Abstract
Background
Birt-Hogg-Dubé (BHD) syndrome is a hereditary hamartoma syndrome that predisposes patients to develop hair follicle tumors, lung cysts, and kidney cancer. Genetic studies of BHD patients have uncovered the causative gene, FLCN, but its function is incompletely understood.
Methods
Mice with conditional alleles of FLCN and/or peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PPARGC1A), a transcriptional coactivator that regulates mitochondrial biogenesis, were crossbred with mice harboring either muscle creatine kinase (CKM) –Cre or myogenin (MYOG) –Cre transgenes to knock out FLCN and/or PPARGC1A in muscle, or cadherin 16 (CDH16)– Cre transgenes to knock out FLCN and/or PPARGC1A in kidney. Real-time polymerase chain reaction, immunoblotting, electron microscopy, and metabolic profiling assay were performed to evaluate mitochondrial biogenesis and function in muscle. Immunoblotting, electron microscopy, and histological analysis were used to investigate expression and the pathological role of PPARGC1A in FLCN-deficient kidney. Real-time polymerase chain reaction, oxygen consumption measurement, and flow cytometry were carried out using a FLCN-null kidney cancer cell line. All statistical analyses were two-sided.
Results
Muscle-targeted FLCN knockout mice underwent a pronounced metabolic shift toward oxidative phosphorylation, including increased mitochondrial biogenesis (FLCN f/f vs FLCN f/f /CKM–Cre: % mitochondrial area mean = 7.8% vs 17.8%; difference = 10.0%; 95% confidence interval = 5.7% to 14.3%; P < .001), and the observed increase in mitochondrial biogenesis was PPARGC1A dependent. Reconstitution of FLCN-null kidney cancer cells with wild-type FLCN suppressed mitochondrial metabolism and PPARGC1A expression. Kidney-targeted PPARGC1A inactivation partially rescued the enlarged kidney phenotype and abrogated the hyperplastic cells observed in the FLCN-deficient kidney.
Conclusion
FLCN deficiency and subsequent increased PPARGC1A expression result in increased mitochondrial function and oxidative metabolism as the source of cellular energy, which may give FLCN-null kidney cells a growth advantage and drive hyperplastic transformation.
Affected individuals with Birt-Hogg-Dubé (BHD) syndrome are at risk for the development of multifocal chromophobe and hybrid oncocytic renal cell carcinomas (1,2). The FLCN gene (alias BHD) is a tumor suppressor (3–6), but the mechanism by which FLCN deficiency causes kidney cancer is not completely understood. FLCN encodes folliculin (FLCN), a 64-kDa protein with no known functional or structural domain. FLCN binds to two FLCN-interacting proteins, FNIP1 and FNIP2, both of which interact with 5’-adenosine monophosphate–activated protein kinase (AMPK) (7–9), and several studies have shown that loss of FLCN modulates activity of the mammalian target of rapamycin (mTOR) pathway (3,4,10–12). AMPK is an important energy sensor in cells that negatively regulates mTOR (13,14). Although the function of FLCN remains to be determined, its involvement in a FNIP1/FNIP2/AMPK complex and the modulation of mTOR activity in response to FLCN deficiency suggest that FLCN may play a role in cellular energy and nutrient sensing through interaction with the AMPK–mTOR pathway, a precedent seen with other tumor suppressors involved in cell metabolism (15–19).
FLCN has been reported to interact with several other pathways in addition to the FLCN and AMPK–mTOR pathways already highlighted. The Drosophila FLCN homolog is required for germline stem cell maintenance through the JAK/STAT pathway (20). FLCN-null embryonic stem cells and the FLCN-null cancer cell line UOK257 show loss of transforming growth factor β–dependent signaling (10,21). Transcriptional activity of TFE3, a member of the MiTF/TFE family, is enhanced by its nuclear localization in FLCN-null cells (22). Although these reports indicate that FLCN might be essential for connecting these important pathways, distinct functions of FLCN remain to be elucidated.
To clarify a potential role of FLCN in metabolism, we conducted metabolic assays using FLCN-deficient muscle, which is an important organ for energy production. We also carried out metabolic assays using a FLCN-null kidney cancer cell line, which was reconstituted with wild-type FLCN. We further evaluated the physiological meaning of metabolic alterations under FLCN deficiency in the development of FLCN-deficient polycystic enlarged kidneys.
Methods and Materials
Mice
The mice carrying FLCN alleles flanked by loxP sites (floxed, f) were generated as previously described (3). Muscle creatine kinase (CKM)– Cre transgenic mice [FVB–Tg(Ckmm–cre)5Khn/J, stock number: 006405] were obtained from Jackson Laboratories. Cadherin 16 (CDH16)–Cre transgenic mice were described previously (3). The mice carrying PPARGC1A alleles flanked by loxP sites (floxed, f) were generous gifts from Dr Bruce Spiegelman at Dana-Farber Cancer Institute, Harvard Medical School (23), and myogenin (MYOG) –Cre transgenic mice were generous gifts from Dr Eric N. Olson at University of Texas Southwestern Medical Center (24). We confirmed that the CKM–Cre, MYOG–Cre, and CDH16–Cre transgenes had no detectable effect on mouse phenotypes. Rapamycin (LC Laboratories, Woburn, MA) was administrated 2mg/kg daily from postnatal day 7 for 2 months. For muscle-specific FLCN knockout mice, conditional FLCN knockout mice (FLCN f/f) were crossbred with CKM–Cre transgenic mice and MYOG–Cre transgenic mice to generate FLCN f/f /CKM–Cre and FLCN f/f /MYOG–Cre mice, respectively. For muscle-specific double knockout of FLCN and PPARGC1A, conditional FLCN (FLCN f/f) and PPARGC1A (PPARGC1A f/f) knockout mice were crossbred with CKM–Cre transgenic mice and MYOG–Cre transgenic mice to generate FLCN f/f /PPARGC1A f/f /CKM–Cre and FLCN f/f /PPARGC1A f/f /MYOG–Cre mice, respectively. For kidney-specific double knockout of FLCN and PPARGC1A, conditional FLCN (FLCN f/f) and PPARGC1A (PPARGC1A f/f) knockout mice were crossbred with CDH16–Cre transgenic mice to generate FLCN f/f / PPARGC1A f/f /CDH16–Cre mice. All mice that were used in these experiments were housed in the National Cancer Institute animal facilities according to the National Cancer Institute Animal Care and Use Committee guidelines. The aged mice were killed by carbon dioxide asphyxiation, and the white portion of the quadriceps muscle or kidney was removed, cut into small pieces, snap frozen in liquid nitrogen, and stored at −80°C for further analysis.
Cell Lines and Transfections
FLCN-null UOK257, a cancer cell line originally derived from a renal tumor of a BHD syndrome patient surgically treated at the Urologic Oncology Branch, National Institutes of Health, Bethesda, Maryland, with written patient permission under National Institutes of Health Institutional Review Board–approved protocol 97-C-0147, was reconstituted with wild-type FLCN gene expression by two sequential lentiviral transductions using the full length rtTA3(Tet-on) gene (Clontech, Mountain View, CA) and the full length FLCN gene under the TRE-tight promoter. Different concentrations of doxycycline were tested, and we confirmed that doxycycline less than 0.5ng/µl had no effect on the UOK257 parental cell line [rtTA3(Tet-on) gene only without FLCN gene]. Generally, doxycycline inducible UOK257 cells were analyzed 12 hours after addition of doxycycline. The C2C12 myoblast cell line was purchased from ATCC (Manassas, VA), transfected with Stealth small interfering RNA for FLCN (MSS278133: UUCAAACGCUGAAUGGACCAGGUUC; Invitrogen, Carlsbad, CA) using RNAi Max (Invitrogen) in the presence of 10% horse serum, and harvested 48 hours later.
FACS Analysis
JC-1 (Biotium, Hayward, CA) and tetramethylrhodamine methyl ester (TMRM) (Invitrogen) were used for membrane potential experiments. For JC-1, cells were stained at 37°C in a 5% carbon dioxide incubator for 15 minutes with JC-1 dye. For TMRM, cells were stained at room temperature for 30 minutes with 20nM TMRM. Cells were trypsinized and resuspended into 10% fetal bovine serum in phosphate-buffered saline (PBS). Stained cells were analyzed with CyAn adenosine diphosphate (Beckman Coulter, Brea, CA), and data analysis was performed with FlowJo 7.6.4 software (FlowJo, Ashland, OR).
Quantitative Real-Time Polymerase Chain Reaction
Total RNA was isolated from the flash frozen white portion of the quadriceps muscle, from cell cultures, and from human kidney specimens (surgically treated at the Urologic Oncology Branch, National Institutes of Health, Bethesda, MD, with written patient permission under National Institutes of Health Institutional Review Board–approved protocol 97-C-0147) using TRIzol reagent (Invitrogen), and total RNA was reverse transcribed to cDNA using a Superscript III reverse transcriptase kit (Invitrogen). Quantitative real-time polymerase chain reaction (PCR) was performed with the 7300 Real-Time PCR System (Applied Biosystems, Foster City, CA) using SYBR Green PCR Master Mix (Fermentas, Glen Burnie, CA). Primer sequences are given in Supplementary Table 1 (available online). Signal intensity obtained from Real-Time PCR System was described in relative units; each value was normalized using 36B4 for muscle tissue experiments and β-actin for other experiments.
