Abstract
Neurons express new gene transcripts and proteins upon receiving synaptic inputs, and these events are essential for achieving proper neuronal wiring, adequate synaptic plasticity, and updatable memory. However, the biological impact of new gene expression on input-specific synaptic potentiation remains largely elusive, in part because the cell biological and biochemical mechanisms for synaptic targeting of newly synthesized proteins has remained obscure. A new study investigating the targeting of the memory related protein Arc from the soma to the synapses teases apart a novel “inverse” synaptic tagging mechanism that enables Arc to specifically target the un-potentiated synapses, thereby helping to maintain the contrast of synaptic weight between strengthened and weak synapses.
Keywords: Arc, CaMKIIβ, activity-dependent gene expression, inverse synaptic tagging, memory trace
Several pioneering studies have suggested the significance of new gene transcription and new protein synthesis in long-term memory formation.1-5 Recently, the critical importance of neurons in which transcription of various neuronal genes such as c-fos is heightened during the memory encoding period was directly tested. Thus, optogenetic6 or chemical manipulations7 to re-active a transcriptionally pre-activated subset of neurons were found to be sufficient to trigger, or to recapitulate at least in part, the memory recall process, as judged from mouse behavioral criteria. Additional works also indicated that reconsolidation and extinction of fear memory are dissociable processes of memory “updating,” which rely upon induction of de novo gene expression in distinct areas of the brain.8-11 Such memory reallocation processes appear to play a crucial role in updating the emotional valence as well as the sensory and contextual information associated with an episodic event.12 In spite of an growing interest in the physiological role of activity-dependent gene expression and a widely recognized role of CREB in this process,1-3,13-16 how differential gene expression across various brain areas contribute to formation of new memory and to determining the boundary between extinction and reconsolidation of memories that were once formed remain as yet unknown. What we clearly need to better understand are the synaptic mechanisms that underlie the assembly and re-assembly (i.e., the dynamic updating of functional connectivity) of activated neurons in which new memory-related genes are induced. This is not only a fundamental challenge in memory research, but also in molecular systems neuroscience in general (Fig. 1).
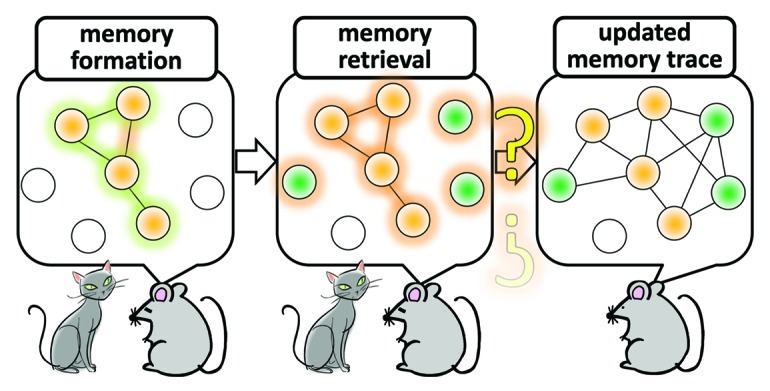
Figure 1. A model of assembly and re-assembly of neurons that are activated during formation and retrieval of memory. Neurons are activated in response to stimuli that trigger memory formation and these activated neurons constitute a functional assembly, or an active neuronal network, in several brain regions. Re-activating this neuronal network is a critical process during memory retrieval. A large amount of evidence indicate that molecular and cellular traces of memory which were produced during memory encoding within the original neuronal network that was assembled in response to the initial memory-inducing stimuli persist in this network. An outstanding question is whether, and if so how, this network of neurons can be “re-assembled” at the cellular and synaptic levels when updating memory after retrieval. Addressing this problem is essential for not only in memory research but also in molecular systems neuroscience.
