Abstract
Sleep improves cognition and is necessary for normal brain plasticity, but the precise cellular and molecular mechanisms mediating these effects are unknown. At the molecular level, experience-dependent synaptic plasticity triggers new gene and protein expression necessary for long-lasting changes in synaptic strength.1 In particular, translation of mRNAs at remodeling synapses is emerging as an important mechanism in persistent forms of synaptic plasticity in vitro and certain forms of memory consolidation.2 We have previously shown that sleep is required for the consolidation of a canonical model of in vivo plasticity (i.e., ocular dominance plasticity [ODP] in the developing cat).3 Using this model, we recently showed that protein synthesis during sleep participates in the consolidation process. We demonstrate that activation of the mammalian target of rapamycin [mTOR] pathway, an important regulator of translation initiation,4 is necessary for sleep-dependent ODP consolidation and that sleep promotes translation (but not transcription) of proteins essential for synaptic plasticity (i.e., ARC and BDNF). Our study thus reveals a previously unknown mechanism operating during sleep that consolidates cortical plasticity in vivo.
Keywords: development, mRNA, ontogeny, plasticity, protein synthesis, translation
Experience-dependent plasticity involves gene expression that is highly regulated at both the transcriptional and translational levels. In particular, regulation at the translational step has become an important mechanism allowing spatial fine-tuning of protein expression and input specific synaptic plasticity, unlike transcription that is confined to the nucleus.1 Translational machinery (ribosomes, mRNA, translation factors) is present in axons and dendrites and allows neurons to adapt their response to environmental stimulation by changing their proteomic profile locally.5 The importance of protein synthesis in the consolidation of synaptic plasticity and memory has long been recognized.6,7 More recently substantial progress has been made in indentifying the molecular mechanisms that regulate activity-dependent protein synthesis. Most steps of translation (initiation, elongation and mRNA sequestration [e.g., mRNA binding proteins]) have been implicated in plasticity-dependent translation regulation.8,9 One critical step involved in this process is the initiation step, mostly controlled by the mTOR pathway.4,10 mTOR, via its direct downstream target, eukaryotic initiation factor 4E (eIF4E)-binding protein 1 (4E-BP1), regulates the translation initiation of 5′ capped mRNA (which comprises most of the mRNA in the cell).10 Previous studies have shown that sleep promotes transcription of mRNA involved in translation regulation,11,12 but whether those factors are activated during sleep is not known. It is also known that sleep, especially deep slow-wave sleep,13,14 is correlated with increased protein synthesis, but to date no specific function has been associated with this phenomenon.
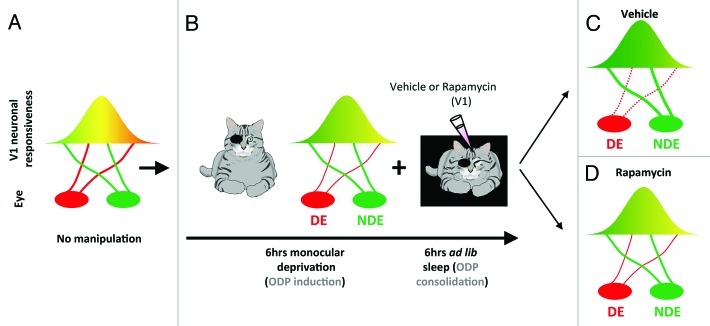
ODP is a classic form of plasticity in vivo that refers to physiological and anatomical changes in visual cortical circuits triggered by transiently blocking patterned vision in one eye (monocular deprivation, Figure 1).15 This type of brain plasticity is considered physiological because in contrast to electrically evoked potentials or tetanic stimulation, it is triggered by alterations of natural sensory input. It also involves various types of synaptic plasticity (Hebbian and non-Hebbian16), many of which require de novo protein synthesis to be consolidated. We have previously shown that ODP can be divided temporally into an induction phase (during waking) and a consolidation phase (during sleep) involving cellular mechanisms of depression and potentiation of the deprived and the non-deprived visual pathways, respectively (Fig. 1).3,17 In our recent study,18 we now show that inhibition of mTOR-dependent protein synthesis during sleep impairs the consolidation