Abstract
The FCGR3A-V158F and FCGR2A-H131R polymorphisms are associated with clinical responses to therapeutic mAbs and with immune thrombocytopenic purpura (ITP). The FCGR2C-ORF/STOP polymorphism, controlling FcγRIIC expression on natural killer cells and therefore FcγRIIC-mediated antibody dependent cell-mediated cytotoxicity, is also associated with ITP. Using a new pyrosequencing assay to determine this polymorphism in a control population, we observed the expected allele frequencies (ORF:12.6%) and percentages of individuals with a single copy (10.0%) or 3 copies (12.1%) of FCGR2C, or with at least one FCGR2C-ORF allele (20.1%). No association of FCGR2C copy number variations with the FCGR3A-V158F or FCGR2A-H131R genotype was detected. More importantly, our results demonstrate a strong and a weaker linkage disequilibrium associating the FCGR2C-ORF allele with the FCGR3A-158V and the FCGR2A-131H allele, respectively.
Keywords: receptors for the Fc portion of IgG, immune thrombocytopenic purpura, linkage disequilibrium
Introduction
The low-affinity receptors for the Fc portion of IgG (FcγRs) are encoded by a 200 kb gene cluster located on chromosome 1 and include FCGR2A, FCGR3A, FCGR2C, FCGR3B and FCGR2B (in following order from the centromere). The FCGR3A and FCGR2A genes display a functional allelic dimorphism resulting in a valine to phenylalanine substitution at position 158 (V158F) of FcγRIIIA and a histidine to arginine substitution at position 131 (H131R) of FcγRIIA. The FcγRIIIA-158V and FcγRIIA-131H allotypes have greater affinity for IgG1 and IgG2, respectively.1,2 On the other hand, the FCGR2C-ORF/STOP polymorphism in exon 3 of the gene controls the expression (ORF allele) or the absence (STOP allele) of FcγRIIC on natural killer (NK) cells.3 Moreover, it has been recently reported that an additional mutation at the splice sites of intron 7 in their FCGR2C-ORF alleles results in another stop codon in ≈20% of the FCGR2C-ORF donors and, therefore, the absence of FcγRIIC expression in these donors.4 We and others have shown that the FCGR3A-V158F and FCGR2A-H131R polymorphisms are associated with clinical responses to therapeutic mAbs such as rituximab,5-7 cetuximab,8 or trastuzumab.9 The better responses observed in patients expressing the FcγRIIIA-158V and FcγRIIA-131H allotypes demonstrate that antibody dependent cell-mediated cytoxicity (ADCC) or other functions exerted by cells expressing these receptors play a critical role in mediating the clinical effects.10 It has been shown that FcγRIIc-expressing NK cells can mediate ADCC to antibody-coated targets,3,4 suggesting that this receptor might also be involved in the anti-tumor responses mediated by cytolytic mAbs.
These polymorphisms of FCGR2A, FCGR3A and FCGR2C have also been reported to be associated with immune-complex-mediated auto-inflammation.11 For instance, an association of the FCGR3A-158V allele with immune thrombocytopenic purpura (ITP) has been repeatedly reported.12-15 The association of FCGR2A-H131R with ITP was also investigated in these studies with conflicting results. No association was found by Foster et al. in a small cohort12 (n = 37) or by Breunis et al. in a larger cohort of Caucasian patients15 (n = 116), but Carcao et al. reported that FCGR2A-131H was associated with ITP in a cohort of 98 Caucasian children.14 The latter result might be explained at least partially by the linkage disequilibrium (LD) between the FCGR3A-V158F and FCGR2A-H131R polymorphisms that we and others reported in Caucasian populations.16,17 This hypothesis is supported by the fact that the FCGR2A-H131R polymorphism, which is not in LD with FCGR3A-V158F in the Asian population,17 was not associated with ITP in a cohort of 104 Asian patients.13 Breunis et al. also reported an association between the FCGR2C-ORF allele and ITP in their Caucasian cohort; however, the possibility that this might result from a LD between the FCGR2C-ORF/STOP and the FCGR3A-V158F polymorphisms was not reported. Overall, the issue of the LD between the FCGR2C-ORF/STOP polymorphism and the FCGR3A-V158F or FCGR2A-H131R polymorphisms has not yet been addressed. This probably results from the difficulty in FCGR2C genotyping, due to the fact that FCGR2C and FCGR2B are identical in this region and that FCGR2C is subject to copy number variations (CNV). To clarify this situation, we developed an approach based on both a paralog ratio test and a pyrosequencing assay for the determination of the FCGR2C-ORF/STOP polymorphism in a control population.