Western Blotting, Immunofluorescence Staining, and Antibodies
Frozen muscle pieces and kidney samples were homogenized in radioimmunoprecipitation assay buffer [20mM tris(hydroxymethyl)aminomethane–hydrogen chloride pH 7.5, 150mM sodium chloride, 1mM ethylenediaminetetraacetic acid, 1.0% Triton X-100, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate] supplemented with Complete protease inhibitor cocktail and PhosStop phosphatase inhibitor cocktail (Roche, Indianapolis, IN). Immunofluorescence staining of human BHD-associated tumor was done as previously reported (3,4). Antibodies used included mouse monoclonal FLCN, which was developed by the Monoclonal Antibody Core at the National Cancer Institute, National Institutes of Health, Bethesda, Maryland (7), at 1:1000 dilution, Myoglobin(FL154) at 1:200 dilution, PPARGC1A (H300, Santa Cruz Biotechnology, Santa Cruz, CA) at 1:200 dilution, COX4 at 1:1000 dilution, Cytochrome c (136F3) at 1:1000 dilution,β-actin (Cell Signaling, Danvers, MA) at 1:1000 dilution, and α-tubulin (Sigma Aldrich, St. Louis, MO) at 1:1000 dilution. Antibody– protein complexes were detected using SuperSignal West pico chemiluminescent substrate (Pierce Biotechnology, Rockford, IL) or Odyssey imager (LI-COR Biotechnology, Lincoln, NE).
Electron Microscopy
Specimens of human BHD-associated kidney cancer and normal kidney were obtained with intraoperative core needle biopsies at the Urologic Oncology Branch, National Institutes of Health, Bethesda, Maryland, with written patient permission under National Institutes of Health Institutional Review Board–approved protocol 97-C-0147, and immediately immersed in 4% formaldehyde/2% glutaraldehyde (Electron Microscopy Sciences, Hartfield, PA)/PBS. For specimens of mouse muscle and kidney, the mice were perfused with 5 units/mL heparin/PBS and 4% formaldehyde/2% glutaraldehyde/PBS using a 27-gauge needle immediately after euthanization. The gastrocnemius muscles or kidneys were removed and immersed in 4% formaldehyde/2% glutaraldehyde/PBS. Small blocks were then cut, osmicated, and dehydrated before embedding. The blocks were sectioned and observed in a Jeol 1200 transmission electron microscope equipped with an XR-100 Charge-coupled device camera (Advanced Microscopy Techniques Corporation, Danvers, MA). Percentage of mitochondrial area was analyzed with Image J (National Institute of Health, Bethesda, MD). One sectional image of muscle was divided into 24 square areas, each of which was 20 µm2, and the ratios of mitochondrial area to muscle area of 24 areas were measured and represented as means and 95% confidence intervals (CIs). The perinuclear area was omitted because of the abundance of mitochondria. At least three pairs of independent sections were analyzed, and similar results were obtained. For percentage of mitochondrial area in kidney cells, the ratios of mitochondrial area to cell area were measured in at least 20 cells and represented as means and 95% confidence intervals.
Metabolite Analyses
Metabolite profiling analyses using mass spectrometry platforms of liquid chromatography (LC-MS) and gas chromatography (GC-MS) were carried out in collaboration with Metabolon, Inc (Durham, NC), and 202 compounds were detected, including linolenate, eicosapentaenoate, docosapentaenoate, docosahexaenoate, dihomo-linolenate, palmitoleate, oleate, linoleate, arachidonate, cis-vaccenate, mead acid, stearidonate, 10-nonadecenoate, palmitate, dihomo-linoleate, 10-heptadecenoate, eicosenoate, stearate, margarate, coenzyme A, acetyl coenzyme A, citrate, malate, flavin adenine dinucleotide, ATP, glucose, glucose 6-phosphate, fructose 6-phosphate, fructose 1,6-biphosphate, 3-phosphoglycerate, 2-phosphoglycerate, phosphoenolpyruvate, pyruvate, lactate, ribose 5-phosphate, and ribulose 5-phosphate. Six quadriceps muscle tissues of FLCN wild-type mice (FLCN f/f) and six quadriceps muscle tissues of FLCN knockout mice (FLCN f/f /CKM–Cre) were prepared for analysis using Metabolon’s proprietary solvent extraction method. The extracted supernatant was split into equal parts for analysis on the GC, LC+, and LC− platforms. Signal intensity obtained through LC-MS and GC-MS was normalized to protein concentration determined by Bradford protein assays and described in relative units. Nicotinamide adenine dinucleotide, reduced (NADH) was measured using a nicotinamide adenine dinucleotide/NADH quantitation kit (Biovision, Milpitas, CA).
Respiratory Capacity of Cancer Cell Line and Isolated Mitochondria From Muscle Tissue
Baseline oxygen consumption ratio of doxycycline-inducible UOK257 cells was measured with the XF96 extracellular flux analyzer (Seahorse Bioscience, North Billerica, MA) using 8.3g/L Dulbecco’s modified Eage medium supplemented with 200mM glutamine, 100mM sodium pyruvate, 25mM D-glucose, 63.3mM sodium chloride, and phenol red. After monitoring baseline respiration, 1 µM oligomycin and 3 µM carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP) were injected into wells of the XF96 cell culture plate. For respiratory capacity measurements of isolated mitochondria from muscle tissues, an XF96 V3 PET plate (Seahorse Bioscience) was coated overnight with 1:15 000 polyethyleneimine solution/assay buffer (137mM potassium chloride, 2mM monopotassium phosphate, 2.5mM magnesium chloride, 20mM HEPES, 0.5mM EGTA, 0.2% fatty acid–free BSA). One microgram of mitochondria isolated from entire hind limbs using the standard Nagarse method (25) was attached to the plate bottom at 936 g for 10 minutes. The plate was warmed at 37°C for 10 minutes and transferred to the Seahorse XF96 analyzer. State 3 respiration (maximum adenosine diphosphate–stimulated oxygen consumption ratio; oxygen consumption ratio under sufficient substrate for mitochondrial complex) of complex I was measured immediately after addition of 5mM glutamate, 5mM malate, and 0.5mM adenosine diphosphate, and complex IV–dependent respiration was measured immediately after addition of 0.5mM tetramethyl phenylenediamine and 3mM ascorbic acid.
Statistical Analysis
Experimental data are summarized as the mean values with 95% confidence intervals (CIs). Statistical analyses were performed using a two-tailed Student’s t test for in vivo data and analysis of variance for in vitro data (SPSS Statistics version 20, IBM, Armonk, NY), and differences were considered to be statistically significant at a value of P less than .05.
Results
To evaluate the potential metabolic role of FLCN, we crossbred mice carrying loxP-flanked FLCN alleles (floxed, f) with muscle creatine kinase (CKM)– Cre transgenic mice, which express Cre recombinase driven by the CKM promoter, thereby deleting FLCN gene sequences, specifically in muscle. Conditional gene expression was necessary because homozygous FLCN knockout mice are embryonic lethal (4). Strikingly, the muscles in the muscle-specific FLCN knockout mice (FLCN f/f /CKM– Cre) were characterized by distinctive red coloring (Figure 1, A), which was indicative of increased expression of mitochondrial components (26,27). Indeed, FLCN-deficient muscle showed evidence of increased mitochondrial biogenesis, including increased mRNA of mitochondrial genes (FLCN f/f vs FLCN f/f /CKM– Cre: n = 6 each; ATP5g mean = 1 vs 2.69, difference = 1.69, 95% CI = 0.64 to 2.73, P = .005; COX5a mean = 1 vs 2.15, difference = 1.15, 95% CI = 0.42 to 1.87, P = .006; CYCS (Cytochrome C) mean = 1 vs 1.89, difference = 0.89, 95% CI = 0.39 to 1.38, P = .003; PDK4 mean = 1 vs 1.79, difference = 0.79, 95% CI = 0.46 to 1.11, P < .001; Ndufs8 mean = 1 vs 1.64, difference = 0.64, 95% CI = 0.42 to 0.86, P < 0.001; UCP3 mean = 1 vs 1.91, difference = 0.91, 95% CI = 0.32 to 1.51, P = .007) (Figure 1, B), increased expression of mitochondrial proteins (Figure 1, C), and increased mitochondrial area per muscle fiber area (FLCN f/f vs FLCN f/f /CKM–Cre: percentage of mitochondrial area mean = 7.8% vs 17.8%, difference = 10.0%, 95% CI = 5.7% to 14.3%, P < .001) (Figure 1, D).
Figure 1.
Mitochondrial biogenesis in FLCN-deficient muscle. A) Mice carrying FLCN alleles flanked by loxP sites (floxed, f) were crossed with CKM (muscle creatine kinase)– Cre transgenic mice to generate muscle-specific FLCN knockout mice (FLCN f/f /CKM– Cre). Pictures show representative images of muscle with indicated genotypes. B) Mitochondrial gene mRNA levels in quadriceps muscle tissue from littermate-matched mice were quantified with the indicated genes by real-time polymerase chain reaction. * Cytochrome C. Mean±95% confidence interval (CI). n=6. Student’s t test (two-sided). C) Lysates of quadriceps muscle from littermate-matched mice were immunoblotted with the indicated antibodies. *Cytochrome C. β-actin served as a loading control. D) Electron microscopy images were obtained from gastrocnemius muscle tissues of littermate matched mice. ×1000 (scale bar: 2 µm) and ×15 000 (scale bar: 500nm) magnified images. Arrows indicate mitochondria. Right bars represent percentage of mitochondrial area. Mean ± 95% confidence interval. Student’s t test (two-sided).