of ODP (Fig. 1). Remarkably, mTOR inhibition during wake does not affect the induction phase of ODP. This first finding is important because it suggests that, at a functional level, protein synthesis necessary for ODP consolidation occurs only during sleep. We also find that mTOR inhibition during sleep impairs both potentiation of the open eye and depression of the deprived eye visual pathways (Fig. 1). This confirms previous findings in in vitro models showing the importance of protein synthesis for stabilization of both weakening (e.g., long-term depression) and strengthening (e.g., long-term potentiation) of synapses and demonstrates the requirement for mTOR during sleep in both processes.19
Figure 1. Protein synthesis is required for sleep-dependent ocular dominance plasticity (ODP). (A) In developing cats with normal vision, most neurons in the primary visual cortex (V1) are binocular (i.e. equally responsive to inputs from either eye, represented as the yellow area). (B) When animals are deprived of patterned visual input in one eye (i.e. monocular deprivation) most neurons in V1 become responsive only to stimulation of the non-deprived eye (NDE). This process is induced very rapidly in awake cats (6 h) and is enhanced/consolidated by subsequent sleep (6 h). To test the role of mTOR in sleep-dependent ODP, visual cortices are infused with vehicle or the selective mTOR inhibitor rapamycin during the post-MD sleep period. (C) Sleep-dependent ODP is intact in the vehicle infused hemispheres and includes a maintenance of depression of the DE visual input (dotted red line) and potentiation of the NDE input (thick red line). (D) Inhibition of protein synthesis in V1 with rapamycin during post-MD sleep blocks sleep-dependent ODP. This reflects inhibition of both plastic changes normally observed after sleep (the weakening of the DE and the strengthening of NDE inputs). This results in a V1 plasticity phenotype that is normally seen after the initial 6 h of monocular deprivation only in awake animal (compare B and D).
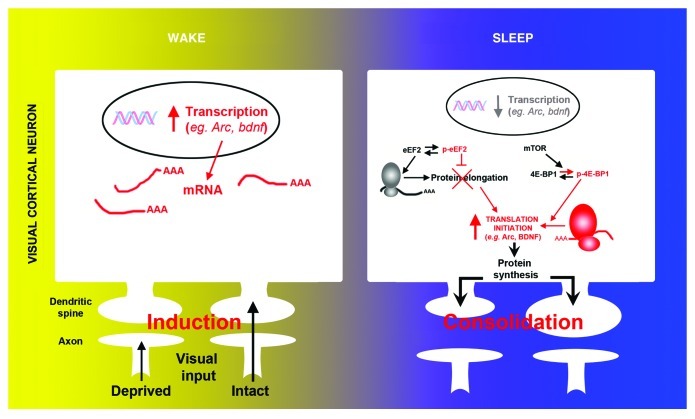
Considerable effort has been focused on identifying mRNAs that undergo rapid translation in response to synaptic activation.20 Most mRNAs identified in neuronal processes (i.e., dendrites and axons) have important functions in synaptic plasticity. This includes plasticity related genes such as arc and bdnf. In our recent report we show that sleep differentially affects transcription and translation of these genes. Using quantitative PCR in visual cortical tissue, we show that the mRNA level of both genes decreased during sleep, as described in studies in adult rodents12 (Fig. 2). However levels of the corresponding proteins transiently increased during the same time period in total protein extracts as well as in synaptic enriched protein fractions (i.e., synaptoneursome) (Fig. 2), demonstrating selective activation of protein synthesis during sleep. In the visual cortex, transcription of the immediate early gene (IEG) arc is triggered by visual input. This suggested that the decreased level of transcripts observed after sleep might also be an indirect consequence of reduced visual input. This was confirmed as transcription of another important IEG, c-fos, decreased in a similar manner during sleep, and both transcripts were similarly reduced by simply blocking binocular vision in an awake animal.
Figure 2. Molecular evidence of protein synthesis regulation during sleep. During wake, the induction of ocular dominance plasticity (monocular deprivation) triggers activity-dependent transcription of selected genes (e.g., arc, bdnf, c-fos) in V1. Subsequent sleep activates a cascade of translational events (increased translation initiation via 4E-BP1 phosphorylation and reduced global elongation via eEF2 phosphorylation) leading to a net increase in translation initiation of subsets of mRNA. Arc and bdnf are two examples of important plasticity-related genes where transcription is decreased and translation is increased during sleep.
