Abstract
Deorphanization of GPR54 receptor a decade ago led to the characterization of the kisspeptin receptor (Kissr) in mammals and the discovery of its major role in the brain control of reproduction. While a single gene encodes for Kissr in eutherian mammals including human, other vertebrates present a variable number of Kissr genes, from none in birds, one or two in teleosts, to three in an amphibian, xenopus. In order to get more insight into the evolution of Kissr gene family, we investigated the presence of Kissr in osteichthyans of key-phylogenetical positions: the coelacanth, a representative of early sarcopterygians, the spotted gar, a non-teleost actinopterygian, and the European eel, a member of an early group of teleosts (elopomorphs). We report the occurrence of three Kissr for the first time in a teleost, the eel. As measured by quantitative RT-PCR, the three eel Kissr were differentially expressed in the brain-pituitary-gonadal axis, and differentially regulated in experimentally matured eels, as compared to prepubertal controls. Subfunctionalisation, as shown by these differences in tissue distribution and regulation, may have represented significant evolutionary constraints for the conservation of multiple Kissr paralogs in this species. Furthermore, we identified four Kissr in both coelacanth and spotted gar genomes, providing the first evidence for the presence of four Kissr in vertebrates. Phylogenetic and syntenic analyses supported the existence of four Kissr paralogs in osteichthyans and allowed to propose a clarified nomenclature of Kissr (Kissr-1 to -4) based on these paralogs. Syntenic analysis suggested that the four Kissr paralogs arose through the two rounds of whole genome duplication (1R and 2R) in early vertebrates, followed by multiple gene loss events in the actinopterygian and sarcopterygian lineages. Due to gene loss there was no impact of the teleost-specific whole genome duplication (3R) on the number of Kissr paralogs in current teleosts.
Introduction
In 1999, a novel G protein-coupled receptor named GPR54 was cloned from the rat brain [1]. GPR54 ligands were later shown to be kisspeptins, previously described as metastasis suppressors, encoded by Kiss1 gene [2]. As soon as this link between kisspeptins and GPR54 was unveiled, major discoveries in the field of reproductive endocrinology were realised. Kisspeptin and its receptor (GPR54/Kissr) emerged as major upstream regulators of the gonadotropic axis in mammals, by their key roles in the control of GnRH, mediation of steroid feedbacks, as well as initiation of puberty [3].
In mammalian species, a single gene, named Kiss1r, encodes for the kisspeptin receptor. To date the only exception is the platypus (Ornithorhynchus anatinus), a non-placental mammal, in which two receptors are present [4]. Contrasting situations are found in other tetrapods, as shown by a lack of Kissr in birds and up to three Kissr paralogous genes in an amphibian species, the xenopus (Xenopus tropicalis) [4]. In teleosts, at least one Kissr is present in all species investigated so far (for review [5]). A second Kissr gene could be evidenced in some species including zebrafish (Danio rerio) [6], goldfish (Carassius auratus) [7], medaka (Oryzias latipes) [4], and striped bass (Morone saxatilis) [8]. However, this second paralog is lacking in the genomes of other teleosts, such as fugu (Takifugu niphobles), tetraodon (Tetraodon nigroviridis) and stickleback (Gasterosteus aculeatus). Concerning cyclostomes, we recently identified one Kissr gene in the sea lamprey (Petromyzon marinus) genome [9]. The homology relationships between the various Kissr and the evolutionary events that led to such diversity are still ambiguous.
Recently the genomes of three osteichthyan species of particular phylogenetic interest have been published: the coelacanth, Latimeria chalumnae, a representative of early sarcopterygians (coelacanth genome project, Broad Institute), the spotted gar, Lepisosteus oculatus, a non-teleost actinopterygian [10], and the European eel, Anguilla anguilla, a member of an early group of teleosts (elopomorphs) [11]. In the present study, we investigated the presence of Kissr in the genome of those relevant species.
We previously initiated the study of Kissr in the eel [9]. Due to its phylogenetical position, the eel may provide insights into ancestral regulatory functions in teleosts [12], the largest group of vertebrates. Furthermore, its striking biological cycle, with a blockade of sexual maturation as long as the reproductive oceanic migration is not performed, makes the eel a powerful model to investigate neuroendocrine mechanisms of puberty [13]. We formerly cloned the cDNA of one Kissr from the European eel brain [9].
In the present study, we report the occurrence of two additional Kissr in the eel, providing the first evidence for three Kissr in a teleost species. We also identified four Kissr in both coelacanth and spotted gar genomes, providing the first evidence for the presence of four Kissr in vertebrate species. Phylogenetic and syntenic analyses allowed us to assess the existence of four Kissr paralogs in osteichthyans and to raise new hypotheses on the origin and evolutionary history of vertebrate Kissr family. These data also let us propose a clarified Kissr nomenclature, based on these four paralogons. Finally, in order to get some insights into the potential process driving the conservation or the loss of multiple Kissr, we focused on the analyses of their tissue distributions and regulations during experimental maturation in the eel.
Materials and Methods
Animals
European eels (Anguilla anguilla) were at the prepubertal “silver” stage, which corresponds to the last continental phase of the eel life cycle, preceding the oceanic reproductive migration. Cloning and tissue distribution were performed using female and male eels purchased from Rungis International Market (Rungis, France) and transferred to MNHN, France. Eel manipulations were performed according to the guidelines of the French Ministry of Agriculture and Fisheries; Veterinary Department for Animal Health and Protection, under the supervision of authorised investigators (agreement N°I-75UPMC-F1-07). Experimental gonadal maturation was performed on farmed female eels at the DTU Aqua research facility at Lyksvad Fishfarm, Vamdrup, Denmark. Eel manipulations and experimental maturation were performed according to the guidelines of Danish Ministry of Food, Agriculture and Fisheries; Danish Veterinary and Food Administration (Approval reference for maturation experimentations: 2010/561-1783 “artificial reproduction of European eel”). Pain, suffering and stress were attempted minimized during transport and rearing throughout the experiments. All eels were anesthetized using benzocaine before sacrifice and whenever needed in relation to treatment.
Identification of Kissr sequences
European eel genome database analysis
The TBLASTN algorithm of the CLC DNA Workbench software (CLC bio, Aarhus, Denmark) was used to retrieve the genomic sequences of three Kissr (named Kissr-1, Kissr-2 and Kissr-3) from the European eel genome database [11]. The peptidic sequences of the previously characterized eel Kissr (named here Kissr-2) [9], of the two zebrafish Kissr, and of the three xenopus Kissr were used as queries.
Coelacanth and spotted gar genomic database analyses
The TBLASTN algorithm (search sensitivity: near exact matches (short)) of the e!ENSEMBL website was used to retrieve the genomic sequences of four coelacanth Kissr and four spotted gar Kissr from their genomic databases, respectively. The peptidic sequences of the three eel Kissr identified in the present study, of the three xenopus Kissr and of the two predicted platypus Kissr were used as queries.
The exons and splicing junctions were predicted using the empirical nucleotidic splicing signatures, i.e. intron begins with “GT” and ends with “AG”. The 7 transmembrane domains were determined using TMHMM software (TMHMM Server v. 2.0).
Phylogenetic analysis
Amino-acid sequences of 51 known or predicted Kissr were first aligned using ClustalW [14], then manually adjusted. The JTT (Jones, Taylor and Thornton) protein substitution matrix of the resulting alignment was determined using ProTest software [15]. Phylogenetic analysis of the Kissr sequence alignment was performed using the maximum likelihood method (RaxML software [16]), with 1,000 bootstrap replicates.
Identity and similarity percentages between two sequences were calculated using EMBOSS Matcher.
Syntenic analysis
The synteny analyses of the eel Kissr-1, Kissr-2, Kissr-3 genomic regions were manually performed using CLC DNA Workbench 6 software and the European eel genome database. The analyses of the neighbouring genes of the four predicted spotted gar Kissr were performed using the preliminary gene annotation of the genome assembly LepOcu1 generated by Ensembl release 67. Synteny maps for the genomic neighbourhoods of the Kissr genes in human, platypus, lizard (Anolis carolinensis), xenopus, zebrafish, medaka, stickleback, tetraodon and coelacanth ( as well as of the corresponding region in chicken (Gallus gallus) were performed using the PhyloView of Genomicus v67.01 web site [17]. The analysis of the neighbouring genes of the four predicted coelacanth Kissr paralogs was completed by the automatic and manual gene annotation of the coelacanth genome.
Cloning of the full-length cDNAs encoding eel Kissr-1 and Kissr-3
Total RNA from eel brain (di-/mes-encephalon) was extracted using Trizol reagent and reverse-transcripted as previously described [9]. 5′ and 3′ UTR genomic sequences of predicted Kissr-1 and Kissr-3 were used to design specific primers (Table S1) in order to amplify by PCR their full coding sequences (CDS). Standard PCRs were performed as follows: an initial step of polymerase activation for 3 min at 94°C; then 40 cycles with 30 s at 94°C for denaturing, 30 s at various temperatures (54°C for Kissr-1 and 66°C for Kissr-3) for annealing, 1 min 30 s at 72°C for primer extension, and a single final extension step of 5 min at 72°C. PCR products were purified with the QUIAquick PCR Purification Kit (Qiagen, Hilden, Germany) and inserted in a pCR™4-TOPO® TA vector provided by the TOPO® TA Cloning® Kit (Invitrogen). The vectors were then transfected in One Shot® TOP10 Chemically Competent E. coli (Invitrogen). After the bacteria containing a vector with insert had grown in miniprep cultures, vectors were extracted and purified using QUIAquick Spin Miniprep Kit (Qiagen, Hilden, Germany). Their inserts were then sequenced at GATC biotech Ltd (Konstanz, Germany). The obtained sequences were submitted to EMBL.
