (AGL2) and tomato (tm1, tm4, tm5, and tm8) genomic DNAs. Isolated cDNA clones were sequenced using the Dye Terminator Cycle DNA Sequencing kit (Perkin-Elmer), and products were separated on an automated sequenator (model 377, Applied Biosystems). Sequence data were analyzed using the GCG sequence analysis program (Genetics Computer Group, Madison, WI).
In Situ Hybridization
For in situ-hybridization experiments, the 3′ ends of all cDNA clones were introduced into the pSPT18 vector (Boehringer Mannheim). Both the antisense and sense single-stranded DIG-labeled RNA probes used in this study were derived from these constructs using the DIG RNA-labeling kit (Boehringer Mannheim). For PrMADS1 and PrMADS2, the plasmids were linearized in the polylinker by digestion with SmaI, and used as a template for synthesizing RNA using T7 polymerase. As a control, a single-stranded DIG-labeled sense probe was synthesized using SP6 polymerase, and the same plasmid was linearized in the polylinker with SalI. For PrMADS3, the antisense probe was synthesized by SP6 from the template linearized with the NdeI restriction enzyme (in the coding region). As a control, the sense probe was synthesized using T7 polymerase from the template linearized with the NotI restriction enzyme (in the coding region). Plant material was fixed, embedded in Paraplast, and prepared for in situ hybridization, according to the method of Jack et al. (1992). Immunological detection of the hybridized probe was carried out using the DIG detection kit (Boehringer Mannheim).
Construction of the Phylogenetic Tree
Alignment of conceptual amino acid sequences was made using the GCG program PILEUP. Phylogenetic trees were constructed as described (Münster et al., 1997), employing the neighbor-joining algorithm, as provided by the program NEIGHBOR of the GCG phylogeny inference package (PHYLIP).
RESULTS
Isolation of the PrMADS1, PrMADS2, and PrMADS3 Genes
Phylogenetic analyses of AGL2-like and AGL6-like proteins showed that the three pine genes PrMADS1, PrMADS2, and PrMADS3 belong to the AGL2 and AGL6 gene subfamilies (Fig. 1) (Ma et al., 1991; Münster et al., 1997). The PrMADS1 protein belongs to the AGL2 subfamily, and is most similar to the AGL2 and AGL4 (Flanagan and Ma, 1994) and AGL3 (Huang et al., 1995) proteins from Arabidopsis. PrMADS2 and PrMADS3, together with MADS-box proteins from other conifers such as DAL1 from spruce (Tandre et al., 1995) and PMADS1 from red pine (Liu and Podila, 1997), belong to the AGL6 subfamily (Ma et al., 1991).
Figure 1.
Phylogenetic tree showing the relationship among AGL2-like and AGL6-like proteins. Pine MADS-domain proteins are indicated by black boxes. MADS-domain proteins from other gymnosperms are indicated by shaded boxes.
Organ- and Tissue-Specific Expression of PrMADS1, PrMADS2, and PrMADS3 Genes in Early Cone-Bud Development
The expression patterns of the genes were first analyzed by RNA gel-blot analysis. Cone sizes selected for this experiment approximately represent three stages of SCB and two stages of PCB development. In situ hybridization was used to determine precise localization of the transcripts in developing SCBs, PCBs, and DSBs. Some of the most informative examples of expression of these genes are shown in Figures 3 and 4.
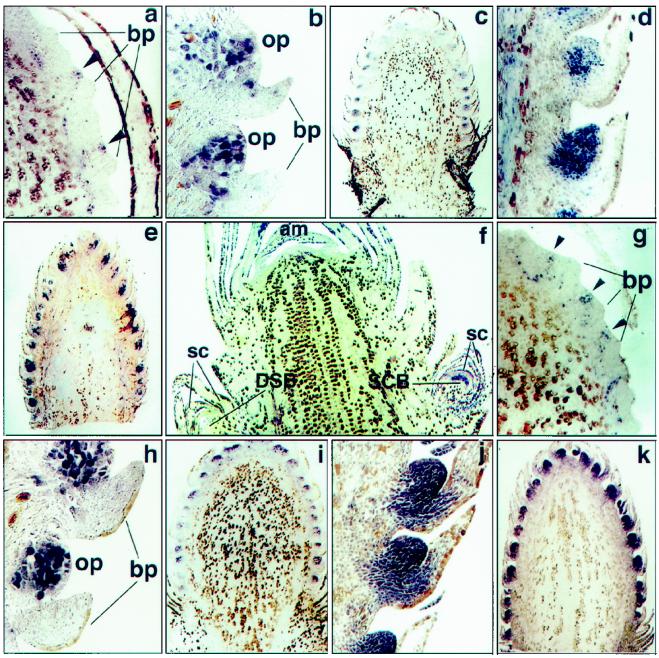
Figure 3.
