Abstract
Gas exchangers fundamentally form by branching morphogenesis (BM), a mechanistically profoundly complex process which derives from coherent expression and regulation of multiple genes that direct cell-to-cell interactions, differentiation, and movements by signaling of various molecular morphogenetic cues at specific times and particular places in the developing organ. Coordinated expression of growth-instructing factors determines sizes and sites where bifurcation occurs, by how much a part elongates before it divides, and the angle at which branching occurs. BM is essentially induced by dualities of factors where through feedback- or feed forward loops agonists/antagonists are activated or repressed. The intricate transactions between the development orchestrating molecular factors determine the ultimate phenotype. From the primeval time when the transformation of unicellular organisms to multicellular ones occurred by systematic accretion of cells, BM has been perpetually conserved. Canonical signalling, transcriptional pathways, and other instructive molecular factors are commonly employed within and across species, tissues, and stages of development. While much still remain to be elucidated and some of what has been reported corroborated and reconciled with rest of existing data, notable progress has in recent times been made in understanding the mechanism of BM. By identifying and characterizing the morphogenetic drivers, and markers and their regulatory dynamics, the elemental underpinnings of BM have been more precisely explained. Broadening these insights will allow more effective diagnostic and therapeutic interventions of developmental abnormalities and pathologies in pre- and postnatal lungs. Conservation of the molecular factors which are involved in the development of the lung (and other branched organs) is a classic example of nature’s astuteness in economically utilizing finite resources. Once purposefully formed, well-tested and tried ways and means are adopted, preserved, and widely used to engineer the most optimal phenotypes. The material and time costs of developing utterly new instruments and routines with every drastic biological change (e.g. adaptation and speciation) are circumvented. This should assure the best possible structures and therefore functions, ensuring survival and evolutionary success.
Keywords: Lung, Development, Tracheal system, Branching morphogenesis, Growth factors
Introduction
"‘Specifically, convergence provides a way to tell features that have important functional significance from features that do not.’ Vogel[1]."
The development of the vertebrate lung was a decisive preparatory event for adaptation to air-breathing and successful transition from water to land, momentous occasions in the evolution and diversification of animal life (e.g. [2-4]). Cost-effective acquisition of molecular oxygen (O2) allowed accomplishment of more energetic lifestyles like flight (e.g. [5-7]) which lead to marked adaptive radiation and speciation (e.g. [8,9]). Motley of conserved genes and molecular factors orchestrated proper transformations from simple to complex body forms by programmed organization and arrangement of cells and tissue components (e.g. [10-18]). Physiological processes (e.g. metabolic-, respiratory-, and ionic exchange) occur across two-dimensional (2D) surfaces which are increased by the three-dimensional (3D) assemblage of organs, tissues, and cells. Branched structures are common in nature and biology in particular. In the later, they include the conductive nervous tissue (e.g. axons and their arborisation), vascular system, vertebrate lungs, secretory glands, tubules of the kidney, and the insectan tracheal system (e.g. [19-24]). Mainly formed to secrete and/or transport materials/fluids or conduct/transfer ions and effects like electric activity, e.g., actions potentials in nervous tissue, branched systems comprise of distinctively hierarchically arranged structural parts (segments = domains) which interconnect in a coherent manner. The overall morphology granted by the frequency and the geometry of bifurcation. In tubular branched systems, depending on the type of organ, endothelial- (in the vascular system) or epithelial cells (in all the other organs) line the internal space (cavity/lumen). The similarities amongst the various branched biological structures underscore existence of shared programmed regulatory mechanisms which involve signaling molecules, transcription factors, and other growth instructing molecular agents which instruct the growth and bifurcation patterns (e.g. [15,16,21,23]). Identification and characterization of the morphogenetic drivers which prompt budding and bifurcation from pre-existent domains is pivotal to determining the control and the regulation of the process of branching morphogenesis (BM). The controlling mechanism of BM involves dualities of morphogenetic factors: an agonist is regulated by an inhibitor and vice versa (e.g. [16,21,25-29]). There is mounting evidence that combinatorial action of various signaling molecules, transcription factor families, and other molecular factors is vital to cell specification, differentiation, and tissue development. For example, interactions between fibroblast growth factor (FGF) signaling and Wnt/β-catenin signaling in the lung mesenchyme positively reinforce each other (e.g. [17]), BMP-4 counteracts the effects of FGF-10 [30]; De Langhe et al. [31,32], mesenchymal FGF signaling is needed for the expression of Wnt-2a and mesenchymal Wnt/β-catenin signalling [33,34], and mesenchymal Wnt/β-catenin signaling is required to sustain expression of FGFR-1 and FGFR-2 and GATA-6 and Nkx2.1 operate in a synergistic way to instruct pulmonary epithelial differentiation and development [35]. Well-coordinated spatiotemporal expressions and repressions of the morphogenetic factors initiate, hone, and optimize the ultimate phenotypes (e.g. [36-38]). For the murine lung, comprehensive gene expression profiling using oligonucleotide-based microarrays showed that ∼ 11,000 genes are expressed throughout the developmental stages of the lung [36].
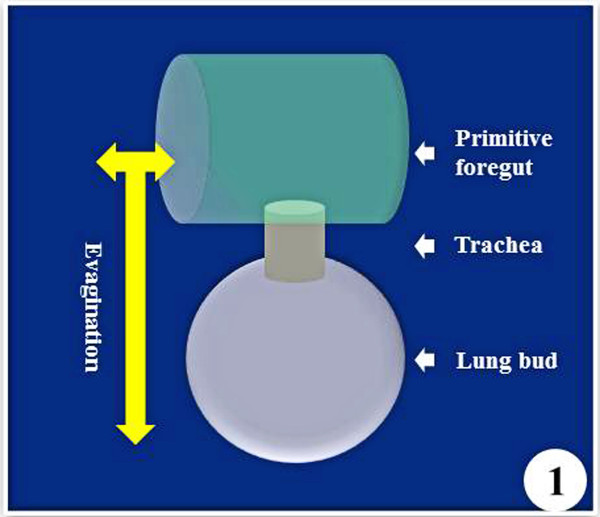
The embryonic development of the vertebrate lung inaugurates with a ventral out-pocketing (evagination) of dedicated (committed) cells from the formative primitive foregut endoderm into the splanchnic mesenchyme (Figure 1) (e.g. [39-42]) and interactions between cells originating from two germ layers - endoderm and mesoderm (e.g. [42-45]). The lung buds elongate and branch to form the trachea and the main bronchi followed by stereotypic branching and budding which produces the conducting airways, leading to the alveolar region of the peripheral lung. Among others, these changes prepare the lung for air-breathing in the postnatal life. The distinctive structural features of lungs are large surface area, intense vascularization (which generates large capillary blood volume), and thin blood-gas barrier (e.g. [46,47]). Large respiratory surface area emanates from intense internal subdivision of the lung, a phenotype fashioned by different morphogenetic cues that are expressed in form of growth factors (GFs) and cytokines, molecular instruments which act as paracrine signals that control cell division and differentiation (e.g. [21,39,48]).
Figure 1 .
The lung bud and subsequently the lung inaugurates by evagination (out-pocketing = diverticulation) of committed endodermal cells of the primitive foregut. Its growth and development is regulated by various molecular morphogenetic factors that are expressed at different times and places in the epithelial, mesenchymal, and mesothelial compartments.
In evolutionary terms, many developmental pathways are conserved in the animal kingdom (e.g. [13,49-55]): the common origin of these processes is strongly implied in the biological past. In the developing lung bud, epithelial cells of the airways and endothelial ones of the vasculature multiply rapidly while the formative structures undergo reiterative branching, producing a highly ordered arrangement (e.g. [45,56]). While a lot still remains to be resolved, the signals and the manner in which they are regulated during BM are becoming clearer (e.g. [16,21,23,40,57]). Detailed insights into the mechanisms of BM will not only increase our understanding of the development of the lung but will further advance techniques of lung engineering and regenerative medicine (e.g. [58,59]), design of artificial organs to replace failed/failing ones (e.g. [60]), and therapeutic interventions, especially during early stages of lung development (e.g. [61-65]). More importantly, insight into the process of BM of one kind of branched organ should permit meaningful understanding of others. For an area that is receiving intense scientific interest and scientific accounts (publications) are appearing prodigiously, occasional critical reviews of the subject matter are necessary to identify what is certain, what is speculative and therefore ambiguous, and where gaps in knowledge exist. Heuristic collation and reconciliation of available information should help highlight areas for further research, helping avert improvident and costly duplication of effort by investigators. Presently, much of the investigative activity of the genetic- and molecular aspects of the development of gas exchangers in particular and branched structures in general is on the mammalian lung (particularly on those of laboratory animals - mainly the mouse and rat) and the tracheal system of insects (especially of Drosophila). Relatively little information exists on the morphogenetic aspects of the development of the avian lung and hardly any is available on the amphibian- and reptilian ones. Holistic understanding of the mechanisms involved in the process of BM of the gas exchangers will only be accomplished once these gaps are closed. Moreover, although instructive in their own right, the shortcomings that are inherent in in vitro studies and those involving genetically manipulated (engineered) animals should be appreciated. A first in comparatively integrating the available data, this account succinctly outlines the process of BM and that of the development of the mammalian- (bronchioalveolar) and avian (parabronchial) lungs as well as that of the insectan tracheal system, the only taxa where meaningful data are presently available.
Branching Morphogenesis (BM)
Branched structures are ubiquitous in nature. They occur at every scale and form of development in both the plant- (e.g. [66,67]) and the animal kingdoms (e.g. [52,67]). The design of branched forms has constantly fascinated biologists, mathematicians, and physicists (e.g. [67-72]). A prototypical developmental process, BM is mechanistically fabricated by few simple iterative genetic subroutines through which complex well-ordered, functionally efficient architecture is engineered [73]. An assemblage described as ‘growth and branching of epithelial buds’ by Saxena and Sariola [74] and ‘creation of branched structures’ by Davies [67], in animal tissues and organs, BM occurs in the lung (e.g. [20,21,25,53,75-79]), glandular organs like the mammary gland, the salivary gland, and the pancreas (e.g. [28,80-83]), the kidney (e.g. [22,84-86]), the tooth [87], the tracheal system of insects (e.g. [23,88,89]), and the vasculature (e.g. [19]). In most cases, the functional units (e.g. secretory or gas exchange units) display distinctive 3D architecture (e.g. [30,79]). Organs that form by BM provide good models for studying and understanding application of the mode of development in animal patterning, cell differentiation, and organ and tissue organization (e.g. [15,45,90,91]). Branched structures form by coordinated spatiotemporal expression of specified morphogenetic cues [25,92].
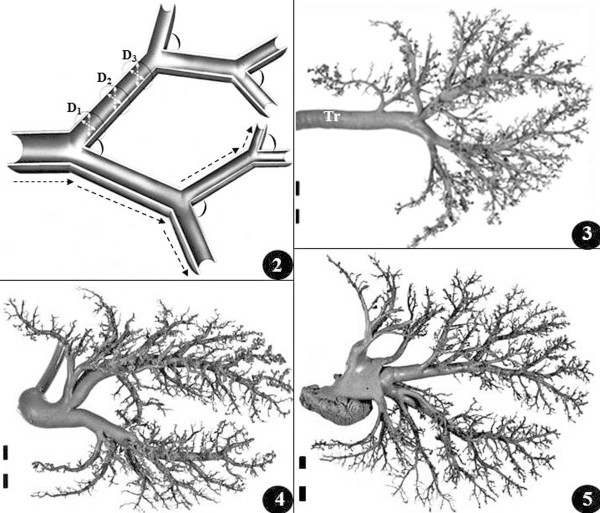
Normal lung development culminates in formation of airways and blood vessels which branch (Figure 2), pattern, and closely relate to each other (Figures 345): this increases the respiratory surface area and reduces the diffusion distance for molecular oxygen (O2) between inhaled air and capillary blood. Also, proper geometries and sizes of the airways and the blood vessels grant optimal (cost-effective) flows of the respiratory fluid media, saving on energy required to transport them through the conduits (e.g. [93]). While the iterating process involved in BM may appear deceptively simple to genetically program, the instructions and the molecular factors that drive it are profoundly intricate (e.g. [20,21,23,94,95]). BM is driven by an assortment of genes and intercellular signaling molecules that include transcriptional factors, soluble peptide growth factors, and insoluble extracellular matrix molecules that are expressed in the right quantities, time, place, and sequence. This determines the points where new branches form, the lengthening of the intervening duct/trunk/stalk before downstream branching occurs, and where groups of cells detach from the epithelium of the main duct to form side branches (secondary budding) (e.g. [16,18,21,25,39,40,79,96-98]). In computer lexicon, a highly specific genomic information flow engine initiates and regulates spatiotemporal expression and transcription of appropriate morphogenetic cues which produce a protocol-based pulmonary architecture by programming and reprogramming branching periodicity and bifurcation angles. Additionally, physicochemical and environmental cues and factors like intraluminal hydraulic pressure (e.g. [21,99,100]), relative hypoxia (e.g. [101]), and calcium concentration (e.g. [102,103]) play important roles in BM. ‘Cross-talk’, i.e., cell-to-cell signaling, especially between the mesenchymal- and the epithelial cells (e.g. [85,91]), is necessary to correctly educe cell-specific developmental pathways that lead to proper lung development and differentiation of various epithelial cell lineages (e.g. [104-106]). In the adult human lung, e.g., after 20–23 bifurcations, a highly ordered system of airways with ~25,000 bronchioles (e.g. [45,107]) gives rise to ~300 to ~600 million alveoli (e.g. [108-110]): a respiratory surface area of ∼ 140 m2 exists [111]. More than 40 different types of cells (pneumocytes) exist in the lung (e.g. [109,112-114]).
Figure 2 .
A generic diagram of branched tubular system. By specifying where signaling molecules, transcription factors, and other molecular factors are localized or expressed, the lengths, the diameters, and the sites where branches form and the angles of bifurcation are specified. Dashed arrows show the lengths of the constituent segments; D1-D3, axial diameters (small dashed arrows) at various points of the length of a part; the arcs show the angles of bifurcation. Figures 345: Latex cast preparations of the airway (Figure 3), the venous (Figure 4), and the arterial- (Figure 5) systems of a mature lung of the pig (Sus scrofa). Product of complex branching process, the systems match to optimize gas exchange between air and blood. Tr, trachea. Scale bars: 1 cm. From Maina and van Gils [115].
Figure 3 .
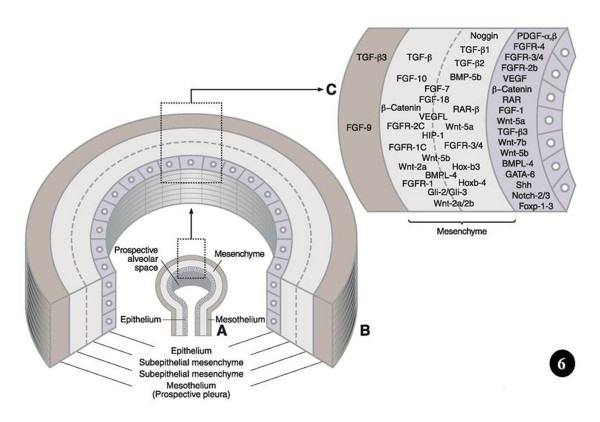
Figure6A-C. Sites where key signaling molecular factors, transcription factors, and other molecular agents are expressed and/or localized to regulate the development and growth of the lung. In the mesenchyme, the areas of expression are not exact.
Figure 4 .
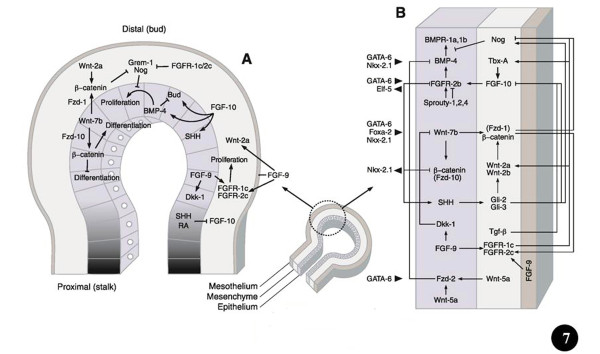
Ways and means by which prominent signaling molecules and transcription factors control lung development. The middle (small) diagram shows the different parts (germ layers) of a lung bud (proximal and distal). Figure 7A: Location of signalling molecules and the pathways by which they control growth, budding, and differentiation of the airway epithelium and surrounding mesenchyme. The shading shows the proximal-distal extent of the lung bud epithelium. Figure 7B: Signaling programs in the distal lung bud bring about mesenchymal- and epithelial cell proliferation as well as airway branching. Details to these processes can be obtained from the text and particularly from the following comprehensive reviews: Metzger and Krasnow [25], Perl and Whitsett [11], Roth-Kleiner and Post [116], Cardoso and Lü [40], Lu and Werb [53], De Langhe and Reynolds [117], Affolter et al. [77], Warburton et al. [21], [15,16,18,29,98]. Modified after Ornitz and Yin [18].
Figure 5 .
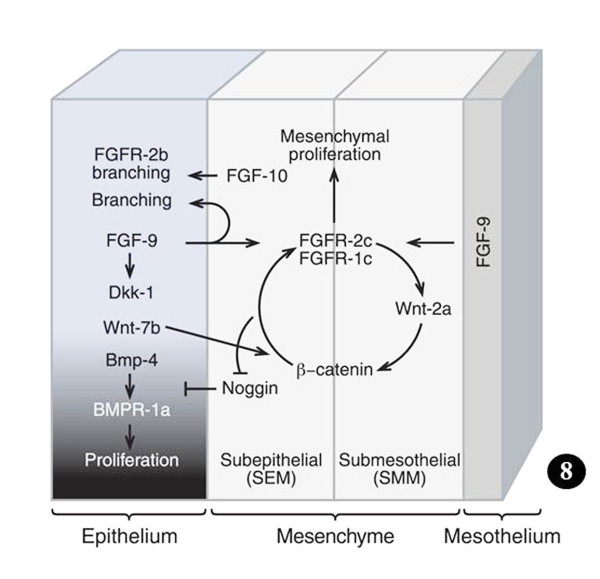
Figure 8: Feed-forward signaling in which FGF-9 controls Wnt-2a expression and mesenchymal Wnt/β-catenin signaling, and in which mesenchymal Wnt/β-catenin signaling is requisite for mesenchymal FGFR expression and mesenchymal responsiveness to FGF-9. In absence of either FGF-9 or canonical Wnt ligands, the mesenchymal FGF-9-Wnt/β-catenin feed-forward network breaks up, occasioning loss of mesenchymal FGFR expression and FGF responsiveness. Wnt/β-catenin signaling has to be preserved in order to induce or maintain FGFR expression, FGF-9 responsiveness, and continuance of FGF-Wnt/β-catenin feed-forward signaling. Loss of mesenchymal FGF-Wnt/β-catenin signaling increases Noggin expression in both the subepithelial and submesothelial mesenchyme. Lack of either pathway decreases mesenchymal proliferation. The change in the density of the shading of the epithelium shows the proximal-distal extent of the lung bud. Details can be acquired from Yin et al. [33], Rajagopal et al. [118], Yi et al. [119] De Langhe et al. [32], and Ornitz and Yin [18]. Modified after Ornitz and Yin [18].
Evolution causes selection pressure to conserve functionally important coding and regulatory pathways (e.g. [120]). The genes that are involved in BM are conserved across animal species (e.g. [23,25,52,67,79]). In computational jargon, these elements are ‘hard-wired’. The genetic instructions that lead to analogous designs have been continued over long evolutionary time scales. By among others Wagner [121,122], Wallace [123], and Mojica et al. [124], such constant and ubiquitous forms, structures, and systems have been termed ‘Bauplans’ (= ‘builder’s plans’ = ‘blue-prints’ = ‘frozen an excellent example of such conserved construct. The transformation of the unicellular organisms (protozoa) to multicellular organisms (Metazoa), which occurred in the Ediacaran era, ~500 to 700 million years ago (mya), is the original branching morphogenetic process (e.g. [125-128]). The genes involved in BM can be traced back to a common origin - a set dedicated to regulating pattern formation [129]. Much of the genomic reconfiguration occurred during the Cambrian Explosion, ~530 million years ago, when massive increase in animal body plans occurred and phenomenal speciation happened (e.g. [9,130]). The genes that encode for BM appear to have developed gradually: Pavlova et al. [75], e.g., showed them to correlate with the expression of different, but interrelating, genomic subgroups, signifying differences in morphogenetic mechanisms at the various stages in evolution of branching tubules. In signaling biology, which entails transduction and transcriptional controls, few canonical developmental programs are exploited more frequently across species, tissues, and stages of elaboration (e.g. [52]). Based on the notion that important regulatory pathways are commonly genetically conserved among species, comparative genomics approach has been used to identify well-conserved controlling factors (e.g. [120,131]). For the development of the lung, among others, the best known genes, molecular factors, and regulatory pathways are the Bone Morphogenetic Proteins (BMPs), the Fibroblast Growth Factors (FGFs), Sonic Hedgehog (Shh), Wnt genes/proteins (Wnts), Transformation Growth Factors (TGFs), Retinoic Acid (RA), Vascular Endothelial Growth Factor (VEGF), and Extracellular Matrix (ECM) component proteins. Most of these instruments have been shown to be involved in the formation of the insectan tracheal system. The morphogenetic factors are succinctly outlined below. The sites of localization and expression of signaling molecules, transcription factors, and other morphogenetic molecular cues, which regulate the development of the lung bud, are shown in Figure 6 and the mechanisms by which they achieve it are outlined in Figures 7 and 8. Table 1 summarizes the areas of expression of some of the lung’s signaling molecules, transcription factors, and other molecular factors and the phenotypes specified by genetic mutations, targeted inhibition, abrogation, blockage or under expression of certain genes and their products (proteins).
Figure 6 .
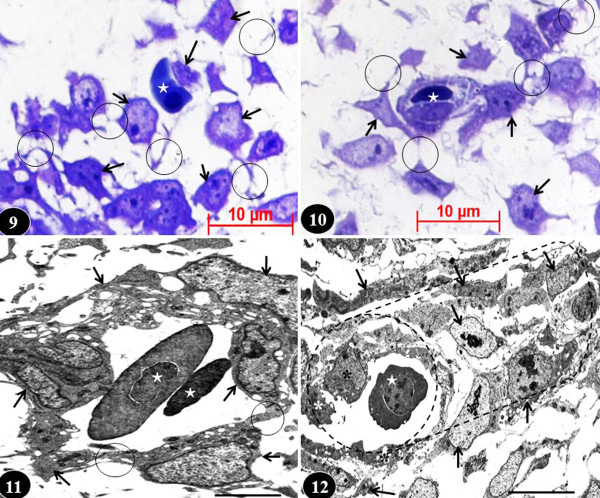
Developing chicken lung. Figure 9: Radical transformation of mesenchymal cells into characteristically stellate angioblasts (arrows) with conspicuous filopodia (circles) (which allow them to move) and into hemopoietic cells, i.e., red blood cells (star), at the fourth day of embryonic lung development. Toluidine stained section. Figure 10: Angioblasts (arrows) moving to surround a red blood cell (star) on the fifth day of embryonic lung’s development to form a blood vessel. Circles, filopodia. Toluidine stained section. Figure 11: Angioblasts (arrows) surrounding red blood cells (stars) in the developing lung on the sixth day of embryonic life forming a blood vessel. Circles, filopodia connecting angioblasts to form the vessel lumen. Figure 12: A blood vessel with a well-defined lumen in the lung of a seven day old embryo. Arrows, angioblasts forming the wall; asterisks, endothelial cells; star, red blood cell; dashed cylindrical shape, orientation of the blood vessel. These changes occur under the influence of VEGF. Figures 11 and 12 are transmission electron micrographs. Scale bars: Figure 11, 10 μm; Figure 12, 15 μm.
Figure 7 .
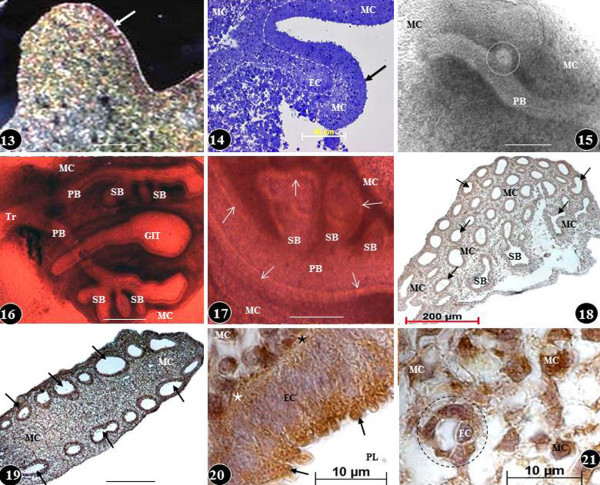
Early development of the chicken lung (Figures 13–17) and expression of basic FGF (bFGF-2) at different stages of development (Figures 18–21). Figures 13 and 14: Lung buds (arrows) on the third- (Figure 13) and fourth days (Figure 14) of embryonic life: initially, the bud comprises of a rather homogenous group of cells (Figure 13) while a little later the epithelial cells (EC) and the mesenchymal cells (MC) reorganize into separate areas (dashed lines). Toluidine blue stained preparations. Scale bars: Figure 13, 100 μm; Figure 14, 50 μm. Figures 15–17: Matrigel cultured lung buds. On the fourth day of embryonic life (Figure 15), a secondary bronchus (dashed circle) can be seen sprouting from a primary bronchus (PB) and at the fifth day of life (Figure 16), extrapulmonary primary bronchi (PB) can be seen continuing into the developing lung and giving rise to secondary bronchi (SB). Tr, trachea; GIT, developing part of the gastrointestinal system. MC, mesenchymal cells. Figure 17: Close-up of developing airways at the sixth day of embryonic life. The epithelium lining the primary bronchus (PB) and the secondary bronchi (SB) can be seen (arrows). Figures 16 and 17 are colour adapted images of black and white microscopic originals. Scale bars: Figure 15, 200 μm; Figure 16, 200 μm; Figure 17, 0.2 mm. Figures 18 and 19: Transverse- (Figure 18) and longitudinal (Figure 19) sections of developing chicken lungs at the sixth day of development showing expression of basic fibroblast growth factor-2 (bFGF-2) in the mesenchymal cells (MC) and in the epithelial cells lining the airways (arrows). SB, secondary bronchi. Scale bar: Figure 19, 100 μm. Figures 20 and 21: Expression of bFGF-2 in the parabronchial epithelial cells (EC) (Figure 20) and in the mesenchymal cells (MC) (Figure 21) of the lung of a seven day old embryo. bFGF is overexpressed in the apical parts of the epithelial cells (arrows) (Figure 20) and in the mesenchymal cells (Figures 20 and 21). PL, parabronchial lumen; stars, basement membrane of epithelial cells; circle, developing blood vessel; EC, endothelial cell. Figure 13 is from Maina [132]; Figure 13 is from Maina [133]; other figures-unpublished.
