Abstract
Podosomes and invadopodia seen in osteoclasts and cancer cells, respectively, are actin-rich membrane protrusions. We recently demonstrated that an adaptor protein, Tks5, which is an established regulator of invadopodia in cancer cells, drives osteoclast-osteoclast fusion as well as osteoclast-cancer cell fusion by generating circumferential podosomes/invadopodia. This finding revealed an unexpected potential of podosomes/invadopodia to act as fusion-competent protrusions. Fusion of biological membranes involves the intricate orchestration of various proteins and lipids. Recent literature suggests the importance of membrane curvature formation in lipid bilayer fusion. In this study, we investigated the expression of Bin-Amphiphysin-Rvs161/167 (BAR) domain superfamily proteins, which have membrane deforming activity, during osteoclastogenesis. We found that IRTKS was specifically induced during osteoclast fusion and interacted with Tks5, suggesting the role of IRTKS in the formation of fusion-competent protrusions via its BAR domain.
Keywords: osteoclast fusion, podosomes, fusion-competent protrusions, membrane deformation, BAR domain, IRTKS
Membrane fusion is a fundamental biological phenomenon ranging from intracellular vesicle fusion and mitochondrial remodelling to virus entry, fertilization, myogenesis, and osteoclastogenesis. These fusion processes are organized by a wide variety of molecules with little conservation among species and cell types, which may reflect the independent evolution of each fusion process.1 Therefore, characterization of every fusion molecule is required for a thorough understanding of molecular mechanisms. On the other hand, some principles seem to be shared by various fusion processes that overcome an energetic barrier caused by apposing and merging two lipid bilayers. In vitro protein-free experiments indicated that for lipid bilayers to fuse, they undergo the following steps: establishment of a close contact between the bilayers to become at least partially dehydrated; formation of a protrusion with high curvature between bilayers to expose an unstable outer leaflet thereby resulting in hemifusion; and fusion pore opening associated with membranes with lateral tension that is typically caused by local or global membrane expansion.2-5 Therefore, “fusion proteins” on biological membranes would promote fusion via lowering intermediate energy barriers that otherwise hinder each step.
As shown by Ohya et al., several categories of proteins such as tethering proteins and Rab GTPases sequentially facilitate each fusion step to merge physiological membrane bilayers efficiently.6 Membrane proteins that can induce high curvature and/or tether the two bilayers are widely used in fusion processes such as mitochondrial fusion,7,8 yeast cell mating9 and epithelial or anchor cell fusion of Caenorhabditis elegans.10,11 One of the best-studied fusion events is synaptic vesicle fusion with the presynaptic plasma membrane at the synapse. Synaptotagmin-1 was shown to trigger soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs)-mediated bilayer fusion by generating highly curved membrane upon binding to Ca2+ 12. In this case, local bending of the lipid bilayers by shallow membrane insertion of the C2 domains of synaptotagmin-1 is thought to be important, because deformation of the flat membrane by another polypeptide called the Bin-Amphiphysin-Rvs161/167 (BAR) domain could rescue a synaptotagmin-1 mutant lacking lipid-deforming activity.13 SNAREs bind to synaptotagmin-1 and form complexes of four helices to tightly pull apposing bilayers.14 Since the shallow membrane insertion of proteins promotes bilayer fusion in vitro,15 it is possible that proteins with amphipathic helices and BAR domain superfamily proteins constitute fusion proteins.
