Abstract
Unlike bones, behavior does not fossilize, so it is hard to infer the evolutionary history of social traits. However, we have shown elsewhere that Bayesian phylogenetic methods allow the investigation of ancestral states and models of evolution of social grouping behaviour in primates. Here, we extend this analysis to another significant aspect of primate social life, which may be subject to different evolutionary pressures—mating systems. We show that mating systems evolved from a polygynandrous state at the root of the phylogeny to the two derived states of harem-polygyny and monogamy. Unlike social organization, where there were no transitions from uni-male groups to pairs, here we found positive transition rates from both polygynous mating states into monogamy. There were no transitions out of monogamy to another mating state. Both derived mating systems evolved late in primate evolution. Nocturnal primates remained solitary foragers while their mating systems evolved from polygynandry to harem-polygyny and monogamy. However, among diurnal primates the derived mating states evolved at the same time as the derived states of social organization.
Keywords: Bayesian phylogenetics, mating systems, monogamy, phylogeny, polygynandry, polygyny, primates
Although rare among mammals, sociality is common among primates.1 In a recent study we showed how group living evolved in primates following a switch to diurnal activity patterns.2 However, primate societies are shaped not only by the groups that animals live in, but also by the mating strategies that they employ. Therefore, for a full understanding of the evolutionary history of primate social systems both need to be investigated.
We have shown previously that for social traits with a strong phylogenetic signal, such as social organization or dispersal patterns, it is possible to infer models of evolution, transition rates between states and ancestral states at nodes on the primate phylogeny.2 Here we use Bayesian phylogenetic methods3 to infer ancestral states and evolutionary pathways for mating systems in primates, complementing our previous investigation of social organization and allowing the comparison of these two features of primate life.
An analysis using Ape4 in R gave a Lambda value of 0.996, not significantly different from 1 (p = 0.679), suggesting a strong phylogenetic signal in the mating system data among primates.
We ran RJ MCMC analyses in BayesTraits5 with mating system classified as both a binary trait (polygyny/monogamy) and a three state trait (harem-polygyny/polygynandry/monogamy). The analysis for the binary classification supports polygyny as the ancestral primate mating system at the root of the phylogeny (polygyny mean probability = 0.974 +/− 0.001; monogamy mean = 0.026 +/− 0.001). For the three state trait the root was polygynandry (polygynandry mean = 0.853 +/− 0.003; harem-polygyny mean = 0.109 +/− 0.002; monogamy mean = 0.038 +/− 0.001).
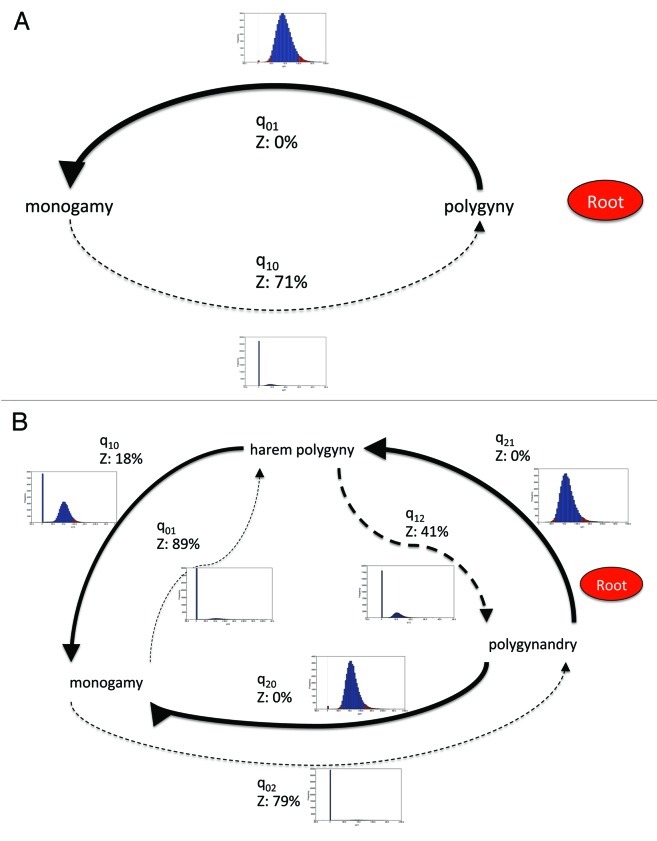
The best fitting model from the RJ procedure (with 71% of the posterior probability distribution) revealed that from polygynous mating at the root of the phylogeny there was a strong transition rate to monogamy, but a zero reverse rate (Fig. 1A). We also split the polygynous mating state into harem-polygyny and polygynandry to understand the dynamics of mating change better.6 From polygynandry at the root of the phylogeny there were strong transition rates into both harem-polygyny and monogamy, and a non-zero but weaker rate from harem-polygyny to monogamy (best fitting model from the RJ procedure, with 33% of the posterior probability distribution). All other rates were zero (Fig. 1B).