Kissr tissue distribution in the eel
Various tissues were collected from eight freshwater female silver European eels to investigate the distribution of Kissr-1, Kissr-2 and Kissr-3 expressions, using qPCR. Eels were sacrificed by decapitation. The following organs were sampled, stored in RNAlater (Ambion-Inc, Austin, TX, USA) and kept frozen at −20°C until RNA extraction: brain, pituitary, ovary, muscle, eye, liver, adipose tissue, kidney, intestine and spleen. The brain was dissected into five parts [18]: olfactory bulbs, telencephalon, di-/mes-encephalon, cerebellum and medulla oblongata. In addition, testes from eight freshwater male silver European eels were also sampled.
Eel experimental maturation
Farmed female silver European eels transferred to seawater (36‰) received weekly injections of salmon pituitary extract for four months to induce vitellogenesis followed by one dihydroxy-progesterone injection to induce final oocyte maturation and ovulation, according to [19]. Analyzes were performed on nine matured eels and seven controls. After sacrifice, anterior brain divided in two parts (olfactory bulbs/telencephalon; di-/mesencephalon), pituitary and ovary were dissected and stored in RNAlater and kept frozen at −20°C until RNA extraction.
Quantitative real-time PCR (qPCR)
Eel Kissr-1, Kissr-2 and Kissr-3 specific primers (Table S1) were designed based on the full-length European eel CDS sequences cloned in this study (Kissr-1 and Kissr-3) and previously [9] (Kissr-2), using Primer3 Software (Whitehead Institute/Massachusetts Institute of Technology, Boston, MA). The Kissr-3 qPCR primers were designed in exon-3, to quantify the expression level of the three Kissr-3 mRNA splicing isoforms at once. To optimize the assays, different annealing temperatures were tested according to the melting temperature (Tm) of primers. To assess their specificity, amplification products were sequenced at GATC Biotech Ltd. Primers for European eel LHβ, mGnRH and reference gene β-actin were as previously designed [18], [20] (Table S1). All primers were purchased from Eurofins (Ebersberg, Germany).
Quantitative assays of eel Kissr-1, Kissr-2, Kissr-3, mGnRH, LH-β and β-actin mRNAs were performed using the LightCycler® System (Roche, Ltd. Basel, Switzerland) with SYBER Green I sequence-unspecific detection as previously described [18], [20], [21]. The qPCR primers are listed in Table S1. The qPCRs were prepared with 4 µl of diluted cDNA template, 2 µl of PCR grade water, 2 µl of SYBR Green master mix and 1 µl of each forward and reverse primer (0.5 pmole each at final concentration). The qPCRs were performed as follows: an initial step of polymerase activation for 10 min at 95°C; then 41 cycles with 15 s at 95°C for denaturing, 5 s at 60°C (Kissr-1, Kissr-2, mGnRH, LH-β and β-actin) or 62°C (Kissr-3) for annealing, 10 s at 72°C for primer extension, 5 s at 83°C to avoid measurement of non-specific annealing; and a single final extension step of 5 min at 72°C. Each qPCR run contained a non-template control (cDNA was substituted by water) for each primer pairs to confirm that reagents were not contaminated. The efficiency of all primers was tested and the specificity of each reaction was assessed by melting curve analysis to ensure the presence of only one product, and by sequencing. Individual tissue samples were then analyzed in duplicate by qPCR. Serial dilutions of cDNA pool of brain tissues were run in duplicate and used as a common standard curve and also included in each run as a calibrator. Normalisation of data was performed using total RNA content for the tissue distribution samples, and using β-actin mRNA level for experimental maturation samples.
Statistical analysis
Results are given as mean ± SEM. Means were compared by Student's t-test using Instat (GraphPad Software Inc., San Diego, Calif., USA).
Results and Discussion
Characterization of three European eel Kissr
Genomic prediction of three eel Kissr genes
We used the deduced peptidic sequence from the previously cloned European eel Kissr cDNA [9] (considered as Kissr-2 in the present study) as a query to retrieve Kissr sequences in the European eel genome. TBLASTN results revealed the presence of three different Kissr genes, called Kissr-1, -2, -3, each made of 5 exons and 4 introns (Fig. S1), that constitutes the conserved structure of Kissr genes. The lengths of exon-2, exon-3 and exon-4 are the same for all three genes with 125 bp, 136 bp and 239 bp, respectively. The exon-1 of Kissr-1, Kissr-2 and Kissr-3 is 235 bp, 214 bp and 229 bp, respectively. The exon-5 is 348 bp, 372 bp and 351 bp, respectively. This leads to predicted Kissr-1, Kissr-2 and Kissr-3 CDS of 1083 bp, 1086 bp and 1080 bp, respectively.
Cloning of eel Kissr cDNAs
Using European eel specific Kissr-1 primers, PCRs performed on brain cDNAs led to a single product. Its sequence (EMBL: HE802271) encompassed partial 5′ and 3′ UTRs of 199 bp and 102 bp, respectively, and a full CDS of 1083 bp. Once translated, the cloned Kissr-1 CDS gives a 360-aa receptor, exhibiting the seven transmembrane domains (TMD) characteristic of the GPCR family (Fig. 1A).
Figure 1. Molecular cloning of eel Kissr-1 and Kissr-3_v1.
Nucleotide and deduced amino-acid sequence of the cDNA encoding the eel Kissr-1 (A) and Kissr-3_v1 (B). Nucleotides (top) are numbered from 5′ to 3′. The amino-acid residues (bottom) are numbered beginning with the first methionine residue in the ORF. The asterisk (*) indicates the stop codon. The predicted seven transmembrane domains (TMD) are underlined and the cysteines involved in a disulphide bridge are shaded in grey.
The previously cloned eel Kissr cDNA [9] is named Kissr-2 in the present study.
Using European eel specific Kissr-3 primers, PCRs performed on brain cDNAs led to the isolation of three different products (EMBL: HE802272, HE802273 and HE802274). Their sequence revealed three Kissr-3 isoforms – named here Kissr-3_v1 (Fig. 1B), Kissr-3_v2 and Kissr-3_v3 (Fig. S2). The partial 5′ and 3′ UTR are 167 and 147 bp for all three isoforms. The Kissr-3_v1 isoform corresponds to the 5 predicted exons from the eel genome. Once translated, it gives a 359-aa receptor exhibiting the seven conserved TMD (Fig. 1B). The Kissr-3_v2 CDS corresponds to the 5 predicted exons, minus the 76 first nucleotides of the exon-2 that are missing (Fig. S2A). The loss of these 76 nucleotides results in a shift of the Kissr-3 reading frame and the occurrence of a first premature termination codon at position 280 pb. Once translated, it gives a 93-aa protein. This shorter protein exhibits a single TMD encoded by exon-1. The Kissr-3_v3 CDS corresponds to exon-1, exon-3, exon-4 and exon-5, while exon-2 is completely missing (Fig. S2B). The loss of exon-2 results in a shift of the Kissr-3 reading frame without the occurrence of any premature termination codon. Once translated, it gives a 361-aa protein, which exhibits only one TMD encoded by exon-1. The presence of the three Kissr-1, -2, and -3 transcripts and of the three Kissr-3 mRNA isoforms was also observed in Japanese eel (Anguilla japonica) brain cDNAs (data not shown).
To date, only one or two Kissr genes had been found in the genome of teleosts (reviewed by [5]), so the eels provide the first evidence of the presence of three Kissr genes in teleosts. Until now three Kissr genes had been found only in an amphibian species, xenopus [4].
In addition to this Kissr gene diversity, European and Japanese eels present three Kissr-3 mRNA isoforms revealing similar alternative splicing in both species. The existence of Kissr mRNA splicing isoforms has been described in two other teleosts, the sole (Solea senegalensis) [22] and the zebrafish [23]. In the sole, two isoforms were identified for the single Kissr (Kissr-2 type) gene present in this species and they differ by the retention of intron III, while in the eel they differ by partial or complete deletion of exon-2 in Kissr-3 [22]. In the zebrafish, five isoforms were identified for Kiss1rb (Kissr-3 type), while no splicing isoform was observed concerning Kiss1ra (Kissr-2 type). Among the five zebrafish Kiss1rb isoforms, one of them, called KRBDP1, resulted from the deletion of exon-2 and corresponds to eel Kissr-3_v3 [23]. Even though the existence of other Kissr mRNA isoforms in the eel cannot be excluded, the three genes and several isoforms described here highlight the Kissr molecular diversity in basal teleosts. They also could reflect an ancestral post-transcriptional regulatory process of Kissr.
Genomic prediction of four coelacanth and four spotted gar Kissr genes
To further assess the Kissr diversity in vertebrates, we investigated the presence of these genes in the genomes of the spotted gar, a non-teleost actinopterygian, and the coelacanth, a basal sarcopterygian, two species of relevant phylogenetical positions. We performed a TBLASTN in both genomes (Ensembl) using the three European eel, the two zebrafish and the three xenopus Kissr proteins as queries. The TBLASTN results revealed the existence of four Kissr genes in both coelacanth and spotted gar genomes, named here Kissr-1, Kissr-2, Kissr-3 and Kissr-4.
Four Kissr genes in the coelacanth genome
The four predicted Kissr genes of coelacanth are made of 5 exons and 4 introns. The predicted CDS and the exon-exon junctions of each putative transcript are shown in Fig. S3. Once translated, the putative transcripts lead to four predicted proteins, i.e. coelacanth Kissr-1, Kissr-2, Kissr-3 and Kissr-4, of 368-aa, 367-aa, 377-aa and 367-aa, respectively (Fig. S3).