BAR domain superfamily proteins are known to induce membrane deformation with the assistance of the actin cytoskeleton.16-18 They include BAR, Extended Fer-CIP homology (EFC)/F-BAR (FCH-BAR), and insulin receptor tyrosine kinase substrate p53 (IRSp53)-missing-in-metastasis (MIM) homology domain (IMD)/I-BAR (inverse BAR), all of which form homo dimers and deform liposomes in vitro.19 Although an increasing number of studies have suggested their critical roles in endocytosis and protrusion formation during cell migration, the function of BAR domain superfamily proteins in membrane fusion has just begun to be investigated.20
Osteoclastogenesis is a process wherein monocyte-derived precursor cells differentiate into multinuclear osteoclasts following the stimulation of macrophage colony-stimulating factor (M-CSF) and receptor activator of NF-κB ligand (RANKL). During osteoclast differentiation, osteoclast precursors fuse with each other to become multinuclear giant cells, with the formation and organization of columnar actin structures called podosomes.21 For osteoclasts to come into close proximity with each other, they need to activate Rho-family GTPases in combination with molecules that rearrange the actin cytoskeleton.22,23 A membrane type 1 matrix metalloproteinase (MT1-MMP)–p130Cas–Rac signaling pathway was recently shown to be essential in this process.24 Cell-cell recognition and tethering are thought to be achieved via cell surface molecules such as E-cadherin,25 vacuolar-type H+-ATPase V0 subunit d2,26 immunoglobulin-like domain containing proteins such as macrophage fusion receptor (MFR),27 and members of the multipass transmembrane protein family such as DC-STAMP28 and OC-STAMP.29 These molecules were shown to be essential for osteoclast fusion, i.e., they can be regarded as bona fide fusion proteins. However, how they facilitate fusion, especially how they cooperate with other fusion proteins, is not known.
We have recently reported that osteoclast fusion and osteoclast-cancer cell fusion is mediated by circumferential podosomes and invadopodia formed in osteoclasts and cancer cells, respectively. Tks5, an indispensable adaptor protein for Src-induced cancer cell invadopodia formation was shown to be a key molecule to drive those fusion processes.30 Although we concluded that Tks5-dependent circumferential podosomes/invadopodia could serve as “fusion-competent protrusions” downstream of phosphatidylinositol 3-kinase, what exactly happens on the fusing membranes remains an enigma. An intriguing hypothesis is that rapid lateral membrane expansion caused by circumferential podosomes/invadopodia provides an unstable bilayer, which would be relaxed by the supply of the membrane from the fusion partner.5 In the course of our studies, we also observed that tiny spindle-like membrane protrusions were generated at the sites of podosome expansion. Penetration of one of those protrusions resulted in osteoclast fusion, which we defined as “filopodia-mediated fusion.”30 Therefore, local bending of the plasma membrane in addition to circumferential podosome formation might drive osteoclast fusion.
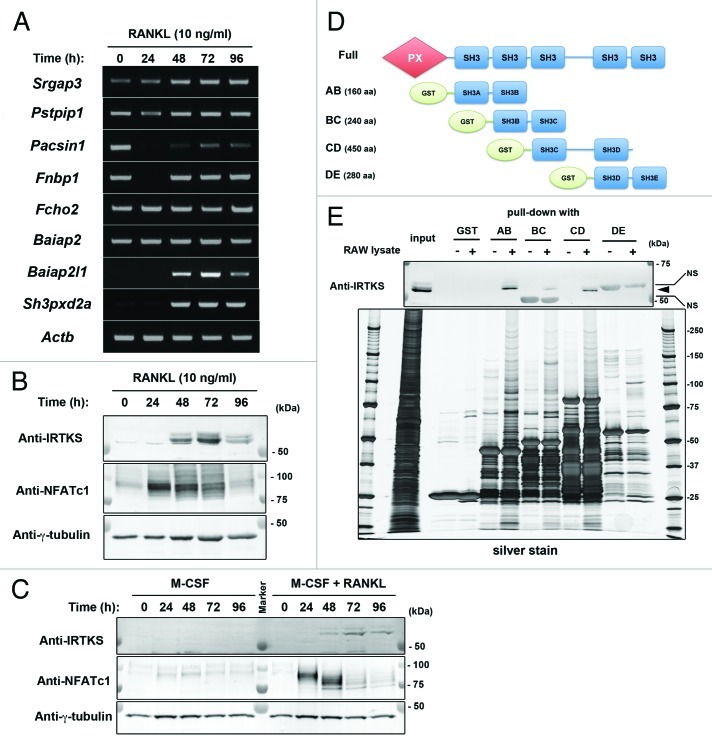