In situ localization of PrMADS1 and PrMADS2 transcripts in longitudinal sections of the developing SCBs, PCBs, and DSBs. Accumulation of PrMADS1 transcripts in: a, differentiating SCB, in the group of cells initiating ovuliferous scale primordia (arrowheads) (magnification ×50); b and c, SCB at stage 2 (magnification ×100 and ×11.25, respectively); and d and e, SCB at stage 3 (magnification ×45 and ×8.3, respectively). Accumulation of PrMADS2 transcripts in: f, SCB differentiating in developing female LSTB (magnification ×21.6); g, SCB in the group of cells initiating ovuliferous scale primordia (arrowheads) (magnification ×50); h and i, SCB at stage 2 (magnification ×100 and ×11.25 respectively); and j and k, SCB at stage 3 (magnification ×45 and ×8.3, respectively). am, Apical meristem; bp, bract primordia; op, ovuliferous scale primordia; and sc, sterile cataphylls.
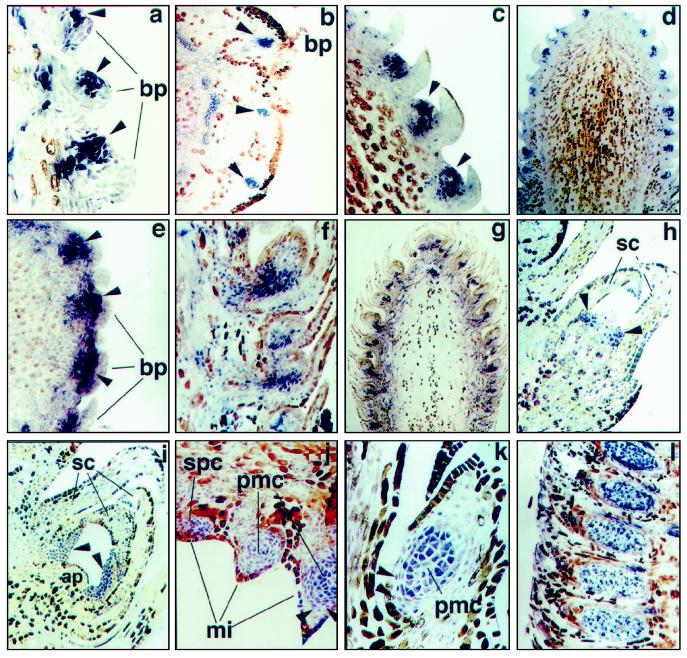
Figure 4.
In situ localization of PrMADS3 transcripts in developing SCBs, PCBs, and DSBs. a, Differentiating SCB. Expression in the group of cells initiating ovuliferous scale primordia is shown by arrowheads (magnification ×42). b, Cross-section of differentiating SCB. Expression in the group of cells initiating ovuliferous scale primordia is shown by arrowheads (magnification ×45). c and d, SCB at stage 2. Ovuliferous scale primordia shown by arrowheads (magnification ×100 and ×11.25, respectively). e, Cross-section of SCB at stage 3. Ovuliferous scale primordia shown by arrowheads (magnification ×55). f and g, SCB at stage 3 (magnification ×45 and ×8.3, respectively). h, DSB with initiating needle primordia (arrowheads) (magnification ×90). i, DSB with developing needle primordia (arrowheads) (magnification ×45). j and k, Differentiated PCB with initiating microsporophylls (stage 1). Two layers of tapetal cells surrounding pollen mother cells are shown by arrowheads (magnification ×45 and ×60, respectively). l, PCB after completion of microsporophyll initiation (stage 2) (magnification ×23.5). All sections except b and e, which are cross-sections, are longitudinal. ap, Apical meristem; bp, bract primordia; mi, microsporophylls; pmc, pollen mother cells; sc, sterile cataphylls; and spc, sporogenous cells.