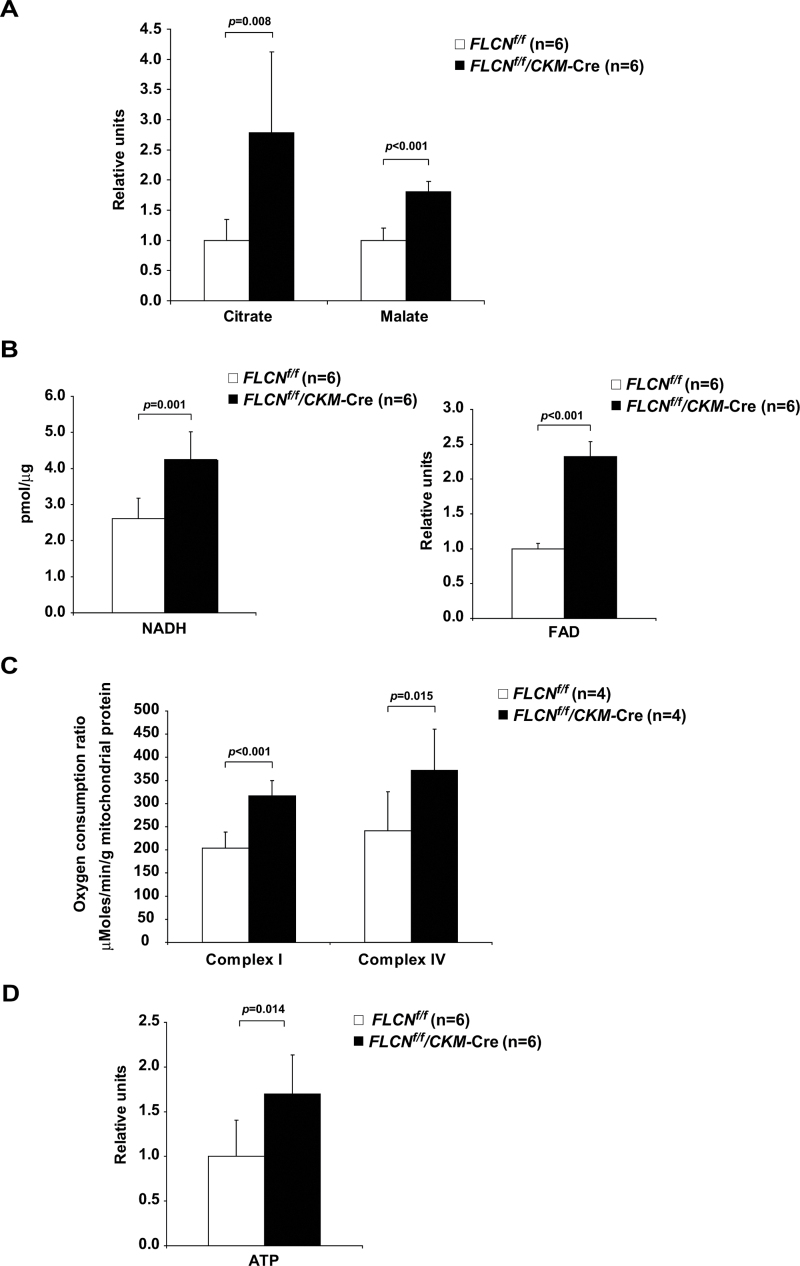
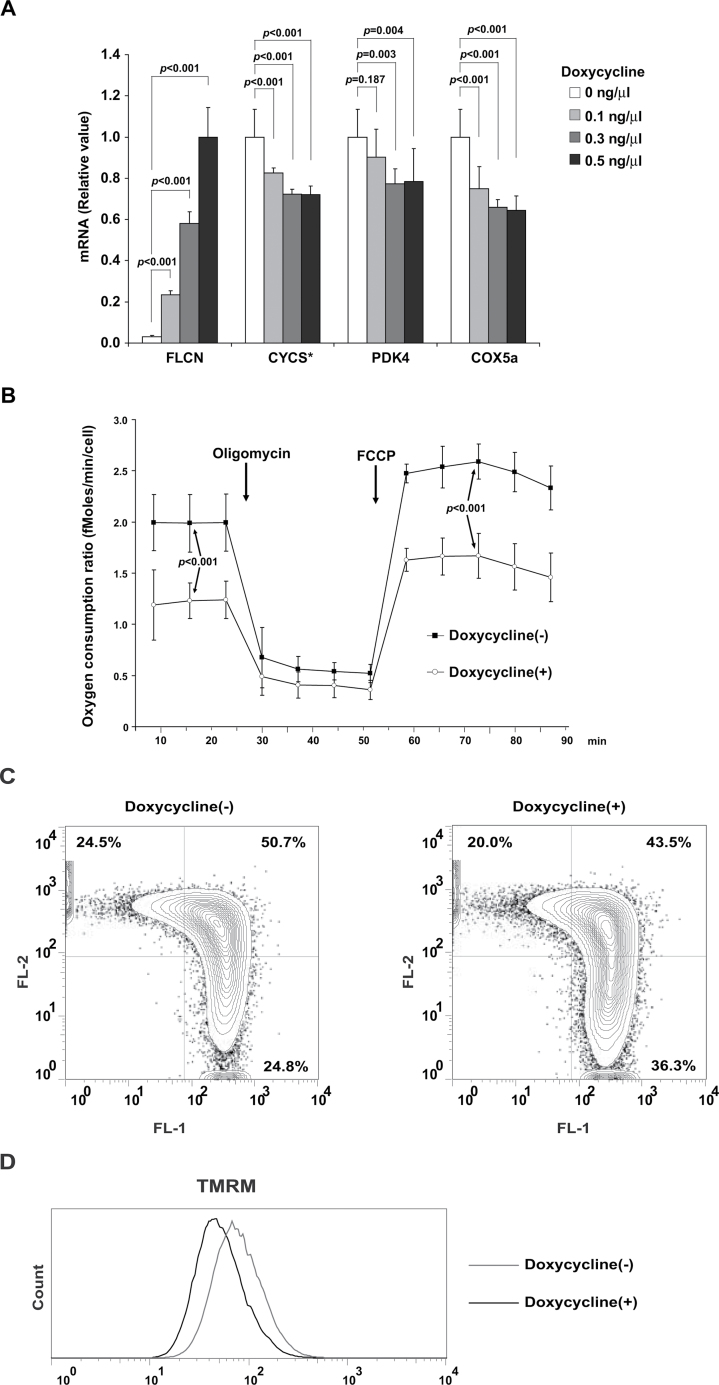
To further investigate mitochondrial function in FLCN-deficient muscle, we conducted metabolite profiling analyses using LC-MS and GC-MS. Nutrients for fatty acid β-oxidation (Table 1), metabolites in the tricarboxylic acid cycle (FLCN f/f vs FLCN f/f /CKM–Cre: n = 6 each; citrate mean = 1 vs 2.78, difference = 1.78, 95% CI = 0.59 to 2.98, P = .008; malate mean = 1 vs 1.80, difference = 0.80, 95% CI = 0.58 to 1.03, P < .001) (Figure 2, A), and coenzymes for the electron transport chain (FLCN f/f vs FLCN f/f /CKM– Cre: n = 6 each; NADH mean = 2.61 vs 4.24 pmol/µg, difference = 1.63 pmol/µg , 95% CI = 0.80 to 2.46 pmol/µg, P = 0.001; flavin adenine dinucleotide mean = 1 vs 2.32, difference = 1.32, 95% CI =1.12 to 1.52, P <0.001) (Figure 2, B) were increased in FLCN-deficient muscle. These results suggest that FLCN-deficient muscle has a higher flux through the tricarboxylic acid cycle to produce abundant cofactors for the mitochondrial electron transport chain. Furthermore, both the respiratory capacity of isolated mitochondria (FLCN f/f vs FLCN f/f /CKM– Cre: n = 4 each; complex I state 3 respiration mean = 203.9 vs 316.8 µMoles/minute/g mitochondrial protein, difference = 112.9 µMoles/minute/g mitochondrial protein, 95% CI =76.0 to 150.0 µMoles/minute/ g mitochondrial protein, P < .001; complex IV–dependent respiration mean = 240.8 vs 372.0 µMoles/minute/g mitochondrial protein, difference = 131.3 µMoles/minute/g mitochondrial protein, 95% CI = 36.5 to 226.0 µMoles/minute/g mitochondrial protein, P = .02) (Figure 2, C) and the amount of ATP in muscle tissue were higher in FLCN-deficient mice compared with FLCN f/f control mice (FLCN f/f vs FLCN f/f /CKM– Cre: n = 6 each; mean = 1 vs 1.70, difference = 0.70, 95% CI = 0.18 to 1.21, P = .02) (Figure 2, D). These results demonstrate that inactivation of FLCN triggers a metabolic shift to mitochondrial oxidative metabolism, resulting in excessive ATP production (Supplementary Table 2 and Supplementary Figure 1, available online). Furthermore, in a doxycycline-inducible, FLCN-null renal tumor cell line model (Supplementary Figure 2, available online), it is noteworthy that the enhanced mitochondrial oxidative functions, including increased expression of mitochondrial genes (Figure 3, A), elevated oxygen consumption ratio (Figure 3, B), and increased membrane potential (Figure 3, C and D) associated with FLCN-deficiency were reversed by doxycycline induction of FLCN expression. Thus, we conclude that FLCN is essential for maintaining a physiologically normal rate of mitochondrial metabolism.
Table 1.
Metabolites related to β-oxidation in FLCN-deficient quadriceps muscle*
| Classification | Metabolite (IUPAC nomenclature)† | KO (n = 6)/ WT (n = 6) | P ‡ |
|---|---|---|---|
| Essential fatty acids | Linolenate (18:3n3 or 6) | 2.406 | .0002 |
| Essential fatty acids | Eicosapentaenoate (20:5n3) | 1.701 | .005 |
| Essential fatty acids | Docosapentaenoate (22:5n3) | 1.605 | .02 |
| Essential fatty acids | Docosahexaenoate (22:6n3) | 1.566 | .02 |
| Essential fatty acids | Dihomo-linolenate (20:3n3 or n6) | 1.407 | .04 |
| Long chain fatty acids | Palmitoleate (16:1n7) | 3.080 | .0006 |
| Long chain fatty acids | Oleate (18:1n9) | 2.289 | .0002 |
| Long chain fatty acids | Linoleate (18:2n6) | 2.225 | .0002 |
| Long chain fatty acids | Arachidonate (20:4n6) | 2.210 | .003 |
| Long chain fatty acids | Cis-vaccenate (18:1n7) | 2.086 | .0004 |
| Long chain fatty acids | Mead acid (20:3n9) | 1.988 | .001 |
| Long chain fatty acids | Stearidonate (18:4n3) | 1.944 | .006 |
| Long chain fatty acids | 10-Nonadecenoate (19:1n9) | 1.930 | .005 |
| Long chain fatty acids | Palmitate (16:0) | 1.923 | .0001 |
| Long chain fatty acids | Dihomo-linoleate (20:2n6) | 1.845 | .01 |
| Long chain fatty acids | 10-Heptadecenoate (17:1n7) | 1.652 | .0001 |
| Long chain fatty acids | Eicosenoate (20:1n9 or 11) | 1.596 | .03 |
| Long chain fatty acids | Stearate (18:0) | 1.447 | .005 |
| Long chain fatty acids | Margarate (17:0) | 1.330 | .02 |
| CoA metabolism | CoA | 1.600 | .0005 |
| CoA metabolism | Acetyl CoA | 3.532 | .0001 |
* Metabolite profiling analyses using mass spectrometry platforms of liquid chromatography and gas chromatopgraphy were carried out with extracts from quadriceps muscle, and knock-out (KO) (FLCN f/f /CKM–Cre)/wild-type (WT) (FLCN f/f) ratios of metabolites related to β-oxidation were obtained. CoA = coenzyme A; IUPAC = International Union of Pure and Applied Chemistry.