Results
FCGR2C genotyping of a Caucasian population using pyrosequencing
Due to its chimeric nature resulting from an unequal crossover between FCGR2B and FCGR2A, analysis of FCGR2C is complex.18 Consequently, in the region of FCGR2C-ORF/STOP polymorphism, FCGR2C is identical to FCGR2B and, except at the level of the FCGR2C-STOP codon, it is not possible to distinguish between FCGR2C and FCGR2B (see Supplemental Material). An additional level of complexity arises from the fact that FCGR2C is subject to CNV, in contrast to FCGR2A and FCGR2B.19,20 The close relationship of both FCGR2B and FCGR2A paralogs with FCGR2C provides the opportunity to use a paralog ratio test21 to evaluate both polymorphisms and the CNV of FCGR2C in a pyrosequencing approach (see Supplemental Material). In our control Caucasian population (Table 1), the percentages of individuals with a single copy (11.1%; n = 21) or 3 copies (12.2%; n = 23) of FCGR2C were close to those reported in the previous studies using multiplex ligation-dependent probe amplification.4,15 It is of note that we identified two individuals with two null alleles corresponding to a total lack of FCGR2C. In our control population, 79.9% of individuals did not express FcγRIIc because they only carry the FCGR2C-STOP allele (149/151) or lack the FCGR2C gene (2/151), whereas 20.1% of them possessed at least one FCGR2C-ORF allele (Table 1) leading to FcγRIIc expression in those who do not have the additional mutation at the splice sites of intron 7.4 We also identified an unusual allele combination of FCGRC-ORF/STOP, with 5 copies of FCGR2C-STOP and one copy of FCGR2C-ORF (Table 1).
Table 1. FCGR2C genotypes in a Caucasian population.
| Number of FCGR2C-ORF | Number of FCGR2C-STOP | Genotypea | |
|---|---|---|---|
| 0 |
0 |
nul/nul |
2 |
| 1 |
STOP |
21 |
|
| 2 |
STOP/STOP |
108 |
|
| 3 |
STOP/STOP/STOP |
20 |
|
|
Total |
|
|
151 (79.9%) |
| 1 |
0 |
ORF |
0 |
| 1 |
ORF/STOP |
27 |
|
| 2 |
ORF/STOP/STOP |
1 |
|
| 5 |
ORF/(STOP)5 |
1 |
|
| 2 |
0 |
ORF/ORF |
6 |
| 1 |
ORF/ORF/STOP |
2 |
|
| 3 |
0 |
ORF/ORF/ORF |
1 |
| |
|
|
|
| Total | 38 (20.1%) |
a Allele frequencies estimated from these genotypes are 12.6 and 87.4 respectively for FCGR2C-ORF and FCGR2C-STOP.
LD between FCGR2C-ORF/STOP, FCGR3A-V158F and FCGR2A-H131R
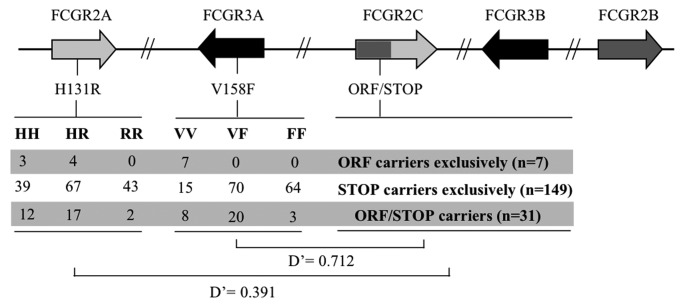
The FCGR3A-V158F and FCGR2A-H131R genotypes of the 189 individuals of our population were therefore analyzed together with the FCGR2C-ORF/STOP data. First, we did not find any association between FCGR2C CNV and FCGR3A-158V/F or FCGR2A-H131R genotypes (data not shown). Second, we analyzed the LD between FCGR2C-ORF/STOP and FCGR3A-V158F or FCGR2A-H131R, without taking the FCGR2C CNV into account. Our results demonstrated the high LD between FCGR2C-ORF/STOP and FCGR3A-V158F: the FCGR2C-ORF allele was strongly associated with the FCGR3A-158V allele (Fig. 1). Indeed, the frequency of the ORF allele was 53.5%, 20.2%, and 4.5% in VV, VF and FF donors, respectively. On the other hand, analysis of the LD between FCGR2C-ORF/STOP and FCGR2A-H131R revealed a lower LD, which tended to associate the FCGR2C-ORF allele with the FCGR2A-131H allele (Fig. 1).
