Abstract
The Cooperative Studies Program Coordinating Center provided the data management, administrative, and statistical support to the Global Enteric Multicenter Study (GEMS). The GEMS study, the largest epidemiological study in the diarrheal disease area among children <5 years of age, was carried out in 4 African countries and 3 Asian countries. Given the geographical and geopolitical differences among the countries, the administration of a centralized data management operation was a major challenge. The sheer volume of the data that were collected, regular transfer of the data to a centralized database, and the cleaning of the same also posed some challenges. This paper outlines the details of the support that the data coordinating center provided and the challenges faced during the course of the study.
The Cooperative Studies Program Coordinating Center at Perry Point, Maryland, is one of 5 coordinating centers under Clinical Sciences Research and Development in the Department of Veterans Affairs, and specializes in providing data management, statistical, and administrative support to VA clinicians in the planning, conduct, and close-out of multisite clinical trials and epidemiological studies. In early 2006, the Center for Vaccine Development (CVD) of the University of Maryland School of Medicine approached the Perry Point Data Coordinating Center (DCC) for data management and other related services for the Global Enteric Multicenter Study (GEMS), an international epidemiological study of diarrhea in children <5 years of age, to utilize the Perry Point DCC's years-long experience in handling large-scale clinical studies.
With support from the Bill & Melinda Gates Foundation, GEMS was carried out in 7 countries: 4 sites in Africa (Basse, The Gambia; Kisumu, Kenya; Bamako, Mali; Manhica, Mozambique) and 3 sites in Asia (Kolkata, India; Mirzapur, Bangladesh; Karachi, Pakistan). GEMS began with a Health Utilization and Attitude Survey (HUAS) in each country, where approximately 1100 households were randomly sampled from either an existing or a newly initiated demographic surveillance system. After the completion of the HUAS, a 3-year case/control study was initiated in each country. In the case/control study, 660 children with moderate-to-severe diarrhea (cases), along with 660 matched children without diarrhea (controls), were recruited in each of 3 age strata (0–11, 12–23, and 24–59 months) in each country. During the 3-year case/control study, a shorter version of the original HUAS (“HUAS-Lite”) was performed 2–3 times per year where 1100 households were randomly sampled from the respective demographic surveillance system for each round completed. The study used a total of 20 case report forms (CRFs). These included 1 CRF for the original HUAS, 1 CRF for the HUAS-Lite, 9 clinical/epidemiological CRFs (5 for cases, 4 for controls), 6 laboratory CRFs, and 3 verbal autopsy CRFs. The interviews for HUAS and for HUAS-Lite rounds and the data collection for the case/control study were conducted by locally employed and trained health workers in each country. To satisfy the need of the health workers who spoke native languages, the CRFs and the informed consent forms were translated and printed in different languages. For example, Mali used CRFs in French, Mozambique in Portuguese, and Kenya in dual languages (Dholuo and English). Bangladesh used informed consent forms in Bengali and Pakistan in Urdu.
PERRY POINT DCC SUPPORT
Once contracted by the CVD and approved for participation by the VA central office, the DCC established a core team for the GEMS. The core team was comprised of a team lead (also a biostatistician), a statistical programmer, a project manager, 2 data managers, and 4 computer assistants. The team lead and the project manager provided the administrative support, the biostatistician and the statistical programmer provided the statistical support, and the data managers and the computer assistants provided the data management support for the study.
The Perry Point DCC provided the following services for the GEMS:
Data Management Support
Implementation of a data flow model where data collection would take place at the individual countries and data would be sent to the DCC periodically where the study master database(s) would be established, maintained, and managed.
Selection of a standardized data management platform that would work seamlessly in 8 different countries in 3 continents (7 participating countries in 2 continents and the United States as the central hub for data management) with a significant variability in technical support and other related logistical support.
Design standardized CRFs that adhere to specifications required by the selected data collection software.
Generate paper CRFs in 4 languages (English, French, Portuguese, and Dholuo).
Generate paper informed consent forms in 3 languages (English, Bengali, and Urdu).
Establish a standardized data transfer protocol between the participating countries and the DCC.
