Abstract
Background
The glycoprotein (G) gene sequences of bovine ephemeral fever virus (BEFV) strains derived from mainland China have not been compared with those of the isolates from other countries or areas. Therefore, the G genes of four BEFV isolates obtained from mainland China were amplified and sequenced. A phylogenetic tree was constructed in order to compare and analyze the genetic relationships of the BEFV isolates derived from mainland China and different countries and areas.
Results
The complete BEFV G gene was successfully amplified and sequenced from four isolates that originated from mainland China. A total of fifty-one BEFV strains were analyzed based on the G gene sequence and were found to be highly conserved. A phylogenetic tree showed that the isolates were grouped into three distinct lineages depending on their source of origin. The antigenic sites of G1, G2 and G3 are conserved among the isolates, except for several substitutions in a few strains.
Conclusions
The phylogenetic relationships of the BEFV isolates that originated from mainland China, Taiwan, Japan, Turkey, Israel and Australia were closely related to their source of origin, while the antigenic sites G1, G2 and G3 are conserved among the BEFV isolates used in this work.
Keywords: Bovine ephemeral fever virus, Glycoprotein gene, Mainland China, Phylogenetic relationship, Variation
Background
Bovine ephemeral fever virus (BEFV) is an arthropod-borne rhabdovirus which belongs to the genus Ephemerovirus in the Rhabdoviridae[1]. Bovine ephemeral fever (BEF), caused by BEFV, is an acute febrile disease in cattle and water buffalo in tropical and subtropical regions of Africa, Asia, Australia and the Middle East. The disease has a considerable economic impact on dairy farming in China. Most infected livestock present with a decrease in the quantity and quality of milk, and lameness or paralysis [2,3].
BEFV is a negative ssRNA genome and viral particles have a bullet-like appearance or tapered shape. In addition, the virus bears spikes on the surface of envelope proteins. Five structural proteins of BEFV have been described, which comprise nucleoprotein (N), surface glycoprotein (G), large RNA-dependent RNA polymerase (L), polymerase associate protein (P) and matrix protein (M) [4-7]. Monoclonal antibody (MAb) studies of the prototype virus indicate that the G protein is the main protective antigen [8]. Four distinct antigenic sites (G1, G2, G3 and G4) on the surface of the G protein have been identified [8-10]. Antigenic site G1 is linear, and G2 and G3 are conformational. G1 reacts only with anti-BEFV antibodies, but the other antigenic sites show cross-reactivity with sera against other related viruses [11]. A blocking enzyme-linked immunoabsorbent assay (ELISA) and two indirect ELISAs for the detection of the antibodies against the G1 site of BEFV have been established [12-14].
BEF was first documented in China in 1955 [15]. The first BEFV strain JB76H was isolated from infected dairy cattle during the 1976 epidemic in mainland China [16]. The disease was reported to be prevalent in twenty-five provinces in mainland China from 1952 to 1991 [11,15]. From 1991 to date, because BEF is not carried out, there is a lack of detailed epidemiological data on the disease in mainland China, except for Henan Province in central China. The major BEF epidemics are shown in Table 1. The data were mainly obtained from the previous reports [11,15], while the data relating to the BEF epidemics in Henan Province from 1983 to 2011 were obtained from our monitoring system.
Table 1.
