Abstract
Antibodies are a unique class of proteins with the ability to adapt their binding sites for high affinity and high specificity to a multitude of antigens. Many analyses have been performed on antibody sequences and structures to elucidate which amino acids have a predominant role in antibody interactions with antigens. These studies have generally not distinguished between amino acids selected for broad antigen specificity in the primary immune response and those selected for high affinity in the secondary immune response. By studying a large data set of affinity matured antibodies derived from in vitro directed evolution experiments, we were able to specifically highlight a subset of amino acids associated with affinity improvements. In a comparison of affinity maturations using either tailored or full amino acid diversification, the tailored approach was found to be at least as effective at improving affinity while requiring fewer mutagenesis libraries than the traditional method. The resulting sequence data also highlight the potential for further reducing amino acid diversity for high affinity binding interactions.
Keywords: antibody, affinity maturation, directed evolution, ScFv, display technology
Introduction
The human immune system has evolved to recognize a vast number of different organic molecules, primarily through the enormous diversity of different binding sites contained within the antibody repertoire. For instance, it is estimated that we synthesize as many as 1010 different antibody sequences in our lifetimes to provide an immune defense against pathogens.1 The route to generating this vast antibody sequence diversity differs according to the stage of the immune response. In the primary immune response, when it is beneficial to generate antibodies to many different antigen specificities, sequence diversity is achieved by the process of V(D)J recombination, which introduces considerable structural diversity into the complementarity-determining region (CDR) loops that bind to antigen.2 In the secondary immune response, antibody affinity is improved by further diversification of antibody sequences, this time by the process of somatic hypermutation, in which the variable regions of the antibody are heavily point-mutated and B cells bearing the highest affinity antibodies, often with multiple CDR mutations, are positively selected.3,4 The primary response, therefore, uses gene recombination to yield generally lower affinity antibodies of broad specificity, whereas the secondary response uses point mutagenesis to yield higher affinity antibodies with singular specificity. As such, the amino acid usage required in CDR loops to generate high affinity in the secondary immune response can differ from that required to generate broad specificity in the primary response.
For the successful application of antibodies in both research and therapy, high affinity is generally a key attribute. For therapy in particular, many antibodies function by stoichiometric blockade of a target protein, so higher affinity enables a longer duration of effect for a given dose of drug. Because of the need for high affinity antibodies, it is useful to understand the amino acid biases in CDR loops that are most appropriate for high affinity antigen interactions. This information is useful because, to improve antibody affinity by mutation, there are practical limitations on the number of variant sequences that can be generated and tested. For example, to generate all possible combinations of amino acid replacements in the antibody CDR loops requires a combinatorial diversity of ~1 × 1078, which vastly exceeds what can be generated in vitro or in vivo (< 1 × 1011). Therefore, if a subset of amino acids can be found that are generally linked to higher affinity binding, then this can help reduce the combinatorial diversity required and improve the efficiency of affinity maturation.
Several studies have aimed to elucidate which amino acids are most prevalent in the CDR loops of naturally-occurring antibodies. The initial approach was to measure CDR amino acid preferences by performing sequence analysis of antibody databases,5-7 but with an increasing number of publicly available antibody:antigen co-crystal structures, these studies then included structural analyses, such as looking for amino acid residues that frequently become buried upon interaction with antigen.8-11 Although not always in complete agreement, these studies highlighted certain amino acids that seem to be over-represented in CDR loops, and therefore are presumed to have a critical role in antigen binding. For instance, most studies were in agreement that tyrosine was a critical CDR residue for binding interactions due to the large side-chain volume and the ability to engage in several different types of bond formations with residues in the antigen interface. This finding was further emphasized in studies using limited antibody diversity in CDR loops, which showed that tyrosine could be responsible for up to 70% of antibody contacts with antigen.12
Due to the different ways in which sequence diversity is generated in vivo during the primary and secondary immune responses, these previous analyses do not necessarily provide information on the amino acid preferences that are specifically linked to higher affinity. One exception was the study of 80 somatic hypermutation variants of six germline antibodies specific to the antigen thyroid peroxidase (TPO), which concluded that certain amino acids such as threonine, serine and glycine were over-represented following affinity maturation.13 The interpretation of these results, however, may be somewhat limited by the relatively small number of mutations included in the analysis and the focus on a single antigen. As an alternative approach, we performed an analysis of a large collection of in-house, unpublished antibody affinity maturation data to specifically assess the different amino acid contributions to affinity. To our knowledge, this analysis of 6095 amino acid replacements arising from the in vitro affinity maturation of 21 different monoclonal antibodies (mAbs) against seven distinct protein antigens is the largest analysis of its kind. The data resulting from the study has allowed us to simplify the affinity maturation process by precisely introducing the amino acid diversity that is preferred in higher affinity antibodies. We refer to this approach as ‘tailored in vitro affinity maturation’ (TiAM). To test the strategy experimentally, we focus on the affinity maturation of two antibodies that bind to the protein antigen inter-cellular adhesion molecule 1 (ICAM-1), a target of therapeutic interest for the prevention of human rhinovirus infections. We report a comparison of libraries in which amino acid diversity is specifically tailored for high affinity vs. those in which complete randomization is employed.