Figure 1. Model of evolution of primate mating systems showing posterior distribution of transition rates between states. A. Monogamy and polygyny. B. Monogamy, harem-polygyny and polygynandry. Thickness of arrows reflects proportion of time the transition rate is not zero. Z denotes a zero transition rate as a proportion of posterior probability distribution. A dashed line denotes a zero transition rate in the RJ derived model. Graphs show posterior probability distribution of each transition rate.
Across the whole posterior probability distribution, for the three state analysis, the mode transition rate out of monogamy was zero, while the transitions from harem-polygyny to monogamy and back to polygynandrous mating were zero for 18% and 41% of the time respectively (Fig. 1B). Transitions from polygynandrous mating to the other two states were never zero.
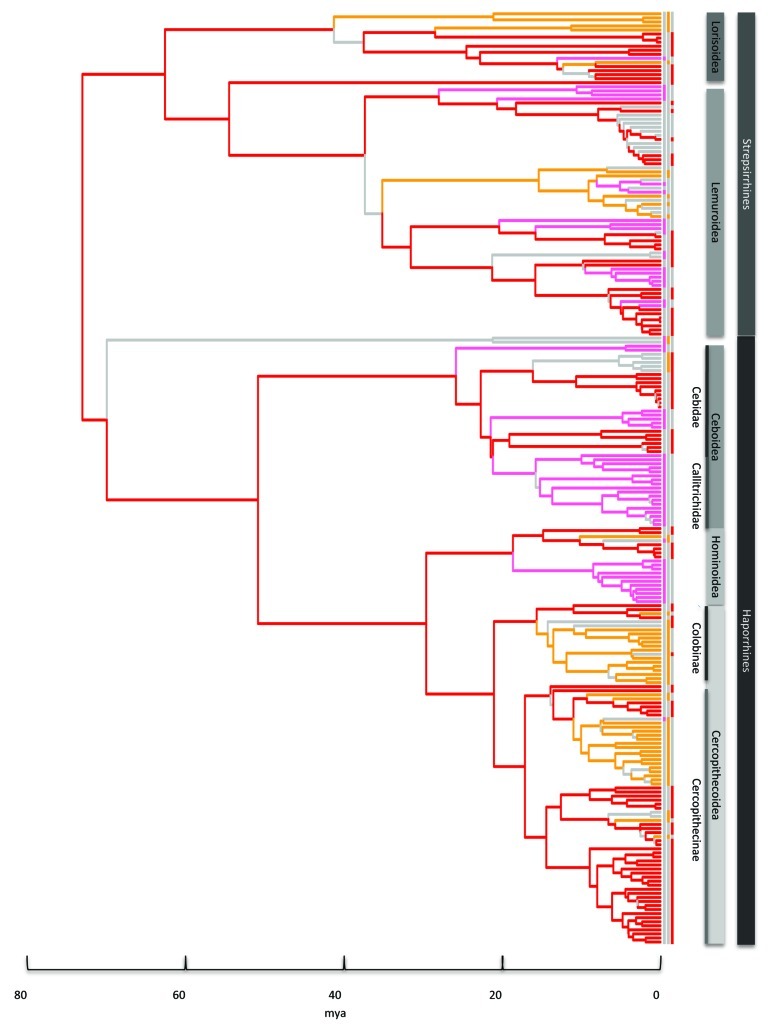
Harem-polygyny evolved earliest among strepsirrhines, first at the root of the Loris sub-family Perodicticinae (~42mya) and later at the root of the Lepilemurs (~36mya) (Fig. 2). Monogamy evolved in a number of Lemur families from ~28mya. Among haplorrhines monogamy emerged first at the root of Callicebus (~26mya), followed by Aotus and the Callitrichids (~22mya) and then Gibbons (~19mya). Harem-polygyny evolved later among Anthropoids, at the root of the Trachypithecus genus (~16mya), and later still at the root of the Cercopithecus genus (~11mya).
Figure 2. Primate phylogeny with ancestral states for mating systems derived from RJ MCMC model of evolution. The tree topology is the maximum clade credibility tree from the 10k Trees Project13 posterior distribution with branch length drawn proportional to time. Branches and tips are colored for polygynandry (red), harem-polygyny (orange) and monogamy (pink) where the combined probability of the state and the branch is greater than or equal to 0.7. Where the combined probability is less than 0.7 the branch is gray.
Like social organization2 mating system data in primates show a strong phylogenetic signal, indicating that it is possible to make inferences about the evolution of this trait across the primate tree. This also suggests that history plays a significant role in the current distribution of both these social traits across species. The model of evolution for primate mating systems is similar to that for primate social organization,2 once multi-male/multi-female groups were established, with transitions to both monogamy and harem-polygyny. The pathways differ in that monogamy was found to evolve from both harem-polygyny and polygynandry.