The BEF epidemics that have occurred in mainland China (mainly from 1955 to 1991)
| Provinces | Years |
|---|---|
| Guangdong |
1955, 1962, 1966, 1971, 1972, 1976, 1977, 1978, 1979, 1983, 1985, 1987, 1988, 1991 |
| Guangxi |
1976, 1982 |
| Hunan |
1955, 1963, 1966, 1978, 1979, 1983–1984, 1987, 1988, 1991 |
| Hubei |
1959, 1964, 1970, 1976, 1983, 1987, 1991 |
| Hainan |
1957-1959, 1967–1969, 1971–1972, 1975–1984, 1985–1989, 1991 |
| Henan |
1949-1982, 1983, 1985, 1989, 1991, 1997, 2004, 2005, 2011 |
| Jiangsu |
1954-1955, 1966, 1976–1977, 1991 |
| Zhejiang |
1955, 1958, 1965, 1971, 1983, 1987, 1988, 1991, 2002 |
| Fujian |
1954, 1955, 1958, 1963, 1966, 1972, 1975 |
| Jiangxi |
1949-1989, 1991 |
| Anhui |
1954, 1955, 1958, 1966, 1970, 1976, 1983, 1987, 1988, 1991 |
| Shandong |
1954, 1955, 1959, 1965, 1966, 1970, 1971, 1976, 1983, 1987, 1991 |
| Shanghai |
1952, 1955, 1958, 1971, 1976, 1983, 1991 |
| Yunnan |
1965, 1973, 1975, 1977, 1978, 1982, 1983, 1986–1989, 1991 |
| Guizhou |
1955, 1957, 1969, 1976, 1983 |
| Sichuan |
1954, 1957, 1962, 1967, 1969, 1976, 1980, 1983, 1986, 1987, 1989 |
| Xizang |
1976-1979, 1985-1989 |
| Shǎnxi |
1953-1960, 1961, 1962, 1966, 1968, 1969, 1975–1978, 1982–1986, 1989, 1991 |
| Gansu |
1956, 1964, 1969, 1975, 1977, 1981, 1986, 1987, 1989 |
| Ningxia |
1957-1989 |
| Shānxi |
1954, 1959, 1991 |
| Beijing |
1956, 1966, 1976 |
| Neimenggu |
1966, 1971, 1974, 1975, 1976 |
| Liaoning |
1985, 1991 |
| Jilin | 1983, 1991 |
Currently, no information is available on the variation in antigenic properties and nucleotide sequences of the G gene of BEFV isolated in mainland China. In this study, the complete G genes of four BEFV strains (LS11, LYC11, JT02L and JB76H) obtained from mainland China were amplified and sequenced. For the first time, the phylogenetic relationships and antigenic variation of the G genes of BEFV, isolated from mainland China, Taiwan, Japan, Turkey, Israel and Australia, were analyzed.
Materials and methods
Virus isolation and identification
The JB76H strain was used in mainland China as the vaccine against BEFV. The BEFV strain JT02L was obtained from an outbreak that occurred in 2002 in Zhejiang Province. The LS11 and LYC11 strains of BEFV were isolated from blood samples of the infected dairy cattle during the 2011 epidemic in Luoyang city, Henan Province. The blood samples were collected in order to monitor BEF, which is required by the foundation item supporting for this work.
Isolation of BEFV was carried out in the brains of suckling mice and baby hamster kidney (BHK-21) cells as described previously [17]. Briefly, the blood collected from infected dairy cattle was mixed with Alsever's solution, and BEFV was concentrated decuple by centrifugation of the blood sample. Subsequently, the BEFV samples were inoculated into the brains of suckling mice and subjected to seven blind passages in suckling mice. Thereafter, BHK-21 cells were inoculated with BEFV extracted from the brains of the 7th passage infected suckling mice. The virus underwent five to ten passages in BHK-21 cells, until cytopathogenic effects (CPE) were observed.
The presence of BEFV was confirmed by reverse-transcription polymerase chain reaction (RT-PCR) as reported previously [18]. The BEFV RNAs were extracted from the infected blood and BHK-21 cells using a QIAamp viral RNA mini kit (Qiagen, Hilden, Germany). For RT-PCR, the primers were 420F (5' AGA GCT TGG TGT GAA TAC 3') and 420R (5' CCA ACC TAC AAC AGC AGA TA 3'). The forward primer 420F was used to reverse-transcribe BEFV RNA to cDNA. Subsequently, a partial fragment of the BEFV G gene was amplified using the primers 420F and 420B. After the initial denaturation at 94°C for 5min, the amplification proceeded through a total of 35 cycles consisting of denaturation at 94°C for 40s, annealing at 46°C for 1min, primer extension at 72°C for 40s and a final extension for 10min at 72°C. The expected DNA fragments were 420 base pairs (bp) in length.
Amplification and sequencing of the BEFV G gene
The complete BEFV G gene was amplified as described in a previous report [19]. The primers were GF (5' ATG TTC AAG GTC CTC ATA ATT ACC 3') and GR (5' TAA TGA TCA AAG AAC CTA TCA TCA C 3'). The amplification procedures were carried out according to the report [19], except for the extension at 68°C for 2 min and the final extension at 68°C for 10 min.
The amplified fragments of the BEFV G gene were purified with an agarose gel DNA purification kit (TaKaRa, Dalian, China) and ligated with the pGEM-T Easy vector. Subsequently, the ligated mixtures were transformed into Escherichia coli DH5α. The plasmids were extracted from positive clones and then sequenced by TaKaRa (Dalian, China). The sequences obtained were deposited in the NCBI GenBank database.