Collectively, these findings are important for the following reasons. First, they demonstrate that transcriptional changes are not necessarily mimicked, at the functional level, by changes of the corresponding proteins. This may be especially true for forms of synaptic plasticity that involve translation in the consolidation process. Second, they suggest that some transcriptional changes ascribed to sleep might be epiphenomena, rather than reflecting an active process. This is important because genome wide screening methods have been extensively used to clarify the role of sleep in brain function11,12,21 and current hypotheses on sleep function are based on changes in transcriptomes.11,22 Of course, more comprehensive studies examining a wider-array of genes, their proteins and activity-dependent transcription mechanisms (e.g., CREB-dependent transcription) are needed. Several studies have shown that sleep (especially REM sleep) may promote rapid gene transcription23,24 and that several important gene groups are transcribed during sleep.11,12 This indicates that while the transcription of some genes is not actively regulated by sleep, this may not be universally true for others.
Interestingly, sleep following monocular deprivation, not only induced translation initiation (via increased phosphorylation of the mTOR target 4E-BP1), it also decreased protein elongation (via phosphorylation of the eukaryote elongation factor 2 [eEF2]) (Fig. 2). This may seem paradoxical (i.e., enhanced initiation and decreased elongation rate), but similar events are triggered during synaptic plasticity in vitro25,26 and in vivo27,28 and may promote the translation of specific subsets of mRNAs (e.g., ARC29) (Fig. 2). This was supported by our results showing that translation of plasticity related genes other than ARC and BDNF, such as αCamKII or GlurI, was not affected by sleep. We further confirmed that the molecular changes observed at the translational level (i.e., increased BDNF and ARC protein expression and translation factors phosphorylation) were specific to sleep as they did not occur in animals instead kept awake after monocular deprivation. Therefore an important and exciting future direction for this research is to discover which genes are actively translated during sleep.
These findings provide a new way to investigate sleep function and raise a number of exciting questions. The first is to determine the exact location of sleep-dependent protein synthesis. Our results suggest that translation mechanisms are activated at synapses because most of the protein changes we detect occur in synaptoneurosomes. But these results have to be confirmed and extended to a wider panel of candidate proteins that are translated in an activity-dependent manner (e.g., MAP1b, tissue-plasminogen activator). Second, our results suggest that specific proteins (e.g., ARC and BDNF) are produced during sleep and not others (e.g., αCaMKII and GlurI). This could be explained by the fact that sleep promotes the translation of specific pools of mRNAs. The underlying mechanisms are likely to be complex. For example, RNA-binding proteins are important translation regulators as they allow mRNA transport while inhibiting their translation. Among them, the cytoplasmic polyadenylation element binding protein (CPEB) and Fragile X mental retardation protein (FMRP) are known to be critical in in vitro models of synaptic plasticity, memory formation and proper brain development.30 Interestingly, it is well known that translation of αCaMKII and GluRI (that are not regulated in our model) depends on CPEB.31-33 Other important mRNA-binding mechanisms, such as microRNAs, have also been shown to be regulated by sleep,34,35 but their function, if any, in sleep-dependent plasticity is unknown. Third, our findings indicate that understanding the synaptic proteomic profile of the sleeping brain will provide key insights into sleep function and neurological disorders. In a wide range of vertebrate species, sleep is maximal during times when the brain is rapidly maturing and highly plastic. Sleep also promotes brain protein synthesis not only postnatally, but prenatally as well.36 These ontogenetic periods are accompanied by a number of important processes that require protein synthesis (e.g., axon guidance, dendritogenesis, and synaptogenesis). It is therefore likely that sleep-dependent protein synthesis plays critical roles in the initial establishment and refinement of developing synaptic circuitry.37 Moreover, it is also likely that this function is retained in some fashion across the lifespan.38
At the clinical level, deregulation in mRNA transport and translation is responsible for mental retardation symptoms in several developmental psychiatric disorders, such as Fragile X syndrome (FXS; mutation of the fmrp gene)39 and most likely Down and Angelman syndromes and autism spectrum disorders.40-42 Genomic screening studies provide strong evidence that sleep promotes RNA trafficking and general translational events.43,44 This in turn suggests that sleep is indeed a preferred time for these processes. Mice with mutations in fmrp (FXS) and Ube3a (involved in Angelman syndrome) genes, both involved in different aspect of translation regulation, have impaired sleep characterized by increased wake periods for example.45,46 Therefore a better understanding of how natural behavioral brain states (sleep vs. wake) participate in general mRNA transport and translation is central to understanding how miscues in translation regulation contribute to neurological diseases.