Figure 8 .
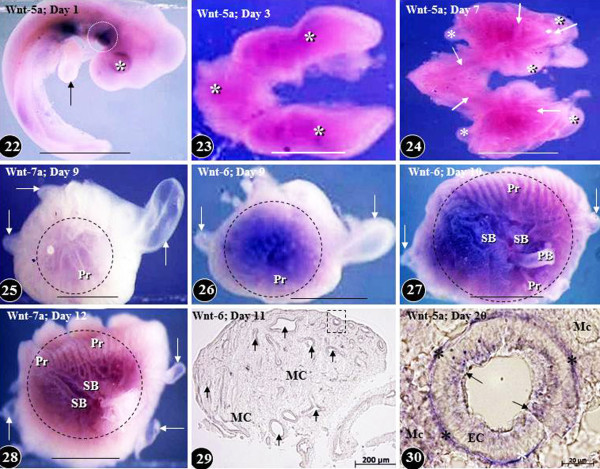
Expression of Wnt signaling molecules in developing chicken embryo (Figure 22) and in the lung at different times of embryonic life (Figures 23–30). Wnt-5a is expressed on the ventral aspect of the body trunk at day two of life (Figures 22). Arrow, developing fore limb bud; asterisk, optic placode; dashed circle, prospective site of the development of the lung and the heart. Scale bar, 1 mm. Figures 23: Diffuse expression of Wnt-5a (asterisks) in the developing lung on day three of life. Scale bar, 1 mm. Figure 24: Expression of Wnt-5a in developing lung on day five of life. Wnt-5a is predominantly localized in the developing airways (arrows) and relatively little of it in the mesenchyme and the air sacs (asterisks). Scale bar, 1 mm. Figures 25–28: Expression of Wnts in developing lungs at different embryonic days of life (areas marked by dashed circles). Figures 25, 26, and 28 show lateral aspects of the lung while Figure 27 is that of a medial one. The Wnts are poorly expressed in the developing air sacs (arrows). PB, primary bronchus; Pr, parabronchi; SB, secondary bronchi. Scale bars, 1 mm. Figure 29: Transverse section of a developing lung at the eleventh day of life showing Wnt-6 expression in the epithelium lining the airways (arrows) and the mesenchyme (MC). An airway like the one enclosed in the dashed square is shown enlarged in Figure 30. Figure 30: Cross-section of a parabronchus showing the expression of Wnt-5a at the 20th day of development. Wnt-5a is highly expressed in the epithelial cells (arrows), the mesenchymal cells (MC), and in the basement membrane of the epithelial cells (stars). Figures 22–28 are from Maina [132]; Figure 30 is from Maina [134]; Figure 29 is from unpublished work of RG Macharia and JN Maina.
Table 1.
Expression patterns of some lung signaling molecules, transcription factors, and other molecular factors and the phenotypes specified by their mutations, targeted inhibition, abrogation, blockage or underexpression
| Signaling molecule | ||||
|---|---|---|---|---|
| |
Gene name |
Expression site |
Phenotype |
References |
| FGF-18 |
Fibroblast growth factor-18 |
Mesenchyme |
Poor alveolization |
Usui et al. [135] |
| FGF-9 |
Fibroblast growth factor-9 |
Epithelium; mesothelium (future pleura) |
Little airway branching; mesenchymal hypoplasia |
Colvin et al. [136]; Yin et al. [17] |
| PDGF-a |
Platelet derived growth factor-a |
Epithelium |
Compromised myoblast and elastin formation; defective alveolization |
Bostrom et al. [137]; Buch et al. [138] |
| Notch-2/3 |
Notch gene homologue-2/3 |
Epithelium |
Defective alveolization |
Xu et al. [139] |
| FGFR-3/4 |
Fibroblast growth factor receptor-3/4 |
Epithelium; mesenchyme |
Deficient production of elastin; lacking alveolization |
Weinstein et al. [140] |
| BMP-4 |
Bone morphogenetic protein-4 |
Epithelium; Mesenchyme |
poor lung development with cystic terminal air spaces |
Bellusci et al. [141]; Weaver et al. [142]; Jang et al. [143] |
| FGFR-2b |
Fibroblast growth factor receptor-2b |
Epithelium |
Lung agenesis; flawed branching; poor epithelial cell differentiation |
De Moerlooze et al. [144]; Shu et al. [145] |
| FGF-10 |
Fibroblast growth factor-10 |
Mesenchyme |
Lung agenesis and different abnormalities |
Sekine et al. [146]; De Moerlooze et al. [144]; [147] |
| WNT-5a |
Wingless-related MMTV integration site-5A |
Mesenchyme; epithelium |
Excessive branching; tracheal deficiency |
Li et al. [148] |
| WNT-7b |
Wingless-related MMTV integration site-7B |
Epithelium |
Vascular scantiness; reduced mesenchymal proliferation |
Shu et al. [149]; Shi et al. [150]; Mucenski et al. [151] |
| TGF-3β |
Transforming growth factor-β3 |
Epithelium; mesothelium |
Anomalous airway branching; poor extent of alveolization |
Kaartinen et al. [152]; Shi et al. [153] |
| Shh |
Sonic hedgehog |
Epithelium |
Poor bronchial branching; hypoplastic lungs; tracheoesophangeal fistula |
Litingtung et al. [154]; Minoo et al. [155] |
| HIP-1 |
Hedgehog interacting protein-1 |
Mesenchyme |
Defective branching |
Chuang and McMahon [156]; Chuang et al. [26] |
| Catnnb-1 |
β-catenin |
Epithelium |
Impaired airway branching |
Mucenski et al.[151]) |
| VEGF |
Vascular endothelial growth factor |
Epithelium; mesenchyme |
VEGF knockout have lethal phenotype; anormal vasculature |
Zeng et al. [157]; Miquerol et al. [158]; Kasahara et al. [159] |
| Nog |
Noggin |
Mesenchyme |
Lobation deficiencies |
Weaver et al. [142] |
|
Transcription factors | ||||
| FOXA-1/2 |
Forkhead box-A1/A2 |
Epithelium |
Hypoplastic lungs; defective branching; poor smooth muscle formation |
Wan et al. [160] |
| FOX-j1 |
Forkhead box-J1 |
Epithelium |
Deficiency of ciliated cells; left-right asymmetry |
Brody et al. [161] |
| FOX-F1a |
Forkhead box-F1a |
Mesenchyme |
Defective branching and lobation; tracheoesophangeal fistula |
Lim et al. [162] |
| HOX-A5 |
Homeobox-A5 |
Mesenchyme |
Tracheal occlusion; poor airway branching |
Aubin et al. [163]; Golpon et al. [164] |
| GATA-6 |
GATA-binding protein-6 |
Epithelium |
Diminished airway branching; defective sacculation |
Yang et al. [165] |
| Gli-2/3 |
GLI-Kruppel family member GLI-2/3 |
Mesenchyme |
Lung agenesis |
Motoyama et al. [166]; van Tuyl and Post [167] |
| Wnt-2/2b |
Wingless-related MMTV integration site-2/2b |
Mesenchyme |
Lung agenesis |
Harris-Johnson et al. [168] |
| NKX-2.1 (TTF-1) |
Nkx homeodomain/thyroid-specific transcription factor |
Epithelium |
Lung agenesis |
Stahlman et al. [169]; Kimura et al. [170] |
| HOX-a5 |
Homeobox-A5 |
Mesenchyme |
Poor airway branching; thickening of the alveolar walls |
Aubin et al. [163] |
| ALK-3 |
Aurora-like kinase |
Epithelium |
Impairment of lung branching; decreased cell proliferation |
Sun et al. [171] |
|
Other molecular factors | ||||
| Tmem 16a |
Transmembrane protein-16a |
Epithelium |
Abnormal tracheal cartilage development |
Rock et al. [172] |
| RARs |
Retinoic receptors |
Epithelium; mesenchyme; mesothelium |
Lung agenesis and hypolasia; reduction in alveolar number; tracheoesophangeal fistula |
Mendelsohn et al. [173]; Dickman et al. [174]; McGowan et al. [175]; Cardoso and Lü [40] |
| Itga-3 |
Integrinα-3 |
Epithelium |
Defective branching |
Kreidberg et al. [176]; De Arcangelis et al. [177] |
| Lama-5 |
Lamininα-5 |
Epithelium; mesothelium |
Defective lobation |
Schuger et al. [178]; Nguyen et al. [179] |
| Lmnb-1 |
Laminin-B1 |
Epithelium; mesenchyme |
Impaired lobation |
Schuger et al. [180]; Vergnes et al. [181] |
| PTHlh | Parathyroid hormone-like peptide | Epithelium | Impaired branching | Rubin et al. [182] |
Molecular aspects of the development of the Mammalian lung
Bone Morphogenetic Proteins (BMPs) and Transforming Growth Factors (TGFs)
Out of the more than 20 family members, some BMPs have been shown to be directly involved in various developmental processes (e.g. [183]). Some like BMP-4, -5, and −7 occur in developing lung (e.g. [184]) where they control cell differentiation and proliferation in the epithelial lung buds. BMP-2 and −4 expressions are induced by FGF signaling [141,185]. Although the actual source of BMP-4 is controversial (e.g. [17,142]), it regulates epithelial proliferation through signaling to BMPR-1a (ALK-3) [171,185]. A recent study by Jang et al. [143], however, indicates that BMP-4 may only be expressed in the lung epithelium. It (BMP-4) has been shown to be involved in proliferation and survival of distal lung epithelial cells and in specifying development of smooth muscle cell precursors (e.g. [30,185-187]) and lung vasculogenesis and angiogenesis (e.g. [141,153,188,189]). BMP-5 is expressed across the embryonic lung mesenchyme and BMP-7 in the lung endoderm [190]. BMP-4 is similarly expressed by the proximate mesenchymal cells but its actual role there is unclear [77]: BMP-4 is an antagonist of FGF-10 [30]. Its expression decreases after the branching process has finished. This suggests that BMP-4 has negligible effect on the branching of the airways [141]. Null mutants of BMP-1 and −7 show no pulmonary defects. Unlike other BMPs, BMP-1 isn’t a member of the TGF-β Superfamily: it has a structure unique from the other BMPs and may be involved in activation of other BMPs [191,192].
Transforming growth factor-beta (TGF-β) family is a group of growth factors (GFs) (cytokines) which instruct lung development and play a pivotal role in instigation and pathogenesis of lung diseases [193-197]. The TGF-β family is part of a superfamily of proteins known as the TGF-β superfamily, which includes inhibins, activin, anti-müllerian hormone, BMP, decapentaplegic, and VG-1 [198,199]. Three isoforms of TGF-β, namely TGF-β1, TGF-β2, and TGF-β3, which utilize two receptors (TGFR-β1 and −2) have been reported in the mouse lung, with TGFR-β-2 being expressed only in the distal airway epithelium at early gestation (E11.5) and in both airway epithelium and mesenchyme from mid-gestation (E14.5) to postnatal day 14 [196]. Lack of TGF-β signaling causes abnormalities in BM and alveolization of the lung [194-196,200-202] while interestingly, excessive amounts of it causes serious hypoplasia in immature lung and fibrosis in the adult one [153]. This may occur from disruption of other peptide GFs which are involved in BM [203]. Chen et al. [196] noted that TGFR-β2 mediates TGF-β signaling and performs different roles in the lung epithelium and mesenchyme by differently controlling specific stages of lung development [196].
Fibroblast Growth Factors (FGFs)
FGFs are multifunctional proteins with a broad variety of mitogenic, regulatory, morphological, and endocrine effects (e.g. [144,204]). Termed ‘pluripotent GFs’ and ‘promiscuous GFs’ (e.g. [205]) due to their multiple actions on many cell types, FGFs are involved in proliferation and differentiation of cells and tissues (e.g. [39,144,206]). They consist of a family of ∼ 23 gene encoding low molecular weight polypeptides with different developmental roles which include further cell growth and migration together with tissue repair, inflammation, angiogenesis, and tumour growth (e.g. [207-210]). They are heparin binding proteins and their interactions with cell-surface associated heparan sulfate proteoglycans have been shown to be required for FGF signal transduction. First isolated in pituitary extracts by Armelin [211], FGFs are the first angiogenetic factors to be sequenced [212]. Six members, namely FGF-1, -2, -7, -9, -10, and −18, are expressed in the lung (e.g. [140,213-216]). FGFs bind and signal via FGF tyrosine kinase receptors (FGFR1-5) which are expressed in the lung (e.g. [140,217,218]). They are mostly expressed in the pulmonary mesenchyme while their receptors are located in the lung epithelium. The exceptions are FGF-1 and -2 [(the acidic FGF (aFGF, FGF-1) and the basic FGF (bFGF, FGF-2, FGF-β)] which are expressed both in the fetal pulmonary epithelium and the mesenchyme [219,220]. FGF signaling, and specifically FGFR-3 and −4, are involved in the regulation of the basement membrane formation in the lung [36,221]. Interestingly, mutation of these two genes occasions failure of terminal lung development [36].
FGF-1 (aFGF) which instructs surfactant protein SP-B mRNA production stimulates epithelial cell proliferation which leads to formation, branching, and cell differentiation in the developing lung [204]. FGF-2 is a highly conserved GF which is generally involved in growth and development of different organs and tissues and induction of the mesoderm (e.g. [222-224]). It is a potent mitogen of the type-II pneumocytes [225] and has been associated with compensatory lung growth after damage from exposure to 95% O2[226]. FGF-2 is, however, an idiosyncratic GF: while it is produced in many cell types as well as in endothelial cells and cardiac myocytes and has been localized in cytoplasm, nucleus, and extracellular membrane (e.g. [227,228]), the actual mechanism by which it is secreted under normal physiological conditions is uncertain: it must have a consensus N-terminal signal sequence for its secretion [229].
Albeit the fact they possess some vascular and hematological defects, FGF-2 knockout mice are morphologically normal [230]. The disseminated expression of FGF-2 in the rat fetal lung, i.e., its localization both in the airway epithelial cells and the extracellular membrane (ECM) [219], resemble the pattern it presents in the avian lung [231]. One of the first genes which are upregulated in response to FGF-2 is Sprouty (Spry-2) [232,233]. Spry-2 negatively regulates FGF signal transduction by limiting or moderating the mitogen-activated protein kinase (MAPK) pathway (e.g. [232,234]): it (FGF-2) determines the site of production and hence the number of branches that develop in a certain domain [79]. FGF-1 and −7 produce different airway arrangements during pulmonary growth and development [204]. While FGF-7 is expressed very early in the mesenchymal cells of the developing lung (at sites where active branching occurs), its receptor (FGFR-2), is expressed only on epithelial cells [235]: it promotes mesenchymal-epithelial cell interactions [236]. According to Tichelaar et al. [237], during the development of the mammalian lung, FGF-7 is a more potent morphogen than FGF-10. In presence of other soluble factors, it (FGF-7) causes the trachea to transdifferentiate into distal lung, a transformation that FGF-10 doesn’t cause [90]: this shows that the developments of trachea and lung are regulated differently. In vitro, BM is disrupted by addition of exogenous FGF-7, antisense oligonucleotides, and neutralizing antibodies (e.g. [236,238]). In the mouse, the epithelial receptor for FGF-7 is the FGFR-2 IIIb isoform [144]: there is a lung phenotype when this isoform (but not another) is deleted. At day 10.5 of gestation, in the mouse lung, FGF-9 is abundantly expressed in the mesothelial cell layer and the epithelium [215]. It disperses to the mesenchyme to activate FGFR-1 signaling [239], ostensibly controlling the expression of mesenchymal genes, including FGF-10 [240]. FGF-9 performs functions of reciprocal epithelial-to-mesenchymal signaling and BM in the lung [17,215,241]. While many distal air spaces form and alveolar epithelial cell differentiation occur, FGF-9 null mice (FGF-9−/−) have severe lung hypoplasia and succumb in the prenatal stage [136,241]. Mouse embryos lacking FGF-9 show mesenchymal hypoplasia, diminished BM, and at the end of gestation, hypoplastic lungs that cannot support life [17]. Signaling concurrently, FGF-9 and Shh control growth and patterning of the pulmonary capillary plexes by regulating the expression of VEGF-A [242]. Together with β-catenin-mediated Wnt signalling, mesenchymal FGF-9 signaling acts in a feed-forward loop which sustains mesenchymal FGF receptivity and mesenchymal Wnt/β-catenin signaling [17]. FGF-9 largely signals to mesenchymal FGF receptors FGFR- 1 and −2 but also has the unique capacity of activating epithelial FGFR signaling [17,33,241,243].
Amongst all the FGFs expressed in the lung, only FGF-10 has been shown to be absolutely necessary to lung development (e.g. [184]). Through an intricate regulatory loop, it (FGF-10) regulates expression of BMP-4 at the growing (terminal) epithelial bud (e.g. [30,184,187]) while in response, BMP-4 [141], TGF-β1 [244], and Shh [245,246] (molecular factors expressed by the lung epithelial cells) limit FGF-10 production in the mesenchyme [39,91,247]. Up regulation of these factors in the very highly proliferative regions of the lung may stop or delay growth, induce quiescence, or promote lung bud maturation. Park et al. [235] attributed the formation of the airways largely to FGF-7 and FGF-10 and very little of it to FGF-2. During early lung development (branching period), FGF-10 is expressed in the mesenchyme at the distal tip of the new lung buds (e.g. [39,235,248,249]). From there, it spreads to activate FGFR-2b in adjacent epithelium, instructing a regular bifurcation pattern [116,187,246,250]. Appropriate spatiotemporal expression of FGF-10 is essential to correct organization of the lung epithelial tubules. Disruption of FGF-10-FGFR-2b signaling as well as overexpression of a dominant negative FGFR-2 in the mouse lung is lethal at birth. It causes multiple organ defects, including agenesis of the lung and termination of the trachea in a blind sac (e.g. [144,249,251]). FGF-10 plays a vital role in maintaining epithelial progenitor cell proliferation as well as co-ordination of alveolar smooth muscle cell formation and vascular development [147,187,251]. Furthermore, it (FGF-10) induces Shh, BMP-4, and Wnt-2 signaling, all of which are necessary for lung development (e.g. [30,146]). In the mouse, removal of FGF-18 gene has no specific effect on lung development [252,253]. However, FGF-18 knockout mice have decreased cell proliferation and alveolar spaces while overexpression causes asymmetric expansion of the conducting airways [135,254]. FGF-18 performs a vital role in lung alveolar development during late embryonic lung development but it is not directly involved in BM [216]. FGF signaling is mostly responsible for regulating mesenchymal proliferation while β-catenin signaling is an obligatory permissive factor for mesenchymal FGF signaling [17].
Wnt growth factors/genes
The Wnt proteins, which are named in reference to the Drosophila gene Wingless and its mouse homolog Integrase-1, are a number of 19 family of secreted glycoproteins, signaling molecules which exert a broad range of important developmental processes (e.g. [42,98,255-257]). They produce morphogenetic effects by binding to cell surface receptor proteins (Frizzled), triggering a multi-step signaling cascade within the cell which allows β-catenin to move into the nucleus where it activates certain genes (e.g. [118,255,258-262]). By means of the canonical pathway, Wnt-2 and Wnt-2b signaling perform crucial and cooperative roles in determining lung endoderm progenitors within the anterior foregut, without affecting the specification of other foregut-derived tissues [42]: embryos lacking Wnt-2/2b expression present complete lung agenesis and don’t express Nkx2.1, the first marker of the lung endoderm. Also, Wnt proteins are profoundly involved in epithelial cell tubulogenesis in organs like lung, kidney, ear, mammary gland, gut, and heart (e.g. [261]). They regulate location and concentration of β-catenin, a protein which complexes with T-cell factor (TCF) in the nucleus: the complex (of β-catenin and TCF) activates the transcription of over 100 genes which perform various functions [263,264]. Wnt-β-catenin signaling is decisive to proper BM [118,265]: it refines the morphogenetic processes that are instructed by other upstream signaling pathways. Mesenchymal Wnt-β-catenin signaling controls FGFR-1 and FGFR-2 expression and consequently determines FGF signaling [33]. Wnt-5a and -7b are both expressed largely in the distal lung bud tip which is the site of most cell proliferation in embryonic lung [145,149,266,267]. Moreover, the signaling pathway regulates local specialization of the epithelium and the mesenchyme and the development of progenitor cell groups (e.g. [33,117]).
During the pseudoglandular stage of lung development, Wnt-2a and Wnt-7b are canonical Wnt ligands that actuate mesenchymal Wnt/β-catenin signaling while FGF-9 is the only ligand that signals to mesenchymal FGF receptors (FGFRs) [17]. Wnt-2 is expressed in the mesenchyme next to the tips of the airway buds [266,268]. This suggests presence of a relationship between Wnt expression and Shh signaling (e.g. [245]). During early lung development, Wnt-5a is expressed in both mesenchymal- and epithelial parts of the branching airways while in the pseudoglandular- and canalicular stages it localizes in the epithelium of the end-bud, with distinctive proximal-distal gradient [148]. Wnt-5a null mice evince increased cell proliferation both in the epithelium and the mesenchyme. This leads to growth of the distal lung, increased branching, and enlargement of the lung [148]. FGF-10, BMP-4, and Shh, which are all profoundly involved in BM, are expressed in Wnt-5a null mice [148]: Wnt-5a therefore acts as an inhibitory regulator of BM. Like the FGF-9 null mice, the only other mutant animal to display severe growth deficiencies [136], Wnt-7b null lungs are noticeably hypoplastic but show signs of normal patterning and cell differentiation [118]. Expressed only in the airway epithelium, with its highest levels occurring at the tips of the branching end-buds [266], Wnt-7b signals to adjacent cells to activate both autocrine and paracrine canonical Wnt signaling cascades. In Wnt-7b null mice, FGF-9 expression remains normal and both Wnt-7b and FGF-9 null mutants present reduced FGF-10 expression in the distal inter-bud region while normal expression in the proximal part of the lung bud occurs [118,241]. Concomitantly, these cascades provoke co-ordinated proliferation of contiguous epithelial- and mesenchymal cells to promote the growth of the organ, with limited changes in cell differentiation and morphogenetic patterning. Wnt-5a, a noncanonical Wnt, may disrupt the function of Wnt-7b by instructing and impeding lung growth [31,148]. Wnt-7b expression is regulated by TTF-1 (thyroid transcription factor-1), GATA-6, and FOXA-2, morphogenetic factors which are critical to proper lung development [169,266]. Mesothelial- and epithelial-derived FGF-9, mesenchymal Wnt-2a, and epithelial Wnt-7b have unique functions in the development of the mouse lung [17]: mesothelial FGF-9 and mesenchymal Wnt-2a are mostly in charge of supporting mesenchymal FGF-Wnt/β-catenin signaling while epithelial FGF-9 mainly affects epithelial branching.
Sonic hedgehog (Shh)
Hedgehog (Hh) is a family of three secreted proteins termed Sonic hedgehog (Shh), Indian hedgehog (Ihh), and Desert hedgehog (Dhh) which play important roles in embryonic development. Amongst the Hh family, Shh, which is one of the morphogens involved in early lung development and is the best studied ligand (e.g. [269]): it (Shh) is expressed in the distal epithelium of the lung for the period of pseudoglandular stage of development. It produces its effects by binding to its receptors, patched-1 (Ptc-1) and Smoothen, transmembrane proteins that exist in contiguous sub-epithelial mesenchyme (e.g. [245,246,270]). Expressed at the tips of the end-buds, Shh negatively controls the distal mesenchyme FGF-10 expression, blocking lung bud extension while upregulating FGF-7 [246,247,271]. The zinc finger Gli genes are transducers of Shh signaling [272]. During the development of the lung, the genes are expressed in overlapping but well-defined areas of the mesenchyme [26,272-275]. Gli-2(−1-) and Gli-3(−1-) double mutant mice die by day 10.5 [166]: the lungs are hypoplastic, the right and left lobes don’t separate, and the tracheo-oesophangeal septum is defective, a phenotype which is similar to that displayed by Shh(−/−)[154] or TTF-1 (Nkx2.1)(−/−) mice [155]. Mice with Gli-3 deficiency are viable but the lung is underdeveloped [273]. In Gli-2 null mutant mice, the tracheobronchial tube is not separated, the right and left lungs are connected, and the growth of the alveolar region is stunted [154,166,273]: the lung forms as one undersized lobe. Gli-1 double mutant mice have severe lung defects which are similar to those of the Shh(−/−) mice, where the lung develops but BM is repressed [154]. Disruption of the membrane-bound Hedgehog interacting protein-1 (HIP-1) results in upregulation of Hh signaling, causing neonatal lethality from respiratory failure [156,276,277]: Hip-1 directly binds mammalian Hh proteins and moderates their signaling. Null mutation of Shh supresses lung epithelial branching [154,271]. In the mouse, conditional knockout of Shh in the lung epithelium generates fewer blood vessels and reduces VEGF expression [269]. Experimentally induced overexpression of Shh in the lung epithelium (using SP-C promoter) intensifies cell proliferation in both the mesenchyme and the epithelium while branching is not affected: it leads to development of superfluous mesenchyme and dearth of alveoli [246]. While FGF-10 doesn’t effect Shh expression, excessive amounts of FGF-7 suppress both Shh expression and signaling [235,247]. Shh- and FGF-9 signals control mesenchymal proliferation in specific submesothelial and subepithelial cellular compartments [241].