† ([Number of carbon atoms] : [Number of double bonds] n [Position of double bond]).
‡ Student’s t test (two-sided).
Figure 2.
Mitochondrial function in FLCN-deficient muscle tissues. A) Relative tricarboxylic acid cycle metabolite values for citrate and malate were obtained in metabolite profiling analyses on quadriceps muscle tissues of indicated genotypes. Mean ± 95% confidence interval. Quadriceps, n = 6. Student’s t test (two-sided). B) Coenzymes for the electron transport chain were measured in quadriceps muscle tissues of indicated genotypes. Nicotinamide adenine dinucleotide, reduced (NADH) was measured using a commercially available kit, and a relative value of flavin adenine dinucleotide was obtained by metabolite profiling analyses. Mean ± 95% confidence interval. Quadriceps, n = 6. Student’s t test (two-sided). C) Respiratory capacity of isolated mitochondria was analyzed in muscle tissues of indicated genotypes. State 3 respiration of complex I– and complex IV–dependent respiration was measured by Seahorse XF96 analyzer using mitochondria isolated from the entire hind limb muscle. Mean ± 95% confidence interval. n = 4. Student’s t test (two-sided). D) Relative ATP amounts were obtained from metabolite profiling analyses on quadriceps muscle tissues of indicated genotypes. Mean ± 95% confidence interval. Quadriceps, n = 6. Student’s t test (two-sided).
Figure 3.
Mitochondrial oxidative function of FLCN-null UOK257 renal cancer cells. A) FLCN-null UOK257 renal cancer cells were reconstituted with wild-type FLCN expression. The mRNA of mitochondrial genes was quantified by real-time polymerase chain reaction in FLCN-null UOK257 cells, which express wild-type FLCN in a doxycycline-dependent manner. * Cytochrome C. Mean ± 95% confidence interval of triplicates. Analysis of variance test. B) Baseline respiration and respiratory capacity were analyzed by Seahorse XF96 analyzer on FLCN-inducible UOK257 cells cultured with or without doxycycline. Oxygen consumption ratio of baseline respiration was measured in completely supplemented media, and oxygen consumption ratio of maximum respiration was measured after the addition of carbonyl cyanide-p-trifluoromethoxyphenylhydrazone. Mean ± 95% confidence interval of six wells. Student’s t test (two-sided). C) Representative flow cytometry analysis of JC-1–stained FLCN inducible UOK257 cells, cultured with or without doxycycline. The FL2+, FL1− fraction (upper left quadrant) represents cells with high membrane potential, whereas the FL2−, FL1+ fraction (lower right quadrant) represents cells with low membrane potential. D) Representative flow cytometry analysis of tetramethylrhodamine methyl ester (TMRM)–stained FLCN-inducible UOK257 cells, cultured with or without doxycycline.
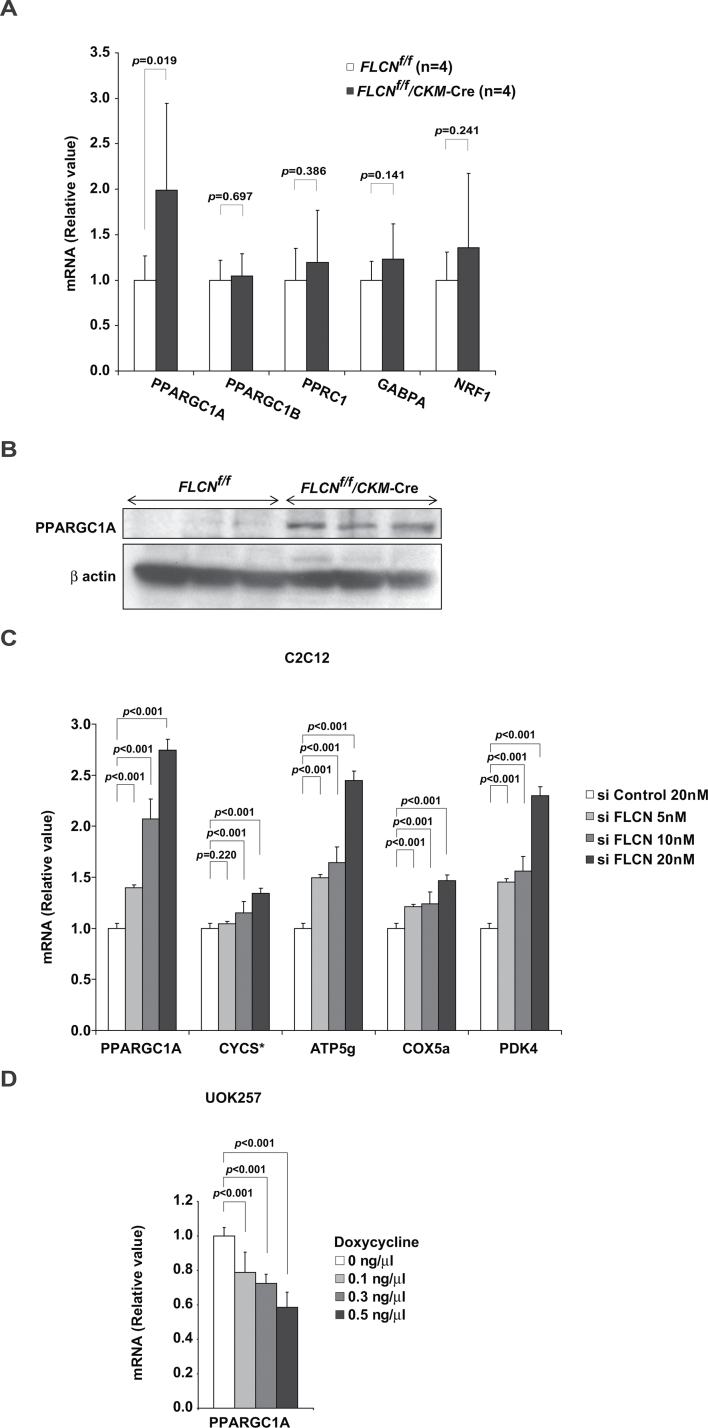
To clarify the molecular mechanism by which FLCN deficiency increases mitochondrial biogenesis and enhances oxidative metabolism, we examined the expression of genes, including transcription factors and coactivators, which had been reported to regulate mitochondrial biogenesis or whose gene-manipulated murine models resulted in a phenotype similar to that found in our FLCN-deficient model that included red-colored muscle and increased mitochondrial biogenesis (26–28). In our screen, we observed increased PPARGC1A mRNA (FLCN f/f vs FLCN f/f /CKM– Cre: n = 4 each; PPARGC1A mean = 1 vs 1.99, difference = 0.99, 95% CI = 0.23 to1.75, P = .02) (Figure 4, A) and increased expression of PPARGC1A protein (Figure 4, B) in FLCN-deficient muscle. Consistent with these results, C2C12 myoblasts showed increased mRNA of PPARGC1A when FLCN was knocked down (Figure 4, C and Supplementary Figure 3, available online). Additionally, FLCN restoration in FLCN-null UOK257 cells suppressed PPARGC1A mRNA (Figure 4, D), suggesting that FLCN modulates PPARGC1A expression levels to produce an adequate level of mitochondrial oxidative phosphorylation for cellular energy needs.
Figure 4.
PPARGC1A expression under FLCN deficiency. A) The mRNA of transcription factor and coactivator was quantified by real-time polymerase chain reaction on quadriceps of FLCN f/f and FLCN f/f /CKM– Cre mice. Mean ± 95% confidence interval. Quadriceps, n = 4. Student’s t test (two-sided). B) Lysates from quadriceps muscle of indicated genotypes were immunoblotted with PPARGC1A antibody. Quadriceps, n = 3. β-actin served as a loading control. C) FLCN expression was knocked down in C2C12 myoblasts using FLCN small interfering RNA (siRNA), and mRNA of indicated genes was quantified with real-time polymerase chain reaction. * Cytochrome C. Mean ± 95% confidence interval of triplicates. Analysis of variance test. D) PPARGC1A mRNA was quantified with real-time polymerase chain reaction on FLCN-inducible UOK257 cells cultured with or without doxycycline. Mean ± 95% confidence interval of triplicates. Analysis of variance test.