Figure 1.LD between FCGR2C-ORF/STOP, FCGR3A-V158F and FCGR2A-H131R. Polymorphisms of interest were localized on the genomic cluster using gene order and orientation annotated in build 37.2 of NCBI. For FCGR3A-V158F and FCGR2A-H131R, number of individual for each genotype is indicated depending on FCGR2C-ORF/STOP polymorphism.
Discussion
We have developed an approach based on both a paralog ratio test and a pyrosequencing assay for the determination of the FCGR2C-ORF/STOP polymorphism in a control population. This paralog ratio test is based on the premise that there are no CNV in FCGR2A and FCGR2B. Niederer HA et al. using a paralog ratio test to determine copy number variation identified a single genomic region (CNR2) containing CNV of FCGR2C, FCGR3A and FCGR2A.22 Frequencies of CNV for this region were respectively 1.3% for 1 copy and 4.5% for 3 copies. It is important that primer pair used to amplify a 279 bp fragment of FCGR2A were localized in 3′-UTR sequence. On the other hand, using multiplex ligation-dependent probe amplification with probes specific for coding regions, Breunis et al. did not observed CNV of FCGR2A and FCGR2B in a large (> 600 individuals) control population.19
In our pyrosequencing approach, we have also used primer pair localized in coding region of FCGR2A and FCGR2B where no CNV has been reported so far. Moreover, if the CNR2 described by Niederer et al. expanded to the coding region of FCGR2A, it will be expected that 4.5% of individuals would carry 3 copies of FCGR2A. In these individuals, those with the frequent FCGR2C-STOP/STOP genotype and carrying two copies of FCGR2B should have a ratio between FCGR2B/FCGR2C-ORF and FCGR2A < to 1 [(2 FCGR2B + 0 FCGR2C-ORF)/3 FCGR2A = 0.66]. Nevertheless, we did not observed such a result in our cohort (all the ratios were > to 1). Therefore, our results in accordance with those of Breunis et al.19 do not substantiate the hypothesis of CNV in the coding region of FCGR2A.
Using a pyrosequencing approach, we evaluated frequencies of FCGR2C-ORF/STOP alleles in a control Caucasian population. These frequencies are concordant with previous results showing that the FCGR2C-ORF allele is present in only half of the 40% of individuals with detectable CD32+ NK cells (i.e., expressing one of the FcγRII).23 The different genotype frequencies observed in our study (Table 1) were mostly close to those previously reported in the Caucasian population by Breunis et al.15 and by van der Heijden et al.4 However, Breunis et al. did not identify individuals with at least 2 copies of FCGR2C-ORF in their control group, whereas 4.3% and 0.5% had two or three copies of the FCGR2C-ORF allele in our population, respectively. Given the allele and CNV frequencies, all the ORF/STOP combinations expected were observed.