Retinoic Acid
Vitamin A (retinol) brings about molecular signaling by the binding of its active metabolite (RA) to a group of heterodimerized TFs (transcription factors) [RA receptors α, β, and γ (RARα, -β, and -γ)] and retinoic-X receptors [α, β, and γ (RXR α, -β, and -γ)] (e.g. [278-280]). After RA binds, the nuclear receptors are activated and attach to their specific response sites in the promoter region of their target genes [281]. RA effects transcription of many genes and development and homeostasis in various organs, including the lung (e.g. [282]). It is expressed very early in lung development and continues throughout the process [283-285]. RAR-β is absent in the distal epithelium during BM but is expressed in the epithelial cells of the proximal- and the medium-sized airways while RAR-γ localizes mostly in the epithelium of the distal end-buds and demonstrates only weak expression in the proximal airway epithelium of the fetal- and adult lungs [282,286]. When RA is lacking during early stages of lung development (e.g. [282]), formation of oesophagotracheal septum is inhibited and the primary lung bud outgrowth doesn’t develop: it leads to lung agenesis or serious lung hypoplasia. Interestingly, upregulation of RA impedes BM while suppressing epithelial cell differentiation [282,286]. RA acts on cell programming and meaningfully instructs their differentiation [287]. Exogenous administration (in vitro) of RA upregulates FOXA-2 and TGFβ-3, two inhibitors of BM [287,288]. If RA signaling is blocked by a pan-RAR antagonist, expression of FGF-10, BMP-4, Shh, TTF-1, and GATA-6 is altered, prompting excessive airway branching [287,288]. Among the RA receptors, only signaling from RAR-β and RAR-γ is implicated in BM [282,286-288]. While RAR-β seems to impede branching, it is incontrovertibly involved in formation and stabilization of the conducting airways [282,286,288]. RA is vital in subdivision of the lung parenchyma [116,175,289]. Lungs of mice with obliterations of RAR-γ have less elastin and fewer alveoli [175] while RAR-α null mutant mice also have fewer alveoli [290]. Overexpression of dominant negative RAR-α in the mouse, just before and during alveolization, causes fewer but larger alveoli to form [291]. RAR-β signaling in the early postnatal period hinders alveolization [290,292]. Endogenous RA controls TGF-β activity in the prospective area where the lung forms, permitting local expression of FGF-10 and induction of lung buds Chen et al. [41].
Extracellular matrix (ECM) component proteins
The ECM performs various roles in regulating cell function (e.g. [85]). It separates tissues, providing mechanical and structural support and/or offering a structure for cells to attach and or move on. Consisting of a collagen scaffold to which glycoproteins like tenasin, laminins, fibronectin, and proteoglycogens attach and intermingle with fibrinous proteins such as fibrillins and elastin, the ECM comprises of the basement membrane and the interstitial matrix (e.g. [293,294]). Signals transduced by α-3β-1 integrin may be involved in stimulating branch formation in the developing lung [176]. Matrix metalloproteinases perform important roles in remodeling the ECM (e.g. [295]). Absence or inhibition of interaction between epithelial cells with the basement membrane occasions failure of either normal lung development or lung injury repair [296]. Dearth of elastin decreases subdivision of the parenchyma in the mouse lung [297]. Elastin is important in alveolarization [298]. Suggestive of a prospective role in airway branching, tenascin-C accumulates in areas where new bronchial branches form [299,300]. Fibronectin expression increases to the highest level during airway branching [301,302]: it localizes in the mesenchyme at the epithelial-mesenchymal interface, commonly at points where airways bifurcate [302,303]. Inhibition of fibronectin matrix accumulation reduces BM [304]. The laminins are large multidomain glycoproteins that include three polypeptide subunits, namely α, β, and γ. Laminin α-1 is critical to lung BM and bronchial smooth muscle cell formation while laminin α-5 is necessary for normal lobulation and alveolization [180,305,306].
Vascular Endothelial Growth Factor (VEGF)
Vascular development entails highly complex, well-coordinated processes which include physicochemical stimulators and inhibitors and various gene regulators and signaling molecules (e.g. [307-309]). It is necessary that during lung development, proper juxtaposition occurs between the alveolar surface and the pulmonary capillary endothelial system, forming the blood-gas barrier. It is axiomatic that the development of the vascular system influences the BM of the airways and alveolization [157,310]. Among other organs, the lung has the greatest expression of VEGF (e.g. [311]). Great progress has been made in identifying the signaling pathways which control endothelial cell differentiation and their assembly into a network of cylindrical (tubular) structures with a lumen (e.g. [48,307,308,312]). Lung mesenchyme isn’t uniform in nature. By using histological benchmarks and molecular markers, it has been divided into a sub-mesothelial zone (SMZ) and a sub-epithelial zone (SEZ) [18,241]: Wnt-2a is expressed in the SMZ while Noggin (Nog) is expressed in the SEZ (e.g. [33,142]). Pulmonary vasculature apparently forms between the two mesenchymal compartments [18,241,242]. In vitro, 3-D gel preparations have shown that as many as 1,000 different genes are expressed or upregulated during endothelial tubulogenesis (e.g. [48,56,307]). With some of them converting to red blood cells and accumulating haemoglobin (Figure 9), particular mesenchymal cells change to blood cell/blood vessel forming cells (angioblasts), sense the environment, and move by means of long filopodia to surround red blood cells (Figure 10). Progressively, the cells aggregate and demarcating a lumen [308] (Figures 1112). Members of the VEGF (e.g. [313,314]) and the angiopoetin and the emprin family (e.g. [315-317]) have been associated with the formation of pulmonary vasculature. Transformation, proliferation, and migration of angioblasts is regulated by the local VEGF-A levels and activation of VEGFR-2 [308].
Figure 9 .
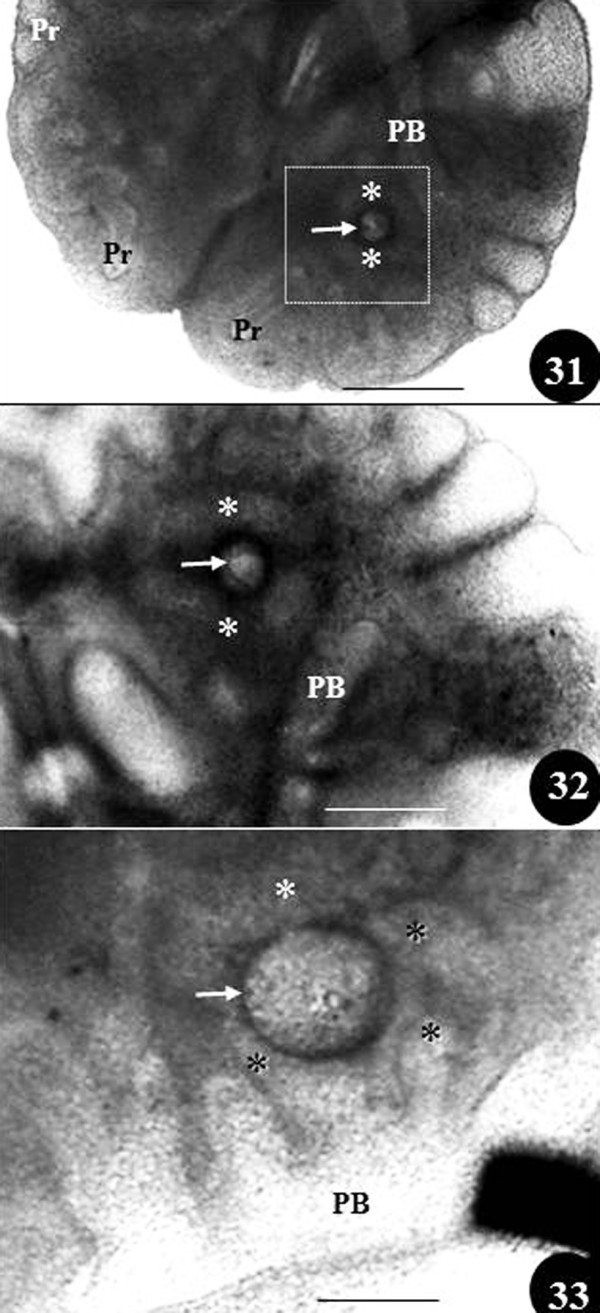
Figure 31–33: Matrigel cultured preparations of developing chicken lungs showing effect of FGF-10 soaked in beads (arrows) on the developing secondary bronchi (asterisks) between days 6 (Figures 31, 32) and 10 (Figure 33) of embryonic life: Figure 32 is an enlarged view of the area enclosed in Figure 31. The secondary bronchi (asterisks) are seen growing towards and surrounding the bead. PB, primary bronchus; Pr, parabronchi. Scale bars: Figure 31, 200 μm; Figure 32, 100 μm; Figure 33, 100 μm. From unpublished work done by JN Maina and B Kramer.
Figure 10 .
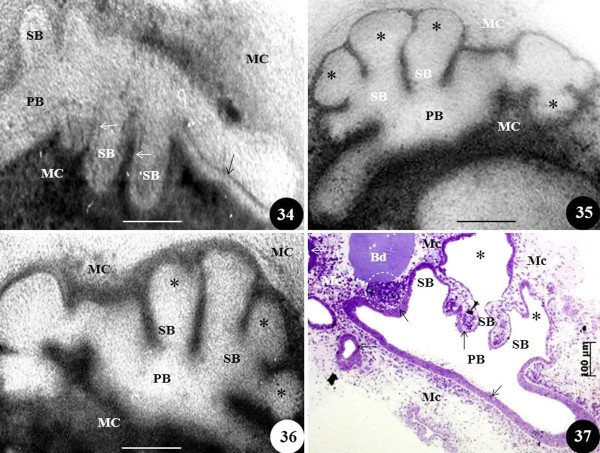
Effect of exposure of excessive FGF-10 on matrigel cultured developing chicken lung. Figure 34 shows normal (control) development of the primary bronchus (PB), the secondary bronchi (SB), and the epithelium lining the airways at day four of embryonic development (arrows). MC, mesenchymal cells. Figures 35 and 36: When exposed to high concentration of exogenous FGF-10 (dissolved in matrigel) at day the fourth day of embryonic life, the primary bronchi (PB) and the secondary bronchi (SB) are aberrantly distended (asterisks) and the epithelium lining the airways is unspecified. Figure 37: A toluidine blue-stained section of a matrigel cultured lung at the fourth day of development showing an FGF-10 soaked bead (Bd) which was placed close to the developing lung (the matrigel also had high concentration of dissolved exogenous FGF-10). The secondary bronchi (SB) were bloated (asterisks) but the epithelium which lines the airways (arrows) was well-defined, particularly that associated with the primary bronchus (PB). MC, mesenchymal cells; asterisks, abnormally swollen distal parts of the secondary bronchi; dashed circle, mesenchymal cell proliferation in the area between the FGF-10 bead and the airway epithelium. Scale bars: 100 μm. From unpublished work by JN Maina and B Kramer.
Figure 11 .
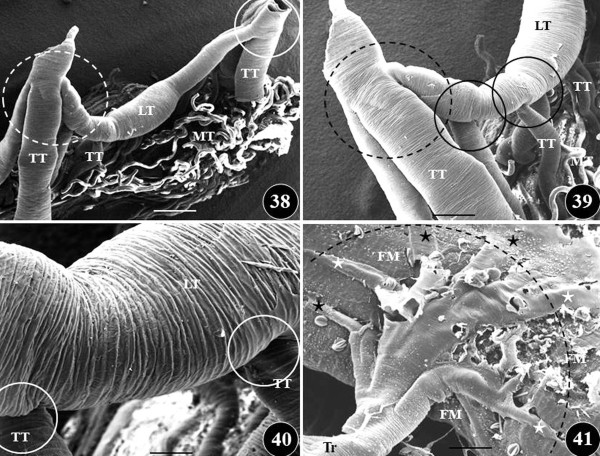
Figures 38–41: Scanning electron microscope views of the branching pattern of the insectan trachea system, from the grasshopper,Chrotogonus senegalensis. Figures 38–40: In some areas, especially close to the spiracles, many branches originate from a parent segment (dashed circles) (Figures 38, 39) while in other cases, single branches (continuous circles) derive from an original part. TT, transverse trachea; LT, longitudinal trachea; MT, Malphigian tubules. Figure 41: Terminal trachea (Tr) giving rise to many terminal tracheoles (stars) which enter the flight muscle (FM). Dashed arch, shows the expansive part of the flight muscle which is supplied with air by a single terminal trachea. Scale bars: Figure 38, 1.5 mm; Figure 39, 1 mm; Figure 40, 0.5 mm; Figure 41, 0.5 mm. From Maina [318].
Figure 12 .
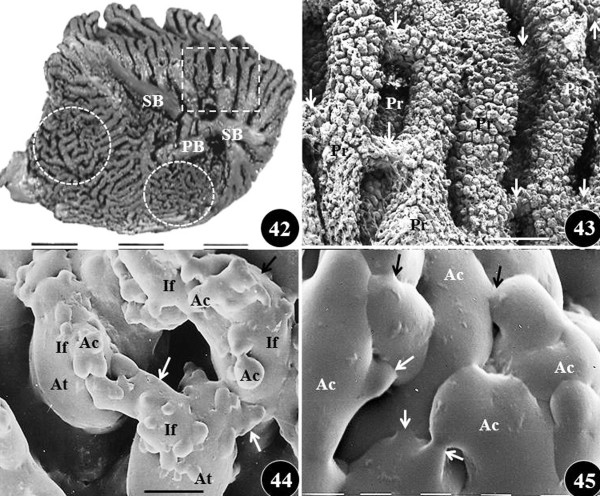
Figures 42–45: Scanning electron microscope views of the branching pattern and anastomoses of the airways in the mature lung of the domestic fowl,Gallus gallusvariantdomesticus. Figure 42: Medial view of latex cast of the lung showing the different sizes, orientations and anastomoses of the tertiary bronchi (parabronchi). Dashed square, paleopulmonic parabronchi which are long and lie parallel to each other; dashed circles, neopulmonic parabronchi which are short and anastomose profusely; PB, primary bronchus; SB secondary bronchi. Figure 43: Close-up of a stack of cast of paleopulmonic parabronchi (Pr) showing areas which they anastomose (arrows). Figure 44: Close-up of cast of a parabronchus showing atria (At) which give rise to infundibula (If) which anastomose (arrows). AC, air capillaries; dashed lines, interatrial septa. Figure 45: Close-up of cast of air capillaries (AC), the terminal gas exchange units, which profusely anastomose (arrows). Scale bars: Figure 42, 1 cm; Figure 43, 1 mm; Figure 44, 0.5 mm; Figure 45, 8 μm.
VEGF is a dimeric, heparin-binding glycoprotein. It is an endothelial cell-specific mitogen which initiates cell proliferation and chemotaxis (e.g. [319-322]). By differential mRNA splicing, the VEGF gene generates at least five protein isoforms (VEGF-122, -145, -164, -188, and −206) which have different affinities for heparan sulfate as well as for the receptors [VEGFR-1, FLT-1 (fetal liver tyrosinase-1), VEGF-2, FLK-1 (fetal liver kinase-1/KDR)], and neuropilin-1 [323-325]. In different organs, angiogenetic response to VEGF varies. This is dependent on the genetic composition of the animal [326]. VEGF-122 doesn’t bind to heparan sulfate and is freely diffusible; VEGF-188 is heparin-binding and is mostly associated with the cell surface and the ECM, while; VEGF-164 has transitional properties (e.g. [327,328]). Presence of various VEGF ligands and receptors shows specific and redundant regulatory pathways of vascular development (e.g. [329,330]). Mice with an inactivated FLK-1 [313] and -II receptors [326] or VEGF gene die in utero from lack of endothelial cells while knockout ones lack yolk-sac blood-islands and organized blood vessels [321,326]. Inactivation of gene encoding for VEGFR-1 leads to increased number of endothelial cells which block the vessel lumen while that of VEGFR-3 produces abnormally organized blood vessels and causes cardiac failure [326]. Precise control through VEGFR-3 signaling is needed to correct vasculoangiogenesis and hematopoiesis [331]. Gene inactivation experiments show that VEGFR-1 utilizes a negative regulatory effect on VEGFR-2, at least during embryogenesis [326]. Lethality with deletion of a single allele shows the importance of VEGF in embryonic vascular growth [332].
During the development of the lung, airway epithelial cells express VEGF and direct it into the subendothelial matrix while the pulmonary endothelial cells synthesize correct receptors (e.g. [333]). VEGF promotes proliferation, cell mediator migration, angioblast differentiation (in the direction of endothelial cell- lineage), and increases vascular permeability [313,334-336]. The functions are mediated by binding of high-affinity cell receptors and matrix binding sites (e.g. [337,338]). VEGF is vital in de novo development of new blood vessels (vasculogenesis) or growth from pre-existing vessels (angiogenesis) (e.g. [189,309,322,323,331,339]). Angiogenesis involves pruning, vessel enlargement, intussusception (vessel splitting), branch remodeling, and extension to form trunks and complex network (e.g. [340,341]). Expression of VEGF gene at the mRNA level is highest in the airway epithelial cells of the lung [342,343], especially in the alveolar type-II epithelial cells [344,345]. Vasculo-epithelial interactions are critical to proper patterning of the airway- and vascular systems (e.g. [346,347]). During the development of the lung, VEGF-A is expressed by the epithelial cells while its primary receptor, VEGFR-2 or FLK-1, is localized in endothelial cells (e.g. [348]). VEGFR-1 and VEGFR-2 expression increases during lung development and accumulates in the pulmonary endothelial cells that lie close to the developing epithelium [116]. HGF (Hepatocyte Growth Factor), a putative endothelial derived factor, mediates reciprocal signaling from the vasculature to the respiratory epithelium [349].
Inhibition of VEGF signaling influences postnatal alveolization [116]. Disruption of the VEGF gene produces mutant embryos with abnormal pulmonary blood vessel development [320,332]. Knockouts for VEGF-A and its two recognized high affinity tyrosine kinase receptors [VEGFR-1 (FLT-1) and VEGFR-2 (KDR/FLK-1)], which are expressed in the primitive vascular endothelium [335,350], die before the lung’s blood capillary plexus forms. Mice overexpressing VEGF in distal epithelial cells present abnormal BM, paucity of acinar buds, impairment of type-I and -II cells, loose mesenchymal mass, and premature development of blood vessels [57,157]. Overexpression of VEGF in the respiratory epithelium leads to excessive vasculogenesis [157,351]. VEGF-188 (which is formed in the pulmonary epithelium, especially by the type-II cells) [352] may mediate the convergence and stabilization of the highly organized blood vessel networks that come to be located in the interalveolar wall. VEGF plays different important roles in the repair and maintenance of blood vessels in different pathologies of the mature lung (e.g. [311]). VEGF-A signaling performs an essential part in facilitating communication between the epithelial, mesenchymal, and endothelial parts of the early mouse embryonic lung [310]. It regulates the expression of BMP-4, mSpry-2, mSpry-4, and Sp-c as well as proliferation of both epithelial and mesenchymal compartments. Lazarus et al. [353] showed that blood vessels are requisite for stereotypic 3D epithelial branching and patterning in the lung. They conjectured that inhibition of normal branching, which ensued from vascular loss caused experimentally by ablative methods, could be partly explained by interruption of spatial expression pattern of the branching mediator FGF-10 and by misregulated expression of the branching regulators Shh and Sprouty-2. Del Moral et al. [310] observed that VEGF pathway is involved in driving epithelial to endothelial communication in embryonic mouse lung morphogenesis: VEGF-164 stimulates mouse embryonic BM in culture and increases the intensity of the index of proliferation in both epithelium and mesenchyme.
Transcription Factors (TFs) and other Growth Factors (GFs)
The platelet-derived growth factor (PDGF) is a potent stimulator of cell motility and growth, especially that of connective tissue cells such as fibroblasts and smooth muscle cells (e.g. [138,354,355]). PDGF and its receptor (PDGFR) are expressed in the lung from the onset of the pseudoglandular stage of development [219,356]. Lack of PDGF introduces pulmonary phenotypes that lack alveolar smooth muscle cells and diminished deposition of elastin fibers [137]: PDGF-A and PDGF-Rα are requisite for alveolization [357]. PDGF-Rα positive cells are largely found in the mesenchyme next to the bronchial end-buds [358]. While no distinct lung branching defects were described in PDGF-Rα null mice by Bostrom et al. [137], secondary subdivision didn’t occur in PDGF-A null ones: they exhibited an emphysematous phenotype [357].
FOX [Foxhead Box (Fox)] TFs, also called hepatocyte nuclear factor-3β (HNF-3β), are expressed in the lung [359-362]: they are known to play an important role during lung morphogenesis. FOXA-1 and −2 are co-expressed in the developing lung epithelium while FOXA-1 is correspondingly expressed in the mesenchyme [363]. Silencing FOXA-1 and −2 disrupts BM in the mouse lung, producing a hypoplastic lung with severe defects in epithelial- and smooth muscle cell differentiation [160]. Overexpression of FOXA-2 impairs airway branching, epithelial cell differentiation, and decreases production of surfactant proteins, SP-A, SP-B, and SP-C [364]. Lungs of transgenic mice overexpressing FOXA-2 also display reduced vasculogenesis, possibly from decreased VEGF production by epithelial cells [364].
The GATA family consists of a group of zinc finger domain transcription factors which recognize DNA motif (AT)GATA(A/G) to regulate target gene expression (e.g. [365-367]): they play important roles in regulating cell differentiation during vertebrate development. In the developing lung, GATA-5 and −6 transcription factors are expressed independently [363]: GATA-6 expression is restricted to the respiratory epithelial cells of the developing lung [166] while that of GATA-5 occurs in the smooth muscle cells of the large airways [366]. Furthermore, corresponding to that of SP-A mRNA [165,166,368,369], among the GATA family of zinc finger domain TFs, GATA-6 is expressed before GATA-5. GATA-6 has been shown to regulate specification, differentiation, and maturation of the pulmonary epithelium, branching morphogenesis, and late epithelial cell differentiation [165,252,368,370,371]. Type-II epithelial cells isolated from adult mice and immortalized MLE-15 cells express TTF-1, GATA-6, and various surfactant protein mRNAs [360]. In vivo, GATA-6 and Nkx2.1 act in synergistic manner, directing pulmonary epithelial differentiation and development [35]. Inhibition of GATA-6 at E6.0 impeded alveolar maturation and also reduced expression of surfactant proteins which are vital to normal pulmonary function. GATA-6 may play a role in lung development essentially because it regulates expression of TTF-1, which is crucial to lung formation [360,372]. During postnatal alveolization, GATA-6 is not expressed in the developing lung [367]. GATA-6 null mice succumb shortly after implantation, i.e., ~5.5 days after conception [373] and chimeric GATA-6 null ones display a pulmonary phenotype with reduced airway branching [370]. GATA-6 overexpression impairs alveolization [367].
Numbered 1–4, the Notch family consists of four proteins which interact with five ligands (Jagged-1 and −2, and Delta-1, 3, -4) which are expressed on the surface of a neighbouring cell [374]. The greatly conserved Notch/Notch-ligand signaling pathway significantly regulates the development of the lung [48,375].
TTF-1, a member of the Nkx-2 family, is involved in lung development [372,376,377]. TTF-1 promoter activity is directed by combinatorial or cooperative actions of HNF-3 [hepatocyte nuclear factor-3; also known as FOXA (forkhead box A)], Sp (specificity protein)-1, Sp-3, GATA-6, and HOXB-3 (homeobox B-3) TFs [372]. At first expressed in the epithelial cells of dividing lungs, with advancing gestation, TTF-1 expression is considerably reduced and restricted to the type-II cells [169]. In the lung, TTF-1 controls the expression of surfactant proteins that are required for lung stability and lung host defence [372]. Lungs of transgenic mice with increased TTF-1 expression display modest alveolization and type-II cell hyperplasia [378]. Indicating a high degree of conservation, the amino acid sequences of TTF-1 from human, rat, mouse, and other species are very similar [372]. TTF-1 null mice exhibit severe deficiencies of the lung’s BM [155,170]: the bronchial tree is undeveloped while the distal parenchyma is lacking. TTF-1 expression can be activated by other TFs such as FOXA-2 (e.g. [378]).
A number of distinct HOX (homeodomain) TFs are expressed in the developing lung as the mouse embryo gets the end of gestation (e.g. [163,164,379,380]). The HOXB-3 and −4 genes are expressed in the mesenchyme of the trachea, bronchi, and distal lung while HOXA-2 and HOXB-5 are confined to the distal lung mesenchyme, specifying their possible role(s) in BM. The HOXA-5 null mice present defective tracheal structure and defective BM, diminished surfactant production, and thickened alveolar walls [163]. GATA-5 and −6 TFs exhibit non-overlapping spatial expression in the developing lung [365]: GATA-6 expression is restricted to the bronchiolar epithelial cells while GATA-5 is expressed in the smooth muscle cells of the large airways [366].
In humans, Homeobox protein Six1 is a protein which is encoded by the Six-1gene (e.g. [381]). It is a member of the six homeodomain family of TFs [382]. Six-1(−1-) lungs are particularly hypoplastic with considerably reduced epithelial branching and augmented mesenchymal cell density [383]: expressed at the distal epithelial tips of the branching airways and also in the proximate distal mesenchyme, Six-1 coordinates Shh-FGF-10 signaling in embryonic lung to ensure correct levels of proliferation and differentiation of epithelial, mesenchymal, and endothelial cells.