Four Kissr genes in the spotted gar genome
The four predicted Kissr genes of spotted gar are also made of 5 exons and 4 introns. Although Kissr-1 exon-4 and Kissr-4 exon-5 are partial due to the current status of the genome, the predicted CDS and the exon-exon junctions of each putative transcript are shown in Fig. S4. Once translated, the putative transcripts lead to four predicted proteins, i.e. spotted gar Kissr-1 to 4 of 318-aa, 362-aa, 382-aa, and 385-aa, respectively (Fig. S4).
In both coelacanth and spotted gar, all predicted Kissr proteins present the typical seven TMD of the GPCR family (Fig. S3 and S4).
These new findings evidence for the first time the potential existence of four Kissr paralogs in vertebrate species, representatives of the sarcopterygian (coelacanth) and the actinopterygian (spotted gar) lineages.
Phylogenetic analysis of the Kissr family
Other genomic database analyses
In addition to the characterisation and/or the prediction of European eel, coelacanth and spotted gar Kissr, we used the ENSEMBL Genome Browser to retrieve the sequences of two Kissr in the medaka genome (GPR54-1, chromosome 17 and GPR54-2, chromosome 9; [4]), and to search for new candidate Kissr genes in the genomes of other vertebrates. We found one putative Kissr gene, named here Kissr-2, in several teleost genomes, stickleback (chromosome group III), tetraodon (chromosome 15-random) and cod (Gadus morhua; scaffold 377), one Kissr in a cyclostome genome, sea lamprey (scaffold GL478157; [9]), and one in the lizard genome (chromosome 2).
Phylogenetic analysis
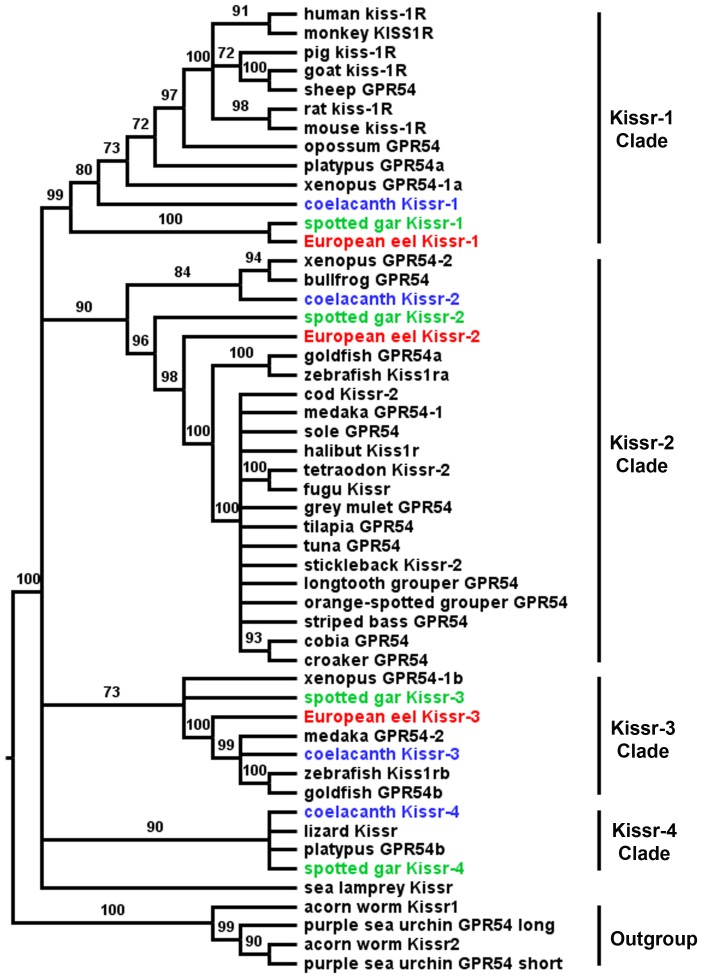
Based on an alignment of 51 Kissr peptidic sequences (Fig. S5), and assuming ambulacrarian (acorn worm, Saccoglossus kowalevskii, and purple sea urchin, Strongylocentrotus purpuratus (Table S2)) Kissr as outgroup, a phylogenetic tree was generated using the Maximum Likelihood method (the list of sequences and accession numbers is provided in Table S2). As shown in Fig. 2, it clusteres the vertebrate Kissr into four main clades, which are supported by significant bootstrap values (99, 90, 73, and 90%, respectively). Based on this analysis and the determination of those four clades, we propose the following new nomenclature of the different Kissr.
Figure 2. Consensus phylogenetic tree of the vertebrate kisspeptin receptors (Kissr).
This phylogenetic tree was constructed based on the amino-acid sequences of Kissr (for the references of each sequence see Table S2) using the Maximum Likelihood method with 1,000 bootstrap replicates. The number shown at each branch node indicates the bootstrap value (%); only values and branching above 70% are indicated. The tree was rooted using the two sequences of the hemichordata acorn worm GPR54-1 and GPR54-2 and the two sequences of echinodermata purple sea urchin Kissr_short and Kissr_long. The European eel Kissr and predicted coelacanth and spotted gar Kissr are coloured in red, blue and green, respectively.
The first clade mainly encompasses sarcopterygian Kissr including all eutherian Kissr, a metatherian Kissr (Monodelphis domestica; opossum GPR54), a prototherian Kissr (platypus GPR54a), a xenopus Kissr (GPR54-1a) and coelacanth Kissr-1. In addition, two actinopterygian Kissr, spotted gar and European eel Kissr-1, branch together at the base of this clade. This is the first evidence of the presence of an ortholog to eutherian Kissr in actinopterygians. Acknowledging the presence of eutherian (including human) Kissr, which have been the first to be discovered, this clade was named Kissr-1 clade. The eel is up to now the only teleost presenting an ortholog (eel Kissr-1) to mammalian Kissr.
The second clade, named Kissr-2, clusters mainly actinopterygian Kissr, i.e. spotted gar Kissr-2 and most of the previously described teleost Kissr including zebrafish Kiss1ra, goldfish GPR54a, medaka GPR54-1 and European eel Kissr-2. This clade also clusters together three sequences from sarcopterygian species, xenopus GPR54-2, bullfrog (Rana catesbeiana) GPR54 and coelacanth Kissr-2.
The third clade, named Kissr-3, clusters two sarcopterygian Kissr (xenopus GPR54-1b and coelacanth Kissr-3) and some actinopterygian Kissr (spotted gar Kissr-3 and a few teleost Kissr: zebrafish Kiss1rb, goldfish GPR54b, medaka GPR54-2 and European eel Kissr-3).
The fourth clade, named Kissr-4, clusters three sarcopterygian Kissr (platypus GPR54b, lizard Kissr and coelacanth Kissr-4) with one actinopterygian Kissr, the spotted gar Kissr-4.
This phylogenetic analysis suggests the existence of four distinct paralogous Kissr in osteichthyans. Furthermore, it shows that each sarcopterygian Kissr is orthologous to actinopterygian Kissr. This is specially highlighted by the relationship between the four coelacanth and spotted gar Kissr.
Syntenic analysis of Kissr genes
In order to test the results obtained with the phylogenetic analysis, we performed a syntenic analysis of the Kissr neighbouring genes, an approach which is applied to determine gene orthology relationships as well as the origin and evolutionary history of gene families. For this analysis (Fig. 3), we considered the following vertebrate representatives: mammals (eutherian: human and prototherian: platypus), bird (chicken), squamate (lizard), amphibian (xenopus), basal sarcopterygian (coelacanth), non-teleost actinopterygian (spotted gar) and teleosts (zebrafish, medaka, stickleback, tetraodon and European eel). As already reported (for review: [5], [24]–[26]), genomic synteny analysis shows that birds do not possess any Kissr gene.
Figure 3. Conserved genomic synteny of osteichthyan Kissr.
Genomic synteny maps comparing the orthologs of Kissr-1 (A), Kissr-2 (B), Kissr-3 (C), Kissr-4 (D) loci and their neighbouring genes. Analyses were performed on the genomes of human (Homo sapiens), platypus (Ornithorhynchus anatinus), lizard (Anolis carolinensis), chicken (Gallus gallus), xenopus (Xenopus (Silurana) tropicalis), coelacanth (Latimeria chalumnae), spotted gar (Lepisosteus oculatus), zebrafish (Danio rerio), medaka (Oryzias latipes), stickleback (Gasterosteus aculeatus), tetraodon (Tetraodon nigroviridis) and European eel (Anguilla anguilla). This map was established using the PhyloView of Genomicus v67.01 web site, manual annotation of European eel genome using CLC DNA Workbench 6 software and the gene annotation of the coelacanth and spotted gar genomic databases (see section 2). Kissr genes are named according to our proposed nomenclature (Kissr-1 to Kissr-4). The other genes are named after their human orthologs according to Human Genome Naming Consortium (HGNC). Orthologs of each gene are shown in the same color. Each of the eight conserved gene families is identified by an exponent number. The direction of arrows indicates the gene orientation, with the position of the gene (in 10−6 base pairs) indicated below. The full gene names and detailed genomic locations are given in Table S3.
The mammalian, amphibian, coelacanth, spotted gar and European eel genes from Kissr-1 clade are positioned in genomic regions containing common loci, including PALM, PTBP1, LPPR3, MED16, ARID3A, WDR18, GRIN3B, C19orf6, GADD45B and DIRAS1, exhibiting well conserved synteny (Fig. 3A). This supports the orthology of the Kissr-1 genes. Syntenic analysis suggests that the other teleost genomes do not contain any Kissr-1 gene, though the above-mentioned neighbouring genes are present in the corresponding genomic regions (Fig. 3A). The eel currently provides a unique example of Kissr-1 ortholog in teleosts.