Results
An analysis of amino acid frequencies during affinity maturation was performed on data arising from the directed evolution of 21 mAbs in vitro. As summarized in Table 1, the data used for the analysis shows that these 21 mAbs comprise a diverse set of antibody sequences representing multiple V-region families and with diverse VH and VL CDR3 lengths. The data in Table 1 also highlights that these 21 mAbs were selected against seven distinct protein antigens covering both secreted proteins and cell surface receptor classes. To focus on sequence changes that were associated with significant improvements in affinity, we only analyzed the variants of these 21 mAbs that improved their affinity at least 5-fold over the wild type, or parental, antibody. Figure 1A shows that in most cases the affinity gains were much greater than five-fold and also shows that there was some variation in the number of improved sequences derived from each mAb. To avoid any skewing of data as a result of this variation in sample size, we divided the data into the 36 individual VH or VL CDR3 loop optimizations, and in each case determined which amino acids were selected for, selected against or were relatively neutral (Table S1). To draw some general conclusions about enrichment of certain amino acids during affinity maturation, we then looked across all 36 CDR optimizations and determined the overall frequency at which each amino acid was selected for, selected against, or was relatively neutral (Fig. 1B). We classified amino acid families by volume and polarity14,15 and selected seven amino acids (single letter code: G, P, S, N, H, L and Y) for being most consistently selected from their respective amino acid families during the affinity maturation process. Since the first amino acid family of ‘neutral and small’ side-chains had a strong propensity to be selected, we chose three representatives (G, P and S) from that group, but none from the ‘special’ group that contains only cysteine because this side-chain was consistently selected against during affinity maturation. As a measure of the consistency of these findings across different CDR loops, the data was separately analyzed for VH CDR3 loops, which constituted 19 of the 36 total loop optimizations and VL CDR3 loops, which constituted the other 17 loop optimizations (Fig. 1C and D, respectively). The findings from the overall analysis and the sub-analyses appeared to be in good correlation, suggesting that the amino acid preferences were broadly relevant to both heavy and light chain CDR3 loop optimizations. The data was also analyzed separately to look at the positional bias of mutations in the affinity matured pool; it was of interest to note that there was clearly an over-representation of mutations toward the middle, or apex, of both VH and VL CDR3 loops (Fig. S1).
Table 1. Characteristics of the antibodies in the affinity maturation sequence analysis data set.
| Antibody (CDRs targeted) | Variant amino acids analyzed | Antigen type | VH Germline | VH CDR3, a.a. length | VL Germline | VL CDR3, a.a. length |
|---|---|---|---|---|---|---|
|
1 (H3) |
53 |
Secreted protein |
VH1 1–69 |
17 |
Vλ1 1b |
11 |
|
2 (H3) |
445 |
Secreted protein |
VH1 1–69 |
19 |
Vλ1 1b |
11 |
|
3 (H3) |
73 |
Secreted protein |
VH1 1–46 |
18 |
Vλ3 3l |
13 |
|
4 (H3) |
75 |
Secreted protein |
VH1 1–69 |
16 |
Vλ1 1b |
11 |
| 5 (H3&L3) |
187 |
Receptor |
VH3 3–23 |
19 |
Vλ3 3r |
11 |
| 6 (H3&L3) |
346 |
Receptor |
VH5 5–51 |
12 |
Vλ3 3l |
9 |
|
7 (H3&L3) |
1232 |
Receptor |
VH1 1–24 |
11 |
Vλ1 1e |
11 |
| 8 (H3&L3) |
266 |
Receptor |
VH3 3–23 |
9 |
Vλ3 3r |
10 |
| 9 (H3&L3) |
394 |
Receptor |
VH1 1–24 |
12 |
Vλ1 1c |
11 |
| 10 (H3&L3) |
341 |
Receptor |
VH3 3–30.5 |
13 |
Vλ3 3l |
10 |
|
11 (H3) |
28 |
Secreted protein |
VH3 3–11 |
10 |
Vκ1 L12 |
9 |
|
12 (H3&L3) |
318 |
Secreted protein |
VH1 1–18 |
10 |