Footnotes
Previously published online: www.landesbioscience.com/journals/cib/article/21010
References
- 1.Cohen S, Greenberg ME. Communication between the synapse and the nucleus in neuronal development, plasticity, and disease. Annu Rev Cell Dev Biol. 2008;24:183–209. doi: 10.1146/annurev.cellbio.24.110707.175235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bramham CR, Wells DG. Dendritic mRNA: transport, translation and function. Nat Rev Neurosci. 2007;8:776–89. doi: 10.1038/nrn2150. [DOI] [PubMed] [Google Scholar]
- 3.Frank MG, Issa NP, Stryker MP. Sleep enhances plasticity in the developing visual cortex. Neuron. 2001;30:275–87. doi: 10.1016/S0896-6273(01)00279-3. [DOI] [PubMed] [Google Scholar]
- 4.Banko JL, Klann E. Cap-dependent translation initiation and memory. Prog Brain Res. 2008;169:59–80. doi: 10.1016/S0079-6123(07)00004-0. [DOI] [PubMed] [Google Scholar]
- 5.Martin KC. Local protein synthesis during axon guidance and synaptic plasticity. Curr Opin Neurobiol. 2004;14:305–10. doi: 10.1016/j.conb.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 6.Hernandez PJ, Abel T. The role of protein synthesis in memory consolidation: progress amid decades of debate. Neurobiol Learn Mem. 2008;89:293–311. doi: 10.1016/j.nlm.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gold PE. Protein synthesis inhibition and memory: formation vs amnesia. Neurobiol Learn Mem. 2008;89:201–11. doi: 10.1016/j.nlm.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Groppo R, Richter JD. Translational control from head to tail. Curr Opin Cell Biol. 2009;21:444–51. doi: 10.1016/j.ceb.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costa-Mattioli M, Sonenberg N. Translational control of gene expression: a molecular switch for memory storage. Prog Brain Res. 2008;169:81–95. doi: 10.1016/S0079-6123(07)00005-2. [DOI] [PubMed] [Google Scholar]
- 10.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–18. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 11.Mackiewicz M, Shockley KR, Romer MA, Galante RJ, Zimmerman JE, Naidoo N, et al. Macromolecule biosynthesis: a key function of sleep. Physiol Genomics. 2007;31:441–57. doi: 10.1152/physiolgenomics.00275.2006. [DOI] [PubMed] [Google Scholar]
- 12.Cirelli C, Gutierrez CM, Tononi G. Extensive and divergent effects of sleep and wakefulness on brain gene expression. Neuron. 2004;41:35–43. doi: 10.1016/S0896-6273(03)00814-6. [DOI] [PubMed] [Google Scholar]
- 13.Ramm P, Smith CT. Rates of cerebral protein synthesis are linked to slow wave sleep in the rat. Physiol Behav. 1990;48:749–53. doi: 10.1016/0031-9384(90)90220-X. [DOI] [PubMed] [Google Scholar]
- 14.Nakanishi H, Sun Y, Nakamura RK, Mori K, Ito M, Suda S, et al. Positive correlations between cerebral protein synthesis rates and deep sleep in Macaca mulatta. Eur J Neurosci. 1997;9:271–9. doi: 10.1111/j.1460-9568.1997.tb01397.x. [DOI] [PubMed] [Google Scholar]
- 15.Wiesel TN, Hubel DH. Comparison of the effects of unilateral and bilateral eye closure on cortical unit responses in kittens. J Neurophysiol. 1965;28:1029–40. doi: 10.1152/jn.1965.28.6.1029. [DOI] [PubMed] [Google Scholar]