As with social organization, once monogamy was established (or pair living, in the case of social organization) there were no further transitions. This may be because the cognitive changes to enable the behavioral co-ordination required for stable monogamy are hard to reverse,7 and also because the factors leading to the evolution of monogamy persisted over time.9 Harem-polygyny also evolved directly from polygynandry, but there were back transitions. In contrast, there were no direct transitions into harem-polygyny from monogamy suggesting that rather than a ‘combination of pairs’10 harem-polygyny can be better described as ‘degraded’ polygynandry. An interesting question for future work is the extent to which this is due to ecological and physiological conditions, such as small female groups and asynchronous estrus, enabling a single male to monopolise mating within a group of females.11,12 Co-evolutionary models could be used to test the factors leading to the emergence of harem-polygyny.
The derived states of mating system and social organization evolved at a similar time across diurnal primate clades such that they matched through most of primate evolutionary history. However, among those primates that remained nocturnal, solitary social organization persisted even when mating systems changed from the ancestral state of polygynandry to the derived states of harem-polygyny or monogamy.
Methods
Model testing was performed across a Bayesian posterior distribution of 10,000 ultrametric primate trees derived from genetic data (version 2 of the 10kTrees Project) to account for uncertainty in the underlying phylogeny.13 We present a maximum clade credibility tree that was inferred from the complete 10kTrees sample using TreeAnnotator.14 Pagel’s Lambda was estimated in R, using the Ape4 and Geiger15 packages. We used a reversible-jump (RJ) Markov chain Monte Carlo (MCMC) analysis in the Multistate procedure of BayesTraits (available from www.evolution.rdg.ac.uk)5,16 to derive the posterior distribution of log-likelihoods, the rate parameters of models of evolution, and trait values at ancestral nodes on the primate phylogeny including the root, where the frequency in the posterior distribution represents the posterior belief in that outcome. Primate mating systems were classified as monogamous (0) harem-polygynous (1) or polygynandrous (2), polymorphic species were coded accordingly, with data taken from the literature (see SI for2). The term monogamy is used in the sense of social monogamy, which may vary from genetic monogamy in a number species.
Footnotes
Previously published online: www.landesbioscience.com/journals/cib/article/20821
References
- 1.Krebs J, Davies N. Behavioral ecology: an evolutionary approach 4th edn, (Wiley-Blackwell, 1997). [Google Scholar]
- 2.Shultz S, Opie C, Atkinson QD. Stepwise evolution of stable sociality in primates. Nature479, 219-222,http://www.nature.com/nature/journal/v479/n7372/abs/nature10601.html - supplementary-information (2011). [DOI] [PubMed]
- 3.Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP. Bayesian inference of phylogeny and its impact on evolutionary biology. Science. 2001;294:2310–4. doi: 10.1126/science.1065889. [DOI] [PubMed] [Google Scholar]
- 4.Paradis E, Claude J, Strimmer K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics. 2004;20:289–90. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 5.Pagel MD, Meade A. Bayesian analysis of correlated evolution of discrete characters by reversible-jump Markov chain Monte Carlo. Am Nat. 2006;167:808–25. doi: 10.1086/503444. [DOI] [PubMed] [Google Scholar]
- 6.Kappeler PM, van Schaik CP. Evolution of primate social systems. Int J Primatol. 2002;23:707–40. doi: 10.1023/A:1015520830318. [DOI] [Google Scholar]
- 7.Shultz S, Dunbar RIM. The evolution of the social brain: anthropoid primates contrast with other vertebrates. Proc Biol Sci. 2007;274:2429–36. doi: 10.1098/rspb.2007.0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Schaik CP, Kappeler PM. in Monogamy: mating strategies and partnerships in birds, humans and other mammals (eds U.H. Reichard & C. Boesch) Ch. 4, 59–80 (Cambridge University Press, 2003). [Google Scholar]
- 9.Opie, C., Atkinson, Q.D., Dunbar, R.I.M. and Shultz, S. The evolution of pair living in primates. (in prep).
- 10.Harcourt AH, Greenberg J. Do gorilla females join males to avoid infanticide? A quantitative model. Anim Behav. 2001;62:905–15. doi: 10.1006/anbe.2001.1835. [DOI] [Google Scholar]
- 11.Dunbar RIM. Primate Social Systems (Chapman & Hall, 1988). [Google Scholar]
- 12.Dunbar RIM. in Primate Males: causes and consequences of variation in group composition (ed Peter M. Kappeler) 259-268 (Cambridge University Press, 2000). [Google Scholar]
- 13.Arnold C, Matthews LJ, Nunn CL. The 10k trees website: a new online resource for primate phylogeny. Evol Anthropol. 2010;19:114–8. doi: 10.1002/evan.20251. [DOI] [Google Scholar]
- 14.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harmon LJ, Weir JT, Brock CD, Glor RE, Challenger W. GEIGER: investigating evolutionary radiations. Bioinformatics. 2008;24:129–31. doi: 10.1093/bioinformatics/btm538. [DOI] [PubMed] [Google Scholar]
- 16.Pagel MD, Meade A, Barker D. Bayesian estimation of ancestral character states on phylogenies. Syst Biol. 2004;53:673–84. doi: 10.1080/10635150490522232. [DOI] [PubMed] [Google Scholar]