The amphibian, coelacanth, spotted gar and teleost genes from Kissr-2 clade are positioned in genomic regions containing common loci, including PTPRF, KDM4A, ST3GAL3, DPH2, ATPV60B, B4GALT2, CCDC24, KIF2C, PTCH2, EIF2B3, ZCCHC11, GLIS1, GADD45A, DIRAS3, LPAR3, ZNF644, PTBP2 and PALMD, exhibiting well conserved synteny (Fig. 3B). This supports the orthology of the Kissr-2 genes. European eel Kissr-2 orthology is only supported by the presence of partial GLIS1 due to the small size of the scaffold. Syntenic analysis suggests that squamate (lizard) and mammalian genomes do not contain any Kissr-2 gene, though the above-mentioned neighbouring genes are present in the corresponding genome regions (Fig. 3B).
The amphibian, teleost (zebrafish, medaka, European eel) and spotted gar Kissr-3 genes are positioned in genomic regions containing common loci including PSAT1, ISCA1, ZCCHC6, GADD45G, DIRAS2, PALM2, LPAR1 and PTBP3, exhibiting conserved synteny (Fig. 3C). This supports the orthology of the Kissr-3 genes. Syntenic analysis suggests that the other teleost and tetrapod genomes do not contain any Kissr-3 gene, though the above-mentioned neighbouring genes are present in the corresponding genome regions. The coelacanth predicted Kissr-3 is split into the scaffolds JH131603.1 and JH131921.1, which are too small to contain any other gene (Fig. 3C).
Platypus and lizard Kissr-4 gene neighbouring regions present only RETN gene in common, due to the small size of the platypus scaffold (Fig. 3D). Assemblage of three scaffolds (JH133705.1, JH132986.1, and JH127844) from coelacanth genome allowed us to reveal a Kissr-4 neighbouring region comprising LPAR2, STXBP2 and EPHX3 genes, with STXBP2 and EPHX3, being also present in lizard and spotted gar Kissr-4 region (Fig. 3D). These data suggest that Kissr-4 genes can be considered as orthologous. Syntenic analysis also suggests that the genomes from placental mammals, amphibian and teleosts do not contain any Kissr-4 gene, though they present in a common region some conserved genes including CAMSAP3, XAB2, STXBP2, RETN, RAB3D, PALM3, NOTCH3, EPHX3, WIZ and LPAR2 (Fig. 3D).
This syntenic analysis of Kissr genes delineated four different conserved genomic regions among osteichthyans. For each conserved genomic region, Kissr genes from various species clustered in the corresponding phylogenetic Kissr clade. Thus, the results of the syntenic analysis fully validated the orthology relationships of the phylogenetic analysis and further supported our proposal for a new nomenclature.
Evolutionary history of Kissr family
Origin of the four Kissr present in basal osteichthyans
So far, the presence of Kissr has been characterised in two non-vertebrate species, the purple sea urchin and the acorn worm (Table S2). This revealed the existence of at least one ancestral Kissr before the vertebrate emergence. The predictions of four Kissr in the genomes of a basal sarcopterygian, the coelacanth, and of an actinopterygian, the spotted gar, together with the results of both phylogenetic and syntenic analyses, enabled us to hypothesise the existence of at least four Kissr paralogous genes in the common osteichthyan ancestor of the sarcopterygian and actinopterygian lineages. A remaining question was to infer whether these four genes resulted from the duplications of one or multiple ancestral Kissr genes.
It is currently admitted that two rounds of whole genome duplication (1R and 2R) occurred in the early vertebrate evolutionary history, resulting in four-fold replicates of the ancestral genome. Even though numerous genomic rearrangements and loss events occurred during the vertebrate radiation, the vestiges of these two successive genome duplications can be revealed in the current vertebrate species by the existence of numerous four-fold repeated regions (tetra-paralogon) carrying paralogous genes [27].
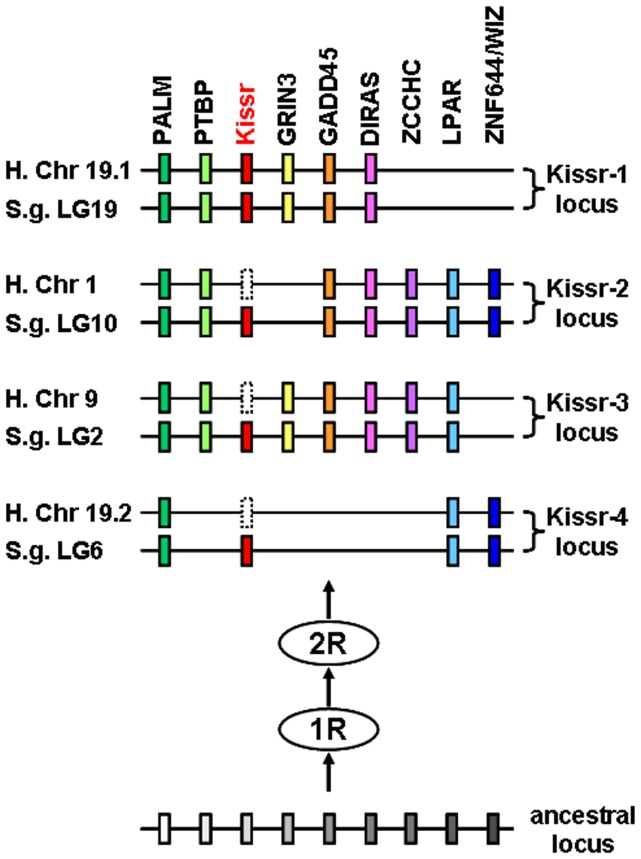
Our syntenic analysis demonstrated that the current osteichthyan Kissr genes are localised in four different genomic regions. In the Fig. 3, in addition to the Kissr genes, we could reveal paralogs from eight gene families present among the four syntenic regions of the osteichthyan representative species. The eight considered families are PALM (4 paralogs), PTBP (3 paralogs), GRIN3 (2 paralogs), GADD45 (3 paralogs), DIRAS (3 paralogs), ZCCHC (2 paralogs), LPAR (3 paralogs) and ZNF644/WIZ (2 paralogs). Fig. 4 focuses on the comparison of these four conserved Kissr syntenic regions in one sarcopterygian, the human, and one actinopterygian, the spotted gar, two species chosen for their genomic assembly in chromosomes and their phylogenetic positions among osteichthyans. Fig. 4 shows that the members of these eight gene families delineate a tetra-paralogon with the four conserved genomic regions of both human and spotted gar. This syntenic observation strongly suggests that the four Kissr resulted from “en-bloc” duplications of a unique ancestral genomic region.
Figure 4. Proposed origin of osteichthyans Kissr loci based on human and spotted gar Kissr tetra-paralogons.
The paralogous genes of each of the eight identified families delineate four tetra-paralogons in both human and spotted gar. This suggests a common origin of the four loci before the two whole genome duplication rounds (1R and 2R) occurred in the early vertebrate history.
Recently, two studies proposed reconstructions of the genomes of vertebrate and chordate ancestors. In the first study, ten proto-chromosomes of the ancestral vertebrate karyotype and their linkage to the corresponding tetra-paralogons in the human genome were hypothesised [28]. The second study proposed a reconstruction into seventeen proto-chromosomes of the last chordate ancestor genome and their linkage to the human tetra-paralogons [29]. Considering these two studies, together with our localisation of the four Kissr syntenic regions in the human genome, we can hypothesise that the corresponding tetra-paralogons resulted from the duplications of one single region localised on the proto-chromosome-A of the vertebrate ancestor and on the proto-chromosome-1 of the last chordate ancestor. From these analyses, we can infer that the four Kissr paralogons may have resulted from the two successive rounds of whole genome duplication (1R and 2R) that occurred in basal vertebrates (Fig. 5).
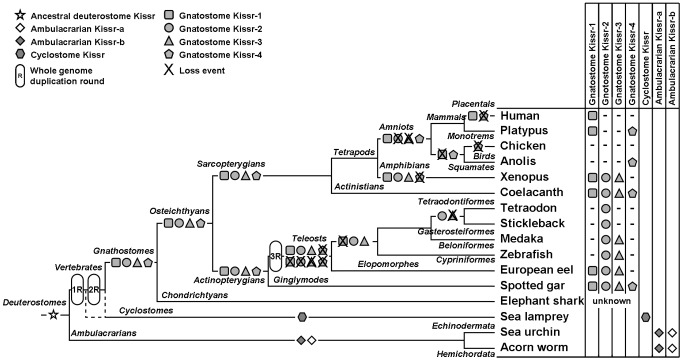
Figure 5. Current status and proposed evolutionary history of Kissr family.
This representation is based on the phylogenetic and synteny analyses. The names of the main phyla are given on the corresponding branches. The names of the current representative species of each phylum are given at the end of the final branches, together with the symbol of the Kissr gene they possess. This hypothesis assumes the presence of the four Kissr paralogs in the osteichthyan lineage resulting from the two rounds of vertebrate whole genome duplication. Multiple subsequent Kissr gene loss events are indicated in the actinopterygian and sarcopterygian lineages.