PrMADS1
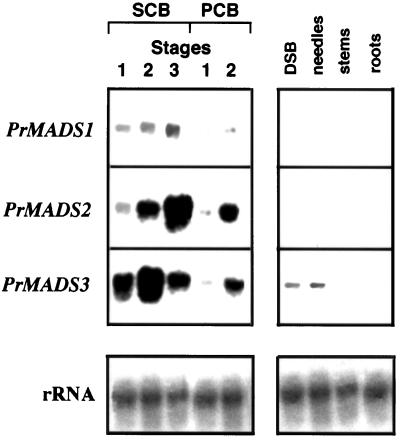
The steady-state levels of PrMADS1 increased markedly during female and male cone development (Fig. 2). Thus, the highest level of accumulation of PrMADS1 transcripts was found at stage 3, and the lowest at stage 1. In differentiated PCBs, expression of PrMADS1 increased at the stage of completion of microsporophyll initiation (stage 2). PrMADS1 transcripts were not detected in vegetative organs, including DSBs, needles, stems, and roots.
Figure 2.
Northern-hybridization expression patterns of the PrMADS1, PrMADS2, and PrMADS3 genes in RNA samples isolated from different-sized immature SCBs, PCBs, and DSBs, needles, stems, and roots.
In situ hybridization showing localization of PrMADS1 transcripts in differentiating SCBs and PCBs is shown in Figure 3. Within SCBs, PrMADS1 was expressed in a group of cells within the ovuliferous scale primordia emergent at the adaxial side of the bract primordia (Fig. 3a). The hybridization signals extended approximately five to six cells deep into the inflorescence and often followed the traces of undifferentiated vasculature, but never reached into maturing vascular cells. The number of cells expressing the PrMADS1 gene increased during cell proliferation within the developing ovuliferous scale primordia (stage 2) (Fig. 3b). Development of ovuliferous scale primordia usually began at the basal part of differentiating SCBs and proceeded acropetally. At this stage, PrMADS1 expression was consistent with ovuliferous scale primordia development, and stronger expression signals were detected in more-developed ovuliferous scale primordia (Fig. 3c). At stage 3, when the fused bract-ovuliferous scale complex formed and become displaced from the cone-bud axis, PrMADS1 was strongly expressed in a large number of cells within the developing ovuliferous scale primordia (Fig. 3d). Expression signals at this stage were equally strong in fused bract-ovuliferous scale complexes located at the base and at the top part of the SCB axis (Fig. 3e).
PrMADS2
Northern-hybridization analyses revealed that PrMADS2 was more strongly expressed in immature SCBs and in PCBs than PrMADS1 (Fig. 2). Stronger levels of expression were detected in more developed cone buds. No expression was found in DSBs, needles, stems, or roots.
In situ hybridization showed that PrMADS2 transcripts were first detected in emergent axillary apices within young female LSTBs. These apices could potentially differentiate as SCBs (Fig. 3f). At this stage, PrMADS2 transcripts accumulated strongly and almost uniformly. No signal was detected at the apical meristem or in the axillary bud, which could differentiate as DSBs (Fig. 3f). Within differentiated SCBs, PrMADS2 transcripts were detected after the bract primordia initiation (stage 1), but before ovuliferous scale primordia had emerged (Fig. 3g). Expression was detected within numerous, conspicuous cell pockets that were regularly spaced peripherally at the adaxial side of the bract primordia (arrowheads). Continued periclinal division of the cells in these basal pockets led to the formation of ovuliferous scale primordia. At stage 2, PrMADS2 expression was strong and almost uniform within ovuliferous scale primordia cells (Fig. 3, h and i). Almost no expression signal was detected at this stage in bract primordia. PrMADS2 transcripts also persisted throughout stage 3, showing strong and uniform accumulation in ovule primordia located at the top and bottom parts of the SCB axis (Fig. 3, j and k).