Vκ1 L12 |
9 |
|
13 (H3&L3) |
102 |
Secreted protein |
VH3 3–32 |
12 |
Vλ2 2a2 |
9 |
|
14 (H3&L3) |
405 |
Secreted protein |
VH1 1–02 |
10 |
Vλ3 3h |
10 |
|
15 (H3&L3) |
216 |
Secreted protein |
VH1 1–02 |
12 |
Vλ1 1e |
10 |
| 16 (H3&L3) |
225 |
Receptor |
VH3 3–23 |
12 |
Vλ2 2a2 |
7 |
| 17 (H3&L3) |
513 |
Receptor |
VH3 3–11 |
7 |
Vλ3 3m |
11 |
|
18 (H3&L3) |
88 |
Receptor |
VH3 3–30.5 |
13 |
Vλ1 1e |
11 |
|
19 (H3&L3) |
310 |
Receptor |
VH3 3–23 |
21 |
Vλ1 1b |
11 |
|
20 (H3&L3) |
178 |
Receptor |
VH3 3–23 |
7 |
Vλ3 3l |
11 |
| 21 (H3&L3) | 300 | Receptor | VH3 3–23 | 8 | Vλ2 2a2 | 11 |
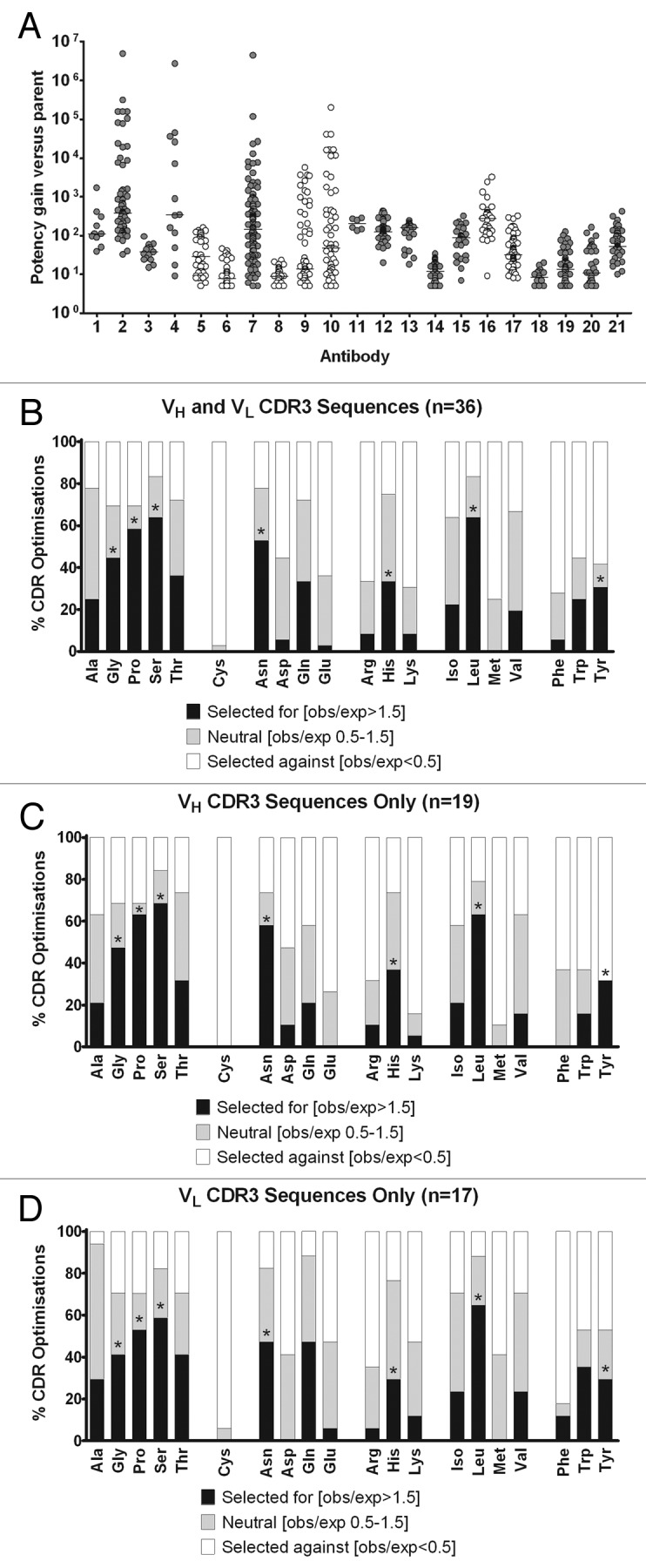
Figure 1. Summary of the amino acid bias observed during the in vitro affinity maturation of 36 individual CDR loops from 21 different monoclonal antibodies (mAbs). (A) shows the range of potency gains for each of the 21 mAbs upon which affinity maturation was performed. The 21 mAbs are grouped according to the antigens they were selected on and data points colored gray or white according to antigen grouping. The affinity matured variant sequences were subsequently included in an analysis for amino acid bias, which is summarized in (B–D). (B) shows the frequency, in the overall data set of VH and VL CDR3 loops, with which each amino acid type was categorised as "selected for" (black bars), "neutral" (light gray bars) or "selected against" (white bars). Amino acids are grouped in six families from left to right, neutral and small, special, polar and relatively small, polar and relatively large, nonpolar and relatively small and nonpolar and relatively large. The representative amino acids from five of the six families that were prioritized for further study on the basis of being "selected for" during the affinity maturation process are highlighted with asterisks. In (C and D), the equivalent data are plotted for VH CDR3 loops alone and VL CDR3 loops alone, respectively.
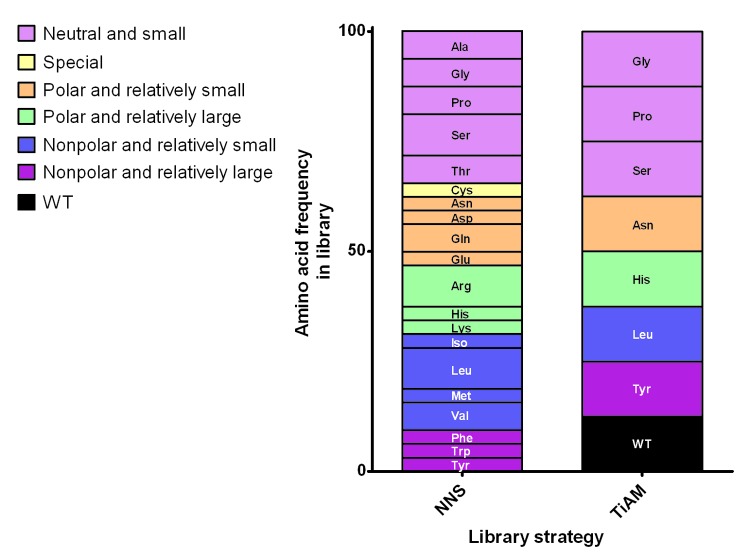
Two antibodies specific for the protein antigen ICAM-1, ICM10064 and ICM10088, were used for the comparison of the TiAM strategy for affinity maturation to the standard, non-tailored approach using NNS codons. Diversified VH and VL CDR3 loop libraries were created for each antibody in scFv format by either a standard approach using the degenerate codon NNS to introduce 32 codon variants representing all 20 amino acids, or the TiAM approach to diversify only the seven amino acids prioritized in the sequence analysis, plus the wild type codon if not already encoded in the set of seven chosen amino acids (Fig. 2). It is worth noting that, to approach full combinatorial diversity, the NNS approach was deliberately limited to stretches of six adjacent positions (326 = 1.1 × 109), whereas the TiAM approach was extended to 12 positions in a CDR loop (712 = 1.4 × 1010; 812 = 6.9 × 1010). We also used the TiAM approach to simultaneously optimize the VH CDR1 and CDR2 loops. Therefore, the NNS method used six libraries to randomize two loops (VH and VL CDR3) in each scFv while the TiAM approach used three libraries to explore four CDR loops (VHCDR1, 2 and 3 and VLCDR3).
Figure 2. Representation of the CDR loop randomization strategies for affinity maturation. In the typical in vitro optimization strategy, blocks of six CDR residues are fully randomized in separate libraries using degenerate NNS codons, producing the amino acid frequencies shown in the graph. In contrast, the TiAM approach uses an equal frequency of eight different codons (encoding the wild type plus the seven preferred amino acid sub-types), allowing up to 12 residues to be mutated in parallel in a single library. Amino acids are grouped in six families according to side-chain properties, neutral and small, special, polar and relatively small, polar and relatively large, nonpolar and relatively small and nonpolar and relatively large.