Phylogenetic analysis of the BEFV G gene sequences
The nucleotide length of the region encoding the entire ectodomain of BEFV G protein is 1527 bp [10]. An alignment of BEFV sequences corresponding to the ectodomain region were carried out using the Clustal W program [20]. The BEFV strains were isolated from mainland China, Taiwan, Japan, Turkey, Israel and Australia (Table 2). The nucleotide and deduced amino acid (aa) sequence homologies among the isolates were analyzed using the MegAlign program of DNAstar. The phylogenetic tree based on the nucleotide sequence (1527 bp) of the analyzed G genes was constructed by the neighbor-jointing method [21] with the Kimura two-parameter model [22]. The reliability of the branching orders was evaluated by the bootstrap test with 1000 replicates [23]. Phylogenetic analyses were conducted using MEGA 5 software [24]. If the nucleotide sequences of several BEFV strains had 100% homology, a representative isolate was used to construct the phylogenetic tree.
Table 2.
Characteristics of BEFV strains used in this study
| Strain | Source | Year collected | Geographical origin | Cluster | Accession No. |
|---|---|---|---|---|---|
| JB76H |
Bovine blood |
1976 |
Beijing, Mainland China |
I |
JQ728557 |
| JT02L |
Bovine blood |
2002 |
Zhejiang, Mainland China |
I |
JQ728558 |
| LS11 |
Bovine blood |
2011 |
Henan, Mainland China |
I |
JQ728559 |
| LYC11 |
Bovine blood |
2011 |
Henan, Mainland China |
I |
JQ728560 |
| YHL |
Bovine blood |
1966 |
Yamaguchi, Japan |
I |
AB462028 |
| Hirado-6 |
Bovine plasma |
1988 |
Nagasaki, Japan |
I |
AB462029 |
|
a Hirado-9 |
Bovine plasma |
1988 |
Nagasaki, Japan |
I |
AB462030 |
| Amakusa-1 |
Bovine blood |
1988 |
Kumamoto, Japan |
I |
AB462031 |
| * a Amakusa-2 |
Bovine blood |
1988 |
Kumamoto, Japan |
I |
AB462032 |
|
b Azuma |
Bovine erythrocyte |
1988 |
Kagoshima, Japan |
I |
AB462033 |
| * b ON-BEF-88-1 |
Bovine white blood cells |
1988 |
Okinawa, Japan |
I |
AB462034 |
| ON-BEF-88-3 |
Bovine white blood cells |
1988 |
Okinawa, Japan |
I |
AB462035 |
| ON-BEF-88-4 |
Bovine white blood cells |
1988 |
Okinawa, Japan |
I |
AB462036 |
| ON-BEF-89-1 |
Bovine white blood cells |
1989 |
Okinawa, Japan |
I |
AB462037 |
| ON-BEF-89-2 |
Bovine white blood cells |
1989 |
Okinawa, Japan |
I |
AB462038 |
| ON-BEF-89-3 |
Bovine white blood cells |
1989 |
Okinawa, Japan |
I |
AB462039 |
| * b Onna3 |
Bovine erythrocyte |
1989 |
Okinawa, Japan |
I |
AB462040 |
| ON-BEF-01-1 |
Bovine white blood cells |
2001 |
Okinawa, Japan |
I |
AB462041 |
|
c ON-BEF-01-2 |
Bovine erythrocyte |
2001 |
Okinawa, Japan |
I |
AB462042 |
| ON-BEF-01-3 |
Bovine erythrocyte |
2001 |
Okinawa, Japan |
I |
AB462043 |
| ON-04-1 |
Bovine blood |
2004 |
Okinawa, Japan |
I |
AB462044 |
| CS1180 |
Bovine blood |
1982 |
Queensland, Australia |
III |
AF058321 |
| CS1647 |
Culicoides brevitarsis |
1984 |
Queensland, Australia |
III |
AF058322 |
| CS1619 |
|
|
Australia |
III |
AF058323 |
| CS42 |
Anopheles bancrofti |
1975 |
Northern Territory, Australia |
III |
AF058324 |
| CS1818 |
Bovine blood |
1970 |
Queensland, Australia |
III |
AF058325 |
|
d BB7721 |
Bovine blood |
1968 |
Queensland, Australia |
III |
AF234533 |
| * d |
|
|
Australia |
III |
NC002526 |
| 1984/TW/TN1 |
Bovine blood |
1984 |
Taiwan |
I |
AY935239 |
| 1996/TW/TN1 |
Bovine blood |
1996 |
Taiwan |
I |
AY935240 |
| TN88128 |
Bovine blood |
1999 |
