Abstract
Background. Although specific human immunodeficiency virus type 1 (HIV-1) drug resistance mutations are well studied, little is known about cumulative amino acid changes, or how regimen and participant characteristics influence these changes.
Methods. In the AIDS Clinical Trials Group randomized study A5202 of treatment-naive HIV-infected participants, cumulative HIV-1 amino acid changes from pretreatment to virologic failure were evaluated in protease and reverse transcriptase (RT) gene sequences.
Results. Among 265 participants with virologic failure, those assigned atazanavir plus ritonavir (ATV/r) did not have significantly more protease changes compared with those assigned efavirenz (EFV) (P ≥ .13). In contrast, participants with virologic failure assigned EFV had more RT changes, including and excluding known resistance codons (P < .001). At pretreatment, lower CD4 cell count, major resistance, more amino acid mixtures (all P < .001), hepatitis C antibody negativity (P = .05), and black race/ethnicity (P = .02) were associated with more HIV-1 amino acid changes.
Conclusions. Virologic failure following EFV-containing treatment was associated with more HIV-1 amino acid changes compared to failure of ATV/r-containing treatment. Furthermore, we show that non–drug resistance mutations occurred more frequently among those failing EFV, the clinical relevance of which warrants further investigation. Pretreatment immunologic status may play a role in viral evolution during treatment, as evidenced by increased amino acid changes among those with lower pretreatment CD4 count.
Clinical Trials Registration. NCT00118898.
More than 5 million individuals are receiving chronic antiretroviral therapy (ART) to suppress the replication of human immunodeficiency virus type 1 (HIV-1). Current US guidelines recommend monitoring of ART with measurement of HIV-1 RNA every 3–6 months to assess virologic response [1]. Virologic suppression with ART results in profound long-term clinical benefits including decrease in immunodeficiency and an increase in life span [2, 3]. Virologic failure reverses each of these benefits, and the recrudescence of viral replication, is associated with the selection of drug-resistant viruses and CD4 cell count decline [4–6]. Upon rebound of viremia on ART, recommendations include reinforcement of adherence and drug resistance testing of plasma HIV-1 to determine the optimal regimen to achieve virologic suppression.
In AIDS Clinical Trials Group (ACTG) Study A5202, treatment-naive participants were randomized to atazanavir plus ritonavir (ATV/r) or efavirenz (EFV) with either tenofovir DF/emtricitabine (TDF/FTC) or abacavir/lamivudine (ABC/3TC). The major drug resistance mutations detected by sequencing and consensus alignment at the time of virologic failure were previously reported, where reverse transcriptase (RT) resistance-associated mutations were found to be more frequent among participants with virologic failure assigned to EFV-containing than ATV/r-containing treatment arms [7–9]. Here we present a thorough assessment of HIV-1 major and nonmajor mutations, and cumulative amino acid changes from pretreatment to virologic failure, to provide additional evidence for selection pressures and evolution of drug resistance in response to the different combinations of antiretrovirals.
METHODS
Study Design
In ACTG A5202, treatment-naive participants were randomized to a blinded nucleoside reverse transcriptase inhibitor (NRTI) combination of TDF/FTC or ABC/3TC with either open-label ATV/r or EFV. Randomization was stratified on the basis of whether participants had low (<100 000 copies/mL) or high (≥100 000 copies/mL) HIV-1 plasma viral load on a single screening test done before study entry. Pretreatment drug resistance screening was allowed but not required during the enrollment period (September 2005–November 2007), except among those with recent HIV-1 infection. Overall, 45% of participants had drug resistance testing prior to screening, and potential participants with evidence of resistance were not eligible for enrollment. Following randomization, plasma HIV-1 RNA level was tested at weeks 4, 16, and 24, and every 12 weeks thereafter. The study's primary efficacy endpoint was time from randomization to virologic failure, defined as confirmed (2 consecutive) HIV-1 RNA levels ≥1000 copies/mL at or after 16 weeks and before 24 weeks, or confirmed ≥200 copies/mL at or after 24 weeks following randomization; confirmation was recommended to be performed within 30 days of initial failure [7–9]. The human subjects committees of all sites approved the A5202 protocol and written informed consent was obtained from all participants in compliance with human experimentation guidelines of the US Department of Health and Human Services.
HIV-1 Sequencing
Participants who experienced protocol-defined virologic failure had pretreatment and failure plasma samples tested for drug resistance at Stanford University by reverse transcriptase-nested polymerase chain reaction/sequencing protocol as previously described [10]. Mutants comprising ≥25% of the population are typically detectable on consensus sequence chromatogram; virologic failure samples were tested in real time and pretreatment samples were tested retrospectively. Major drug resistance mutations were defined based on the International AIDS Society–USA Spring 2008 guidelines [11], with the addition of T69D, L74I, and G190C/E/Q/T/V for RT and L24I, F53L, I54V/A/T/S, G73C/S/T/A, and N88D for protease as specified by the A5202 protocol.
General Statistical Considerations
Analyses were restricted to participants with virologic failure who had HIV sequence data at both pretreatment and virologic failure, including those with retrospectively identified pretreatment resistance mutations. Treatment comparisons were analyzed per the randomized treatment assignment, regardless of ART status at the time of virologic failure. Reported P values are 2-sided. Analyses were performed with SAS software, version 9.
Individual Mutations
Within HIV protease and RT, each nonconsensus mutation that occurred in 2 or more participants was tested for discordance from pretreatment to virologic failure with an exact McNemar test. Discordance was evaluated pooled over all the virologic failures, and also within ATV/r-containing and EFV-containing arms. Amino acid mixture mutations were counted as individual mutations (eg, M184IV → M184I and M184V, and M184MV counted the same as M184V). Benjamini and Hochberg's false discovery rate (FDR) multiple comparison adjustment was applied separately to prespecified mutations (RT: major; protease: major plus ATV minor) [11] and unspecified mutations, with statistical significance defined as FDR-adjusted P ≤ .05.