- 16.Tropea D, Van Wart A, Sur M. Molecular mechanisms of experience-dependent plasticity in visual cortex. Philos Trans R Soc Lond B Biol Sci. 2009;364:341–55. doi: 10.1098/rstb.2008.0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aton SJ, Seibt J, Dumoulin M, Jha SK, Steinmetz N, Coleman T, et al. Mechanisms of sleep-dependent consolidation of cortical plasticity. Neuron. 2009;61:454–66. doi: 10.1016/j.neuron.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seibt J, Dumoulin MC, Aton SJ, Coleman T, Watson A, Naidoo N, et al. Protein synthesis during sleep consolidates cortical plasticity in vivo. Curr Biol. 2012;22:676–82. doi: 10.1016/j.cub.2012.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klann E, Dever TE. Biochemical mechanisms for translational regulation in synaptic plasticity. Nat Rev Neurosci. 2004;5:931–42. doi: 10.1038/nrn1557. [DOI] [PubMed] [Google Scholar]
- 20.Cajigas IJ, Tushev G, Will TJ, Tom Dieck S, Fuerst N, Schuman EM. The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron. 2012;74:453–66. doi: 10.1016/j.neuron.2012.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maret S, Dorsaz S, Gurcel L, Pradervand S, Petit B, Pfister C, et al. Homer1a is a core brain molecular correlate of sleep loss. Proc Natl Acad Sci U S A. 2007;104:20090–5. doi: 10.1073/pnas.0710131104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tononi G, Cirelli C. Sleep and synaptic homeostasis: a hypothesis. Brain Res Bull. 2003;62:143–50. doi: 10.1016/j.brainresbull.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 23.Ribeiro S, Mello CV, Velho T, Gardner TJ, Jarvis ED, Pavlides C. Induction of hippocampal long-term potentiation during waking leads to increased extrahippocampal zif-268 expression during ensuing rapid-eye-movement sleep. J Neurosci. 2002;22:10914–23. doi: 10.1523/JNEUROSCI.22-24-10914.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ribeiro S, Goyal V, Mello CV, Pavlides C. Brain gene expression during REM sleep depends on prior waking experience. Learn Mem. 1999;6:500–8. doi: 10.1101/lm.6.5.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weatherill DB, McCamphill PK, Pethoukov E, Dunn TW, Fan X, Sossin WS. Compartment-specific, differential regulation of eukaryotic elongation factor 2 and its kinase within Aplysia sensory neurons. J Neurochem. 2011;117:841–55. doi: 10.1111/j.1471-4159.2011.07251.x. [DOI] [PubMed] [Google Scholar]
- 26.Scheetz AJ, Nairn AC, Constantine-Paton M. NMDA receptor-mediated control of protein synthesis at developing synapses. Nat Neurosci. 2000;3:211–6. doi: 10.1038/72915. [DOI] [PubMed] [Google Scholar]
- 27.Belelovsky K, Elkobi A, Kaphzan H, Nairn AC, Rosenblum K. A molecular switch for translational control in taste memory consolidation. Eur J Neurosci. 2005;22:2560–8. doi: 10.1111/j.1460-9568.2005.04428.x. [DOI] [PubMed] [Google Scholar]
- 28.Kanhema T, Dagestad G, Panja D, Tiron A, Messaoudi E, Håvik B, et al. Dual regulation of translation initiation and peptide chain elongation during BDNF-induced LTP in vivo: evidence for compartment-specific translation control. J Neurochem. 2006;99:1328–37. doi: 10.1111/j.1471-4159.2006.04158.x. [DOI] [PubMed] [Google Scholar]
- 29.Park S, Park JM, Kim S, Kim J-A, Shepherd JD, Smith-Hicks CL, et al. Elongation factor 2 and fragile X mental retardation protein control the dynamic translation of Arc/Arg3.1 essential for mGluR-LTD. Neuron. 2008;59:70–83. doi: 10.1016/j.neuron.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin KC, Zukin RS. RNA trafficking and local protein synthesis in dendrites: an overview. J Neurosci. 2006;26:7131–4. doi: 10.1523/JNEUROSCI.1801-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang Y-S, Jung M-Y, Sarkissian M, Richter JD. N-methyl-D-aspartate receptor signaling results in Aurora kinase-catalyzed CPEB phosphorylation and alpha CaMKII mRNA polyadenylation at synapses. EMBO J. 2002;21:2139–48. doi: 10.1093/emboj/21.9.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu L, Wells D, Tay J, Mendis D, Abbott MA, Barnitt A, et al. CPEB-mediated cytoplasmic polyadenylation and the regulation of experience-dependent translation of alpha-CaMKII mRNA at synapses. Neuron. 1998;21:1129–39. doi: 10.1016/S0896-6273(00)80630-3. [DOI] [PubMed] [Google Scholar]