Design a data quality control (QC) protocol where “data QC” reports would be generated and sent to the participating countries periodically.
Design a data accountability protocol where “missing CRF” reports would be generated and sent to the sites periodically based on an “expected CRF” algorithm.
Establish a study-specific numbering protocol for the HUAS households, case/control children, and their laboratory samples.
Establish a data management handbook with instructions for CRF completion, addressing the QC and missing form reports, etc.
Build and maintain close contacts between the data management workgroups in the participating countries and the data management group at the DCC using conference calls, emails, etc.
Administrative Support
Implementation and tracking of “CRF request forms” from each participating country.
Printing and shipping of CRFs for each country for the entire duration of the study.
Tracking of all regulatory documentation from each participating countries, including institutional review board/ethics board approvals, Federal Wide Assurance numbers, translation certificates, etc.
Setup of regular meetings between the CVD core group and the DCC.
Maintain required minimum staff to ensure execution of the DCC support.
Participate in annual or other meetings during the study.
Statistical Support
Generate weekly tables on eligibility and enrollment.
Generate monthly aggregate tables on variables (either original or constructed) as requested by the CVD core group.
Generate analytic datasets for analysis purposes as requested by the CVD core group.
Perform statistical analysis based on an established statistical analysis plan.
Participate in statistical workgroup meetings.
DATA COLLECTION TOOL DATAFAX: AN OVERVIEW
DataFax was chosen as the data collection tool for GEMS by the data management group from DCC. The primary objective of DataFax is to automate the collection and processing of paper case report forms, and ultimately improving the timeliness and quality of the study database. The specific design objectives are as follows:
Use simple technology in the clinical sites. Clinical sites (or participating countries) can send CRFs to the DCC in TIFF or PDF format using either an ordinary fax machine (from any standard G3 fax machine), an Internet fax machine, email, or secure file transfer protocol (SFTP). Data can be added via raw data entry into data screens (from paper CRFs), or by importing ASCII data files (eg, from a central laboratory).
Computerize the receipt, logging, and filing of CRFs. When data are submitted to DataFax, the software reads information embedded in barcodes on the individual CRF pages and routes them to the appropriate study database. Each CRF image is assigned a fax ID and is placed in a queue for validation. Once validation is complete, all images and data records are stored electronically in a secure location on optical or magnetic disc.
Automatically generate an initial data record. DataFax reads data boxes (Xs, handwritten numbers, and visual analog scales) to create an initial data record as the starting point for the clinical review and data validation process.
Provide split screen review of CRFs and the corresponding data records. All CRFs and initial data records are reviewed on screen to complete data entry, make corrections, and flag problems (eg, missing data).
Automate the QC process. Problems detected on the CRFs received by the DCC are flagged using QC notes (electronic sticky notes), which are automatically formatted into standard QC reports for transmission by fax or email to the clinical sites.
Automate work flow management. CRFs are stamped with a validation level at each CRF review and data processing stage.
DataFax does not work with arbitrary CRFs. Study CRFs have to be designed by the DCC to adhere to DataFax specifications, which include the following:
Bar coding must be placed at the top of each CRF page to identify the study, the CRF plate (page), and optionally the sequence (or visit) number.
All pages must be US letter or A4 size and oriented vertically (portrait) not horizontally (landscape). The DCC selected standard US letter size for the GEMS study.
All boxes designed for numerical data and spacing must conform to DataFax standards.
Faxed/scanned/emailed CRFs are automatically indexed upon receipt (by study number, CRF page number, and optionally by visit number) from barcodes printed at the top of each CRF page. The remaining fields on each CRF page are processed by the intelligent character recognition (ICR) software, which reads numeric, date, check, choice, and visual analog scale fields to create an initial data record ready for subsequent validation by the data management staff. Text fields are not read by the ICR software and must be entered manually when the record is validated.
Each newly received CRF page and the corresponding data record created by the ICR software are reviewed by data management staff using the DataFax validation tool. Data management staff flag any CRF problems (eg, missing data), using pop-up QC notes, during validation. Lookup tables provide standardization of queries to be sent to clinical investigators. Preprogrammed edit checks will detect inconsistencies within forms, across forms, across visits, and even across study participants, if necessary.