HIV-1 Cumulative Amino Acid Changes
HIV-1 cumulative changes from pretreatment to virologic failure were defined as a weighted percentage of amino acid changes; a codon was assigned change = 0 if the amino acid remained the same (eg, K103K → K103K), change = 1 if there was a complete amino acid mutation (eg, K103K → K103N), and change = 0.5 if there was a mixture of the pretreatment amino acid with an emergent mutation (eg, K103K → K103K/N). Codons with missing information were excluded from the calculation. This measure is based on the information theory concept of Hamming distance [12]. Reverse transcriptase codons (1–230) and protease codons (1–99) were analyzed for amino acid changes separately and combined; changes were also evaluated when including and excluding major drug resistance mutation codons, to provide a measure of polymorphic and compensatory changes associated with virologic failure.
Regimen comparisons of cumulative amino acid changes were evaluated with a stratified Mann-Whitney test. During the course of the study, the data safety monitoring board (DSMB) recommended premature unblinding of the NRTIs for participants in the screening plasma HIV-1 RNA ≥100 000 stratum. Consequently, NRTI comparisons are presented separately for the <100 000 and ≥100 000 screening plasma HIV-1 RNA strata, and virologic failures within ≥100 000 stratum were restricted to those reviewed at the time of the DSMB recommendation (February 2008); otherwise, all follow-up through study completion was analyzed.
Associations between covariates and the amount of HIV-1 amino acid changes were evaluated with univariate and multivariable linear regression. The following pretreatment covariates were evaluated: assigned ATV/r vs EFV, assigned ABC/3TC vs TDF/FTC, sex, race/ethnicity, age, history of intravenous drug use, hepatitis C antibody status, having undergone drug resistance screening, presence of major drug resistance mutations, number of codons containing amino acid mixtures, plasma HIV-1 RNA level, and CD4 cell count. Covariates measured proximal and prior to virologic failure genotyping included participant-recall antiretroviral adherence and medication status in the preceding week [13], modification of assigned treatment per clinical ARV treatment records, visit weeks until initial virologic failure, plasma HIV-1 RNA level, HIV-1 RNA ever <200 copies/mL prior to virologic failure, and change in plasma HIV-1 RNA level from pretreatment.
Covariate associations with P value ≤.1 in the univariate model, and assigned treatment factors, were considered for a multivariable regression model chosen via backward selection, with a P value ≤.05 required to remain in the model. All 2-way interactions among covariates in the resulting model were tested. The measure of HIV-1 weighted cumulative amino acid changes was square-root transformed to stabilize the variance of these Poisson-like data (not true Poisson due to mixture weighting).
RESULTS
Data Availability
Among the 269 confirmed virologic failures in study A5202, HIV-1 sequence results were available at both pretreatment and failure for 265 participants (98.5%), of the 4 remaining virologic failures with unavailable results, for 2 the samples did not pass quality assurance, and the other 2 each had a sample that did not amplify. Missing codon information due to trimming of sequences was unusual, with the exception of early protease codons 1–12, and the latter codons of RT.
Characteristics of Participants Who Experienced Virologic Failure
Of the 265 participants with sequence results at pretreatment and failure, 254 (96%) had HIV subtype B virus; the other subtypes were A1 (n = 4), AG (n = 3), F2 (n = 2), C (n = 1), and D (n = 1). These 265 participants with virologic failure were predominantly male (78%); the most common race/ethnicities were black non-Hispanic (48%), white non-Hispanic (32%), and Hispanic (17%). Median (Q1, Q3) age was 35 (29, 43) years. Twenty-four (9%) were hepatitis C antibody positive, 8% self-reported intravenous drug use, and 42% had resistance testing prior to study screening.
Among participants with virologic failure, 154 (58% of 265) were assigned to receive ABC/3TC (83 with ATV/r, 71 to EFV), and 111 (42% of 265) to receive TDF/FTC (57 with ATV/r, 54 to EFV). At pretreatment, the median (Q1, Q3) plasma HIV-1 RNA and CD4 count were 4.7 (4.4, 5.3) log10 copies/mL and 188 (38, 330) cells/μL, respectively (Table 1).
Table 1.
Participant and HIV-1 Disease Characteristics Among A5202 Participants With Virologic Failure With HIV-1 Genotype Results
| EFV + TDF/FTC (n = 54) | EFV + ABC/3TC (n = 71) | ATV/r + TDF/FTC (n = 57) | ATV/r + ABC/3TC (n = 83) | Total (N = 265) | ||
|---|---|---|---|---|---|---|
| Pretreatment characteristics | ||||||
| Sex | Male | 45 (83%) | 60 (85%) | 38 (67%) | 64 (77%) | 207 (78%) |
| Age (y) | Median (Q1, Q3) | 39 (30, 43) | 32 (27, 40) | 39 (30, 45) | 35 (30, 42) | 35 (29, 43) |
| Race/ethnicitya | Black non-Hispanic | 26 (48%) | 31 (44%) | 34 (60%) | 36 (43%) | 127 (48%) |
| White non-Hispanic | 18 (33%) | 27 (38%) | 15 (26%) | 25 (30%) | 85 (32%) | |
| Hispanic | 10 (19%) | 10 (14%) | 8 (14%) | 18 (22%) | 46 (17%) | |