PrMADS3
Northern hybridization of PrMADS3 showed the opposite expression pattern to that of PrMADS1 and PrMADS2. This gene was strongly expressed at early stages of SCB development (stages 1 and 2), but expression decreased during further SCB development.
Whereas PrMADS3 showed a different pattern of expression in immature SCBs, its expression pattern in differentiating PCBs was similar to those of PrMADS1 and PrMADS2. The expression signal was strongest at the completion of microsporophyll initiation (stage 2). In contrast to PrMADS1 and PrMADS2, PrMADS3 was expressed in vegetative tissues. A low level of expression was detected in vegetative buds and fully elongated needles.
In situ hybridization analysis showing the expression of PrMADS3 at different stages of development of ovuliferous scale primordia within the same differentiated SCB is shown in Figure 4a. PrMADS3 transcript was first detected in SCBs at the stage when the bract primordia were well defined, but before ovuliferous scale primordia emerged. The signal was much stronger in more developed ovuliferous scale primordia located at the basal part of the SCB axis (Fig. 4a). Cross-sections of a SCB showed regularly spaced cell groups within the ovuliferous scale primordia expressing PrMADS3 (Fig. 4b). In immature pine cone buds, strong, nonspecific binding of sense (control) and antisense probes to phloem and vascular cells was sometimes observed, suggesting that the positive expression signals seen in these cells may have been artificial. At stage 2, expression signals were concentrated mainly in the ovuliferous scale primordia, with very low expression found in bract primordia (Fig. 4, c–e). At later stages (stage 3) of SCB differentiation, the number of cells expressing PrMADS3 did not correlate with the number of cells proliferating within the developing ovule primordia. Thus, the RNA signal at this stage was no longer uniform within ovule primordia, and transcripts were detected mostly in the pedicels at the base of the ovule primordia (Fig. 4, f and g).
Unlike PrMADS1 and PrMADS2, PrMADS3 transcripts were detected in differentiated DSBs. Low levels of PrMADS3 transcripts were found in groups of cells within the peripheral zone of DSBs (Fig. 4h). Continued periclinal division of these cells gave rise to needle primordia. Uniform accumulation of PrMADS3 transcripts in two of the three to five needle primordia surrounding the apex is shown in Figure 4i.
Within differentiated PCBs, PrMADS1, PrMADS2, and PrMADS3 showed identical patterns of expression at different stages of development. Expression of PrMADS2 is shown in Figure 4, j to l. At stage 1, low levels of microsporophyll PrMADS2 transcript were concentrated within initiating microsporophylls, in a group of cells initiating sporogenous cells (Fig. 4j). In more developed microsporophylls, expression signals were found in pollen mother cells and in two layers of tapetal cells surrounding the microsporangium (Fig. 4, j and k). Much stronger levels of expression were detected at stage 2 in developed pollen mother cells, when microsporophyll initiation was complete (Fig. 4l).
DISCUSSION
PrMADS1, PrMADS2, and PrMADS3 as Part of the Regulatory Hierarchy Controlling Early Stages of Flowering in Monterey Pine
Three MADS-box genes were isolated and characterized from Monterey pine. Analysis of the PrMADS2 expression pattern suggests that PrMADS2 may be involved in several aspects of flower development in Monterey pine. PrMADS2 was detected for the first time, to our knowledge, within LSTBs in emergent axillary buds, which had begun to separate from the inflorescence meristem, but before the emergence of organ primordia. Expression of PrMADS2 was specific to SCBs and PCBs, since no signal was detected in initiating vegetative cones or in inflorescence meristem. The fact that PrMADS2 expression began after the SCB and PCB meristems emerged suggests that PrMADS2 is unlikely to be required for initiation of these meristems. However, it may be involved in some aspects of development after initiation, such as growth or maintenance of the floral meristem, or preparation for the establishment of organ identity. This suggests a possible role of PrMADS2 as a gymnosperm homolog of the members of the family of intermediate genes (such as AGL2/4, AGL6, and AGL9). These genes possibly act as mediators between the floral meristem and floral organ-identity genes, being expressed later than the onset of the flower meristem-identity genes, but before the flower organ-identity genes. This places the PrMADS2 gene upstream of PrMADS1 and PrMADS3 in the regulatory hierarchy of cone-bud development.