Our investigations showed a high LD between FCGR2C-ORF/STOP and FCGR3A-V158F. Despite the reported association of FCGR3A, FCGR2A and FCGR2C polymorphisms with several autoimmune diseases, their LD has not been extensively studied. Using flow cytometry, Steward-Akers et al. showed that the FcγRIIIA-158F/F genotype was over-represented in CD32neg rheumatoid arthritis patients as compared with CD32pos patients, suggesting a LD between FCGR3A-158F and FCGR2C-STOP alleles.22 On the other hand, Breunis et al. reported that they did not observe an absolute linkage when the FcγRIIIa-158V data were combined with the FCGR2C-ORF allele and FCGR2C-386C/-120T promoter haplotype.15 Second, we found a lower LD between FCGR2C-ORF allele and the FCGR2A-131H allele. This low LD between FCGR2C-ORF/STOP and FCGR2A-H131R could explain why Breunis et al. did not find an association between ITP and FCGR2A-H131R, whereas they found an association of ITP with both the V158F and the ORF/STOP polymorphisms. Moreover, the conclusion that the FCGR2C ORF/STOP polymorphism predisposes to ITP, as proposed by Breunis et al., should be re-evaluated in light of its LD with FCGR3A-V158F.
The importance of FCGR3A-V158F, FCGR2A-H131R and FCGR2C-ORF/STOP polymorphisms is well established in several autoimmune diseases11 and in response to therapeutic antibodies.5 In accordance with our previous report,17 this study strongly supports the notion that the different LD between these polymorphisms should be systematically taken into account in these types of study.
Materials and Methods
DNA samples
A total of 189 Caucasian DNA samples (CHRU of Tours, regular collection approved by the Ministry of Health, DC-2008–308) were initially extracted using the QIAamp DNA Blood Mini Kit (Qiagen) from peripheral blood of healthy donors.
PCR amplification
A 110 pb region common to FCGR2A, FCGR2B and FCGR2C and spanning the site of the FCGR2C-ORF/STOP polymorphism was amplified by polymerase chain reaction (PCR) (Fig. S1). The primer sequences used were Biot-2abci2e3-S 5′-CTCTGCCCCTCAG-3′ and 2abce3-AS 5′-TGTCAGAGTCACACAGT-3′. The forward primer was 5′-biotinylated to allow single-strand DNA template isolation for the pyrosequencing reaction. Each PCR mix contained 50 ng genomic DNA, 10 pmol of each primer, and 0.3 μL Taq polymerase (Eurobio) in a total volume of 55 μL. Cycling was performed in a Bio-Rad iCycler as follows: 94°C for 5 min, 35 cycles of 94°C for 1 min, 62°C for 30 sec, 72°C for 30 sec and a final extension at 72°C for 2 min. Successful and specific amplification of the region of interest was verified by visualizing 5 μL of the PCR product in an 8% acrylamide gel.
Pyrosequencing
Preparation of the single stranded DNA template for pyrosequencing was performed using the PSQ Vacuum Prep Tool (Biotage) according to the manufacturer’s instructions: 45 μL of biotinylated PCR product was immobilized on streptavidin-coated Sepharose high-performance beads and processed to obtain single stranded DNA using the PSQ 96 Sample Preparation kit (Biotage). The template was incubated with 1 μL of 10 pmol sequencing primer (pyro2-AS: 5′- TGGAGCACGTTGATCCAC-3′) at 80°C for 2 min on a PSQ 96 plate. The sequencing reaction of the complementary strand was automatically performed on a PSQ 96MA instrument (Biotage) at room temperature using PyroGold reagents (Fig. S2).
FCGR3A-V158F and FCGR2A-H131R genotyping
FCGR3A-V158F (rs 396991) and FCGR2A-H131R (rs 1801274) polymorphisms were genotyped as previously described.5
Statistical analyses
LD between polymorphisms was estimated using Haploview.24 The association between FCGR2C CNV and FCGR3A-158V/F or FCGR2A-H131R genotypes was tested using a CHI 2 test.
Supplementary Material
Acknowledgments
This work was supported by the Institut National du Cancer and Canceropôle Grand Ouest (Mab Impact), and the Fondation Langlois. Julien Lejeune is granted by the Région Centre.