| Hepatitis C antibody | Positive | 9 (17%) | 3 (4%) | 6 (11%) | 6 (7%) | 24 (9%) |
| Screening HIV-1 RNA stratum | ≥100 000 | 22 (41%) | 33 (46%) | 28 (49%) | 48 (58%) | 131 (49%) |
| Plasma HIV-1 RNA (log10 copies/mL) | Median (Q1, Q3) | 4.7 (4.3, 5.0) | 4.8 (4.2, 5.5) | 4.6 (4.3, 4.9) | 4.8 (4.5, 5.5) | 4.7 (4.4, 5.3) |
| CD4+ T-cell count (cells/μL) | Median (Q1, Q3) | 197 (51, 308) | 185 (20, 324) | 253 (79, 360) | 90 (32, 320) | 188 (38, 330) |
| Genotype before screening | Genotyped | 21 (39%) | 33 (46%) | 16 (28%) | 40 (48%) | 110 (42%) |
| Major resistance mutations | Yes | 7 (13%) | 8 (11%) | 3 (5%) | 7 (8%) | 25 (9%) |
| Number of codons with amino acid mixtures | Mean (SD) | 2.4 (2.4) | 2.1 (1.9) | 1.7 (1.5) | 2.1 (1.7) | 2.1 (1.9) |
| Median (Q1, Q3) | 2 (0, 4) | 2 (0, 4) | 2 (1, 3) | 2 (1, 3) | 2 (1, 3) | |
| Closest subtype cladeb | Subtype B | 51 (94%) | 68 (96%) | 56 (98%) | 79 (95%) | 254 (96%) |
| At the time of virologic failure | ||||||
| Plasma HIV-1 RNA (log10 copies/mL) | Median (Q1, Q3) | 3.6 (2.8, 4.3) | 3.9 (3.2, 4.7) | 3.7 (2.8, 4.5) | 3.5 (2.8, 4.3) | 3.6 (2.9, 4.5) |
| Major resistance mutationsc | Yes | 34 (63%) | 48 (68%) | 7 (12%) | 19 (23%) | … |
| K103N and M184V/I | Both | 8 (15%) | 17 (24%) | 0 (0%) | 2 (2%) | … |
| K103N without M184V/I | 16 (30%) | 17 (24%) | 0 (0%) | 0 (0%) | … | |
| M184V/I without K103N | 3 (6%) | 9 (13%) | 5 (9%) | 12 (14%) | … | |
| Antiretroviral adherence self-reportd | 100% adherent | 37 (69%) | 42 (60%) | 33 (58%) | 52 (63%) | 164 (62%) |
| <100% adherent | 7 (13%) | 9 (13%) | 13 (23%) | 17 (21%) | 46 (17%) | |
| Not taking any antiretroviral treatment | 10 (19%) | 19 (27%) | 11 (19%) | 13 (16%) | 53 (20%) | |
| Days between initial failure and genotype specimens | Median (Q1, Q3) | 28 (17, 56) | 28 (14, 62) | 35 (21, 80) | 28 (19, 57) | 28 (19, 62) |
| Assigned antiretroviral treatment status | No modification | 38 (70%) | 42 (59%) | 47 (82%) | 55 (66%) | 182 (69%) |
| Restricted to ≥100 000 screening stratum | Virologic failure after DSMB recommendation | 11/22 (50%) | 8/33 (24%) | 13/28 (46%) | 16/48 (33%) | 48/131 (37%) |
Data are presented as no. (%) unless otherwise specified.
Abbreviations: ABC/3TC, abacavir/lamivudine; ATV/r, atazanavir/ritonavir; DSMB, data safety and monitoring board; EFV, efavirenz; HIV-1, human immunodeficiency virus type 1; Rx, treatment; TDF/FTC, tenofovir DF/emtricitabine.
a n = 7 have other race/ethnicity.
b Other observed subtypes included: A1 (n = 4), AG (n = 3), F2 (n = 2), C (n = 1), and D (n = 1).
c Includes participants with and without pretreatment resistance.
d Self-report 1 week recall, most recent report prior to virologic failure genotype sample, n = 2 with missing data.
Pretreatment Major Mutations Among Participants With Virologic Failure
Retrospective genotyping of pretreatment samples among virologic failures identified 25 (9%) with major resistance mutations at study entry, 15 and 10 in the EFV and ATV/r arms, respectively (Table 1). Among these 25, 6 had major protease inhibitor (PI) mutations, 15 had major nonnucleoside reverse transcriptase inhibitor (NNRTI) mutations, and 9 had major NRTI mutations; 1 had pretreatment resistance mutations in 2 drug classes (PI and NNRTI), and 2 had major mutations in all 3 evaluated drug classes. Notably, all the observed major NRTI mutations at pretreatment were the thymidine analogue–associated mutations (TAMs) M41L and L210W; no other major consensus NRTI mutations (eg, M184V/I) were detected. Among EFV failures, the time to virologic failure was shorter for those with pretreatment resistance, with a median (Q1, Q3) weeks elapsed from randomization to virologic failure of 24 (16, 36) vs 36 (24, 84) weeks for 15 with and 110 without pretreatment resistance, respectively (Mann-Whitney P = .04); among ATV/r failures, these numbers were 36 (24, 84) and 36 (24, 72) weeks for 10 and 130 with and without pretreatment major resistance, respectively (P = .99).
There was a trend toward differences in the frequency of NNRTI pretreatment major resistance mutations based on the assigned drug class; 9% (11/125) in the EFV arms had NNRTI-related mutations at pretreatment, compared with 3% (4/140) in the ATV/r arms (Fisher exact P = .06). Pretreatment major NRTI mutations were identified in 2% (2/125) in the EFV arms compared with 5% (7/140) in the ATV/r arms (P = .18). Pretreatment major protease mutations were present for 2 of 125 assigned to EFV and 4 of 140 assigned to ATV/r (P = .7).
Emergence of Individual Mutations at Virologic Failure
Presence or absence of each mutation was compared between pretreatment and failure sequences from each participant, including all prespecified and unspecified mutations. Among ATV/r virologic failures, no significant increase in frequency from pretreatment to failure was observed for any specific protease mutation (adjusted for multiple testing, all FDR P = 1.0, all unadjusted P > .22). For the ATV/r arms, the only mutations significantly more frequent at failure than pretreatment were M184V (FDR P = .001) and M184I (FDR P = .047), 12 of 13 and 4 of 7 emergent M184V and M184I, respectively, occurred in the ABC/3TC arm.