SCBs become anatomically differentiated with the initiation of bract primordia (stage 1). Ovuliferous scale primordia then arise from the periclinally dividing group of hypodermal cells located on the side of the bract primordia (stage 2). Expression of all three pine MADS-box genes in this group of cells at this stage suggests their possible role in ovule primordia initiation. In emergent ovuliferous scale primordia, PrMADS3 expresses abundantly and shows a much higher level of expression than PrMADS1 and PrMADS2. However, later in SCB development, when bract and ovuliferous scale primordia have developed and become displaced from the cone-bud axis, expression of PrMADS3 begins to wane and is restricted primarily to the basal part of the ovule primordia. Expression of PrMADS1 and PrMADS2 at this stage remains elevated and almost uniform within developing ovule primordia. This suggests that the PrMADS1 and PrMADS2 genes may also play a role in establishing and elaborating ovule morphology, whereas the role of PrMADS3 is probably restricted to initiation and very early stages of development of the ovule primordia.
In differentiated PCBs, all three genes showed similar expression in a group of cells initiating sporogenous cells within initiating microsporophylls (stage 1). Later, when microsporophyll initiation was complete, PrMADS1, PrMADS2, and PrMADS3 transcripts were detected in developed pollen mother cells (stage 2). This expression pattern suggests the involvement of all three genes in the initiation and early stages of male organ development.
Expression of the PrMADS3 gene in emergent needle primordia in differentiated vegetative buds and in fully elongated needles suggests a more universal function of PrMADS3, which also plays a role at the vegetative stage of radiata pine development.
Correlation between Structure and Function of PrMADS1, PrMADS2, and PrMADS3, and Their Angiosperm Homologs
It has been shown that the subfamily structure of the MADS-box gene family largely reflects the phylogenetic relationship between MADS-box gene family members (Theissen et al., 1996).
Phylogenetic tree analyses showed that PrMADS1 belongs to the subfamily of AGL2-like proteins and showed highest homology to a AGL2 protein of unknown function. Expression of the AGL2 gene is not restricted to certain whorls, and it is abundantly and ubiquitiously expressed during early and intermediate stages of flower development (stages 2–8). Expression of AGL2 was also found in developing ovules and, after fertilization, in developing embryos (Flanagan and Ma, 1994; Savidge et al., 1995). The snapdragon gene DEFH49 is expressed mainly in carpels, but also in petals and stamens (Davies and Schwarz-Sommer, 1994). The OM1 transcript of the orchid Aranda debora was found only in mature flowers (Lu et al., 1993). Six different AGL2-like genes reported from maize (Cacharrón et al., 1995) have diverse expression patterns, ranging from early onset of expression in developing spikelet primordia (ZMM6) to late expression restricted to the developing gynoecial ridge and silk of the female inflorescence (ZMM7). The expression of ZMM8 is restricted to stamens and gynoecium, as well as palea and lemma. A similar expression pattern was found for the OSMAD1 gene (Chung et al., 1994). The AGL3 gene, however, expresses in leaves, stems, and flowers (Huang et al., 1995).
The PrMADS2 and PrMADS3 proteins belong to the subfamily of AGL6 proteins comprising AGL6, AGL13, ZAG3, ZAG5, and DAL1. The expression pattern of PrMADS2, however, at early stages of SCB meristem initiation was similar to that of AGL2 expressed at stage 2, after the floral meristem has emerged, but before any floral organ primordia emerge. Within the AGL6-like subfamily, PrMADS2 showed very strong homology and a similar expression pattern to the PMADS1 gene from red pine (Liu and Podila, 1997), suggesting that these two genes are orthologs. Within angiosperms, the PrMADS2 gene is most similar to the AGL13 gene from Arabidopsis, which is expressed in the ovule and in the style (Rounsley et al., 1995).
