Abstract
In many animal species, traits associated with male fitness evolve rapidly. Intersexual conflict and male-male competition have been suggested to drive this rapid evolution. These fast evolutionary dynamics result in elevated rates of amino acid replacement and modification of gene expression attributes. Gene acquisition is another mechanism that might contribute to fitness differences among males. However, empirical evidence of fitness effects associated with newly evolved genes is scarce. The Sdic multigene family originated within the last 5.4 myr in the lineage that leads to D. melanogaster and encodes a sperm dynein intermediate chain presumably involved in sperm motility. The silencing of the Sdic multigene family, followed by the screening of relevant phenotypes, supports the role of the Sdic multigene family in sperm competition. The case of the Sdic multigene family illustrates the flexibility of genetic networks in incorporating lineage-specific gene novelties that can trigger an evolutionary arms race between males.
Keywords: Drosophila, chromosome engineering, fitness variation, genotype-phenotype map, male fertility, newly evolved genes, sperm competition
The gene Sperm dynein intermediate chain (Sdic) is located at the cytological location 19C1 of the X chromosome of D. melanogaster but is absent in its closest relatives (Fig. 1A).1,2 This gene was discovered during the characterization of one of its adjacent genes, short wing (sw) [aka Cdic], which encodes a cytoplasmic dynein intermediate chain.3 In fact, the structure of the gene Sdic includes exonic sequences from the gene sw and from its other flanking gene Annexin X (AnnX). Additionally, the gene Sdic possesses one newly evolved exon and regulatory sequences.4 Its chimeric nature and its presence as a tandem array of multiple copies suggested an evolutionary scenario associated with several consecutive segmental duplications.4-6 Early functional analysis of one of the Sdic copies, Sdic1, indicated that its expression is confined to testis with its encoded protein being present in seminal vesicles and maturing spermatocytes, especially along their tails.4 Given the expression profile of Sdic1 and its structural relationship with sw (only the exons of the parental gene sw are present in the transcript of Sdic), it was proposed that the SDIC protein is a sperm-specific axonemal dynein intermediate chain and therefore putatively relevant to the fertility of males. Genes associated with reproduction are potential targets of sexual selection, e.g., through male-male competition.7 In the case of the Sdic multigene family, sexual selection may adopt the form of sperm competition, thus explaining the rapid evolution of Sdic. This notion would be consistent with the multiple rearrangements this region has undergone in a very short evolutionary period and with unusually low levels of nucleotide variation.8,9 However, empirical evidence of the phenotypic effect associated with the Sdic multigene family, and therefore of its presumably adaptive role, was still lacking.
Figure 1. (A) Phylogeny of D. melanogaster (mel), its closest relatives (rel: D. simulans, D. sechellia, and D. mauritiana), and the outgroup species D. yakuba (yak). The Sperm dynein intermediate chain (Sdic) multigene family is only present in D. melanogaster and therefore it must have been originated after its lineage branched off from that leading to its closest relatives 5.4 mya.36 (B) Details of the molecular organization of the Sdic multigene family at 19C1 of the X chromosome. Although four copies are annotated in FlyBase,25 our computational analyses revealed the existence of additional haplotypes that denote the existence of further copies, in good agreement with earlier estimates (see main text). The presence of additional copies of the gene Sdic could conflict with models previously proposed for the generation of the multigene family.6
In a recent report, we knocked out the whole Sdic multigene family at 19C1.10 Four copies of Sdic are annotated in the most recent release of the D. melanogaster genome. For at least two of these copies, Sdic1 and Sdic3, there is unambiguous evidence of their expression in adult males.4,10 We induced the deletion of the Sdic multigene family through an ectopic recombination event between two flanking FRT-bearing transposable elements. Since the deletion of the Sdic multigene family also spans the essential gene sw,10-12 a sw transgene was introduced into the genome of the engineered stocks, i.e., those without the Sdic multigene family. The transgene recapitulates the endogenous expression of the gene sw in whole males10 and in testis (Fig. 2). The absence of the Sdic multigene family did not result in obvious sperm malformations or significantly diminished fertility. However, the silencing of the Sdic multigene family did lead to an impairment of the competence of the sperm in experimental settings involving consecutive matings between one female and two individual males.13 This phenotype is likely to be important for fitness because polyandry is well documented in natural populations of D. melanogaster.14-18 The type of assay performed compares the ability of the sperm from males with and without the Sdic multigene family (the experimental males) to displace or inactivate the sperm of a reference wild-type male, which results in differences in the fraction of the progeny fathered by each male. The experimental males can be first or second to mate relative to the reference male depending on the assay. In the assay in which the experimental males were second to mate, we detected that the fraction of progeny fathered by the knockout experimental male was significantly reduced compared with that of the experimental male carrying the Sdic multigene family. The molecular mechanisms whereby this diminished competence appears remain unknown. It is possible that mobility of the sperm, which has been proposed to affect fertilization efficiency,19 had been subtly altered. This altered mobility can also affect sperm utilization upon contact with particular accessory gland proteins, some of which have been shown to influence the outcome of sperm competition due to their involvement in sperm storage and displacement.20 Likewise, there could be an altered interaction with the proteins secreted by the spermathecal secretory cells in females21 or a combination of any of these scenarios. The phenotype detected supports that the Sdic multigene family indeed has a measurable impact on male fertility in spite of its recent origin.