Following multiple rounds of affinity selections by phage display, individual scFv proteins were screened in a high-throughput affinity assay. Twenty-eight scFv variants of ICM10064 and 72 scFv variants of ICM10088 demonstrated improved affinity over their respective parent scFv in this assay and each was purified for IC50 analysis. In the ICM10064 lineage, the VLCDR3 libraries from both the TiAM and standard NNS approaches produced hits that showed affinity improvements of up to 134-fold and 115-fold, respectively (Figs. 3A and 5A). Differences in variance of the two populations were, however, not significant, suggesting that the two mutagenesis strategies were equally efficient at the task of affinity maturation. For the ICM10088 lineage, affinity improvements were observed from all the CDR loop libraries (Fig. 3B), including multiple variants from VHCDR1, 2 and 3 (Tables S2A and S2B), but again the most highly improved variants were isolated from the VLCDR3 libraries, with affinity improvements over ICM10088 of up to 25-fold and 81-fold, respectively, for the NNS and TiAM strategies (Figs. 3Band 5B). In this case, the TiAM population was more significantly improved than the NNS population (p = 0.0012). The full dose titration curves and IC50 values for most highly improved antibodies from each lineage are shown in Figure 4.
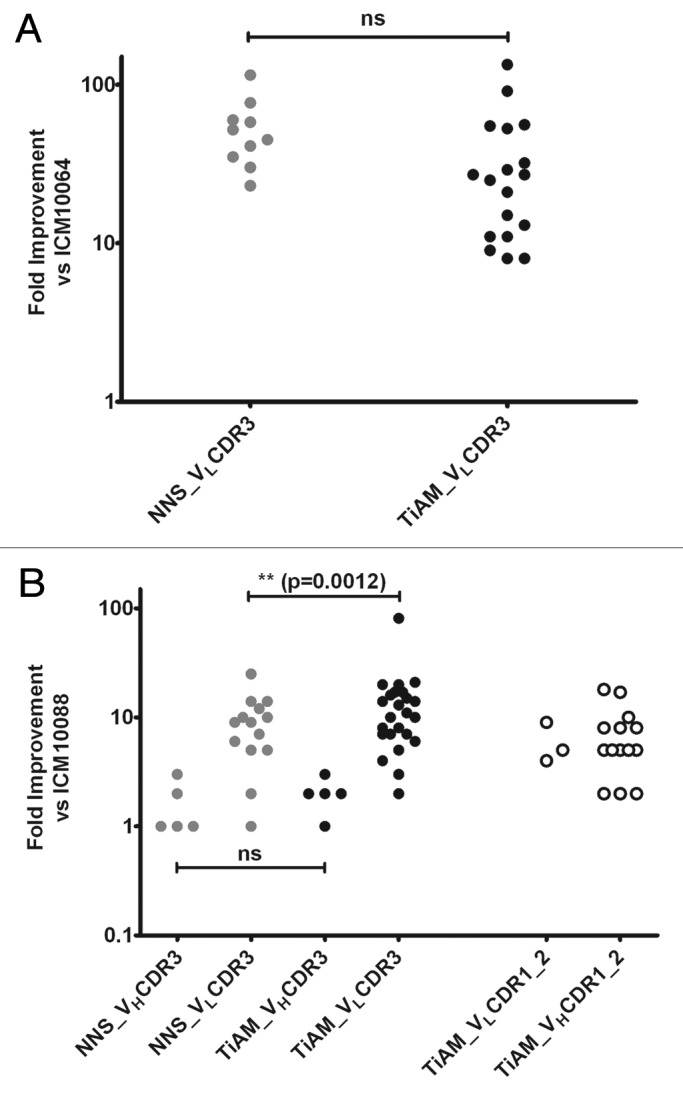
Figure 3. Comparison of affinity gains from the standard NNS optimization approach and the TiAM method. (A) The affinity improvement over the ICM10064 parent antibody for all optimized VLCDR3 variants from the TiAM and NNS methods, respectively. (B) The affinity improvement over the ICM10088 parent antibody for all optimized VHCDR3 and VLCDR3 variants from the TiAM and NNS methods. Also plotted are the improvements conferred by performing the TiAM approach in VLCDR 1 and 2 and VHCDR 1 and 2.
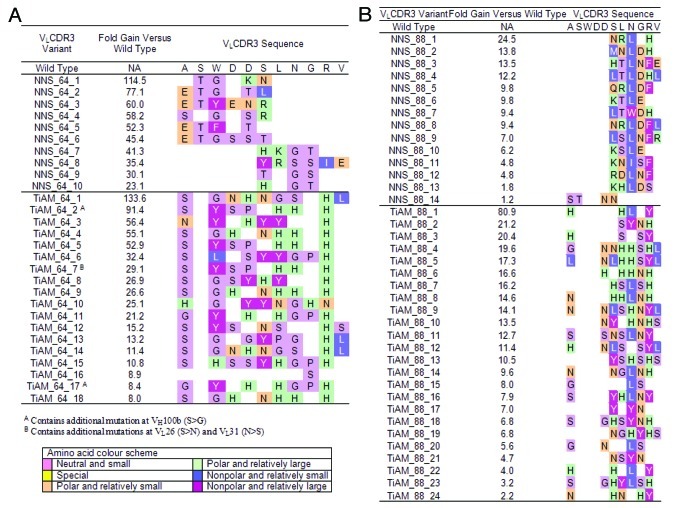
Figure 5. Sequence summary for variants which have undergone TiAM and NNS optimization of VLCDR3 loops in antibodies ICM10064 and ICM10088. For lineage ICM10064 (A) and ICM10088 (B), the wild type VLCDR3 loop sequence is shown at the top and for each variant the changes incorporated during the affinity maturation process are highlighted. Amino acids are colored according to side-chain properties, as detailed in the legend, and the improvement in affinity measured for each individual variant is recorded.