- 33.Pavlopoulos E, Trifilieff P, Chevaleyre V, Fioriti L, Zairis S, Pagano A, et al. Neuralized1 activates CPEB3: a function for nonproteolytic ubiquitin in synaptic plasticity and memory storage. Cell. 2011;147:1369–83. doi: 10.1016/j.cell.2011.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davis CJ, Clinton JM, Taishi P, Bohnet SG, Honn KA, Krueger JM. MicroRNA 132 alters sleep and varies with time in brain. J Appl Physiol. 2011;111:665–72. doi: 10.1152/japplphysiol.00517.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davis CJ, Bohnet SG, Meyerson JM, Krueger JM. Sleep loss changes microRNA levels in the brain: a possible mechanism for state-dependent translational regulation. Neurosci Lett. 2007;422:68–73. doi: 10.1016/j.neulet.2007.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Czikk MJ, Sweeley JC, Homan JH, Milley JR, Richardson BS. Cerebral leucine uptake and protein synthesis in the near-term ovine fetus: relation to fetal behavioral state. Am J Physiol Regul Integr Comp Physiol. 2003;284:R200–7. doi: 10.1152/ajpregu.00190.2002. [DOI] [PubMed] [Google Scholar]
- 37.Swanger SA, Bassell GJ. Making and breaking synapses through local mRNA regulation. Curr Opin Genet Dev. 2011;21:414–21. doi: 10.1016/j.gde.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frank MG. The function(s) of sleep. In: Foundations of Psychiatric Sleep Medicine. Cambridge University Press, Cambridge; 2010; pp 59-78. [Google Scholar]
- 39.Bassell GJ, Warren ST. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60:201–14. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Troca-Marín JA, Alves-Sampaio A, Montesinos ML. Deregulated mTOR-mediated translation in intellectual disability. Prog Neurobiol. 2012;96:268–82. doi: 10.1016/j.pneurobio.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 41.Hoeffer CA, Klann E. mTOR signaling: at the crossroads of plasticity, memory and disease. Trends Neurosci. 2010;33:67–75. doi: 10.1016/j.tins.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Greer PL, Hanayama R, Bloodgood BL, Mardinly AR, Lipton DM, Flavell SW, et al. The Angelman Syndrome protein Ube3A regulates synapse development by ubiquitinating arc. Cell. 2010;140:704–16. doi: 10.1016/j.cell.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mackiewicz M, Zimmerman JE, Shockley KR, Churchill GA, Pack AI. What are microarrays teaching us about sleep? Trends Mol Med. 2009;15:79–87. doi: 10.1016/j.molmed.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cirelli C. The genetic and molecular regulation of sleep: from fruit flies to humans. Nat Rev Neurosci. 2009;10:549–60. doi: 10.1038/nrn2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bushey D, Tononi G, Cirelli C. The Drosophila fragile X mental retardation gene regulates sleep need. J Neurosci. 2009;29:1948–61. doi: 10.1523/JNEUROSCI.4830-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Colas D, Wagstaff J, Fort P, Salvert D, Sarda N. Sleep disturbances in Ube3a maternal-deficient mice modeling Angelman syndrome. Neurobiol Dis. 2005;20:471–8. doi: 10.1016/j.nbd.2005.04.003. [DOI] [PubMed] [Google Scholar]