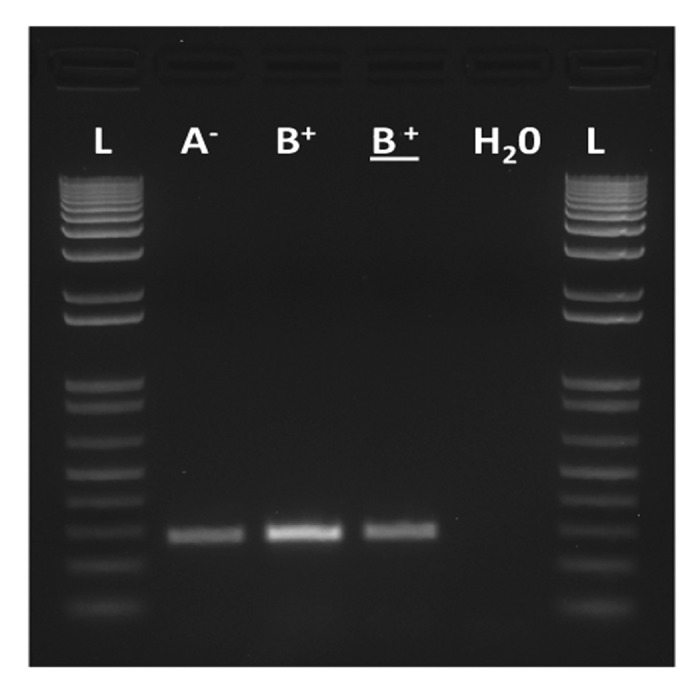
Figure 2. Molecular verification by RT-PCR of the proper expression of the sw transgene in testis of males with (A-) and without Sdic (B+). The lane of the underlined B+ corresponds to males carrying the endogenous copy of sw but not the transgene. H2O, negative control (no cDNA was added); L, ladder. Primers and methods are as described.10
Several important aspects of our findings warrant further clarification. First, the phenotype observed as a consequence of the silencing of the Sdic multigene family does not seem to diminish sperm motility qualitatively in non-competitive conditions. One explanation is that at least one other gene is performing this function. An alternative explanation is the presence of at least one additional functional copy of the gene Sdic outside 19C1 on the X chromosome, which would have not been knocked out by our silencing approach. Computational analysis using raw sequences absent in the main assembly of the reference strain of D. melanogaster y1; cn bw1 sp1 22 supported the existence of additional haplotypes that must belong to other copies of the gene Sdic (Fig. 1B). This result would be in good agreement with previous estimates based on Southern blot experiments,4 and the restriction profile of BACs (Berkeley Drosophila Genome Project; unpublished results) and P1 phages (J.M. Ranz, unpublished results). In situ hybridization experiments on mitotic chromosomes rejected this scenario, eliminating the possibility that extra copies outside the cluster of 19C1 on the X chromosome exist, e.g., in poorly annotated regions of the D. melanogaster genome like the Y chromosome. Another possibility is that the gene sw, which is also known to be involved in sperm differentiation,23 may encode protein isoforms functionally equivalent to the SDIC protein. Although multiple SW protein isoforms exist, they all differ significantly from the SDIC protein at both termini.4,5 Recently, the first axonemal dynein intermediate chain gene has been characterized in D. melanogaster.24 The gene Dic61B exists as a single copy, is expressed in testis, and shows homology to axonemal dynein intermediate chain encoding genes from other organisms, including that of the gene DNAI1 in humans. Importantly, when the gene Dic61B is knockout, sperm individualization fails. Since the gene Dic61B is found in D. simulans, we hypothesize that it is the truly indispensable axonemal intermediate chain encoding gene, while Sdic may be still evolving its function affecting the competence of the sperm of D. melanogaster in a more subtle manner.