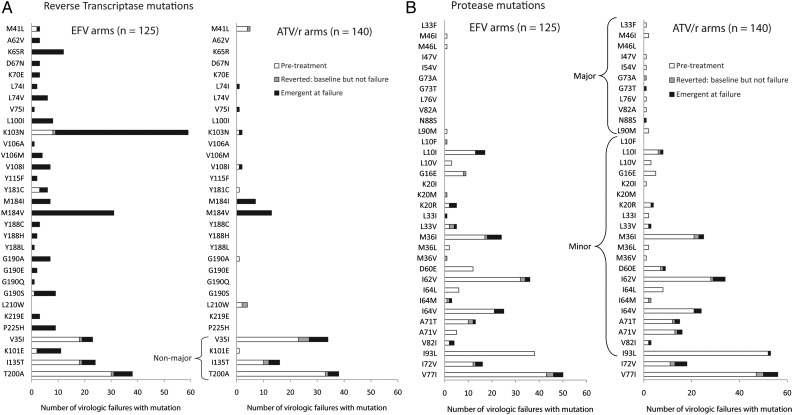
Among all virologic failures, no unknown (ie, nonmajor) RT mutations were more frequent at failure (adjusted for multiple testing, all FDR P > .58). Ignoring multiple testing, there was a trend toward higher frequency at failure compared to pretreatment for 3 nonmajor RT mutations: E6K (6 emergent at failure), K101E (9), and T200A (11) (all unadjusted P ≤ .031). When evaluated among all failures, particular major RT drug resistance mutations were significantly more frequent at failure than pretreatment, including K65R, L100I, K103N, V108I, M184I, M184V, G190A, G190S, and P225H (all adjusted FDR P ≤ .042), and a trend for L74V (FDR P = .075). Figure 1 presents all observed major RT and protease mutations and minor ATV/r mutations, plus additional mutations emergent in 5 or more virologic failures within the EFV or ATV/r-containing arms.
Figure 1.
Reverse transcriptase and protease mutations among participants with virologic failure. A, All observed major reverse transcriptase and 151 complex mutations are presented, plus nonmajor mutations emergent in 5 or more participants with virologic failure in either group. V35I, K101E, I135T, and T200A are nonmajor. B, All observed major protease (L33F-L90M) and atazanavir/ritonavir (ATV/r) minor mutations (L10F-I93L) are presented, plus additional mutations emergent in 5 or more participants with virologic failure in either group (I72V and V77I). The one participant with G73T at failure had G73A at pretreatment. Amino acid mixtures were separated into individual mutations; mutations are ordered by codon within major, minor, and additional mutations. Abbreviation: EFV, efavirenz.
Mutation Patterns Among Participants With Virologic Failure Without Major Pretreatment Mutations
Excluding the 25 participants (15 assigned to EFV, 10 assigned to ATV/r) with major pretreatment resistance mutation(s), 68 of 110 in the EFV arms had major drug resistance mutations at virologic failure, 59 (87% of 68) of whom had at least 1 of K103N or M184V/I, and 67 (99% of 68) of whom had 1 or more of K103N, M184V/I, K65R, or G190A/C/E/Q/S/T/V. Additionally, in the EFV arms, all 35 of 110 with NRTI major resistance mutation(s) at failure also had NNRTI major resistance mutations (10 TDF/FTC arm, 25 ABC/3TC arm). In the ATV/r arms, among participants with virologic failure without pretreatment resistance, 17 of 130 had major emergent mutations at failure, and all but 1 (16/17) had M184V/I; the 16 with M184V/I accounted for all ATV/r failures with NRTI-associated mutations. Only 1 failure assigned to ATV/r had a major emergent protease resistance mutation at failure (N88S, with M184V).
Cumulative Amino Acid Changes Among Participants With Virologic Failure
The amount of cumulative protease amino acid changes was not significantly different between ATV/r-containing and EFV-containing arms (P = .50 in TDF/FTC arm, P = .13 in ABC/3TC arm). However, there were more RT amino acid changes with EFV compared to ATV/r virologic failures, with either TDF/FTC or ABC/3TC, whether including and excluding major drug resistance codons (P values <.001, Table 2).
Table 2.
HIV-1 Amino Acid Changes From Pretreatment to Virologic Failure, Comparing Efavirenz vs Atazanavir/Ritonavir Among Participants With Virologic Failure With Pretreatment and Failure Sequences
| With TDF/FTC |
With ABC/3TC |
||||
|---|---|---|---|---|---|
| EFV (n = 54) | ATV/r (n = 57) | EFV (n = 71) | ATV/r (n = 83) | ||
| Protease changes (%) | Median (Q1, Q3) | 0.51 (0, 1.16) | 0.57 (0, 1.15) | 0.51 (0, 1.52) | 0.51 (0, 0.72) |
| Absence of any protease AA change | n (%) | 23 (43%) | 20 (35%) | 28 (39%) | 37 (45%) |
| P = .5 | P = .13 | ||||
| RT changes (%) | Median (Q1, Q3) | 1.09 (0.65, 1.96) | 0.43 (0.22, 0.65) | 1.12 (0.43, 2.17) | 0.43 (0.22, 0.87) |
| Absence of any RT AA change | n (%) | 4 (7%) | 12 (21%) | 6 (8%) | 18 (22%) |
| P < .001 | P < .001 | ||||
| RT changes (%), excluding codons associated with major resistancea | Median (Q1, Q3) | 0.71 (0.24, 1.19) | 0.24 (0.24, 0.48) | 0.71 (0.24, 1.19) | 0.48 (0, 0.86) |
| P < .001 | P < .001 | ||||
Total n = 265. Changes indicates the percentage of amino acid codons changed from pretreatment to virologic failure, with mixture changes assigned weight = 0.5 change.
Abbreviations: AA, amino acid; ABC/3TC, abacavir/lamivudine; ATV/r, atazanavir/ritonavir; EFV, efavirenz; HIV-1, human immunodeficiency virus type 1; RT, reverse transcriptase; TDF/FTC, tenofovir DF/emtricitabine.
a Twenty RT and 16 protease major mutation codons excluded; RT 62, 75, 77, and 116 from the 151 complex were included since no participants had Q151M in this study.
When virologic failures assigned to EFV were categorized into 3 predefined resistance groups (major resistance at pretreatment, n = 15; emergent major resistance at virologic failure, n = 68; no major resistance at pretreatment or failure, n = 42), differences between groups were observed in the amount of RT amino acid changes when excluding codons associated with major drug resistance (Kruskal-Wallis P = .001). EFV virologic failures with major emergent resistance mutations had more nonmajor RT amino acid changes compared to participants with no major resistance mutations at pretreatment or failure, with median changes 1.0% vs 0.5%, respectively (95% confidence interval for difference, 0.2%–0.7%, P < .001); the median nonmajor RT changes was 1.0% among those with pretreatment major resistance.