Among the genes whose expression was upregulated (> 1.3-fold increase) in RANKL-stimulated RAW264.7 macrophages, we selected genes known to have a potential activity in membrane protrusion formation.30,31 These included Srgap3, Pstpip1, Pacsin1, Fnbp1, Fcho2, Baiap2 and Baiap2l1. We then confirmed RANKL-dependent upregulation of their expression by reverse transcription–polymerase chain reaction (RT-PCR), and found that insulin receptor tyrosine kinase substrate (IRTKS) (encoded by Baiap2l1) was specifically upregulated in response to RANKL as was Tks5 (encoded by Sh3pxd2a) (Fig. 1A and Table 1). The protein expression of IRTKS was also detected in response to RANKL stimulation with its maximum at 72 h in both RAW264.7 macrophages (Fig. 1B) and in primary bone marrow macrophages (Fig. 1C). Furthermore, IRTKS was shown to interact with Tks5 by a pull-down assay using the truncated Tks5 mutants (Fig. 1D and E). The Tks5 mutants containing either the first two SH3 domains (“AB”) or the middle two SH3 domains (“CD”) bound to IRTKS, suggesting that IRTKS interacted with Tks5 at two different sites. Therefore, IRTKS might function in concert with Tks5 to generate fusion-competent protrusions. However, reduced expression of IRTKS did not show a reproducible effect on osteoclast fusion (data not shown). This might be caused by redundancy of other BAR superfamily proteins in osteoclasts such as Slit-Robo GTPase activating protein 3 (srGAP3, encoded by Srgap3) and/or IRSp53 (encoded by Baiap2), which are expressed when filopodia-like membrane protrusions are formed32,33 (Fig. 1A).
Figure 1. IRTKS is induced in response to RANKL and binds to Tks5. (A) RAW264.7 macrophages were subjected to RT-PCR analysis using indicated primers. (B and C) RAW264.7 macrophages cultured in the presence of RANKL (10 ng/ml) for the indicated times (B) or mouse bone marrow–derived macrophages cultured in the presence of M-CSF (10 ng/ml) with or without RANKL (10 ng/ml) for the indicated times (C) were subjected to immunoblot analysis with antibodies against IRTKS, NFATc1 or γ-tubulin (loading control). The blot with anti-NFATc1 and anti-γ-tubulin were reused from our previous work (©Oikawa, T et al., 2012. Originally published in The Journal of Cell Biology. doi:10.1083/jcb.201111116). (D) Schematic representation of the Tks5 constructs used for the pull-down assay. PX, phox homology domain. SH3, Src homology 3 domain. GST, glutathione S-transferase. (E) RANKL-stimulated RAW264.7 cell lysates were mixed with the GST-SH3 domains of Tks5 immobilized on beads; the bound proteins were subjected to SDS-PAGE and silver staining or to immunoblot analysis with anti-IRTKS antibody. An arrowhead indicates the bands of IRTKS. NS, non-specific.
IRTKS has been shown to regulate plasma membrane deformation as well as actin cytoskeleton remodelling. IRTKS induces short filopodia-like protrusions,34 and is also known to regulate pedestal formation during infection of enterohemorrhagic E. coli.35 Therefore, IRTKS might result in filopodia-mediated osteoclast fusion by generating highly curved membrane protrusions with bundled actin at circumferential podosomes.
Finally, we searched in silico for the putative NFATc1 binding sequences [(A/T)GGAAA(A/N)(A/T/C)N]36 on the promoter regions of Baiap2l1 and Sh3pxd2a. Both Baiap2l1 and Sh3pxd2a had conserved NFATc1 binding sequences at 2208 bp and 1162/1174 bp upstream of the initiation codon, respectively, supporting their specific expression depending on RANKL-NFATc1 signaling (Fig. 2).

Figure 2. Potential NFATc1 binding sites in the Baiap2l1, BAIAP2L1, Sh3pxd2a or SH3PXD2A promoter region: Gene IDs were obtained from the NCBI Gene database. Consensus NFATc1 binding sequences conserved between mouse and human are underlined.
In conclusion, we identified a novel candidate molecule in osteoclast fusion, IRTKS, which was specifically induced in response to RANKL and predicted to mediate local membrane destabilization through its membrane deforming activity. Future study should be aimed to understand how osteoclast-specific molecules such as IRTKS and Tks5 regulate membrane fusion in in vitro reconstitution systems and in vivo.