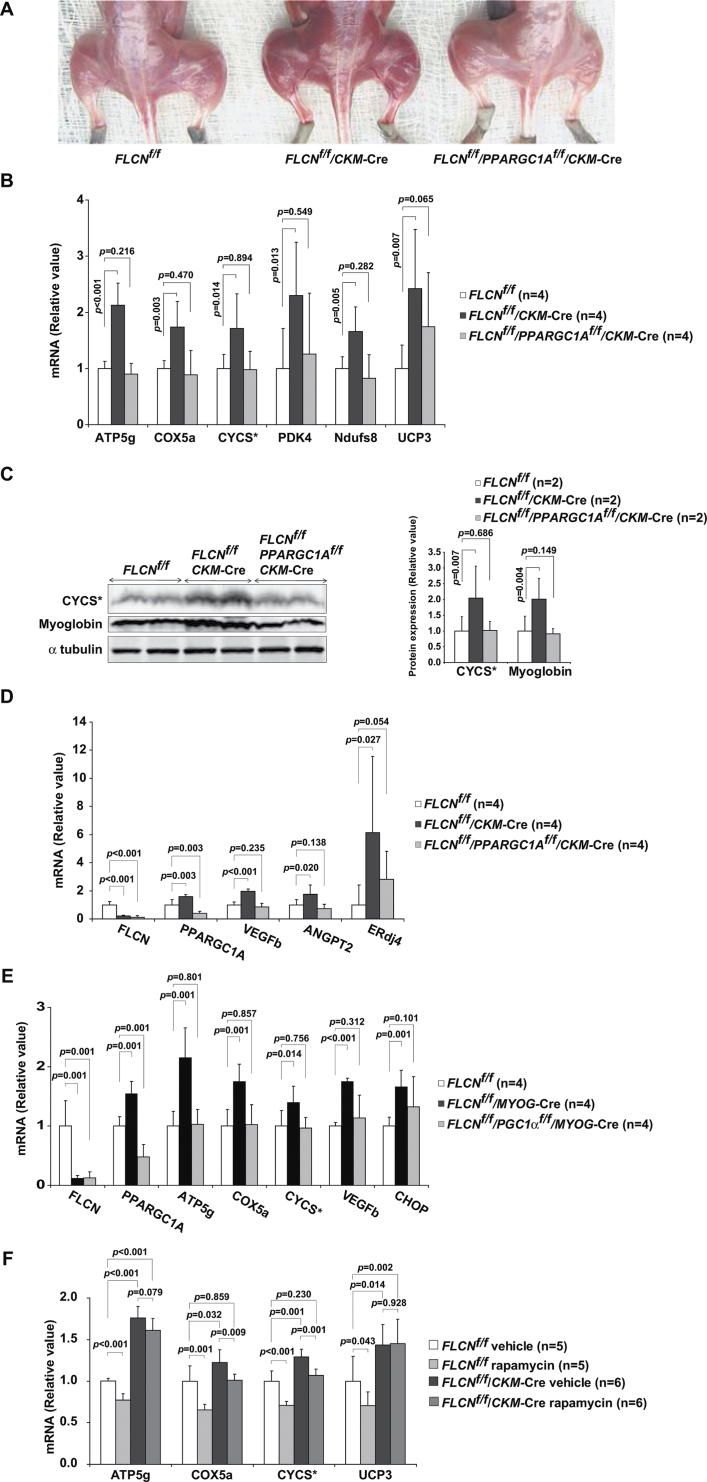
PPARGC1A is a key regulator of mitochondrial function, oxidative metabolism, and energy homeostasis in a variety of tissues (29–34). Notably, muscle-specific PPARGC1A transgenic mice have been shown to develop red-colored muscle with increased mitochondrial biogenesis (27), and PPARGC1A-null mice are characterized by decreased mitochondrial biogenesis and an inability to increase ATP levels (32). Moreover, in vitro bioenergetic analyses have revealed that overexpression of PPARGC1A is associated with an increased membrane potential and respiratory capacity (34–37). Phenotypes of our mouse model in which FLCN inactivation is directed to muscle and FLCN-null in vitro systems are consistent with those of PPARGC1A overexpressing cells and these findings led us to crossbreed muscle-specific FLCN knockout mice with muscle-specific PPARGC1A knockout mice to see whether increased mitochondrial biogenesis under FLCN deficiency is caused by upregulated PPARGC1A. Notably, increased mitochondrial biogenesis, which was associated with FLCN deletion in mouse muscle, was completely reversed by crossbreeding that resulted in deletion of PPARGC1A alleles (Figure 5, A–C). These data strongly support the concept that upregulation of mitochondrial biogenesis caused by FLCN deficiency is PPARGC1A dependent, underscoring a role for FLCN as a physiological negative regulator of PPARGC1A.
Figure 5.
PPARGC1A inactivation in FLCN-deficient muscle. A) FLCN f/f /CKM– Cre mice were crossbred with mice carrying PPARGC1A alleles flanked by loxP sites (floxed, f), and FLCN f/f /PPARGC1A f/f /CKM– Cre mice were generated. A photograph shows representative images of muscle from mice with FLCN f/f, FLCN f/f /CKM– Cre and FLCN f/f /PPARGC1A f/f /CKM– Cre genotypes. B) The mRNA of mitochondrial genes was quantified with real-time polymerase chain reaction on quadriceps samples obtained from FLCN f/f, FLCN f/f /CKM– Cre and FLCN f/f /PPARGC1A f/f /CKM– Cre mice. * Cytochrome C. Mean ± 95% confidence interval. Quadriceps, n = 4. Student’s t test (two-sided). C) Lysates from quadriceps muscle tissues were immunoblotted with the indicated antibodies. α-tubulin served as a loading control. Protein levels in left panel were quantified by Li-Cor Odyssey imager as shown in right panel. * Cytochrome C. Mean ± 95% confidence interval. n = 2. Student’s t test (two-sided). D) The mRNA of angiogenic factors (VEGFb and ANGPT2) and unfolded protein response gene (ERdj4) was quantified with real-time polymerase chain reaction in quadriceps muscle tissues of indicated genotypes. Mean ± 95% confidence interval. Quadriceps, n = 4. Student’s t test (two-sided). E) A different muscle-specific Cre transgenic strain, myogenin (MYOG)– Cre transgenic mice, was crossbred with mice carrying floxed alleles of FLCN and PPARGC1A. The mRNA of PPARGC1A, mitochondrial genes (ATP5g, COX5a, and CYCS), angiogenic factor (VEGFb) and unfolded protein response gene (CHOP) was quantified with real-time polymerase chain reaction on quadriceps samples obtained from FLCN f/f, FLCN f/f /MYOG– Cre and FLCN f/f /PPARGC1A f/f /MYOG– Cre mice. * Cytochrome C. Mean ± 95% confidence interval. Quadriceps, n = 4. Student’s t test (two-sided). F) Mice were treated with rapamycin 2mg/kg daily from postnatal day 7 for 2 months. The mRNA of the indicated genes was quantified with real-time polymerase chain reaction. * Cytochrome C. Mean ± 95% confidence interval. Quadriceps, n = 5 for FLCN f/f group and n = 6 for FLCN f/f /CKM– Cre group. Student’s t test (two-sided).
To date, few studies have evaluated the role of PPARGC1A in cancer development; therefore, the contribution of PPARGC1A to tumor progression (38,39) or inhibition is unclear (40). In addition to its role in energy homeostasis, PPARGC1A has reported functions that include suppression of reactive oxygen species (41), regulation of angiogenesis (42), and induction of unfolded protein response that mitigates endoplasmic reticulum stress (43), all of which may play a role in tumor development. Both of our muscle-directed FLCN knockout mouse models showed increased mRNA of angiogenic factors such as VEGFb and ANGPT2 and unfolded protein response genes such as ERdj4 and CHOP (FLCN f/f vs FLCN f/f /CKM–Cre: n = 4 each; VEGFb mean = 1 vs 1.97, difference = 0.97, 95% CI = 0.76 to 1.17, P < .001; ANGPT2 mean = 1 vs 1.75, difference = 0.75, 95% CI = 0.17 to 1.33, P = .02; ERdj4 mean = 1 vs 6.13, difference = 5.13, 95% CI = 0.82 to 9.44, P = .03; and FLCN f/f vs FLCN f/f /MYOG–Cre: n = 4 each; VEGFb mean = 1 vs 1.75, difference = 0.75, 95% CI = 0.69 to 0.82, P < .001; CHOP mean = 1 vs 1.66, difference = 0.66, 95% CI = 0.42 to 0.91, P = .001) (Figure 5, D and E). Notably, the increased mRNA of these genes was reversed when both FLCN and PPARGC1A were deleted. These results suggest that FLCN-deficient tissues may have increased angiogenesis and upregulated unfolded protein responses through PPARGC1A. Thus, PPARGC1A may contribute to the development of kidney cancer by enhancing angiogenesis and suppressing endoplasmic reticulum stress during FLCN deficiency.
We previously reported that highly proliferating cystic kidneys in kidney-specific FLCN knockout mice have upregulation of the mTOR and MEK/Erk pathways and that rapamycin treatment statistically significantly reduced the kidney size and extended the lifespan of these mice (3). mTOR has been found to control mitochondrial biogenesis by modulating the interaction between PPARGC1A and YY1, a transcriptional factor whose motif is highly enriched in the promoter region of oxidative phosphorylation genes (28). However, although we treated our muscle-specific FLCN knockout mice with rapamycin, rapamycin was more effective in suppressing expression of mitochondrial genes in the control mouse group (FLCN f/f vehicle vs FLCN f/f rapamycin: n = 5 each; ATP5g mean = 1 vs 0.77, difference = 0.23, 95% CI = 0.16 to 0.30, P < .001; UCP3 mean = 1 vs 0.70, difference = 0.30, 95% CI = 0.01 to 0.58, P = .04) than in the muscle-specific FLCN knockout mouse group (FLCN f/f /CKM– Cre vehicle vs FLCN f/f /CKM–Cre rapamycin: n = 6 each; ATP5g mean = 1.76 vs 1.61, P = .72; UCP3 mean=1.44 vs 1.45, P = .84) (Figure 5, F). This suggests that the increased mitochondrial biogenesis in FLCN-deficient muscle might be due to upregulated PPARGC1A itself rather than enhanced interaction of PPARGC1A-YY1 by mTOR. Notably, mTORC1 was not activated in FLCN-deficient muscle (data not shown).