Within the <100 000 and ≥100 000 copies/mL screening viral load strata and separately by third drug (EFV or ATV/r), the measure of cumulative RT amino acid changes was not significantly different comparing those with virologic failure by NRTI treatment assignment (P > .2, Table 3). With ATV/r, there were more protease amino acid changes in those assigned TDF/FTC than ABC/3TC among the screening plasma HIV-1 RNA <100 000 copies/mL stratum (median: 1.0% vs 0.0%, P = .006), but no significant difference was observed in the ≥100 000 screening stratum.
Table 3.
HIV-1 Amino Acid Changesa From Pretreatment to Virologic Failure, Comparing Tenofovir/Emtricitabine vs Abacavir/Lamivudine Virologic Failures
| With EFV |
With ATV/r |
||||
|---|---|---|---|---|---|
| TDF/FTC (n = 32) | ABC/3TC (n = 38) | TDF/FTC (n = 29) | ABC/3TC (n = 35) | ||
| A: Screening plasma HIV-1 RNA<100 000 copies/mL (n = 134) | |||||
| Protease changes (%) | Median (Q1, Q3) | 0.51 (0, 1.35) | 0.51 (0, 1.01) | 1.01 (0, 1.52) | 0.00 (0.00, 0.58) |
| Absence of any protease AA change | n (%) | 12 (38%) | 17 (45%) | 8 (28%) | 18 (51%) |
| P = .5 | P = .006 | ||||
| RT changes (%) | Median (Q1, Q3) | 1.09 (0.65, 1.96) | 0.87 (0.43, 1.74) | 0.22 (0, 0.65) | 0.22 (0, 0.79) |
| Absence of any RT AA change | n (%) | 2 (6%) | 6 (16%) | 8 (28%) | 11 (31%) |
| P = .2 | P = .9 | ||||
| With EFV |
With ATV/r |
||||
| TDF/FTC (n = 11) | ABC/3TC (n = 25) | TDF/FTC (n = 15) | ABC/3TC (n = 32) | ||
| B: Screening plasma HIV-1 RNA ≥100 000 copies/mL, limited through the time of the DSMB recommendations (n = 83) | |||||
| Protease changes (%) | Median (Q1, Q3) | 0 (0, 1.52) | 0.51 (0, 1.01) | 0 (0, 1.01) | 0.51 (0, 1.01) |
| Absence of any protease AA change | n (%) | 7 (64%) | 10 (40%) | 8 (53%) | 11 (34%) |
| P = .3 | P = .3 | ||||
| RT changes (%) | Median (Q1, Q3) | 1.75 (0.22, 2.39) | 1.96 (1.12, 2.39) | 0.43 (0.22, 0.65) | 0.54 (0.22, 0.87) |
| Absence of any RT AA change | n (%) | 2 (18%) | 0 (0%) | 2 (13%) | 4 (13%) |
| P = .2 | P = .4 | ||||
Abbreviations: AA, amino acid; ABC/3TC, abacavir/lamivudine; ATV/r, atazanavir/ritonavir; DSMB, data safety and monitoring board; EFV, efavirenz; HIV-1, human immunodeficiency virus type 1; RT, reverse transcriptase; TDF/FTC, tenofovir DF/emtricitabine.
a Indicates the percentage of amino acid codons changed from pretreatment to virologic failure, with mixture changes assigned weight = 0.5 change.
Among 265 virologic failures, the median (Q1, Q3) percent of amino acid changes was 0.63 (0.30, 1.25); 9% had no amino acid changes. A multivariable regression model of cumulative amino acid changes showed that efavirenz treatment (compared to ATV/r), lower pretreatment CD4, presence of pretreatment major drug resistance mutations, increased pretreatment amino acid mixture codons (all P < .001), hepatitis C antibody negativity (P = .05), and black non-Hispanic race/ethnicity (compared to white or Hispanic, P = .02) were each associated with significantly more HIV-1 amino acid changes (protease and RT combined). In contrast, the 20% who reported not taking any antiretroviral treatment proximal to the sample collected at time of virologic failure for genotyping had fewer amino acid changes (P = .003, Table 4). The above covariates in the multivariable model accounted for 40% of the variability in amino acid changes (adjusted R2). The following covariates were not significantly associated with amino acid changes among virologic failures: assigned NRTI (P = .95), sex (P = .80), age (P = .18), history of intravenous drug use (P = .34), presence vs absence of pretreatment resistance screening (P = .39), and HIV-1 RNA level at the time of virologic failure genotype sample (P = .78). Pretreatment viral load and timing of virologic failure were associated with cumulative amino acid change in univariate analysis but were not significant in the multivariable model. Amino acid changes are presented with respect to time elapsed between pretreatment and virologic failure for each regimen, delineated by CD4 cell count group (Figure 2A) and the presence of pretreatment or emergent drug resistance mutations (Figure 2B).
Table 4.
Covariates Associated With HIV-1 Amino Acid Changes From Pretreatment to Virologic Failurea
| Covariate | R2 (%) | Univariate Models |
Multivariable Model |
||||
|---|---|---|---|---|---|---|---|
| Regression Coefficient | SE | P Value | Regression Coefficient | SE | P Value | ||
| Assigned ATV/r vs EFV | 14 | −0.32 | 0.05 | <.001 | −0.31 | 0.04 | <.001 |
| Assigned ABC/3TC vs TDF/FTC | 0 | −0.003 | 0.05 | .95 | |||
| Pretreatment resistance mutations: major vs no major | 6 | 0.33 | 0.09 | <.001 | 0.27 | 0.07 | <.001 |
| Pretreatment number of codons with amino acid mixtures | 14 | 0.09 | 0.01 | <.001 | 0.06 | 0.01 | <.001 |
| Pretreatment plasma HIV-1 RNA (log10 cp/mL) | 3 | 0.10 | 0.03 | .005 | |||
| Pretreatment CD4 count (per 50 cells/μL increase) | 12 | −0.04 | 0.01 | <.001 | −0.03 | 0.01 | <.001 |
| Race/ethnicity: Black non-Hispanic vs Hispanic or white | 2 | 0.12 | 0.05 | .020 | 0.10 | 0.04 | 0.02 |
| Pretreatment hepatitis C antibody status: positive vs negative | 2 | −0.18 | 0.09 | .041 | −0.14 | 0.07 | 0.05 |
| Timing of virologic failure (per 4 visit weeks) | 2 | −0.006 | 0.003 | .017 | |||
| HIV-1 RNA <200 copies/mL prior to virologic failure genotype; yes vs no | 1 | −0.11 | 0.06 | .055 | |||
| Change in HIV-1 RNA from pretreatment to virologic failure (per 1.0 log10 copies/mL increase) | 1 | −0.04 | 0.02 | .097 | |||
| Self-report antiretroviral treatment status prior to virologic failure: | |||||||
| Not taking antiretroviral treatment vs taking | 4 | −0.20 | 0.06 | .002 | −0.16 | 0.05 | 0.003 |
| Modified the assigned regimen prior to failure vs no prior modification | 2 | −0.12 | 0.06 | .033 | |||
Abbreviations: ABC/3TC, abacavir/lamivudine; ATV/r, atazanavir/ritonavir; EFV, efavirenz; HIV-1, human immunodeficiency virus type 1; R2, the percentage of variability in the outcome variable accounted for by the given covariate; SE, standard error; TDF/FTC, tenofovir DF/emtricitabine.