Summary of molecular regulatory processes in lung development
FGF-10: Signals through FGFR-2b. Through instructing Sp-C expression and downregulating expression of BMP-4, FGF-10 regulates differentiation of epithelial cells. Regulatory molecules like other FGFs, Shhs, β-catenin (Catnb), and TGF-βs cross-talk with FGF-10-FGFR-2b to tweak lung development: positive regulators of FGF-10 include FOXf-1, Tbx-4, and Tbx-5. Shh inhibits FGF-10 expression but via Gli-3 also controls FOXf-1 availability (Figure 7). By upregulating BMP-4, FGF-10 may influence parabronchial smooth muscle cell development. FGF-7: Expressed in the mesenchyme during late stages of lung development and its receptor (FGFR-2) only in the epithelium. Lungs of FGF-7−/− mutant mice are normal. This suggests its redundancy in lung development. FGF-7 activation of FGFR-2b has been shown to regulate interferon-mediated gene expression in adult airway epithelial cell cultures. FGF-9: Signals through FGFR-1 and −2. Signaling from the epithelium to the subepithelial mesenchyme, FGF-9 sustains Shh signaling. In a feed-forward loop which maintains mesenchymal FGF sensitivity and mesenchymal Wnt/β-catenin signaling, mesenchymal FGF-9 signaling interacts with β-catenin mediated Wnt-signaling (Figure 8). FGF-9 (and probably Wnt-7) are two known ligands that can specifically signal from mesothelial (FGF-9) and epithelial cells (FGF-9 and Wnt-7b) to lung mesenchymal FGFRs to control lung development. Regulating mesenchymal proliferation and Wnt-2a expression, mesothelial FGF-9 signals mesenchymal FGFR-1c and FGFR-2c while epithelial FGF-9 predominantly instructs epithelial branching. Overexpression of FGF-9 promptly stops branching morphogenesis (BM). In the submesothelial region, i.e., distal to the source of Shh, FGF-9 induces FGF-10 expression which may promote lengthening of the airways. FGFR-2c: Wnt/ β-catenin signaling is requisite to activate and sustain expression of FGFR-2c. Sprouty family of genes is one of the key inducible negative regulators of FGFR-2c: FGFR-2b signaling induces expression of Spry-2, a RTK modulator which negatively controls FGF signaling, i.e., it inhibits morphogenesis. The positive feedback loop between FGFR-1, FGFR-2, Wnt-2a, and β-catenin (in the mesenchymal cell compartment) countenances input from FGF- and Wnt signaling systems to modulate the output of the whole system, thus coordinating mesenchymal- and epithelial growths. TGFβ: Controls lung development through two receptors, TGFRβ-1 and -II, which work in series. TGFβ ligands bind to their associated receptors on the cell surface and activate downstream Smad proteins which translocate into the nucleus and modulate target gene expression. β-integrin and thrombospondin are involved in regulating release of TGFβ mature peptide. BMP: Bind to heteromeric complexes of BMP serine/threonine kinase types-1 and -II receptors to activate intracellular signaling pathway. BMP-4 signals to BMPR-1A (ALK 3). Mesenchymal Pod-1 (Tcf-21) and epithelial Wnt signaling regulate BMP-4 which is a well-known target for FGF-10. BMP-4 is believed to control (i.e., to be an antagonist) of FGF-mediated lung bud growth. It probably inhibits distal lung budding through autocrine signaling from the epithelium and can also promote budding in a paracrine manner through unclear mesenchymal signaling. Expression of BMP-4 is controlled by TTF-1. Shh: Binds to patched (Ptc), a transmembrane protein, and releases its inhibitory effect on downstream smoothened (Smo), a G-protein coupled transmembrane bridging receptor, leading to activation of cubitus interruptus (Ci). Shh induces Gli gene (Gli-1 and −3) expression which encode transcription factors (TFs) which work downstream of Shh, suppressing FGF-10 expression (Figure 7). Mesenchymal Ptc, Gli-2, and Gli-3 are downregulated in Shh knockdown lung. By directing Hip expression, Shh inhibits FGF-10 expression. Gli: Gli-1, -2, and −3, the three vertebrate Ci gene orthologues, are zinc-finger transcription effectors of the Shh signaling pathway. GATA: GATA-6 binds to and activates transcription of TTF-1 (Nkx2.1) gene. It also activates expression of different genes involved in respiratory epithelial cell differentiation, including SP-A and SP-C. Hip-1: Binds to hedgehog (HH) proteins, moderating HH signaling. Conforming to the expression domains of Ptc-1, Hip-1 is transcriptionally activated in response to HH signaling. Hip-1 and Ptch-1 have redundant roles of instructing airway branching. Wnt: Wnt signals are transduced through seven transmembrane-type Wnt receptors encoded by frizzled (Fzd) genes to activate the canonical β-catenin-TCF-, the JNK- or the intracellular Ca2+-releasing noncanonical pathways. Wnt-2a and Wnt-7b are the canonical Wnt-ligands that activate mesenchymal Wnt/β-catenin signaling. Mesenchymal FGF signaling is required for expression of Wnt-2a and for mesenchymal Wnt/β-catenin signaling which is vital to sustaining the expression of FGFR-1 and FGFR-2. Wnt-7b- and FGF-9 null mutants exhibit diminished FGF-10 expression. Wnt-5a may antagonize Wnt-7b function of inhibiting lung growth (Figure 7). Receptor internalization-dependent and -independent mechanisms are regulated by Wnt-5a though distinctive pathways. TTF-1, GATA-6, and FOXA-2 TFs, which are critical to lung morphogenesis, regulate Wnt-7b expression (Figure 7). TGF: A tyrosine kinase receptor which transfers epidermal growth factor (EGF), TGF signals into the cell. Through EGFR, EGFs positively modulate early lung BM and cytodifferentiation. VEGF: VEGF-A, -B, -C, -D, and placental growth factor (PGF) are the VEGF family members. They signal through the cognate receptors - Fetal Liver Kinase-1 [Flk-1/KDR (VEGFR-2), Fetal Liver Tyrosinase-1 [Flt-1 (VEGFR-1)], and Flt-4 (VEGFR-3). Flk-1 positively regulates the VEGF-A signals while Flt-1 negatively regulates the signals. Binding of VEGF-C to VEGFR-3 controls VEGFR-2 signaling. Spatiotemporal expression of Flk-1 and Flt-1 regulates the vascular endothelial cell proliferation and differentiation, inducing vasculogenesis and angiogenesis. VEGF signaling through VEGFR-C occurs synergistically with VEGF-A. VEGF-A induces upregulation of BMP-4 and Sp-C expression. Transcription of VEGF is regulated by hypoxia-inducible TF-1 (HIF-1α) and -2α. RA: Its signals are mediated by its nuclear receptors of the steroid hormone receptor superfamily namely retinoic acid receptors (RARs) which include -α, -β, -γ, and δ (i.e., RARα, -β, γ, and δ) and retinoic X receptors α, -β, and -γ (i.e., RXRα, -β, and -γ) which translocate to the nucleus, where they effect gene transcription in target cells. Airway bifurcation is only influenced by the RARα and the RARγ receptors. Together with Tbx genes (in chicks), RA impedes expression and alters distribution of FGF-10 and BMP-4, which must be downregulated in order for BM to happen. Exogenous administration of RA upregulates FOXA-2 and TGFβ-3, two inhibitors of BM. Details are given in the text and can be found mostly in the following detailed reviews: Metzger and Krasnow [25], Perl and Whitsett [11], Roth-Kleiner and Post [116], Cardoso and Lü [40], Lu and Werb [53], De Langhe and Reynolds [117], Affolter et al. [77], Warburton et al. [21], [15,16,18,29,98].
Molecular aspects of the development of the Avian Lung (Al)
Birds evolved from reptilian stock following mammals (e.g. [8]). Their respiratory system, the parabronchial lung and the air sac system, is remarkably different from the brochioalveolar one of mammals (e.g. [107,132,134,384,385]). Among the air-breathing vertebrates, structurally, the avian lung is purportedly the most complex (e.g. [134,386]) and functionally efficient (e.g. [387-390]) gas exchanger. While the structure of the avian lung has been studied for a long time, e.g., since Coitier [391], compared to the mammalian lung, the genetic and the molecular aspects of its development have been less well-studied. Some of the studies are those by Goldin and Opperman [392] who examined stimulation of DNA synthesis in embryonic chick lung and that of the trachea by the epidermal growth factor (EGF); Chen et al. [393] examined expression and distribution of cell-to-cell adhesion molecules (fibronectin and laminin) on the embryonic chick lung cells; using lectin probes and cationic dyes, Gallanger [394] studied the process of BM; Muraoka et al. [395] examined expression of nuclear factor-kappa-β on epithelial growth and branching of the airways in embryonic chick lung; using tissue recombination experiments, Sakiyama et al. [396] studied the effect of the Tbx-4-FGF-10 system on the separation of the lung bud from the oesophagus and showed that the formation of the airways and the air sacs was caused by region-specific mesenchymal properties and HOXb genes which were expressed in the proximity of the ventral-distal tips of the lung; Stabellini et al. [397] evaluated the roles of polyamines and TGF-β1 on the branching of the airways; Sakiyama et al. [398] found region-specific expression of HOXB-5 to 9 genes, BMP-2, BMP-4, Wnt-5a, and Wnt-11 in the developing respiratory tract of the avian lung; Maina [133,399,400,441] microscopically studied the development of the chicken lung (Figures 13–17) and Maina et al. [231] (Figures 18–21) showed that FGF-2 is expressed and remains upregulated in the epithelial- and mesenchymal cells from very early- to late stages of lung development. In an unpublished study (RG Macharia and JN Maina), it has been observed that Wnt proteins are expressed in the embryo (Figure 22) and at different times and parts of the developing chicken lung (Figures 23–30): together with other morphogenetic factors, the Wnts appear to contribute to the development of the intricate airway- and vascular systems of the avian lung; Miura et al. [401] observed that the development of the air sacs (‘cysts’ as they called them) occurred because of differences in the diffusion of FGF-10 between the dorsal- and the ventral parts of the lung: they attributed the higher dispersal coefficient of the morphogen in the ventral region to relatively loose tissue/cell arrangement in the mesenchyme and the lower one in the dorsal region to greater expression of heparan sulphate proteoglycan (HSPG) which locks in FGF-10: this observation supports the assertion made by, e.g., Kutejova et al. [92] that during lung development, signaling gradient regulates differential gene expression in a concentration-dependent manner; Moura et al. [29] showed that in the embryonic chick lung, expression of FGF-10, FGFR-1 to −4, and Spry-1 was similar to that in the mammalian lung and FGFR inhibition (with FGF receptor agonist SU5402) caused impairment of secondary bronchi (SB) and abnormal lung growth with swollen SB: by in vitro tissue culture study (JN Maina and B Kramer, unpublished observations), it was noted that while FGF-10 influences lung development (Figures 31–33), exposure to excessive exogenous level of FGF-10 (dissolved in matrigel and/or soaked in beads) gives similar phenotype, i.e., abnormal (cystic) SB, at expense of the mesenchymal space (Figures 34–37).
Molecular aspects of development of the insectan tracheal system
Best formed in insects, tracheal respiration has evolved in various animal taxa which include the Onychophora (Peripatus), Solifugae, Phalangidae, some Acarina, Myriapoda, and Chilopoda. The bodies of the tracheates are suffused by air-filled tubes, the trachea. The insectan tracheal system is structurally and functionally remarkable both for its structural design (e.g. [318,402]) and functional efficiency (e.g. [403]). The circulatory- and the respiratory (tracheal) systems are totally disengaged [404,405]: the former plays no meaningful role in gas exchange. Entering through the spiracular openings, oxygen diffuses from the atmosphere to reach the target tissues and cells (e.g. [318,402,406]). The tracheal system in Drosophila larvae has afforded a suitable and convenient model for studying the molecular aspects of the development of branched structures [24]. The ‘external factors’ which drive the development of the tracheal system includes the metabolic levels and degrees of hypoxia in the different parts of the body (e.g. [403,407]). Genetic screening of Drosophila larvae (e.g. [408-410]) showed that in excess of 200 patterning and morphogenesis genes are involved in the formation of the tracheal system [23]. Some of the genes are involved in the early stages of the development of the tracheal network (primary formation) while others come into effect late to initiate secondary tracheal development. By means of distinct ectodermal placodes (consisting of ~80 cells each) which form on the lateral aspects of the left- and right sides of the embryo, in D. melanogaster, trachea start to form at mid-embryogenesis [25,89,408,409,411]. The placodes express the gene (transcription factor) Trachealess [406,411,412] which as per the name, without it, no trachea form. The gene codes for Helix-Loop-Helix-Period Arnt Single-Minded (bHLH-PAS) TF which sequentially regulates transcription of downstream genes that mediate tracheal development [406,411,413]. Each of the placodes invaginates (in- pockets) into the body and then gradually penetrates organs and tissues [24,412,414,415]. The tracheal system develops by BM (e.g. [23,24,89,409,410,416-428]) where the first two branching levels (primary- and secondary trachea) display a stereotypical morphology (Figures 38–40) while the smaller terminal branches (tracheoles), which are very thin extensions, branch profusely (Figure 41), in many tissues possibly contacting practically every single cell in the body.
The repeating pattern of the primary- and the secondary tracheal branches (e.g. [318,402]) shows that an exacting morphogenetic program is controls their development [21,23,25,418]. By means of transcriptional regulation of ‘Trachealess’, all tracheal cells express a Drosophila ortholog of the mammalian fibroblast growth factor receptor (FGFR) ‘Breathless (Btl)’ (e.g. [417,429-432]). External to the trachea, in the target tissues, the ligand for this receptor, Branchless (Bnl) (Drosophila ortholog of FGF) acts as a chemoattractant of the migrating cells (e.g. [89,418,431]): budding tracheal branches that express Btl migrate towards masses of cells expressing the ligand Bnl. When tracheal cells have reached Bnl-positive cluster of cells, Bnl expression turns-off in that cluster and is instantaneously turned-on a short distance in the path of the advancing branch [88,89]. Quantitatively spatially regulated Btl activity directs tracheal cell migration [431]. High concentration of Bnl provokes expression of pointed (pnt) (a transcription factor) and Sprouty (antagonist of Btl) at the tip of the branch [433]: pointed causes the tip of the tracheal branch to split (forming secondary branches) and Sprouty restricts branching to the tip by inhibiting branching further along the tracheal branch. Expression of Bnl is regulated by the prevailing tissue O2-levels during early larval stages of development [434]. When FGF signaling is extremely upregulated in the entire tracheal tree (by overexpression of a constitutively active form of Btl), many ectopic terminal tracheal branches develop [418]. In Drosophila, DWnt-2 gene is involved in tracheal development while wingless (Wg) influences both the developing epidermis and the trachea [51]. DWnt-2 is expressed near the tracheal cells in a pattern different from the Wg one but is also transduced through the canonical Wnt pathway [51]: when the two genes (DWnt-2 and Wg) are deleted, the phenotype is identical or very similar to one observed when the Wnt-pathway is shut down. Drosophila Frizzled-2 (Dfz-2) has been identified as a Wg receptor [435]. In Drosophila, tracheas don’t start functioning until after fluid in them is cleared at embryonic stage 17 [436].
Concluding remarks
Insects evolved ~350 million years ago (mya) (e.g. [437]), mammals ~200 mya [8], and birds ~160 mya (e.g. [438]). While their respiratory systems form essentially by branching morphogenesis, a process mechanistically driven by broadly highly conserved genes and molecular factors, in certain important aspects, structural and functional differences exist in the respective gas exchangers, namely the tracheal system, and the bronchioalveolar-, and the parabronchial lungs. For example, in contrast to the dichotomous pattern of branching of particularly the airways in the mammalian lung (e.g. [93,108,115,439]) (Figures 2345), in insects, depending on the part of the tracheal system, many as well as single branches stem from original segments (Figures 38–41) and it is even arguable whether the tracheoles, the terminal air conduits (Figure 41), anastomose (e.g. [318,402]): in the avian lung, three morphologically distinctive tiers of airways, namely the primary-, the secondary- and the tertiary bronchi (parabronchi) exist [134,386] and at the parabronchial-, the atrial-, the infundibular, and the air capillary levels, anastomoses occur liberally (Figures 42–45). A fundamental question is why and how actions of congruently conserved genes and molecular factors consequence in different morphologies. Ostensibly, the variations may emanate from spatiotemporal and qualitative and quantitative differences in the patterns of expression of the morphogenetic drivers which specify how long parts lengthen before branching occurs and how branches form. Furthermore, external signals (e.g. hypoxia) may locally modify the morphogenetic programs and regulate upstream and downstream gene expression pathways. Few evolutionally conserved signal transduction pathways are reiteratively exploited during metazoan development [55]. In order to allow more robust systems for regulating a broad spectrum of signaling responses by a limited number of signalling pathways, signals may be integrated at specific connections (nodes) of ‘crosstalk’ between pathways to permit more integrated operations. In nature, economy of structural design (e.g. [440]), which allows systems to perform more by using less, is ubiquitous innovative strategy. More studies are required to identify signaling connections or dedicated protein molecular complexes which may be involved in the integration of the signaling pathways into complex cell signaling networks. It would explain the operational dynamics of genes and molecular factors that lead to different phenotypes. Molecular recursive signalling processes may be hardwired in developmental programs themselves and may be used to craft structural refinements within limits inherent in the conserved systems.
Ethical
The author’s own research investigations were approved by the University of Witwatersrand’s Animal Ethics Screening Committee.
Competing interests
The author declares that concerning this manuscript there are no competing interests.
Acknowledgement
The preparation of this manuscript was funded by the National Research Foundation (NRF). I thank the two involved referees for their suggestions which greatly improved the manuscript.
References
- Vogel S. In: Principles of animal design: the optimization and symmorphosis debate. Weibel ER, Taylor CR, Bolis L, editor. Cambridge University Press, Cambridge; 1998. Convergence as an analytical tool in evaluating design; pp. 13–20. [Google Scholar]
- Schmalhausen II. The origin of terrestrial vertebrates. Academic, London; 1967. [Google Scholar]
- Little C. The terrestrial invasion: an ecophysiological approach to the origins of land animals. Cambridge University Press, Cambridge; 1990. [Google Scholar]
- Maina JN. The gas exchangers: structure, function, and evolution of the respiratory processes. Springer, Heidelberg; 1998. [Google Scholar]
- Ellington CE. Limitations of animal flight performance. J Exp Biol. 1999;160:71–91. [Google Scholar]
- Nudds RL, Bryant DM. The energy cost of short flights in birds. J Exp Biol. 2000;203:1561–1582. doi: 10.1242/jeb.203.10.1561. [DOI] [PubMed] [Google Scholar]
- Tobalske BW, Hedrik TL, Dial KP, Biewener AA. Comparative power curves in bird flight. Nature. 2003;421:363–366. doi: 10.1038/nature01284. [DOI] [PubMed] [Google Scholar]
- Pough FH, Heiser JB, McFarland WN. Vertebrate life. 3d. Macmillan Publishing Company, New York; 1989. [Google Scholar]
- Jessop NM. General zoology, 6th edtn. McGraw-Hill Inc., New York; 1995. [Google Scholar]
- Gudas L, Sporn MB, Roberts AB. In: The retinoids. Sporn MB, Roberts AB, Goodman DS, editor. Raven, New York; 1994. Cellular biology and biochemistry of retinoids; pp. 443–520. [Google Scholar]
- Perl AKT, Whitsett TA. Molecular mechanisms controlling lung morphogenesis. Clin Genet. 2001;56:14–27. doi: 10.1034/j.1399-0004.1999.560103.x. [DOI] [PubMed] [Google Scholar]
- Hall BK. Evo-devo: evolutionary developmental mechnisms. Int J Dev Biol. 2003;47:491–495. [PubMed] [Google Scholar]
- Kimura J, Deutsch GH. Key mechanisms of early lung development. Pediatr Dev Pathol. 2007;10:335–347. doi: 10.2350/07-06-0290.1. [DOI] [PubMed] [Google Scholar]
- Onodera T, Sakai T, Chi-feng Hsu J, Matsumoto K, Chiorini J, Yamada KM. Btbd-7 regulates epithelial cell dynamics and branching morphogenesis. Science. 2010;329:562–565. doi: 10.1126/science.1191880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjorevski N, Nelson CM. Branch formation during organ development. WIRES Syst Biol Med. 2010;2: . doi: 10.1002/wsbm.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlins EL. The building blocks of mammalian lung development. Dev Dyn. 2011;240:463–476. doi: 10.1002/dvdy.22482. [DOI] [PubMed] [Google Scholar]
- Yin Y, Wang F, Ornitz DM. Mesothelial- and epithelial-derived FGF-9 have distinct functions in the regulation of lung development. Development. 2011;138:3169–3177. doi: 10.1242/dev.065110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz DM, Yin Y. Networks regulating development of the lower respiratory tract. Cold Spring Harb Perspect Biol. 2012;4:a008318. doi: 10.1101/cshperspect.a008318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble F, Fleury V, Pries A, Corvol P, Eichmann A, Reneman RS. Control of arterial branching morphogenesis in embryogenesis: go with the flow. Cardiovasc Res. 2005;65:619–628. doi: 10.1016/j.cardiores.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Yates LL, Schnatwinkel C, Murdoch JN, Bogani D, Formstone CJ. et al. The PCP genes Celsr-1 and Vangl-2 are required for normal lung branching morphogenesis. Hum Mol Genet. 2010;19:2251–2267. doi: 10.1093/hmg/ddq104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton D, El-Hashash A, Carraro G, Tiozzo C, Sala F, Rogers O, De Langhe S, Kemp PJ. et al. Lung organogenesis. Curr Top Dev Biol. 2010;90:73–158. doi: 10.1016/S0070-2153(10)90003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner OB, Bush KT, Nigam KB, Yamaguchi Y, Xu D. et al. Stage dependent regulation of mammary ductal branching by heparin sulfate and HGF-cMet signaling. Dev Biol. 2011;355:394–403. doi: 10.1016/j.ydbio.2011.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghabrial AS, Levi BP, Krasnow MA. A systematic screen for tube morphogenesis and branching genes in the Drosophila tracheal system. PLoS Genet. 2011;7(7):e1002087. doi: 10.1371/journal.pgen.1002087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama R, Andrew DJ. Drosophila as a model for epithelial tube formation. Dev Dyn. 2012;241:119–135. doi: 10.1002/dvdy.22775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger RJ, Krasnow MA. Genetic control of branching morphogenesis. Science. 1999;284:1635–1659. doi: 10.1126/science.284.5420.1635. [DOI] [PubMed] [Google Scholar]
- Chuang PT, Kawcak T, McMahon AP. Feedback control of mammalian Hedgehog signaling by the Hedgehog-binding protein, Hip-1, modulates FGF signaling during branching morphogenesis of the lung. Genes Dev. 2003;17:342–347. doi: 10.1101/gad.1026303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torday JS, Rehan VK. Up-regulation of fetal lung parathyroid hormone-related protein gene regulatory network down-regulates the sonic hedgehog/Wnt/β-catenin gene regulatory network. Pediatr Res. 2006;60:382–388. doi: 10.1203/01.pdr.0000238326.42590.03. [DOI] [PubMed] [Google Scholar]
- Nelson CM, VanDujin MM, Inman JL, Fletcher DA, Bissell MJ. Tissue geometry determines sites of mammary branching morphogenesis in organotypic cultures. Science. 2006;314:298–3000. doi: 10.1126/science.1131000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura RS, Coutinho-Borges JP, Pacheco AP, Damota PO, Correia-Pinto J. FGF signaling pathway in the developing chick lung: expression and inhibition studies. PLoS One. 2011;11(3):e17660. doi: 10.1371/journal.pone.0017660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver M, Dunn NR, Hogan BL. BMP-4 and FGF-10 play opposing roles during lung bud morphogenesis. Development. 2000;127:2695–2704. doi: 10.1242/dev.127.12.2695. [DOI] [PubMed] [Google Scholar]
- De Langhe SP, Sala FG, Del Moral PM, Fairbanks TJ, Yamada KM. et al. Dickkopf-1 (DKK1) reveals that fibronectin is a major target of Wnt signaling in branching morphogenesis of the mouse embryonic lung. Dev Biol. 2005;277:316–331. doi: 10.1016/j.ydbio.2004.09.023. [DOI] [PubMed] [Google Scholar]
- De Langhe SP, Carraro G, Tefft D, Li C, Xu X, Chai Y, Minoo P, Hajihosseini MM. et al. Formation and differentiation of multiple mesenchymal lineages during lung development is regulated by β-catenin signaling. PLoS One. 2008;2:e1516. doi: 10.1371/journal.pone.0001516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, White AC, Huh SH, Hilton MJ, Kanazawa H, Long F, Ornitz DM. An FGT-Wnt gene regulatory network controls lung mesenchyme development. Dev Biol. 2008;319:426–436. doi: 10.1016/j.ydbio.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Domyan ET, Lewandoski M, Sun X. Fibroblast growth factor-9 signaling inhibits airway smooth muscle differentiation in mouse lung. Dev Dyn. 2009;238:123–137. doi: 10.1002/dvdy.21831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Rath N, Hannenhalli S, Wang Z, Cappola T, Kimura S, Atochina-Vasserman E. et al. GATA and Nkx factors synergistically regulate tissue-specific gene expression and development in vivo. Development. 2007;134:189–198. doi: 10.1242/dev.02720. [DOI] [PubMed] [Google Scholar]
- Mariani TJ, Reed JJ, Shapiro SD. Expression profiling of the developing mouse lung insights into the establishment of the extracellular matrix. Am J Respir Cell Mol Biol. 2002;26:541–548. doi: 10.1165/ajrcmb.26.5.2001-00080c. [DOI] [PubMed] [Google Scholar]
- Torday JS, Rehan VK. Deconvoluting lung evolution using functional/comparative genomics. Am J Respir Cell Mol Biol. 2004;31:8–12. doi: 10.1165/rcmb.2004-0019TR. [DOI] [PubMed] [Google Scholar]
- Torday JS, Rehan VK, Hicks JW, Wang T, Maina J. et al. Deconvoluting lung evolution: from phenotypes to gene therapy. Int Comp Biol. 2007;47:601–609. doi: 10.1093/icb/icm069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso WN. Lung morphogenesis revisted: old facts, current ideas. Dev Dyn. 2000;219:121–130. doi: 10.1002/1097-0177(2000)9999:9999<::aid-dvdy1053>3.3.co;2-8. [DOI] [PubMed] [Google Scholar]
- Cardoso WV, Lü J. Regulation of early lung morphogenesis: questions, facts and controversies. Development. 2006;133:1611–1624. doi: 10.1242/dev.02310. [DOI] [PubMed] [Google Scholar]
- Chen F, Desai TJ, Qian J, Niederreither K, Lü J, Cardoso WV. Inhibition of TGF-β signaling by endogenous retinoic acid is essential for primary lung bud induction. Development. 2007;134:2969–2979. doi: 10.1242/dev.006221. [DOI] [PubMed] [Google Scholar]
- Goss AM, Tian Y, Tsukiyama T, Cohen ED, Zhou D, Lu MM, Yamaguchi TP, Morrisey EE. Wnt2/2b and β-catenin signaling are necessary and sufficient to specify lung progenitors in the foregut. Dev Cell. 2009;17:290–298. doi: 10.1016/j.devcel.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett BP, Binge CD, Gitlin JD. Mechanisms of gene expression and cell fate determination in the developing pulmonary epithelium. Annu Rev Physiol. 1996;58:51–71. doi: 10.1146/annurev.ph.58.030196.000411. [DOI] [PubMed] [Google Scholar]
- Chinoy MR. Lung growth and development. Front Biosci. 2003;8:392–415. doi: 10.2741/974. [DOI] [PubMed] [Google Scholar]
- Schittny J, Burri PH. In: Lung development and regeneration. Massaro DJ, Massaro GC, Chambon P, editor. Marcel Dekker Inc, New York; 2004. Morphogenesis of the mammalian lung: Aspects of structure and extracellular matrix; pp. 275–316. [Google Scholar]
- Gehr P, Mwangi DK, Amman A, Maloiy GMO, Taylor CR, Weibel ER. Design of the mammalian respiratory system. V. Scaling morphometric diffusing capacity to body mass: wild and domestic animals. Respir Physiol. 1981;44:41–86. doi: 10.1016/0034-5687(81)90077-3. [DOI] [PubMed] [Google Scholar]
- Maina JN, West JB. Thin and strong! the bioengineering dilemma in the structural and functional design of the blood-gas barrier. Physiol Rev. 2005;85:811–844. doi: 10.1152/physrev.00022.2004. [DOI] [PubMed] [Google Scholar]
- van Tuyl MV, Del Riccio V, Post M. In: Lung development and regeneration. Massaro DJ, Massaro GC, Chambon P, editor. Marcel Dekker, New York; 2004. Lung branching morphogenesis: potential for regeneration of small conducting airways; pp. 355–393. [Google Scholar]
- Sidow A. Diversification of the Wnt gene family on the ancestral lineage of vertebrates. Proc Natl Acad Sci. 1992;89:5098–5102. doi: 10.1073/pnas.89.11.5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody T. The interactive fly. 1995. www.sdbonline.org/fly/vdevlhom/pcyclina.htm.