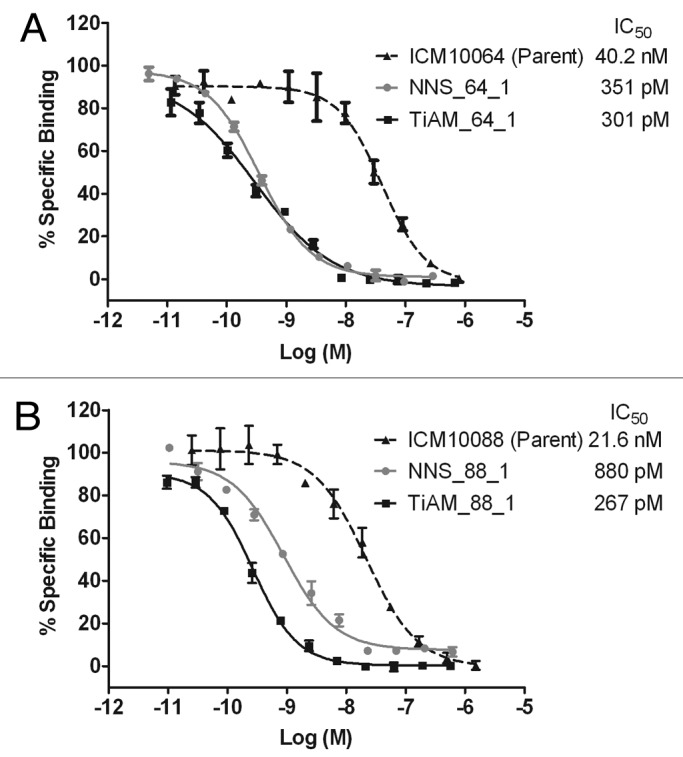
Figure 4. Dose titration analysis of lead antibodies comparing the affinity gains of antibodies from the biased library strategy vs. the standard optimization approach. For lineage ICM10064 (A), the most potent scFv antibodies from the NNS and TiAM approaches respectively, were NNS_64_1 and TiAM_64_1. In the ICM10088 lineage (B), the most potent scFv antibodies from the NNS and TiAM approaches, respectively, were NNS_88_1 and TiAM_88_1. Full dose titration curves, with standard error bars, are plotted and calculated IC50 values are shown for all antibodies.
Analysis of the VLCDR3 sequence changes associated with improved affinity (Fig. 5) shows that different sequence solutions are found for each antibody lineage, with ICM10088 mutations being found predominantly at the C-terminus of the loop, while ICM10064 mutations are dispersed throughout. This is despite the two parent antibodies sharing 96% sequence identity in the VL region and having identical VLCDR3 loop sequences. Comparing the variant sequences from the NNS and TiAM approaches reveals that, for lineage ICM10064, there is a general selection in both approaches for small neutral side-chains (G, P, S and T) with relatively few small, hydrophobic residues (Fig. 6). In contrast, the ICM10088 lineage shows a greater prevalence of hydrophobic (mainly L) and aromatic (F and Y) side-chains in both groups, with strong selection at positions 6, 8 and 10 in the loop (Fig. 5B). The location of mutations within the loops also looks similar between the different strategies, especially in ICM10088, where positions 3 and 4 were never found mutated, while positions 6 to 11 were frequently changed. One notable difference, however, was that many of the ICM10088 TiAM variants, including the highest affinity antibody (TiAM_88_1), combined changes at the C-terminus of the loop with a mutation at the first residue.
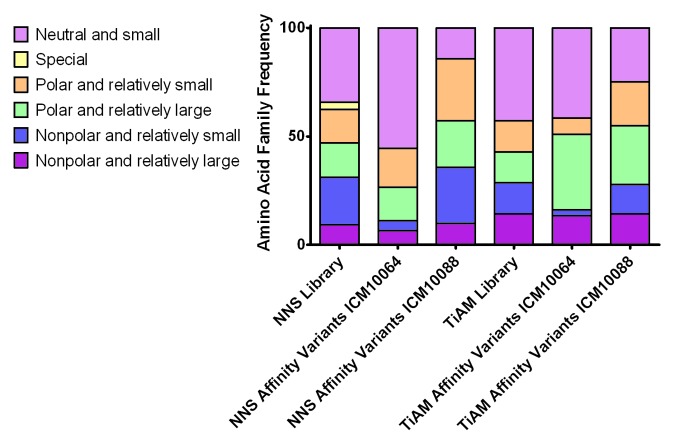
Figure 6. Amino acid frequency in TiAM and NNS libraries before and after selection for improved affinity. The amino acid distribution before library selection was calculated based on the respective library designs and the distribution after affinity selection was calculated from the CDR loop sequences of the affinity matured ICM10064 and ICM10088 variants. Amino acid families are colored according to the key.
With the exception of the ‘special’ amino acid family, containing only cysteine, no families were lost from the libraries during the affinity maturation process (Fig. 6). One consistent feature of the two TiAM affinity maturations was the increase in prevalence of the amino acid histidine, which increased from 16% prevalence in the initial libraries to 35% and 27% in the affinity matured ICM10064 and ICM10088 variant populations, respectively.
Discussion
This study investigates whether an analysis of amino acid preferences in the CDR loops of in vitro affinity matured antibodies can yield useful data to inform future affinity maturation strategies. The analysis revealed that a subset of seven amino acids were over-represented during affinity maturations and this information was used to create tailored libraries to test the hypothesis that this subset were particularly well-suited for affinity maturation. Overall, this tailoring strategy performed at least as effectively as the standard approach of full amino acid randomization, and in some cases could outperform the standard method, while requiring fewer libraries to be built.