Materials and Methods
Antibodies
The following antibodies were used for immunoblot analysis: mouse monoclonal anti-IRTKS (sc-100680) and anti-NFATc1 (sc-7294) (Santa Cruz Biotechnology); mouse monoclonal anti–γ-tubulin (Sigma).
Cell Culture
RAW264.7 macrophages were cultured under 5% CO2 at 37°C in α-minimum essential medium (α-MEM) supplemented with 10% fetal bovine serum (FBS) and penicillin-streptomycin. For stromal cell-free osteoclast formation, nonadherent cells were plated at a density of 1 × 106 per well (bone marrow) or 3 × 106 per well (spleen) in 6-well plates and were cultured for 3 d in α-MEM supplemented with 10% FBS and recombinant human M-CSF (R&D Systems) at 10 ng/ml. The resulting M-CSF–dependent macrophages were used as osteoclast precursors. Osteoclast differentiation was induced by culture for 3 to 4 d in the presence of recombinant human M-CSF and recombinant mouse RANKL (R&D Systems), each at 10 ng/ml.
RT-PCR Analysis
Total RNA was extracted from cells with the use of the Trizol reagent (Invitrogen), and portions (0.5 μg) of the RNA were subjected to reverse transcription with SuperScriptII polymerase (Invitrogen). The primers used to detect specific transcripts by subsequent PCR are listed in Table 1.
Table 1. List of primer pairs used for RT-PCR.
| Gene symbol | Sequence 5′-3′ |
|---|---|
|
Srgap3 |
TCGTGGATGACCAAAATGAGC |
| |
TTCGTCACTGGTGTGAGGCT |
|
Pstpip1 |
CTGCAGTTCCGAGATGCCTT |
| |
GATATTTTGCCTGTACACTCTTT |
|
Pacsin1 |
ATGTCTGGCTCCTACGATGAG |
| |
GCCAGCCTTTCCTCCTTACAA |
|
Fnbp1 |
GATCAGTTTGACAACTTGGAAA |
| |
CTGGAAGATGTTGGGGATGTG |
|
Fcho2 |
CTACACAAAGAGAAATAGAAAAG |
| |
AGTAGAAATGGTTCTCTTTGGT |
|
Baiap2 |
ATGTCGCTTTCACGCTCGGA |
| |
CTTGGAATGGTAAGCAGCAGA |
|
Baiap2l1 |
CTCACGGAGAACACGTACCG |
| |
CTGCATGTGATAGTAGTGTATG |
|
Sh3pxd2a |
CAGTCCTGCATTTCTGACGG |
| |
GTTTTTCACGGCAAGGGACA |
|
Actb |
CTCTTTGATGTCACGCACGAT |
| GTGGGCCGCTCTAGGCACCAA |
Acknowledgments
This work was supported by the Promotion of Environmental Improvement for Independence of Young Researchers, “Kanrinmaru Project,” of the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan, a Grant-in-Aid for Young Scientists (A) (23689020) from MEXT and Takeda Science Foundation.