a The outcome measure is the square-root transform of the percentage of amino acid codons changed from pretreatment to virologic failure among protease and reverse transcriptase, with mixture changes assigned weight = 0.5 change. Treatment assignment and covariates with univariate P ≤ .10 were included in the table. Sex, age, intravenous drug use, resistance screening status, and plasma HIV-1 RNA level at virologic failure genotyping were not significantly associated with cumulative amino acid changes. N = 251 were included in the models, 7 were of other or missing race/ethnicity, 5 were missing hepatitis C antibody status, and 2 were missing self-report treatment status.
Figure 2.
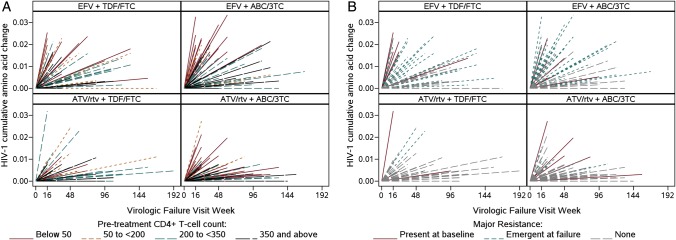
Human immunodeficiency virus type 1 (HIV-1) cumulative amino acid changes over time to virologic failure. Each participant with virologic failure is represented by a line from (0,0) to time of virologic failure on the horizontal axis and HIV-1–weighted cumulative amino acid changes on the vertical axis, with mixture changes assigned weight = 0.5 change. A, Among participants with virologic failure, lower pretreatment CD4 count (cells/μL) was associated with more HIV-1 amino acid changes. B, Many of the HIV-1 amino acid changes occurred within participants with virologic failure who had major resistance mutations present prior to treatment or emergent at failure, particularly with efavirenz (EFV). Abbreviations: 3TC, lamivudine; ABC, abacavir; ATV/rtv, atazanavir/ritonavir; FTC, emtricitabine; TDF. tenofovir DF.
DISCUSSION
Transmitted and acquired drug resistance mutations and the evolution of drug resistance are associated with clinical and immunologic progression, and constrain the effectiveness of ART. Viral rebound during treatment is associated with emergent resistance and changes in the RT and protease. Here we presented changes in HIV-1 consensus sequence from pretreatment to protocol-defined virologic failure from a large, randomized clinical trial of once-daily regimens.
Strikingly, there were remarkably few changes among consensus protease amino acids, regardless of treatment assignment; changes were not significantly different between ATV/r and EFV failures, even though only the former drug would be expected to exert selective pressure on protease. This finding is consistent with the absence of consensus protease inhibitor resistance mutations, described only in small numbers of virologic failures enrolled in other clinical trials including RTV-boosted ATV [14–16]. The paucity of protease drug resistance mutation may be explained by reduced drug exposure, due to the variable pharmacokinetics of ATV, or poor adherence [17, 18]. Yet in this study, absence of amino acid changes in the protease was not significantly associated with <100% adherence or being off ART near the time of virologic failure (data not shown). The near absence of major protease resistance at virologic failure and minimal cumulative changes in protease and RT is a consistent outcome among those failing RTV-boosted protease inhibitor-containing regimens [14–16, 19, 20].
Among EFV virologic failures, there were more frequent changes in RT, including and excluding amino acid positions associated with major antiretroviral resistance mutations that increase drug resistance and the fitness of resistant virus [21]. Amino acid changes at virologic failure were observed at greater frequency in association with pretreatment major resistance mutations and pretreatment amino acid mixtures. Measures of increased amino acid mixtures in RT are associated with more advanced disease, lower CD4 cell count, and longer duration of infection [22, 23]. Among participants with virologic failure, we also found that lower pretreatment CD4 cell count and higher pretreatment HIV-1 viral load were associated with more amino acid changes. Surprisingly, the CD4 count association with HIV-1 amino acid changes was stronger than the viral load association, providing some evidence for immunologic control of virus evolution during treatment.
Race/ethnicity and hepatitis C coinfection were associated with HIV-1 amino acid changes in RT and protease. Black non-Hispanics who experienced virologic failure in this study had more HIV-1 amino acid changes compared with whites or Hispanics with virologic failure. Hepatitis C antibody–positive participants experiencing HIV-1 virologic failure (n = 24) had marginally fewer changes. Finally, in this study, participants not taking ART proximal to virologic failure had significantly fewer amino acid changes compared to those taking treatment. Sensitivity analyses evaluating protease and RT changes separately show that the covariates significantly associated with cumulative amino acid changes were also associated with RT changes. The directionality of covariate associations was similar for protease and RT, but only amino acid mixtures were significantly associated with protease changes, perhaps because protease changes were not as frequent.