To date, the exact timing of the 2R occurrence is uncertain and the impact of this event on the cyclostomes is still debated [30], [31]. In our study, neither phylogenetic nor syntenic analyses enabled us to specifically relate the sea lamprey Kissr to one of the four osteichthyan Kissr. The currently available data also led to a polytomy of the four Kissr clades and did not cluster them into two major clades, each of them divided in two sub-clades, which would have reflected the successive 1R and 2R. In the future, increasing number of characterised Kissr sequences, especially from Kissr-3 and Kissr-4 clades, may bring sufficient new information to resolve this polytomy. Though kisspeptin genes have been characterised in chondrichthyans, it could be of particular interest to investigate the presence of its receptor in this lineage to further assess Kissr history in early gnathostomes.
A subsequent history of losses
Our study suggests that four Kissr paralogs would have been present in ancestral gnathostomes, resulting from 1R and 2R. It also shows that these four paralogons are still present in two early emerged osteichthyans, a sarcopterygian, the coelacanth and an actinopterygian, the spotted gar. All other vertebrate species investigated so far possess less Kissr genes (from 3 to none) indicating multiple events of Kissr losses in both the sarcopterygian and actinopterygian lineages (Fig. 5).
In tetrapods (sarcopterygians), Kissr-4 would have been lost in amphibians, Kissr-1, -2 and -3 being present in the xenopus, while in amniotes the losses would have first concerned Kissr-2 and Kissr-3, Kissr-1 and Kissr-4 being present in a prototherian mammal (Fig. 5). Further alternative losses occurred in amniotes, with only Kissr-1 remaining in eutherian mammals, but only Kissr-4 in squamates (lizard) (Fig 5). Finally, an additional loss would have led to the complete absence of Kissr in birds.
In teleosts (actinopterygians), a third round of whole genome duplication (3R) is supposed to have occurred specifically in the early history of this group. The 3R is usually considered as one of the main factors that drove the large radiation and adaptative success of the teleost lineage [11], [32]. As four kissr were present in basal actinopterygians, the teleost-specific 3R implied the potential existence of up to eight Kissr genes in the early teleost history. However, our results show that the largest number of Kissr exhibited by current teleosts is three in the eel, and that each of them is orthologous to one of the coelacanth and spotted gar Kissr. This indicates that 3R did not impact the number of Kissr in teleosts, suggesting an early loss of teleost-specific duplicated Kissr genes, before the emergence of the elopomorphs (Fig. 5). Apart from the eel, only one or two Kissr genes have been described so far in teleosts, indicating additional loss events after the emergence of the elopomorphs (Fig. 5).
The occurrence of these many independent loss events may have led to the current situation of Kissr in the various vertebrate lineages. Indeed, some species seem to be more conservative than others, and it is of particular interest to clarify what may have driven the conservation or the loss of Kissr.
Conservation of multiple Kissr: the example of the eel
As shown by the present study, the eel is one of the most conservative species among current vertebrates, as three different Kissr have been retained. Conservation of multiple Kissr may reflect evolutionary processes such as neo- or sub-functionalisation. The comparative analyses of eel Kissr peptidic sequences and eel Kissr tissue distribution and regulation may constitute the first steps in the understanding of such processes.
Comparison of eel Kissr peptidic sequences
The analysis of the peptidic sequences deduced from the three cloned eel Kissr cDNAs revealed a conserved disulfide bridge between cysteines at positions 112/192 for Kissr-1, 105/185 for Kissr-2 and 110/190 for Kissr-3. The seven TMD of each receptor comprise 23 aa, except for TMD3 of Kissr-1 and Kissr-2 which comprise 18 aa and 19 aa, respectively. The pairwise comparison of the three peptidic sequences revealed 60.5% identity for Kissr-1/Kissr-2, 63.3% for Kissr-1/Kissr-3, and 63.4% for Kissr-2/Kissr-3. These low identities are mostly due to differences between the N-terminal extracellular domains (28.6% to 40.5% identity) and between the C-terminal intracellular domains (43.6% to 58.3% identity) (Fig. S6). Differences within the N-terminal domains could reflect variations in ligand binding properties, while those in the C-terminal domain may correspond to differences in G protein association properties and activation of intracellular signalling pathways. Future studies, including recombinant receptors and characterization of endogenous ligands, will aim at further investigating the potential differences in the structure/function of three eel Kissr.
Differential tissue distribution of the three eel Kissr
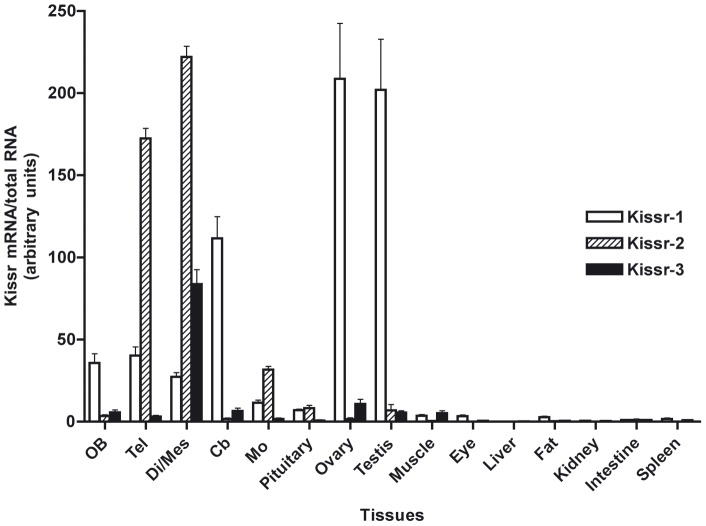
Specific qPCR were developped for each eel Kissr and applied to the analysis of their respective tissue distribution (Fig. 6).
Figure 6. Tissue distribution of the expression of the three eel Kissr mRNAs.
Olfactory bulbs (OB), telencephalon (Tel), di-/mes-encephalon (Di/Mes), cerebellum (Cb), medulla oblongata (MO), pituitary, ovary, testis, muscle, eye, liver, adipose tissue (Fat), kidney, intestine and spleen. The relative expression of each Kissr mRNA was normalised to the amount of total RNA. Each bar represents mean ± SEM from 8 individuals.
Kissr-1 mRNA was expressed in all parts of the brain and in the pituitary. Its highest expression level was found in the gonads, in both ovary and testis. Low Kissr-1 mRNA levels were measured in muscle, retina and fat tissue, while its expression was close to the limit of detection in the other peripheral tissues investigated (liver, kidney, intestine, and spleen).
Kissr-2 mRNA was also expressed in the brain and pituitary, with the highest transcript levels in the telencephalic and di-/mes-encephalic areas, in agreement with our previous study [9]. A low expression of Kissr-2 mRNA was measured in the testis, while its expression in the ovary and all the other peripheral organs was close to the limit of detection.
Kissr-3 mRNA was highly expressed in the di-/mes-encephalic area and to a much lesser extent in the other parts of the brain. Transcript level was close to the limit of detection in the pituitary. Low expression of Kissr-3 mRNA was measured in the gonads, ovary and testis, as well as in the muscle, while its expression in the other peripheral organs was close to the limit of detection.
The three eel Kissr thus appear mainly expressed in the eel Brain-Pituitary-Gonad (BPG) axis, which highlights the potential involvement of the kisspeptin system in the eel reproductive function. Considering the phylogenetic position of the eel, this tissue distribution could reflect an ancestral role of the kisspeptin system in reproduction, which would have been largely conserved across vertebrate evolution. The three Kissr transcripts are all highly expressed in the eel brain, but with various relative levels according to brain regions. Both Kissr-1 and Kissr-2 are expressed in the pituitary, where they may mediate direct kisspeptin effects as previously investigated [9]. Kissr-1 is also highly expressed in both the ovary and testis. Thus, the three eel Kissr present a differential distribution among BPG axis that suggests differential putative roles in the control of eel reproduction and implies a potential sub-functionalisation of the three receptors.
In the other teleosts, only Kissr-2 and for some species Kissr-3 have been described so far. As in the eel, they are expressed in the BPG axis (grey mullet, Mugil cephalus [33]; zebrafish [6], [34]; fathead minnow, Pimephales promelas [35]; tilapia, Oreochromis niloticus [36], [37]; goldfish [7]; fugu [38]). In xenopus, which presents the three orthologs of eel Kissr [4], all three receptors are expressed in the brain, while only GPR54-1a (kissr-1) and GPR54-2 (kissr-2) mRNA are detected in the pituitary, similarly to the eel. In placental mammals, only Kissr-1 ortholog is present and expressed in the brain, pituitary (human [2], [39]; mouse [40]) and ovary (rat [41]; human and marmoset [42]), where a role in local control of ovarian functions has been suggested.
Differential regulation of the three eel Kissr by experimental maturation
Gonadal development in the silver eel is blocked at a pre-pubertal stage due to a deficient production of pituitary gonadotropins, which results from a dual brain blockade: lack of stimulation by GnRH and direct inhibition by dopamine (for review: [43]). Experimental gonadal maturation was induced in female silver eels according to classical hormonal treatments [19]. The expressions of the three Kissr were analyzed by qPCR in the BPG axis, which are the major sites of Kissr expression in the eel.
In the brain (Fig. 7), Kissr-1 transcript level was significantly up-regulated in matured eels as compared to control ones (x3.73, P<0.0001 in olfactory bulbs/telencephalon and x7.15, P<0.0001 in di-/mes-encephalon), while no changes were recorded for Kissr-2 nor Kissr-3. In the pituitary (Fig. 7), Kissr-1 and Kissr-2 transcript levels were significantly down-regulated in matured eels compared to controls (x0.42; P<0.05 and x0.14; P<0.01, respectively), while Kissr-3 remained at the limit of detection. In the ovary (Fig. 7), Kissr-3 transcript level was significantly down-regulated in matured eels (x 0.39; P = 0.01), while Kissr-1 transcript levels did no change significantly and Kissr-2 remained at the limit of detection.