Comparing the current analysis to previously published ones such as the global survey of CDR amino acid preferences in large antibody databases5-7 or side-chains in antibody interfaces,8-11 it is clear that prior studies were not designed to focus on affinity maturation per se. Amino acid preferences highlighted in such studies would therefore include contributions from both V(D)J recombination in the primary immune response and from somatic hypermaturation of sequences in the secondary response. In addition, there would have been more subtle evolutionary biases from ancestral features not necessarily related to antigen binding affinity and also inherent DNA biases in CDR loops that allow for preferential incorporation of mutations at certain hotspots during somatic hypermutation.4 In essence, prior analyses have not distinguished between amino acids that are important for creating a diversity of antigen binding sites and those required for the affinity maturation of an antibody toward a single antigen. In the former case, CDR loop flexibility is important to enable binding to the greatest possible diversity of antigenic shapes, whereas in the latter case the CDR loops are required to adopt a single, preferred conformation with the highest possible complementarity to their cognate antigen.
Analyzing naturally hypermutated sequences does allow one to focus on mutations appropriate for high affinity;13 however the nature of somatic hypermutation leads to amino acid changes via single point mutations within a codon, so the resulting amino acids tend to be inherently linked to the starting codon rather than a full exploration of the optimal amino acid.4 In contrast, the analysis described here was based on a large number of sequences arising from a controlled mutagenesis and affinity selection procedure that explored all possible amino acid replacements. The NNS randomization approach used for mutagenesis yields predictable diversity, in terms of the location of mutations and the relative amino acid frequencies in the starting library, which makes it possible to normalize for any inherent biases in the process and, therefore, draw meaningful conclusions about the affinity contributions of individual amino acid types. We also excluded positions that did not change during maturation from the analysis to avoid any biases caused by ancestral sequence features that could have confounded the interpretation.
The conclusion of this new analysis was that certain amino acids representing different families of side-chains,14,15 were over-represented during affinity maturation. Since the whole family of ‘neutral and small’ side-chains had a strong propensity to be selected, we chose three representatives (G, P and S) from that group for further analysis to reflect the observed bias toward that family, but only one representative from other families. The finding that cysteine was strongly under-represented in the affinity matured pool would have been predicted because of the likely folding and solubility problems associated with inserting a single, unpaired cysteine residue into an exposed loop. The significance of the amino acid enrichment data was underlined by a comparison between restricted analyses of VHCDR3 and VLCDR3 loops. Despite these loops normally being very different in both sequence and length, there was strong similarity in the selection of the amino acids in both loop types (Fig. 1). It was of interest to note that a separate positional analysis demonstrated a preference for affinity-enhancing mutations at the apices of both the VHCDR3 and VLCDR3 loops, which is in agreement with previous studies on positional bias of contact residues in CDR loops.16 This may be of interest to others planning directed evolution studies on antibody CDR loops.
The selected amino acids in this study (G, P, S, N, H, L and Y) show some overlap with the amino acids Y, A, D and S, which were selected in a previous study as being able to confer binding to multiple different antigens when introduced into the CDR loops of a single antibody framework.12 In further studies by the same group, the tetranomial diversity was further reduced to a binomial diversity (Y, S) in the CDR loops and the resulting antibodies were still able to perform well in interactions with a protein antigen.17 The main focus of the previous work by Fellouse et al., however, was to test the ability of limited diversity libraries to encode antibodies to any given antigen rather than to improve the affinity of an existing antibody. An unrelated study attempted to reduce the number of mutations required for the engineering of a high affinity antibody by exploring a restricted amino acid diversity of nine amino acids.18 These nine were chosen arbitrarily as representing different side-chain chemistries, rather than based on any analysis, and the second stage of the affinity maturation introduced further amino acid diversity, making it difficult to conclude whether the original tailored diversity had been effective at conferring high affinity binding.
It is worth noting that in our study all seven of the prioritized amino acids were evident in the mutated antibodies following affinity maturation (Figs. 5 and Figure 6), suggesting that they may all actively contribute to the affinity gain. There were, however, some minor variations in frequency among the representatives of the different amino acid families. Worth noting is the apparent enrichment of histidine in the TiAM libraries for both ICM10064 and ICM10088, which suggests that further refinement of the chosen diversity could be explored. It is also evident that within the neutral and small amino acid family, glycine and serine are more enriched than proline during affinity maturation, which might suggest that two or even just one representative from this family are sufficient. The advantage of further restricting the amino acid diversity would be to enable more positions to be mutated in a single library, potentially offering a greater chance of finding synergistic combinations of mutations within the variant pool. This needs to be balanced, however, against the risk that insufficient diversity may prevent some antibodies from finding a solution for high affinity binding.
In terms of overall effectiveness, the tailored approach was seen to be comparable to the full randomization (i.e., NNS) mutagenesis method as measured by the range of affinities isolated. In fact, for the lineage ICM10088, the highest affinity variant TiAM1 (81-fold improved to 267 pM) came from the tailored approach and combined mutations at both the N-terminal and C-terminal ends of the VLCDR3 loop. The ability to select for this combination was enabled by the greater number of positions that could be explored within the limited amino acid subset. It would be of great interest to further explore limiting the diversity and expanding the number of mutated positions for the affinity maturation of additional antibodies. It should be emphasized that the TiAM approach using seven amino acid diversity enabled the exploration of four individual CDR loops in just three libraries, whereas the NNS method required six libraries just to explore two CDR loops, and both approaches delivered comparable affinity gains.
In summary, this study shows the benefit of using a precise antibody sequence analysis to inform further affinity maturations and has led to meaningful conclusions on the relative importance of different side-chains in conferring high affinity binding to antigen. A step toward proving the utility of this approach was made by showing equal, and in some cases superior, effectiveness of tailored diversity vs. full randomization and it is expected that the efficiency of the tailoring strategy could be further enhanced in future studies. Despite the increased synthesis cost of oligonucleotides encoding precise amino acid diversity, there are now novel ways to tailor diversity using conventional synthesis19 and, ultimately, the tailored approach has the possibility to find synergistic sequence combinations not accessible with standard methods.