Taiwan |
I |
AF208840 |
|
e 2001/TW/TN1 |
Bovine blood |
2001 |
Taiwan |
I |
AY935241 |
| * c 2001/TW/TN2 |
Bovine blood |
2001 |
Taiwan |
I |
AY954451 |
| 2001/TW/TN3 |
Bovine blood |
2001 |
Taiwan |
I |
AY954452 |
| * e 2001/TW/TN4 |
Bovine blood |
2001 |
Taiwan |
I |
AY954453 |
| * e 2001/TW/TN5 |
Bovine blood |
2001 |
Taiwan |
I |
AY954454 |
| * e 2001/TW/TN6 |
Bovine blood |
2001 |
Taiwan |
I |
AY954455 |
| * c 2001/TW/TN7 |
Bovine blood |
2001 |
Taiwan |
I |
AY954456 |
| 2001/TW/TN8 |
Bovine blood |
2001 |
Taiwan |
I |
AY954457 |
| 2001/TW/TN9 |
Bovine blood |
2001 |
Taiwan |
I |
AY954458 |
| 2001/TW/TN10 |
Bovine blood |
2001 |
Taiwan |
I |
AY954459 |
| * e 2001/TW/TN11 |
Bovine blood |
2001 |
Taiwan |
I |
AY954460 |
| TN-2004-124 |
Bovine blood |
2004 |
Taiwan |
I |
AY818194 |
| 2008/TR/CP62 |
Bovine blood |
2008 |
Turkey |
II |
GQ229451 |
| 2008/TR/CP77 |
Bovine blood |
2008 |
Turkey |
II |
GQ229452 |
| ISR00 |
Bovine blood |
2000 |
Israel |
II |
JN833630 |
| ISR01 |
Bovine blood |
2001 |
Israel |
II |
JN833631 |
| ISR04 |
Bovine blood |
2004 |
Israel |
II |
JN833632 |
| ISR10/1 |
Bovine blood |
2010 |
Israel |
II |
JN833633 |
| ISR10/2 |
Bovine blood |
2010 |
Israel |
II |
JN833634 |
| ISR10/3 | Bovine blood | 2010 | Israel | II | JN833635 |
Amino acid sequence variation of the antigenic sites of BEFV G protein
The aa sequences corresponding to the antigenic sites G1, G2 and G3 have been determined previously [8,10]. The sites G1 and G2 are located at the residues 487–503 and 168–189, respectively. The conformational site G3 is located at residues 49–63, 215–231 and 262–271. The aa sequences deduced from BEFV G genes were aligned, and the variations in the aa corresponding to the sites G1, G2 and G3 were analyzed. The representative BEFV strains used were isolated at different times or from different countries and areas.
Results
Virus isolation and identification
The DNA fragments of 420 bp were amplified from blood samples of the infected dairy cattle by RT-PCR. It was confirmed by sequence analysis that the gene fragments represented part of the BEFV G gene indicating that the disease shown in the cattle was in fact BEF.
From the outbreak of BEF in Luoyang in 2011, infected blood was collected from dairy cattle in the Songxian and Yichuanxian areas, and two BEFV strains, designated as LS11 and LYC11, were isolated by intracerebral inoculation of suckling mice and in BHK-21 cells. The infected suckling mice showed paralysis and stiffness in their hind legs on the second to third day after inoculation and died during 12–24 hours post-morbidity. The infected BHK-21 cells showed specific CPE. The specific DNA fragments of 420 bp were also amplified from the LS11 and LYC11 strains.
Amplification and sequencing of the BEFV G gene
Complete BEFV G genes (1872 bp) were successfully amplified and sequenced from the JB76H, JT02L, LS11 and LYC11 strains. The G gene sequences of LS11, LYC11, JT02L and JB76H isolates have been assigned the accession numbers JX564637, JX564638, JX564639 and JX564640, respectively, in the GenBank database.
Phylogenetic analysis of the BEFV G gene sequences
The G gene sequences of the other forty-seven BEFV isolates were obtained from the GenBank database. A total of fifty-one BEFV isolates were used in this study (Table 2), and forty-one representative strains were used to produce Figures 1 and 2.
Figure 1.