- Llimargas M, Lawrence PA. Seven Wnt homologues in Drosophila: a case study of the developing trachea. PNAS. 2001;98:14487–14492. doi: 10.1073/pnas.251304398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JA. Do different branching epithelia use a conserved developmental mechanism? Bioassays. 2002;24:937–948. doi: 10.1002/bies.10161. [DOI] [PubMed] [Google Scholar]
- Lu P, Werb Z. Patterning mechanisms of branched organs. Science. 2008;322:1506–1509. doi: 10.1126/science.1162783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen R, Gouar ML, Pechmann M, Poulin F, Bolognesi R. et al. Conservation, loss and redeployment of Wnt ligands in protostomes: implications for understanding the evolution of segment formation. BMC Evol Biol. 2010;10:374. doi: 10.1186/1471-2148-10-374. http://www.biomedicalcentral.com/1471-2148/10/374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancratov R, DasGupta R. Postgenomic technologies targeting the Wnt signaling network. Interdispl Rev Syst Biology Med. 2011;3:649–665. doi: 10.1002/wsbm.140. [DOI] [PubMed] [Google Scholar]
- Alsberg E, Moore K, Huang S, Polte T, Inger DE. In: Lung development and regeneration. Massaro DJ, Massaro GC, Chambon P, editor. Marcel Dekker, New York; 2004. The mechanical and cytoskeletal basis of lung morphogenesis; pp. 247–274. [Google Scholar]
- van Tuyl MV, Liu J, Wang J, Kuliszewski M, Tibboel D, Post M. Role of oxygen and vascular development in epithelial branching morphogenesis of the developing mouse lung. Am J Physiol Lung Cell Mol Physiol. 2005;288:167–178. doi: 10.1152/ajplung.00185.2004. [DOI] [PubMed] [Google Scholar]
- Nichols JE, Niles JA, Cortiella J. Design and development of tissue engineered lung: Progress and challenges. Organogenesis. 2009;5:57–61. doi: 10.4161/org.5.2.8564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TH, Calle EA, Zhao L, Lee EJ, Gui L. et al. Tissue-engineered lungs for in vitro implanatation. Science. 2010;329:538–541. doi: 10.1126/science.1189345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade CF, Wong AP, Waddell TK, Keshvjee S, Liu M. Cell-based tissue engineering for lung regeneration. Am J Physiol. 2007;292:L510–L518. doi: 10.1152/ajplung.00175.2006. [DOI] [PubMed] [Google Scholar]
- Carmeliet P. Gene therapy: stimulating angiogenesis or angioma-genesis. Nat Med. 2000;6:1102–1103. doi: 10.1038/80430. [DOI] [PubMed] [Google Scholar]
- Kumar VH, Ryan RM. Growth factors in the fetal- and neonatal lung. Front Biosci. 2004;9:464–480. doi: 10.2741/1245. [DOI] [PubMed] [Google Scholar]
- Cao L, Mooney DJ. Spatiotemporal control over growth factor signaling for therapeutic neovasculalization. Adv Drug Deliv Rev. 2007;59:1340–1350. doi: 10.1016/j.addr.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torday JS, Rehan VK. The evolutionary continuum from lung development to homeostatsis and repair. Am J Physiol Lung Cell Mol Physiol. 2007;292:L608–L611. doi: 10.1152/ajplung.00379.2006. [DOI] [PubMed] [Google Scholar]
- Torday JS, Rehan VK. A cell-molecular approach predicts vertebrate evolution. Mol Biol Evol. 2011;28:2973–2981. doi: 10.1093/molbev/msr134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleury V, Gouyet JF, Léonetti M. (eds) Branching in nature: dynamics and morphogenesis of branching structures, from cell to river networks. Springer, Berlin; 2001. [Google Scholar]
- Davies JA. Branching morphogenesis. Landes Biosience, Austin; 2006. [Google Scholar]
- Mandelbrot BB. The fractal geometry of nature. Freeman, San Francisco; 1983. [Google Scholar]
- Gross LJ, Miura RM. Some mathematical questions in biology. Lectures on mathematics in the life sciences. American Mathematical Society, Society for Industrial and Applied Mathematics. 1986;18:179. [Google Scholar]
- Steen LA. The science of patterns. Science. 1988;240:611–616. doi: 10.1126/science.240.4852.611. [DOI] [PubMed] [Google Scholar]
- Keith D. Mathematics: The science of patterns - the search for order in life, mind and the Universe. Scientific American Paperback Library, Philadelphia; 1996. [Google Scholar]
- Hartmann D, Miura T. Mathematical analysis of a free-boundary model for lung branching morphogenesis. Math Med Biol. 2007;24:209–224. doi: 10.1093/imammb/dql029. [DOI] [PubMed] [Google Scholar]
- Jamie D. Branching morphogenesis. Springer, Berlin; 2006. [Google Scholar]
- Saxena L, Sariola H. Early organogenesis of the kidney. Pediatr Nephrol. 1987;1:385–392. doi: 10.1007/BF00849241. [DOI] [PubMed] [Google Scholar]
- Pavlova A, Stuart RO, Pohl M, Nigam SK. Evolution of gene expression patterns in a model of branching morphogenesis. Am J Physiol. 1999;277:F650–F663. doi: 10.1152/ajprenal.1999.277.4.F650. [DOI] [PubMed] [Google Scholar]
- Affolter M, Bellusci S, Itoh B, Thiery JB, Web Z. Tube or not tube: remodeling epithelial tissues via branching morphogenesis. Dev Cell. 2003;4:1–20. doi: 10.1016/s1534-5807(02)00410-0. [DOI] [PubMed] [Google Scholar]
- Affolter M, Zeller R, Caussinus E. Tissue remodelling through branching morphogenesis. Nature Rev. 2009;10:831–842. doi: 10.1038/nrm2797. [DOI] [PubMed] [Google Scholar]
- Chuang PT, McMahon AP. Branching morphogenesis of the lung: new molecular insights into an old problem. Trends Cell Biol. 2003;13:86–91. doi: 10.1016/s0962-8924(02)00031-4. [DOI] [PubMed] [Google Scholar]
- Metzger RJ, Klein OD, Martin GR, Krasnow MA. The branching programme of the mouse lung development. Nature, Lond. 2008;453:745–750. doi: 10.1038/nature07005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaskoll T, Zhou YM, Chai Y, Makarenkova HP, Collinson JM. et al. Embryonic submandibular gland morphogenesis: Stage-specific protein localization of FGFs, BMPs, Pax-6 and Pax-9 in normal mice and abnormal SMG phenotypes in FgfR-2-IIIc(+)/Delta), BMP-7(−/−) and Pax-6(−/−) mice. Cells Tissues Organs. 2002;170:83–98. doi: 10.1159/000046183. [DOI] [PubMed] [Google Scholar]
- Villasenor A, Chong DC, Henkemeyer M, Cleaver O. Epithelial dynamics of pancreatic branching morphogenesis. Development. 2010;137:4295–4305. doi: 10.1242/dev.052993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otso H, Fujimori S, Schmidt-Ullrich R, Hartmann C, Theslefft I, Mikkola ML. Ectodysplasin and Wnt pathways are required for salivary gland branching morphogenesis. Development. 2011;138:2681–2691. doi: 10.1242/dev.057711. [DOI] [PubMed] [Google Scholar]
- McDermont KM, Liu BY, Tisty TD, Pazour GJ. Primary cilia regulate branching morphogenesis during mammary gland development. Curr Biol. 2010;20:731–737. doi: 10.1016/j.cub.2010.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl M, Stuart RO, Sakurai H, Nigam SK. Branching morphogenesis during kidney development. Annu Rev Physiol. 2000;62:595–620. doi: 10.1146/annurev.physiol.62.1.595. [DOI] [PubMed] [Google Scholar]
- Hsu JC, Yamada KM. Salivary gland branching morphogenesis – recent progress and future opportunities. Int J Oral Sci. 2010;2:117–126. doi: 10.4248/IJOS10042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah MM, Sakurai H, Gallegos TF, Sweeney DE, Bush KT. et al. Growth factor-dependent branching of the ureteric bud is modulated by selective 6-O sulfation of heparin sulfate. Dev Biol. 2011;356:19–27. doi: 10.1016/j.ydbio.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith DB, Cutler LS, Kollar EJ. The correlation of temporal regulation of glycosaminoglycan synthesis with morphogenetic events in mouse tooth development. Arch Oral Biol. 1992;37:623–628. doi: 10.1016/0003-9969(92)90124-q. [DOI] [PubMed] [Google Scholar]
- Ghabrial AS, Krasnow MA. Social interactions among epithelial cells during tracheal branching morphogenesis. Nature, Lond. 2006;441:746–749. doi: 10.1038/nature04829. [DOI] [PubMed] [Google Scholar]
- Ghabrial A, Luschnig S, Metzstein MM, Krasnow MA. Branching morphogenesis of the Drosophila and tracheal system. Annu Rev Cell Dev Biol. 2003;19:623–647. doi: 10.1146/annurev.cellbio.19.031403.160043. [DOI] [PubMed] [Google Scholar]
- Shannon JM, Gebb SA, Nielsen LD. Induction of alveolar type-II cell differentiation in embryonic tracheal epithelium in mesenchyme-free culture. Development. 1998;126:1675–1688. doi: 10.1242/dev.126.8.1675. [DOI] [PubMed] [Google Scholar]
- Hogan BL. Morphogenesis. Cell. 1999;96:225–233. doi: 10.1016/s0092-8674(00)80562-0. [DOI] [PubMed] [Google Scholar]
- Kutejova E, Briscoe J, Kicheva A. Temporal dynamics of patterning by morphogen gradients. Curr Opin Genet Dev. 2009;19:315–322. doi: 10.1016/j.gde.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Horsfield K, Thurbeck A. Relation between diameter and flow in the bronchial tree. Bull Math Biol. 1981;43:681–691. doi: 10.1007/BF02458417. [DOI] [PubMed] [Google Scholar]
- Miura T. Modeling lung branching morphogenesis. Curr Top Deb Biol. 2008;81:291–319. doi: 10.1016/S0070-2153(07)81010-6. [DOI] [PubMed] [Google Scholar]
- Morrisey EE, Hogan BLM. Preparing for the first breath: genetic and cellular mechanisms in lung development. Dev Cell. 2010;18:8–23. doi: 10.1016/j.devcel.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J, Csete M. Rules of engagement. Nature, Lond. 2007;446:860. doi: 10.1038/446860a. [DOI] [PubMed] [Google Scholar]
- Warburton D. Developmental biology: order in the lung. Nature, Lond. 2008;453:733–735. doi: 10.1038/453733a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusse R. Wnt signaling. Cold Spring Harb Perspect Biol. 2012;4:a011163. doi: 10.1101/cshperspect.a011163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant D, Mostov K. Development: inflationary pressures. Nature, Lond. 2007;449:549–550. doi: 10.1038/449549a. [DOI] [PubMed] [Google Scholar]
- Cartwright JHE, Piro O, Tuval I. Fluid dynamics in developmental biology: moving fluids that shape ontogeny. HFSP J. 2009;3:77–93. doi: 10.2976/1.3043738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remesal A, Pedraz C, Feliciano LS, Ludena D. Pulmonary expression of vascular endothelial growth factor (VEGF) and alveolar septation in a newborn rat model exposed to acute hypoxia and recovered under conditions of air or hyperoxia. Histol Histopathol. 2009;24:325–330. doi: 10.14670/HH-24.325. [DOI] [PubMed] [Google Scholar]
- Finney BA, Del Moral PM, Wilkinson WJ, Cayzac S, Cole M. et al. Regulation of mouse lung development by extracellular calcium-sensing receptor. Can J Physiol. 2008;586:6007–6019. doi: 10.1113/jphysiol.2008.161687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson SM. The extracellular Ca 2+-sensing receptor branches out - a new role in lung morphogenesis. J Physiol. 2008;586:5847–5848. doi: 10.1113/jphysiol.2008.166405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoo P, King RJ. Epithelial-mesenchymal interaction in lung development. Annu Rev Physiol. 1994;56:13–45. doi: 10.1146/annurev.ph.56.030194.000305. [DOI] [PubMed] [Google Scholar]
- Torday JS, Rehan VK. Cell-cell signaling drives the evolution of complex traits: introduction- lung evo-devo. Integr Comp Biol. 2009;49:142–154. doi: 10.1093/icb/icp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torday JS, Rehan VK. Lung evolution as a cipher for physiology. Physiol Genomics. 2009;38:1–6. doi: 10.1152/physiolgenomics.90411.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weibel ER. The pathways for oxygen: structure and function in the mammalian respiratory system. Harvard University Press, Harvard; 1984. [Google Scholar]
- Weibel ER. Morphometry of the human lung. Springer, Berlin; 1963. [Google Scholar]
- Pinkerton KE, Joad JP. The mammalian respiratory system and critical windows of exposure for children’s health. Environ Health Perspect. 2000;108:457–462. doi: 10.1289/ehp.00108s3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochs M, Nyengaard JR, Jung A, Knudsen L, Voigt M. et al. The number of alveoli in the human lung. Am J Respir Crit Care Med. 2004;169:120–124. doi: 10.1164/rccm.200308-1107OC. [DOI] [PubMed] [Google Scholar]
- Gehr P, Bachofen M, Weibel ER. The normal human lung: ultrastructure and morphometric estimation of diffusion capacity. Respir Physiol. 1978;32:121–140. doi: 10.1016/0034-5687(78)90104-4. [DOI] [PubMed] [Google Scholar]
- Breeze RG, Wheeldon EB. The cells of the pulmonary airways. Am Rev Respir Dis. 1977;116:705–717. doi: 10.1164/arrd.1977.116.4.705. [DOI] [PubMed] [Google Scholar]
- Cardoso WV, Whitsett JA. Resident cellular components of the lung: developmental aspects. Proc Am Thorac Soc. 2008;5:767–771. doi: 10.1513/pats.200803-026HR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine A. Breathing life into the lung stem cell field. Cell Stem Cell. 2009;4:468–469. doi: 10.1016/j.stem.2009.05.012. [DOI] [PubMed] [Google Scholar]
- Maina JN, van Gils P. Morphometric characterization of the airway and vascular systems of the lung of the domestic pig, Sus scrofa: comparison of the airway, arterial, and venous systems. Comp Biochem Physiol. 2001;130A:781–798. doi: 10.1016/s1095-6433(01)00411-1. [DOI] [PubMed] [Google Scholar]
- Roth-Kleiner M, Post M. Similarities and dissimilarities of branching and septation during lung development. Pediatr Pulmonol. 2005;40:113–134. doi: 10.1002/ppul.20252. [DOI] [PubMed] [Google Scholar]
- De Langhe SP, Reynolds SD. Wnt signaling in lung organogenesis. Organogenesis. 2008;4:100–108. doi: 10.4161/org.4.2.5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal J, Carroll TJ, Guseh JS, Bores SA, Blank LJ, Anderson WJ. et al. Wnt-7b stimulates embryonic lung growth by co-ordinately increasing the replication of epithelium and mesenchyme. Development. 2008;135:1625–1634. doi: 10.1242/dev.015495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi L, Domyan ET, Lewandoski M, Sun X. Fibroblast growth factor-9 signaling inhibits airway smooth muscle differentiation in mouse lung. Dev Dyn. 2009;238:123–137. doi: 10.1002/dvdy.21831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panneerselvam P, Bawankar P, Kulkarni S, Patankar S. In silico prediction of evolutionarily conserved GC-rich elements associated with antigenic proteins of Plasmodium falciparum. Evol Bioinformatics. 2011;7:235–255. doi: 10.4137/EBO.S8162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GP. The origin of morphological characters and the biological basis of homology. Evolution. 1989;43:1157–1171. doi: 10.1111/j.1558-5646.1989.tb02566.x. [DOI] [PubMed] [Google Scholar]
- Wagner GP. Complexity matters. Science. 1998;279:1158–1159. doi: 10.1126/science.279.5354.1158. [DOI] [PubMed] [Google Scholar]
- Wallace A. Animal body plans. Cambridge University Press, Cambridge (England); 1997. [Google Scholar]
- Mojica NS, Navarro J, Marijuán PC, Lahoz-Beltra R. Cellular "bauplans": evolving unicellular forms by means of Julia sets and Pickover biomorphs. Biosystems. 2009;98:19–30. doi: 10.1016/j.biosystems.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Salazar-Cludad I, Jernvall J, Newman SA. Mechanisms of pattern formation in development and evolution. Development. 2003;130:2027–2037. doi: 10.1242/dev.00425. [DOI] [PubMed] [Google Scholar]
- Knoll AH. Life on a young planet: The first three billion years of evolution on Earth. Princeton University Press, Princeton; 2004. [Google Scholar]
- Schmidt-Rhaesa A. The evolution of organ systems. Oxford University Press, Oxford; 2007. [Google Scholar]
- Jacob W, Schierwater B. Changing hydrozoan bauplans by silencing Hox-like genes. PLoS One. 2007;2(8):e694. doi: 10.1371/journal.pone.0000694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB, Grenier J, Weatherbee SD. From DNA to diversity: molecular genetics and the evolution of animal design. Blackwell, Cambridge; 2001. [Google Scholar]
- Valentine JW, Jablonski D, Erwin DH. Fossils, molecules and embryos: new perspectives on the Cambrian Explosion. Development. 1999;126:851–859. doi: 10.1242/dev.126.5.851. [DOI] [PubMed] [Google Scholar]
- Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- Maina JN. Bioengineering aspects in the design of gas exchangers: comparative evolutionary, morphological, functional, and molecular perspectives. Springer, Heidelberg, Berlin; 2011. [Google Scholar]
- Maina JN. A systematic study of the development of the airway (bronchial) system of the avian lung from days 3 to 26 of embryogenesis: A transmission electron microscopic study on the domestic fowl, Gallus gallus variant domesticus. Tissue Cell. 2003;35:375–391. doi: 10.1016/s0040-8166(03)00058-2. [DOI] [PubMed] [Google Scholar]
- Maina JN. The lung-air sac system of birds: development, structure, and function. Springer- Verlag, Heidelberg; 2005. [DOI] [PubMed] [Google Scholar]
- Usui H, Shibayama M, Ohbayashi N, Konishi M, Takada S. et al. FGF-18 is required for embryonic lung alveolar development. Biochem Biophy Res Commun. 2004;322:887–892. doi: 10.1016/j.bbrc.2004.07.198. [DOI] [PubMed] [Google Scholar]
- Colvin JS, White AC, Pratt SJ, Ornitz DM. Lung hypoplasia and neonatal death in FGF9- null mice identify this gene as an essential regulator of lung mesenchyme. Development. 2001;128:2095–2106. doi: 10.1242/dev.128.11.2095. [DOI] [PubMed] [Google Scholar]
- Bostrom H, Gritli-Linde A, Betsholz C. PDGF-A/PDGF alpha-receptor signaling is required for lung growth and the formation of alveoli but not for early lung branching morphogenesis. Dev Dyn. 2002;223:155–162. doi: 10.1002/dvdy.1225. [DOI] [PubMed] [Google Scholar]
- Buch S, Jassal D, Cannigia I, Edelson J, Han R, Liu J, Tanswell K, Post M. Ontogeny and regulation of platelet-derived growth factor gene expression in distal fetal lung epithelial cells. Am J Respir Cell Mol Biol. 1994;11:251–261. doi: 10.1165/ajrcmb.11.3.8086163. [DOI] [PubMed] [Google Scholar]
- Xu K, Nieuwenhuis E, Cohen BL, Wang W, Canty AJ, Danska JS. et al. Lunatic Fringe- mediated Notch signaling is required for lung alveogenesis. Am J Physiol Lung Cell Mol Physiol. 2009;298:L45–L56. doi: 10.1152/ajplung.90550.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein M, Xu X, Ohyama K, Deng CX. FGFR-3 and FGFR-4 function coorperatively to direct alveogenesis in the murine lung. Development. 1998;125:3615–3623. doi: 10.1242/dev.125.18.3615. [DOI] [PubMed] [Google Scholar]
- Bellusci S, Henderson R, Winnier G, Oikawa T, Hogan BL. Evidence from normal expression and targeted mis-expression that bone morphogenetic protein-4 (BMP-4) plays a role in mouse embryonic lung morphogenesis. Development. 1996;122:1693–1702. doi: 10.1242/dev.122.6.1693. [DOI] [PubMed] [Google Scholar]
- Weaver M, Batts L, Hogan BL. Tissue interactions pattern the mesenchyme of embryonic mouse lung. Dev Biol. 2003;258:169–194. doi: 10.1016/s0012-1606(03)00117-9. [DOI] [PubMed] [Google Scholar]
- Jang CW, Gao L, Dickinson ME, Behringer RR. BMP-4 directed nuclear cyan fluorescent protein provides a tool for live imaging and reveals cellular resolution of BMP-4 expression patterns during embryogenesis. Int J Dev Biol. 2010;54:931–938. doi: 10.1387/ijdb.092911cj. [DOI] [PubMed] [Google Scholar]
- De Moerlooze L, Spencer-Dene B, Revest J, Hajihosseini M, Rosewell I, Dickson C. An important role for the IIIb isoform of fibroblast growth factor receptor-2 (FGFR-2) in mesenchymal-epithelial signaling during mouse organogenesis. Development. 2000;127:483–492. doi: 10.1242/dev.127.3.483. [DOI] [PubMed] [Google Scholar]
- Shu W, Guttentag S, Wang Z, Andril T, Ballad P, Lu MM, Piccolo S, Birchmeir S. et al. Wnt/ β-catenin signaling acts upstream of N-myc, BMP4, and FGF signaling to regulate proximal-distal patterning in the lung. Dev Biol. 2005;283:226–239. doi: 10.1016/j.ydbio.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Sekine K, Ohuchi H, Fujiwara M, Yamasaki M, Yoshizawa T. et al. FGF-10 is essential for limb and lung development. Nat Genet. 1999;21:138–141. doi: 10.1038/5096. [DOI] [PubMed] [Google Scholar]
- Min H, Danilenko DM, Scully SA, Bolon B, Ring BD, Tarpley JE, DeRose M, Simonet WS. FGF-10 is required for both limb and lung development and exhibits striking functional similarities to Drosophila branchless. Genes Dev. 1998;12:3156–3161. doi: 10.1101/gad.12.20.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Xiao J, Hormi K, Borok Z, Minoo P. Wnt-5a participates in distal lung morphogenesis. Dev Biol. 2002;248:68–81. doi: 10.1006/dbio.2002.0729. [DOI] [PubMed] [Google Scholar]
- Shu W, Jiang YQ, Lu MM, Morissey EE. Wnt-7b regulates mesenchymal proliferation and vascular development in the lung. Development. 2002;129:4831–4842. doi: 10.1242/dev.129.20.4831. [DOI] [PubMed] [Google Scholar]
- Shi W, Heisterkamp N, Groffen J, Zhao J, Warburton D, Kaartinen V. TGF-3 beta null mutation does not abrogate fetal lung maturation in vivo by glucocorticoids. Am J Physiol. 1999;277:L1205–L1213. doi: 10.1152/ajplung.1999.277.6.L1205. [DOI] [PubMed] [Google Scholar]
- Mucenski ML, Wert SE, Nation JM, Loudy DE, Huelsken J, Birchmeir W. et al. β-catenin is required for specification of proximal/distal cell fate during lung morphogenesis. J Biol Chem. 2003;278:40231–40238. doi: 10.1074/jbc.M305892200. [DOI] [PubMed] [Google Scholar]
- Kaartinen V, Voncken KW, Shuler C, Warburton D, Bu D, Heisterkamp N, Groffen J. Abnormmal lung development and cleft palate in mice lacking TGF-beta 3 indicates defects of epithelial-mesenchymal interaction. Nat Genet. 1995;11:415–421. doi: 10.1038/ng1295-415. [DOI] [PubMed] [Google Scholar]
- Shi W, Xu J, Warburton D. Development, repair and fibrosis: What is common and why it matters. Respirology. 2009;14:656–665. doi: 10.1111/j.1440-1843.2009.01565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litingtung Y, Lei L, Westphal H, Chiang C. Sonic hedgehog is essential to foregut development. Nat Genet. 1998;20:58–61. doi: 10.1038/1717. [DOI] [PubMed] [Google Scholar]
- Minoo P, Su G, Drum H, Bringas P, Kimura S. Defects in tracheo-oesophangeal and lung morphogenesis in Nkx2.1(−/−) mouse embryos. Dev Biol. 1999;209:60–71. doi: 10.1006/dbio.1999.9234. [DOI] [PubMed] [Google Scholar]
- Chuang PT, McMahon AP. Vertebrate Hedgehog signaling modulated by induction of a hedgehog-binding protein. Nature, Lond. 1999;397:617–621. doi: 10.1038/17611. [DOI] [PubMed] [Google Scholar]