Materials and Methods
Data analysis
The sequence data of variant antibodies derived from the in vitro affinity maturation of 21 different human mAbs, using methods described previously,20 was included in the initial analysis. Each parental antibody had been subjected to VHCDR3 or VLCDR3 randomization in blocks of 6–7 consecutive CDR loop positions by degenerate codon (NNS) mutagenesis, followed by phage or ribosome display selections for improved affinity.20 For each individual CDR optimization, CDR loop variants were then assayed for improved affinity vs. the original parent antibody in an assay such as that described.21 Only amino acid replacements that improved the affinity of the antibody by at least five-fold were included in the analysis. For each CDR optimization, the total frequency of each amino acid at a mutated position was calculated to generate an observed amino acid frequency (Fobs). To minimize bias caused by the parental CDR sequence, any positions that remained unchanged during affinity maturation were excluded and all amino acid replacements were treated equally regardless of the potency increase they conferred, whether located in a VH or VL CDR3 loop, their position within the loop and whether accompanied by other nearby replacements. To normalize for the known bias introduced by the NNS randomization codon, Fobs was divided by the expected amino acid frequency (Fexp), which was calculated based on the frequency of each amino acid codon provided by the degenerate NNS triplet. Fobs/Fexp ratios greater than 1 indicated an amino acid type that was preferentially selected for during affinity maturation, whereas Fobs/Fexp ratios less than 1 indicated an amino acid type selected against. Using these criteria, it was possible to calculate Fobs/Fexp values for all 20 amino acids across 36 CDR loop optimizations, based on the analysis of 6,095 amino acid replacements. For analysis of positional bias within the CDR3 loop, the frequency at which a given position was found to be mutated in the affinity matured pool (observed frequency) was divided by the frequency at which the residue was mutated in the library designs for each parent antibody (expected frequency).
Library construction and in vitro selection
For NNS-based mutagenesis, oligonucleotides were designed to randomize stretches of six adjacent amino acids within regions encoding the CDR3 loops of the antibodies.22 Multiple, separate libraries were constructed to span the entire CDR3 loop. For tailored mutagenesis, trinucleotide-based oligonucleotides23 were designed to introduce the selected amino acids into regions of up to 12 adjacent positions within a loop. In the tailored approach, the CDR1 and CDR2 loops of the heavy and light chain were also diversified in separate libraries. These oligonucleotides were synthesized at Ella Biotech. All library construction was performed by oligonucleotide-directed mutagenesis, using well-established protocols24 in the pCantab6 phage display vector.25 Phage display selections for improved affinity were performed according to the published methods.26 For a fair comparison of the NNS and tailored library approaches, identical selection conditions were used for each library.
Expression and purification of individual scFvs
For preparation of scFvs, plasmids containing scFv genes were cultured in E. coli, expression induced by isopropyl-D-thiogalactoside, and scFv proteins were isolated from the periplasm by osmotic shock followed by capture of the C-terminal His-tag by Ni2-nitrilotriacetic acid chromatography.27 For IgG expression, the VH and VL chains of selected scFvs were cloned into human IgG expression vectors, and expressed and purified as described.28
Analysis of scFv affinity by epitope competition assay
For affinity screening of variant scFv from the library selections, an HTRF® assay was used to measure the inhibition of the biotinylated parent antibody binding to europium-chelate-labeled ICAM-1 by the scFv or IgG antibody samples, as described for other antibody:antigen interactions.21 Biotinylated parent antibody was bound to streptavidin XLent! (CIS BioInternational, 611SAXAC) by preincubation in the dark for 30 min at room temperature. After preincubation, the biotinylated antibody/streptavidin mix was added to a 384-well black Optiplate (Perkin Elmer, 6005529). This was followed by the addition of diluted test antibody sample and diluted europium-chelate-labeled ICAM-1. Unlabelled parent antibody was used as a positive control for inhibition. After 1 h of incubation in the dark, time-resolved fluorescence at 620 nm and 665 nm was read using an Envision 2101 reader (Perkin Elmer). Data were analyzed by using PRISM software (GraphPad).
Supplementary Material
Acknowledgments
We thank the DNA Chemistry, Tissue Culture and High-Throughput Expression teams at MedImmune for their invaluable support of this work.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/mabs/article/21728
References
- 1.Lesk AM. Introduction to Protein Science. Oxford: Oxford University Press, 2004. [Google Scholar]
- 2.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–81. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 3.Kim S, Davis M, Sinn E, Patten P, Hood L. Antibody diversity: somatic hypermutation of rearranged VH genes. Cell. 1981;27:573–81. doi: 10.1016/0092-8674(81)90399-8. [DOI] [PubMed] [Google Scholar]
- 4.Neuberger MS, Milstein C. Somatic hypermutation. Curr Opin Immunol. 1995;7:248–54. doi: 10.1016/0952-7915(95)80010-7. [DOI] [PubMed] [Google Scholar]
- 5.Kabat EA, Wu TT, Bilofsky H. Unusual distributions of amino acids in complementarity-determining (hypervariable) segments of heavy and light chains of immunoglobulins and their possible roles in specificity of antibody-combining sites. J Biol Chem. 1977;252:6609–16. [PubMed] [Google Scholar]