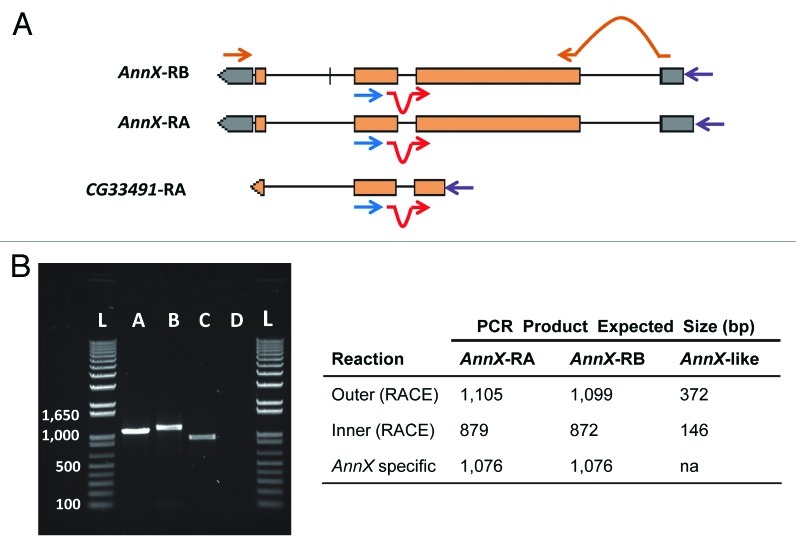
A second important aspect is the phenotype detected in the absence of the Sdic multigene family. In recent releases of the D. melanogaster genome assembly, the residual stretches of the parental gene AnnX still part of the Sdic gene structure have been annotated as independent transcriptional units (CG33491, CG33496, CG33487, and CG33498; collectively AnnX-like hereafter).25 Evidence of the expression of these genes stemmed from an RNA-seq experiment,26 which indicated that these AnnX-like genes reach their peak of expression in 5-d-old males. These transcriptional units are part of the deleted chromosomal stretch. If these AnnX-like genes can somehow affect male fertility, it is unclear whether the reduced sperm competence observed results partially or entirely from their deletion as opposed to the deletion of the gene copies of Sdic. 5′ RACE experiments using 5-d-old males did not find evidence of transcription of the AnnX-like gene upstream Sdic (Fig. 3), in good agreement with the absence of supporting ESTs for these genes.27 Collectively, these results point to a potential artifact in the current functional annotation of the AnnX-like genes.
Figure 3. Functional test of the expression of AnnX-like genes. (a) Outline of the strategy followed. 5′ RACE experiments were performed to detect the presence of the putative transcripts corresponding to the AnnX-like genes in whole body and testis of 5-d-old males. The experiments were done following the instructions of the kit from Epicenter ExactSTART Eukaryotic mRNA 5′-&3′-RACE. Primers used: outer primer (blue; 5′GGATCCAAGTAGGGCTGTCA3′), located on the third exon of AnnX and the second exon of AnnX-like (only CG33491 is shown); inner primer (red; 5′GACTGGACGCACTCAACTATGG3′), located at the junction of the second and third exons; and a 5′ primer from the kit used (purple). The sequences of the outer and inner primers are complementary to those of the two AnnX transcripts and to that putatively transcribed from the AnnX-like genes according to FlyBase.25AnnX transcript specific primers (brown) were used for RT-PCR experiments with the only purpose to test for the quality of the cDNA and the presence of genomic DNA. (B) Results for the expression profiling experiments in testis. Left, 5′ RACE products amplified from cDNAs of total RNA from males carrying the wild-type configuration for the Sdic multigene family (B+). Lanes: 1) PCR product using AnnX specific primers; 2) PCR product using the outer primer and the 5′ primer; 3) PCR product using the inner primer and the 5′ primer; 4) H2O, negative control (cDNA but no primers added); L) ladder. Right, summary table with the expected size of the PCR products for the two transcripts of AnnX and for the putative transcript of AnnX-like genes. Evidence for the presence of the two AnnX transcripts was detected but not for the putative transcript of AnnX-like genes. Experiments using total RNA from whole bodies as starting material shown identical results (not shown).