A QC report program formats all QC notes for each clinical site into a clear, compact report identifying all outstanding CRF problems and clarification requests. Missing pages and overdue visits may be included in QC reports. Each QC report may also include a scheduling summary for all participants at the clinical site including, entry date, date of last visit, and target date for the next scheduled visit. The QC reports may be faxed and/or emailed to clinical sites at scheduled times.
Corrected CRFs, re-sent from the clinical sites, are automatically identified on arrival for revalidation, entry of corrected fields, and resolution of QC notes using the DataFax validation tool. All versions of each CRF page (and all versions of the corresponding data records) are retained for subsequent review, but only 1 version of each CRF page is flagged as the primary (good) copy and linked to the primary data record.
Full journaling identifies all changes made to the database by user, date, and time. A QC database tracks all data clarification queries by problem type (eg, missing data, illegal value) and current status (resolved or outstanding). Audit trail reports show all changes made to the database, at data record and individual data field levels by user, date, and time, including history of QC notes.
DATA TRANSFER PROTOCOL
The data flow model that was adopted for GEMS was as follows:
Data were collected in the field in each participating country using paper CRFs in the appropriate language with appropriate barcodes.
Completed CRFs were scanned and saved as TIFF or PDF files using a scanner with prespecified resolution settings (each country purchased a scanner locally based on the supplied specifications by the DCC) to ensure readability by the DataFax software.
- The TIFF and PDF files were electronically transferred to the DCC at an agreed-upon interval.
- Three different transfer platforms were used for GEMS:
- Email: In the beginning of the study, the participating countries were requested to send the scanned files as attachments via emails, but the size limitations for the attached documents as outlined by the VA exchange email server (5 MB) made this mode very time consuming and difficult to track as it was requiring the sites to send multiple emails to be within the acceptable file size limit.
- Microsoft Groove: Collaborative workspaces were created for each country where the sites could post their scanned files for DCC to retrieve from. Each country's folder structure in their respective workspaces was set up by the DCC staff to ensure ease of posting and retrieval of files. DCC had access to all of these workspaces, but each country's access was restricted to its own workspace only. DCC staff also regulated/controlled access to these workspaces by the site staff. DCC staff deleted the posted files regularly once the files were retrieved from the workspaces to keep the workspace synchronization times under control. These workspaces were working very well for data transfer until a decision was made by the GEMS executive committee to use these workspaces as archives for the submitted CRFs. At the end of the second year of the study the size of each workspace exceeded the size limit of Groove (2 GB), which made the workspaces unusable.
- SFTP server: As an alternative solution, accounts were created on an SFTP server accessible by each participating country for data transfer. The screenshot in Figure 1 shows an account for one of the participating countries.
- The scanned files, when received at the DCC, were routed to the respective form queues based on the barcodes placed on top of each page of each CRF.
- For the ease of data management for GEMS, 4 separate databases (and thus 4 form queues) were maintained—1 for HUAS (included data from original HUAS and the HUAS-Lite rounds), 1 for case registration (CRF 2), 1 for the rest of the case/control study (included all the clinical/epidemiological and the laboratory CRFs), and 1 for the verbal autopsy data.
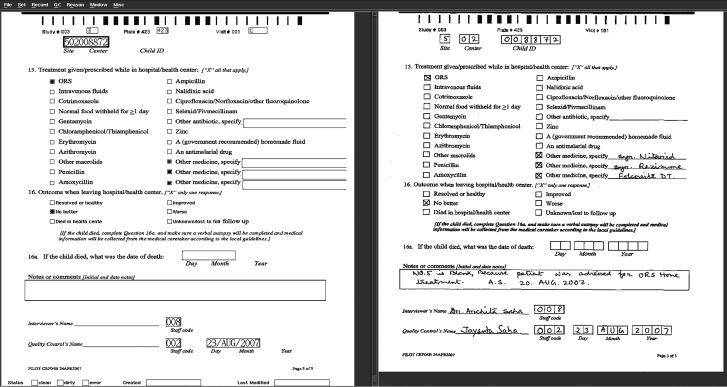
Before submission to the respective master databases, the data management group at the DCC validated the forms on a split screen. The right half of the screen displays the actual scanned image of a CRF page and the left half of the screen displays the software's ICR interpretation of the same page (Figure 2).