- Zeng X, Wert SE, Federici R, Peters KG, Whitsett JA. VEGF enhances pulmonary vasculogenesis and disrupts lung morphogenesis in vivo. Dev Dyn. 1998;211:215–227. doi: 10.1002/(SICI)1097-0177(199803)211:3<215::AID-AJA3>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Miquerol L, Gertsenstein M, Harpal K, Rossant J, Nagy A. Multiple developmental roles of VEGF suggested by a LacZ tagged allele. Dev Biol. 1999;212:307–322. doi: 10.1006/dbio.1999.9355. [DOI] [PubMed] [Google Scholar]
- Kasahara Y, Tuder RM, Taraseviciene-Stewart L, Le Cras TD, Abman S, Hirth PK. et al. Inhibition of VEGF receptors causes lung cell apoptosis and emphysema. J Clin Invest. 2000;106:1311–1319. doi: 10.1172/JCI10259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan H, Dingle S, Xu Y, Besnard V, Kaestner KH. et al. Compensatory roles of FOXA-1 and FOXA-2 during lung morphogenesis. J Biol Chem. 2005;280:13809–13816. doi: 10.1074/jbc.M414122200. [DOI] [PubMed] [Google Scholar]
- Brody SL, Yan XH, Wuerffel MK, Song SK, Shapiri SD. Ciliogenesis and left-right axis defects in forkhead factor HFH-4-null mice. Am J Respir Cell Mol Biol. 2000;23:45–51. doi: 10.1165/ajrcmb.23.1.4070. [DOI] [PubMed] [Google Scholar]
- Lim L, Kalinichenko VV, Whitsett JA, Costa RH. Fusion of lung lobes and vessels in mouse embryos heterozygous for the forkhead box-f1 targeted allele. Am J Physiol Lung Cell Mol Physiol. 2002;282:L1012–L1022. doi: 10.1152/ajplung.00371.2001. [DOI] [PubMed] [Google Scholar]
- Aubin J, Lemeieux M, Tremblay M, Berard J, Jeannotte L. Early postnatal lethality in Hoxa-5 mutant mice is attributable to respiratory tract defects. Dev Biol. 1997;192:432–445. doi: 10.1006/dbio.1997.8746. [DOI] [PubMed] [Google Scholar]
- Golpon HA, Geraci MW, Miller HL, Miller GJ, Tuder RM, Voelkel NF. HOX genes in human lungs: altered expression in primary pulmonary hypertension and emphysema. Am J Pathol. 2001;158:955–966. doi: 10.1016/S0002-9440(10)64042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Min M, Zhang L, Whitsett JA, Morrisey EE. GATA-6 regulates differentiation of distal lung epithelium. Development. 2002;129:2223–2246. doi: 10.1242/dev.129.9.2233. [DOI] [PubMed] [Google Scholar]
- Motoyama J, Liu J, Mo R, Ding Q, Post M, Hui CC. Essential function of Gli-2 and Gli-3 in the formation of lung, trachea and oesophagus. Nat Genet. 1998;20:54–57. doi: 10.1038/1711. [DOI] [PubMed] [Google Scholar]
- van Tuyl MV, Post M. From fruitflies to mammals: mechanisms of signaling via the Sonic hedgehog pathway in lung development. Respir Res. 2000;1:30–35. doi: 10.1186/rr9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris-Johnson KS, Domyan ET, Vezina CM, Sun X. β-catenin promotes respiratory progenitor identity in mopuse foregut. Proc Natl Acad Sci, USA. 2009;106:16287–16292. doi: 10.1073/pnas.0902274106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahlman MT, Gray ME, Whitsett JA. Expression of thyroid transcription factor-1 (TTF-1) in fetal and neonatal human lung. J Histochem Cytochem. 1996;44:673–678. doi: 10.1177/44.7.8675988. [DOI] [PubMed] [Google Scholar]
- Kimura S, Hara Y, Pineau T, Fernadez-Salguero P, Fox CH. et al. The T/ebp null mouse: thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid gland, lung, ventral forebrain, and pituitary. Genes Dev. 1996;10:60–69. doi: 10.1101/gad.10.1.60. [DOI] [PubMed] [Google Scholar]
- Sun J, Chen H, Chen C, Whitsett JA, Mishina Y, Bringas P, Ma JC, Warburton D, Shi W. Prenatal lung epithelial cell-specific abrogation of ALK-3 bone morphogenetic protein signaling causes neonatal respiratory distress by disrupting distal airway formation. Am J Pathol. 2008;172:571–582. doi: 10.2353/ajpath.2008.070286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock JR, Futtner CR, Harfe BD. The transmebrane protein TMEM-16A is required for normal development of the murine trachea. Dev Biol. 2008;321:141–149. doi: 10.1016/j.ydbio.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Mendelsohn C, Lohnes D, Decimo D, Lufkin T, LeMeuer M, Chambon P, Mark M. Function of the retinoic acid recptors (RARs) during development (II ). Multiple abnormalities at various statges of organogenesis in RAR double mutants. Development. 1994;120:2749–2771. doi: 10.1242/dev.120.10.2749. [DOI] [PubMed] [Google Scholar]
- Dickman ED, Thaller C, Smith SM. Temporally regulated retinoic acid depletion produces specific neural crest, ocular, and nervous system defects. Development. 1997;124:311–3121. doi: 10.1242/dev.124.16.3111. [DOI] [PubMed] [Google Scholar]
- McGowan S, Jackson SK, Jenkins-Moore M, Dai HM, Chambon P, Snyder JM. Mice bearing deletions of retinoic acid receptors demonstrate reduced lung elastin and alveolar numbers. Am J Respir Cell Mol Biol. 2000;23:162–167. doi: 10.1165/ajrcmb.23.2.3904. [DOI] [PubMed] [Google Scholar]
- Kreidberg JA, Donovan MJ, Goldstein SL, Rennke H, Shepherd J, Jones RC. et al. Alpha-3 β-1 integrin has a crucial role in kidney and lung organogenesis. Development. 1996;122:3537–3547. doi: 10.1242/dev.122.11.3537. [DOI] [PubMed] [Google Scholar]
- De Arcangelis A, Mark M, Kreidberg J, Sorokin L, Georges-Labouesse E. Synergistic activities of alpha-3 and alpha-6 integrins are required during apical ectodermal ridge formation and organogenesis in the mouse. Development. 1999;126:3957–3968. doi: 10.1242/dev.126.17.3957. [DOI] [PubMed] [Google Scholar]
- Schuger L, Skubitz AP, O’Shea KS, Chang JF, Varani J. Identification of laminin domains involved in branching morphogenesis: effects of anti-laminin monoclonal antibodies on mouse embryonic lung development. Dev Biol. 1991;146:531–541. doi: 10.1016/0012-1606(91)90254-z. [DOI] [PubMed] [Google Scholar]
- Nguyen NM, Milner JH, Pierce RA, Senior RM. Laminin alpha 5 is required for lobar septation and visceral pleural basement membrane formation in the developing mouse lung. Dev Biol. 2002;246:231–244. doi: 10.1006/dbio.2002.0658. [DOI] [PubMed] [Google Scholar]
- Schuger L, Skubitz AP, Zhang J, Sorokin L, He L. Laminin alpha-1 chain synthesis in the mouse developing lung: requirement for epithelial-mesenchymal contact and possible role in bronchial smooth muscle development. J Cell Biol. 1997;139:553–562. doi: 10.1083/jcb.139.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergnes L, Peterfy M, Bergo MO, Young SG, Reue K. Laminin B-1 is required for mouse development and nuclear integrity. Proc Natl Acad Sci, USA. 2004;101:10428–10433. doi: 10.1073/pnas.0401424101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin LP, Kovacs CS, De Paepe ME, Tsai SW, Torday JS, Kronenberg HM. Arrested pulmonary alveolar cytodifferentiation and defective surfactant synthesis in mice missing the gene for parathyroid hormone-related protein. Dev Dyn. 2004;230:278–289. doi: 10.1002/dvdy.20058. [DOI] [PubMed] [Google Scholar]
- Hogan BL. Bone morphogenetic proteins: Multifunctional regulators of vertebrate development. Genes Dev. 1996;10:1580–1594. doi: 10.1101/gad.10.13.1580. [DOI] [PubMed] [Google Scholar]
- Hyatt BA, Shangguan X, Shannon JM. BMP-4 modulates fibroblast growth factor- mediated induction of proximal and distal lung differentiation in mouse embryonic tracheal epithelium in mesenchyme free culture. Dev Dyn. 2002;225:153–165. doi: 10.1002/dvdy.10145. [DOI] [PubMed] [Google Scholar]
- Eblangie MC, Reedy M, Oliver T, Mishina Y, Hogan BL. Evidence that autocrine signaling through BMPR-1a regulates the proliferation, survival and morphogenetic behaviour of distal lung epithelial cells. Dev Biol. 2006;291:67–82. doi: 10.1016/j.ydbio.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Jeffery TK, Upton PD, Trembath RC, Morrell NW. BMP-4 inhibits proliferation and promotes myocyte differentiation of lung fibroblasts via Smad-1 and JNK pathways. Am J Physiol Lung Cell Mol Physiol. 2004;288:L370–L378. doi: 10.1152/ajplung.00242.2004. [DOI] [PubMed] [Google Scholar]
- Mailleux AA, Kelly R, Veltmaat JM, Langhe SP, Zaffran S. et al. FGF-10 expression identifies parabronchial smooth muscle cell progenitors and is required for their entry into the smooth muscle cell lineage. Development. 2005;132:2157–2166. doi: 10.1242/dev.01795. [DOI] [PubMed] [Google Scholar]
- Kurz H. Physiology of angiogenesis. J Neurooncol. 2000;50:17–35. doi: 10.1023/a:1006485716743. [DOI] [PubMed] [Google Scholar]
- Patan S. Vasculogenesis and angiogenesis. Cancer Treat Res. 2004;117:3–32. doi: 10.1007/978-1-4419-8871-3_1. [DOI] [PubMed] [Google Scholar]
- Vukićević S, Sampath KT, editor. Bone morphogenetic proteins: regeneration of bone and beyond. Birkhäuser Verlag, Basel; 2004. [Google Scholar]
- Shore EM, Cook AL, Hahn GV, Kaplan FS, Wozney JM. et al. BMP-1 sublocalization on human chromosome 8: molecular anatomy and orthopaedic implications. Clin Orthop Relat Res. 1995;311:199–209. [PubMed] [Google Scholar]
- Park JO, Pan J, Möhrlen F, Marcus-Oliver S, Johnsen R. et al. Characterization of astacin family of metalloproteases in C.elegans. BMC Dev Biol. 2010;10:14. doi: 10.1186/1471-213X-10-14. http://biomedcentral.com/1471-213X/10/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korfhagen TR, Swantz RJ, Wert SE, McCarty JM, Kerlakian CB, Glasser SW, Whitsett JA. Respiratory epithelial cell expression of human transforming growth factor-alpha induces lung fibrosis in transgenic mice. J Clin Invest. 1994;93:1691–1699. doi: 10.1172/JCI117152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Tseu I, Wang J, Tanswell K, Post M. Transforming growth factor beta-2, but not beta-1 and beta-3, is critical for early rat lung branching. Dev Dyn. 2000;180:242–257. doi: 10.1002/(SICI)1097-0177(200004)217:4<343::AID-DVDY2>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Bartram U, Speer CP. The role of transforming growth factor beta in lung development and disease. Chest. 2004;125:754–765. doi: 10.1378/chest.125.2.754. [DOI] [PubMed] [Google Scholar]
- Chen H, Zhuang F, Liu YH, Xu B, Del Moral B. et al. TGF-beta receptor-II in epithelial versus mesenchyme plays distinct roles in the developing lung. Eur Respir J. 2008;32:285–295. doi: 10.1183/09031936.00165407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirakihara T, Horiguchi K, Miyazawa K, Ehata S, Shibata T, Morita I, Miyazono K, Saitoh M. TGF-β relualates isoform switching of FGF receptors and epithelial-mesenchymal transition. EMBO J. 2011;30:783–795. doi: 10.1038/emboj.2010.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signaling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- Zhou L, Dey CR, Wert SE, Whitsett JA. Arrested lung morphogenesis in transgenic mice bearing an SP-C-TGF-beta 1 chimeric gene. Dev Biol. 1996;175:227–238. doi: 10.1006/dbio.1996.0110. [DOI] [PubMed] [Google Scholar]
- Zhao J, Bu D, Lee M, Slavkin HC, Hall FL, Warburton D. Abrogation of transforming growth factor-beta type-II receptor stimulates embryonic mouse lung branching morphogenesis in culture. Dev Biol. 1996;180:242–257. doi: 10.1006/dbio.1996.0298. [DOI] [PubMed] [Google Scholar]
- Zeng X, Gray M, Stahlman MT, Whitsett JA. TGF-beta1 perturbs vascular development and inhibits epithelial differentiation in fetal lung in vivo. Dev Dyn. 2001;221:289–301. doi: 10.1002/dvdy.1140. [DOI] [PubMed] [Google Scholar]
- Zhao J, Sime PJ, Bringas P Jr, Gauldie J, Warburton D. Epithelium-specific adenoviral transfer of a dominant-negative mutant TGF-beta type-II receptor stimulates embryonic lung branching morphogenesis in culture and potentiates EGF and PDGF- A. Mech Dev. 1998;72:89–100. doi: 10.1016/s0925-4773(98)00019-7. [DOI] [PubMed] [Google Scholar]
- Cardoso WV, Itoh A, Nogawa H, Mason I, Brody JS. FGF-1 and FGF-7 induce distinct patterns of growth and differentiation in embryonic lung epithelium. Dev Dyn. 1997;208:398–405. doi: 10.1002/(SICI)1097-0177(199703)208:3<398::AID-AJA10>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Green PJ, Walsh FS, Doherty P. Promiscuity of fibroblast growth factor receptors. Bioessays. 1996;18:639–646. doi: 10.1002/bies.950180807. [DOI] [PubMed] [Google Scholar]
- Ornitz DM, Itoh N. Fibroblast growth factors. Genome Biol. 2001;2:REVIEWS3005. doi: 10.1186/gb-2001-2-3-reviews3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gospodarowicz D. In: The fibroblast growth factor family. Baird A, Klagsbrun M, editor. New York Academy of Science, New York; 1991. Biological activities of fibroblast growth factors; pp. 1–8. [DOI] [PubMed] [Google Scholar]
- Desai TJ, Cardoso WV. Growth factors in lung development and disease: friends or foe? Respir Res. 2002;3:2. doi: 10.1186/rr169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoine M, WirzW TCG, Mavituna M, Emans N. et al. Expression pattern of fibroblast growth factors (FGFs), their receptors and antagonists in primary endothelial cells and vascular smooth muscle cells. Growth Factors. 2005;23:87–95. doi: 10.1080/08977190500096004. [DOI] [PubMed] [Google Scholar]
- Kumar VH, Lakshminrusimha S, El Abiad MT, Chess PR, Ryan RM. Growth factors in lung development. Adv Clin Chem. 2005;40:261–316. doi: 10.1016/s0065-2423(05)40007-4. [DOI] [PubMed] [Google Scholar]
- Armelin HA. Pituitary extracts and steroid hormones in the control of 3 T3 cell growth. Proc Natl Acad Sci, USA. 1973;70:2702–2706. doi: 10.1073/pnas.70.9.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klagsbrun M. The fibroblast growth factor family: structural and biological properties. Prog Growth Factor Res. 1989;1:207–235. doi: 10.1016/0955-2235(89)90012-4. [DOI] [PubMed] [Google Scholar]
- González AM, Buscaglia M, Ong M, Baird A. Distribution of fibroblast growth factor in the 18-day rat fetus: localization in the basement laminas of diverse tissues. J Cell Biol. 1990;110:753–765. doi: 10.1083/jcb.110.3.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YM, Spirito P, Yu ZX, Biro S, Sasse J. et al. Acidic fibroblast growth factor in the developing rat embryo. J Cell Biol. 1991;114:1261–1273. doi: 10.1083/jcb.114.6.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin JS, Feldman B, Nadeau JH, Goldfarb M, Ornitz DM. Genomic rganization and embryonic expression of the mouse fibroblast growth factor 9 gene. Dev Dyn. 1999;216:72–88. doi: 10.1002/(SICI)1097-0177(199909)216:1<72::AID-DVDY9>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Haque T, Nakada S, Hamdy RC. A review of FGF-18: its expression, signaling pathways and possible functions during embryogenesis and post-natal development. Histol Histopathol. 2007;22:97–105. doi: 10.14670/HH-22.97. [DOI] [PubMed] [Google Scholar]
- Powell PP, Wang CC, Horinouchi H, Shephard K, Jacobson M. et al. Differential expression of fibroblast growth factor receptors 1 to 4 and ligand genes in late fetal and early postnatal rat lung. Am J Respir Cell Mol Biol. 1998;19:563–572. doi: 10.1165/ajrcmb.19.4.2994. [DOI] [PubMed] [Google Scholar]
- Sleeman M, Fraser J, McDonald M, Yuan S, White D. et al. Identification of a new fibroblast growth factor receptor, FGFR-5. Gene. 2001;271:171–182. doi: 10.1016/s0378-1119(01)00518-2. [DOI] [PubMed] [Google Scholar]
- Han RNN, Liu J, Tanswell AK, Post M. Expression of basic fibroblast growth factor and receptor: immunolocalization studies in developing rat fetal lung. Pediatr Res. 1992;31:435–440. doi: 10.1203/00006450-199205000-00004. [DOI] [PubMed] [Google Scholar]
- González AM, Hill DJ, Logan A, Maher PA, Baird A. Distribution of fibroblast growth factor (FGF-2) and FGF receptor-1 messanger RNA expression and protein presence in the mid- trimester human fetus. Pediatr Res. 1996;39:375–385. doi: 10.1203/00006450-199603000-00001. [DOI] [PubMed] [Google Scholar]
- Li X, Chen Y, Scheele S, Arman E, Haffner-Krausz R, Ekblom P, Lonai P. Fibroblast growth factor signaling and basement membrane assembly are connected during epithelial morphogenesis of the embroid body. J Cell Biol. 2001;153:811–822. doi: 10.1083/jcb.153.4.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler DS, Melton DA. Vertebrate embryonic induction: mesodermal and neural patterning. Science. 1994;266:595–604. doi: 10.1126/science.7939714. [DOI] [PubMed] [Google Scholar]
- Bikfalvi A, Klein S, Guiseppe P, Rifkin D. Biological roles of fibroblast growth factor-2. Endocrine Rev. 1997;18:26–45. doi: 10.1210/edrv.18.1.0292. [DOI] [PubMed] [Google Scholar]
- Le ACN, Musil L. FGF signaling in chick lens development. Development. 2001;233:394–411. doi: 10.1006/dbio.2001.0194. [DOI] [PubMed] [Google Scholar]
- Tanswell AK, Buch S, Liu M, Post M. In: Chronic lung disease of early infancy. Brand RD, Coalson J, editor. Marcel Dekker, New York; 1999. Factors mediating cell growth in lung injury; pp. 493–534. [Google Scholar]
- Jankov RP, Luo X, Campbell A, Belcastro R, Cabacungan J. et al. Fibroblast growth factor receptor-1 and neonatal compensatory lung growth after exposure to 95% oxygen. Am J Respir Crit Care Med. 2003;167:1554–1561. doi: 10.1164/rccm.200207-662OC. [DOI] [PubMed] [Google Scholar]
- Vlodavsky I, Folkman J, Sullivan R, Fridman R, Ishai MR. et al. Endothelial cell derived basic fibroblast growth factor: synthesis and deposition into subendothelial matrix. Proc Natl Acad Sci, USA. 1987;84:2292–2296. doi: 10.1073/pnas.84.8.2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speir E, Tanner V, Gonzalez MA, Farris J, Baird A, Casscells W. Acidic and basic fibroblast growth factors in adult rat heart myocytes. Circ Res. 1992;71:251–259. doi: 10.1161/01.res.71.2.251. [DOI] [PubMed] [Google Scholar]
- Abraham JA, Mergia A, Wang JL, Tumolo A, Friedman KA. et al. Nucleotide sequence of a bovine cDNA clone encoding the angiogenetic protein, basic fibroblast growth factor. Science. 1986;233:545–548. doi: 10.1126/science.2425435. [DOI] [PubMed] [Google Scholar]
- Zhou M, Sutliff RL, Paul RJ, Lorenz JN, Hoying JB. et al. Fibroblast growth factor-2 control of vascular tone. Nature (Med) Lond. 1998;4:201–207. doi: 10.1038/nm0298-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maina JN, Madan AK, Alison B. Expression of fibroblast growth factor-2 (FGF-2) in early stages (days 3–11) of the development of the avian lung, Gallus gallus variant domesticus. J Anat. 2003;203:505–512. doi: 10.1046/j.1469-7580.2003.00236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tefft JD, Lee M, Smith S, Leinwand M, Zhao J. et al. Conserved function of mSpry-2. a murine homolog of Drosophila sprouty, which negatively modulates respiratory organogenesis. Curr Biol. 1999;9:219–222. doi: 10.1016/s0960-9822(99)80094-3. [DOI] [PubMed] [Google Scholar]
- Mailleux AA, Tefft D, Ndiaye D, Ito N, Thiery JP. et al. Evidence that Sprouty-2 functions as an inhibitor of mouse embryonic lung growth and morphogenesis. Mech Dev. 2001;102:81–94. doi: 10.1016/s0925-4773(01)00286-6. [DOI] [PubMed] [Google Scholar]
- Tefft JD, Lee M, Smith S, Crowe DL, Bellusci S, Warburton D. mSprouty-2 inhibits FGF-10 activated MAP kinase by differentially binding upstream target proteins. Am J Physiol Lung Cell Mol Physiol. 2002;283:L700–L706. doi: 10.1152/ajplung.00372.2001. [DOI] [PubMed] [Google Scholar]
- Park WY, Miranda B, Lebeche D, Hashimoto G, Cardoso WV. FGF-10 is a chemotactic factor for distal epithelial buds during lung development. Dev Biol. 1998;201:125–134. doi: 10.1006/dbio.1998.8994. [DOI] [PubMed] [Google Scholar]
- Post M, Souza P, Liu J, Tseu I, Wang J, Kuliswewski M, Tanswell AK. Keratinocyte growth factor and its receptor are involved in regulating early lung branching. Development. 1996;122:3107–3115. doi: 10.1242/dev.122.10.3107. [DOI] [PubMed] [Google Scholar]
- Tichelaar JW, Lu W, Whitsett JA. Conditional expression of fibroblast growth factor-7 in the developing and mature lung. J Biol Chem. 2000;275:111858–111864. doi: 10.1074/jbc.275.16.11858. [DOI] [PubMed] [Google Scholar]
- Shiratori M, Oshika E, Ung LP, Sigh G, Shinozuka H. et al. Keratinocyte growth factor and embryonic rat lung morphogenesis. Am J Respir Cell Mol Biol. 1996;15:328–338. doi: 10.1165/ajrcmb.15.3.8810636. [DOI] [PubMed] [Google Scholar]
- Szebenyi G, Fallon JF. Fibroblast growth factors as multifunctional signaling factors. Int Rev Cytology. 1999;185:45–106. doi: 10.1016/s0074-7696(08)60149-7. [DOI] [PubMed] [Google Scholar]
- Arman E, Haffner-Krausz R, Gorivodsky M, Lonai P. FGF-2 is required for limb outgrowth and lung branching morphogenesis. Proc Nat Acad Sci. 1999;96:11895–11899. doi: 10.1073/pnas.96.21.11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AC, Xu J, Yin Y, Smith C, Schmid G, Ornitz DM. FGF-9 and Shh signaling co- ordinate lung growth and development through regulation of distinct mesenchymal domains. Development. 2006;133:1507–1517. doi: 10.1242/dev.02313. [DOI] [PubMed] [Google Scholar]
- White CR, Lavine KJ, Ornitz DM. FGF-9 and Shh regulate mesenchymal VEGF-A expression and development of the pulmonary capillary network. Development. 2007;134:3743–3752. doi: 10.1242/dev.004879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Moral PM, De Langhe SP, Sala FG, Veltmaat JM, Tefft D. et al. Differential role of FGF-9 on epithelium and mesenchyme in mouse embryonic lung. Dev Biol. 2006;293:77–89. doi: 10.1016/j.ydbio.2006.01.020. [DOI] [PubMed] [Google Scholar]
- Serra R, Moses HL. pRb is necessary for inhibition of N-myc expression by TGF-beta-1 in embryonic lung organ cultures. Development. 1995;121:3057–3066. doi: 10.1242/dev.121.9.3057. [DOI] [PubMed] [Google Scholar]
- Bellusci S, Furuta Y, Rush MG, Handerson R, Winnier G, Hogan BL. Involvement of sonic hedgehog (Shh) in mouse embryonic lung growth and morphogenesis. Development. 1996;124:53–63. doi: 10.1242/dev.124.1.53. [DOI] [PubMed] [Google Scholar]
- Bellusci S, Grindley J, Emoto H, Itoh N, Hogan BL. Fibroblast growth factor-10 (FGF-10) and branching morphogenesis in the embryonic mouse lung. Development. 1997;124:4867–4878. doi: 10.1242/dev.124.23.4867. [DOI] [PubMed] [Google Scholar]
- Lebeche D, Malpel S, Cardoso WV. Fibroblast growth factor interactions in the developing lung. Mech Dev. 1999;86:125–136. doi: 10.1016/s0925-4773(99)00124-0. [DOI] [PubMed] [Google Scholar]
- Kaplan F. Molecular determinants of fetal lung organogenesis. Mol Genet Metab. 2000;71:321–341. doi: 10.1006/mgme.2000.3040. [DOI] [PubMed] [Google Scholar]