The acquisition of new genes with the potential to affect male fitness is widespread across taxa.28-30 Recently originated genes can become stable components of the genome if they contribute to differences in reproductive success.7 Empirical evidence of the impact of newly evolved genes on male fertility has only been documented in very few cases.31-35 Our functional characterization of the Sdic multigene family precisely illustrates the genomic consequences of the arms race established among males, which continuously, and in a very short evolutionary time, reshapes the genetic network that underlies male reproductive traits.
Acknowledgments
This work is supported by NSF grant MCB-1157876 to J.R.
Glossary
Abbreviations:
- AnnX
Annexin X
- EST
expressed sequence tag
- mya
million years ago
- RACE
Rapid Amplification of cDNA End
- sw
short wing
- Sdic
Sperm dynein intermediate chain
Footnotes
Previously published online: www.landesbioscience.com/journals/cib/article/21136
References
- 1.Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, et al. Drosophila 12 Genomes Consortium Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–18. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- 2.Ponce R. The recent origin of the Sdic gene cluster in the melanogaster subgroup. Genetica. 2009;135:415–8. doi: 10.1007/s10709-008-9288-0. [DOI] [PubMed] [Google Scholar]
- 3.Nurminsky DI, Nurminskaya MV, Benevolenskaya EV, Shevelyov YY, Hartl DL, Gvozdev VA. Cytoplasmic dynein intermediate-chain isoforms with different targeting properties created by tissue-specific alternative splicing. Mol Cell Biol. 1998;18:6816–25. doi: 10.1128/mcb.18.11.6816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nurminsky DI, Nurminskaya MV, De Aguiar D, Hartl DL. Selective sweep of a newly evolved sperm-specific gene in Drosophila. Nature. 1998;396:572–5. doi: 10.1038/25126. [DOI] [PubMed] [Google Scholar]
- 5.Ranz JM, Ponce AR, Hartl DL, Nurminsky D. Origin and evolution of a new gene expressed in the Drosophila sperm axoneme. Genetica. 2003;118:233–44. doi: 10.1023/A:1024186516554. [DOI] [PubMed] [Google Scholar]
- 6.Ponce R, Hartl DL. The evolution of the novel Sdic gene cluster in Drosophila melanogaster. Gene. 2006;376:174–83. doi: 10.1016/j.gene.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 7.Singh RS, Artieri CG. Male sex drive and the maintenance of sex: evidence from Drosophila. J Hered. 2010;101(Suppl 1):S100–6. doi: 10.1093/jhered/esq006. [DOI] [PubMed] [Google Scholar]
- 8.Nurminsky D, Aguiar DD, Bustamante CD, Hartl DL. Chromosomal effects of rapid gene evolution in Drosophila melanogaster. Science. 2001;291:128–30. doi: 10.1126/science.291.5501.128. [DOI] [PubMed] [Google Scholar]
- 9.Kulathinal RJ, Sawyer SA, Bustamante CD, Nurminsky D, Ponce R, Ranz JM, et al. Selective sweep in the evolution of a new sperm-specific gene in Drosophila In: Selective Sweep. Nurminsky D (ed), Austin, Texas: Kluwer Academic/Plenum Publishers, 2004. [Google Scholar]
- 10.Yeh SD, Do T, Chan C, Cordova A, Carranza F, Yamamoto EA, et al. Functional evidence that a recently evolved Drosophila sperm-specific gene boosts sperm competition. Proc Natl Acad Sci USA. 2012;109:2043–8. doi: 10.1073/pnas.1121327109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindsley DL, Zimm GG. The genome of Drosophila melanogaster San Diego: Academic Press, 1992. [Google Scholar]
- 12.Boylan KL, Hays TS. The gene for the intermediate chain subunit of cytoplasmic dynein is essential in Drosophila. Genetics. 2002;162:1211–20. doi: 10.1093/genetics/162.3.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clark AG, Aguadé M, Prout T, Harshman LG, Langley CH. Variation in sperm displacement and its association with accessory gland protein loci in Drosophila melanogaster. Genetics. 1995;139:189–201. doi: 10.1093/genetics/139.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milkmann R, Zeitler RR. Concurrent multiple paternity in natural and laboratory populations of Drosophila melanogaster. Genetics. 1974;78:1191–3. doi: 10.1093/genetics/78.4.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parker GA. Sperm competition and its evolutionary consequences in the insects. Biol Rev Camb Philos Soc. 1970;45:525–67. doi: 10.1111/j.1469-185X.1970.tb01176.x. [DOI] [Google Scholar]