Hamming distance is a measure of cumulative amino acid changes that ignores the interdependence structure between amino acid changes. Some or perhaps many of the amino acid changes are not independent; particular amino acid substitutions may be occurring in reaction to other substitutions, especially those within close structural proximity [24]. However, Hamming distance is straightforward to calculate and interpret, does not make model-based assumptions about HIV-1 genetic change, and demonstrated an accumulation of nonmajor mutations in the presence of major RT mutations under the selective pressure of EFV. To our knowledge, no previous studies have evaluated this amino acid change metric. Our application was adapted from a nucleic acid distance metric presented previously [25]. Although we analyzed nonsynonymous substitutions that result in amino acid changes, models predict a role for synonymous and nonsynonymous point mutations and recombination in the emergence of drug resistance [26, 27].
The treatment comparisons presented here are observational comparisons among participants who experienced virologic failure, within a clinical trial where participants were randomly assigned to 1 of 4 first-line treatment options. Participants were analyzed according to the initially assigned regimen in order to evaluate resistance and HIV amino acid outcomes with respect to the first-line treatment strategy, regardless of changes or discontinuation of therapy prior to virologic failure. Previously reported A5202 results provide evidence that participants with higher pretreatment RNA or lower CD4 cell counts were at an increased risk for virologic failure with ABC/3TC compared to TDF/FTC [7], thus, it was not unexpected that virologic failures assigned to ABC/3TC treatment had lower CD4 counts and higher plasma HIV-1 RNA levels at pretreatment in this analysis among failures. A complication of study A5202 is that resistance testing before treatment initiation was performed in only 45% of participants. At study outset in 2005, pretreatment resistance testing was not yet the standard of care in the United States; as a result, there was heterogeneity among the sites regarding whether resistance testing was done. Today, testing treatment-naive patients for resistance prior to initiating therapy is strongly recommended [1].
Drug resistance mutations acquired during treatment may be due to decreased adherence, diminished drug levels, or both. Nevertheless, the frequency of emergent drug resistance mutations and amino acid changes in RT was dramatically higher among EFV compared to ATV/r virologic failures. This suggests that following virologic failure with ATV/r, changing to an alternative RTV-boosted PI or NNRTI-based regimen with adherence counseling and continued NRTI treatment should be effective in achieving virologic suppression. In contrast, effective treatment following EFV virologic failure should include a change to a boosted PI and consideration of alternative nucleosides and drug classes. The first-line choice of a boosted protease or NNRTI regimen, pretreatment levels of amino acid mixtures or viral diversity, CD4 cell count, and HIV-1 viral load may each impact the selection of mutations and drug resistance at virologic failure. In addition to genotyping upon virologic failure, cumulative amino acid changes from pretreatment to failure may provide insight into drug exposure, adherence, and reasons for virologic failure. Additionally, changes in amino acids may predict risks for second-line virologic failure and optimize continued antiretroviral treatment.
Notes
Acknowledgments. We thank the patients for their participation in this study; Michael Wantman for data quality assurance and database programming; and Courtney Ashton for lab data coordination.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases (award number U01AI068636, along with the previous grant number for the ACTG Central Group [AI38858] and the SDMC [AI68634]). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health. This work was also supported in part by the General Clinical Research Center Units funded by the National Center for Research Resources. The study medications were provided by Abbott Pharmaceuticals, Bristol-Myers Squibb, Gilead Sciences, and GlaxoSmithKline.
Potential conflicts of interest. E. S. D. has received research support from Abbott Laboratories, Merck Laboratories, Pfizer, Gilead, and ViiV and has served as an advisor or consultant for Bristol Myers Squibb, Gilead Sciences, Merck Laboratories, and ViiV. P. E. S. has received research support from Bristol-Myers Squibb, Gilead, GlaxoSmithKline, Merck Laboratories, and Janssen, and consulting fees from Bristol-Myers Squibb, Gilead Sciences, GlaxoSmithKline, and Merck Laboratories. A. C. C. has received past research support from Tibotec, Koronis, Merck Laboratories, Schering-Plough, Boehringer-Ingelheim, and Gilead Sciences and consulting fees from GlaxoSmithKline, Pfizer, and Merck Laboratories, and has personal equity and a family member with equity in Bristol-Myers Squibb, Abbott Laboratories, Johnson & Johnson, and Pfizer. M. A. F. has received grant support from Abbott Laboratories, Bionor Immuno AS, Cytheris, GlaxoSmithKline, Pfizer, and Tibotec, and grant support, lecture, and consulting fees from Merck Laboratories. C. T. is a paid member of a data monitoring committee for a Tibotec hepatitis C drug. D. K. has received research support from Pfizer, Roche Molecular Diagnostics, Bristol-Myers Squibb, GlaxoSmithKline, and Gilead Sciences. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Panel on Antiretroviral Guidelines for Adults and Adolescents. Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents. Department of Health and Human Services; 2011. pp. 1–166. 10 January http://www.aidsinfo.nih.gov/ContentFiles/AdultandAdolescentGL.pdf. Accessed 19 August 2011. [Google Scholar]
- 2.Kitahata MM, Gange SJ, Abraham AG, et al. Effect of early versus deferred antiretroviral therapy for HIV on survival. N Engl J Med. 2009;360:1815–26. doi: 10.1056/NEJMoa0807252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antiretroviral Therapy Cohort Collaboration. Life expectancy of individuals on combination antiretroviral therapy in high-income countries: a collaborative analysis of 14 cohort studies. Lancet. 2008;372:293–9. doi: 10.1016/S0140-6736(08)61113-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gupta R, Hill A, Sawyer AW, Pillay D. Emergence of drug resistance in HIV type 1-infected patients after receipt of first-line highly active antiretroviral therapy: a systematic review of clinical trials. Clin Infect Dis. 2008;47:712–22. doi: 10.1086/590943. [DOI] [PubMed] [Google Scholar]