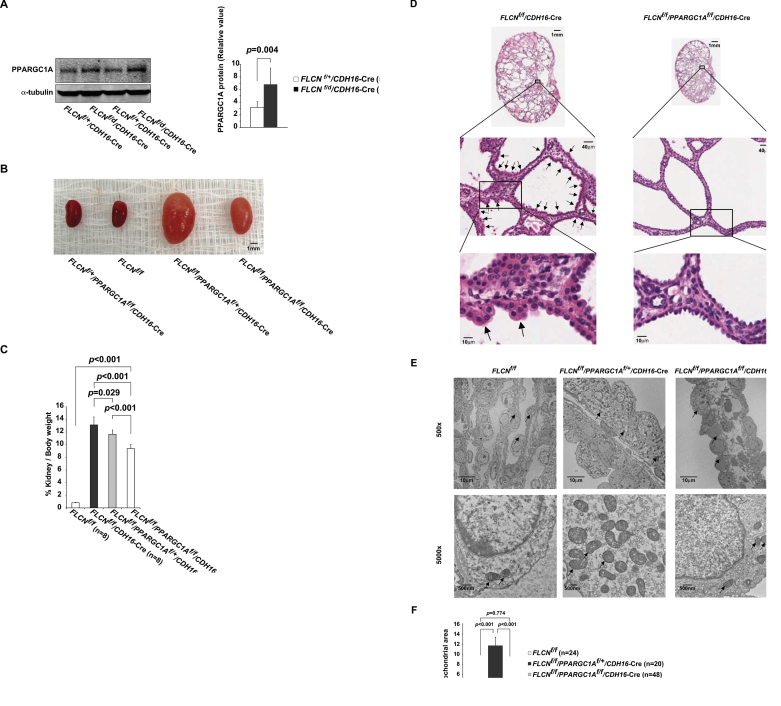
To investigate a potential role of PPARGC1A in the pathogenesis of human BHD-associated kidney cancer, we examined PPARGC1A mRNA and observed high mRNA levels in BHD-associated kidney cancer relative to normal kidney (normal kidney from BHD patients, n = 4 vs BHD-associated kidney cancer, n = 5: mean = 1.02 vs 4.45, difference = 3.44, 95% CI = 2.13 to 4.75, P < .001) (Figure 6, A). In support of this result, we observed elevated protein levels of PPARGC1A in FLCN-deficient mouse tumors compared with adjacent normal kidney (Figure 6, B). Additionally, we observed increased COX4 staining (Figure 6, C), a downstream target of PPARGC1A, and tumor cells filled with increased numbers of mitochondria in human BHD-associated kidney cancer (Figure 6, D). These results indicate that upregulated PPARGC1A and subsequent increased numbers of mitochondria may play an important role in the growth of FLCN-null tumor cells. We previously reported that kidney-specific FLCN knockout mice develop highly proliferating cystic kidneys by 3 weeks of age (3). In fact, we observed increased PPARGC1A protein in FLCN-deficient kidneys (FLCN f/f vs FLCN f/f /CDH16– Cre: n = 2 each; PPARGC1A mean = 3.17 vs 6.82, difference = 3.65, 95% CI =2.71 to 4.59, P = .004) (Figure 7, A), and this observation led us to crossbreed kidney-targeted FLCN knockout mice with kidney-targeted PPARGC1A knockout mice. Surprisingly, inactivation of PPARGC1A alleles in this mouse model partially reduced the size of the enlarged kidneys (FLCN f/f /CDH16– Cre vs FLCN f/f /PPARGC1A f/f /CDH16– Cre: n = 8 each; mean of kidney to body weight ratio = 13.13% vs 9.40%, difference = 3.73%, 95% CI = 2.44% to 5.02%, P < .001) (Figure 7, B and C). To clarify the underlying mechanism by which inactivation of PPARGC1A reduced the size of FLCN-deficient kidneys, we examined the histology of FLCN/PPARGC1A doubly inactivated kidneys. Previously, we observed the presence of hyperplastic cells with enlarged eosinophilic, granular cytoplasm in FLCN-deficient kidneys (3). Interestingly, we did not observe any hyperplastic cells in the FLCN/PPARGC1A doubly inactivated kidneys (Figure 7, D). Furthermore, by electron microscopy we observed increased numbers of mitochondria in FLCN-deficient kidneys (FLCN f/f vs FLCN f/f /PPARGC1A f/+ /CDH16– Cre: n = 24 vs n = 20 cells; mean of percentage of mitochondrial area = 3.44% vs 11.70%, difference = 8.26%, 95% CI = 6.46% to 10.06%, P < .001), and this phenotype was reversed by inactivation of PPARGC1A (FLCN f/f /PPARGC1A f/f /CDH16–Cre) (Figure 7, E and F), consistent with our results in the FLCN-deficient muscle and in vitro systems.
Figure 6.
PPARGC1A expression in human Birt-Hogg-Dubé (BHD)–associated kidney tumor and FLCN-deficient mouse kidney tumor. A) PPARGC1A mRNA in human BHD-associated kidney cancer was quantified with real-time polymerase chain reaction. Bars represent mean ± 95% confidence interval. Normal kidney from sporadic cases (n = 5) vs normal kidney from BHD patients (n = 4) vs BHD-associated kidney cancer (n = 5). Student’s t test (two-sided). B) Lysates of FLCN-deficient mouse kidney tumors, solid tumors that developed in heterozygous FLCN knockout mice at 18–24 months of age, were immunoblotted with the indicated antibodies. β-actin served as a loading control. Normal mouse kidney (n = 4) vs FLCN-deficient mouse kidney tumor (n = 5). C) Human BHD-associated kidney cancer and adjacent normal kidney were stained with an antibody to COX4, a downstream target of PPARGC1A. Green = COX4; Blue = 4’,6-diamidino-2-phenylindole. Right panel shows hematoxylin and eosin (H&E) staining of the corresponding area, and inserts show higher magnification images. Dotted lines indicate the border between normal kidney and kidney cancer. Four samples of human BHD-associated kidney cancer were stained and similar results were obtained. (Scale bar: 100 µm in low magnified image, 10 µm in high magnified image.) D) Electron microscopy images were obtained from renal medulla of normal kidney from patients with non-BHD (sporadic) and BHD-associated kidney cancer. ×500 (scale bar: 10µm) and ×5000 (scale bar: 500nm) magnified images. Arrows indicate mitochondria.
Figure 7.
Pathological significance of upregulated PPARGC1A in FLCN-deficient kidney. A) Lysates of kidney from mice at 5 days of age with indicated genotypes were immunoblotted with the PPAGC1A antibody. α-tubulin served as a loading control. CDH16– Cre = cadherin 16–Cre. Protein level in left panel was quantified by Li-Cor Odyssey imager as shown in right panel. Mean ± 95% confidence interval. n = 2. Student’s t test (two-sided). B) FLCN f/f /CDH16– Cre mice were crossbred with mice carrying PPARGC1A alleles flanked by loxP sites (floxed, f), and FLCN f/f /PPARGC1A f/f / CDH16– Cre mice were generated. Images show representative kidneys of each genotype at 14 days of age. Scale bar: 1mm. C) Ratios of kidney to body weight of indicated genotypes were measured at 21 days of age. Mean ± 95% confidence interval of eight mice per genotype. Student’s t test (two-sided). D) Representative hematoxylin- and eosin-stained tissues of enlarged kidneys from indicated genotypes. Arrows indicate hyperplastic cells in FLCN-deficient kidney (FLCN f/f / CDH16– Cre). Scale bars: 1mm, 40 µm, 10 µm. E) Representative electron microscopy images were obtained from kidney tissues. ×500 (scale bar: 10 µm) and ×5000 (scale bar: 500nm) magnified images. Arrows indicate mitochondria. F) Percentage of mitochondrial area per cell was quantified for the indicated genotypes. n = 24 cells for FLCN f/f, n = 20 cells for FLCN f/f /PPARGC1A f/+ / CDH16– Cre, n = 48 cells for FLCN f/f /PPARGC1A f/f /CDH16– Cre. Mean ± 95% confidence interval. Student’s t test (two-sided).
Discussion
In this study, we report an important role of FLCN in mitochondrial oxidative metabolism. FLCN-deficient muscle in a mouse model displays a distinctive red color and enhanced mitochondrial oxidative metabolism. PPARGC1A expression was high in FLCN-deficient muscle, and inactivation of PPARGC1A in muscle completely reversed the elevated mitochondrial biogenesis. The UOK257 FLCN-null kidney cancer cell line displayed increased mitochondrial function and an elevated level of PPARGC1A mRNA, which were reversed by reconstitution with wild-type FLCN. Human BHD-associated kidney cancer also displayed an elevated level of PPARGC1A mRNA. Furthermore, inactivation of PPARGC1A in a kidney-targeted FLCN-deficient mouse model partially rescued the highly proliferating cystic kidney phenotype. A partial reduction in the size of FLCN-deficient kidneys and complete disappearance of hyperplastic cells that were observed in the FLCN-deficient kidney model (3) underscore the pathological significance of upregulated PPARGC1A in FLCN-null kidney cells.
Although the details of FLCN function have not been completely elucidated, studies have shown that the FLCN pathway interacts with a number of important metabolic pathways, including the AMPK-mTOR (3,4,10–12), TFE3 (22), and HIF–VEGF (44) pathways. In the current study, we demonstrate that FLCN plays a role in mitochondrial homeostasis, further supporting prior work implicating FLCN in metabolism.
In FLCN-deficient muscle, we did not observe the activation of mTORC1, whereas we previously observed the activation of mTORC1 in FLCN-deficient kidney models (3,4). mTORC1 integrates a variety of environmental cues, including energy, nutrient, and growth factors, and, in addition to these inputs, mTORC1 activity in muscle tissues is affected by exercise (45). Therefore, our observation that mTORC1 was not activated in FLCN-deficient muscle may be due to the particular organ context. The precise mechanism by which FLCN deficiency dysregulates the mTOR pathway will require further study.