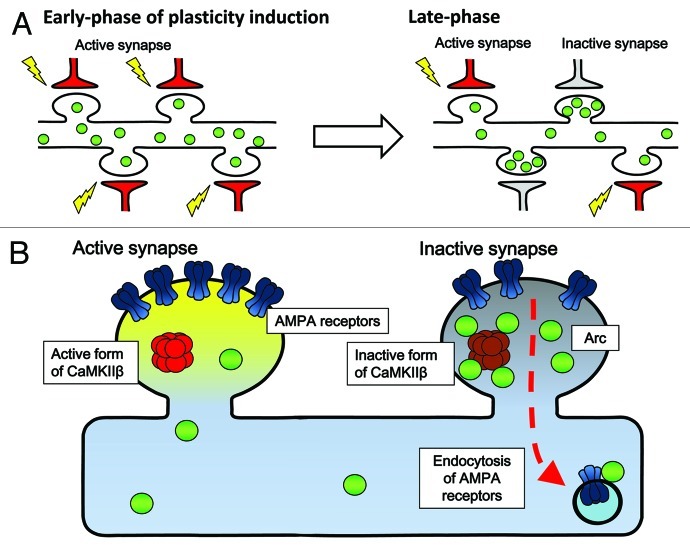
In parallel to in vivo memory studies in mammals, it has been clearly demonstrated in many experimental settings, such as brain slices and dissociated neuronal cultures, that long-lasting changes in synaptic transmission efficacy and structures also require new synthesis of transcripts and proteins.3,17 This may be especially true during a particular time window after receiving plasticity-inducing stimuli when newly induced plasticity-related proteins may functionally interact with synaptic tags.18 However, to date, it remains unclear how specific protein products derived from newly transcribed genes may mediate alteration of neuronal synaptic efficacy. An even larger mystery is to understand how newly-expressed plasticity-related gene products selectively target the very synapses that need to be modulated. In an attempt to address this question, Okuno et al.19 focused on the activity-regulated neuron-specific gene product Arc.20-25 Arc protein induction has been correlated with ongoing cognitive activity in the hippocampus26,27 and in the cortex,5 and its absence caused severe memory disorders.28 When the dynamics of Arc protein’s targeting to the synapses was investigated, a privileged accumulation of Arc was unexpectedly found in non-potentiated, weak, synapses (Fig. 2A). Furthermore, synaptic levels of induced Arc were negatively correlated with the surface expression of glutamate receptors at these synapses during the late phase of potentiation. The critical molecular beacon that physically attracted Arc to weak synapses turned out to be the calmodulin-unbound form of the β subunit of Ca2+/Calmodulin kinase II (CaMKIIβ), a well-known molecular player implicated in synaptic plasticity and memory formation (Fig. 2B). Because the number of glutamate receptors directly determines the efficacy of synaptic connections between neurons in the brain, these results demonstrate that one critical role of Arc may be to keep weak synapses weak, while allowing strong, essential synapses to remain strong and capable of memory storage. These results provide a novel framework for an “inverse synaptic tagging,” which may subserve memory consolidation (and initially block memory updating) by preventing undesired synaptic enhancement at weak synapses, while sparing potentiated synapses. This finding presents new mechanistic insights on how the contrast between strong and weak synapses may be maintained during long-term synaptic plasticity, and shed light on the fundamental role of new gene expression and of the guided targeting of new protein products to synapses as a molecular basis of memory weight allocation at individual synapses within an activated neuronal network.29
Figure 2. A model for Arc's targeting to weak synapses and its net effect on glutamate receptor clearance. (A) Selective accumulation activity-induced Arc protein in inactive synapses during late-phase synaptic plasticity. Upon receiving plasticity-inducing synaptic inputs, Arc is newly synthesized in the cell body and delivered to the dendrites (left). Arc is then gradually lost from active synapses, but in contrast, accumulates in inactive synapses during a following period (right). This Arc accumulation in the inactive synapses relies upon selective interaction with the inactive form of CaMKIIβ. (B) Selective Arc-CaMKIIβ interaction in inactive synapses favors the removal of AMPA receptors from inactive synapses neighboring the active, potentiated synapses.