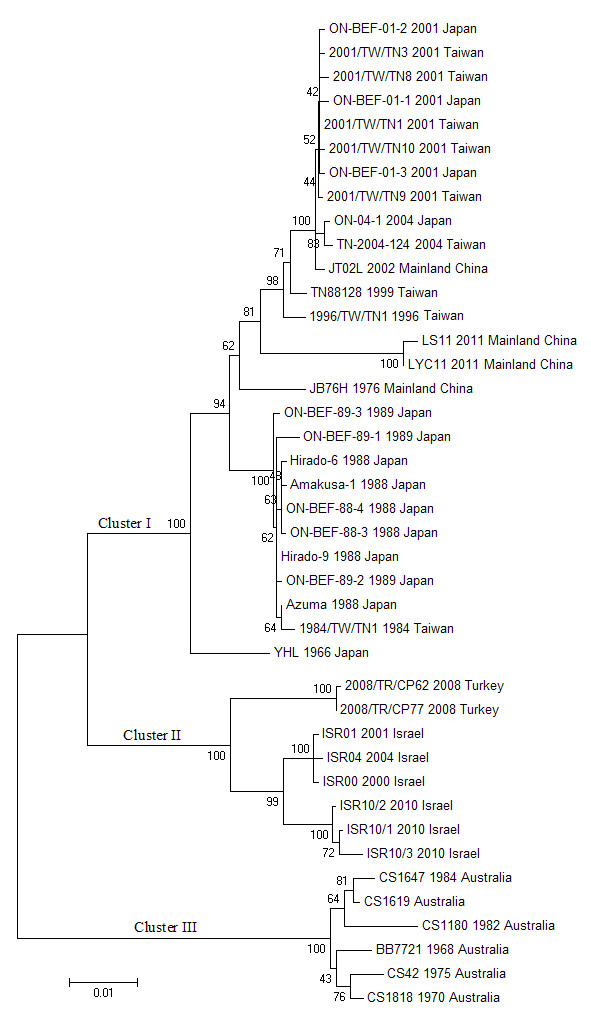
Phylogenetic profiles of the BEFV isolates based on the comparison of the G gene sequences. The source, year of isolation and geographical origin of each strain are indicated in the tree. The scale represents 1% sequence divergence. A total of forty-one BEFV isolates were used to construct the phylogenetic tree.
Figure 2.
Alignment of the aa sequences corresponding to the antigenic sites of G1 G2 and G3 of BEFV G protein. The residues differing from the sequences of the majority are denoted.
All nucleotide and deduced aa sequences corresponding to the ectodomain region of BEFV G protein were highly conserved among the BEFV isolates obtained from mainland China, Taiwan, Japan, Turkey, Israel and Australia. The identities of the nucleotide sequences were between 89.3% and 99.9%, and those of the aa sequences were between 94.5% and 100%.
Forty-one BEFV isolates were grouped into three distinct lineages (Figure 1). Cluster I contained the strains isolated from mainland China, Taiwan and Japan. The Turkish and Israeli isolates were grouped into cluster II, and Australian strains were placed in the independent cluster III.
Amino acid sequence variation of the antigenic sites of BEFV G protein
As shown in Figure 2, the antigenic sites of G1, G2 and G3 were highly conserved among the BEFV isolates obtained from mainland China and Taiwan, except for six aa substitutions in the JB76H, 1984/TW/TN1 and 2001/TW/TN10 strains. In the G3 site of the JB76H strain, two residues at positions 53 and 263 were substituted from N to K and from E to G respectively. Three substitutions, at positions 220 (E to A), 271 (Q to R) and 499 (S to N), were found in the 1984/TW/TN1 strain. There was a residue change at position 215 (K to N) in the 2001/TW/TN10 isolate. The residues Q at 271 and S at 499 were substituted by R and N in the Japanese strains, except for the ON-04-1, ON-BEF-01-1, ON-BEF-01-2, ON-BEF-01-3 and YHL isolates. Two substitutions at positions 170 (T to N) and 223 (E to D) were detected in the YHL strain. An additional substitution was observed at position 215 (K to T) in the ON-BEF-89-3 isolate. The three antigenic sites were completely conserved in the strains isolated from Turkey and Israel. Among the eight isolates, only two amino acids at positions 223 and 503 were substituted from E to D and from K to T, respectively. In the antigenic site G1 of Australian isolates, the residue was N at position 499, differing from the majority of sequences, which contained S. The substitutions were found at positions 223–224 (ET to DK) in the G3 site of the six Australian isolates. An additional substitution was observed at position 218 (R to K) in the CS1647, CS1619 and CS1180 isolates. Two additional aa changes were found at positions 215 (K to T) and 263 (E to K) in the CS1818 and CS42 isolates.