In 2010, Klomp et al. reported increased mRNA expression of PPARGC1A in human BHD-associated kidney tumors (46). In the current study, we have established PPARGC1A as a key molecule that confers enhanced mitochondrial oxidative metabolism on FLCN-deficient cells. Although aerobic glycolysis (known as the “Warburg effect”) is thought to be the dominant metabolic pathway active in cancer cells, Otto Warburg originally developed his theory based on the efficiency of ATP production from one molecule of glucose through mitochondrial oxidative phosphorylation (n = 29–30 ATP) compared with glycolysis (n = 2 ATP) (47,48), which is a requirement for proliferating cells to maximize energy production (18). Our data indicating that upregulation of PPARGC1A in response to FLCN deficiency contributes to hyperplastic cell formation in the FLCN-deficient mouse kidneys strongly support the idea that mitochondria are important organelles that can fuel tumor suppressor–null cells to initiate tumorigenesis or acquire a growth advantage driving tumor progression.
Numerous electron microscopy studies have demonstrated increased numbers of mitochondria in sporadic chromophobe renal cell carcinoma and renal oncocytoma compared with normal kidney (49–51), and expression profiling analyses of both sporadic (52–56) and familial (46) chromophobe renal cell carcinoma and renal oncocytoma revealed the increased expression of mitochondrial genes or transcriptional regulators for mitochondrial components. For many years, these observations have raised questions among researchers about the underlying mechanisms by which mitochondria increase in these types of tumors. On the other hand, genetic studies of sporadic chromophobe renal cell carcinoma and renal oncocytoma have reported the rearrangement of mitochondrial DNA in these types of tumors (57–60). Taken together, these findings have led researchers to speculate that increased mitochondrial expression might be a compensatory consequence of impaired mitochondrial function. However, our findings that loss of FLCN enhances ATP production and mitochondrial respiratory function support the concept that the increased number of mitochondria observed in BHD-associated kidney cancer is a primary consequence of upregulated PPARGC1A resulting from FLCN deficiency and that FLCN-null cells augment mitochondrial oxidative metabolism by increasing mitochondrial number. Although it is not know whether the FLCN pathway is dysregulated in sporadic chromophobe renal cell carcinoma and renal oncocytoma, increased numbers of mitochondria in these tumor types would suggest the potential for attenuation of a pathway involving FLCN.
In mouse kidneys in which FLCN and PPARGC1A were both inactivated, we no longer detected the eosinophilic, hypertrophic cells with increased numbers of mitochondria that were observed in the FLCN-deficient mouse kidneys. Therefore, it is possible that the increased expression of PPARGC1A and its target genes leading to excess mitochondrial ATP production under FLCN deficiency may contribute to tumorigenesis and/or tumor progression. However, one obvious limitation of this study is that the kidney-targeted FLCN knockout mice die due to renal failure around 3 weeks of age without progression of the hyperplastic cells to malignancy (3). To address this limitation, we have recently developed another FLCN knockout mouse model in which one FLCN allele is deleted in the germline. Subsequent loss of heterozygosity of the remaining wild-type FLCN allele in kidney cells leads to development of chromophobe and hybrid oncocytic tumors at 18–24 months of age, thereby mimicking the human BHD-associated renal tumor paradigm (4). Although this in vivo tumor model of BHD requires a longer time frame, it would be interesting to see the consequence of PPARGC1A inactivation on tumor formation in this heterozygous FLCN knockout mouse model.
In addition to its role in mitochondrial homeostasis, PPARGC1A regulates angiogenesis (42), neutralizes reactive oxygen species, and induces expression of unfolded protein response proteins that mitigate endoplasmic reticulum stress (43). Because these functions may be important for cancer cells to grow, the PPARGC1A pathway could be a potential therapeutic target in patients with BHD-associated kidney cancer.
Our findings support a role for tumor suppressor FLCN as a physiologically important negative regulator of PPARGC1A, a key regulator of energy homeostasis, and demonstrate that loss of FLCN leads to increased mitochondrial oxidative metabolism with subsequent energy overproduction. Furthermore, PPARGC1A enhances the development of hyperplastic cells and enlarged cystic kidneys in the kidney-targeted FLCN-null mouse model, suggesting that upregulation of PPARGC1A and subsequent increased mitochondrial oxidative metabolism in FLCN-null kidney cells may contribute to renal tumor initiation and/or progression in BHD syndrome. These findings provide a foundation for the management of BHD patients and the development of targeted therapeutic approaches for patients with BHD-associated kidney cancer.
Funding
This work was supported by the Intramural Research Program of the National Institutes of Health (NIH) , National Cancer Institute, Center for Cancer Research. This project was funded in part with federal funds from the Frederick National Laboratory for Cancer Research, NIH, under contract HHSN261200800001E.
Supplementary Material
H. Hasumi, M. Baba, L. S. Schmidt, and W. M. Linehan conceived the project, analyzed the data, and wrote the manuscript. L.S. Schmidt organized mice projects, and M. Baba provided FLCN knockout mice. Y. Hasumi, Y. Huang, and H. Oh performed mice dissection, quantitative real-time PCR, and immuno blotting. M. Baba and R. M. Hughes generated a UOK257 doxycycline- inducible cell line. H. Oh and M. E. Klein performed PCR genotyping of mice. S. Takikita performed mice metabolic assays. K. Nagashima performed electron microscopy studies. W. M. Linehan decided to submit the manuscript. All the authors discussed the results and commented on the manuscript.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. NCI-Frederick is accredited by AAALAC International and follows the Public Health Service Policy for the Care and Use of Laboratory Animals. Animal care was provided in accordance with the procedures outlined in the National Research Council’s Guide for Care and Use of Laboratory Animals.
The authors thank Dr Bruce Spiegelman for distributing PPARGC1A knockout mice; Dr Eric N. Olson for distributing myogenin (MYOG)– Cre transgenic mice; Dr Tracey A. Rouault and Dr Toshimitsu Kawate for critical review of the manuscript; Dr Oksana Gavrilova, Dr Toren Finkel, Dr Samuel W. Cushman, Dr Karin G. Stenkula, Dr Marcella Fulco, and Dr Andrew D. Patterson for helpful discussions; Dr Celia Quijano and Dr Sonia Cortassa for technical support for isolated mitochondrial assay; Dr Diana C. Haines for pathological diagnosis; Protein Expression Laboratory, SAIC-Frederick, Inc for generating lentiviral constructs; and Louise Cromwell for excellent technical support with the mouse studies.
References
- 1. Linehan WM, Srinivasan R, Schmidt LS. The genetic basis of kidney cancer: a metabolic disease Nat Rev Urol. 2010. 7(5):277–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Linehan WM, Pinto PA, Srinivasan R, et al. Identification of the genes for kidney cancer: opportunity for disease-specific targeted therapeutics Clin Cancer Res. 2007. 13(2 Pt 2):671s–679s [DOI] [PubMed] [Google Scholar]
- 3. Baba M, Furihata M, Hong SB, et al. Kidney-targeted Birt-Hogg-Dube gene inactivation in a mouse model: Erk1/2 and Akt-mTOR activation, cell hyperproliferation, and polycystic kidneys J Natl Cancer Inst. 2008. 100(2):140–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hasumi Y, Baba M, Ajima R, et al. Homozygous loss of BHD causes early embryonic lethality and kidney tumor development with activation of mTORC1 and mTORC2 Proc Natl Acad Sci U S A. 2009. 106(44):18722–18727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nickerson ML, Warren MB, Toro JR, et al. Mutations in a novel gene lead to kidney tumors, lung wall defects, and benign tumors of the hair follicle in patients with the Birt-Hogg-Dube syndrome Cancer Cell. 2002. 2(2):157–164 [DOI] [PubMed] [Google Scholar]
- 6. Vocke CD, Yang Y, Pavlovich CP, et al. High frequency of somatic frameshift BHD gene mutations in Birt-Hogg-Dube-associated renal tumors J Natl Cancer Inst. 2005. 97(12):931–935 [DOI] [PubMed] [Google Scholar]
- 7. Baba M, Hong SB, Sharma N, et al. Folliculin encoded by the BHD gene interacts with a binding protein, FNIP1, and AMPK, and is involved in AMPK and mTOR signaling Proc Natl Acad Sci U S A. 2006. 103(42):15552–15557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hasumi H, Baba M, Hong SB, et al. Identification and characterization of a novel folliculin-interacting protein FNIP2 Gene. 2008. 415(1–2):60–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Takagi Y, Kobayashi T, Shiono M, et al. Interaction of folliculin (Birt-Hogg-Dube gene product) with a novel Fnip1-like (FnipL/Fnip2) protein Oncogene. 2008. 27(40):5339–5347 [DOI] [PubMed] [Google Scholar]
- 10. Cash TP, Gruber JJ, Hartman TR, Henske EP, Simon MC. Loss of the Birt-Hogg-Dube tumor suppressor results in apoptotic resistance due to aberrant TGFbeta-mediated transcription Oncogene. 2011. 30(22):2534–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hartman TR, Nicolas E, Klein-Szanto A, et al. The role of the Birt-Hogg-Dube protein in mTOR activation and renal tumorigenesis Oncogene. 2009. 28(13):1594–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hudon V, Sabourin S, Dydensborg AB, et al. Renal tumour suppressor function of the Birt-Hogg-Dube syndrome gene product folliculin J Med Genet. 2010. 47(3):182–189 [DOI] [PubMed] [Google Scholar]