- 16.Griffiths RC, McKechnie SW, McKenzie JA. Multiple mating and sperm displacement in natural populations of Drosophila melanogaster. Theor Appl Genet. 1982;62:89–96. doi: 10.1007/BF00276292. [DOI] [PubMed] [Google Scholar]
- 17.Gromko MH, Sheehan K, Richmond RC. Random mating in two species of Drosophila. Am Nat. 1980;115:467–79. doi: 10.1086/283574. [DOI] [Google Scholar]
- 18.Lefevre G, Jr., Jonsson UB. Sperm transfer, storage, displacement, and utilization in Drosophila melanogaster. Genetics. 1962;47:1719–36. doi: 10.1093/genetics/47.12.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Birkhead TR, Martínez JG, Burke T, Froman DP. Sperm mobility determines the outcome of sperm competition in the domestic fowl. Proc Biol Sci. 1999;266:1759–64. doi: 10.1098/rspb.1999.0843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ravi Ram K, Wolfner MF. Seminal influences: Drosophila Acps and the molecular interplay between males and females during reproduction. Integr Comp Biol. 2007;47:427–45. doi: 10.1093/icb/icm046. [DOI] [PubMed] [Google Scholar]
- 21.Schnakenberg SL, Matias WR, Siegal ML. Sperm-storage defects and live birth in Drosophila females lacking spermathecal secretory cells. PLoS Biol. 2011;9:e1001192. doi: 10.1371/journal.pbio.1001192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–95. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 23.Ghosh-Roy A, Desai BS, Ray K. Dynein light chain 1 regulates dynamin-mediated F-actin assembly during sperm individualization in Drosophila. Mol Biol Cell. 2005;16:3107–16. doi: 10.1091/mbc.E05-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fatima R. Drosophila Dynein intermediate chain gene, Dic61B, is required for spermatogenesis. PLoS ONE. 2011;6:e27822. doi: 10.1371/journal.pone.0027822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S, et al. FlyBase Consortium. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 2009;37(Database issue):D555–9. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2010 doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhan Z, Ren J, Zhang Y, Zhao R, Yang S, Wang W. Evolution of alternative splicing in newly evolved genes of Drosophila. Gene. 2011;470:1–6. doi: 10.1016/j.gene.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 28.Betrán E, Thornton K, Long M. Retroposed new genes out of the X in Drosophila. Genome Res. 2002;12:1854–9. doi: 10.1101/gr.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marques AC, Dupanloup I, Vinckenbosch N, Reymond A, Kaessmann H. Emergence of young human genes after a burst of retroposition in primates. PLoS Biol. 2005;3:e357. doi: 10.1371/journal.pbio.0030357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krzywinska E, Krzywinski J. Analysis of expression in the Anopheles gambiae developing testes reveals rapidly evolving lineage-specific genes in mosquitoes. BMC Genomics. 2009;10:300. doi: 10.1186/1471-2164-10-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loppin B, Lepetit D, Dorus S, Couble P, Karr TL. Origin and neofunctionalization of a Drosophila paternal effect gene essential for zygote viability. Curr Biol. 2005;15:87–93. doi: 10.1016/j.cub.2004.12.071. [DOI] [PubMed] [Google Scholar]
- 32.Kalamegham R, Sturgill D, Siegfried E, Oliver B. Drosophila mojoless, a retroposed GSK-3, has functionally diverged to acquire an essential role in male fertility. Mol Biol Evol. 2007;24:732–42. doi: 10.1093/molbev/msl201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dai H, Chen Y, Chen S, Mao Q, Kennedy D, Landback P, et al. The evolution of courtship behaviors through the origination of a new gene in Drosophila. Proc Natl Acad Sci USA. 2008;105:7478–83. doi: 10.1073/pnas.0800693105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ding Y, Zhao L, Yang S, Jiang Y, Chen Y, Zhao R, et al. A young Drosophila duplicate gene plays essential roles in spermatogenesis by regulating several Y-linked male fertility genes. PLoS Genet. 2010;6:e1001255. doi: 10.1371/journal.pgen.1001255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heinen TJ, Staubach F, Häming D, Tautz D. Emergence of a new gene from an intergenic region. Curr Biol. 2009;19:1527–31. doi: 10.1016/j.cub.2009.07.049. [DOI] [PubMed] [Google Scholar]
- 36.Tamura K, Subramanian S, Kumar S. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol Biol Evol. 2004;21:36–44. doi: 10.1093/molbev/msg236. [DOI] [PubMed] [Google Scholar]