Based on the discrepancies/errors/illegible entries identified during split-screen validation, QC notes are added and reports were generated and sent back to the countries for corrections/explanations.
The data management workgroup at each country rescanned the corrected pages of the CRFs and posted them in an appropriate folder on the SFTP server for the DCC to retrieve. Once received at the DCC, the CRF pages were revalidated before being submitted to the respective databases.
Once the data were cleaned, the datasets were shared with each country and the CVD core group intermittently.
After the final data lock, entire datasets from the respective countries were sent for final analysis.
Figure 1.
Structure of a typical FTP (file transfer protocol) account.
Figure 2.
Split-screen validation in DataFax.
NUMBERING PROTOCOL
For GEMS, a unique identification number was assigned to each household surveyed for the HUAS (original and the Lite rounds), to each child enrolled as a case or control for the case/control study, and to each laboratory sample that was collected from each child enrolled. To achieve this task, a numbering protocol was established by the DCC and the details are given below:
- HUAS
- ◦ ID: 5 numbers divided into 2 sections
- ▪ Section 1: Position 1 = Country # (1–7, 9 possible)
- ▪ Section 2: Positions 2 to 5 = Survey # (0001–9999)
- ▪ Entire ID numbers are preprinted on CRFs
- ▪ Child ID numbers assigned sequentially within each country
- Ex. 10001 = Country 1, Sequential # 0001
- HUAS Lite
- ◦ ID: 7 numbers divided into 2 sections
- ▪ Section 1: Position 1 = Country # (1–7, 9 possible)
- ▪ Section 2: Positions 2 to 7 = Survey # (000001–999999)
- ▪ Entire ID numbers are preprinted on CRFs
- ▪ Child ID numbers assigned sequentially within each country
- Ex. 1000001 = Country 1, Sequential # 000001
- CASES AND CONTROLS
- ◦ ID: 9 numbers divided into 3 sections
- ▪ Section 1: Position 1 = Site # (1–7, 9 possible)
- ▪ Section 2: Positions 2 to 3 = Center # (99 possible for each country)
- ▪ Section 3: Positions 4 to 9 = Sequential # (000001–999999)
- ◦ Case ID number transcribed from the Registration Log (CRF 2)
- ▪ Sequential numbering from 000001 to 899999
- ▪ Ex. 101000001 = Country 1, Center 01, Sequential # 000001
- ▪ Ex. 102000025 = Country 1, Center 02, Sequential # 000025
- ◦ Control Patient ID
- ▪ Sequential numbering from 900001 to 999999
- ▪ Ex. 101900001 = Country 1, Center 01, Sequential # 900001
- ▪ Ex. 202900050 = Country 2, Center 02, Sequential # 900050
- SPECIMEN IDs
- ◦ ID: 6 numbers divided into 2 sections
- ▪ Section 1: Position 1 = Country # (1–7)
- ▪ Section 2: Positions 2 to 6 = Sequential numbering from 00001 to 99999
- ▪ Ex. 100001 = Country 1, Sequential # 00001
DCC CHALLENGES
The following sections outline the challenges that DCC experienced and the lessons learned at different phases of GEMS.
One of the major challenges that the data management group experienced was to identify and implement a data management platform that would work seamlessly in 8 different countries (7 participating countries and the US as the centralized data hub). DataFax served very efficiently as a centrally managed data management system supporting standardized CRFs with a standardized data transfer protocol for the GEMS. DCC supplied the technical specifications of scanners, which were purchased/installed/managed locally at each country and used to scan and generate TIFF or PDF files with a specified resolution setting for validation purposes at the DCC. The other major challenge the data management team at DCC faced was the use of different language CRFs in some countries. India, Bangladesh, Pakistan, and The Gambia used CRFs in English but Mali, Kenya, and Mozambique used CRFs in the languages spoken in those countries. The challenge was to maintain the English CRF format (length, number of questions on each page, location of boxes for responses on each question) intact on each page of a given CRF while the translations in respective languages (French for Mali, Portuguese for Mozambique, and Dholuo for Kenya) were overlaid either in place of corresponding English questions or in combination. The validation screens at the DCC were maintained in English for standardization purposes.