Figure 7. Regulations of the three eel Kissr expressions during experimental maturation.
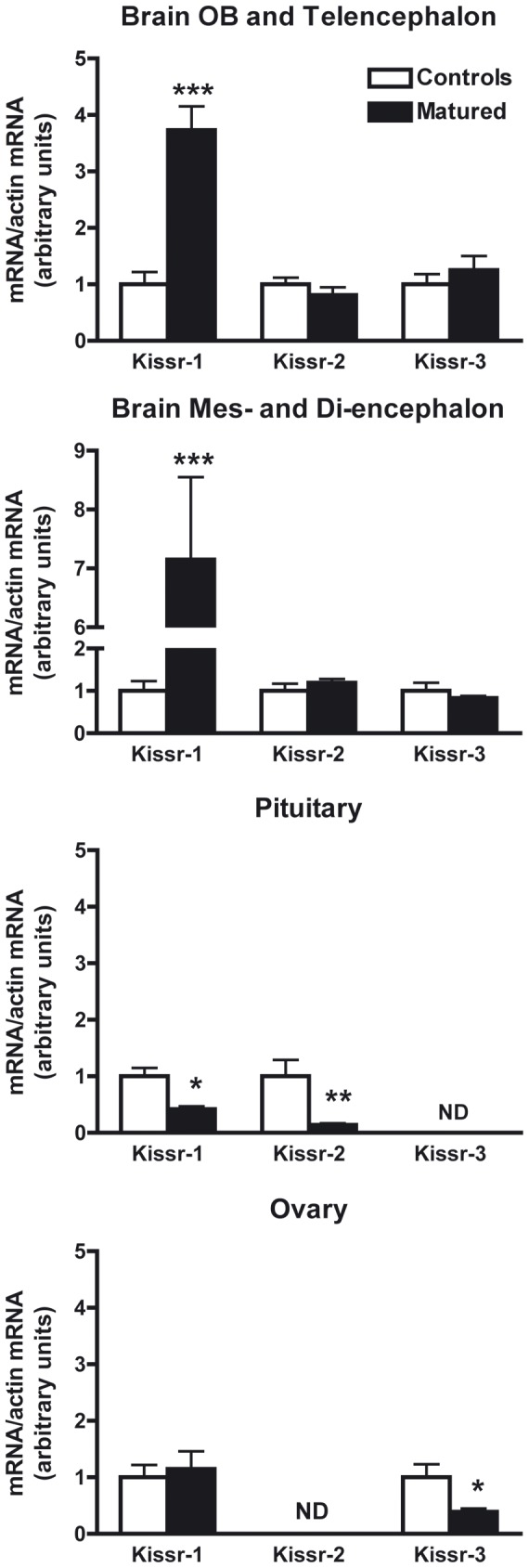
The relative expression of each Kissr mRNA was normalised to ß-actin mRNA. Each bar represents mean ± SEM from 7 control and 9 matured female eels. Significant difference between control and matured groups: *P<0.05, **P<0.01 and *** P<0.001 (Student's t test). N.D. = Non detectable.
We also analyzed by qPCR the expression of brain mGnRH and pituitary LHß. Significant increases in mRNA levels for brain mGnRH (x5.6; P<0.001) and pituitary LHß (x206; P<0.0001) were measured in matured eels, as compared to control eels (data not shown) in agreement with our previous studies [20], [44], [45].
A parallel study was performed on the regulation of the three Kissr transcript levels in experimentally matured Japanese female eels. Eels were treated according to Jeng et al. [46]. As for the European eel, a differential regulation was evidenced. This was shown by selective increase in brain Kissr-1, decreases in pituitary Kissr-1 and Kissr-2 and decrease in ovarian Kissr-3 transcripts, in matured eels as compared to controls (data not shown).
Those results clearly evidence a differential regulation of the three eel Kissr, with receptor- and tissue- specific variations. The selective up-regulation of Kissr-1 expression in the brain suggests that Kissr-1 may contribute to enhance kisspeptin stimulatory control of GnRH at puberty in the eel. Similarly, brain Kissr-1 expression increases at the onset of puberty in mammals (rodent [47]; monkey [48]). This role may have shifted to Kissr-2 in other teleosts, which lack Kissr-1 ortholog. For instance, parallel variations in the brain expressions of Kissr-2 ortholog and GnRH were observed during sexual maturation/puberty in several teleosts (cobia, Rachycentron canadum [49]; grey mullet [33]; fathead minnow [35]; Nile tilapia [36]). We recently revealed an unexpected inhibition by kisspeptins of LHβ expression in eel pituitary cells in vitro, providing the first evidence of an inhibitory role of kisspeptin on gonadotropic function [9]. In the present study, we observed a down-regulation of eel Kissr-1 and Kissr-2 in the pituitary, while LHβ expression largely increased. This suggests that the inhibitory control of kisspeptin on LHβ expression could be removed during the sexual maturation by the down regulation of its receptors in the pituitary. Kissr-3 was down-regulated in the ovary implying a potential involvement of this receptor in ovarian function. Future studies should aim at clarifying the role of kisspeptin system at gonadal level in teleosts.
The regulation of the expression of the three Kissr in the BPG tissues reinforces the hypothesis of a conserved ancestral role in vertebrate reproductive function. In addition to their differential distribution in the BPG axis, their differential regulations further strengthen the potential sub-functionalisation of the three Kissr paralogs for the control of eel reproductive function.
Our previous studies, including castration experiments and steroid treatments, demonstrated that the activation of mGnRH and LH in matured eel result from a positive feedback by sex steroids [44], [45], [50], [51]. Increasing data support a role of brain kisspeptin system in the mediation of steroid feedbacks on GnRH neurons in mammals (for review: [52], [53]) and recently in teleosts (medaka [54]; goldfish [55]; zebrafish [34]). Future studies will aim at investigating the role of sex steroids in the differential regulation of Kissr paralogs in the eel.
Conclusions
In conclusion, our study provides the first evidence of multiple Kissr paralogs in basal osteichthyans, with the cloning of three Kissr in a basal teleost, the eel, and the prediction of four Kissr in a non-teleost actinopterygian, the spotted gar, and four Kissr in a basal sarcopterygian, the coelacanth. Phylogenetic and syntenic analyses support the existence of four Kissr paralogs in osteichthyans, leading to the proposal of a new, simplified classification and nomenclature (Kissr-1 to 4). The four Kissr paralogs may have arisen during the two rounds of whole genome duplication (1R and 2R) in early vertebrates, followed by multiple gene loss events in the various groups of actinopterygian and sarcopterygian lineages. In particular, no impact of teleost-specific 3R can be recorded on the number of Kissr paralogs in current teleosts. Sub-functionalisation of the three eel Kissr, as shown by differences in their sequences, tissue distributions and regulations during sexual maturation, may have represented significant evolutionary constraints for the conservation of multiple Kissr paralogs in this species.
Supporting Information
Three eel Kissr gene sequences. Genomic sequences of the eel Kissr-1 extracted from the scaffold 90.1 (A), Kissr-2 extracted from the scaffold 3158.1 (B) and Kissr-3 extracted from the scaffold 1832.1 (C) (European eel genome [11]). Nucleotides are numbered from 5′ to 3′. The five exons of each gene are shaded in grey.
(DOC)
Molecular cloning of eel Kissr-3 splicing variants Kissr-3_v2 and Kissr-3_v3. Nucleotide and deduced amino-acid sequence of the cDNAs encoding the eel Kissr-3_v2 (A) and Kissr-3_v3 (B). Nucleotides (top) are numbered from 5′ to 3′. The amino-acid residues (bottom) are numbered beginning with the first methionine residue in the ORF. The asterisk (*) indicates the stop codon. The predicted transmembrane domains (TMD) are underlined.
(DOC)
Prediction of four Kissr CDS from the coelacanth genome. Nucleotide and deduced amino-acid sequences of the CDS encoding the coelacanth Kissr-1 (A), Kissr-2 (B), Kissr-3 (C) and Kissr-4 (D). Nucleotides (top) are numbered from 5′ to 3′. The amino-acid residues (bottom) are numbered beginning with the first methionine residue in the ORF. The asterisk (*) indicates the stop codon. The predicted transmembrane domains (TMD) are underlined. The exon-exon junctions are represented by two nucleotides coloured in red.
(DOC)
Prediction of four Kissr CDS from the spotted gar genome. Nucleotide and deduced amino-acid sequences of the CDS encoding the spotted gar Kissr-1 (A), Kissr-2 (B), Kissr-3 (C) and Kissr-4 (D). Nucleotides (top) are numbered from 5′ to 3′. The amino-acid residues (bottom) are numbered beginning with the first methionine residue in the ORF. The asterisk (*) indicates the stop codon. The predicted transmembrane domains (TMD) are underlined. The exon-exon junctions are represented by two nucleotides coloured in red.
(DOC)
Alignment of the amino-acid sequences of 51 Kissr used for the phylogenetic analysis. The amino-acid sequences were aligned by ClustalW and manually adjusted. The identical amino-acid residues between sequences are shaded in black and the similar (with similar physico-chemical properties) amino-acid residues are shaded in grey. Sequence references are listed in Table S2.
(TIF)
Alignment of the deduced amino-acid sequences of the three eel Kissr. The entire amino-acid sequences were aligned by ClustalW and manually adjusted. The identical amino-acid residues between the three sequences are shaded in black and the similar (with similar physico-chemical properties) amino-acid residues are shaded in grey.
(TIF)
European eel gene specific primers. Specific primers (F for Forward and R for Reverse) were designed for PCR and qPCR amplifications. Kissr, kisspeptin receptor; LHβ, Luteinizing Hormone β subunit; mGnRH, mammalian Gonadotrophin Releasing Hormone.