- 5.Deeks SG, Gange SJ, Kitahata MM, et al. Trends in multidrug treatment failure and subsequent mortality among antiretroviral therapy-experienced patients with HIV infection in North America. Clin Infect Dis. 2009;49:1582–90. doi: 10.1086/644768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hogg RS, Bangsberg DR, Lima VD, et al. Emergence of drug resistance is associated with an increased risk of death among patients first starting HAART. PLoS Med. 2006;3:e356. doi: 10.1371/journal.pmed.0030356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sax PE, Tierney C, Collier AC, et al. Abacavir-lamivudine versus tenofovir-emtricitabine for initial HIV-1 therapy. N Engl J Med. 2009;361:2230–40. doi: 10.1056/NEJMoa0906768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daar ES, Tierney C, Fischl MA, et al. Atazanavir plus ritonavir or efavirenz as part of a 3-drug regimen for initial treatment of HIV-1. Ann Intern Med. 2011;154:445–56. doi: 10.1059/0003-4819-154-7-201104050-00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sax PE, Tierney C, Collier AC, et al. Abacavir/lamivudine versus tenofovir DF/emtricitabine as part of combination regimens for initial treatment of HIV: Final results. J Infect Dis. 2011;204:1191–201. doi: 10.1093/infdis/jir505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winters MA, Coolley KL, Girard YA, et al. A 6-basepair insert in the reverse transcriptase gene of human immunodeficiency virus type 1 confers resistance to multiple nucleoside inhibitors. J Clin Invest. 1998;102:1769–75. doi: 10.1172/JCI4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson VA, Brun-Vezinet F, Clotet B, et al. Update of the drug resistance mutations in HIV-1. Top HIV Med. 2008;16:138–45. [PubMed] [Google Scholar]
- 12.Hamming RW. Error detecting and error correcting codes. Bell System Technical Journal. 1950;29:147–60. [Google Scholar]
- 13.Chesney MA, Ickovics JR, Chambers DB, et al. Self-reported adherence to antiretroviral medications among participants in HIV clinical trials: the AACTG adherence instruments. Patient Care Committee and Adherence Working Group of the Outcomes Committee of the Adult AIDS Clinical Trials Group (AACTG) AIDS Care. 2000;12:255–66. doi: 10.1080/09540120050042891. [DOI] [PubMed] [Google Scholar]
- 14.Ross LL, Weinberg WG, DeJesus E, et al. Impact of low abundance HIV variants on response to ritonavir-boosted atazanavir or fosamprenavir given once daily with tenofovir/emtricitabine in antiretroviral-naive HIV-infected patients. AIDS Res Hum Retroviruses. 2010;26:407–17. doi: 10.1089/aid.2009.0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ulbricht KU, Behrens GM, Stoll M, et al. A multicenter, open labeled, randomized, phase III study comparing lopinavir/ritonavir plus atazanavir to lopinavir/ritonavir plus zidovudine and lamivudine in naive HIV-1-infected patients: 48-week analysis of the LORAN trial. Open AIDS J. 2011;5:44–50. doi: 10.2174/1874613601105010044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilkin TJ, McKinnon JE, DiRienzo AG, et al. Regimen simplification to atazanavir-ritonavir alone as maintenance antiretroviral therapy: final 48-week clinical and virologic outcomes. J Infect Dis. 2009;199:866–71. doi: 10.1086/597119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colombo S, Buclin T, Cavassini M, et al. Population pharmacokinetics of atazanavir in patients with human immunodeficiency virus infection. Antimicrob Agents Chemother. 2006;50:3801–8. doi: 10.1128/AAC.00098-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ray JE, Marriott D, Bloch MT, McLachlan AJ. Therapeutic drug monitoring of atazanavir: Surveillance of pharmacotherapy in the clinic. Br J Clin Pharmacol. 2005;60:291–9. doi: 10.1111/j.1365-2125.2005.02413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Molina JM, Andrade-Villanueva J, Echevarria J, et al. Once-daily atazanavir/ritonavir versus twice-daily lopinavir/ritonavir, each in combination with tenofovir and emtricitabine, for management of antiretroviral-naive HIV-1-infected patients: 48 week efficacy and safety results of the CASTLE study. Lancet. 2008;372:646–55. doi: 10.1016/S0140-6736(08)61081-8. [DOI] [PubMed] [Google Scholar]
- 20.Riddler SA, Haubrich R, DiRienzo AG, et al. Class-sparing regimens for initial treatment of HIV-1 infection. N Engl J Med. 2008;358:2095–106. doi: 10.1056/NEJMoa074609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhee SY, Kantor R, Katzenstein DA, et al. HIV-1 pol mutation frequency by subtype and treatment experience: Extension of the HIVseq program to seven non-B subtypes. AIDS. 2006;20:643–51. doi: 10.1097/01.aids.0000216363.36786.2b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park SY, Love TM, Nelson J, Thurston SW, Perelson AS, Lee HY. Designing a genome-based HIV incidence assay with high sensitivity and specificity. AIDS. 2011;25:F13–9. doi: 10.1097/QAD.0b013e328349f089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cousins MM, Laeyendecker O, Beauchamp G, et al. Use of a high resolution melting (HRM) assay to compare gag, pol, and env diversity in adults with different stages of HIV infection. PLoS One. 2011;6:e27211. doi: 10.1371/journal.pone.0027211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dahirel V, Shekhar K, Pereyra F, et al. From the cover: coordinate linkage of HIV evolution reveals regions of immunological vulnerability. Proc Natl Acad Sci U S A. 2011;108:11530–5. doi: 10.1073/pnas.1105315108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kantor R, Wantman M, Katzenstein D, et al. Viral shedding and drug resistance in plasma and genital compartments among viremic, multi-drug-experienced HIV-infected men and women. Presented at the 15th Conference on Retroviruses and Opportunistic Infections; 3–6 February 2008; Boston, MA. [Google Scholar]
- 26.Carvajal-Rodriguez A, Crandall KA, Posada D. Recombination favors the evolution of drug resistance in HIV-1 during antiretroviral therapy. Infect Genet Evol. 2007;7:476–83. doi: 10.1016/j.meegid.2007.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Althaus CL, Bonhoeffer S. Stochastic interplay between mutation and recombination during the acquisition of drug resistance mutations in human immunodeficiency virus type 1. J Virol. 2005;79:13572–8. doi: 10.1128/JVI.79.21.13572-13578.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Among the 269 confirmed virologic failures in study A5202, HIV-1 sequence results were available at both pretreatment and failure for 265 participants (98.5%), of the 4 remaining virologic failures with unavailable results, for 2 the samples did not pass quality assurance, and the other 2 each had a sample that did not amplify. Missing codon information due to trimming of sequences was unusual, with the exception of early protease codons 1–12, and the latter codons of RT.