Discussion
The clinical signs, morbidity and mortality associated with current cases of BEF are different from those of BEF cases reported before 2000. The current disease cases showed more severe symptoms, and the morbidity and mortality have increased significantly. Luoyang city, in Henan Province, central China, is an epidemic area for BEF, and there have been eight BEF epidemics in the area from 1983 to 2011. The three BEF epizootics in 2011, 2005 and 2004 caused considerable economic loss to dairy cattle farming. During the latest an outbreak, which occurred in 2011, the infected dairy cattle showed a sudden onset of fever, stiffness, and nasal and ocular discharges. Moreover, difficulty in breathing and shortness of breath were the most obvious clinical symptoms shown by the infected dairy cattle. Some of the severe cases died between 6 and 12 hours after infection. The morbidity was about thirty percent, and the mortality rate was about five percent. However, the morbidity was from ten to twenty percent and the mortality was lower than one percent before 2000. During the 2004 BEF epizootic, about 12,200 dairy cattle were affected, of which, 2,198 cases died. Similar data were obtained in the 2005 epizootic. In the 2011 outbreak, the infected and dead dairy cattle rose to about 32,051 and 5,360, respectively. Certainly, the numbers of dairy cattle were increased from 44,000 in 2004 and 2005 to 107,300 in 2011 (http://henan.people.com.cn/news/2011/08/02/558227.html). The high feeding density of dairy cattle and the suffocation caused by the BEF may be the leading reasons for the high mortalities.
The phylogenetic relationships of the G gene sequence of BEFV isolated in Japan, Taiwan, Turkey, Israel and Australia had been analyzed previously [25,26]. To date, the genetic relationships of BEFV derived from mainland China and those from other countries or areas have not been studied. In order to clarify the variation in the BEFV G gene with time and location, the G genes of four BEFV strains (LS11, LYC11, JT02L and JB76H) isolated from mainland China were amplified and sequenced. The G gene sequences of the three field isolates was repeatedly amplified and sequenced from infected blood, suckling brain and BHK-21 cells. The results showed that no change was found in the nucleotide sequences, indicating that the adaptation to suckling mice and BHK-21 cells through low passages had no significant effect on the nucleotide sequences of the BEFV G gene. However, it is worth noting that only one G gene sequence of the JB76H strain was used, because the original samples could not be obtained. It was unclear whether the extensive passages in BHK-21 cells affected variation in the G gene sequence of JB76H isolate.
The nucleotide and deduced aa sequences of the region encoding the ectodomain of BEFV G protein were well conserved among the BEFV isolates. In particular, the strains that originated from mainland China, Taiwan and Japan had higher identities. The corresponding sequences of the isolates derived from Turkey and Israel were highly conserved. However, the identities of the sequences were slightly lower among Australian isolates and other strains.
The phylogenetic relationships of the sequences of the ectodomain region of the BEFV G gene were analyzed in this work. The analysis revealed that the clusters of the BEFV isolates were closely related with geographical location. The strains derived from oriental areas (mainland China, Taiwan and Japan) had a close relationship. Turkish and Israeli isolates were grouped into one cluster, which had a close relationship. The Australian isolates were grouped into an independent cluster, and had a distant relationship with the Asian strains. The results revealed that the phylogenetic relationships among the BEFV isolates were closely interrelated with geographical location. Close genetic relationships among BEFV strains can be deduced if the isolates originate from adjacent areas. Similarly, the BEFV isolates derived from widely separated regions have distant genetic relationships. This may indicate that BEFV circulates in neighboring region for a long time.
The clusters of the isolates were also chronologically related. In cluster I, the JT02L strain clustered with other East Asian isolates from 2001–2004, suggesting that the same BEF outbreak spread through mainland China, Taiwan and Japan across the borders. The LS11 and LYC11 strains slightly diverged from the isolates from previous epizootics in East Asia, which indicated that the new BEFV possibly invaded mainland China from a neighboring area via infected vectors carried on the seasonal wind over a long distance or the import of live cattle. In fact, some evidence has shown that both winds and animal transport have an important role in trans-boundary transmission of BEFV [25,26]. The oldest Chinese mainland vaccine strain, JB76H, and the oldest Japanese strain, YHL, sat separately. The Japanese and Taiwanese isolates from 1984–1989 clustered together. Similar results were obtained in the clusters II and III.