- 6.Lea S, Stuart D. Analysis of antigenic surfaces of proteins. FASEB J. 1995;9:87–93. doi: 10.1096/fasebj.9.1.7821764. [DOI] [PubMed] [Google Scholar]
- 7.Collis AV, Brouwer AP, Martin AC. Analysis of the antigen combining site: correlations between length and sequence composition of the hypervariable loops and the nature of the antigen. J Mol Biol. 2003;325:337–54. doi: 10.1016/S0022-2836(02)01222-6. [DOI] [PubMed] [Google Scholar]
- 8.Padlan EA. On the nature of antibody combining sites: unusual structural features that may confer on these sites an enhanced capacity for binding ligands. Proteins. 1990;7:112–24. doi: 10.1002/prot.340070203. [DOI] [PubMed] [Google Scholar]
- 9.Mian IS, Bradwell AR, Olson AJ. Structure, function and properties of antibody binding sites. J Mol Biol. 1991;217:133–51. doi: 10.1016/0022-2836(91)90617-F. [DOI] [PubMed] [Google Scholar]
- 10.Davies DR, Cohen GH. Interactions of protein antigens with antibodies. Proc Natl Acad Sci U S A. 1996;93:7–12. doi: 10.1073/pnas.93.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lo Conte L, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J Mol Biol. 1999;285:2177–98. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 12.Fellouse FA, Wiesmann C, Sidhu SS. Synthetic antibodies from a four-amino-acid code: a dominant role for tyrosine in antigen recognition. Proc Natl Acad Sci U S A. 2004;101:12467–72. doi: 10.1073/pnas.0401786101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.David MP, Asprer JJ, Ibana JS, Concepcion GP, Padlan EA. A study of the structural correlates of affinity maturation: antibody affinity as a function of chemical interactions, structural plasticity and stability. Mol Immunol. 2007;44:1342–51. doi: 10.1016/j.molimm.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 14.Zhang J. Rates of conservative and radical nonsynonymous nucleotide substitutions in mammalian nuclear genes. J Mol Evol. 2000;50:56–68. doi: 10.1007/s002399910007. [DOI] [PubMed] [Google Scholar]
- 15.Dagan T, Talmor Y, Graur D. Ratios of radical to conservative amino acid replacement are affected by mutational and compositional factors and may not be indicative of positive Darwinian selection. Mol Biol Evol. 2002;19:1022–5. doi: 10.1093/oxfordjournals.molbev.a004161. [DOI] [PubMed] [Google Scholar]
- 16.MacCallum RM, Martin AC, Thornton JM. Antibody-antigen interactions: contact analysis and binding site topography. J Mol Biol. 1996;262:732–45. doi: 10.1006/jmbi.1996.0548. [DOI] [PubMed] [Google Scholar]
- 17.Fellouse FA, Li B, Compaan DM, Peden AA, Hymowitz SG, Sidhu SS. Molecular recognition by a binary code. J Mol Biol. 2005;348:1153–62. doi: 10.1016/j.jmb.2005.03.041. [DOI] [PubMed] [Google Scholar]
- 18.Rajpal A, Beyaz N, Haber L, Cappuccilli G, Yee H, Bhatt RR, et al. A general method for greatly improving the affinity of antibodies by using combinatorial libraries. Proc Natl Acad Sci U S A. 2005;102:8466–71. doi: 10.1073/pnas.0503543102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang L, Gao H, Zhu X, Wang X, Zhou M, Jiang R. Construction of “small-intelligent” focused mutagenesis libraries using well-designed combinatorial degenerate primers. Biotechniques. 2012;52:149–58. doi: 10.2144/000113820. [DOI] [PubMed] [Google Scholar]
- 20.Thom G, Cockroft AC, Buchanan AG, Candotti CJ, Cohen ES, Lowne D, et al. Probing a protein-protein interaction by in vitro evolution. Proc Natl Acad Sci U S A. 2006;103:7619–24. doi: 10.1073/pnas.0602341103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lowe DC, Gerhardt S, Ward A, Hargreaves D, Anderson M, Ferraro F, et al. Engineering a high-affinity anti-IL-15 antibody: crystal structure reveals an α-helix in VH CDR3 as key component of paratope. J Mol Biol. 2011;406:160–75. doi: 10.1016/j.jmb.2010.12.017. [DOI] [PubMed] [Google Scholar]
- 22.Barbas CF, 3rd, Hu D, Dunlop N, Sawyer L, Cababa D, Hendry RM, et al. In vitro evolution of a neutralizing human antibody to human immunodeficiency virus type 1 to enhance affinity and broaden strain cross-reactivity. Proc Natl Acad Sci U S A. 1994;91:3809–13. doi: 10.1073/pnas.91.9.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kayushin A, Korosteleva M, Miroshnikov A, Kosch W, Zubov D, Piel N, et al. A new approach to the synthesis of trinucleotide phosphoramidites - synthons for the generation of oligonucleotide/peptide libraries. Nucleosides Nucleotides. 1999;18:1531–3. doi: 10.1080/07328319908044778. [DOI] [Google Scholar]
- 24.Baker KP, Edwards BM, Main SH, Choi GH, Wager RE, Halpern WG, et al. Generation and characterization of LymphoStat-B, a human monoclonal antibody that antagonizes the bioactivities of B lymphocyte stimulator. Arthritis Rheum. 2003;48:3253–65. doi: 10.1002/art.11299. [DOI] [PubMed] [Google Scholar]
- 25.McCafferty J, Fitzgerald KJ, Earnshaw J, Chiswell DJ, Link J, Smith R, et al. Selection and rapid purification of murine antibody fragments that bind a transition-state analog by phage display. Appl Biochem Biotechnol. 1994;47:157–71, discussion 171-3. doi: 10.1007/BF02787932. [DOI] [PubMed] [Google Scholar]
- 26.Hawkins RE, Russell SJ, Winter G. Selection of phage antibodies by binding affinity. Mimicking affinity maturation. J Mol Biol. 1992;226:889–96. doi: 10.1016/0022-2836(92)90639-2. [DOI] [PubMed] [Google Scholar]
- 27.Vaughan TJ, Williams AJ, Pritchard K, Osbourn JK, Pope AR, Earnshaw JC, et al. Human antibodies with sub-nanomolar affinities isolated from a large non-immunized phage display library. Nat Biotechnol. 1996;14:309–14. doi: 10.1038/nbt0396-309. [DOI] [PubMed] [Google Scholar]
- 28.Persic L, Roberts A, Wilton J, Cattaneo A, Bradbury A, Hoogenboom HR. An integrated vector system for the eukaryotic expression of antibodies or their fragments after selection from phage display libraries. Gene. 1997;187:9–18. doi: 10.1016/S0378-1119(96)00628-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.