Footnotes
Previously published online: www.landesbioscience.com/journals/cib/article/21252
References
- 1.Chen EH, Grote E, Mohler W, Vignery A. Cell-cell fusion. FEBS Lett. 2007;581:2181–93. doi: 10.1016/j.febslet.2007.03.033. [DOI] [PubMed] [Google Scholar]
- 2.Yang L, Huang HW. A rhombohedral phase of lipid containing a membrane fusion intermediate structure. Biophys J. 2003;84:1808–17. doi: 10.1016/S0006-3495(03)74988-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kozlovsky Y, Kozlov MM. Stalk model of membrane fusion: solution of energy crisis. Biophys J. 2002;82:882–95. doi: 10.1016/S0006-3495(02)75450-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shillcock JC, Lipowsky R. Tension-induced fusion of bilayer membranes and vesicles. Nat Mater. 2005;4:225–8. doi: 10.1038/nmat1333. [DOI] [PubMed] [Google Scholar]
- 5.Kozlov MM, McMahon HT, Chernomordik LV. Protein-driven membrane stresses in fusion and fission. Trends Biochem Sci. 2010;35:699–706. doi: 10.1016/j.tibs.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohya T, Miaczynska M, Coskun U, Lommer B, Runge A, Drechsel D, et al. Reconstitution of Rab- and SNARE-dependent membrane fusion by synthetic endosomes. Nature. 2009;459:1091–7. doi: 10.1038/nature08107. [DOI] [PubMed] [Google Scholar]
- 7.Hales KG, Fuller MT. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell. 1997;90:121–9. doi: 10.1016/S0092-8674(00)80319-0. [DOI] [PubMed] [Google Scholar]
- 8.Koshiba T, Detmer SA, Kaiser JT, Chen H, McCaffery JM, Chan DC. Structural basis of mitochondrial tethering by mitofusin complexes. Science. 2004;305:858–62. doi: 10.1126/science.1099793. [DOI] [PubMed] [Google Scholar]
- 9.Heiman MG, Walter P. Prm1p, a pheromone-regulated multispanning membrane protein, facilitates plasma membrane fusion during yeast mating. J Cell Biol. 2000;151:719–30. doi: 10.1083/jcb.151.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mohler WA, Shemer G, del Campo JJ, Valansi C, Opoku-Serebuoh E, Scranton V, et al. The type I membrane protein EFF-1 is essential for developmental cell fusion. Dev Cell. 2002;2:355–62. doi: 10.1016/S1534-5807(02)00129-6. [DOI] [PubMed] [Google Scholar]
- 11.Sapir A, Choi J, Leikina E, Avinoam O, Valansi C, Chernomordik LV, et al. AFF-1, a FOS-1-regulated fusogen, mediates fusion of the anchor cell in C. elegans. Dev Cell. 2007;12:683–98. doi: 10.1016/j.devcel.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martens S, Kozlov MM, McMahon HT. How synaptotagmin promotes membrane fusion. Science. 2007;316:1205–8. doi: 10.1126/science.1142614. [DOI] [PubMed] [Google Scholar]
- 13.Hui E, Johnson CP, Yao J, Dunning FM, Chapman ER. Synaptotagmin-mediated bending of the target membrane is a critical step in Ca(2+)-regulated fusion. Cell. 2009;138:709–21. doi: 10.1016/j.cell.2009.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hernandez JM, Stein A, Behrmann E, Riedel D, Cypionka A, Farsi Z, et al. Membrane fusion intermediates via directional and full assembly of the SNARE complex. Science. 2012;336:1581–4. doi: 10.1126/science.1221976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chernomordik LV, Kozlov MM. Mechanics of membrane fusion. Nat Struct Mol Biol. 2008;15:675–83. doi: 10.1038/nsmb.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Itoh T, De Camilli P. BAR, F-BAR (EFC) and ENTH/ANTH domains in the regulation of membrane-cytosol interfaces and membrane curvature. Biochim Biophys Acta 2006; 1761:897-912. [DOI] [PubMed]
- 17.Takenawa T. Phosphoinositide-binding interface proteins involved in shaping cell membranes. Proc Jpn Acad, Ser B, Phys Biol Sci. 2010;86:509–23. doi: 10.2183/pjab.86.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suetsugu S, Gautreau A. Synergistic BAR-NPF interactions in actin-driven membrane remodeling. Trends Cell Biol. 2012;22:141–50. doi: 10.1016/j.tcb.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 19.Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, et al. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–9. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 20.Richard JP, Leikina E, Langen R, Henne WM, Popova M, Balla T, et al. Intracellular curvature-generating proteins in cell-to-cell fusion. Biochem J. 2011;440:185–93. doi: 10.1042/BJ20111243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saltel F, Chabadel A, Bonnelye E, Jurdic P. Actin cytoskeletal organisation in osteoclasts: a model to decipher transmigration and matrix degradation. Eur J Cell Biol. 2008;87:459–68. doi: 10.1016/j.ejcb.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 22.Brazier H, Pawlak G, Vives V, Blangy A. The Rho GTPase Wrch1 regulates osteoclast precursor adhesion and migration. Int J Biochem Cell Biol. 2009;41:1391–401. doi: 10.1016/j.biocel.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 23.Leung R, Wang Y, Cuddy K, Sun C, Magalhaes J, Grynpas M, et al. Filamin A regulates monocyte migration through Rho small GTPases during osteoclastogenesis. J Bone Miner Res. 2010;25:1077–91. doi: 10.1359/jbmr.091114. [DOI] [PubMed] [Google Scholar]
- 24.Gonzalo P, Guadamillas MC, Hernández-Riquer MV, Pollán A, Grande-García A, Bartolomé RA, et al. MT1-MMP is required for myeloid cell fusion via regulation of Rac1 signaling. Dev Cell. 2010;18:77–89. doi: 10.1016/j.devcel.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mbalaviele G, Chen H, Boyce BF, Mundy GR, Yoneda T. The role of cadherin in the generation of multinucleated osteoclasts from mononuclear precursors in murine marrow. J Clin Invest. 1995;95:2757–65. doi: 10.1172/JCI117979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee SH, Rho J, Jeong D, Sul JY, Kim T, Kim N, et al. v-ATPase V0 subunit d2-deficient mice exhibit impaired osteoclast fusion and increased bone formation. Nat Med. 2006;12:1403–9. doi: 10.1038/nm1514. [DOI] [PubMed] [Google Scholar]
- 27.Saginario C, Sterling H, Beckers C, Kobayashi R, Solimena M, Ullu E, et al. MFR, a putative receptor mediating the fusion of macrophages. Mol Cell Biol. 1998;18:6213–23. doi: 10.1128/mcb.18.11.6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yagi M, Miyamoto T, Sawatani Y, Iwamoto K, Hosogane N, Fujita N, et al. DC-STAMP is essential for cell-cell fusion in osteoclasts and foreign body giant cells. J Exp Med. 2005;202:345–51. doi: 10.1084/jem.20050645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang M, Birnbaum MJ, MacKay CA, Mason-Savas A, Thompson B, Odgren PR. Osteoclast stimulatory transmembrane protein (OC-STAMP), a novel protein induced by RANKL that promotes osteoclast differentiation. J Cell Physiol. 2008;215:497–505. doi: 10.1002/jcp.21331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oikawa T, Oyama M, Kozuka-Hata H, Uehara S, Udagawa N, Saya H, et al. Tks5-dependent formation of circumferential podosomes/invadopodia mediates cell-cell fusion. J Cell Biol. 2012;197:553–68. doi: 10.1083/jcb.201111116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suetsugu S, Toyooka K, Senju Y. Subcellular membrane curvature mediated by the BAR domain superfamily proteins. Semin Cell Dev Biol. 2010;21:340–9. doi: 10.1016/j.semcdb.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 32.Carlson BR, Lloyd KE, Kruszewski A, Kim IH, Rodriguiz RM, Heindel C, et al. WRP/srGAP3 facilitates the initiation of spine development by an inverse F-BAR domain, and its loss impairs long-term memory. J Neurosci. 2011;31:2447–60. doi: 10.1523/JNEUROSCI.4433-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krugmann S, Jordens I, Gevaert K, Driessens M, Vandekerckhove J, Hall A. Cdc42 induces filopodia by promoting the formation of an IRSp53:Mena complex. Curr Biol. 2001;11:1645–55. doi: 10.1016/S0960-9822(01)00506-1. [DOI] [PubMed] [Google Scholar]
- 34.Millard TH, Dawson J, Machesky LM. Characterisation of IRTKS, a novel IRSp53/MIM family actin regulator with distinct filament bundling properties. J Cell Sci. 2007;120:1663–72. doi: 10.1242/jcs.001776. [DOI] [PubMed] [Google Scholar]
- 35.Vingadassalom D, Kazlauskas A, Skehan B, Cheng HC, Magoun L, Robbins D, et al. Insulin receptor tyrosine kinase substrate links the E. coli O157:H7 actin assembly effectors Tir and EspF(U) during pedestal formation. Proc Natl Acad Sci USA. 2009;106:6754–9. doi: 10.1073/pnas.0809131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rao A, Luo C, Hogan PG. Transcription factors of the NFAT family: regulation and function. Annu Rev Immunol. 1997;15:707–47. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]