Unlike the PrMADS1 and PrMADS2 genes, PrMADS3 transcripts were detected in the group of cells initiating needle primordia in differentiated DSBs and in fully elongated needles. This pattern of expression is similar to most of the members of the AGL6 subfamily. The AGL6 gene is expressed in inflorescence stems, all four flower whorls, and ovules (M. Yanofsky, personal communication). ZAG3 expresses in carpels and in sterile organs of male and female inflorescences (Mena et al., 1995). ZAG5 seems to be an ortholog of AGL6, and expresses in stems, flowers, and ovules (Mena et al., 1995). The fact that PrMADS3 has strong homology to the DAL1 gene (identity between proteins is 97%) and has a similar pattern of expression suggests that these two genes are orthologs. Both genes are expressed in SCBs and PCBs, as well as in developing vegetative shoots, although the PrMADS3 gene has a much lower level of expression in developing vegetative shoots than DAL1, which has an equal level of expression in vegetative and reproductive shoots (Tandre et al., 1995).
Our results demonstrate that molecular mechanisms of flowering, and the gene families involved at different stages of flower initiation, are not novel to the angiosperm lineage, but are a conserved ancestral property of all seed plants. Analysis of the PrMADS1, PrMADS2, and PrMADS3 target genes, as well as other meristem and organ-identity gene families from various gymnosperms, may be valuable in developing a more complete description of the molecular events controlling flowering in conifers, and in further understanding how these genes have been recruited during the evolutionary development of flowering in general.
ACKNOWLEDGMENTS
We thank Dr. Detlef Weigel (The Salk Institute, San Diego, CA), Dr. Marty Yanofsky (University of California, San Diego), and Dr. Elena Alvarez-Buylla (University of Mexico, Mexico City) for very helpful discussions and critical reading of the manuscript; Dr. George Coupland (John Innes Centre, Norwich, UK) for helpful discussions; Dr. Hong Ma (Cold Spring Harbor, New York); Dr. Marty Yanofsky and Dr. Detlef Weigel for communicating unpublished results; Dr. Günter Theissen and Dr. Jan Kim (Max Plank Institut für Züchtungsforshung, Germany) for construction of the phylogenetic tree; Dr. Derek Harrison (University of Victoria, Canada) for many helpful comments; and Ms. Corinna Lange for assistance in preparation of the manuscript.
Abbreviations:
- DIG
digoxigenin
- DSB
dwarf shoot bud
- LSTB
long-shoot terminal bud
- PCB
pollen-cone bud
- SCB
seedcone bud
LITERATURE CITED
- Bradley D, Ratcliffe O, Vincent C, Carpenter R, Coen E. Inflorescence commitment and architecture in Arabidopsis. Science. 1997;275:80–83. doi: 10.1126/science.275.5296.80. [DOI] [PubMed] [Google Scholar]
- Cacharrón J, Fischer A, Saedler H, Theissen G. Expression patterns of MADS-box genes as studied by in situhybridization. Maize Genetics Cooperation Newsletter. 1995;69:37–38. [Google Scholar]
- Chung Y-Y, Kim S-R, Finkel D, Yanofsky MF, An G. Early flowering and reduced apical dominance result from ectopic expression of a rice MADS-box gene. Plant Mol Biol. 1994;26:657–665. doi: 10.1007/BF00013751. [DOI] [PubMed] [Google Scholar]
- Davies B, Schwarz-Sommer Z. Control of floral organ identity by homeotic MADS-box transcription factors. In: Novel L, editor. Results and Problems in Cell Differentiation. Plant Promoters and Transcription Factors. Berlin: Springer-Verlag; 1994. pp. 235–258. [DOI] [PubMed] [Google Scholar]
- Flanagan CA, Ma H. Spatially and temporally regulated expression of the MADS-box gene AGL2 in wild-type and mutant Arabidopsisflowers. Plant Mol Biol. 1994;26:581–595. doi: 10.1007/BF00013745. [DOI] [PubMed] [Google Scholar]
- Harrison D, Slee M. Long shoot terminal bud development and the differentiation of pollen- and seed-cone buds in Pinus caribaeva var. hondurensis. Can J For Res. 1992;22:1656–1668. [Google Scholar]