(XLS)
References of the Kissr sequences used in the phylogenetic analysis.
(XLS)
Names, references and locations of the genes used in the synteny analysis.
(XLS)
Acknowledgments
We thank Dr. B. Quérat (CNRS, Paris, France) for his helpful advices and discussions concerning phylogeny and synteny. We are grateful to Dr C. Henkel (ZF-screen, Leiden, Netherlands) for his work on the European eel genome, P. Lauesen (Billund Aquaculture Service, Billund, Denmark), M. Krüger-Johnsen (DTU aqua, Copenhagen, Denmark) and C. Graver (Danish Eel Farmer's Association, Ribe, Denmark) for performing eel maturation experiment, and S. Baloche (CNRS, Paris, France) for her technical assistance. Special thanks to L. Hardman and C. Atkinson (European Programme Erasmus, Keele University, Keele, UK/MNHN, Paris, France) for English correction. We also thank Eric Ryckelynck and his team from Nodaiwa (Paris, France) for their kind cooperation.
Funding Statement
JP is a recipient of a PhD fellowship from the Ministry of Research and Education. This work was supported by grants from the National Research Agency, PUBERTEEL number ANR-08-BLAN-0173 to SRJ, CFC, KR and SD, and from the European Community, 7th Framework Programme, PRO-EEL number 245257 to AGL, RD, GT, JT and SD. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Lee DK, Nguyen T, O'Neill GP, Cheng R, Liu Y, et al. (1999) Discovery of a receptor related to the galanin receptors. FEBS Lett 446: 103–107. [DOI] [PubMed] [Google Scholar]
- 2. Kotani M, Detheux M, Vandenbogaerde A, Communi D, Vanderwinden JM, et al. (2001) The metastasis suppressor gene KiSS-1 encodes kisspeptins, the natural ligands of the orphan G protein-coupled receptor GPR54. J Biol Chem 276: 34631–34636. [DOI] [PubMed] [Google Scholar]
- 3. Roa J, Navarro VM, Tena-Sempere M (2011) Kisspeptins in reproductive biology: consensus knowledge and recent developments. Biol Reprod 85: 650–660. [DOI] [PubMed] [Google Scholar]
- 4. Lee YR, Tsunekawa K, Moon MJ, Um HN, Hwang JI, et al. (2009) Molecular evolution of multiple forms of kisspeptins and GPR54 receptors in vertebrates. Endocrinology 150: 2837–2846. [DOI] [PubMed] [Google Scholar]
- 5. Tena-Sempere M, Felip A, Gomez A, Zanuy S, Carrillo M (2012) Comparative insights of the kisspeptin/kisspeptin receptor system: lessons from non-mammalian vertebrates. Gen Comp Endocrinol 175: 234–243. [DOI] [PubMed] [Google Scholar]
- 6. Biran J, Ben-Dor S, Levavi-Sivan B (2008) Molecular identification and functional characterization of the kisspeptin/kisspeptin receptor system in lower vertebrates. Biol Reprod 79: 776–786. [DOI] [PubMed] [Google Scholar]
- 7. Li S, Zhang Y, Liu Y, Huang X, Huang W, et al. (2009) Structural and functional multiplicity of the kisspeptin/GPR54 system in goldfish (Carassius auratus). J Endocrinol 201: 407–418. [DOI] [PubMed] [Google Scholar]
- 8. Zmora N, Stubblefield J, Zulperi Z, Biran J, Levavi-Sivan B, et al. (2012) Differential and Gonad Stage-Dependent Roles of Kisspeptin1 and Kisspeptin2 in Reproduction in the Modern Teleosts, Morone Species. Biol Reprod [DOI] [PubMed] [Google Scholar]
- 9. Pasquier J, Lafont AG, Leprince J, Vaudry H, Rousseau K, et al. (2011) First evidence for a direct inhibitory effect of kisspeptins on LH expression in the eel, Anguilla anguilla. Gen Comp Endocrinol 173: 216–225. [DOI] [PubMed] [Google Scholar]
- 10. Amores A, Catchen J, Ferrara A, Fontenot Q, Postlethwait JH (2011) Genome evolution and meiotic maps by massively parallel DNA sequencing: spotted gar, an outgroup for the teleost genome duplication. Genetics 188: 799–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Henkel CV, Burgerhout E, de Wijze DL, Dirks RP, Minegishi Y, et al. (2012) Primitive duplicate Hox clusters in the European eel's genome. PLoS One 7: e32231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pasqualini C, Weltzien FA, Vidal B, Baloche S, Rouget C, et al. (2009) Two distinct dopamine D2 receptor genes in the European eel: molecular characterization, tissue-specific transcription, and regulation by sex steroids. Endocrinology 150: 1377–1392. [DOI] [PubMed] [Google Scholar]
- 13.Dufour S, Burzawa-Gerard E, Le Belle N, Sbaihi M, Vidal B (2003) Reproductive endocrinology of the European eel, Anguilla anguilla. In: Aida K, Tsukamoto K, Yamauchi K, editors. Eel Biology. Tokyo: Springer. pp. 373–383.
- 14. Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Abascal F, Zardoya R, Posada D (2005) ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21: 2104–2105. [DOI] [PubMed] [Google Scholar]
- 16. Stamatakis A, Ott M (2008) Efficient computation of the phylogenetic likelihood function on multi-gene alignments and multi-core architectures. Philos Trans R Soc Lond B Biol Sci 363: 3977–3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Muffato M, Louis A, Poisnel CE, Roest Crollius H (2010) Genomicus: a database and a browser to study gene synteny in modern and ancestral genomes. Bioinformatics 26: 1119–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sébert ME, Legros C, Weltzien FA, Malpaux B, Chemineau P, et al. (2008) Melatonin activates brain dopaminergic systems in the eel with an inhibitory impact on reproductive function. J Neuroendocrinol 20: 917–929. [DOI] [PubMed] [Google Scholar]
- 19. Ohta H, Kagawa H, Tanaka H, Okuzawa K, Hirose K (1996) Changes in fertilization and hatching rates with time after ovulation induced by 17, 20β-dihydroxy-4-pregnen-3-one in the Japanese eel, Anguilla japonica. Aquaculture 139: 291–301. [Google Scholar]
- 20. Aroua S, Weltzien FA, Le Belle N, Dufour S (2007) Development of real-time RT-PCR assays for eel gonadotropins and their application to the comparison of in vivo and in vitro effects of sex steroids. Gen Comp Endocrinol 153: 333–343. [DOI] [PubMed] [Google Scholar]
- 21. Weltzien FA, Pasqualini C, Le Belle N, Vidal B, Vernier P, et al. (2005) Brain expression of tyrosine hydroxylase and its regulation by steroid hormones in the European eel quantified by real-time PCR. Ann NY Acad Sci 1040: 518–520. [DOI] [PubMed] [Google Scholar]
- 22. Mechaly AS, Vinas J, Piferrer F (2009) Identification of two isoforms of the Kisspeptin-1 receptor (kiss1r) generated by alternative splicing in a modern teleost, the Senegalese sole (Solea senegalensis). Biol Reprod 80: 60–69. [DOI] [PubMed] [Google Scholar]
- 23. Onuma TA, Duan C (2012) Duplicated Kiss1 receptor genes in zebrafish: distinct gene expression patterns, different ligand selectivity, and a novel nuclear isoform with transactivating activity. FASEB J 26: 2941–2950. [DOI] [PubMed] [Google Scholar]
- 24. Akazome Y, Kanda S, Okubo K, Oka Y (2010) Functional and evolutionary insights into vertebrate kisspeptin systems from studies of fish brain. J Fish Biol 76: 161–182. [DOI] [PubMed] [Google Scholar]
- 25. Um HN, Han JM, Hwang JI, Hong SI, Vaudry H, et al. (2010) Molecular coevolution of kisspeptins and their receptors from fish to mammals. Ann N Y Acad Sci 1200: 67–74. [DOI] [PubMed] [Google Scholar]
- 26. Kim DK, Cho EB, Moon MJ, Park S, Hwang JI, et al. (2012) Molecular Coevolution of Neuropeptides Gonadotropin-Releasing Hormone and Kisspeptin with their Cognate G Protein-Coupled Receptors. Front Neurosci 6: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dehal P, Boore JL (2005) Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol 3: e314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Putnam NH, Butts T, Ferrier DE, Furlong RF, Hellsten U, et al. (2008) The amphioxus genome and the evolution of the chordate karyotype. Nature 453: 1064–1071. [DOI] [PubMed] [Google Scholar]
- 29. Nakatani Y, Takeda H, Kohara Y, Morishita S (2007) Reconstruction of the vertebrate ancestral genome reveals dynamic genome reorganization in early vertebrates. Genome Res 17: 1254–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kuraku S (2008) Insights into cyclostome phylogenomics: pre-2R or post-2R. Zoolog Sci 25: 960–968. [DOI] [PubMed] [Google Scholar]
- 31. Kuraku S, Meyer A, Kuratani S (2009) Timing of genome duplications relative to the origin of the vertebrates: did cyclostomes diverge before or after? Mol Biol Evol 26: 47–59. [DOI] [PubMed] [Google Scholar]
- 32. Amores A, Force A, Yan YL, Joly L, Amemiya C, et al. (1998) Zebrafish hox clusters and vertebrate genome evolution. Science 282: 1711–1714. [DOI] [PubMed] [Google Scholar]