The variation in the aa sequences of the antigenic sites G1, G2 and G3 of the BEFV isolates was analyzed. The mentioned aa sequences of the three field strains obtained from mainland China corresponded identically with those of the Japanese isolates from 2001–2004 and the Taiwanese strains except for the 1984/TW/TN1 and 2001/TW/TN10. The other Japanese isolates from 1988–1989 had the same aa sequences mentioned above except for a substitution in ON-BEF-89-3 strain. No residues were changed among the isolates derived from Turkey and Israel. Three to five substitutions were found in the antigenic sites of G1 and G3 of Australian isolates compared with the residues of the Chinese mainland strains. These results indicated that the antigenic sites G1, G2 and G3 of BEFV isolates that related closely in place or time were highly conserved.
Conclusions
The sequences of the ectodomain region of the BEFV G gene were analyzed. The BEFV strains were isolated from mainland China, Taiwan, Japan, Turkey, Israel and Australia. The nucleotide and deduced aa sequences were well conserved among the isolates. A phylogenetic tree based on the nucleotide sequences was constructed, and the isolates were grouped into three clusters. The variations in the aa sequences of the antigenic sites G1, G2 and G3 of BEFV G protein were analyzed. The results showed that the phylogenetic relationships of the isolates were closely related to their geographical and chronological sources.
Abbreviations
BEFV: Bovine ephemeral fever virus; BEF: Bovine ephemeral fever; G: Glycoprotein; NCBI: National center for biotechnology information; ELISA: Enzyme-linked immunoabsorbent assay; RT-PCR: Reverse-transcription polymerase chain reaction.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
FZ carried out the studies and drafted the manuscript. CQ gave the polish to the language of the manuscript. The two authors read and approved the final paper.
Contributor Information
Fuying Zheng, Email: zfycaas@126.com.
Changqing Qiu, Email: cqqiu@126.com.
Acknowledgements
This work was supported by grants from BRF080305 and the Director General Foundation of Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences (2007). We appreciate professor Zhijie Zhao of the Center for Animal Disease Control and Prevention of Luoyang City for his sincere assistance.
References
- Wunner WH, Calisher CH, Dietzgen RG, Jackson AO, Kitajima EW, Lafon M, Leong JC, Nichol S, Peters D, Smith JS, Walker PJ. In: Virus Taxonomy. Murphy FA, Fauquet CM, Bishop DHL, Ghabrial SA, Jarvis AW, Martelli GP, Mayo MA, Summers MD, editor. Vienna: Sixth Report of the International Committee on Taxonomy of Viruses. Springer-Verlag; 1995. Rhabdoviridae; pp. 275–288. [Google Scholar]
- Hsieh YC, Chen SH, Chou CC, Hsiao HW, Chen SZ, Lee YF, Liu HJ. Bovine ephemeral fever in Taiwan (2001–2002) J Vet Med Sci. 2005;67:411–416. doi: 10.1292/jvms.67.411. [DOI] [PubMed] [Google Scholar]
- Walker PJ. Bovine ephemeral fever in Australia and the world. Currt Top Microbiol Immunol. 2005;292:57–80. doi: 10.1007/3-540-27485-5_4. [DOI] [PubMed] [Google Scholar]
- Walker PJ, Keren AB, Daisy HC, Denise LD, Doolan, YongHong W. Proteins of bovine ephemeral fever virus. J Gen Virol. 1991;72:67–74. doi: 10.1099/0022-1317-72-1-67. [DOI] [PubMed] [Google Scholar]
- Walker PJ, Byme KA, Riding GA, Cowley JA, Wang Y, McWilliam SM. The genome of bovine ephemeral fever rhabdovirus contains two related glycoprotein genes. Virology. 1992;191:49–61. doi: 10.1016/0042-6822(92)90165-L. [DOI] [PubMed] [Google Scholar]
- Walker PJ, Wang Y, Cowley JA, McWilliam SM, Prehaud CJ. Structural and antigenic analysis of the nucleoptotein of bovine ephemeral fever rhabdovirus. J Gen Virol. 1994;75:1889–1899. doi: 10.1099/0022-1317-75-8-1889. [DOI] [PubMed] [Google Scholar]
- Dhillon J, Cowley JA, Wang Y, Walker PJ. RNA polymerase (L) gene and genome terminal sequences of ephemeroviruses bovine ephemeral fever virus and Adelaide River virus indicate a close relationship to vesiculovirus. Virus Res. 2000;70:87–95. doi: 10.1016/S0168-1702(00)00215-X. [DOI] [PubMed] [Google Scholar]
- Cybinski DH, Walker PJ, Byrne KA, Zakrzewski H. Mapping of antigenic sites on the bovine ephemeral fever virus glycoprotein using monoclonal antibodies. J Gen Virol. 1990;71:2065–2072. doi: 10.1099/0022-1317-71-9-2065. [DOI] [PubMed] [Google Scholar]
- Cybinski DH, Davis SS, Zakrzewski H. Antigenic variation of the bovine ephemeral fever virus glycoprotein. Arch Virol. 1992;124:211–224. doi: 10.1007/BF01309803. [DOI] [PubMed] [Google Scholar]
- Kongsuwan K, Cybinski DH, Cooper J, Walker PJ. Location of neutralizing epitopes on the G protein of bovine ephemeral fever rhabdovirus. J Gen Virol. 1998;79:2573–2581. doi: 10.1099/0022-1317-79-11-2573. [DOI] [PubMed] [Google Scholar]
- Jin H. Express of bovine ephemeral fever virus transmembrane glycoprotein G in recombinant vaccinia virus and baculovirus. 2001. Doctoral Paper (in chinese).