Glossary
Abbreviations:
- ADCC
antibody-dependent cell-mediated cytoxicity
- CNV
copy number variations
- FcγR
Receptor for the Fc portion of IgG
- ITP
immune thrombocytopenic purpura
- LD
linkage disequilibrium
- NK
natural killer
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental material may be found here: www.landesbioscience.com/journals/mabs/article/22287
Footnotes
Previously published online: www.landesbioscience.com/journals/mabs/article/22287
References
- 1.Dall’Ozzo S, Tartas S, Paintaud G, Cartron G, Colombat P, Bardos P, et al. Rituximab-dependent cytotoxicity by natural killer cells: influence of FCGR3A polymorphism on the concentration-effect relationship. Cancer Res. 2004;64:4664–9. doi: 10.1158/0008-5472.CAN-03-2862. [DOI] [PubMed] [Google Scholar]
- 2.Bruhns P, Iannascoli B, England P, Mancardi DA, Fernandez N, Jorieux S, et al. Specificity and affinity of human Fcgamma receptors and their polymorphic variants for human IgG subclasses. Blood. 2009;113:3716–25. doi: 10.1182/blood-2008-09-179754. [DOI] [PubMed] [Google Scholar]
- 3.Metes D, Ernst LK, Chambers WH, Sulica A, Herberman RB, Morel PA. Expression of functional CD32 molecules on human NK cells is determined by an allelic polymorphism of the FcgammaRIIC gene. Blood. 1998;91:2369–80. [PubMed] [Google Scholar]
- 4.van der Heijden J, Breunis WB, Geissler J, de Boer M, van den Berg TK, Kuijpers TW. Phenotypic variation in IgG receptors by nonclassical FCGR2C alleles. J Immunol. 2012;188:1318–24. doi: 10.4049/jimmunol.1003945. [DOI] [PubMed] [Google Scholar]
- 5.Cartron G, Dacheux L, Salles G, Solal-Celigny P, Bardos P, Colombat P, et al. Therapeutic activity of humanized anti-CD20 monoclonal antibody and polymorphism in IgG Fc receptor FcgammaRIIIa gene. Blood. 2002;99:754–8. doi: 10.1182/blood.V99.3.754. [DOI] [PubMed] [Google Scholar]
- 6.Weng WK, Levy R. Two immunoglobulin G fragment C receptor polymorphisms independently predict response to rituximab in patients with follicular lymphoma. J Clin Oncol. 2003;21:3940–7. doi: 10.1200/JCO.2003.05.013. [DOI] [PubMed] [Google Scholar]
- 7.Treon SP, Hansen M, Branagan AR, Verselis S, Emmanouilides C, Kimby E, et al. Polymorphisms in FcgammaRIIIA (CD16) receptor expression are associated with clinical response to rituximab in Waldenström’s macroglobulinemia. J Clin Oncol. 2005;23:474–81. doi: 10.1200/JCO.2005.06.059. [DOI] [PubMed] [Google Scholar]
- 8.Bibeau F, Lopez-Crapez E, Di Fiore F, Thezenas S, Ychou M, Blanchard F, et al. Impact of Fc{gamma}RIIa-Fc{gamma}RIIIa polymorphisms and KRAS mutations on the clinical outcome of patients with metastatic colorectal cancer treated with cetuximab plus irinotecan. J Clin Oncol. 2009;27:1122–9. doi: 10.1200/JCO.2008.18.0463. [DOI] [PubMed] [Google Scholar]
- 9.Musolino A, Naldi N, Bortesi B, Pezzuolo D, Capelletti M, Missale G, et al. Immunoglobulin G fragment C receptor polymorphisms and clinical efficacy of trastuzumab-based therapy in patients with HER-2/neu-positive metastatic breast cancer. J Clin Oncol. 2008;26:1789–96. doi: 10.1200/JCO.2007.14.8957. [DOI] [PubMed] [Google Scholar]
- 10.Congy-Jolivet N, Bolzec A, Ternant D, Ohresser M, Watier H, Thibault G. Fc gamma RIIIa expression is not increased on natural killer cells expressing the Fc gamma RIIIa-158V allotype. Cancer Res. 2008;68:976–80. doi: 10.1158/0008-5472.CAN-07-6523. [DOI] [PubMed] [Google Scholar]
- 11.Nimmerjahn F, Ravetch JV. Fcgamma receptors as regulators of immune responses. Nat Rev Immunol. 2008;8:34–47. doi: 10.1038/nri2206. [DOI] [PubMed] [Google Scholar]