The validation process also posed some challenges to the DCC staff. Because DataFax uses an ICR technology to recognize “X” and numerals, it did not recognize handwritten notes. The computer assistants at the DCC needed to manually type the handwritten responses. Even though the number of handwritten responses was limited, illegible handwriting (often in languages other than English) posed challenges.
Other challenges, related to the data management processes that the DCC staff encountered, are outlined below:
Printing and shipping of CRFs to ensure smooth conduct of data collection in each participating country: Because a paper-based data management system was chosen for GEMS, an enormous amount of CRFs needed to be printed and shipped to the participating countries. Since DataFax was very restrictive about the paper size, the decision was made to print all the CRFs in US standard letter size. Country-specific mailing times and customs issues posed additional challenges toward maintaining the timeline. Moreover, receipt of CRF requests from the countries in a timely manner was also a crucial element in maintaining the timeline.
- CRF submission issues:
- Not using the correct/recommended settings for the scanner by the participating countries during scanning of the paper CRFs (this posed challenges in reading the CRFs correctly by the DataFax software);
- Not including the file submission tracking log with each submission of CRFs;
- Not following the suggested naming conventions of the submitted files.
- Data cleaning challenges:
- Overlooking some of the identified errors on a QC report (in these situations the QC reports with the same errors were sent back repeatedly for correction);
- Not following the “Resubmission of CRFs with Corrections” protocol while sending back the corrected CRFs to the DCC;
- Long-overdue CRFs.
To overcome some of these challenges, the DCC data management staff scheduled numerous conference calls with each participating country to enforce various standardized protocols that DCC established for GEMS and also to help the site personnel to understand the contents of the QC reports.
SOME DATA ON THE SCALE OF GEMS
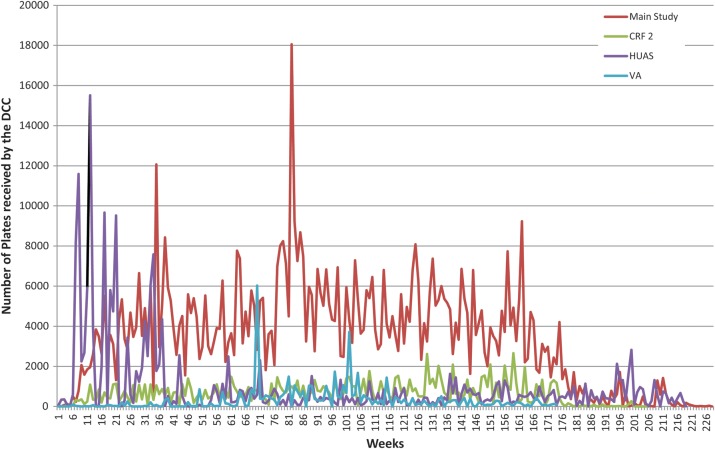
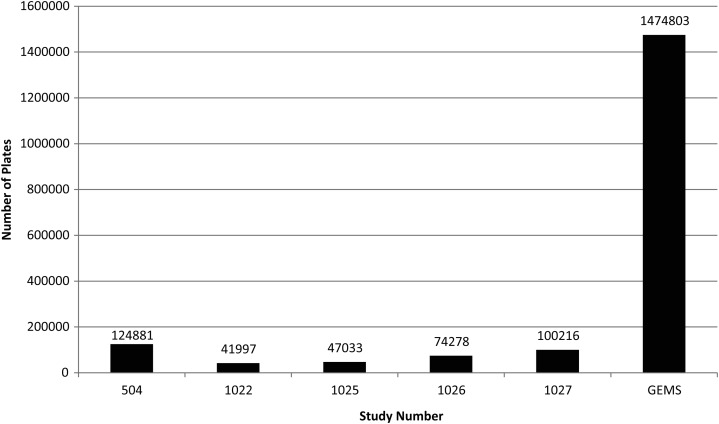
GEMS is the largest international, epidemiological study executed in the diarrheal disease area in Asia and in sub-Saharan Africa. GEMS amassed a huge volume of data. In the original round of HUAS, a total of approximately 7000 households were surveyed. On average, each country completed 6 rounds of HUAS-Lite surveys during the 3-year case/control study, with the exception of 2 countries. In total, 30 000–35 000 households were surveyed during the HUAS-Lite rounds. For the case/control study, approximately 27 000 children were enrolled in the 7 participating countries. In total, the DCC validation team has processed about 1.5 million pages of CRFs for GEMS. Figure 3 illustrates the volume of CRFs received over the first 130 weeks of the study and Figure 4 provides a perspective of GEMS' scale compared to other studies conducted at DCC.