- 13. Inoki K, Corradetti MN, Guan KL. Dysregulation of the TSC-mTOR pathway in human disease Nat Genet. 2005. 37(1):19–24 [DOI] [PubMed] [Google Scholar]
- 14. Shaw RJ, Bardeesy N, Manning BD, et al. The LKB1 tumor suppressor negatively regulates mTOR signaling Cancer Cell. 2004. 6(1):91–99 [DOI] [PubMed] [Google Scholar]
- 15. Crino PB, Nathanson KL, Henske EP. The tuberous sclerosis complex N Engl J Med. 2006. 355(13):1345–1356 [DOI] [PubMed] [Google Scholar]
- 16. Goncharova EA, Goncharov DA, Li H, et al. mTORC2 is required for proliferation and survival of TSC2-null cells Mol Cell Biol. 2011. 31(12):2484–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jones RG, Thompson CB. Tumor suppressors and cell metabolism: a recipe for cancer growth Genes Dev. 2009. 23(5):537–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation Science. 2009. 324(5930):1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yeung RS. Multiple roles of the tuberous sclerosis complex genes Genes Chromosomes Cancer. 2003. 38(4):368–375 [DOI] [PubMed] [Google Scholar]
- 20. Singh SR, Zhen W, Zheng Z, et al. The Drosophila homolog of the human tumor suppressor gene BHD interacts with the JAK-STAT and Dpp signaling pathways in regulating male germline stem cell maintenance Oncogene. 2006. 25(44):5933–5941 [DOI] [PubMed] [Google Scholar]
- 21.Hong SB, Oh H, Valera VA, et al. Tumor suppressor FLCN inhibits tumorigenesis of a FLCN-null renal cancer cell line and regulates expression of key molecules in TGF-beta signaling. Mol Cancer. 2010;9:160. doi: 10.1186/1476-4598-9-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hong SB, Oh H, Valera VA, Baba M, Schmidt LS, Linehan WM. Inactivation of the FLCN tumor suppressor gene induces TFE3 transcriptional activity by increasing its nuclear localization PLoS One. 2010. 5(12):e15793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Handschin C, Chin S, Li P, et al. Skeletal muscle fiber-type switching, exercise intolerance, and myopathy in PGC-1alpha muscle-specific knock-out animals J Biol Chem. 2007. 282(41):30014–30021 [DOI] [PubMed] [Google Scholar]
- 24. Li S, Czubryt MP, McAnally J, et al. Requirement for serum response factor for skeletal muscle growth and maturation revealed by tissue-specific gene deletion in mice Proc Natl Acad Sci U S A. 2005. 102(4):1082–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bhattacharya SK, Thakar JH, Johnson PL, Shanklin DR. Isolation of skeletal muscle mitochondria from hamsters using an ionic medium containing ethylenediaminetetraacetic acid and nagarse Anal Biochem. 1991. 192(2):344–349 [DOI] [PubMed] [Google Scholar]
- 26. Arany Z, Lebrasseur N, Morris C, et al. The transcriptional coactivator PGC-1beta drives the formation of oxidative type IIX fibers in skeletal muscle Cell Metab. 2007. 5(1):35–46 [DOI] [PubMed] [Google Scholar]
- 27. Lin J, Wu H, Tarr PT, et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres Nature. 2002. 418(6899):797–801 [DOI] [PubMed] [Google Scholar]
- 28. Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex Nature. 2007. 450(7170):736–740 [DOI] [PubMed] [Google Scholar]
- 29. Arany Z, He H, Lin J, et al. Transcriptional coactivator PGC-1 alpha controls the energy state and contractile function of cardiac muscle Cell Metab. 2005. 1(4):259–271 [DOI] [PubMed] [Google Scholar]
- 30. Fernandez-Marcos PJ, Auwerx J. Regulation of PGC-1alpha, a nodal regulator of mitochondrial biogenesis Am J Clin Nutr. 2011. 93(4):884S–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Handschin C, Chin S, Li P, et al. Skeletal muscle fiber-type switching, exercise intolerance, and myopathy in PGC-1alpha muscle-specific knock-out animals J Biol Chem. 2007. 282(41):30014–30021 [DOI] [PubMed] [Google Scholar]
- 32. Lin J, Wu PH, Tarr PT, Lindenberg KS, St-Pierre J, Zhang CY, et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice Cell. 2004. 119(1):121–135 [DOI] [PubMed] [Google Scholar]
- 33. Michael LF, Wu Z, Cheatham RB, et al. Restoration of insulin-sensitive glucose transporter (GLUT4) gene expression in muscle cells by the transcriptional coactivator PGC-1 Proc Natl Acad Sci U S A. 2001. 98(7):3820–3825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wu Z, Puigserver P, Andersson U, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1 Cell. 1999. 98(1):115–124 [DOI] [PubMed] [Google Scholar]
- 35. Puigserver P, Rhee J, Lin J, et al. Cytokine stimulation of energy expenditure through p38 MAP kinase activation of PPARgamma coactivator-1 Mol Cell. 2001. 8(5):971–982 [DOI] [PubMed] [Google Scholar]
- 36. Spiegelman BM. Transcriptional control of mitochondrial energy metabolism through the PGC1 coactivators Novartis Found Symp. 2007. 287 60–69 [PubMed] [Google Scholar]
- 37. St-Pierre J, Lin J, Krauss S, et al. Bioenergetic analysis of peroxisome proliferator-activated receptor gamma coactivators 1alpha and 1beta (PGC-1alpha and PGC-1beta) in muscle cells J Biol Chem. 2003. 278(29):26597–26603 [DOI] [PubMed] [Google Scholar]
- 38. Bhalla K, Hwang BJ, Dewi RE, et al. PGC1alpha promotes tumor growth by inducing gene expression programs supporting lipogenesis Cancer Res. 2011. 71(21):6888–6898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shiota M, Yokomizo A, Tada Y, et al. Peroxisome proliferator-activated receptor gamma coactivator-1alpha interacts with the androgen receptor (AR) and promotes prostate cancer cell growth by activating the AR Mol Endocrinol. 2010. 24(1):114–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. D'Errico I, Salvatore L, Murzilli S, et al. Peroxisome proliferator-activated receptor-gamma coactivator 1-alpha (PGC1alpha) is a metabolic regulator of intestinal epithelial cell fate Proc Natl Acad Sci U S A. 2011. 108(16):6603–6608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. St-Pierre J, Drori S, Uldry M, et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators Cell. 2006. 127(2):397–408 [DOI] [PubMed] [Google Scholar]
- 42. Arany Z, Foo SY, Ma Y, et al. HIF-independent regulation of VEGF and angiogenesis by the transcriptional coactivator PGC-1alpha Nature. 2008. 451(7181):1008–1012 [DOI] [PubMed] [Google Scholar]
- 43. Wu J, Ruas JL, Estall JL, et al. The unfolded protein response mediates adaptation to exercise in skeletal muscle through a PGC-1alpha/ATF6alpha complex Cell Metab. 2011. 13(2):160–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Preston RS, Philp A, Claessens T, et al. Absence of the Birt-Hogg-Dube gene product is associated with increased hypoxia-inducible factor transcriptional activity and a loss of metabolic flexibility Oncogene. 2011. 30(10):1159–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Laplante M, Sabatini DM. mTOR signaling in growth control and disease Cell. 2012. 149(2):274–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Klomp JA, Petillo D, Niemi NM, et al. Birt-Hogg-Dube renal tumors are genetically distinct from other renal neoplasias and are associated with up-regulation of mitochondrial gene expression BMC Med Genomics. 2010. 3:59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Warburg O, Wind F, Negelein E. The metabolism of tumors in the body J Gen Physiol. 1927. 8(6):519–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Warburg O. On the origin of cancer cells Science. 1956. 123(3191):309–314 [DOI] [PubMed] [Google Scholar]
- 49. Kuroda N, Toi M, Hiroi M, Shuin T, Enzan H. Review of renal oncocytoma with focus on clinical and pathobiological aspects Histol Histopathol. 2003. 18(3):935–942 [DOI] [PubMed] [Google Scholar]
- 50. Moreno SM, Benitez IA, Martinez Gonzalez MA. Ultrastructural studies in a series of 18 cases of chromophobe renal cell carcinoma Ultrastruct Pathol. 2005. 29(5):377–387 [DOI] [PubMed] [Google Scholar]
- 51. Nagashima Y, Mitsuya T, Shioi KI, et al. Renal oncocytosis Pathol Int. 2005. 55(4):210–5 [DOI] [PubMed] [Google Scholar]
- 52. Higgins JP, Shinghal R, Gill H, et al. Gene expression patterns in renal cell carcinoma assessed by complementary DNA microarray Am J Pathol. 2003. 162(3):925–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Osunkoya AO, Yin-Goen Q, Phan JH, et al. Diagnostic biomarkers for renal cell carcinoma: selection using novel bioinformatics systems for microarray data analysis Hum Pathol 2009. 40(12):1671–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schuetz AN, Yin-Goen Q, Amin MB. Molecular classification of renal tumors by gene expression profiling J Mol Diagn. 2005. 7(2):206–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Takahashi M, Yang XJ, Sugimura J, et al. Molecular subclassification of kidney tumors and the discovery of new diagnostic markers Oncogene. 2003. 22(43):6810–6818 [DOI] [PubMed] [Google Scholar]
- 56. Young AN, Amin MB, Moreno CS, et al. Expression profiling of renal epithelial neoplasms: a method for tumor classification and discovery of diagnostic molecular markers Am J Pathol. 2001. 158(5):1639–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mayr JA, Meierhofer D, Zimmermann F, et al. Loss of complex I due to mitochondrial DNA mutations in renal oncocytoma Clin Cancer Res. 2008. 14(8):2270–2275 [DOI] [PubMed] [Google Scholar]
- 58. Gasparre G, Hervouet E, de Laplanche E, et al. Clonal expansion of mutated mitochondrial DNA is associated with tumor formation and complex I deficiency in the benign renal oncocytoma Hum Mol Genet. 2008. 17(7):986–995 [DOI] [PubMed] [Google Scholar]
- 59. Kovacs A, Storkel S, Thoenes W, Kovacs G. Mitochondrial and chromosomal DNA alterations in human chromophobe renal cell carcinomas J Pathol. 1992. 167(3):273–277 [DOI] [PubMed] [Google Scholar]
- 60. Welter C, Kovacs G, Seitz G, Blin N. Alteration of mitochondrial DNA in human oncocytomas Genes Chromosomes Cancer. 1989. 1(1):79–82 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.