- 12.Foster CB, Zhu S, Erichsen HC, Lehrnbecher T, Hart ES, Choi E, et al. Early Chronic ITP Study Group Polymorphisms in inflammatory cytokines and Fcgamma receptors in childhood chronic immune thrombocytopenic purpura: a pilot study. Br J Haematol. 2001;113:596–9. doi: 10.1046/j.1365-2141.2001.02807.x. [DOI] [PubMed] [Google Scholar]
- 13.Fujimoto TT, Inoue M, Shimomura T, Fujimura K. Involvement of Fc gamma receptor polymorphism in the therapeutic response of idiopathic thrombocytopenic purpura. Br J Haematol. 2001;115:125–30. doi: 10.1046/j.1365-2141.2001.03109.x. [DOI] [PubMed] [Google Scholar]
- 14.Carcao MD, Blanchette VS, Wakefield CD, Stephens D, Ellis J, Matheson K, et al. Fcgamma receptor IIa and IIIa polymorphisms in childhood immune thrombocytopenic purpura. Br J Haematol. 2003;120:135–41. doi: 10.1046/j.1365-2141.2003.04033.x. [DOI] [PubMed] [Google Scholar]
- 15.Breunis WB, van Mirre E, Bruin M, Geissler J, de Boer M, Peters M, et al. Copy number variation of the activating FCGR2C gene predisposes to idiopathic thrombocytopenic purpura. Blood. 2008;111:1029–38. doi: 10.1182/blood-2007-03-079913. [DOI] [PubMed] [Google Scholar]
- 16.Lehrnbecher T, Foster CB, Zhu S, Leitman SF, Goldin LR, Huppi K, et al. Variant genotypes of the low-affinity Fcgamma receptors in two control populations and a review of low-affinity Fcgamma receptor polymorphisms in control and disease populations. Blood. 1999;94:4220–32. [PubMed] [Google Scholar]
- 17.Lejeune J, Thibault G, Ternant D, Cartron G, Watier H, Ohresser M. Evidence for linkage disequilibrium between Fcgamma RIIIa-V158F and Fcgamma RIIa-H131R polymorphisms in white patients, and for an Fcgamma RIIIa-restricted influence on the response to therapeutic antibodies. J Clin Oncol. 2008;26:5489–91, author reply 5491-2. doi: 10.1200/JCO.2008.19.4118. [DOI] [PubMed] [Google Scholar]
- 18.Warmerdam PA, Nabben NM, van de Graaf SA, van de Winkel JG, Capel PJ. The human low affinity immunoglobulin G Fc receptor IIC gene is a result of an unequal crossover event. J Biol Chem. 1993;268:7346–9. [PubMed] [Google Scholar]
- 19.Breunis WB, van Mirre E, Geissler J, Laddach N, Wolbink G, van der Schoot E, et al. Copy number variation at the FCGR locus includes FCGR3A, FCGR2C and FCGR3B but not FCGR2A and FCGR2B. Hum Mutat. 2009;30:E640–50. doi: 10.1002/humu.20997. [DOI] [PubMed] [Google Scholar]
- 20.Bournazos S, Woof JM, Hart SP, Dransfield I. Functional and clinical consequences of Fc receptor polymorphic and copy number variants. Clin Exp Immunol. 2009;157:244–54. doi: 10.1111/j.1365-2249.2009.03980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hollox EJ, Detering JC, Dehnugara T. An integrated approach for measuring copy number variation at the FCGR3 (CD16) locus. Hum Mutat. 2009;30:477–84. doi: 10.1002/humu.20911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niederer HA, Willcocks LC, Rayner TF, Yang W, Lau YL, Williams TN, et al. Copy number, linkage disequilibrium and disease association in the FCGR locus. Hum Mol Genet. 2010;19:3282–94. doi: 10.1093/hmg/ddq216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stewart-Akers AM, Cunningham A, Wasko MC, Morel PA. Fc gamma R expression on NK cells influences disease severity in rheumatoid arthritis. Genes Immun. 2004;5:521–9. doi: 10.1038/sj.gene.6364121. [DOI] [PubMed] [Google Scholar]
- 24.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.