Figure 3.
The number of case report forms (CRFs) received in the 4 databases (main study, CRF2, HUAS, and Verbal Autopsy) of Global Enteric Multicenter Study. Abbreviations: CRF, case report forms; HUAS, Health Utilization and Attitude Survey; VA, Verbal Autopsy; DCC, Perry Point Data Coordinating Center.
Figure 4.
The number of plates received for Global Enteric Multicenter Study versus other studies completed by the Data Coordinating Center.
DISCUSSION
The GEMS project was an enormous undertaking from the DCC perspective, not only because of its scale but also for its involvement with multiple countries from 3 continents with very diverse cultural, social, and technological backgrounds. It was a valuable experience for the DCC staff, who learned from dealing with a group of talented, diligent, and focused individuals from the 7 countries. DCC staff was successful in building up close and mutually respectful working relationships with the data management staff from all the participating countries. The use of standardized case report forms for data collection, utilization of standardized data management software, and the application of numerous standardized data management procedures all helped the data management staff at the DCC to successfully collect and clean a huge amount of data in a timely fashion.
Arguably, perhaps the most onerous challenge for DCC was to print and ship enormous amounts of paper CRFs on a tight schedule to ensure continuity in data collection at the sites. An electronic data management/transfer system with electronic CRFs could have eliminated this huge and expensive logistical challenge. On the other hand, at least initially, the variability in availability of high-speed Internet technology, the varying degrees of expertise of local staff in some countries to manage a sophisticated system, and the challenge of implementing and managing an electronic system remotely were some of the reasons for DCC to go with a more conventional system.
As the data coordinating center for large cooperative clinical trials sponsored by the Department of Veterans Affairs, over the years our unit has gained wide recognition for the large and complex datasets that we have managed. Yet GEMS is unique in regards to the number of plates that the DCC staff processed over the duration of the study, surpassing all previous experience. Indeed, in comparison with other large studies completed at the DCC, GEMS was almost 10 times larger in regards to the volume of data. We believe that the insights and experiences that we have described in this paper should be helpful to other research consortia undertaking projects that generate enormous datasets and that must transfer those data expeditiously from field sites (including some very rural sites) in multiple developing countries to a central data coordinating center.
Notes
Acknowledgments. The enormous task of implementing and conducting the study from the Perry Point Data Coordinating Center (DCC) perspective would not have been possible without the talented and hardworking DCC staff. The DCC team was comprised of the following individuals in addition to the first author as the team lead and the biostatistician: project managers—Barbara Yndo, Karen Hessler, Kristy Tomlin; data managers—Steven Berkey, Christina Carty; statistical programmer—Rebecca Horney; computer assistants—Ellen Sterrett, Christopher Crayton, Veronica Debity, Julie Lowe, Carla Davis, Tangra Cole.
Financial support. This work was supported by the Bill & Melinda Gates Foundation (grant number 38874).
Supplement sponsorship. This article was published as part of the supplement entitled “The Global Enteric Multicenter Study (GEMS),” sponsored by the Bill & Melinda Gates Foundation.
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.