- Lü J, Izvolsky KI, Qian J, Cardoso WV. Identification and FGF-10 targets in the embryonic lung epithelium during bud morphogenesis. J Biol Chem. 2005;280:4834–4841. doi: 10.1074/jbc.M410714200. [DOI] [PubMed] [Google Scholar]
- Orr-Urtreger A, Bedford MT, Burakova T, Arman E, Zimmer Y. et al. Developmental localization of the splicing alternatives of fibroblast growth factor receptor-2 (FGFR-2) Dev Biol. 1993;158:475–486. doi: 10.1006/dbio.1993.1205. [DOI] [PubMed] [Google Scholar]
- Ramasamy SK, Mailleux AA, Gupte VV, Mata F, Sala FG. et al. FGF-10 dosage is critical for amplification of epithelial cells progenitors and for the formation of multiple mesenchymal lineages during lung development. Dev Biol. 2007;307:237–247. doi: 10.1016/j.ydbio.2007.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Morrisey EE, Whitsett JA. GATA-6 is required for maturation of the lung in late gestation. Am J Physiol Lung Cell Mol Physiol. 2002;283:L468–L475. doi: 10.1152/ajplung.00044.2002. [DOI] [PubMed] [Google Scholar]
- Ohbayashi N, Shibayama M, Kurotaki Y, Imanishi M, Fujimori T. et al. FGF-18 is required for normal cell proliferation and differentiation during osteogenesis and chondrogenesis. Genes Dev. 2002;16:870–879. doi: 10.1101/gad.965702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitsett JA, Clark JC, Picard L, Tichelaar JW, Wert SE. et al. Fibroblast growth factor 18 influences proximal programming during lung morphogenesis. J Biol Chem. 2002;277:22743–22749. doi: 10.1074/jbc.M202253200. [DOI] [PubMed] [Google Scholar]
- Nusse R, Varmus HE. Wnt genes. Cell. 1992;69:1073–1087. doi: 10.1016/0092-8674(92)90630-u. [DOI] [PubMed] [Google Scholar]
- Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- Gordon MD, Nusse R. Wnt signaling: multiple pathways, multiple receptors, and multiple transcription factors. J Biol Chem. 2006;281:22429–22433. doi: 10.1074/jbc.R600015200. [DOI] [PubMed] [Google Scholar]
- Ilyas M. Wnt signaling and the mechanistic basis of tumor development. J Pathol. 2005;205:130–144. doi: 10.1002/path.1692. [DOI] [PubMed] [Google Scholar]
- Bartscherer K, Boutros M. Regulation of Wnt protein secretion and its role in gradient formation. EMBO Rep. 2008;9:977–982. doi: 10.1038/embor.2008.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirams GR, Bryne M, King JR. A multiple time scale analysis of a mathematical model of the Wnt/β-catenin signaling pathway. J Math Biol. 2010;60:131–160. doi: 10.1007/s00285-009-0262-y. [DOI] [PubMed] [Google Scholar]
- Miller RK, McCrea PD. Wnt to build a tube: contributions of Wnt signaling to epithelial tubulogenesis. Dev Dyn. 2010;239:77–93. doi: 10.1002/dvdy.22059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A, Yamamoto H, Sakane H, Koyama H, Kikuchi A. Wnt-5a regulates distinct signaling pathways by binding to frizzled-2. EMBO J. 2010;29:41–54. doi: 10.1038/emboj.2009.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadigan KM, Nusse R. Wnt signaling: a common theme in animal development. Genes Dev. 1997;11:1106–1115. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- Wodarz A, Nusse R. Wnt signaling in development. Annu Rev Cell Dev Biol. 1998;14:59–88. doi: 10.1146/annurev.cellbio.14.1.59. [DOI] [PubMed] [Google Scholar]
- Moon RT. Wnt/β-catenin pathway. Sci STKE cm1. 2005; : . doi: 10.1126/stke.2712005cm1. [DOI] [PubMed] [Google Scholar]
- Weidenfeld J, Shu W, Zhang L, Millar SE, Morissey EE. Wnt-7b promoter is regulated by TTF-1, GATA6, and FOXA-2 in lung epithelium. J Biol Chem. 2002;277:21061–21070. doi: 10.1074/jbc.M111702200. [DOI] [PubMed] [Google Scholar]
- Wang Z, Shu W, Lu MM, Morissey EE. Wnt-7b activates canonical signaling in epithelial and vascular smooth muscle cells through interactions with Fzd-1, fzd-10, and LRP5. Mol Cell Biol. 2005;25:5022–5030. doi: 10.1128/MCB.25.12.5022-5030.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lako M, Stracha T, Bullen P, Wilson DI, Robson SC, Lindsay S. Isolation, characterization and embryonic expression of Wnt-11, a gene which maps to 11 g13.5 and has possible roles the development of skeleton, kidney, and lung. Gene. 1998;219:101–110. doi: 10.1016/s0378-1119(98)00393-x. [DOI] [PubMed] [Google Scholar]
- Miller LA, Wert SE, Clark JC, Xu Y, Perl AK, Whitsett JA. Role of Sonic hedgehog in patterning of tracheal-bronchial cartilage and the peripheral lung. Dev Dyn. 2004;231:57–71. doi: 10.1002/dvdy.20105. [DOI] [PubMed] [Google Scholar]
- Miller LA, Wert SE, Whitsett JA. Immunolocalization of sonic hedgehog (Shh) in developing mouse lung. J Histochem Cytochem. 2001;49:1593–1604. doi: 10.1177/002215540104901213. [DOI] [PubMed] [Google Scholar]
- Pepicelli CV, Lewis P, McMahon A. Sonic hedgehog regulates branching morphogenesis in the mammalian lung. Curr Biol. 1998;8:1083–1086. doi: 10.1016/s0960-9822(98)70446-4. [DOI] [PubMed] [Google Scholar]
- Hui CC, Slusarski D, Platt KA, Holmgren R, Joyner AL. Expression of three mouse homologs of the Drosophila segment polarity gene cubitus interruptus, GLI, GLI-2, and GLI-3, in ectoderm- and mesoderm-derived tissue suggests multiple roles during postimplantation development. Dev Biol. 1994;162:402–413. doi: 10.1006/dbio.1994.1097. [DOI] [PubMed] [Google Scholar]
- Grindley JC, Bellusci S, Perkins D, Hogan BL. Evidence for the involvement of the Gli gene family in embryonic mouse lung development. Dev Biol. 1997;188:337–348. doi: 10.1006/dbio.1997.8644. [DOI] [PubMed] [Google Scholar]
- Whitsett J. A lungful of transcription factors. Nat Genet. 1998;20:7–8. doi: 10.1038/1654. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Nishizaki Y, Hui C, Nakafuku M, Kondoh H. Regulation of Gli-2 and Gli-3 primary mediators of Shh signaling. Development. 1999;126:3915–3924. doi: 10.1242/dev.126.17.3915. [DOI] [PubMed] [Google Scholar]
- Goodrich LV, Johnson RL, Milenkovic L, McMahon JA, Scott MP. Conservation of the hedgehog/patched signaling pathway from flies to mice: induction of a mouse patched gene by hedgehog genes. Genes Dev. 1996;10:301–312. doi: 10.1101/gad.10.3.301. [DOI] [PubMed] [Google Scholar]
- Kawahira H, Ma NH, Tzanakakis ES, McMahon AP, Chuang PT, Hebrock M. Combined activities of hedgehog signaling inhibitors regulate pancreas development. Development. 2003;130:4871–4879. doi: 10.1242/dev.00653. [DOI] [PubMed] [Google Scholar]
- Chambon P. A decade of molecular biology of retinoic acid receptors. FASEB J. 1996;10:940–954. [PubMed] [Google Scholar]
- Marlétaz F, Holland LZ, Laudet V, Schubert M. Retinoic acid signaling and the evolution of chordates. Int J Biol Sci. 2006;2:38–47. doi: 10.7150/ijbs.2.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodosiou M, Laudet V, Schubert M. From carrot to clinic: an overview of the retinoic acid signaling pathway. Cell Mol Life Sci. 2010;67:1423–1445. doi: 10.1007/s00018-010-0268-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastien J, Rochette-Egly C. Nuclear retinoid receptors and transcription of retinoid-target genes. Gene. 2004;32:1–16. doi: 10.1016/j.gene.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Mollard R, Ghyselinck NB, Wendling O, Chambon P, Mark M. Stage-dependent responses of the developing lung to retinoic acid signaling. Int J Dev Biol. 2000;44:457–462. [PubMed] [Google Scholar]
- Cardoso WV, Williams MC, Mitsialis SA, Joyce-Brady M, Rishi AK, Brody JS. Retinoic acid induces changes in the pattern of airway branching and alters epithelial cell differentiation in the developing lung in vitro. Am J Respir Cell Mol Biol. 1995;12:464–476. doi: 10.1165/ajrcmb.12.5.7742011. [DOI] [PubMed] [Google Scholar]
- Malpel S, Mendelsohn C, Cardoso WV. Regulation of retinoic acid signaling during lung morphogenesis. Developent. 2000;127:3057–3067. doi: 10.1242/dev.127.14.3057. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Suzuki T, Kaneko C, Darnel AD, Moriya T. et al. Retinoid receptors in the developing human lung. Clin Sci. 2002;103:613–621. doi: 10.1042/cs1030613. [DOI] [PubMed] [Google Scholar]
- Mollard R, Viville S, Ward SJ, Decimo D, Chambon P, Delle P. Tissue-specific expression of retinoic acid receptor isoform transcripts in mouse embryo. Mech Dev. 2000;94:223–232. doi: 10.1016/s0925-4773(00)00303-8. [DOI] [PubMed] [Google Scholar]
- Wongtrakool C, Malpel S, Gerenstein J, Sedita J, Ramirez MI. et al. Down-regulation of retinoic acid receptor alpha signaling is required for sacculation and type-I cell formation in the developing lung. J Biol Chem. 2003;278:46911–46918. doi: 10.1074/jbc.M307977200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazaud C, Dolle P, Rossant J, Mollard R. Retinoic acid signaling regulates murine bronchial tubule formation. Mech Dev. 2003;120:691–700. doi: 10.1016/s0925-4773(03)00048-0. [DOI] [PubMed] [Google Scholar]
- Dirami G, Massaro GD, Clerch LB, Ryan US, Reczek PR, Massaro D. Lung retinol storing cells synthesize and secrete retinoic acid, an inducer of alveolus formation. Am J Physiol Lung Cell Mol Physiol. 2004;286:249–256. doi: 10.1152/ajplung.00140.2003. [DOI] [PubMed] [Google Scholar]
- Massaro GD, Massaro D, Chan WY, Clerch LB, Ghyselinck N. et al. Retinoic acid receptor- beta: an endogenous inhibitor of the perinatal formation of pulmonary alveoli. Physiol Genomics. 2000;4:51–57. doi: 10.1152/physiolgenomics.2000.4.1.51. [DOI] [PubMed] [Google Scholar]
- Yang L, Naltner A, Yan C. Overexpression of dominant negative retinoic acid receptor alpha causes alveolar abnormality in transgenic neonatal lungs. Endocrinology. 2003;144:3004–3011. doi: 10.1210/en.2002-0121. [DOI] [PubMed] [Google Scholar]
- Massaro GD, Massaro D, Chambon P. Retinoic acid receptor-alpha regulates pulmonary alveolus formation in mice after, but not during, perinatal period. Am J Physiol Lung Cell Mol Physiol. 2003;284:431–433. doi: 10.1152/ajplung.00245.2002. [DOI] [PubMed] [Google Scholar]
- Leblond CP, Inoue S. Structure, composistion, and assembly of basement membrane. Am J Anat. 1989;185:367–390. doi: 10.1002/aja.1001850403. [DOI] [PubMed] [Google Scholar]
- Crouch EC, Martin GR, Brody JS, Laurie GW. In: The lung: scientific foundations, 2nd edtn. Crystal RD, West JB, Weibel ER, Barnes PJ, editor. Lippincott-Raven Publishers, Philadelphia; 1997. Basement membranes; pp. 769–791. [Google Scholar]
- Hass TL. Endothelial cell regulation of matrix metalloproteinases. Can J Physiol Pharmacol. 2005;83:1–7. doi: 10.1139/y04-120. [DOI] [PubMed] [Google Scholar]
- Hilfer SR. Morphogenesis of the lung: control of embryonic and fetal branching. Annu Rev Physiol. 1996;58:93–113. doi: 10.1146/annurev.ph.58.030196.000521. [DOI] [PubMed] [Google Scholar]
- Wendel DP, Taylor DG, Albertine KH, Keating MT, Li DY. Impaired distal airway development in mice lacking elastin. Am J Respir Cell Mol Biol. 2000;23:320–326. doi: 10.1165/ajrcmb.23.3.3906. [DOI] [PubMed] [Google Scholar]
- Burri PH, Weibel ER. In: Development of the lung. Hodson WA, editor. Marcel Dekker Inc, New York; 1977. Ultrastructure and morphometry of the developing lung; pp. 215–268. [Google Scholar]
- Zhao Y, Young SL. Tenascin in rat lung development: in situ localization and cellular sources. Am J Physiol. 1995;269:482–491. doi: 10.1152/ajplung.1995.269.4.L482. [DOI] [PubMed] [Google Scholar]
- Kaarteenaho-Wiik R, Kinnula V, Herva R, Pääkkö P, Pöllänen R, Soini Y. Distribution and mRNA expression of tenascin-C in developing human lung. Am J Respir Cell Mol Biol. 2001;25:341–346. doi: 10.1165/ajrcmb.25.3.4460. [DOI] [PubMed] [Google Scholar]
- Snyder JM, O’Brien JA, Rodgers HF. Localization and accumulation of fibronectin in rabbit fetal lung tissue. Differentation. 1987;34:32–39. doi: 10.1111/j.1432-0436.1987.tb00048.x. [DOI] [PubMed] [Google Scholar]
- Roman J, McDonald JA. Expression of fibronectin, the integrin alpha-5, and alpha-smooth muscle actin in the heart, and lung development. Am J Respir Cell Mol Biol. 1992;6:472–480. doi: 10.1165/ajrcmb/6.5.472. [DOI] [PubMed] [Google Scholar]
- Sakai T, Larsen M, Yamada KM. Fibronectin requirement in branching morphogenesis. Nature, Lond. 2003;423:876–881. doi: 10.1038/nature01712. [DOI] [PubMed] [Google Scholar]
- Roman J, Crouch EC, McDonald JA. Reagents that inhibit fibronectin matrix assembly of cultured cells also inhibit lung branching morphogenesis in vitro. Implications for lung development, injury, and repair. Chest. 1991;99(Suppl):20–21. [PubMed] [Google Scholar]
- Schuger L, O’Shea KS, Rheinheimer J, Varani J. Laminin in lung development: effects of anti-laminin antibody in murine lung morphogenesis. Dev Biol. 1990;137:26–32. doi: 10.1016/0012-1606(90)90004-3. [DOI] [PubMed] [Google Scholar]
- Coraux C, Meneguzzi G, Rousselle P, Puchelle E, Gaillard D. Distribution of laminin-5, integrin receptors, and branching morphogenesis during human fetal lung development. Dev Dyn. 2002;225:176–185. doi: 10.1002/dvdy.10147. [DOI] [PubMed] [Google Scholar]
- Gerritsen ME, Soriano R, Yang S, Zlot C, Ingle G. et al. Branching out: a molecular fingerprint of endothelial differentiation into tube-like structures generated by affymetrix oligonucleotide arrays. Microcirculation. 2003;10:63–81. doi: 10.1038/sj.mn.7800170. [DOI] [PubMed] [Google Scholar]
- Gerhardt H, Betsholtz C. How do endothelia cells orientate? EXS. 2005;94:3–15. doi: 10.1007/3-7643-7311-3_1. [DOI] [PubMed] [Google Scholar]
- Gerhardt H. VEGF and endothelial guidance in angiogenic sprouting. Organogenesis. 2008;4:241–246. doi: 10.4161/org.4.4.7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Moral PM, Sala FG, Tefft D, Shi W, Keshet E. et al. VEGF-A signaling through Flk-1 is a critical facilitator of early embryonic lung epithelial to endothelial cross-talk and branching morphogenesis. Dev Biol. 2006;290:177–188. doi: 10.1016/j.ydbio.2005.11.022. [DOI] [PubMed] [Google Scholar]
- Voelkel NF, Vandvier RW, Tuder RM. Vascular endothelial growth factor in the lung. Am J Physiol Lung Cell Mol Physiol. 2006;290:L209–L221. doi: 10.1152/ajplung.00185.2005. [DOI] [PubMed] [Google Scholar]
- Taichman DB, Loomes KM, Schachtner SK, Guttentag S, Vu C. et al. Notch-1 and jagged- 1 expression by the developing pulmonary vasculature. Dev Dyn. 2002;225:166–175. doi: 10.1002/dvdy.10146. [DOI] [PubMed] [Google Scholar]
- Ferrara N. Vascular endothelial growth factor and the regulation of angiogenesis. Recent Prog Horm Res. 2000;55:15–35. [PubMed] [Google Scholar]
- D’Angio CT, Maniscalco WM. The role of vascular growth factors in hyperoxia-induced injury to the developing lung. Front Biosci. 2002;7:1609–1623. doi: 10.2741/A865. [DOI] [PubMed] [Google Scholar]
- Dumont DJ, Gradwohl G, Fong GH, Puri MC, Gertsenstein M. et al. Dominant-negative and targeted null mutations in the endothelial receptor tyrosine kinase, tek, reveal a critical role in vasculogenesis of the embryo. Genes Dev. 1994;8:1897–1909. doi: 10.1101/gad.8.16.1897. [DOI] [PubMed] [Google Scholar]
- Hall SM, Hislop AA, Harworth SG. Origin, differentiation, and maturation of human pulmonary veins. Am J Respir Cell Mol Biol. 2002;26:333–340. doi: 10.1165/ajrcmb.26.3.4698. [DOI] [PubMed] [Google Scholar]
- Shigetomo F, Sako K, Noda K, Zhang J, Minami M, Mochizuki N. Angiopoietin-1/Tie2 receptor signaling in vascular quiescence and angiogenesis. Histol Histopathol. 2010;25:387–396. doi: 10.14670/HH-25.387. [DOI] [PubMed] [Google Scholar]
- Maina JN. A scanning and transmission electron microscopic study of the tracheal air-sac system in a grasshopper (Chrotogonus senegalensis, Kraus)- (Orthoptera: Acrididae: Pygomorphinae) Anat Rec. 1989;223:393–405. doi: 10.1002/ar.1092230408. [DOI] [PubMed] [Google Scholar]
- Fong GH, Rossant J, Gertsenstein M, Breitman BL. Role of the flt-1 receptor tyrosine kinase in regulating the assembly of vascular endothelium. Nature, Lond. 1995;376:66–70. doi: 10.1038/376066a0. [DOI] [PubMed] [Google Scholar]
- Ferrara N, Carver-Moore K, Chen H, Dowd M, Lu L. et al. Heterozygous embryonic lethality induced by targeted inactivation of the VEGF gene. Nature, Lond. 1996;380:439–442. doi: 10.1038/380439a0. [DOI] [PubMed] [Google Scholar]
- Shalaby F, Ho J, Stanford WL, Fischer KD, Schuh AC. et al. A requirement for flk-1 in primitive and definitive hematopoiesis and vasculogenesis. Cell. 1997;89:981–990. doi: 10.1016/s0092-8674(00)80283-4. [DOI] [PubMed] [Google Scholar]
- Ferrara N. Molecular and biological properties of vascular endothelial growth factor. J Mol Med. 1999;77:527–543. doi: 10.1007/s001099900019. [DOI] [PubMed] [Google Scholar]
- Ferrara N, Houck K, Jakeman L, Leung DW. Molecular and biological properties of the vascular endothelial growth factor family of proteins. Endocr Rev. 1992;13:18–32. doi: 10.1210/edrv-13-1-18. [DOI] [PubMed] [Google Scholar]
- Shima DT, Kuroka M, Deustsch U, Ng YS, Adamis AP, D’Amore PA. The mouse gene for vascular endothelial growth factor: genomic structure, definition of the transcriptional unit and characterization of transcriptional and post-transcriptional regulatory sequences. J Biol Chem. 1996;271:3877–3883. doi: 10.1074/jbc.271.7.3877. [DOI] [PubMed] [Google Scholar]
- Larrivée B, Karston A. Signaling pathways induced by vascular endothelial growth factor. Int J Mol Med. 2000;5:447–545. doi: 10.3892/ijmm.5.5.447. [DOI] [PubMed] [Google Scholar]
- Matsumoto T, Claesson-Welsh L. VEGF receptor signal transduction. Sci STKE. 2001;112:RE21. doi: 10.1126/stke.2001.112.re21. [DOI] [PubMed] [Google Scholar]
- Park JE, Keller GA, Ferrara N. The vascular endothelial growth factor (VEGF) isoforms: Differential deposition in the subepithelial extracellular matrix and bioactivity of extracellular matrix bound VEGF. Mol Biol Cell. 1993;4:1317–1326. doi: 10.1091/mbc.4.12.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrara N, Davis-Smyth T. The biology of vascular endothelial growth factor. Endocr Rev. 1997;18:4–25. doi: 10.1210/edrv.18.1.0287. [DOI] [PubMed] [Google Scholar]
- Ng YS, Rohan R, Sunday ME, Demello DE, D’Amore PA. Differential expression of VEGF isoforms in mouse during development and in the adult. Dev Dyn. 2001;220:112–121. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1093>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Tomanek RJ, Holifield JS, Reiter RS, Sandra A, Lin JJC. Role of VEGF family members and receptors in coronary vessel formation. Dev Dyn. 2002;225:233–240. doi: 10.1002/dvdy.10158. [DOI] [PubMed] [Google Scholar]
- Hamada K, Oike Y, Takakura N, Ito Y, Jussila L, Dumont DJ, Alitalo K, Suda T. VEGF-C signaling pathways through VEGFR-2 and VEGFR-3 in vasculo-angiogenesis and hematopoiesis. Blood. 2010;96:3793–3800. [PubMed] [Google Scholar]
- Carmeliet P, Ferreira V, Breier G, Pollefeyt S, Kieckens L. et al. Abnormal bloodvessel development and lethality in embryos lacking a single VEGF allele. Nature, Lond. 1996;380:435–439. doi: 10.1038/380435a0. [DOI] [PubMed] [Google Scholar]
- Acarregui MJ, Penisten ST, Goss KL, Ramirez K, Snyder JM. Vascular endothelial growth factor gene expression in human fetal lung in vitro. Am J Respir Cell Mol Biol. 1999;20:14–23. doi: 10.1165/ajrcmb.20.1.3251. [DOI] [PubMed] [Google Scholar]
- Breier G, Albrecht U, Sterrer S, Risau W. Expression of vascular endothelial growth factor during embryonic angiogenesis and endothelial cell differentiation. Development. 1992;114:521–532. doi: 10.1242/dev.114.2.521. [DOI] [PubMed] [Google Scholar]
- Risau W. Mechanisms of angiogenesis. Nature, Lond. 1997;386:671–674. doi: 10.1038/386671a0. [DOI] [PubMed] [Google Scholar]
- Kane RJ, Crystal RG. Compartmentalization of vascular endothelial growth factor to the epithelial surface of the lung. Mol Med. 2001;7:240–246. [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi TP, Dumont DJ, Conlon RA, Breitman MI, Rossant J. Flk-1, and flt-1-related receptor tyrosine kinase is an early marker for endothelial cell precursors. Development. 1993;111:489–498. doi: 10.1242/dev.118.2.489. [DOI] [PubMed] [Google Scholar]
- Soker S, Gollamudi-Payne S, Fidder H, Charmaheli H, Klagsbrun M. Inhibition of vascular endothelial growth factor (VEGF) induced endothelial cell proliferation by a peptide corresponding to the exon 7-encoded domain of VEGF165. J Biol Chem. 1997;272:31582–31588. doi: 10.1074/jbc.272.50.31582. [DOI] [PubMed] [Google Scholar]
- Neufeld G, Cohen T, Gengrinovitch S, Poltorak Z. Vascular endothelial growth factor VEGF and its receptors. FASEB J. 1999;13:9–22. [PubMed] [Google Scholar]
- Burri PH, Hlushchuk R, Djonov V. Intussusceptive angiogenesis: its emergence, its characteristics, and its significance. Dev Dyn. 2004;231:474–488. doi: 10.1002/dvdy.20184. [DOI] [PubMed] [Google Scholar]
- Makanya AN, Stauffer D, Ribatti D, Burri PH, Djonov V. Microvascular growth, development, and remodelling in the embryonic avian kidney: interplay between sprouting and intussusceptive angiogenic mechanisms. Microsc Res Tech. 2005;66:275–288. doi: 10.1002/jemt.20169. [DOI] [PubMed] [Google Scholar]
- Berse B, Brown LF, van de Water L, Dvorak HF, Senger DR. Vascular permeability factor (vascular endothelial growth factor) gene is expressed differentially in normal tissues, macrophages, and tumours. Mol Biol Cell. 1992;3:211–220. doi: 10.1091/mbc.3.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monacci WT, Merrill MJ, Oldfield EH. Expression of vascular permeability factor/vascular endothelial growth factor in normal rat tissues. Am J Physiol. 1993;264:C995–C1002. doi: 10.1152/ajpcell.1993.264.4.C995. [DOI] [PubMed] [Google Scholar]
- Tuder RM, Flook BE, Voelkel NF. Increased gene expression of VEGF and the VEGF receptors KDR/Flk and Flt in lungs exposed to acute or to chronic hypoxia: modulation of gene expression by nitric oxide. J Clin Invest. 1995;95:1798–1807. doi: 10.1172/JCI117858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt AJ, Amin SB, Chess PR, Watkins RH, Maniscalco WM. Expression of vascular endothelial factor and Flk-1 in developing and glucocorticoid-treated mouse lung. Pediatr Res. 2000;47:606–613. doi: 10.1203/00006450-200005000-00009. [DOI] [PubMed] [Google Scholar]
- De Mello DE, Sawyer D, Galvin N, Reid LM. Early fetal development of lung vasculature. Am J Respir Cell Mol Biol. 1997;16:568–581. doi: 10.1165/ajrcmb.16.5.9160839. [DOI] [PubMed] [Google Scholar]