- 33. Nocillado JN, Levavi-Sivan B, Carrick F, Elizur A (2007) Temporal expression of G-protein-coupled receptor 54 (GPR54), gonadotropin-releasing hormones (GnRH), and dopamine receptor D2 (drd2) in pubertal female grey mullet, Mugil cephalus. Gen Comp Endocrinol 150: 278–287. [DOI] [PubMed] [Google Scholar]
- 34. Servili A, Le Page Y, Leprince J, Caraty A, Escobar S, et al. (2011) Organization of two independent kisspeptin systems derived from evolutionary-ancient kiss genes in the brain of zebrafish. Endocrinology 152: 1527–1540. [DOI] [PubMed] [Google Scholar]
- 35. Filby AL, van Aerle R, Duitman J, Tyler CR (2008) The kisspeptin/gonadotropin-releasing hormone pathway and molecular signaling of puberty in fish. Biol Reprod 78: 278–289. [DOI] [PubMed] [Google Scholar]
- 36. Martinez-Chavez CC, Minghetti M, Migaud H (2008) GPR54 and rGnRH I gene expression during the onset of puberty in Nile tilapia. Gen Comp Endocrinol 156: 224–233. [DOI] [PubMed] [Google Scholar]
- 37. Parhar IS (2005) GnRH and gpcr: laser-captured single cell gene profiling. Fish Physiol Biochem 31: 153–156. [DOI] [PubMed] [Google Scholar]
- 38. Shahjahan M, Motohashi E, Doi H, Ando H (2010) Elevation of Kiss2 and its receptor gene expression in the brain and pituitary of grass puffer during the spawning season. Gen Comp Endocrinol 169: 48–57. [DOI] [PubMed] [Google Scholar]
- 39. Muir AI, Chamberlain L, Elshourbagy NA, Michalovich D, Moore DJ, et al. (2001) AXOR12, a novel human G protein-coupled receptor, activated by the peptide KiSS-1. J Biol Chem 276: 28969–28975. [DOI] [PubMed] [Google Scholar]
- 40. Funes S, Hedrick JA, Vassileva G, Markowitz L, Abbondanzo S, et al. (2003) The KiSS-1 receptor GPR54 is essential for the development of the murine reproductive system. Biochem Biophys Res Commun 312: 1357–1363. [DOI] [PubMed] [Google Scholar]
- 41. Castellano JM, Gaytan M, Roa J, Vigo E, Navarro VM, et al. (2006) Expression of KiSS-1 in rat ovary: putative local regulator of ovulation? Endocrinology 147: 4852–4862. [DOI] [PubMed] [Google Scholar]
- 42. Gaytan F, Gaytan M, Castellano JM, Romero M, Roa J, et al. (2009) KiSS-1 in the mammalian ovary: distribution of kisspeptin in human and marmoset and alterations in KiSS-1 mRNA levels in a rat model of ovulatory dysfunction. Am J Physiol Endocrinol Metab 296: E520–531. [DOI] [PubMed] [Google Scholar]
- 43. Dufour S, Sebert ME, Weltzien FA, Rousseau K, Pasqualini C (2010) Neuroendocrine control by dopamine of teleost reproduction. J Fish Biol 76: 129–160. [DOI] [PubMed] [Google Scholar]
- 44. Dufour S, Le Belle N, Baloche S, Fontaine YA (1989) Positive feedback control by the gonads on gonadotropin (GTH) and gonadoliberin (GnRH) levels in experimentally matured female silver eels, Anguilla anguilla. Fish Physiology and Biochemistry 7: 157–162. [DOI] [PubMed] [Google Scholar]
- 45. Montero M, Le Belle N, King JA, Millar RP, Dufour S (1995) Differential regulation of the two forms of gonadotropin-releasing hormone (mGnRH and cGnRH-II) by sex steroids in the European female silver eel (Anguilla anguilla). Neuroendocrinology 61: 525–535. [DOI] [PubMed] [Google Scholar]
- 46. Jeng SR, Pasquier J, Yueh WS, Chen GR, Lee YH, et al. (2012) Differential regulation of the expression of cytochrome P450 aromatase, estrogen and androgen receptor subtypes in the brain-pituitary-ovarian axis of the Japanese eel (Anguilla japonica) reveals steroid dependent and independent mechanisms. Gen Comp Endocrinol 175: 163–172. [DOI] [PubMed] [Google Scholar]
- 48. Shahab M, Mastronardi C, Seminara SB, Crowley WF, Ojeda SR, et al. (2005) Increased hypothalamic GPR54 signaling: a potential mechanism for initiation of puberty in primates. Proc Natl Acad Sci U S A 102: 2129–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mohamed JS, Benninghoff AD, Holt GJ, Khan IA (2007) Developmental expression of the G protein-coupled receptor 54 and three GnRH mRNAs in the teleost fish cobia. J Mol Endocrinol 38: 235–244. [DOI] [PubMed] [Google Scholar]
- 50. Dufour S, Montero M, Le Belle N, Bassompierre M, King JA, et al. (1993) Differential distribution and response to experimental sexual maturation of two forms of brain gonadotropin-releasing hormone (GnRH) in the European eel, Anguilla anguilla. Fish Physiology and Biochemistry 11: 99–106. [DOI] [PubMed] [Google Scholar]
- 51. Schmitz M, Aroua S, Vidal B, Le Belle N, Elie P, et al. (2005) Differential regulation of luteinizing hormone and follicle-stimulating hormone expression during ovarian development and under sexual steroid feedback in the European eel. Neuroendocrinology 81: 107–119. [DOI] [PubMed] [Google Scholar]
- 52. Lehman MN, Coolen LM, Goodman RL (2010) Minireview: kisspeptin/neurokinin B/dynorphin (KNDy) cells of the arcuate nucleus: a central node in the control of gonadotropin-releasing hormone secretion. Endocrinology 151: 3479–3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Garcia-Galiano D, van Ingen Schenau D, Leon S, Krajnc-Franken MA, Manfredi-Lozano M, et al. (2011) Kisspeptin signaling is indispensable for neurokinin B, but not glutamate, stimulation of gonadotropin secretion in mice. Endocrinology 153: 316–328. [DOI] [PubMed] [Google Scholar]
- 54. Kanda S, Akazome Y, Matsunaga T, Yamamoto N, Yamada S, et al. (2008) Identification of KiSS-1 product kisspeptin and steroid-sensitive sexually dimorphic kisspeptin neurons in medaka (oryzias latipes). Endocrinology 149: 2467–2476. [DOI] [PubMed] [Google Scholar]
- 55. Kanda S, Karigo T, Oka Y (2012) Steroid sensitive kiss2 neurones in the goldfish: evolutionary insights into the duplicate kisspeptin gene-expressing neurones. J Neuroendocrinol [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Three eel Kissr gene sequences. Genomic sequences of the eel Kissr-1 extracted from the scaffold 90.1 (A), Kissr-2 extracted from the scaffold 3158.1 (B) and Kissr-3 extracted from the scaffold 1832.1 (C) (European eel genome [11]). Nucleotides are numbered from 5′ to 3′. The five exons of each gene are shaded in grey.
(DOC)
Molecular cloning of eel Kissr-3 splicing variants Kissr-3_v2 and Kissr-3_v3. Nucleotide and deduced amino-acid sequence of the cDNAs encoding the eel Kissr-3_v2 (A) and Kissr-3_v3 (B). Nucleotides (top) are numbered from 5′ to 3′. The amino-acid residues (bottom) are numbered beginning with the first methionine residue in the ORF. The asterisk (*) indicates the stop codon. The predicted transmembrane domains (TMD) are underlined.
(DOC)
Prediction of four Kissr CDS from the coelacanth genome. Nucleotide and deduced amino-acid sequences of the CDS encoding the coelacanth Kissr-1 (A), Kissr-2 (B), Kissr-3 (C) and Kissr-4 (D). Nucleotides (top) are numbered from 5′ to 3′. The amino-acid residues (bottom) are numbered beginning with the first methionine residue in the ORF. The asterisk (*) indicates the stop codon. The predicted transmembrane domains (TMD) are underlined. The exon-exon junctions are represented by two nucleotides coloured in red.
(DOC)
Prediction of four Kissr CDS from the spotted gar genome. Nucleotide and deduced amino-acid sequences of the CDS encoding the spotted gar Kissr-1 (A), Kissr-2 (B), Kissr-3 (C) and Kissr-4 (D). Nucleotides (top) are numbered from 5′ to 3′. The amino-acid residues (bottom) are numbered beginning with the first methionine residue in the ORF. The asterisk (*) indicates the stop codon. The predicted transmembrane domains (TMD) are underlined. The exon-exon junctions are represented by two nucleotides coloured in red.
(DOC)
Alignment of the amino-acid sequences of 51 Kissr used for the phylogenetic analysis. The amino-acid sequences were aligned by ClustalW and manually adjusted. The identical amino-acid residues between sequences are shaded in black and the similar (with similar physico-chemical properties) amino-acid residues are shaded in grey. Sequence references are listed in Table S2.
(TIF)
Alignment of the deduced amino-acid sequences of the three eel Kissr. The entire amino-acid sequences were aligned by ClustalW and manually adjusted. The identical amino-acid residues between the three sequences are shaded in black and the similar (with similar physico-chemical properties) amino-acid residues are shaded in grey.
(TIF)
European eel gene specific primers. Specific primers (F for Forward and R for Reverse) were designed for PCR and qPCR amplifications. Kissr, kisspeptin receptor; LHβ, Luteinizing Hormone β subunit; mGnRH, mammalian Gonadotrophin Releasing Hormone.
(XLS)
References of the Kissr sequences used in the phylogenetic analysis.
(XLS)
Names, references and locations of the genes used in the synteny analysis.
(XLS)