Acknowledgments
We apologize to the many authors whom we could not cite due to space limitations. This work was supported in part by grants-in-aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan and the Japan Society for Promotion of Science (H.O., H.B.) and a CREST investigatorship (H.B.).
Footnotes
These authors contributed equally to this manuscript.
Previously published online: www.landesbioscience.com/journals/cib/article/20853
References
- 1.Bito H, Deisseroth K, Tsien RW. CREB phosphorylation and dephosphorylation: a Ca(2+)- and stimulus duration-dependent switch for hippocampal gene expression. Cell. 1996;87:1203–14. doi: 10.1016/S0092-8674(00)81816-4. [DOI] [PubMed] [Google Scholar]
- 2.Kida S, Josselyn SA, Peña de Ortiz S, Kogan JH, Chevere I, Masushige S, et al. CREB required for the stability of new and reactivated fear memories. Nat Neurosci. 2002;5:348–55. doi: 10.1038/nn819. [DOI] [PubMed] [Google Scholar]
- 3.Kandel ER, Squire LR. Neuroscience: breaking down scientific barriers to the study of brain and mind. Science. 2000;290:1113–20. doi: 10.1126/science.290.5494.1113. [DOI] [PubMed] [Google Scholar]
- 4.McGaugh JL. Memory--a century of consolidation. Science. 2000;287:248–51. doi: 10.1126/science.287.5451.248. [DOI] [PubMed] [Google Scholar]
- 5.Tse D, Takeuchi T, Kakeyama M, Kajii Y, Okuno H, Tohyama C, et al. Schema-dependent gene activation and memory encoding in neocortex. Science. 2011;333:891–5. doi: 10.1126/science.1205274. [DOI] [PubMed] [Google Scholar]
- 6.Liu X, Ramirez S, Pang PT, Puryear CB, Govindarajan A, Deisseroth K, et al. Optogenetic stimulation of a hippocampal engram activates fear memory recall. Nature. 2012;484:381–5. doi: 10.1038/nature11028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garner AR, Rowland DC, Hwang SY, Baumgaertel K, Roth BL, Kentros C, et al. Generation of a synthetic memory trace. Science. 2012;335:1513–6. doi: 10.1126/science.1214985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nader K, Schafe GE, Le Doux JE. Fear memories require protein synthesis in the amygdala for reconsolidation after retrieval. Nature. 2000;406:722–6. doi: 10.1038/35021052. [DOI] [PubMed] [Google Scholar]
- 9.Mamiya N, Fukushima H, Suzuki A, Matsuyama Z, Homma S, Frankland PW, et al. Brain region-specific gene expression activation required for reconsolidation and extinction of contextual fear memory. J Neurosci. 2009;29:402–13. doi: 10.1523/JNEUROSCI.4639-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suzuki A, Josselyn SA, Frankland PW, Masushige S, Silva AJ, Kida S. Memory reconsolidation and extinction have distinct temporal and biochemical signatures. J Neurosci. 2004;24:4787–95. doi: 10.1523/JNEUROSCI.5491-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim R, Moki R, Kida S. Molecular mechanisms for the destabilization and restabilization of reactivated spatial memory in the Morris water maze. Mol Brain. 2011;4:9. doi: 10.1186/1756-6606-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monfils MH, Cowansage KK, Klann E, LeDoux JE. Extinction-reconsolidation boundaries: key to persistent attenuation of fear memories. Science. 2009;324:951–5. doi: 10.1126/science.1167975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bito H, Deisseroth K, Tsien RW. Ca2+-dependent regulation in neuronal gene expression. Curr Opin Neurobiol. 1997;7:419–29. doi: 10.1016/S0959-4388(97)80072-4. [DOI] [PubMed] [Google Scholar]