- Zakrzewski H, Cybinski DH, Walker PJ. A blocking ELISA for the detection of specific antibodies to bovine ephemeral fever virus. J Immunol Methods. 1992;151:287–289. doi: 10.1016/0022-1759(92)90129-h. [DOI] [PubMed] [Google Scholar]
- Zheng FY, Lin GZ, Qiu CQ, Zhou JZ, Cao XA, Gong XW. Serological detection of bovine ephemeral fever virus using an indirect ELISA based on antigenic site G1 expressed in Pichia pastoris. Vet J. 2010;185:211–215. doi: 10.1016/j.tvjl.2009.06.002. [DOI] [PubMed] [Google Scholar]
- Zheng FY, Lin GZ, Qiu CQ, Zhou JZ, Cao XA, Gong XW. Developmeng and application of G1-ELISA for detection of antibodies against bovine ephemeral fever virus. Res Vet Sci. 2009;87:211–212. doi: 10.1016/j.rvsc.2009.03.010. [DOI] [PubMed] [Google Scholar]
- Bai WB, Jiang CL, Davis SS. Preliminary observations on the epidemiology of bovine ephemeral fever in China. Trop Anim Hlth Prod. 1991;23:22–26. doi: 10.1007/BF02361265. [DOI] [PubMed] [Google Scholar]
- Bai WB, Tian FL, Wang C, Jiang CL, Zhang ZG. Preliminary studies of the complement fixation test to confirm the diagnosis of bovine ephemeral fever. Aust J Biol Sci. 1987;40:137–141. [PubMed] [Google Scholar]
- Zheng FY, Lin GZ, Qiu CQ, Zhou JZ, Cao XA, Gong XW. Isolation and characterization of a field strain of bovine ephemeral fever virus in China. J Anim Vet Adv. 2009;8:1478–1483. [Google Scholar]
- Zheng FY, Lin GZ, Qiu CQ, Yuan KZ, Song JY. Expression and antigenic characterization of the epitope-G1 of bovine ephemeral fever virus glycoprotein in Pichia pastoris. Virol Sin. 2007;22:347–352. doi: 10.1007/s12250-007-0031-2. [DOI] [Google Scholar]
- Wang FI, Hsu AM, Huang KJ. Bovine ephemeral fever in Taiwan. J Vet Diagn Invest. 2001;13:462–467. doi: 10.1177/104063870101300602. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Aizawa M, Takayoshi K, Kokuba T, Yanase T, Shirafuji H, Tsuda T, Yamakawa M. Phylogenetic relationships of the G gene sequence of bovine ephemeral fever virus isolated in Japan, Taiwan and Australia. Vet Microbiol. 2009;137:217–223. doi: 10.1016/j.vetmic.2009.01.021. [DOI] [PubMed] [Google Scholar]
- Aziz-Boaron O, Klausner Z, Hasoksuz M, Shenkar J, Gafni O, Gelman B, David D, Klement E. Circulation of bovine ephemeral fever in the Middle East-Strong evidence for transmission by winds and animal transport. Vet Microbiol. 2012;158:300–307. doi: 10.1016/j.vetmic.2012.03.003. [DOI] [PubMed] [Google Scholar]