- Huang H, Tudor M, Weiss C, Hu Y, Ma H. The Arabidopsis MADS-box gene AGL3is widely expressed and encodes a sequence-specific DNA-binding protein. Plant Mol Biol. 1995;28:549–567. doi: 10.1007/BF00020401. [DOI] [PubMed] [Google Scholar]
- Jack T, Brockmann LL, Meyerowitz EM. The homeotic gene APETALA3 of Arabidopsis thalianaencodes a MADS-box and is expressed in petals and stamens. Cell. 1992;89:683–697. doi: 10.1016/0092-8674(92)90144-2. [DOI] [PubMed] [Google Scholar]
- Kempin SA, Savidge B, Yanofsky MF. Molecular basis of the cauliflower phenotype in Arabidopsis. Science. 1995;267:522–525. doi: 10.1126/science.7824951. [DOI] [PubMed] [Google Scholar]
- Liu J-J, Podila GK. Characterization of a MADS-box gene (accession no. Y09611) from immature female cones of red pine (PGR 97-032) Plant Physiol. 1997;113:665. [Google Scholar]
- Lu Z-X, Wu M, Loh C-S, Yeong C-Y, Goh C-J. Nucleotide sequence of a flower-specific MADS-box cDNA clone from orchid. Plant Mol Biol. 1993;23:901–904. doi: 10.1007/BF00021545. [DOI] [PubMed] [Google Scholar]
- Ma H. The unfolding drama of flower development: recent results from genetic and molecular analyses. Genes Dev. 1994;8:745–756. doi: 10.1101/gad.8.7.745. [DOI] [PubMed] [Google Scholar]
- Ma H, Yanofsky MF, Meyerowitz EM. AGL1-AGL6, an Arabidopsisgene family with similarity to floral homeotic and transcription factor genes. Genes Dev. 1991;5:484–495. doi: 10.1101/gad.5.3.484. [DOI] [PubMed] [Google Scholar]
- Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky ME. Molecular characterization of the Arabidopsis floral homeotic gene APETALA. Nature. 1992;360:273–277. doi: 10.1038/360273a0. [DOI] [PubMed] [Google Scholar]
- Mena M, Mandel MA, Lerner DR, Yanofsky MF, Schmidt RJ. A characterization of the MADS-box gene family in maize. Plant J. 1995;8:845–854. doi: 10.1046/j.1365-313x.1995.8060845.x. [DOI] [PubMed] [Google Scholar]
- Mizukami Y, Ma H. Plant Cell. 1997;9:393–408. doi: 10.1105/tpc.9.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münster T, Pahnke J, Di Rosa A, Kim JT, Matin W, Saedler H, Theissen G. Floral homeotic genes were recruited from homologous MADS-box genes preexisting in the common ancestor of ferns and seed plants. Proc Natl Acad Sci USA. 1997;94:2415–2420. doi: 10.1073/pnas.94.6.2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rounsley S, Ditta G, Yanofsky MF. Diverse roles of MADS-box genes in Arabidopsisdevelopment. Plant Cell. 1995;7:1259–1269. doi: 10.1105/tpc.7.8.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savidge B, Rounsley SD, Yanofsky MF. Temporal relationship between the transcription of two ArabidopsisMADS-box genes and the floral organ-identity genes. Plant Cell. 1995;7:721–733. doi: 10.1105/tpc.7.6.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon S, Meeks-Wagner DR. A mutation in the Arabidopsis TFL1gene affects inflorescence meristem development. Plant Cell. 1991;3:877–892. doi: 10.1105/tpc.3.9.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tandre K, Albert VA, Sundas A, Engström P. Conifer homologues to genes that control floral development in angiosperms. Plant Mol Biol. 1995;27:69–87. doi: 10.1007/BF00019179. [DOI] [PubMed] [Google Scholar]
- Theissen G, Kim JT, Saedler H. Classification and phylogeny of the MADS-box multigene family suggest defined roles of MADS-box gene families in the morphological evolution of eukaryotes. J Mol Evol. 1996;43:484–516. doi: 10.1007/BF02337521. [DOI] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM. LEAFY controls floral meristem-identity in Arabidopsis. Cell. 1992;69:843–859. doi: 10.1016/0092-8674(92)90295-n. [DOI] [PubMed] [Google Scholar]
- Yanofsky MF, Ma H, Bowman JL, Drews GN, Feldmann KA, Meyerowitz EM. The protein encoded by the Arabidopsis homeotic gene agamousresembles transcription factors. Nature. 1990;346:35–39. doi: 10.1038/346035a0. [DOI] [PubMed] [Google Scholar]