- 14.Nonaka M. A Janus-like role of CREB protein: enhancement of synaptic property in mature neurons and suppression of synaptogenesis and reduced network synchrony in early development. J Neurosci. 2009;29:6389–91. doi: 10.1523/JNEUROSCI.1309-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silva AJ, Kogan JH, Frankland PW, Kida S. CREB and memory. Annu Rev Neurosci. 1998;21:127–48. doi: 10.1146/annurev.neuro.21.1.127. [DOI] [PubMed] [Google Scholar]
- 16.Silva AJ, Zhou Y, Rogerson T, Shobe J, Balaji J. Molecular and cellular approaches to memory allocation in neural circuits. Science. 2009;326:391–5. doi: 10.1126/science.1174519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bliss TV, Collingridge GL. A synaptic model of memory: long-term potentiation in the hippocampus. Nature. 1993;361:31–9. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- 18.Frey U, Morris RG. Synaptic tagging and long-term potentiation. Nature. 1997;385:533–6. doi: 10.1038/385533a0. [DOI] [PubMed] [Google Scholar]
- 19.Okuno H, Akashi K, Ishii Y, Yagishita-Kyo N, Suzuki K, Nonaka M, et al. An inverse synaptic tagging of inactive synapses via dynamic interaction of Arc/Arg3.1 with CaMKIIβ. Cell. 2012;149:886–98. doi: 10.1016/j.cell.2012.02.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Link W, Konietzko U, Kauselmann G, Krug M, Schwanke B, Frey U, et al. Somatodendritic expression of an immediate early gene is regulated by synaptic activity. Proc Natl Acad Sci U S A. 1995;92:5734–8. doi: 10.1073/pnas.92.12.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lyford GL, Yamagata K, Kaufmann WE, Barnes CA, Sanders LK, Copeland NG, et al. Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron. 1995;14:433–45. doi: 10.1016/0896-6273(95)90299-6. [DOI] [PubMed] [Google Scholar]
- 22.Chowdhury S, Shepherd JD, Okuno H, Lyford G, Petralia RS, Plath N, et al. Arc/Arg3.1 interacts with the endocytic machinery to regulate AMPA receptor trafficking. Neuron. 2006;52:445–59. doi: 10.1016/j.neuron.2006.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawashima T, Okuno H, Nonaka M, Adachi-Morishima A, Kyo N, Okamura M, et al. Synaptic activity-responsive element in the Arc/Arg3.1 promoter essential for synapse-to-nucleus signaling in activated neurons. Proc Natl Acad Sci U S A. 2009;106:316–21. doi: 10.1073/pnas.0806518106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inoue M, Yagishita-Kyo N, Nonaka M, Kawashima T, Okuno H, Bito H. Synaptic activity-responsive element (SARE): A unique genomic structure with an unusual sensitivity to neuronal activity. Commun Integr Biol. 2010;3:443–6. doi: 10.4161/cib.3.5.12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okuno H. Regulation and function of immediate-early genes in the brain: beyond neuronal activity markers. Neurosci Res. 2011;69:175–86. doi: 10.1016/j.neures.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 26.Guzowski JF, McNaughton BL, Barnes CA, Worley PF. Environment-specific expression of the immediate-early gene Arc in hippocampal neuronal ensembles. Nat Neurosci. 1999;2:1120–4. doi: 10.1038/16046. [DOI] [PubMed] [Google Scholar]
- 27.Ramírez-Amaya V, Vazdarjanova A, Mikhael D, Rosi S, Worley PF, Barnes CA. Spatial exploration-induced Arc mRNA and protein expression: evidence for selective, network-specific reactivation. J Neurosci. 2005;25:1761–8. doi: 10.1523/JNEUROSCI.4342-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plath N, Ohana O, Dammermann B, Errington ML, Schmitz D, Gross C, et al. Arc/Arg3.1 is essential for the consolidation of synaptic plasticity and memories. Neuron. 2006;52:437–44. doi: 10.1016/j.neuron.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 29.Bito H. The chemical biology of synapses and neuronal circuits. Nat Chem Biol. 2010;6:560–3. doi: 10.1038/nchembio.408. [DOI] [PubMed] [Google Scholar]