- Schachtner SK, Wang Y, Scott-Baldwin H. Qualitative and quantitative analysis of embryonic pulmonary vessel formation. Am J Respir Cell Mol Biol. 2000;13:328–335. doi: 10.1165/ajrcmb.22.2.3766. [DOI] [PubMed] [Google Scholar]
- Greenberg JM, Thompson FY, Brooks SK, Shannon JM, McCormick-Shannon K. et al. Mesenchymal expression of vascular endothelial growth factors D and A defines vascular patterning in developing lung. Dev Dyn. 2002;224:144–153. doi: 10.1002/dvdy.10095. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Yun EJ, Gerber HP, Ferrara N, Whitsett JA, Vu TH. Epithelial-vascular cross talk mediated by VEGF-A and HGF signaling directs primary septae formation during distal lung morphogenensis. Dev Biol. 2007;308:44–53. doi: 10.1016/j.ydbio.2007.04.042. [DOI] [PubMed] [Google Scholar]
- Millauer B, Wizigmann-Voos S, Schnurch H, Martinez R, Moller NP. et al. High affinity EGF binding and developmental expression suggest Flk-1 as a major regulator of vasculogenesis and angiogenesis. Cell. 1993;72:835–846. doi: 10.1016/0092-8674(93)90573-9. [DOI] [PubMed] [Google Scholar]
- Akeson AL, Greenberg JM, Cameron JE, Thompson FY, Brooks SK. et al. Temporal and spatial regulation of VEGF-A controls vascular patterning in the embryonic lung. Dev Biol. 2003;264:443–455. doi: 10.1016/j.ydbio.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Maniscalco WM, Watkins RH, Finkelstein JN, Campbell MH. Vascular endothelial growth factor mRNA increases in alveolar epithelial cells during recovery from oxygen injury. Am J Respir Cell Mol Biol. 1995;13:377–386. doi: 10.1165/ajrcmb.13.4.7546767. [DOI] [PubMed] [Google Scholar]
- Lazarus A, Del-Moral PM, Ilovich O, Mishani E, Warburton D, Keshet E. A perfusion- independent role of blood vessels determining branching stereotypy of lung airways. Development. 2011;138:2359–2368. doi: 10.1242/dev.060723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldin CM, Westermark B. Mechanism of action and in vivo role of platelet-derived growth factor. Physiol Rev. 1999;79:1283–1316. doi: 10.1152/physrev.1999.79.4.1283. [DOI] [PubMed] [Google Scholar]
- Betsholtz C, Karlson L, Lindahl P. Developmental roles of platelet-derived growth factors. Bioessays. 2001;23:57–71. doi: 10.1002/bies.1069. [DOI] [PubMed] [Google Scholar]
- Han RN, Liu J, Tanswell AK, Post M. Ontogeny of platelet-derived growth factor receptor in fetal rat lung. Microsc Res Tech. 1993;26:381–388. doi: 10.1002/jemt.1070260506. [DOI] [PubMed] [Google Scholar]
- Boström H, Willetts K, Pekny M, Levéen P, Lindahl P. et al. PDGF-A signaling is a critical event in lung alveolar myofibroblast development and alveogenesis. Cell. 1996;85:863–873. doi: 10.1016/s0092-8674(00)81270-2. [DOI] [PubMed] [Google Scholar]
- Lindahl P, Karlsson L, Hellstrom M, Gebre-Medhin S, Willets K. et al. Alveogenesis failure in PDGF-A-deficient mice is coupled to lack of distal spreading of alveolar smooth muscle cell progenitors during lung development. Development. 1997;124:3943–3953. doi: 10.1242/dev.124.20.3943. [DOI] [PubMed] [Google Scholar]
- Ikeda K, Shaw-White JR, Wert SE, Whitsett JA. Hepatocyte nuclear factor-3 activates transcription of thyroid transcription factor-1 in respiratory epithelial cells. Mol Cell Biol. 1996;16:3626–3636. doi: 10.1128/mcb.16.7.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw-White JR, Bruno MD, Whitsett JA. GATA-6 activates transcription of thyroid transcription factor-1. J Biol Chem. 1999;274:2658–2664. doi: 10.1074/jbc.274.5.2658. [DOI] [PubMed] [Google Scholar]
- Tichelaar JW, Lim L, Costa RH, Whitsett JA. HNF-3/forkhead homologue-4 influences lung morphogenesis and respiratory epithelial cell differentiation in vivo. Dev Biol. 1999;213:405–417. doi: 10.1006/dbio.1999.9380. [DOI] [PubMed] [Google Scholar]
- Patient RK, McGhee JD. The GATA family (vertebrates and invertebrates) Curr Opin Genet Dev. 2002;12:416–422. doi: 10.1016/s0959-437x(02)00319-2. [DOI] [PubMed] [Google Scholar]
- Costa RH, Kalinichenko VV, Lim L. Transcription factors in mouse lung development and function. Am J Physiol Lung Cell Mol Physiol. 2001;280:L823–L838. doi: 10.1152/ajplung.2001.280.5.L823. [DOI] [PubMed] [Google Scholar]
- Zhou L, Dey CR, Wert SE, Yan C, Costa RH, Whitsett JA. Hepatocyte nuclear factor-3 beta limits cellular diversity in the developing respiratory epithelium and alters lung morphogenesis in vivo. Dev Dyn. 1997;210:305–314. doi: 10.1002/(SICI)1097-0177(199711)210:3<305::AID-AJA10>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Morrisey EE, Ip HS, Lu MM, Parmacek MS. GATA-6: a zinc finger transcription factor that is expressed in multiple cell lineages derived from lateral mesoderm. Dev Biol. 1996;177:309–322. doi: 10.1006/dbio.1996.0165. [DOI] [PubMed] [Google Scholar]
- Morrisey EE, Ip HS, Lu MM, Parmacek MS. GATA-5: a transcriptional activator expressed in a novel temporally-restricted pattern during embryonic development. Dev Biol. 1997;183:21–36. doi: 10.1006/dbio.1996.8485. [DOI] [PubMed] [Google Scholar]
- Liu C, Ikegami M, Stahlman MT, Dey CR, Whitsett JA. Inhibition of alveolarization and altered pulmonary mechanics in mice expressing GATA-6. Am J Physiol Lung Cell Mol Physiol. 2003;285:1246–1254. doi: 10.1152/ajplung.00443.2002. [DOI] [PubMed] [Google Scholar]
- Bruno MD, Korfhagen TR, Liu C, Morrisey EE, Whitsett JA. GATA-6 activates transcription of surfactant protein A. J Biol Chem. 2000;275:1043–1049. doi: 10.1074/jbc.275.2.1043. [DOI] [PubMed] [Google Scholar]
- Liu Z, Xu J, Colvin JS, Ornitz DM. Coordination of chondrogenesis and osteogenesis by fibroblast growth factor-18. Genes Dev. 2002;16:859–869. doi: 10.1101/gad.965602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keijer R, van Tuyl M, Meijers C, Post M, Tibboel D, Grosveld F, Koutsourakis M. The transcription factor GATA-6 is essential for branching morphogenesis and epithelial cell differentiation during fetal pulmonary development. Development. 2001;128:503–511. doi: 10.1242/dev.128.4.503. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Goss AM, Cohen ED, Kadzik R, Lepore JJ, Muthukumaraswamy K, Yang J. et al. A GATA-6-Wnt pathway required for epithelial stem cell development and airway regeneration. Nat Genet. 2008;40:862–870. doi: 10.1038/ng.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggaram V. Thyroid transcription factor-1 (TTF-1/Nkx2.1/TITF1) gene regulation in the lung. Clin Sci. 2009;116:27–35. doi: 10.1042/CS20080068. [DOI] [PubMed] [Google Scholar]
- Koutsourakis M, Langeveld A, Patient R, Beddington R, Grosveld F. The transcription factor GATA-6 is essential for early extraembryonic development. Development. 1999;126:723–732. [PubMed] [Google Scholar]
- Radtke F, Fasnacht N, Macdonald HR. Notch signaling in the immune syatem. Immunity. 2010;32:14–27. doi: 10.1016/j.immuni.2010.01.004. [DOI] [PubMed] [Google Scholar]
- Kong Y, Glickman J, Subramaniam M, Shahsafaei A, Allameni KP. et al. Functional diversity of notch family genes in fetal lung development. Am J Physiol Lung Cell Mol Physiol. 2004;286:L1075–L1083. doi: 10.1152/ajplung.00438.2002. [DOI] [PubMed] [Google Scholar]
- Guazzi S, Price M, Damante FMG, Mattei MG, Di Lauro R. Thyroid nuclear factor-1 (TTF-1) contains a homeodomain and displays a novel DNA binding specificity. EMBO J. 1990;9:3631–3639. doi: 10.1002/j.1460-2075.1990.tb07574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaro D, Price M, de Felice M, Di Lauro R. The transcription factor TTF-1 is expressed at the onset of thyroid and lung morphogenesis and in restricted regions of the foetal brain. Development. 1991;113:1093–1104. doi: 10.1242/dev.113.4.1093. [DOI] [PubMed] [Google Scholar]
- Wert SE, Dey CR, Blair PA, Kimura S, Whitsett JA. Increased expression of thyroid transcription factor-1 (TTF-1) in respiratory epithelial cells inhibits alveolarization and causes pulmonary inflammation. Dev Biol. 2002;242:75–87. doi: 10.1006/dbio.2001.0540. [DOI] [PubMed] [Google Scholar]
- Bogue CW, Lou LJ, Vasavada H, Wilson CM, Jacobs HC. Expression of Hoxb genes in the developing mouse foregut and lung. Am J Respir Cell Biol. 1996;15:163–171. doi: 10.1165/ajrcmb.15.2.8703472. [DOI] [PubMed] [Google Scholar]
- Volpe MV, Martin A, Vosatka RJ, Mazzoni CL, Nielsen HC. Hoxb-5 expression in the developing mouse lung suggests a role in branching morphogenesis and epithelial cell fate. Histochem Cell Biol. 1997;108:495–504. doi: 10.1007/s004180050190. [DOI] [PubMed] [Google Scholar]
- Boucher CA, Carey N, Edwards YH, Siciliano MJ, Johnson KJ. Cloning of the human Six-1 gene and its assignment to chromosome 14. Genomics. 1996;33:140–142. doi: 10.1006/geno.1996.0172. [DOI] [PubMed] [Google Scholar]
- Ford HL, Landesman-Bollag E, Dacwag CS, Stukenerg PT, Pardee AB, Seldin DC. Cell cycle- regulated phosphorylation of the human Six-1 homeodomain protein. J Biol Chem. 2000;275:22245–22254. doi: 10.1074/jbc.M002446200. [DOI] [PubMed] [Google Scholar]
- El-Hashash AH, Al Alam D, Turcatel G, Rogers O, Li X. et al. Six1 transcription factor is critical for coordination of epithelial, mesenchymal and vascular morphogenesis in the mammalian lung. Dev Biol. 2011;353:242–258. doi: 10.1016/j.ydbio.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maina JN. Development, structure and function of a novel respiratory organ, the lung-air sac system of birds: to go where no other vertebrate has gone. Biol Rev. 2006;81:545–579. doi: 10.1017/S1464793106007111. [DOI] [PubMed] [Google Scholar]
- Maina JN. Functional morphology of the avian respiratory system, the lung-air sac system: efficiency built on complexity. Ostrich. 2008;79:117–132. [Google Scholar]
- McLelland J. In: Form and function in birds. King AS, McLelland J, editor. Academic, London; 1989. Anatomy of the lungs and air sacs; pp. 221–279. [Google Scholar]
- Scheid P. Mechanisms of gas exchange in bird lungs. Rev Physiol Biochem Pharmacol. 1979;86:137–186. doi: 10.1007/BFb0031533. [DOI] [PubMed] [Google Scholar]
- Seller TJ. Bird respiration. I and II. CRC Press, Boca Raton; 1987. [Google Scholar]
- Powell FL, Scheid P. In: Form and function of the avian lung. King AS, McLelland J, editor. Academic, London; 1989. Physiology of gas exchange in the avian respiratory system; pp. 393–437. [Google Scholar]
- Fedde MR. Relationship of structure and function of the avian respiratory system to disease susceptibility. Poult Sci. 1998;77:1130–1138. doi: 10.1093/ps/77.8.1130. [DOI] [PubMed] [Google Scholar]
- Coitier V. Anatomia avium. , Nuremberg, Germany; 1573. [Google Scholar]
- Goldin GV, Opperman LA. Induction of supernumerary tracheal buds and the stimulation of DNA synthesis in the embryonic chick lung and trachea by epidermal growth factor. J Embryol Exp Morphol. 1980;60:235–243. [PubMed] [Google Scholar]
- Chen WT, Chen JM, Mueller SC. Coupled expression and colocalization of 140 K cell adhesion molecules, fibronectin, and laminin during morphogenesis and cytodifferentiation of chick lung cells. J Cell Biol. 1986;103:1073–1090. doi: 10.1083/jcb.103.3.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallanger BC. Branching morphogenesis in the avian lung: electron microscopic studies using cataionic dyes. J Embryol Exp Morphol. 1986;94:189–205. [PubMed] [Google Scholar]
- Muraoka RS, Bushdid PB, Brantley DM, Yull FE, Kerr LD. Mesenchymal expression of nuclear factor-kappaβ inhibits epithelial growth and branching in the embryonic chick lung. Dev Biol. 2000;225:322–338. doi: 10.1006/dbio.2000.9824. [DOI] [PubMed] [Google Scholar]
- Sakiyama J-I, Yamagishi A, Kuroiwa A. Tbx-Fgf-10 system controls lung bud formation during chicken embryonic development. Development. 2003;130:1225–1234. doi: 10.1242/dev.00345. [DOI] [PubMed] [Google Scholar]
- Stabellini G, Locci P, Calvitti M, Evangelisti R, Marinucci L, Bodo M. et al. Epithelial- mesenchymal interactions and lung branching morphogenesis, role of polyamines and transforming growth factor beta-1. Eur J Histochem. 2001;45:151–162. [PubMed] [Google Scholar]
- Sakiyama J, Yokouchi Y, Kuroiwa A. Coordinated expression of Hoxb genes and signaling molecules during development of the chick. Dev Biol. 2000;227:12–27. doi: 10.1006/dbio.2000.9880. [DOI] [PubMed] [Google Scholar]
- Maina JN. Developmental dynamics of the bronchial (airway)- and air sac systems of the avian respiratory system from days 3 to 26 of life: A scanning electron microscopic study of the domestic fowl, Gallus gallus variant domesticus. Anat Embryol. 2003;207:119–134. doi: 10.1007/s00429-003-0333-6. [DOI] [PubMed] [Google Scholar]
- Maina JN. A systematic study of hematopoiesis, vasculogenesis, and angiogensis in the developing avian lung, Gallus gallus variant domesticus. Tissue Cell. 2004;36:307–322. doi: 10.1016/j.tice.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Miura T, Hartmann D, Kinboshi M, Komada M, Ishibashi M, Shiota K. The cyst-branch difference in developing chick lung results from a different morphogen diffusion coefficient. Mech Dev. 2009;126:160–172. doi: 10.1016/j.mod.2008.11.006. [DOI] [PubMed] [Google Scholar]
- Meyer EP. Corrosion casts as a method for investigation of the insect tracheal sytem. Cell Tissue Res. 1989;256:1–6. [Google Scholar]
- Steen JB. Comparative physiology of respiratory mechanisms. Academic, London; 1971. [Google Scholar]
- Romero NM, Dekanty A, Wappner P. Cellular and developmental adaptations to hypoxia:a Drosophila perspective. Methods Enzymol. 2007;435:123–144. doi: 10.1016/S0076-6879(07)35007-6. [DOI] [PubMed] [Google Scholar]
- Kaiser A, Klok CJ, Socha JJ, Lee WK, Quinlan MC, Harrison JF. Increase in tracheal investment with beetle size supports hypothesis of oxygen limitation on insect gigantism. Proc Natl Acad Sci, USA. 2007;104:13198–13203. doi: 10.1073/pnas.0611544104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S, Chavez C, Andrew DJ. Trachealess (Trh) regulates all tracheal genes during Drosophila embryogenesis. Dev Biol. 2011;360:160–172. doi: 10.1016/j.ydbio.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centanin L, Gorr TA, Wappner P. Tracheal remodeling in response to hypoxia. J Insect Physiol. 2010;56:447–454. doi: 10.1016/j.jinsphys.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning G, Krasnow MA. In: The development of Drosophila melanogaster. Bate M, Martinez-Arias A, editor. Cold Spring Harbor Press (Cold Spring Harbor), New York; 1993. Development of the Drosophila tracheal system; pp. 609–685. [Google Scholar]
- Samakovlis C, Hacohen N, Manning G, Sutherland DC, Guillemin K, Krasnow MA. Development of the Drosophila tracheal system occurs by a series of morphologically distinct but genetically coupled branching events. Development. 1996;122:1395–1407. doi: 10.1242/dev.122.5.1395. [DOI] [PubMed] [Google Scholar]
- Samakovlis C, Manning G, Steneberg P, Hacohen N, Cantera R, Krasnow MA. Genetic control of epithelial tube fusion during Drosophila tracheal development. Development. 1996b;122:3531–3531. doi: 10.1242/dev.122.11.3531. [DOI] [PubMed] [Google Scholar]
- Wilk R, Weizman I, Shilo BZ. Trachealess encodes a bHLH-PAS protein that is an inducer of tracheal cell fates in Drosophila. Genes Dev. 1996;10:93–102. doi: 10.1101/gad.10.1.93. [DOI] [PubMed] [Google Scholar]
- Zhou S, Degan S, Potts EN, Forster WM, Sunday M. NPAS3 is a trachealess homolog critical for lung development and homeostasis. Proc Natl Acad Sci, USA. 2009;106:11691–11696. doi: 10.1073/pnas.0902426106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Issac DD, Andrew DJ. Tubulogenesis in Drosophila: a requirement for the tracheates gene product. Genes Dev. 1996;10:103–117. doi: 10.1101/gad.10.1.103. [DOI] [PubMed] [Google Scholar]
- Vincent S, Ruberte E, Grieder NC, Chen CK, Haerry T. et al. DPP controls tracheal cell migration along the dorsoventral body axis of the Drosophila embryo. Development. 1997;124:2741–2750. doi: 10.1242/dev.124.14.2741. [DOI] [PubMed] [Google Scholar]
- Wappner P, Gabay L, Shilo BZ. Interactions between the EGF receptor and DPP pathways establish distinct cell fates in the tracheal placodes. Development. 1997;124:4707–4716. doi: 10.1242/dev.124.22.4707. [DOI] [PubMed] [Google Scholar]
- Reichman-Fried M, Dickson B, Hafen E, Shilo BZ. Elucidation of the role of Breathless, a Drosophila FGF receptor homolog, in tracheal cell migration. Genes Dev. 1994;8:428–439. doi: 10.1101/gad.8.4.428. [DOI] [PubMed] [Google Scholar]
- Reichman-Fried M, Shilo BZ. Breathless, a Drosophila FGF receptor homolog, is required for the insect of tracheal cell migration and tracheole formation. Mech Dev. 1995;52:265–273. doi: 10.1016/0925-4773(95)00407-r. [DOI] [PubMed] [Google Scholar]
- Sutherland D, Samakovlis C, Krasnow MA. Branchless encodes a Drosophila FGF homolog that controls tracheal cell migration and the pattern of branching. Cell. 1996;87:1091–1101. doi: 10.1016/s0092-8674(00)81803-6. [DOI] [PubMed] [Google Scholar]
- Hacohen N, Kramer S, Sutherland D, Hiromi Y, Krasnow M. Sprouty encodes a novel antagonist of FGF signaling that patterns apical branching of the Drosophila airways. Cell. 1998;92:253–263. doi: 10.1016/s0092-8674(00)80919-8. [DOI] [PubMed] [Google Scholar]
- Sato M, Kornberg TB. FGF is an essential mitogen and chemoattractant fo the air sacs of the Drosophila tracheal system. Dev Cell. 2002;3:195–207. doi: 10.1016/s1534-5807(02)00202-2. [DOI] [PubMed] [Google Scholar]
- Ribeiro C, Neumann M, Affolter M. Genetic control of cell intercalation during tracheal morphogenesis in Drosophila. Curr Biol. 2004;14:2197–2207. doi: 10.1016/j.cub.2004.11.056. [DOI] [PubMed] [Google Scholar]
- Myat MM, Lightfoot H, Wang P, Andrew DJ. A molecular link between FGF and Dpp signaling in branch-specific migration of the Drosophila trachea. Dev Biol. 2005;281:38–52. doi: 10.1016/j.ydbio.2005.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl M, Schuh R, Adryan B. Identification of FGF-dependent genes in the Drosophila tracheal system. Gene Expr Patterns. 2007;1-2:202–209. doi: 10.1016/j.modgep.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Weaver M, Krasnow MA. Dual origin of tissue-specific progenitor cells in Drosophila tracheal remodelling. Science. 2008;321:1496–1499. doi: 10.1126/science.1158712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson LE, Yu M, Nelson KS, Laprise P, Tepass U, Beitel GJ. Drosophila convoluted/dALS is an essential gene required for tracheal tube morphogenesis and apical matrix organization. Genetics. 2009;181:1281–1290. doi: 10.1534/genetics.108.099531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer NT, Moberg KH. Regulation of Drosophila embryonic tracheogenesis by dVHL and hypoxia. Dev Biol. 2009;329:294–305. doi: 10.1016/j.ydbio.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson KS, Furuse M, Beitel GJ. The Drosophila Claudin Kune-kune is required for septate junction organization and tracheal tube size control. Genetics. 2010;185:831–839. doi: 10.1534/genetics.110.114959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz JG, Ceulemans H, Caussinus E, Baietti MF, Affolter M. et al. Drosophila syndecan regulates tracheal cell migration by stabilizing Robo levels. EMBO Rep. 2011;12:1039–1046. doi: 10.1038/embor.2011.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazer L, Shilo BZ. The Drosophila FGF-R homolog is expressed in the embryonic tracheal system and appears to be required for directed tracheal cell extension. Genes Dev. 1991;5:697–705. doi: 10.1101/gad.5.4.697. [DOI] [PubMed] [Google Scholar]
- Klämbt C, Glazer L, Shilo BZ. Breathless, a Drosophila FGF receptor homolog, is essential for migration of tracheal and specific midline flial cells. Genes Dev. 1992;6:1668–1678. doi: 10.1101/gad.6.9.1668. [DOI] [PubMed] [Google Scholar]
- Lee T, Hacohen N, Krasnow M, Montell DJ. Regulated Breathless receptor tyrosine kinase activity required to pattern cell migration and branching in the Drosophila tracheal system. Genes Dev. 1996;10:2912–2921. doi: 10.1101/gad.10.22.2912. [DOI] [PubMed] [Google Scholar]
- Centanin L, Dekanty A, Romero N, Irisarri M, Gorr TA, Wappner P. Cell autonomy of HIF effects in Drosophila: tracheal cells sense hypoxia and induce terminal branch sprouting. Dev Cell. 2008;14:547–558. doi: 10.1016/j.devcel.2008.01.020. [DOI] [PubMed] [Google Scholar]
- Scholz H, Deatrock J, Klaes A, Klämbt C. Genetic dissection of Pointed, a Drosophila gene encoding two ETS-related proteins. Genetics. 1993;135:455–468. doi: 10.1093/genetics/135.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarecki J, Johnson E, Krasnow MA. Oxygen regulation of air way branching in Drosophila is mediated by Branchless FGF. Cell. 1999;99:211–220. doi: 10.1016/s0092-8674(00)81652-9. [DOI] [PubMed] [Google Scholar]
- Tomlinson A, Strapps WR. Linking Frizzled and Wnt signalling in Drosophila development. Development. 1997;124:4515–4521. doi: 10.1242/dev.124.22.4515. [DOI] [PubMed] [Google Scholar]
- Tsarouhas V, Senti KA, Jayaram SA, Tiklova K, Hemphala JC. et al. Sequential pulses of apical epithelial secretion and endocytosis drive airway maturation in Drosophila. Dev Cell. 2007;13:214–225. doi: 10.1016/j.devcel.2007.06.008. [DOI] [PubMed] [Google Scholar]
- Grimaldi D, Engel MS. Evolution of the insects. Cambridge University, New York; 2005. [Google Scholar]
- Ostrom JH. Archaeopteryx and the origin of birds. Biol J Linn Soc. 1976;8:91–182. [Google Scholar]
- Horsfield K. Diameters, generations, and orders of branches in the bronchial tree. J Appl Physiol. 1990;68:457–461. doi: 10.1152/jappl.1990.68.2.457. [DOI] [PubMed] [Google Scholar]
- Weibel ER. In: Principles of animal design: the optimization and symmmorphosis debate. Weibel ER, Taylor CR, Bolis L, editor. Cambridge University Press, Cambridge; 1998. Symmorphosis and optimization of biological design: introduction and questions; pp. 1–10. [Google Scholar]
- Maina JN. Morphogenesis of the laminated tripartite cytoarchitectural design of the blood-gas barrier of the avian lung: A systematic electron microscopic study of the domestic fowl, Gallus gallus variant domesticus. Tissue Cell. 2004;36:129–139. doi: 10.1016/j.tice.2003.11.002. [DOI] [PubMed] [Google Scholar]